Abstract
Cancer cachexia is a debilitating metabolic syndrome accounting for fatigue, an impairment of normal activities, loss of muscle mass associated with body weight loss eventually leading to death in majority of patients with advanced disease. Cachexia patients undergoing skeletal muscle atrophy show consistent activation of the SCF ubiquitin ligase (F-BOX) family member Atrogin-1 (also known as MAFBx/FBXO32) alongside the activation of the muscle ring finger protein1 (MuRF1). Other lesser known F-BOX family members are also emerging as key players supporting muscle wasting pathways. Recent work highlights a spectrum of different cancer signaling mechanisms impacting F-BOX family members that feed forward muscle atrophy related genes during cachexia. These novel players provide unique opportunities to block cachexia induced skeletal muscle atrophy by therapeutically targeting the SCF protein ligases. Conversely, strategies that induce the production of proteins may be helpful to counter the effects of these F-BOX proteins. Through this review, we bring forward some novel targets that promote atrogin-1 signaling in cachexia and muscle wasting and highlight newer therapeutic opportunities that can help in the better management of patients with this devastating and fatal disorder.
Keywords: Cancer Cachexia, Muscle Wasting, Atrogin-1, F-BOX, CRM1
1. Introduction
Cachexia is a devastating syndrome that is observed in the majority of end stage cancer patients. Primary symptoms of cancer cachexia comprise of progressive loss in weight and exhaustion of host skeletal muscle tissue as well as adipose tissue reserves [1]. Cachexia should be assumed in case an uncontrolled weight loss is found to be more than 5% weight preceding the occurrence of symptoms of disease in any given 6 months’ time [1]. Traditional management methods, such as 5-HT3 antagonists [2], appetite stimulants and nutrient supplementation [3], or COX-2 inhibitors and antioxidants [4] have been unsuccessful in reversing the metabolic anomalies commonly observed in cancer associated cachexia. At present the management of cachexia is one of the biggest challenges in cancer treatment. The syndrome has psychosocial impact on the individual and requires multi-pronged strategies that carry considerable financial cost to the patient as well as to the healthcare system [5,6].
Cachexia not only directly increases the morbidity and mortality, it also aggravates the side effects of chemotherapy and reduces the overall quality of life that is often times considered the major and direct cause of morbidity of a large proportion (>40%) of cancer patients. Individuals with upper gastrointestinal tumors have the highest rate of developing cachexia associated complications [7]. It is more acute in certain incurable malignancies such as pancreatic cancer where >80% of patients show extensive cachexia at the time of disease identification, considering the detection criterion i.e. weight loss >5% over 6 months span, weight loss > 2% in the host BMI <20 kg/m2, or indications of sarcopenia that is linked with weight > 2% [8]. The heterogeneity of advanced pancreatic tumors and the complex cues arising from multitude signaling pathways creates a microenvironment ideal for cachexia development [9]. In advanced cancers with cachexia the production of the tumor necrosis factor (TNF)-α and cytokines that are pro-inflammatory is most commonly observed [10,11]. The physical cues involve loss of appetite, chronic weight loss, chronic pain, disturbances in GI function as a direct consequence of tumor invasion and adverse effects of chemotherapy [12]. These chemical and physical signals render an environment conducive for disuse and untenable for proper muscle function leading to wasting.
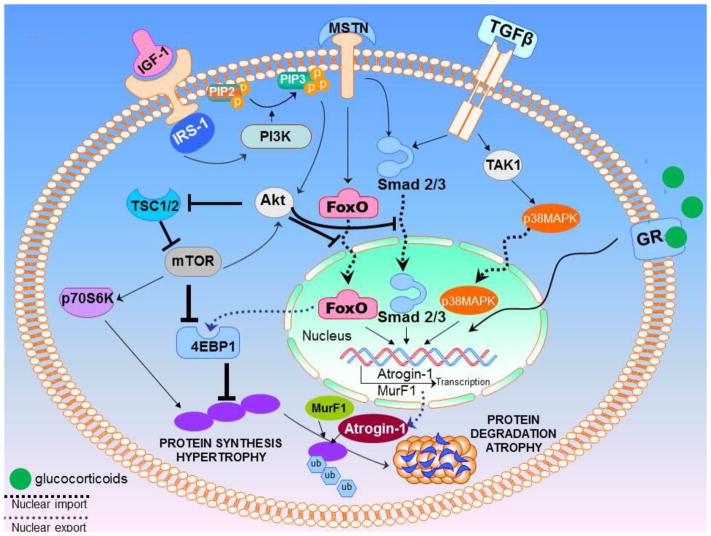
Muscle development results from a very complex crosstalk and the concerted interaction of multitude receptor driven signaling pathways that include IGF-1, IRS-1, PI3K, PIP3, PDK1, Akt, and mTOR among others (some summarized in Figure 1 and reviewed in [13]). On the contrary, muscle atrophy or wasting is mainly regulated by the E-3 ubiquitin ligases particularly MAFBx/FBXO32/Atrogin-1 (topic of this review). Sarcopenia is another form of muscle wasting, however, this term is more commonly, if not exclusively, used to describe skeletal muscle wasting during the aging process [14]. The number of fibers in a muscle and the size of fibers remain relatively stable from puberty until the fifth decade of life, at which point a noticeable decline in muscle fiber number and size begins [15]. Highlighting its overlapping role with cachexia, sarcopenia is directly associated to the poor prognosis of certain cancers such as non-small cell lung cancer [16,17], pancreatic [18], urothelial [19] esophageal [20] and many other malignancies. In line with cancer cachexia associated muscle wasting mechanisms, in sarcopenia, the ubiquitin proteasome assembly is also the main participant guiding muscle protein turnover, and this system holds a pivotal role in controlling muscle size [21]. MAFBx/FBXO32/Atrogin-1 together with MuRF-1 are again considered as chief regulators of ubiquitin-driven protein breakdown in skeletal muscle during sarcopenia (Figure 1 summarizes some of the signaling pathways and their cross talk that drives muscle hypertrophy and atrophy). Therefore, deeper understanding of the pathways driving the activation of these E3 ubiquitin ligase are expected to help in the development of newer and effective strategies for the treatment of a disorder as devastating and fatal as cachexia.
Figure 1. Summary schema of muscle hypertrophy and dystrophy signaling.
Muscle wasting processes are regulated by a number of receptor mediated signaling pathways. The binding of IGF-1 to its receptor results in the activation of insulin receptor substrate-1 (IRS-1), which promotes the activation of phosphatidylinositol-3-kinase (PI3K). The PI3K in-turn is responsible for the addition of a phosphate group to the intramembranous phosphoinositide-(4,5)-biphosphate (PIP2) to result in phosphoinositide-(3,4,5)-triphosphate (PIP3). IGF-1 receptor mediated activation of protein synthesis is PI3K dependent. The serine/threonine kinase Akt, binds PIP3 on the membrane through an N-terminal pleckstrin homology (PH) domain and is activated by phosphoinositide-dependent kinase-1 (PDK1). The recruitment of Akt to the membrane and the activation of Akt through PDK1 promotes Akt release into cytosol to activate mTOR and other downstream effectors. mTOR can also signal upstream to further activate Akt. Akt acts on mTOR by TSC1 and TSC2, which in turn increases mTOR activity. Myostatin (MSTN) and TGF-β when activated, bind to their respective receptors and activate the Smads (Smad2/3) as well as p38 MAPK signaling. MSTN can activate FOXO directly or through the activation of smad, while TGF-β mostly signals through the Smad2/3 alone. p38 MAPK is activated by TAK1 downstream of the activin and TGF-β receptors, and it can regulate the activity of various transcription factors to control gene expression related to muscle function. Under stress, the glucocorticoids act on the muscle antagonistic to the actions of insulin, resulting in the enhancement of protein degradation, reduced protein synthesis, and marked reduction in insulin-stimulated glucose uptake. Both genetic and epigenetic mechanisms have been found to be at play in muscles that in turn regulate or are regulated through protein and glucose signaling through modulation of the insulin/IGF-1 pathway. MuRF1 possesses a GRE in its proximal promoter, and its regulation by GCs depends on GR homodimerization. In contrast, Atrogin-1 is activated through an indirect mechanism, involving competitive binding of GR with AKT leads to the downstream activation of FOXO1/3, transcriptional regulators of MAFbx, thus, indirectly inducing MAFbx expression.
