Abstract
Left-right (LR) asymmetry is a biologically conserved property in living organisms that can be observed in the asymmetrical arrangement of organs and tissues and in tissue morphogenesis, such as the directional looping of the gastrointestinal tract and heart. The expression of LR asymmetry in embryonic tissues can be appreciated in biased cell alignment. Previously an in vitro chirality assay was reported by patterning multiple cells on microscale defined geometries and quantified the cell phenotype–dependent LR asymmetry, or cell chirality. However, morphology and chirality of individual cells on micropatterned surfaces has not been well characterized. Here, a Python-based algorithm was developed to identify and quantify immunofluorescence stained individual epithelial cells on multicellular patterns. This approach not only produces results similar to the image intensity gradient-based method reported previously, but also can capture properties of single cells such as area and aspect ratio. We also found that cell nuclei exhibited biased alignment. Around 35% cells were misaligned and were typically smaller and less elongated. This new imaging analysis approach is an effective tool for measuring single cell chirality inside multicellular structures and can potentially help unveil biophysical mechanisms underlying cellular chiral bias both in vitro and in vivo.
Keywords: Cell Alignment, Nucleus Alignment, Micropatterning, Intensity Gradient, Cell Morphology
INTRODUCTION
Left-right (LR) asymmetry, or chirality, is a well conserved phenomenon in the nature, from dextral and sinistral helices in the shells of snails17, LR asymmetry of the internal organs and the central nervous system of mammals11, 14, to the significant right handed bias in the rotation of climbing plants during twining6. Deviations of normal asymmetry often lead to birth defects such as situs ambiguous, where one or two the internal organs are in mirrored position31. In developmental biology, using animal models such as chick and mouse, several models have been proposed to explain the establishment of LR asymmetry, such as the primary cilium theory15 and voltage gradient theory11, 12. While animal models provide many associations with signaling pathways related to LR asymmetry, well-controlled in vitro approaches and automated chirality characterization are necessary to illustrate intracellular biophysical mechanisms underlying the LR breaking in embryonic development.
Recently, several studies have shown that chirality occurs at the cellular level. With live cell fluorescence imaging, Xu et al. demonstrated that neutrophils exhibited biased migration towards the left side of the centrosome-nucleus axis29. Wan et al. used a cell micropatterning technique, analyzed still phase contrast images of patterned cells, and found that cells have an innate, distinct chiral bias, which is dependent of cell phenotype and mediated by actin24–26. The scale of chiral bias of epithelial cells was shown to depend on cell-cell adhesion28. Cells lost their distinct chirality when exposed to carbon nanotubes due to oxidative stress, suggesting a potential application of cell chirality as a biomarker for detecting cell toxicity18. Chen et al. reported a similar cell chiral alignment with vascular mesenchymal cells at micropatterned boundaries3. On the embryonic level, Taniguchi et al. showed that chiral biases in cell shape regulate asymmetric looping of the Drosophila hindgut by analyzing the angle of cell edges in epithelial sheets8, 21. The authors illustrated that wild-type hindgut epithelial cells have a left-handed bias and demonstrated that the left-handed rotation of the hindgut was caused simply by cell-shape chirality through computer simulation.
Collective cell chiral behavior requires automated, detailed analyses on the cellular level. Previously Wan et al. developed a simple MatLab-based automated cell chirality detection method25, based on the detection of the intensity gradient of phase contrast images of the cells9. The cell has a contour brighter than the interior region. The spatial intensity gradients, determined pixel-by-pixel, were used to define local dominant direction of sub-regions (typically squares) of an arbitrary size from the image, using an accumulator scheme. The other method of chirality analysis was reported by Taniguchi et al., in which the orientations of edges of all cells were measured. The biased angle was defined as the angle between the edge and the vertical (“anterior-posterior”) axis in the image. In the Drosophila epithelial hindgut, cell alignment was determined to bias towards the left side21.
None of these methods, however, are based on individual cells and as a result analysis of cell-cell and cell-boundary interactions is not possible, although they are important for this collective cell phenomenon. In this study, we aim to develop an individual cell-based approach to quantify the chirality of each cell on micropatterned geometries. We believe that such high content analysis will provide deep insights of the biophysical mechanisms of collective chiral morphogenesis on micropatterned surfaces as well as in LR symmetry breaking during embryonic development.
