Abstract
AIM: To investigate alpha-fetoprotein (AFP) mRNA expression in BEL-7404 human hepatoma cells and the effect of L-4-oxalysine (OXL) on the expression.
METHODS: Bel-7404 human hepatoma cells were maintained in RPMI 1640 media. Human AFP cDNA probe was labelled with digoxigenin-11-dUTP by the random primer labelling method. The expression of AFP mRNA in Bel-7404 cells was determined by an in situ hybridization technique with digoxigenin-labelled human AFP cDNA probe. The positive intensities of AFP mRNA in cells were analyzed by microspectrophotometer and expressed as absorbance at 470 nm. For the experiment with OXL, cells were incubated with various concentrations of the agent for 72 h.
RESULTS: Essentially all the hepatoma cells contained AFP mRNA in the cytoplasm, although in various amounts. The specificity of the hybridization reaction was confirmed by control experiments in which the use of Rnase-treated BEL-7404 cells, non-AFP producing cells (HL-60 human leukemia cells) or a nonspecific cDNA probe resulted in negative hybridization. When the cells were treated with OXL (25, 50 mg/L), the content of AFP mRNA in the cytoplasm was decreased with the inhibition percentages of 34.3% and 70.1%, respectively (P < 0.05).
CONCLUSION: AFP mRNA was expressed in BEL-7404 human hepatoma cells and OXL suppressed AFP mRNA expression in the cells.
Keywords: oxalysine; liver neoplasms; fetoprotein; tumor cell, cultured; RNA, messenger; gene expression; in situ hybridization; immunohistochemistry
INTRODUCTION
Serum alpha-fetoprotein (AFP) has been widely detected as a marker for primary hepatocellular carcinoma (PHC). However, the relationship between AFP and PHC is still unclear. We found recently that AFP directly stimulated the growth of mouse ascites hepatoma-22 cells and inhibited the immune responses[1-4]. It is suggested that AFP contributes the generation and development of PHC and is an important target of anti-hepatoma drugs[5-7]. L-4-oxalysine (OXL) is a natural product isolated from a new species of Streptomyces roseo viridofuscus n. sp. in China. Previous studies indicated that OXL exhibited marked antiproliferative activity against several animal tumors. The antimetastatic influence of OXL was also detected in mice bearing Lewis lung carcinoma[8-10]. OXL also exhibited immunoregulatory activity[11]. Preliminary clinical studies suggested that oral treatment with OXL induced an improvement in the symptoms of PHC patients, and no serious side effects were observed[12]. Recently, our laboratory also found that OXL antagonized the biological activities of AFP[13]. AFP content in human BEL-7404 hepatoma cells and cultured media was obviously decreased after the OXL treatment[14]. It is inferred that OXL has anti-AFP activities. In this report, an in situ- hybridization (ISH) technique was used to study the level of AFP mRNA expression in BEL-7404 human hepatoma cells. The effect of OXL on AFP mRNA expression was also observed.
MATERIALS AND METHODS
Cell culture
A human hepatoma cell line, BEL-7404, was maintained in RPMI 1640 media (Gibco) supplemented with 10% calf serum, 100 kU/L of penicillin and 100 mg/L of streptomycin, at 37 °C, 5% CO2 and 100% humidity. The RPMI 1640 media was replaced with fresh media every three to four days. For in vitro experiment with OXL, cells at a density of 5 × 106 cells/L were grown on circular coverslip in each well of 24-well culture plate. Twenty-four hours later, various concentrations of OXL (Department of Antibiotics of this Institute) were added, and cells were again incubated for 72 h. Control group contained cells alone. After incubation, adhesive cells were directly used for ISH assay.
Labelling of probes
The probes used in this experiment are shown in Table 1[15,16]. Recombinant plasmids pHAF-2 containing human AFP cDNA and phalb-7 containing human serum albumin (HSA) cDNA were kind gifts of Drs Yoshitake Hayashi and Kyoskuke Ohta at University of Kobe, Japan. Plasmids were grown in bulk in Escherichia coli- HB101, extracted by the alkaline procedure, purified by phenol and two ethanol precipitations. Plasmids were digested with restrictive enzymes (Promega). The digests were then electrophoresed in 1% preparative agarose gels to separate the purified inserted gene sequences from the residual linearized plasmid band[17].