2. F-BOX proteins and their multifaceted roles in cancer
Cellular homeostasis is governed by the coordinated biosynthesis and breakdown of critical regulatory proteins. There are multitude pathways that regulate protein synthesis. However, protein breakdown is controlled mainly by the ubiquitin proteasome system that comprises of activation of ubiquitin by E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, following which the E3 ubiquitin ligase identifies the specific protein to be ubiquitinated and promotes the translocation of ubiquitin from the E2 enzyme on to target protein leading to 26S proteasome mediated degradation. Among the known E3 ubiquitin ligases, the SCF (SKP1-CUL1-F-box protein) E3 ligases are the largest and most diverse family that directly causes the turnover/degradation of many critical cellular proteins. Highlighting their critical role in protein biology, aberrations in the expression, function and regulation of SCF E3 ligases have been directly or indirectly been associated with various human diseases, including the majority of cancers [22].
There are two E1 ubiquitin-activating enzymes encoded by the genome of higher vertebrates [23]. On the contrary, more than thirty eight E2 ubiquitin-conjugating enzymes and approximately 700 different distinct E3 ubiquitin ligases are found in humans [24]. The largest among the E3 ubiquitin ligases is the SCF family, and is universally conserved across a broad spectrum of hierarchical organisms ranging from single cell eukaryotes to humans. The E3 ubiquitin ligases are composed of four different protein components that directly coordinate the function and structure of the protein: (1) adaptor component called SKP1, (2) the scaffold portion termed Cullin-1 (CUL1), (3) the F-BOX component that is responsible for substrate-recognition and (4) RBX1 or RBX2 RING components. This structural arrangement is conserved in most SCF E3 ligases i.e. CUL1-SKP1 interaction alongside the F-box protein at the N-terminus and the C-terminus binding of the RING protein RBX1/RBX2 [25]. The F-BOX protein is largely responsible for the precise substrate recognition and specificity of SCF E3 ligases through recognition of phosphorylated target proteins [26]. On the other hand the CUL1-RBX1/CUL1-RBX2 is responsible for the fundamental ligase action and catalyzes the relocation of the ubiquitin enzyme from the E2 to the substrate [27]. By guiding a precise and timely turnover of myriad important cellular proteins, the SCF E3 ligases play pivotal part in multitude number of key molecular events such as cell division, signal transduction, DNA replication and also guide the developmental pathways [28,29]. The observation that SCF E3 ligases are aberrantly expressed in majority of malignancies makes them attractive therapeutic candidates for targeted drug development. The F-BOX proteins directly attach to substrates, using various domains, including Leucine Rich Repeats, WD40 motifs, and other domains, which are also the basis for their categorization into sub-families, Fbxl, Fbxw, and Fbxo, respectively. This holoenzyme thus brings a substrate and charged ubiquitin in close proximity, enabling post-translational modification. FBP/Skp1 hetero-dimers can be dissociated from Cul1 by an exchange factor, Cand1, allowing for ‘recycling’ of SCF complexes, and diversifying the cell’s ubiquitinating capacity [30,31,32].
F-BOX proteins have been shown to regulate pathways associated with muscle wasting. In the forthcoming paragraphs we will individually discuss the different F-box family members and their role in muscle development and wasting. We will also review some novel areas in the biology of F-BOX proteins and how these can be approached to block cachexia in cancer.
3. Role of F-BOX in cancer cachexia and muscle wasting
Mass in skeletal muscle is maintained by the coordinated yet opposing processes of constant protein production and proteolysis. In cachexia, this balance shifts in favor of enhanced breakdown leading to a swift loss of muscle mass through breakdown [33]. During wasting the patients muscle protein is quickly mobilized through a series of cellular mechanisms that are parallel and cross linked [34] and work in tandem to the activation of the ubiquitin proteasome pathway [35]. Chief among the ubiquitin proteasome components activated during muscle atrophy is Atrogin-1/MAFbx/FBXO32 belonging to the F-BOX family [36]. Atrogin-1 induction is considered the primary event during the atrophy, and its activation occurs prior to the loss of muscle weight. Transgenic animal models that are null for atrogin-1 show only limited muscle atrophy, highlighting the critical role of atrogin-1 in targeting important muscle protein(s) for breakdown [36]. In parallel, considerable number of investigations has consistently demonstrated the activation of atrogin-1 in dying tissues such as failing heart tissue [37], in uterus during postpartum [38], and also in the terminal stromal tumor tissue that is responsive to treatments such as imatinib [39].
Extensive investigations over the last 15 years have made atrogin-1 as one of the most studied muscle atrophy gene [40]. Given that it is specifically activated in muscles undergoing atrophy, its expression is considered as a biomarker for cachexia [41]. Atrogin-1 expression and nuclear localization (necessary for its cachexia promoting transcriptional regulation activity on genes) is influenced by many key players associated with the tumor microenvironment. Noted among them is the FOXO transcription that is considered the direct promoter of Atrogin-1 expression in a nuclear localization dependent manner [42]. Other notable player in the process includes TNF-α that has been demonstrated to promote atrogin-1 expression [43]. Patients, particularly those with pancreatic cancer associated cachexia show consistent up-regulation in the levels of TNF-α in the serum [44]. TNF-α has also been shown to be activated in pro-inflammatory response [45] through activities such as chronic smoking [46] causing impairment in the lipogenesis processes and ultimately affecting adipocyte differentiation [47]. TNF-α has been shown to drive reactive oxygen species (ROS) production that in turn can promote pro-survival transcription factors such as nuclear factor κB (NF-κB) resulting in atrogin-1 expression [48]. Conversely, TNF-α can block myogenesis by down-regulating MyoD transcription that also involves NF-κB activation as shown in cellular systems [49]. Most importantly, TNF-α also promotes exportin 1/chromosome maintenance region 1 (XPO1/CRM1) nuclear export activity [50] that has been linked to the enhancement in the activities of Atrogin-1 promoting cytokines such as IL-15 [51]. MuRF1 is another ubiquitin ligase activated during muscle atrophy and acts in tandem with Atrogin-1. MurF-1 was discovered as a muscle-specific RING finger protein that attaches to the large sarcomeric protein titin through its kinase domain [52,53]. Later on two additional highly homologous RING finger proteins, named MuRF2 and MuRF3 (62% and 77% homology to MuRF1 respectively) were also identified [52]. All the three MuRFs are encoded by different genes that belong to the subgroup of the TRIM superfamily [54,55]. Nevertheless, MuRF1 is the sole player regulating muscle loss and despite the homology the roles of the additional two murf family members is not clear in the muscle wasting process. Therefore, targeting MurF1 through small molecule inhibitor could be an attractive therapeutic strategy to prevent muscle wasting and certainly requires further attention.
3.1. Role of other F-BOX proteins in regulating cancer associated muscle wasting signaling
The loss of F-BOX family members has been linked to better prognosis and therapy response in some malignancies. For example in an elegant study by Malyukova et al., it was demonstrated that mutation causing functional inactivation of FBXW7, are associated with improved prognosis and early onset response to glucocorticoid treatment in childhood T-cell acute lymphoblastic leukemia (T-ALL) [56]. The authors demonstrated that FBXW7 causes glucocorticoid receptor α (GRα) ubiquitinylation dependent proteasomal degradation that is through glycogen synthase kinase 3 β-mediated phosphorylation. Their studies clearly show that FBXW7 inactivation can cause elevated GRα levels, causing enhanced response to glucocorticoids. On the contrary, reduction in FBXW7 promoted glucocorticoid sensitivity that was independent of the chemotherapy response in T-ALL suggesting that inactivation of FBXW7 could become synthetic lethal approach during chemotherapy. In osteosarcoma model, the loss of FBXW7 has been shown to regress tumors and is an independent prognostic marker [57]. Collectively, the above studies clearly show the importance of additional F-BOX family members in different muscle wasting signaling. Below we discuss the pathways that regulate these important proteins and highlight the therapeutic opportunities that can be harnessed from these signaling mechanisms that can be used to impact muscle wasting.