MATERIALS AND METHODS
Microcontact Printing
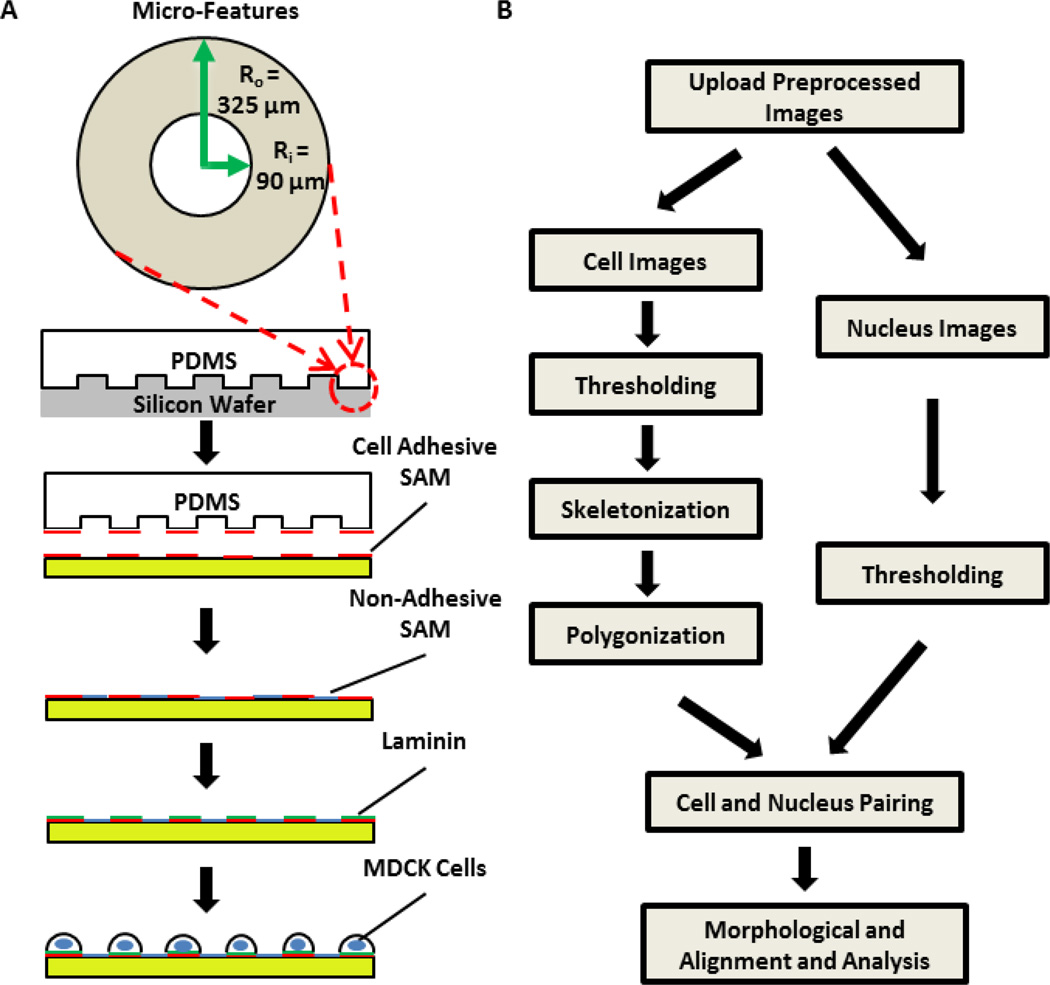
Microcontact printing was performed using polydimethylsiloxane (PDMS) stamps and self-assembled monolayers (SAMs) as previously described23, 25. The procedure is highlighted in Figure 1A. Micropatterned rings have an inner diameter of 90 µm and a width of 235 µm.
Figure 1.
Schematic of micro-contact printing of cells for measuring cell chirality (A) and the flow chart of imaging processing (B). (A) An array of ring micro-features was copied from a silicon master wafer (fabricated through lithography) onto PDMS elastomeric stamps by casting with pre-polymers on the mold. Cell adhesive self-assembled monolayers (SAMs) were transferred from the PDMS stamp onto a gold coated glass slide, which was subsequently treated with a non-adhesive SAM. Laminin was then adsorbed to the adhesive SAM layer and cells seeded. After cells attached, extra cells were washed. Cells were cultured till confluency before fixation and immunofluorescence imaging. (B) Fluorescence images of cells and nuclei were first pre-processed to reduce noises. Cell images (left) were then thresholded to remove artifacts, skeletonized, and polygonalized by identifying nodal points and associated edges in the images, while nuclei images (right) are simply thresholded. Cells and nuclei in the images were paired based upon distance of centroids. Paired cells and nuclei are then analyzed for morphological features and alignment angles.
Cell Culture
Madin-Darby canine kidney (MDCK) epithelial cells were maintained in the Dulbecco's modified Eagle's medium (DMEM) with High Glucose (Life Technologies) with 10% fetal bovine serum (FBS), 1mM sodium pyruvate, and 1% penicillin-streptomycin. Cells were passaged using Trypsin-EDTA and seeded onto patterned surfaces. Once attached, extra cells are removed by washing with culture medium.
Immunofluorescence Imaging
Micropatterned cells were cultured to confluency over 24–36 hours. Cells were fixed with 4% paraformaldehyde and stained for tight junctions with ZO-1 antibody and nuclei with 4',6-diamidino-2-phenylindole (DAPI). Phase contrast and fluorescence images of cells and nuclei were taken for morphological and alignment analyses. The algorithm is described below and also schematically outlined in Figure 1B.
Image Pre-Processing
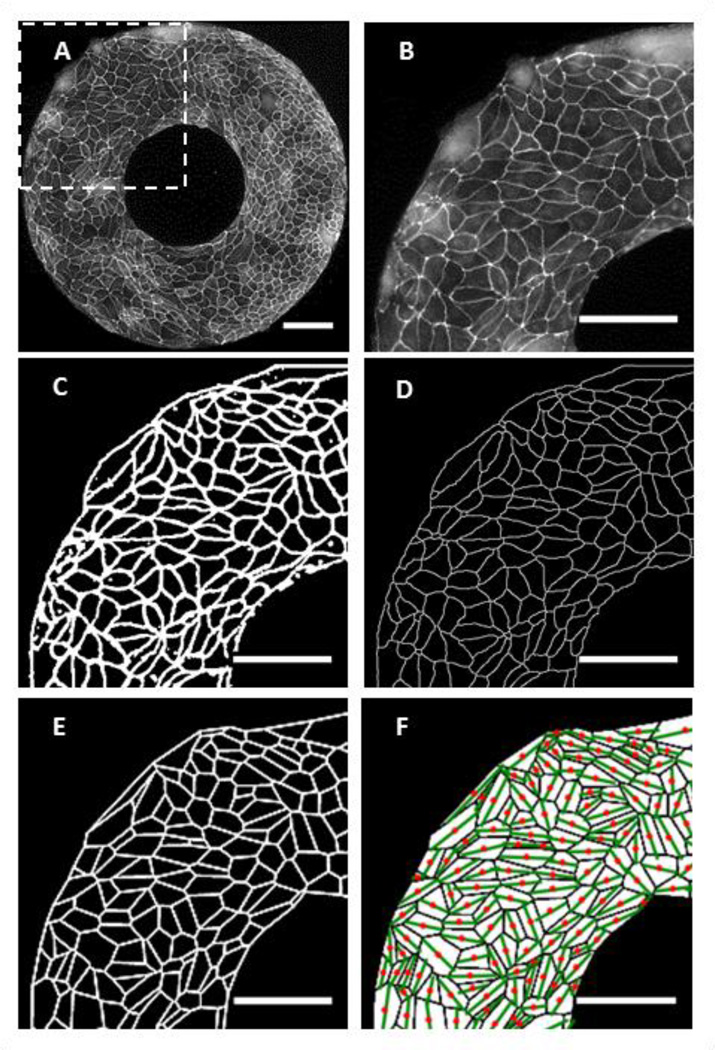
Image processing steps are highlighted visually in Figure 2. Using ImageJ, ZO-1 images of cells (Figure 2A–B) were adjusted for brightness to highlight cells. To remove smooth continuous backgrounds of the fluorescence image, background subtraction from ImageJ was used with a rolling ball radius of 20 pixels, as previously reported2. Images were smoothed with a medium filter of a 3 pixel width. The images were then binarized (Figure 2C) and for discontinuities using the brush tool. Finally, the images were inverted to a black background with white features and a despeckling algorithm was used to remove remaining artifacts such as small dots. Nuclei images were run through a similar protocol.