Table 1.
Probes used in this experiments
| Plasmids | Carrier | Gene contented | Inserted | Restrictive enzyme | Resistance |
| pHAF-2 | pBR322 | AFP | 900 bp | Pst I + Hap II | Tetracycline |
| phalb-7 | pBR322 | Albumin | 727 bp | Pst I + Hind III | Tetracycline |
A 900 bp Pst I-Hap II fragment from the plasmid pHAF-2 and a 727 bp Pst I-Hind III fragment from the plasmid phalb-7 were labelled with digoxigenin (Dig)11-dUTP by random primer labelling method with Dig DNA labelling kit (Boehringer Mannheim Company) and used as AFP and albumin probes, respectively [18]. Briefly, the cDNA was denatured by heating for 5 min at 100 °C and then quickly chilled on ice. The following reagents were added to an Eppendorf tube on ice: 4 μL freshly denatured cDNA (1 μg), 2 μL hexanucleotide mixture, 2 μL- dNTP labelling mixture, 11 μL- sterile water and 1 μL Klenow enzyme, mixed and incubated at 37 °C for at least 1 h (usually incubated 2 h-3 h). Ten μL of 2g/L yeast tRNA (Sigma) was added and the probes were precipitated with the addition of 4 μL of 3 mol/L NaAc (pH 5.2) and 3 volumes of 95% prechilled ethanol (-20 °C) at -70 °C for at least 30 min. The supernatant was discarded by centrifugation and the probes were stored in 50 μL of TE (10mmol/L Tris HCl, 1 mmol/L EDTA, pH 8.0) at -20 °C. The yield of labelled probes in this reaction was 250 ng (5 ng/μL). To test the sensitivity of each probe, dot-blot hybridization was carried out with Dig-labelled cDNA probe. The size of the probes was 50 to 250 bp as estimated by polyacrylamide gel electrophoresis.
In situ hybridization (ISH)
ISH was done essentially according to the procedure of Breborowicz et al[19] with some modifications. Briefly, adhesive cells were fixed in 4% paraformaldehyde for 5 min-8 min at room temperature. The coverslips were serially washed with the following solutions at room temperature: 0.05 mol/L Tris-buffered saline (TBS, pH 7.2) three times, 5 min each; 100 mmol/ L glycine once, 15 min; TBS three times, 5 min each; 0.4% Triton X100 in TBS once, 15min; and TBS three times, 5min each. The coverslips were then treated with 1 mg/L of proteinase K (Sigma) in 20 mmol/L Tris-HCl (pH 7.4) and 2 mmol/L CaCl2 for 15 min at 37 °C, washed with TBS three times for 5min each, air dried, postfixed in 4% paraformaldehyde for 5 min at room temperature, and washed with TBS three times for 5 min each. The coverslips were finally washed with 2 × SSC (1 × SSC: 150 mmol/L NaCl, 15 mmol/L sodium citrate) or treated with RNase (Sigma,100 mg/L in 2 × SSC) for 30 min at 37 °C.