4. Regulation of Atrogenes in cancer
The F-BOX family member atrogin-1 and other atrophy gene Murf1 (collectively called atrogenes) are under the influence of important cancer driver pathways (some summarized in Figure 1). The chief among them include FOXO3a transcription factors, Smad 2,3, glucocorticoid receptors (GR) and p38MAPK that promote the expression of atrogin-1 and MurF1. Cancer associated increase in circulating glucocorticoids (GC) has been very well studied for their roles in promoting muscle wasting through the promotion of atrogin-1 and MurF1 [58]. The glucocorticoid receptor (GR) are considered mandatory for GC induced atrophy as muscle-specific knock down of GR (GR−/−) in mice makes them resistant to GC induced muscle atrophy. GR mediated activation of FOXO proteins can directly promote the transcription of atrogenes. Additionally, GC is recognized to inhibit PI3/Akt signaling resulting in the activation of FOXO and the inhibition of mTOR pathways. On the contrary the proliferator-activated receptor-gamma co-activator 1 alpha (PGC1-α) and cEBP1 are two key negative regulators of atrogin-1. Other novel players such as non-coding RNAs (micro RNAs) which can regulate these atrogenes epigenetically are emerging to play critical roles in the biology of atrogin-1 and murf-1 thereby impacting muscle atrophy during cachexia and are detailed below.
4.1. Atrogin-1 regulation by FOXO proteins
The role of FOXO proteins in muscle wasting is quite extensively studied. The earliest evidence came from studies by Sandri and colleagues who showed that in cultured myotubes undergoing atrophy, the PI3K/AKT signaling is suppressed, leading to activation of FOXO transcription factors and atrogin-1 induction. On the contrary atrogin-1 expression was found to be decreased either through the treatment with IGF-1 or through AKT overexpression that was mediated through the down-regulation of FOXO3 [59]. In the same study, constitutively active FOXO3 was found to act on the atrogin-1 promoter region resulting in atrogin-1 transcription and causing significant atrophy of myotubes and muscle fibers. On the other hand in the presence of dominant negative Foxo constructs, the authors observed lack of atrogin-1 induction even in the presence of glucocorticoids. In another study, IGF-1 was shown to impact PI3K/Akt/mTOR pathway and blocking nuclear translocation of Foxo3 resulting in the lack of atrogin-1 or MurF1 gene activation and thereby preventing muscle atrophy [60]. Conversely, PGC1-α protein is recognized to prevent muscle atrophy by masking the effect of FOXO protein on Atrogin-1[61]. Gene over-expression of PGC1-α in mice resulted in lower decrease in muscle fiber diameter and a minimal induction of atrogenes than in control mice in a mechanism involving inhibition of FOXO protein. These initial findings led to the enhanced research interest towards understanding the role of PGC1-α in preventing muscle wasting some of which are detailed below.
4.2. PGC1-α mediated regulation of atrogin-1 and its consequence on cachexia
A limited number of proteins have been studied that directly suppress the expression of atrogin-1 and among them is PGC1-α. Genome-wide transcriptional analyses point to the elevation of PGC1-α transcript that was linked to mitochondrial function in cancer cell models [62]. Its expression is directly correlated to wt-p53 function in lung cancer models as mutant or null lung tumors show lack of PGC1-α along with de-regulation in mitochondrial oxidative phosphorylation and biogenesis[63]. Earliest studies showed that changes in the redox status and induction of the atrogenes alongside the activation of autophagy pathways and the ensuing muscle disuse could be reverted through forced PGC1-α over-expression [64]. Given its crucial role in preventing muscle disuse, it is not surprising to note that cancer microenvironment has evolved ways to keep PGC1-α function in check. For example tumor microenvironment associated enhancement in TNF-α or activities such as chronic smoking have been shown to block optimal PGC1-α expression leading to muscle dystrophy [65]. Complementing these findings, the insulin growth factor 1 (IGF1) is known to promote PGC1-α and prevent muscle atrophy through atrogin-1 and murf-1 down-regulation [66]. Conversely, during resistance training, IGF1 can also promote muscle hypertrophy by regulating PGC-1α isoform [67]. Highlighting its universal role in disease biology, it has been demonstrated that in a cardiac model the exportin 1 (XPO1) nuclear export function is indispensable for non-pathological (hypertrophy) cardiac gene activation through IGF1 [68]. Most significantly, another form of PGC1-α, the NT-PGC-1α with similar functions possesses a nuclear exclusion sequence (NES) and is targeted by XPO1 mediated nuclear export [69]. These findings point to the fact that restoring PGC1-α expression/nuclear localization may abrogate atrogin-1 that in principle can target cachexia associated muscle wasting.
4.3. C/EBP1-β mediated regulation of the F-Box proteins
CCAAT-enhancer-binding proteins (or C/EBPs) are a family of transcription factors composed of six members, named from C/EBPα to C/EBPζ [70]. C/EBPs can regulate the expression of diverse set of genes by binding to their promoter sequences. Among them the C/EBP-β is a downstream target of Ras signaling and observations point to critical pro-oncogenic functions for C/EBP-β in human malignancies [71,72,73]. Studies indicate that C/EBP-β and δ may be involved in muscle wasting as the expression of both these TFs is found elevated in rat model undergoing sepsis [74]. This was revealed during an analysis of the effect of glucocorticoid receptor antagonist RU38486 on different downstream effector genes in sepsis and wasting muscles showed down-regulation in C/EBP-β and δ. Most importantly the promoter regions of multitude number of genes belonging to ubiquitin-proteasome pathway (including F-BOX family members especially Atrogin-1) carry putative C/EBP binding sites [74]. Further support came from studies where it was shown that dexamethasone treatment causes up-regulation of C/EBP-β and δ along with their DNA binding capacity in atrophying cultured muscle cells in vitro as well as in vivo rat models of muscle wasting [75]
Selective siRNA silencing reveals that among the different C/EBPs, only C/EBP-β DNA binding activity (supershift assay) was identified to be up-regulated and in dexamathasone exposed myotubes the nuclear cofactor p300 interacted specifically with C/EBPβ sparing C/EBP-δ in [76]. These studies indicate that dexamethasone-associated atrogin-1 enhancement and the resultant protein turnover, and atrophy in cultured myotubes is quite significantly regulated by C/EBP-β. More significantly, highlighting a muscle specific function, siRNA against C/EBPβ prevented Atrogin-1 expression that was restricted to myoblasts. These findings clearly point to the critical role played by C/EBPβ in glucocorticoid-promoted MuRF1 expression that could specifically depend on the differentiated status of muscle cells.
4.4. MicroRNA mediated regulation of Atrogin-1
microRNAs (miRNA) are short non-coding RNAs that can regulate many different genes through binding to the 3’UTR of target mRNAs. Aberrant miRNA expression has been recognized to play multi-faceted roles in the biology of cancers. Recently, considerable attention has been paid on deciphering the regulatory mechanisms of miRNAs in the biology of cachexia and muscle wasting [77]. For example, Wada and colleagues demonstrated that miR-23a suppresses the translation of different atrogenes [78]. Their study showed that over-expression of miR-23a could protect against muscle atrophy in vitro as well as suppress GC-induced skeletal muscle atrophy. Supporting these studies, recently another group demonstrated similar effects of exosomal miR-23 against atrogin-1 and murf-1 mediated muscle atrophy through the attenuation of nuclear factor of activated T cells 3 (NFATc3) and calcineurin signaling [79]. In view of these additional findings, Xu and colleagues investigated the impact of FOXO regulatory miR-486 on muscle atrophy genes [80]. Transfection of miR-486 mimic into primary cultures of myotubes was found to block dexamethasone-associated protein degradation with no influence on protein synthesis through inhibition of FOXO1 protein translation. Aside from in vitro work the group also demonstrated that introduction of miR-486 into muscles through electroporation could regress E-3 ligase expression resulting in enhancement in muscle mass. On the contrary, miR-1 was shown to enhance dexamethasone mediated muscle atrophy through inactivation of HSP70 in a mechanism involving activation of FOXO3 [81]. In cardiac hypertrophy models, forced expression of miR-19a/b was shown to induce hypertrophy in rat neonatal cardiomyocytes through simultaneous inhibition of atrogin-1 and MuRF-1 [82]. Recently, miR-182 has also been demonstrated to play preventive role against muscle atrophy through inhibition of FOXO3 mediated activation of atrogin-1 and MurF-1. There are a number of additional studies that point to the critical roles of miRNAs in the regulation of muscle atrophy genes. In a deep sequencing profiling approach, LPS or saline injection in Pig muscle led to the identification of five miRNAs (ssc-miR-146a-5p, ssc-miR-221-5p, ssc-miR-148b-3p, ssc-miR-215 and ssc-miR-192) that were found to have a role in the regulation of muscle wasting genes [83]. Collectively, these data certainly support the premise that a single or very few miRNAs can act on multiple targets (here atrogin-1 and murf-1) to significantly impact biological outcome resulting in protection against complex processes such as muscle atrophy.