Figure 2.
Representative images during cellular image processing. Scale bars = 100 µm. (A) Tight junction ZO-1 stained image of MDCK cells. (B) A highlighted region as indicated by the white box in (A). (C) The cell image is preprocessed to correct for discontinuities, highlight edges, and to remove artifacts. (D) The preprocessed image is skeletonized with cell edges having only a single pixel width. (E) A polygonization image is generated by identifying nodes and edges with a nearest neighbor approach in order to simplify analysis. (F) Cell alignment is indicated by green lines over cell centroids (red dots) on all labeled cells.
Algorithm Development and Pipeline
The Python-based algorithm was developed to further segment and analyze cells and nuclei in the pre-processed images for quantifying both morphological and chiral features. Pre-processed cell images were first doubled in size. The images were then further run through a watershed algorithm and skeletonized, using the SimpleITK library for Python 2.7.613 (Figure 2D). Images were then converted into a binary array and run through a nearest neighbor protocol, using the Numpy library16. This protocol analyzes all the elements in the array and identifies all the “on” pixels, i.e., those that have a value of 1 rather than 0, which are designated as “off” pixels. These “on” pixels are further grouped in order to find pixels with more than two neighbors, which are termed as “nodes”. Nodes are associated with each other through pixels connecting them and paired to designate edges. Edges were simplified to straight edges. OpenCV, an image drawing library, can be utilized to generate the polygonized image for visualization or storage1, which consists of nodes and straight-line approximation of edges. From these saved polygonized images, individual cells and their edges could be easily identified through a connected component protocol in the SimpleTIK library for determining morphological features and alignment angles as described below (Figure 2E).
Pre-processed nuclei images were simply processed through thresholding. Images were analyzed for chiral and morphological features. Cells and nuclei are paired. Cells that either had multiple nuclei or no nuclei were filtered out. Potential cell objects with no nuclei are likely regions that are not occupied by individual cells, while objects with multiple nuclei may represent proliferating cells or improperly segmented cells. By pairing cells and nuclei, we ensure that the objects analyzed are indeed the individual cells and their single nuclei.
Determination of Chiral and Morphological Features
The major morphological features for both cells and nuclei were calculated using the regionprops function in the scikit-image library22. These features include cell and nucleus label area, centroid, major and minor axes, and perimeter. From these parameters, additional morphological features, such as alignment angle, aspect ratio and shape indices were also derived (Figure 2F).
Chiral Alignment Definitions
Alignment angles that were between −90° and −5° were binned as Clockwise (CW), whereas angles that were between 5° and 90° were binned as Counterclockwise (CCW). Angles that were between −5° and 5° are binned as Non-Chiral (NC). All graphs were with respect to the percentage of cells or nuclei.
Comparison with Previous Approaches
The current approach was compared with previous approaches for the bias in angles. The first approach was an intensity gradient approach, reported by Wan et al., in which images were broken into sub-regions and the local intensity magnitudes and orientations were calculated. The dominant orientation was determined in each sub-region and the overall distribution bias was calculated using a von Moses distribution25. The second approach was an edge-based method, reported by Taniguchi et al., in which the angles of the edges were considered21. This concept was modified to calculate angles in cylindrical coordinates in this study. Angles could either be biased left (−90° to −5°) or biased right (5° to 90°), and the bias of the image was calculated using a cumulative distribution function.
Statistical Analysis
All data is presented as mean ± standard error of mean. Alignment bias was determined using a Chi-squared test. The variation of cellular and nuclear morphological parameters with the radial distances was assessed using a one-way analysis of variance (ANOVA). Finally, differences between properly aligned and improperly aligned cells at various radial distances were determined using a two-way ANOVA. Fisher’s LSD post hoc tests were performed at a 95% confidence interval to determine statistical significance.