The hybridization mixture contained 50% deionized formamide, 5 × SSC, 10% dextran sulfate (Sigma), 5 × Denhardt’s solution, 2% sodium dodecyl sulfate, 100 mg/L of salmon sperm DNA (Sigma) denatured at 100 °C and 25 mg/L-50 mg/L of Dig-labelled probe denatured at 100 °C. 100×Denhardt’s solution contained 2% Ficoll 400 (Sigma), 2% polyvinylpyrollidone (Sigma) and 2% bovine serum albumin (BSA, Sigma). Two hundred μL of the hybridization mixture was added to each well of 24-well plate. Cells were incubated in a humidified atmosphere for 18 h at 37 °C. The coverslips were washed at 37 °C in 4 × SSC three times for 5 min each, and then sequentially immersed in 2 × SSC, 1 × SSC, 0.5 × SSC and 0.1 × SSC at 37 °C for 30 min each. The coverslips were then washed in TBS containing 1% BSA and 0.4% Triton X-100 for 30 min at room temperature . Sheep anti-Dig antibody conjugated to alkaline phosphatase (AP, Boehringer Mannheim Company) was diluted 1: 500 with TBS containing 1% BSA and 0.4% Triton X-100, and applied to specimens. Cells were incubated for 2 h at 37 °C, then washed in buffer I (100 mmol/L Tris-HCl, 100 mmol/L NaCl, 10 mmol/L MgCl2, pH 8.0) and II (100 mmol/L Tris-HCl, 100mmol/ L NaCl, 50mmol/L MgCl2, pH 9.5), respectively, for 10min each at room temperature. Development reagent contained 33 μL of nitroblue tetrazolium salt (NBT, 75 μg/L in 70% dimethylformamide) and 25 μL of 5-bromo 4 chloro 3 indocyl phosphate ((BCIP, X-phosphate, 50 μg/L in dime thylformamide) in 7.5 mL of buffer II. Cells were incubated in the color solution at 37 °C for up to 4 h in the dark. Color development was periodically checked and reaction was stopped by washing the coverslips for 5 min in TE at room temperature. The coverslips were rinsed well in distilled water, air-dried, cleared in xylene and mounted with glycerin jelly.
The following controls were performed. Just before the hybridization, cells were incubated with 2 × SSC containing RNase 100 mg/L for 30 min at 37 °C, then were processed for ISH as above; Diglabelled AFP cDNA probe was replaced by Diglabelled albumin cDNA probe; Nonhepatoma cells (human leukemia HL-60 cells) were incubated with hybridization buffer containing the Diglabelled AFP cDNA probe. The positive intensities of AFP mRNA in hepatoma cells were analyzed by microspectrophotometer (Leitz MPV-3) and expressed as absorbancy at 470nm (A470).
Statistical analysis
The statistical significance of differences was evaluated using analysis of variance (ANOVA).
RESULTS
Detection of AFP mRNA by ISH
It was demonstrated with dot-blot hybridization that the sensitivity of the Diglabelled probes was 1.0pg. When Diglabelled AFP cDNA probe was used, purple grains were present in the cytoplasm of BEL-7404 human hepatoma cells, with fewer grains seen in cell nuclei. The number of grains in the cytoplasma varied, but essentially all the hepatoma cells, including those undergoing mitotic division, were considered to contain AFP mRNA-(Figure 1).
Figure 1.
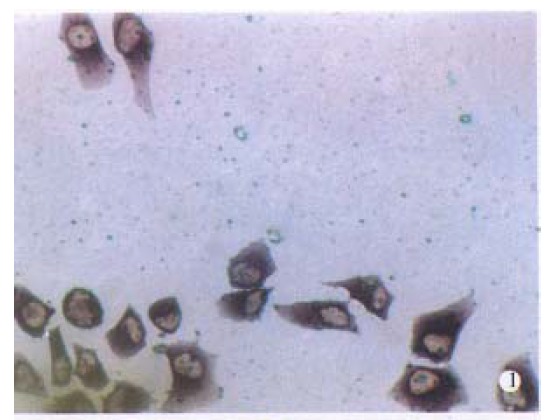
Detection of alpha-fetoprotein (AFP) mRNA in human BEL-7404 hepatoma cells. In A, BEL-7404 cells were hybridized in situ with digoxigenin (Dig)-labelled AFP cDNA probes. Reaction products were visible in almost all cells.
In order to establish the specificity of hybridization, BEL-7404 cells were incubated with Diglabelled albumin cDNA probe under the same hybridization conditions. No accumulation of grains over the cells was observed (Figure 2). Pretreatment of BEL-7404 cells with RNase abolished the formation of grains. Other cell lines not producing AFP, such as HL-60 human leukemia cells, also gave negative results.
Figure 2.