5. Nuclear export mediated F-BOX protein regulation in cancer cachexia and muscle wasting
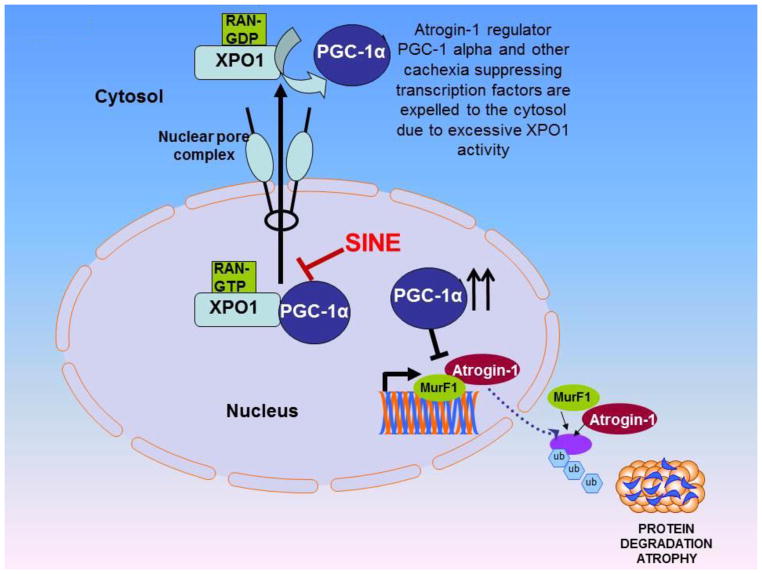
Nuclear protein transport plays an important role in various cellular processes by controlling the activity of genome surveillance TFs and TSPs in a compartmentalization dependent fashion [84]. The nuclear exporter protein exportin 1 (XPO1) is the major exporter of most of the nuclear residing proteins [85]. However, cancer associated enhancement of XPO1 results in excessive efflux of TFs and TSPs to the cytoplasmic compartment leading to their functional inactivation [86]. As described above, most of the cachexia mediators including atrogin-1, PGC-1α and IGF 1 undergo XPO1 mediated nuclear export. Their mis-localization during cachexia could be associated to the enhanced functionality of TNF-α that results in the promotion of XPO1 nuclear export activity. It is not surprising to note that TNF-α mediated production of iNOS can also suppress total albumin expression (a common consequence in cachexia) in a mechanism involving phosphorylation of C/EBP beta that enhances its nuclear export expression through XPO1 [87]. With this multitude of independent evidences from diverse models, it is quite logical to speculate that disturbed nuclear export may promote expulsion of cachexia preventing proteins leading to their functional inactivation (Summarized in Figure 2). Given its significant role in the biology of cachexia, targeting XPO1 could become an attractive therapeutic strategy to make inroads against this debilitating disorder. Earlier we were the first to show that XPO1 inhibition by specific inhibitor of nuclear export (SINE) Selinexor can re-align TSPs in the nucleus of cells causing growth inhibition and suppression of tumors in NHL and PDAC [88]. More striking is the observation that certain F-BOX family members can be retained in the cancer cell nucleus upon Selinexor treatment. Among the list of F-BOX Selinexor targets that our group has shown to be retained in the nucleus include Fbw7[89], FBXL5, FXO33 and related family members [90]. In these studies using epithelial-to-mesenchymal (EMT) harboring snail transduced human mammary epithelial cells (HMLE-Snails) we demonstrated that reversal of mesenchymal phenotype by selinexor directly resulted from nuclear FBXL5 [90]. Drug induced nuclear retention of FBXL5 was found to cause nuclear degradation of EMT promoter snail alongside the suppression of additional EMT markers [90]. Gene expression array and pathway analysis demonstrated differential regulation of additional F-BOX family members that was concurrent with the down-regulation of snail network [90]. Most interestingly, Selinexor could cause a complete loss in tumor growth of ras and snail transduced HMLEs (HMLER-Snail) cells when grown as sub-cutaneous xenograft [90]. Findings from our laboratory are supported by additional groups who have demonstrated nuclear retention of other F-BOX family members FBXL14 as well as FBXL5 by un-related nuclear export inhibitor Leptomycin B (LMB) in breast tumor models [91,92]. Such studies certainly point to the role of nuclear transport in the regulation of F-BOX protein family. Building on these encouraging results our laboratory is currently evaluating the impact of PGC1-α nuclear retention on atrogin-1 and MurF-1 in muscle cell models. Our recent preliminary findings demonstrate that SINE can induce the nuclear localization of PGC1-α in C2C12 muscle cells leading to the suppression of both atrogin-1 and murf1 (unpublished work). Additionally Selinexor can also suppress the transcription of atrogin-1 and murf1 (Figure 3). Our ongoing work reveals that Selinexor can synergize with Dexamethasone (DEX) where we observed superior anti-cancer activity in the combination (apoptosis, viability) in lymphoma models [93]. More striking is the observation that the combination treatment led to the reduction in atrogin-1 expression in the muscle tissue in the Selinexor-DEX combination treated mice (Selinexor at 10 mg/kg twice a week for three weeks in the presence of DEX 10 mg/kg) (unpublished work). These highly significant and novel results clearly support the use of Selinexor in combination with DEX in that not only do we see enhancement in the activity of Selinexor but also observe cachexia preventing effects that are a direct consequence of DEX. These are clinically relevant findings since the combination therapy can lower the dexamethasone dose required in any combination regimen. These findings certainly point to the fact that targeting the nuclear protein export can significantly impact atrophy signaling and warrants further investigations.
Figure 2. Targeting muscle wasting at the nuclear pore.
In cancer, excessive activity of CRM1 results in the functional inactivation of important tumor suppressor proteins (TSPs) and cachexia preventing transcription factors (TFs). Inhibition of nuclear exporter protein Exportin1 (XPO1) by specific inhibitor of nuclear export (SINE) can induce nuclear accumulation of PGC-1 alpha and additional transcription factors that suppress cachexia causing atrogenes.
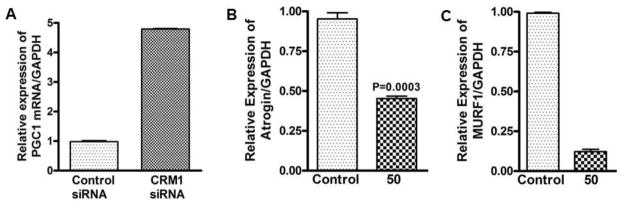
Figure 3. Role of nuclear exporter in muscle wasting (proof of concept).
[A] siRNA silencing of nuclear exporter caused re-expression of PGC1-α in human mammary epithelial cells. Snail transduced Human mammary epithelial cells (HMLE-Snail) grown in duplicate were exposed to either control siRNA or CRM1 siRNA for 72 hrs (for two passages). At the end of the treatment period RNA from each treatment condition was isolated and subjected to RT-PCR analysis. [B] MiaPaCa-2 pancreatic cancer cell lines grown in duplicate in six well plates (50,000 cells per well) were exposed to 50 nM Selinexor (a specific inhibitor of nuclear export for 24 hrs) and RNA was quantified using RT-PCR for [B] Atrogin-1 and [C] MurF1 (p<0.001) normalized GAPDH.
6. Atrogin-1 targeted treatment of cancer cachexia and muscle wasting
Given the significant role of Atrogin-1 in muscle wasting, attempts have been made to develop small molecule targeted drugs against this and other critical F-box proteins. Pharmaceutical strategies were supported by evidence of the beneficial effect of proteasome inhibition on preventing muscle atrophy. For example, in a rat model, Tawa Jr and colleagues demonstrated that the enhanced proteolysis in sepsis is due to a proteasome-dependent pathway, and inhibition of proteasome function by peptide aldehyde inhibitors of the proteasome, N-acetyl-leucyl-leucyl-norleucinal (LLN), CBZ-leucyl-leucyl-leucinal (MG132) resulted in the suppression of proteolysis in incubated rat skeletal muscles [94]. In another study, Fisher and colleagues demonstrated that rats treated with the proteasome inhibitor N-benzyloxycarbonyl-Ile-Glu-(O-t-butyl)-Ala-leucinal (PSI) caused reduction in the myofibrillar muscle protein breakdown by chemically induced sepsis [95]. Combaret et al, showed that the xanthine derivative Pentoxifylline (PTX) could prevent muscle atrophy and suppress increased protein breakdown in Yoshida sarcoma-bearing rats by inhibiting the activation of a non-lysosomal, Ca(2+)-independent proteolytic pathway. In this study, PTX was shown to block the ubiquitin pathway, by inhibiting the 14-kDa ubiquitin conjugating enzyme E2, and the C2 20S proteasome subunit in rat muscles [96]. Indirect approaches to prevent cachexia through ghrelin antagonist have shown pre-clinical success as well. Anamorelin (ANAM), an oral mimetic of ghrelin, enhances body weight and anabolic hormone levels in healthy subjects and these studies have led to their testing in a cachexia preventive setting. In rat models, ANAM showed statistically significant agonistic binding on the ghrelin receptor, and stimulated GH release in vitro. ANAM has been shown to significantly and dose-dependently increase food intake and restore body weight alongside increasing GH and IGF-1 levels across different species tested. In a double-blind, placebo-controlled, crossover study in 16 patients, ANAM was shown to significantly increase body weight compared with placebo [97]. More significantly, preliminary studies also show signs of synergistic interaction between ANAM and specific inhibitors of nuclear export against cancer cells that is concurrent with the suppression of atrogin-1 and murf1. These initial findings are quite encouraging and certainly point to the possibility of rationally combining Selinexor (a Phase II) drug with ANAM (currently in Phase III) to tame in muscle wasting associated with cachexia.