RESULTS
Current Approach Validation
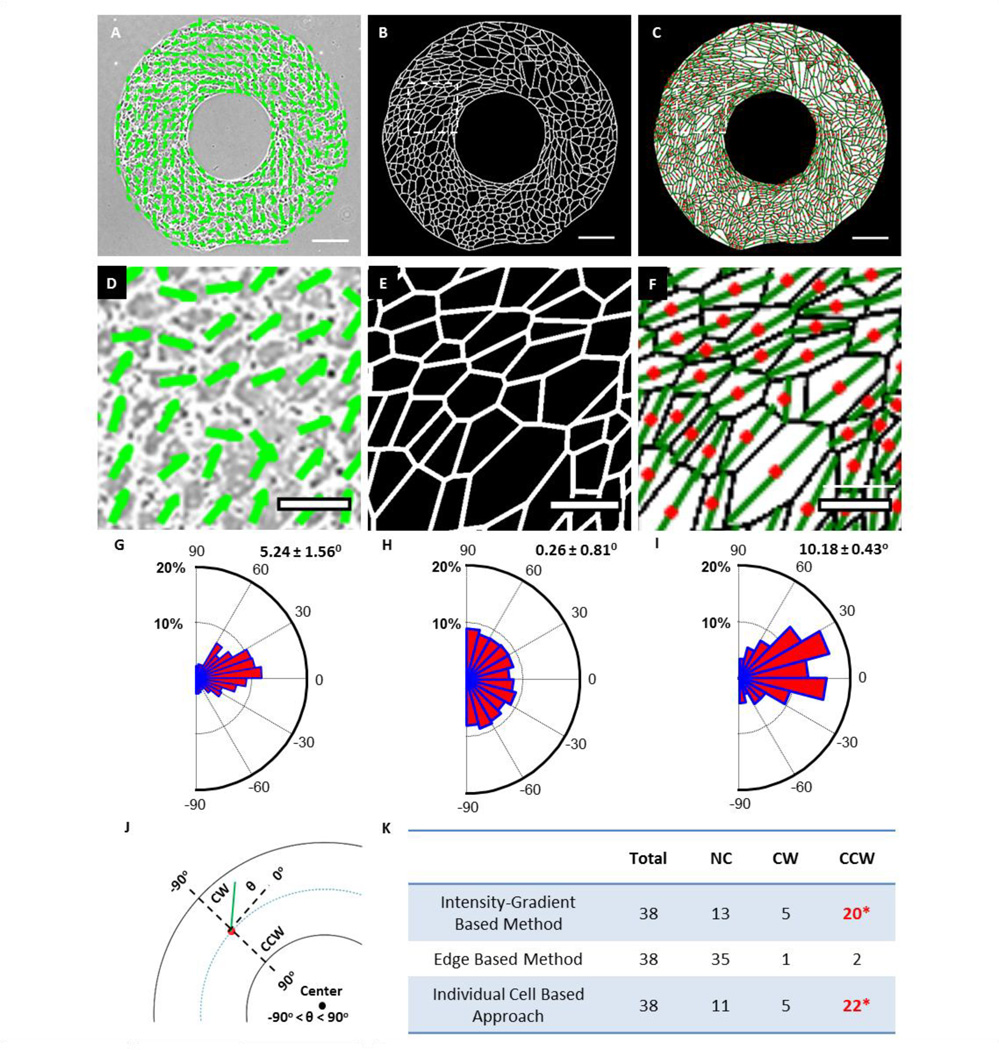
We first compared our results to those of previously published methods. With the intensity gradient-based analysis, the local dominant direction of cell alignment was determined as short green lines for each square sub-region, as seen in Figure 3A, D & G. As the size of sub-region can be chosen more or less arbitrarily, the percentage of calculated CCW rings ranges from 52.6% (20 out of 38) for a 35 µm by 35 µm sub-region, as reported in Figure 3K, to 47.4% (18 out of 38) for a 45 µm by 45 µm sub-region (data not shown). With the edge-based method, the chirality was determined by comparing CW and CCW aligned edges, as highlighted in Figure 3B, E & H. With this method, we failed to detect any significance in chiral alignment in these rings. Finally, with our individual cell-based analysis, demonstrated in Figure 3C, F & I, 57.9% of the rings were CCW, which is comparable to the results from the intensity gradient-based method in Figure 3K. The average alignment angles, as defined in Figure 3J, were 5.24 ± 1.6° (mean ± standard error) for the intensity-gradient technique (Figure 3G) and 10.2 ± 0.4° for the individual cell method (Figure 3I). The distribution of alignment angles and significance of bias for each image, using the individual cell-based method, is demonstrated in Supplementary Material, Table S1.
Figure 3.
Comparison of individual cell based approach with two previously reported approaches. Scale bars = 100 µm. (A–C) Analysis output images from the intensity-gradient method, edge-based method, and the current individual cell based approach, respectively. (D–F) Zoom-in images as indicated by white dashed line boxes in (A–C). (G–I) Rose diagrams of biased angles from 38 rings, according to the definition in (J), where alignment angle θ (green line) is valued relative to the circumferential direction. Red dot stands for cell centroids. (K) The calculated cell chirality of micropatterned rings based on the three methods mention above. *Significantly biased among chiral rings.
Although polygonization is often preferred for analyzing epithelial layer due to its great simplification, it may cause in inaccuracy in determining cell chirality. To evaluate the possible influence, we also performed the analysis without polygonization, we found that it produced similar results (Figure S1).
Nucleus Analysis Demonstrates Chirality
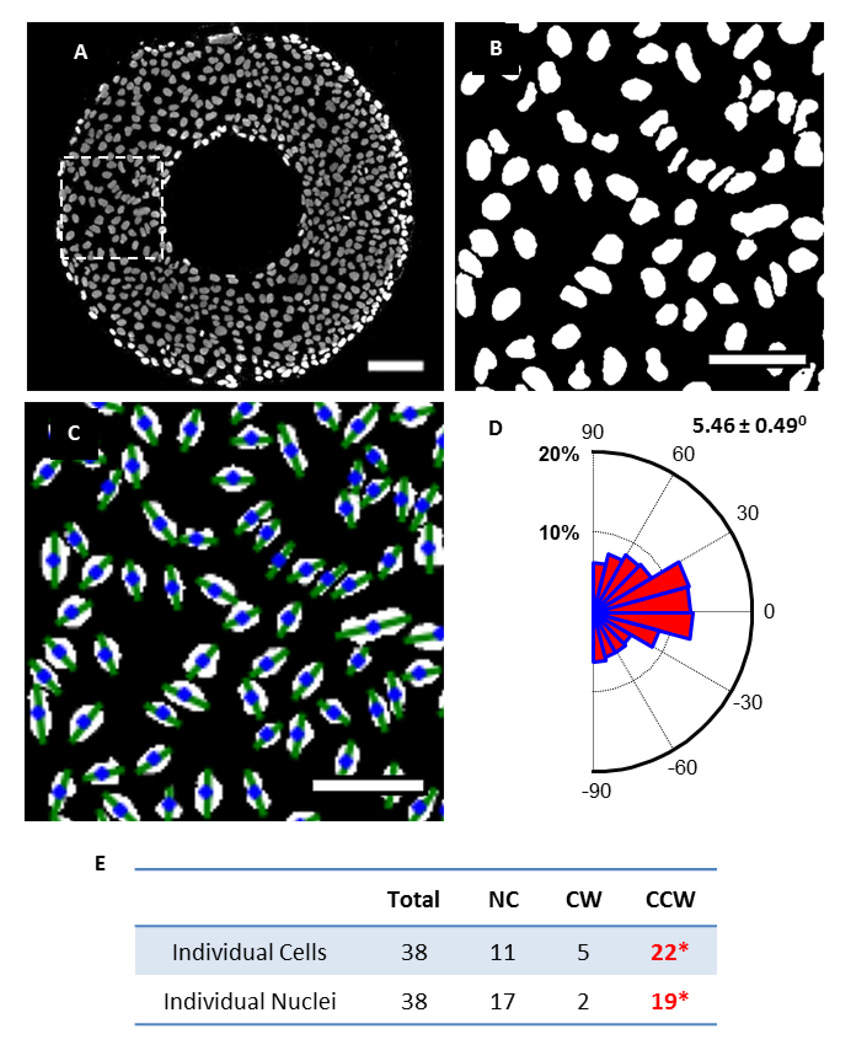
Nuclei also showed a biased alignment (Figure 4). The nuclei image (Figure 4A) corresponds to the ring in Figure 3, and the small dashed line box indicates the region of interest. Figure 4B–C highlights the thresholded image, and the labeled image with alignment angles for individual nuclei. The nuclei have a bias towards CCW alignments with a positive average angle of 5.5 ± 0.5° (Figure 4D). When compared to individual cell analysis, the nuclear alignment can also be used to determine the chirality of multicellular rings, as both demonstrate a similar bias, with 57.9% (22 out of 38) of the cellular rings demonstrated a cellular CCW bias, and 50% (19 out of 38) having a nuclear CCW bias. The distribution of alignment angles and significance of bias of nuclei for each image is demonstrated in Table S2.