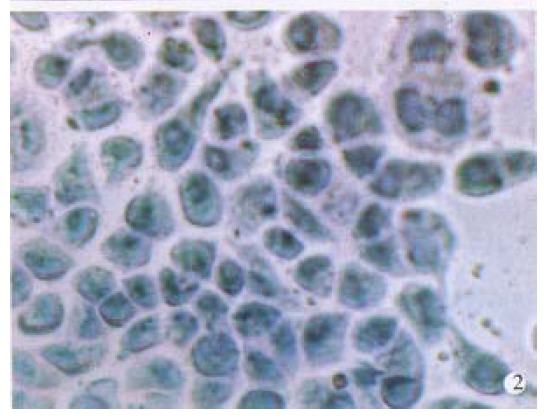
Same as in Figure 1, except that the Diglabelled albumin cDNA was used as probes. Cells were counterstained with hematoxylin. × 400
Influence of OXL on AFP mRNA expression in BEL-7404 human hepatoma cells
Hepatoma cells were incubated with OXL 25, 50 mg/L. Cell viability was greater than 95% using trypan blue exclusion. Seventy-two hours later, the change in AFP mRNA content in hepatoma cells during OXL treatment was determined by ISH. It was found that AFP mRNA content in hepatoma cells was significantly decreased by OXL; the effect of higher concentration was more obvious. It is suggested that OXL inhibits AFP mRNA expression in hepatoma cells (Table 2).
Table 2.
Influence of L-4-oxalysine (OXL) on AFP mRNA expression in BEL-7404 human hepatoma cells
| Treatment | Concentration (mg/L) | Positive intensities of AFP mRNA (A470 ± S) | Inhibition (%) |
| Control | 0.67 ± 0.08 | ||
| OXL | 25 | 0.44 ± 0.06a | 34.3 |
| OXL | 50 | 0.20 ± 0.05b | 70.1 |
BEL-7404 human hepatoma cells (5 × 106 cells/L) were cultivated for 72 h in absence or presence of various concentrations of OXL. The results were expressed as absorbance at 470 nm (A470). The percentage of inhibition was calculated in the cultures in absence of OXL.
P < 0.05;
P < 0.01.
DISCUSSION
Various amounts of AFP were present in almost all the BEL-7404 hepatoma cells shown by the avidin-biotin-peroxidase complex (ABC) method[14]. We got the similar result in our study. The specificity of the hybridization reaction was confirmed by control experiments in which the use of RNase-treated BEL-7404 cells, non-AFP-producing cells or a nonspecific cDNA probe resulted in negative hybridization. We presume that ISH method has the following advantages: It permits to study the expression of genes qualitatively and quantitatively in an individual cell; it is not necessary to purify RNA, fewer instrument and equipment are needed, and only a small quantity of specimen is required; the samples can be preserved for a long time and can be reviewed if necessary; it needs only a small amount of DNA probes; and Dig-labelled probes cause no radioactive contamination.
It has been known that the synthesis of AFP is often elevated to a significant level in association with development of PHC. Hence, the in situ detection of AFP mRNA may help in histopathological diagnosis of PHC which does not secrete AFP in amounts detectable by immunological means. Moreover, ISH also provides a very useful tool for investigating the effect of some drugs on AFP mRNA expression[20]. Present results indicated that OXL suppressed, to a certain extent, AFP mRNA expression in hepatoma cells. Taking our investigation on the whole, it is strongly inferred that OXL exhibits significant anti-AFP activities, which may be one of the mechanisms of anti-tumor action of OXL. Such findings could also lead to the development of new anti-PHC drugs based on AFP target.
ACKNOWLEDGMENTS
The plasmids containing human AFP cDNA and albumin cDNA were kindly provided by Drs Yoshitake Hayashi and Kyosuke Ohta at University of Kobe, Japan. We sincerely thank Dr Wang Yao, Australian National University, Australia; Dr Zhang Da-Bing, Shanghai Institute of Plant Physiology, Chinese Academy of Sciences, Shanghai; and Dr Xiang Zheng Hua, the Second Millitary Medical University, Shanghai for their assistance.