7. Natural products targeting F-BOX family proteins and their impact on cachexia and muscle wasting
A number of non-toxic natural products have been evaluated for their beneficial effects against cachexia and muscle wasting. The turmeric compound curcumin has been studied for its activity against atrophy genes. Curcumin and the anti-oxidant compound N-acetylcysteine (NAC) were shown to inhibit the activation of NF-κB during inflammation in skeletal muscle and thereby protected muscles against loss of mass or wasting [98]. In another study highlighting the mechanism of action, curcumin mediated down regulation of Atrogin-1 and MurF1 was shown to be due in part to inhibitor upstream players such as p38 and NF-kappaB in mice models of LPS induced muscle wasting [99]. Similarly, Siddiqui and colleagues have demonstrated attenuation of muscle proteolysis and muscle wasting through MurF1 and atrogin-1 down-regulation in mice models [100]. Very recently Ono and colleagues showed suppression of curcumin can prevent skeletal muscle destruction in type 1 diabetic mice by inhibiting protein ubiquitination [101]. It should be noted that curcumin also suppresses nuclear export function by targeting CRM1 making it an attractive agent that could impact muscle wasting signaling at different fronts [102]. It would be worthwhile to evaluate the impact of curcumin on PGC1-α and some of the other atrogin-1 regulators described above.
The red wine polyphenol resveratrol has also been investigated for its protective role against ubiquitin pathways. Earliest evidence came from cardiac models in which resveratrol was shown to prevent atrophy through the inhibition of NF-κB (p65) [103,104]. Guided by these initial findings a number of newer investigations have shed light on the multi-faceted effects of resveratrol on muscle wasting genes. Resveratrol has been shown to prevent dexamethasone induced atrophy through the inhibition of both atrogin-1 and murf-1 in a mechanism involving SIRT1[105]. These findings are not restricted to a specific type of muscle wasting model and can be replicated under different conditions. For example, Wang and colleagues have demonstrated that resveratrol can effectively counteract TNF-α promoted muscle protein breakdown and reverses decreasing expression of FoxO1, mTOR, p70S6K, Akt and 4E-BP1, but applies no effect on FOXO3a protein [106]. Given the pleiotropic nature of resveratrol, its potential as an inhibitor of other F-BOX family proteins has also been investigated. For example, it has been demonstrated that the activation of SIRT1 by resveratrol can lead to cellular amassing of β-TrCP E3 ligase via protein breakdown [107]. The anti-cancer potential of resveratrol is well recognized and the newer findings on atrophy suppressive effects certainly support its use as a non-toxic complementary approach in the management of cachexia.
Green tea polyphenols are well known for their cancer preventive effects in cellular and animal tumor models [108,109]. A number of studies have demonstrated the proteasome inhibitory effects of green tea polyphenol EGCG and its related analogs [110,111] and these observations led to the exploration of their potential against F-BOX family members and also for their muscle atrophy preventive mechanisms of action. This becomes especially relevant given that EGCG is capable of down-regulating SKP2 in breast cancer cell lines indicating that it can impact E-3 ubiquitin ligases [112]. As anticipated, EGCG was shown to suppress NF-κB and its related downstream molecules such as atrogenes leading the authors to conclude that it could be an ideal agent to be included in combination therapeutic strategy against the tumor-induced muscle atrophy [113]. Other catechins have also been shown to suppress muscle atrophy by blocking atrogenes [114]. Collectively, there is ample evidence that diet derived natural products and their derivatives especially polyphenols are promising agents that can be used in the management of this deadly syndrome by blocking the F-BOX protein atrogin-1.
8. Conclusions
Cancer cachexia associated muscle wasting is a devastating syndrome that is observed in majority of end stage cancer patients. Cachexia patients undergoing muscle wasting demonstrate a very specific activation of atrogin-1 belonging to the F-BOX family. As such the F-BOX proteins are emerging to play pivotal role in the biology of cancer. As reviewed here, cancer associated de-regulations in F-BOX proteins have a central role in cachexia directly through atrogin-1 or indirectly through the activation of pathways that promote cachexia and muscle wasting signaling. Certain key players are emerging to play a crucial role in the regulation of F-Box family members that includes the nuclear protein transporter protein CRM1 or XPO1. Deeper research on these interactions may lead to the identification of newer therapeutic approaches against cancer cachexia and muscle wasting. Nevertheless, several challenges remain given the diverse nature of these syndromes. Therefore, the scale of the complexity of the problem calls for equally complex solutions. The design of therapies should be holistic enough to simultaneously attack cancer and cachexia symptoms. As discussed in this review, preliminary results, although small show that nuclear transport inhibition could be one a novel avenue that can harnessed to possibly target cancer and associated cachexia and muscle wasting. Similarly miRNAs have to be selectively and carefully evaluated for their multi-targeted effects on overlapping signaling in cancer and atrophy. Such strategies are anticipated to improve the life quality in end stage cancer patients who suffer from this devastating syndrome.