Figure 4.
Nucleus chirality on micropatterned surfaces. (A) Nuclei image corresponding to cell image in Fig. 3. (scale bar = 100 µm). (B) Thresholded nucleus image of the region of interest as indicated by white dashed line box in (A) (scale bar = 25 µm). (C) Alignment angles (green lines) plotted over nucleus centroids (blue dots) (scale bar = 25 µm). (D) Rose diagram of nucleus alignment angle distribution showing a counterclockwise (CCW) bias found on 38 rings. (E) Comparison of individual cell and individual nucleus chirality on micropatterned surfaces. *Significantly biased among chiral rings.
Cells and nuclei were then analyzed for correlations between their respective alignment angles and morphological features (Figure S2). There was no strong correlation between cellular and nuclear aspect ratios. The areas, major axes, and alignment angles were highly correlated between cells and nuclei (R2 > 0.5).
Variation of Cellular and Nuclear Morphologies Over Radial Distance
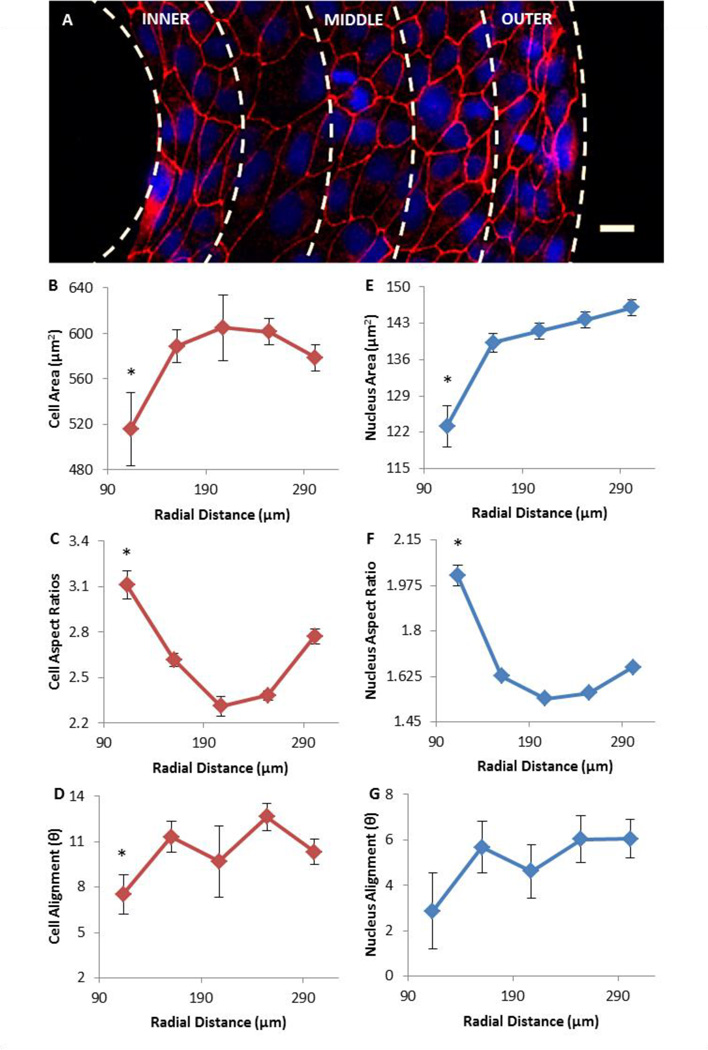
We next analyzed how alignment and morphology change between the inner and outer boundary of the ring patterns. Because CCW was dominant (57.9% of rings), only CCW rings were considered in this analysis (22 rings in total). Figure 5A highlights a region of the ring from the inner boundary to the outer boundary. The cells and nuclei are smallest at the inner boundary and cells are largest in the middle region of the ring, as confirmed in quantification in Figure 5B & E. The area of cells and nuclei at the inner boundary is 516 ± 30 µm2 and 123 ± 3 µm2 respectively. While cells reach a maximal size of 605 ± 11 µm2 in the middle of the ring, nuclei grow larger as they approach the outer boundary to 146 ± 2 µm2. The aspect ratios for cells and nuclei have a similar trend, in which they are largest at the inner boundary, 3.11 ± 0.09 and 2.02 ± 0.03 respectively, and smallest in the middle, 2.31 ± 0.05 and 1.54 ± 0.01 respectively, as demonstrated in Figure 5C and Figure 5F. Alignment angles show a general trend of increasing from the inner boundary towards the outer boundary as seen in Figure 5D and Figure 5G. Further, both cell and nuclei alignment angles share the same alignment trend and appear to have a CCW bias at all radial positions. All morphological features (Figure 5B–F) were found to be significantly different between inner and outer boundaries. In terms of alignment angles, a significant difference between two boundaries was only observed for the cells, but not for the nuclei.