Footnotes
Project supported by the National Natural Science Foundation of China, No.39570824
References
- 1.Wang XW, Xu B. Effect of alpha-fetoprotein on the growth of mouse ascites hepatoma-22 cells in vitro. Acta Oncol Sin. 1995;5(4):229–231. [Google Scholar]
- 2.Wang XW, Xu B. Influence of alpha-fetoprotein on the growth of tumor cells in vitro. Chin J Cancer Res. 1997;9(2):79–82. [Google Scholar]
- 3.Wang XW, Xu B. Effect of alpha-fetoprotein (AFP) on splenocyte prolifera-tion of mice bearing ascites hepatoma-22 in vitro. Shanghai J Immunol. 1995;15(6):327–329. [Google Scholar]
- 4.Wang XW, Xu B. Effect of human alpha-fetoprotein on immunological func-tions in mice bearing ascites hepatoma-22 in vitro. Shanghai J Immunol. 1997;17(4):224–226. [Google Scholar]
- 5.Wang XW, Xu B. Effect of some drugs on alpha-fetoprotein and treatment of primary hepatocellular carcinoma. Chin Oncol. 1995;4(12):24–26. [Google Scholar]
- 6.Wang XW, Xu B. Several new targets of antitumor agents. Zhongguo Yaoli Xuebao. 1997;18:289–292. [PubMed] [Google Scholar]
- 7.Wang XW, Xu B. Alpha-fetoprotein, primary hepatocellular carcinoma and anticancer drugs. In: Cao SR, ed. New theory and technology of oncology. Shanghai: Shanghai Science and Technology Education Press. 1997:485–497. [Google Scholar]
- 8.Wang XW, Xu B. L-4-oxalysine, a new antitumor agent of natural origin. Med Chem Res. 1996;6(4):225–232. [Google Scholar]
- 9.Wang XW, Xu B. Mechanisms of antitumor action of L-4-oxalysine, a new natural product. Med Chem Res. 1996;6(4):233–247. [Google Scholar]
- 10.Wang XW, Xu B. Antitumor and immunological activities of oxalysine. Chin J Oncol. 1997;19(2):115–117. [PubMed] [Google Scholar]
- 11.Wang XW, Xu B. Immunoregulatory activities of L-4-oxalysine: an in vitro study. Methods Find Exp Clin Pharmacol. 1997;19:437–442. [PubMed] [Google Scholar]
- 12.Wang XW, Xu B. L-4-oxalysine: Its antitumor activities and mechanisms of action. Drugs Fut. 1996;21(7):727–731. [Google Scholar]
- 13.Wang XW, Xu B. Anti-alpha-fetoprotein activity of L-4-oxalysine. Asia Pa-cific J Pharmacol. 1996;11(2):25–28. [Google Scholar]
- 14.Wang XW, Xu B. Effect of L-4-oxalysine on alpha-fetoprotein gene expres-sion in human BEL-7404 hepatoma cells. Asia Pacific J Pharmacol. 1997;12(1-2):37–40. [Google Scholar]
- 15.Urano Y, Sakai M, Watanabe K, Tamaoki T. Tandem arrangement of the albumin and alpha-fetoprotein genes in the human genome. Gene. 1984;32:255–261. doi: 10.1016/0378-1119(84)90001-5. [DOI] [PubMed] [Google Scholar]
- 16.Morinaga T, Sakai M, Wegmann TG, Tamaoki T. Primary structures of human alpha-fetoprotein and its mRNA. Proc Natl Acad Sci USA. 1983;80:4604–4608. doi: 10.1073/pnas.80.15.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch EF, Mantiatis T. Molecular cloning: A laboratory mannual. New York: Cold Spring Hober Laboratory Press. 1989:304–316. [Google Scholar]
- 18.Furuta Y, Shinohara T, Sano K, Meguro M, Nagashima K. In situ hybridisation with digoxigenin-labelled DNA probes for detection of viral genomes. J Clin Pathol. 1990;43:806–809. doi: 10.1136/jcp.43.10.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breborowicz J, Tamaoki T. Detection of messenger RNAs of alpha-fetoprotein and albumin in a human hepatoma cell line by in situ hybridization. Cancer Res. 1985;45:1730–1736. [PubMed] [Google Scholar]
- 20.Wang XW, Xu B. Research advances of research of alpha-fetoprotein gene expression and the expression regulation. Acta Oncol Sin. 1996;6(4):281–284. [Google Scholar]