Acknowledgments
Work in the lab of ASA and RMM is supported by NIH R21 grant 1R21CA188818-01A1, SKY Foundation, James H Thie Foundation, QNRF and Perri Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Fearon KC, Voss AC, Hustead DS. Definition of cancer cachexia: effect of weight loss, reduced food intake, and systemic inflammation on functional status and prognosis. Am J Clin Nutr. 2006;83:1345–50. doi: 10.1093/ajcn/83.6.1345. [DOI] [PubMed] [Google Scholar]
- 2.Kast RE, Foley KF. Cancer chemotherapy and cachexia: mirtazapine and olanzapine are 5-HT3 antagonists with good antinausea effects. Eur J Cancer Care (Engl ) 2007;16:351–4. doi: 10.1111/j.1365-2354.2006.00760.x. [DOI] [PubMed] [Google Scholar]
- 3.Ottery FD. Supportive nutrition to prevent cachexia and improve quality of life. Semin Oncol. 1995;22:98–111. [PubMed] [Google Scholar]
- 4.Mantovani G, Madeddu C. Cyclooxygenase-2 inhibitors and antioxidants in the treatment of cachexia. Curr Opin Support Palliat Care. 2008;2:275–81. doi: 10.1097/spc.0b013e32830f47e4. [DOI] [PubMed] [Google Scholar]
- 5.Hopkinson JB. Psychosocial impact of cancer cachexia. J Cachexia Sarcopenia Muscle. 2014;5:89–94. doi: 10.1007/s13539-014-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopkinson JB. The emotional aspects of cancer anorexia. Curr Opin Support Palliat Care. 2010;4:254–8. doi: 10.1097/SPC.0b013e32833ef813. [DOI] [PubMed] [Google Scholar]
- 7.Ozola, Zalite I, Zykus R, Francisco, Gonzalez M, Saygili F, Pukitis A, Gaujoux S, Charnley RM, Lyadov V. Influence of cachexia and sarcopenia on survival in pancreatic ductal adenocarcinoma: a systematic review. Pancreatology. 2015;15:19–24. doi: 10.1016/j.pan.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Johns N, Hatakeyama S, Stephens NA, Degen M, Degen S, Frieauff W, Lambert C, Ross JA, Roubenoff R, Glass DJ, Jacobi C, Fearon KC. Clinical classification of cancer cachexia: phenotypic correlates in human skeletal muscle. PLoS One. 2014;9:e83618. doi: 10.1371/journal.pone.0083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller TC, Burmeister MA, Bachmann J, Martignoni ME. Cachexia and pancreatic cancer: are there treatment options? World J Gastroenterol. 2014;20:9361–73. doi: 10.3748/wjg.v20.i28.9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martignoni ME, Dimitriu C, Bachmann J, Krakowski-Rosen H, Ketterer K, Kinscherf R, Friess H. Liver macrophages contribute to pancreatic cancer-related cachexia. Oncol Rep. 2009;21:363–9. [PubMed] [Google Scholar]
- 11.Barber MD, Fearon KC, Ross JA. Relationship of serum levels of interleukin-6, soluble interleukin-6 receptor and tumour necrosis factor receptors to the acute-phase protein response in advanced pancreatic cancer. Clin Sci (Lond) 1999;96:83–7. [PubMed] [Google Scholar]
- 12.Fearon KC, Baracos VE. Cachexia in pancreatic cancer: new treatment options and measures of success. HPB (Oxford) 2010;12:323–4. doi: 10.1111/j.1477-2574.2010.00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran T, Andersen R, Sherman SP, Pyle AD. Insights into skeletal muscle development and applications in regenerative medicine. Int Rev Cell Mol Biol. 2013;300:51–83. doi: 10.1016/B978-0-12-405210-9.00002-3. [DOI] [PubMed] [Google Scholar]
- 14.Tarantino U, Scimeca M, Piccirilli E, Tancredi V, Baldi J, Gasbarra E, Bonanno E. Sarcopenia: a histological and immunohistochemical study on age-related muscle impairment. Aging Clin Exp Res. 2015;27(Suppl 1):51–60. doi: 10.1007/s40520-015-0427-z. [DOI] [PubMed] [Google Scholar]
- 15.Miljkovic N, Lim JY, Miljkovic I, Frontera WR. Aging of skeletal muscle fibers. Ann Rehabil Med. 2015;39:155–62. doi: 10.5535/arm.2015.39.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Go SI, Park MJ, Song HN, Kang MH, Park HJ, Jeon KN, Kim SH, Kim MJ, Kang JH, Lee GW. Sarcopenia and inflammation are independent predictors of survival in male patients newly diagnosed with small cell lung cancer. Support Care Cancer. 2015 doi: 10.1007/s00520-015-2997-x. [DOI] [PubMed] [Google Scholar]
- 17.Kim EY, Kim YS, Park I, Ahn HK, Cho EK, Jeong YM. Prognostic Significance of CT-Determined Sarcopenia in Patients with Small-Cell Lung Cancer. J Thorac Oncol. 2015 doi: 10.1097/JTO.0000000000000690. [DOI] [PubMed] [Google Scholar]
- 18.Rollins KE, Tewari N, Ackner A, Awwad A, Madhusudan S, Macdonald IA, Fearon KC, Lobo DN. The impact of sarcopenia and myosteatosis on outcomes of unresectable pancreatic cancer or distal cholangiocarcinoma. Clin Nutr. 2015 doi: 10.1016/j.clnu.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Taguchi S, Akamatsu N, Nakagawa T, Gonoi W, Kanatani A, Miyazaki H, Fujimura T, Fukuhara H, Kume H, Homma Y. Sarcopenia Evaluated Using the Skeletal Muscle Index Is a Significant Prognostic Factor for Metastatic Urothelial Carcinoma. Clin Genitourin Cancer. 2015 doi: 10.1016/j.clgc.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Harada K, Ida S, Baba Y, Ishimoto T, Kosumi K, Tokunaga R, Izumi D, Ohuchi M, Nakamura K, Kiyozumi Y, Imamura Y, Iwatsuki M, Iwagami S, Miyamoto Y, Sakamoto Y, Yoshida N, Watanabe M, Baba H. Prognostic and clinical impact of sarcopenia in esophageal squamous cell carcinoma. Dis Esophagus. 2015 doi: 10.1111/dote.12381. [DOI] [PubMed] [Google Scholar]
- 21.Altun M, Besche HC, Overkleeft HS, Piccirillo R, Edelmann MJ, Kessler BM, Goldberg AL, Ulfhake B. Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J Biol Chem. 2010;285:39597–608. doi: 10.1074/jbc.M110.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–47. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 26.Willems AR, Goh T, Taylor L, Chernushevich I, Shevchenko A, Tyers M. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos Trans R Soc Lond B Biol Sci. 1999;354:1533–50. doi: 10.1098/rstb.1999.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, Pan ZQ. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–93. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–67. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 29.Deshaies RJ. Structural biology: Corralling a protein-degradation regulator. Nature. 2014;512:145–6. doi: 10.1038/nature13644. [DOI] [PubMed] [Google Scholar]
- 30.Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan SO, Deshaies RJ. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–15. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun. 2013;4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zemla A, Thomas Y, Kedziora S, Knebel A, Wood NT, Rabut G, Kurz T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- 33.Price SR, Du JD, Bailey JL, Mitch WE. Molecular mechanisms regulating protein turnover in muscle. Am J Kidney Dis. 2001;37:S112–S114. doi: 10.1053/ajkd.2001.20764. [DOI] [PubMed] [Google Scholar]
- 34.Attaix D, Mosoni L, Dardevet D, Combaret L, Mirand PP, Grizard J. Altered responses in skeletal muscle protein turnover during aging in anabolic and catabolic periods. Int J Biochem Cell Biol. 2005;37:1962–73. doi: 10.1016/j.biocel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Attaix D, Combaret L, Bechet D, Taillandier D. Role of the ubiquitin-proteasome pathway in muscle atrophy in cachexia. Curr Opin Support Palliat Care. 2008;2:262–6. doi: 10.1097/spc.0b013e3283196ac2. [DOI] [PubMed] [Google Scholar]
- 36.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–8. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 37.Adams V, Linke A, Wisloff U, Doring C, Erbs S, Krankel N, Witt CC, Labeit S, Muller-Werdan U, Schuler G, Hambrecht R. Myocardial expression of Murf-1 and MAFbx after induction of chronic heart failure: Effect on myocardial contractility. Cardiovasc Res. 2007;73:120–9. doi: 10.1016/j.cardiores.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 38.Bdolah Y, Segal A, Tanksale P, Karumanchi SA, Lecker SH. Atrophy-related ubiquitin ligases atrogin-1 and MuRF-1 are associated with uterine smooth muscle involution in the postpartum period. Am J Physiol Regul Integr Comp Physiol. 2007;292:R971–R976. doi: 10.1152/ajpregu.00617.2006. [DOI] [PubMed] [Google Scholar]
- 39.Frolov A, Chahwan S, Ochs M, Arnoletti JP, Pan ZZ, Favorova O, Fletcher J, von Mehren M, Eisenberg B, Godwin AK. Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol Cancer Ther. 2003;2:699–709. [PubMed] [Google Scholar]
- 40.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–5. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tisdale MJ. Molecular pathways leading to cancer cachexia. Physiology (Bethesda ) 2005;20:340–8. doi: 10.1152/physiol.00019.2005. [DOI] [PubMed] [Google Scholar]