Figure 5.
Morphological analyses of cells and nuclei with CCW bias. Error bars represent standard errors. (A) Image of a region of fluorescence labeled cells (Red: ZO-1 for tight junction) and nuclei (Blue: DAPI) on a CCW ring. White dashed lines indicate 5 regions over which data has been averaged (scale bar = 25 µm). (B–D) Average cell area, aspect ratio and alignment angle over radial distance of ring. (E–G) Average area, aspect ratio and alignment angle of cell nuclei over radial distance.
Misaligned Cells Are Morphologically Distinct
It was then asked whether cells that are misaligned in culture are morphologically distinct from the properly aligned cells. For this purpose, we analyzed individual cells on CCW rings (22 rings in total). Any cell that had an alignment angle that was defined as a CCW was taken to be a properly aligned cell and any cell that had a CW alignment angle was defined as a misaligned cell.
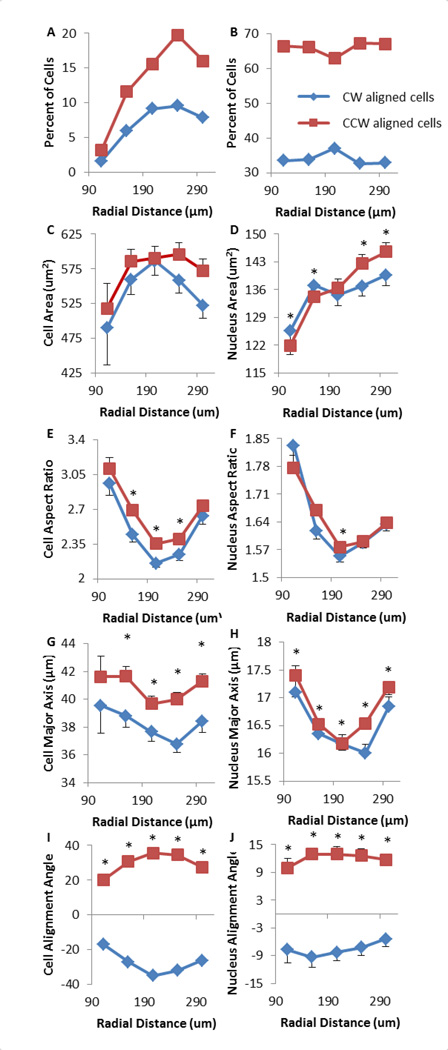
The percentage of misaligned cells is around 33–34% near two boundaries, and is slightly higher (37%) in the middle region (Figure 6A–B). The misaligned cells tend to be slightly smaller than (though not significantly different from) properly aligned cells (Figure 6C). This divergence in size is most significant as the cells begin to approach the outer boundary at 522 ± 19 µm2 for misaligned cells against 573 ± 14 µm2 for properly aligned ones. This trend is shared by the nuclei in Figure 6D, which demonstrate that misaligned cells have significantly smaller nuclei at the outer boundary (140 ± 3 µm2 versus 146 ± 2 µm2). The aspect ratio of misaligned cells is smaller in the interior regions, while it is similar at boundaries as shown in Figure 6E, and the trend is not obvious for the nuclei (Figure 6F). Similarly, the major axis length of the cell is significantly smaller for misaligned cells (Figure 6G–H), while that of the nucleus is similar between two groups. Finally, as expected, alignment angles are similar between properly aligned and misaligned cells, albeit it on opposite sides of 0°, with a larger magnitude found in the interior region compared to two boundaries (Figure 6I). It was also noted that nuclei align themselves similarly to the cells in Figure 6I as trends between aligned and misaligned cells are conserved in the nuclei. Taken together, this data suggested that misaligned cells are typically smaller and less elongated, and have a slightly higher occurrence rate in the interior region than at the boundaries.
Figure 6.
Misaligned cells (i.e., clockwise (CW) aligned cells on CCW rings) demonstrate altered morphology on micropatterned surfaces. Red squares represent properly aligned cells (i.e., CCW aligned cells on CCW rings) and nuclei, while blue diamonds are for misaligned cells. Error bars are standard errors. (A) Percentage of properly and misaligned cells over radial distance, with respect to the total number of cells on an entire patterned ring. (B) Percentage of properly and misaligned cells with respect to the number in each region. (C) Misaligned cells demonstrate smaller average areas. (D) Nuclei of misaligned cells are smaller near the outer border. (E) Misaligned cells show smaller aspect ratios. (F) Aspect ratios of nuclei of properly aligned vs. misaligned cells. (G) Misaligned cells have a shorter major axis. (H) The nucleus of misaligned cells is shorter in its major axis. (I) Cellular alignment angles of misaligned cells are opposite of those of properly aligned cells. (J) Nuclear alignment angles of misaligned cells are opposite to those of properly aligned cells.
DISCUSSION
In this study, a Python-based image processing algorithm has been developed for analyzing the chiral alignment of individual cells and nuclei, using an object labeling technique. We show that this new approach can produce similar but more reliable results compared to the previously reported intensity gradient-based method. In addition, cell nuclei are found to have a similar chirality as the cells. Finally, misaligned cells tend to be smaller and less elongated, when compared to properly aligned cells. Benefiting from the large amount of data retrieved from this algorithm, this system provides a unique insight into chiral morphogenesis of epithelial cells.
Current Approach Accurate and Output Intensive
The presented approach was demonstrated to be highly accurate in both visual validation and when compared to previous approaches, as seen in Figures 2–3. Results were similar to those generated by the intensity gradient based analysis, which have been used in several studies18, 25, 28, and the only slight deviation was in the final count of CCW against NC (non-chiral) rings.
When compared to the edge-based method, the individual cell-based method was more sensitive to the chiral alignment of the cells. One explanation is that the individual cell in vitro is defined to align along its major axis, which is closely associated with the direction of cell elongation, or the direction of longer edges. As a result, the orientation of short edges, which are typically not along the direction of the cell, is weighed less in the individual-cell based analysis. Therefore, it is much more sensitive when compared to the edge-based analysis in which all edges is treated equally.
The edge-based method worked for the images of in vivo gut development8, 21 because the cell morphology is quite different from what was observed on micropatterned surfaces. For gut development, the lengths of cell edges are all similar in a cell, and the direction of the entire cell is determined by the orientation of cell edges, which can be potentially captured by the edge-based method. As a result, the chirality of the cell can be measured as the chiral alignment of edges. For this case, the individual cell-based method should work as well, if not better.
It is worthy to point out that the current approach has several advantages over the intensity-gradient based method. First, this new approach considers the actual morphologies of individual cells rather than regions of the image9, which may need to be tuned for a particular cell type based on cell size and each sub-region can contain portions of multiple cells rather than an individual cell. Further, as seen in Figure 4, this approach is able to also analyze cell nuclei, which the previous approaches analyzed are unable to do. Finally, this approach allows for high-content analysis, as it generates a large amount of data for studying cell-cell and cell-boundary interactions and enables the exploration of biophysical mechanisms of this collective chiral morphogenic process of epithelial cells.
Nuclei Exhibit Chiral Alignments Similar to the Cells
Many studies demonstrated that when cells are aligned on topographical features, the nuclei also tend to take on those alignments4, 7, 19, 27. Here we provided the first evidence that the nuclei also exhibit chiral alignment similar to the cells (Figure 4). These results suggest that the nuclei take on the alignments of the cells and may represent an alternative avenue for the quantification of cell chirality. One possible explanation for the consistent alignment of cells and nuclei is that geometric constraints induced cytoskeletal reorganization that move and reposition the nuclei (e.g. in the astrocyte studies of Dupin et al.5). Another explanation is that forces transmitted between cells through adherens and tight junctions cause a remodeling of the cytoskeleton, which forces nuclei to align10. Because the biased alignment is transmitted from the cell to its nucleus, and the nucleus is not as flexible as the cells, not all nuclei will take on the proper alignment of cells30. This may explain why cell nuclei are typically less biasedly aligned compared to the cells.
Significance of Nucleus Alignment as a Chiral Measure
Cell alignment has been identified as a chiral measure and used for several studies18, 25, 28. As the nucleus tends to align themselves with the cells4, 30, the chiral nuclear alignment is partially expected. However, it has never been proved until now. As nuclear staining or even genetic nuclear labeling is relatively easy to perform, nuclear alignment as a chiral measure potentially has a wide application. In particular, we believe that the identification of nuclear alignment as an additional chiral measure will benefit the LR asymmetry research of embryonic development. For instance, for in vivo 3D studies, it will be much easier to dertimine the nuclear orientation than to measure the shape of cell membrane. Determining the contour of individal cells in 3D cellular structures requires specific labeling and high resolution 3D imaging. Therefore, it will be relatively difficult to implement, especially for 4D time-lapse imaging of embryonic development. Nuclear analysis will be preferred for such applications.
Misaligned Cells are Morphologically Different
Misaligned cells, which take on CW orientations on CCW rings, exhibit divergent morphologies, as shown in Figure 6. Further, the associated nuclei of misaligned cells were also demonstrated to be morphologically distinct. Overall, misaligned cells tend to be smaller and less elongated. When cells are smaller and less polarized, the chirality will not be observed as easily, as the establishment of chirality, LR polarity, requires the proper polarization (front-back polarity) of the cell.
We also noticed that the difference in cellular and nuclear area between misaligned and properly aligned cells were largest at a region close to the outer boundary. This is likely because at the outer boundary, larger cells tend to polarize on the boundary and therefore are able to establish a proper chirality, while smaller cells cannot. In the interior region between the boundaries, even large cells will have a higher chance to experience misalignment, since these cells are far away from the boundaries. As a result, the difference in cell morphology can be easily appreciated on cell boundaries.
Limitations
The current approach has been demonstrated to be highly accurate and produce large amounts of data. The ability to track individual cell migration in a monolayer is a significant advantage for possible applications of this new method, but there are a few drawbacks that are worth a discussion. First, it is computationally expensive compared to other approaches, such as the intensity gradient-based method. Second, this method requires pre-processing of images in order to be analyzed. Ideally, a completely automated algorithm is desired for high-throughput analysis. But this issue can be potentially resolved with the implementation of machine-learning algorithms20. Finally, the current iteration of this approach requires cells that express cell-cell adhesion proteins for analysis. However, we can use confocal imaging of membrane staining for other cell types that do not express these junctions.
Conclusion
In this study, we demonstrated that individual cell and nucleus based analysis represents a robust technique that utilizes object morphologies to determine chiral alignment of patterned cells. Similar but more robust results were obtained, compared to the well-established intensity gradient-based analysis. We found that cell nuclei took on chiral alignments that were similar to cells, highlighting the potential of utilizing nuclei to quantify cell chirality. Interestingly, misaligned cells tended towards smaller and less elongated morphologies compared to properly aligned cells, and this difference was evidently observed on the outer boundary. Overall, the current approach is a step forward towards more thorough analyses of collective cell chiral morphogenesis, and it represents the first that allows researchers to investigate the correlation between chiral and morphological features.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank National Institutes of Health, National Science Foundation, American Heart Association, and March of Dimes for funding Support. Leo Q. Wan is a Pew Scholar in Biomedical Sciences, supported by the Pew Charitable Trusts.
ABBREVIATIONS
- (CW)
Clockwise
- (CCW)
Counterclockwise
- (NC)
Non-chiral
Footnotes
Disclosure
All authors state that they have no conflicts of interest.