- 42.Clavel S, Siffroi-Fernandez S, Coldefy AS, Boulukos K, Pisani DF, Derijard B. Regulation of the intracellular localization of Foxo3a by stress-activated protein kinase signaling pathways in skeletal muscle cells. Mol Cell Biol. 2010;30:470–80. doi: 10.1128/MCB.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Llovera M, Garcia-Martinez C, Lopez-Soriano J, Agell N, Lopez-Soriano FJ, Garcia I, Argiles JM. Protein turnover in skeletal muscle of tumour-bearing transgenic mice overexpressing the soluble TNF receptor-1. Cancer Lett. 1998;130:19–27. doi: 10.1016/s0304-3835(98)00137-2. [DOI] [PubMed] [Google Scholar]
- 44.Karayiannakis AJ, Syrigos KN, Polychronidis A, Pitiakoudis M, Bounovas A, Simopoulos K. Serum levels of tumor necrosis factor-alpha and nutritional status in pancreatic cancer patients. Anticancer Res. 2001;21:1355–8. [PubMed] [Google Scholar]
- 45.Duell EJ, Casella DP, Burk RD, Kelsey KT, Holly EA. Inflammation, genetic polymorphisms in proinflammatory genes TNF-A, RANTES, and CCR5, and risk of pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:726–31. doi: 10.1158/1055-9965.EPI-05-0797. [DOI] [PubMed] [Google Scholar]
- 46.Pessina GP, Paulesu L, Corradeschi F, Luzzi E, Tanzini M, Aldinucci C, Di Stefano A, Bocci V. Chronic cigarette smoking enhances spontaneous release of tumour necrosis factor-alpha from alveolar macrophages of rats. Mediators Inflamm. 1993;2:423–8. doi: 10.1155/S0962935193000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cawthorn WP, Heyd F, Hegyi K, Sethi JK. Tumour necrosis factor-alpha inhibits adipogenesis via a beta-catenin/TCF4(TCF7L2)-dependent pathway. Cell Death Differ. 2007;14:1361–73. doi: 10.1038/sj.cdd.4402127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llovera M, Garcia-Martinez C, Agell N, Lopez-Soriano FJ, Argiles JM. TNF can directly induce the expression of ubiquitin-dependent proteolytic system in rat soleus muscles. Biochem Biophys Res Commun. 1997;230:238–41. doi: 10.1006/bbrc.1996.5827. [DOI] [PubMed] [Google Scholar]
- 49.Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–6. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 50.Ouyang S, Hsuchou H, Kastin AJ, Pan W. TNF stimulates nuclear export and secretion of IL-15 by acting on CRM1 and ARF6. PLoS One. 2013;8:e69356. doi: 10.1371/journal.pone.0069356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Hernandez PL, Hernanz-Macias A, Gomez-Candela C, Grande-Aragon C, Feliu-Batlle J, Castro-Carpeno J, Martinez-Munoz I, Zurita-Rosa L, Villarino-Sanz M, Prados-Sanchez C, Sanchez Garcia-Giron J. Serum interleukin-15 levels in cancer patients with cachexia. Oncol Rep. 2012;28:1443–52. doi: 10.3892/or.2012.1928. [DOI] [PubMed] [Google Scholar]
- 52.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol. 2001;306:717–26. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 53.Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab. 2014;307:E469–E484. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J Cell Biol. 2002;157:125–36. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McElhinny AS, Perry CN, Witt CC, Labeit S, Gregorio CC. Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J Cell Sci. 2004;117:3175–88. doi: 10.1242/jcs.01158. [DOI] [PubMed] [Google Scholar]
- 56.Malyukova A, Brown S, Papa R, O’Brien R, Giles J, Trahair TN, Dalla Pozza L, Sutton R, Liu T, Haber M, Norris MD, Lock RB, Sangfelt O, Marshall GM. FBXW7 regulates glucocorticoid response in T-cell acute lymphoblastic leukaemia by targeting the glucocorticoid receptor for degradation. Leukemia. 2013;27:1053–62. doi: 10.1038/leu.2012.361. [DOI] [PubMed] [Google Scholar]
- 57.Li Z, Xiao J, Hu K, Wang G, Li M, Zhang J, Cheng G. FBXW7 acts as an independent prognostic marker and inhibits tumor growth in human osteosarcoma. Int J Mol Sci. 2015;16:2294–306. doi: 10.3390/ijms16022294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bodine SC, Furlow JD. Glucocorticoids and Skeletal Muscle. Adv Exp Med Biol. 2015;872:145–76. doi: 10.1007/978-1-4939-2895-8_7. [DOI] [PubMed] [Google Scholar]
- 59.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280:2737–44. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 61.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–5. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R, Sneddon S, Hulit J, Howell A, Lisanti MP. Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle. 2012;11:4390–401. doi: 10.4161/cc.22777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taguchi A, Delgado O, Celiktas M, Katayama H, Wang H, Gazdar AF, Hanash SM. Proteomic signatures associated with p53 mutational status in lung adenocarcinoma. Proteomics. 2014;14:2750–9. doi: 10.1002/pmic.201400378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cannavino J, Brocca L, Sandri M, Bottinelli R, Pellegrino MA. PGC1-alpha over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. J Physiol. 2014;592:4575–89. doi: 10.1113/jphysiol.2014.275545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang K, Wagner PD, Breen EC. TNF-alpha-mediated reduction in PGC-1alpha may impair skeletal muscle function after cigarette smoke exposure. J Cell Physiol. 2010;222:320–7. doi: 10.1002/jcp.21955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280:2737–44. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 67.Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, Lanza IR, Rasbach KA, Okutsu M, Nair KS, Yan Z, Leinwand LA, Spiegelman BM. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–31. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrison BC, Roberts CR, Hood DB, Sweeney M, Gould JM, Bush EW, McKinsey TA. The CRM1 nuclear export receptor controls pathological cardiac gene expression. Mol Cell Biol. 2004;24:10636–49. doi: 10.1128/MCB.24.24.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang JS, Huypens P, Zhang Y, Black C, Kralli A, Gettys TW. Regulation of NT-PGC-1alpha subcellular localization and function by protein kinase A-dependent modulation of nuclear export by CRM1. J Biol Chem. 2010;285:18039–50. doi: 10.1074/jbc.M109.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–75. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shuman JD, Sebastian T, Kaldis P, Copeland TD, Zhu S, Smart RC, Johnson PF. Cell cycle-dependent phosphorylation of C/EBPbeta mediates oncogenic cooperativity between C/EBPbeta and H-RasV12. Mol Cell Biol. 2004;24:7380–91. doi: 10.1128/MCB.24.17.7380-7391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sebastian T, Malik R, Thomas S, Sage J, Johnson PF. C/EBPbeta cooperates with RB:E2F to implement Ras(V12)-induced cellular senescence. EMBO J. 2005;24:3301–12. doi: 10.1038/sj.emboj.7600789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sebastian T, Johnson PF. Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle. 2006;5:953–7. doi: 10.4161/cc.5.9.2733. [DOI] [PubMed] [Google Scholar]
- 74.Penner G, Gang G, Sun X, Wray C, Hasselgren PO. C/EBP DNA-binding activity is upregulated by a glucocorticoid-dependent mechanism in septic muscle. Am J Physiol Regul Integr Comp Physiol. 2002;282:R439–R444. doi: 10.1152/ajpregu.00512.2001. [DOI] [PubMed] [Google Scholar]
- 75.Yang H, Mammen J, Wei W, Menconi M, Evenson A, Fareed M, Petkova V, Hasselgren PO. Expression and activity of C/EBPbeta and delta are upregulated by dexamethasone in skeletal muscle. J Cell Physiol. 2005;204:219–26. doi: 10.1002/jcp.20278. [DOI] [PubMed] [Google Scholar]
- 76.Gonnella P, Alamdari N, Tizio S, Aversa Z, Petkova V, Hasselgren PO. C/EBPbeta regulates dexamethasone-induced muscle cell atrophy and expression of atrogin-1 and MuRF1. J Cell Biochem. 2011;112:1737–48. doi: 10.1002/jcb.23093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma M, Juvvuna PK, Kukreti H, McFarlane C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front Physiol. 2014;5:239. doi: 10.3389/fphys.2014.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wada S, Kato Y, Okutsu M, Miyaki S, Suzuki K, Yan Z, Schiaffino S, Asahara H, Ushida T, Akimoto T. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem. 2011;286:38456–65. doi: 10.1074/jbc.M111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hudson MB, Woodworth-Hobbs ME, Zheng B, Rahnert JA, Blount MA, Gooch JL, Searles CD, Price SR. miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol. 2014;306:C551–C558. doi: 10.1152/ajpcell.00266.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu J, Li R, Workeneh B, Dong Y, Wang X, Hu Z. Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 2012;82:401–11. doi: 10.1038/ki.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kukreti H, Amuthavalli K, Harikumar A, Sathiyamoorthy S, Feng PZ, Anantharaj R, Tan SL, Lokireddy S, Bonala S, Sriram S, McFarlane C, Kambadur R, Sharma M. Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J Biol Chem. 2013;288:6663–78. doi: 10.1074/jbc.M112.390369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song DW, Ryu JY, Kim JO, Kwon EJ, Kim do H. The miR-19a/b family positively regulates cardiomyocyte hypertrophy by targeting atrogin-1 and MuRF-1. Biochem J. 2014;457:151–62. doi: 10.1042/BJ20130833. [DOI] [PubMed] [Google Scholar]
- 83.Zhang J, Fu SL, Liu Y, Liu YL, Wang WJ. Analysis of MicroRNA Expression Profiles in Weaned Pig Skeletal Muscle after Lipopolysaccharide Challenge. Int J Mol Sci. 2015;16:22438–55. doi: 10.3390/ijms160922438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–49. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azmi AS. The evolving role of nuclear transporters in cancer. Semin Cancer Biol. 2014;27:1–2. doi: 10.1016/j.semcancer.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 87.Buck M, Zhang L, Halasz NA, Hunter T, Chojkier M. Nuclear export of phosphorylated C/EBPbeta mediates the inhibition of albumin expression by TNF-alpha. EMBO J. 2001;20:6712–23. doi: 10.1093/emboj/20.23.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Azmi AS, Aboukameel A, Bao B, Sarkar FH, Philip PA, Kauffman M, Shacham S, Mohammad RM. Selective inhibitors of nuclear export block pancreatic cancer cell proliferation and reduce tumor growth in mice. Gastroenterology. 2013;144:447–56. doi: 10.1053/j.gastro.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao J, Azmi AS, Aboukameel A, Kauffman M, Shacham S, Abou-Samra AB, Mohammad RM. Nuclear retention of Fbw7 by specific inhibitors of nuclear export leads to Notch1 degradation in pancreatic cancer. Oncotarget. 2014;5:3444–54. doi: 10.18632/oncotarget.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Azmi AS, Muqbil I, Wu J, Aboukameel A, Senapedis W, Baloglu E, Bollig-Fischer A, Dyson G, Kauffman M, Landesman Y, Shacham S, Philip PA, Mohammad RM. Targeting the Nuclear Export Protein XPO1/CRM1 Reverses Epithelial to Mesenchymal Transition. Sci Rep. 2015;5:16077. doi: 10.1038/srep16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, Diaz VM. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014;42:1079–94. doi: 10.1093/nar/gkt935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia JM, Montserrat-Sentis B, Baulida J, Bonilla F, de Herreros AG, Diaz VM. The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J Biol Chem. 2010;285:3794–805. doi: 10.1074/jbc.M109.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Azmi Asfar S, Aboukameel Amro, Carlson Robert O, Elloul Sivan, Shacham Sharon, Kauffman Michael, Frenkel Ran, Mohammad Ramzi M. Abstract 1756: Preclinical activity in non-Hodgkin’s lymphoma of Selinexor a selective inhibitor of nuclear export (SINE) is enhanced through combination with standard-of-care therapies. Cancer Research. 2015;75:1756. [Google Scholar]
- 94.Tawa NE, Jr, Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fischer D, Gang G, Pritts T, Hasselgren PO. Sepsis-induced muscle proteolysis is prevented by a proteasome inhibitor in vivo. Biochem Biophys Res Commun. 2000;270:215–21. doi: 10.1006/bbrc.2000.2398. [DOI] [PubMed] [Google Scholar]
- 96.Combaret L, Ralliere C, Taillandier D, Tanaka K, Attaix D. Manipulation of the ubiquitin-proteasome pathway in cachexia: pentoxifylline suppresses the activation of 20S and 26S proteasomes in muscles from tumor-bearing rats. Mol Biol Rep. 1999;26:95–101. doi: 10.1023/a:1006955832323. [DOI] [PubMed] [Google Scholar]
- 97.Garcia JM, Polvino WJ. Pharmacodynamic hormonal effects of anamorelin, a novel oral ghrelin mimetic and growth hormone secretagogue in healthy volunteers. Growth Horm IGF Res. 2009;19:267–73. doi: 10.1016/j.ghir.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 98.Farid M, Reid MB, Li YP, Gerken E, Durham WJ. Effects of dietary curcumin or N-acetylcysteine on NF-kappaB activity and contractile performance in ambulatory and unloaded murine soleus. Nutr Metab (Lond) 2005;2:20. doi: 10.1186/1743-7075-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin B, Li YP. Curcumin prevents lipopolysaccharide-induced atrogin-1/MAFbx upregulation and muscle mass loss. J Cell Biochem. 2007;100:960–9. doi: 10.1002/jcb.21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siddiqui RA, Hassan S, Harvey KA, Rasool T, Das T, Mukerji P, DeMichele S. Attenuation of proteolysis and muscle wasting by curcumin c3 complex in MAC16 colon tumour-bearing mice. Br J Nutr. 2009;102:967–75. doi: 10.1017/S0007114509345250. [DOI] [PubMed] [Google Scholar]
- 101.Shuto T, Ono T, Ohira Y, Shimasaki S, Mizunoe S, Watanabe K, Suico MA, Koga T, Sato T, Morino S, Sato K, Kai H. Curcumin decreases toll-like receptor-2 gene expression and function in human monocytes and neutrophils. Biochem Biophys Res Commun. 2010;398:647–52. doi: 10.1016/j.bbrc.2010.06.126. [DOI] [PubMed] [Google Scholar]
- 102.Niu M, Wu S, Mao L, Yang Y. CRM1 is a cellular target of curcumin: new insights for the myriad of biological effects of an ancient spice. Traffic. 2013;14:1042–52. doi: 10.1111/tra.12090. [DOI] [PubMed] [Google Scholar]
- 103.Shadfar S, Couch ME, McKinney KA, Weinstein LJ, Yin X, Rodriguez JE, Guttridge DC, Willis M. Oral resveratrol therapy inhibits cancer-induced skeletal muscle and cardiac atrophy in vivo. Nutr Cancer. 2011;63:749–62. doi: 10.1080/01635581.2011.563032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paffett ML, Lucas SN, Campen MJ. Resveratrol reverses monocrotaline-induced pulmonary vascular and cardiac dysfunction: a potential role for atrogin-1 in smooth muscle. Vascul Pharmacol. 2012;56:64–73. doi: 10.1016/j.vph.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castillero E, Alamdari N, Lecker SH, Hasselgren PO. Suppression of atrogin-1 and MuRF1 prevents dexamethasone-induced atrophy of cultured myotubes. Metabolism. 2013;62:1495–502. doi: 10.1016/j.metabol.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 106.Wang DT, Yin Y, Yang YJ, Lv PJ, Shi Y, Lu L, Wei LB. Resveratrol prevents TNF-alpha-induced muscle atrophy via regulation of Akt/mTOR/FoxO1 signaling in C2C12 myotubes. Int Immunopharmacol. 2014;19:206–13. doi: 10.1016/j.intimp.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 107.Woo SR, Byun JG, Kim YH, Park ER, Joo HY, Yun M, Shin HJ, Kim SH, Shen YN, Park JE, Park GH, Lee KH. SIRT1 suppresses cellular accumulation of beta-TrCP E3 ligase via protein degradation. Biochem Biophys Res Commun. 2013;441:831–7. doi: 10.1016/j.bbrc.2013.10.146. [DOI] [PubMed] [Google Scholar]
- 108.Boehm K, Borrelli F, Ernst E, Habacher G, Hung SK, Milazzo S, Horneber M. Green tea (Camellia sinensis) for the prevention of cancer. Cochrane Database Syst Rev. 2009:CD005004. doi: 10.1002/14651858.CD005004.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boehm K. Green tea for the prevention of cancer. J Evid Based Med. 2010;3:53. doi: 10.1111/j.1756-5391.2010.01063.x. [DOI] [PubMed] [Google Scholar]
- 110.Dou QP, Landis-Piwowar KR, Chen D, Huo C, Wan SB, Chan TH. Green tea polyphenols as a natural tumour cell proteasome inhibitor. Inflammopharmacology. 2008;16:208–12. doi: 10.1007/s10787-008-8017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen M, Chan TH, Dou QP. Targeting tumor ubiquitin-proteasome pathway with polyphenols for chemosensitization. Anticancer Agents Med Chem. 2012;12:891–901. doi: 10.2174/187152012802649978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang HC, Way TD, Lin CL, Lin JK. EGCG stabilizes p27kip1 in E2-stimulated MCF-7 cells through down-regulation of the Skp2 protein. Endocrinology. 2008;149:5972–83. doi: 10.1210/en.2008-0408. [DOI] [PubMed] [Google Scholar]
- 113.Wang H, Lai YJ, Chan YL, Li TL, Wu CJ. Epigallocatechin-3-gallate effectively attenuates skeletal muscle atrophy caused by cancer cachexia. Cancer Lett. 2011;305:40–9. doi: 10.1016/j.canlet.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 114.Hemdan DI, Hirasaka K, Nakao R, Kohno S, Kagawa S, Abe T, Harada-Sukeno A, Okumura Y, Nakaya Y, Terao J, Nikawa T. Polyphenols prevent clinorotation-induced expression of atrogenes in mouse C2C12 skeletal myotubes. J Med Invest. 2009;56:26–32. doi: 10.2152/jmi.56.26. [DOI] [PubMed] [Google Scholar]