REFERENCES
- 1.Bradski G. The OpenCV Library (2000) Dr. Dobb's Journal of Software Tools. 2000 [Google Scholar]
- 2.Brigaud I, Duteyrat JL, Chlasta J, Le Bail S, Couderc JL, Grammont M. Transforming Growth Factor beta/activin signalling induces epithelial cell flattening during Drosophila oogenesis. Biol Open. 2015 doi: 10.1242/bio.201410785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen TH, Hsu JJ, Zhao X, Guo C, Wong MN, Huang Y, Li Z, Garfinkel A, Ho CM, Tintut Y, Demer LL. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ Res. 2012;110:551–559. doi: 10.1161/CIRCRESAHA.111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284:274–282. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 5.Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards W, Moles AT, Franks P. The global trend in plant twining direction. Global Ecology and Biogeography. 2007;16:795–800. [Google Scholar]
- 7.Freytes DO, Wan LQ, Vunjak-Novakovic G. Geometry and force control of cell function. Journal of Cellular Biochemistry. 2009;108:1047–1058. doi: 10.1002/jcb.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatori R, Ando T, Sasamura T, Nakazawa N, Nakamura M, Taniguchi K, Hozumi S, Kikuta J, Ishii M, Matsuno K. Left-right asymmetry is formed in individual cells by intrinsic cell chirality. Mech Dev. 2014;133:146–162. doi: 10.1016/j.mod.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Karlon WJ, Hsu PP, Li S, Chien S, McCulloch AD, Omens JH. Measurement of orientation and distribution of cellular alignment and cytoskeletal organization. Ann Biomed Eng. 1999;27:712–720. doi: 10.1114/1.226. [DOI] [PubMed] [Google Scholar]
- 10.Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Annu Rev Biomed Eng. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]
- 11.Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell. 2002;111:77–89. doi: 10.1016/s0092-8674(02)00939-x. [DOI] [PubMed] [Google Scholar]
- 13.Lowekamp BC, Chen DT, Ibáñez L, Blezek D. The Design of SimpleITK. Frontiers in neuroinformatics. 2013;7 doi: 10.3389/fninf.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercola M, Levin M. Left-right asymmetry determination in vertebrates. Annu Rev Cell Dev Biol. 2001;17:779–805. doi: 10.1146/annurev.cellbio.17.1.779. [DOI] [PubMed] [Google Scholar]
- 15.Okada Y, Takeda S, Tanaka Y, Izpisua Belmonte JC, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Oliphant TE. A guide to NumPy. Trelgol Publishing USA; 2006. [Google Scholar]
- 17.Shibazaki Y, Shimizu M, Kuroda R. Body handedness is directed by genetically determined cytoskeletal dynamics in the early embryo. Curr Biol. 2004;14:1462–1467. doi: 10.1016/j.cub.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Singh AV, Mehta KK, Worley K, Dordick JS, Kane RS, Wan LQ. Carbon nanotube-induced loss of multicellular chirality on micropatterned substrate is mediated by oxidative stress. ACS Nano. 2014;8:2196–2205. doi: 10.1021/nn405253d. [DOI] [PubMed] [Google Scholar]
- 19.Singh AV, Raymond M, Pace F, Certo A, Zuidema JM, McKay CA, Gilbert RJ, Lu XL, Wan LQ. Astrocytes increase ATP exocytosis mediated calcium signaling in response to microgroove structures. Sci Rep. 2015;5:7847. doi: 10.1038/srep07847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommer C, Straehle C, Kothe U, Hamprecht FA. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on. IEEE; 2011. ilastik: Interactive learning and segmentation toolkit; pp. 230–233. [Google Scholar]
- 21.Taniguchi K, Maeda R, Ando T, Okumura T, Nakazawa N, Hatori R, Nakamura M, Hozumi S, Fujiwara H, Matsuno K. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science. 2011;333:339–341. doi: 10.1126/science.1200940. [DOI] [PubMed] [Google Scholar]
- 22.Van Der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan LQ, Kang SM, Eng G, Grayson WL, Lu XL, Huo B, Gimble J, Guo XE, Mow VC, Vunjak-Novakovic G. Geometric control of human stem cell morphology and differentiation. Integrative Biology. 2010;2:346–353. doi: 10.1039/c0ib00016g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan LQ, Ronaldson K, Guirguis M, Vunjak-Novakovic G. Micropatterning of cells reveals chiral morphogenesis. Stem cell research & therapy. 2013;4:24. doi: 10.1186/scrt172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan LQ, Ronaldson K, Park M, Taylor G, Zhang Y, Gimble JM, Vunjak-Novakovic G. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc Natl Acad Sci U S A. 2011;108:12295–12300. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan LQ, Vunjak-Novakovic G. Micropatterning chiral morphogenesis. Communicative & integrative biology. 2011;4:745–748. doi: 10.4161/cib.17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worley K, Certo A, Wan LQ. Geometry–Force Control of Stem Cell Fate. BioNanoScience. 2013;3:43–51. [Google Scholar]
- 28.Worley KE, Shieh D, Wan LQ. Inhibition of Cell-Cell Adhesion Impairs Directional Epithelial Migration on Micropatterned Surfaces. Integrative Biology. 2015 doi: 10.1039/c5ib00073d. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Van Keymeulen A, Wakida NM, Carlton P, Berns MW, Bourne HR. Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci U S A. 2007;104:9296–9300. doi: 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamauchi K, Yang M, Jiang P, Yamamoto N, Xu M, Amoh Y, Tsuji K, Bouvet M, Tsuchiya H, Tomita K, Moossa AR, Hoffman RM. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 2005;65:4246–4252. doi: 10.1158/0008-5472.CAN-05-0069. [DOI] [PubMed] [Google Scholar]
- 31.Yokoyama T, Copeland NG, Jenkins NA, Montgomery CA, Elder FF, Overbeek PA. Reversal of left-right asymmetry: a situs inversus mutation. Science. 1993;260:679–682. doi: 10.1126/science.8480178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.