Abstract
Tumour cells often acquire the ability to escape cell death, a key event leading to the development of cancer. In almost half of all human cancers, the capability to induce cell death is reduced by the mutation and inactivation of p53, a tumour suppressor protein that is a central regulator of apoptosis. As a result, there is a crucial need to identify different cell death pathways that could be targeted in malignancies lacking p53. p73, the closely related p53 family member, can regulate many p53 target genes and therefore some of the same cellular responses as p53. Unlike p53, however, p73 is seldom mutated in cancer, making it an attractive, alternative death effector to target. We report here the ability of p73 to upregulate the expression of the A2B receptor, a recently characterized p53 target that effectively promotes cell death in response to extracellular adenosine—a metabolite that accumulates during various forms of cellular stress. Importantly, we show that p73-dependent stimulation of A2B signalling markedly enhances apoptosis in cancer cells that are devoid of p53. This mode of death is caspase- and puma-dependent, and can be prevented by the overexpression of anti-apoptotic Bcl-XL. Moreover, treatment of p53-null cancer cells with the chemotherapeutic drug adriamycin (doxorubicin) induces A2B in a p73-dependent manner and, in combination with an A2B agonist, substantially enhances apoptotic death. We therefore propose an alternate and distinct p53-independent pathway to stimulate programmed cell death involving p73-mediated engagement of adenosine signalling.
Introduction
The prominent tumour suppressor p53 actively protects against cellular transformation by curbing undesirable propagation of damaged cells. One of the ways in which p53 functions in this context is by transactivating genes that execute programmed cell death in response to harmful cellular stresses such as genomic damage or oncogene activation.1 However, over half of all human cancers contain mutations in the p53 gene2 and others have perturbations in pathways required for p53 activation,3 limiting the benefit of targeting p53 in these tumour settings. Hence, the discovery of other factors that can promote cell death in a p53-independent manner is of great interest and utmost importance in targeting such cancers.
p73, a paralogue of p53 from the same family of transcription factors, is now known to have important roles in tumorigenesis.4 Owing to alternative promoter usage and C-terminal alternative splicing, p73 exists in multiple isoforms. Transcription of the gene from the upstream P1 promoter gives rise to the transactivating isoforms (TAp73), while transcripts generated from the internal P2 promoter are amino-terminally truncated and lack the transactivation domain (ΔNp73).5, 6 Functionally, the TAp73 isoforms closely mimic p53 in the ability to stimulate transcription of death genes and to trigger programmed cell death, and have been shown to be bona fide tumour suppressors.4, 7 However, unlike p53, TAp73 is rarely found to be mutated in cancers,8, 9 making it an attractive target for therapeutic intervention. Indeed, previous studies have indicated that TAp73 can be activated in cancer to promote tumour cell death and cause tumour regression.10, 11
Extracellular adenosine, the backbone of ATP, is a natural signalling molecule that activates four cell-surface, G-protein-coupled adenosine receptors. These receptors called A1, A2A, A2B and A3 signal a diverse range of responses in a variety of cells.12 For example, adenosine has been shown to regulate neuro-transmission, immune response, vascular permeability and cell viability.12, 13 A1, A2A and A3 respond to physiological levels of adenosine in the nanomolar range.12 By contrast, A2B is only fully activated by micromolar levels of adenosine that are normally associated with cells under stressed conditions.12 This can occur under conditions of metabolic stress such as hypoxia or upon treatment with cytotoxic drugs.13 Moreover, it has been shown that the extracellular space of many solid tumours has unusually high levels of adenosine.14, 15, 16 Although it is considered that this arises through the enhanced metabolic stress associated with tumour development, the role of adenosine in this context is not entirely clear.
We previously identified ADORA2B (the gene that encodes A2B) to be a target of p53 that functions to prime cells to die in response to the accumulation of extracellular adenosine.13 It was therefore of great interest to investigate whether p73 could also regulate ADORA2B expression and activate programmed cell death via this pathway in a manner independent of p53. Here, we demonstrate the ability of TAp73 to stimulate ADORA2B expression and function, which enhances p73-dependent cell death in response to chemotherapy. These findings reveal an alternative, p53-independent cell death pathway that could potentially be exploited for therapeutic intervention.
Results
ADORA2B is a novel p73 target gene
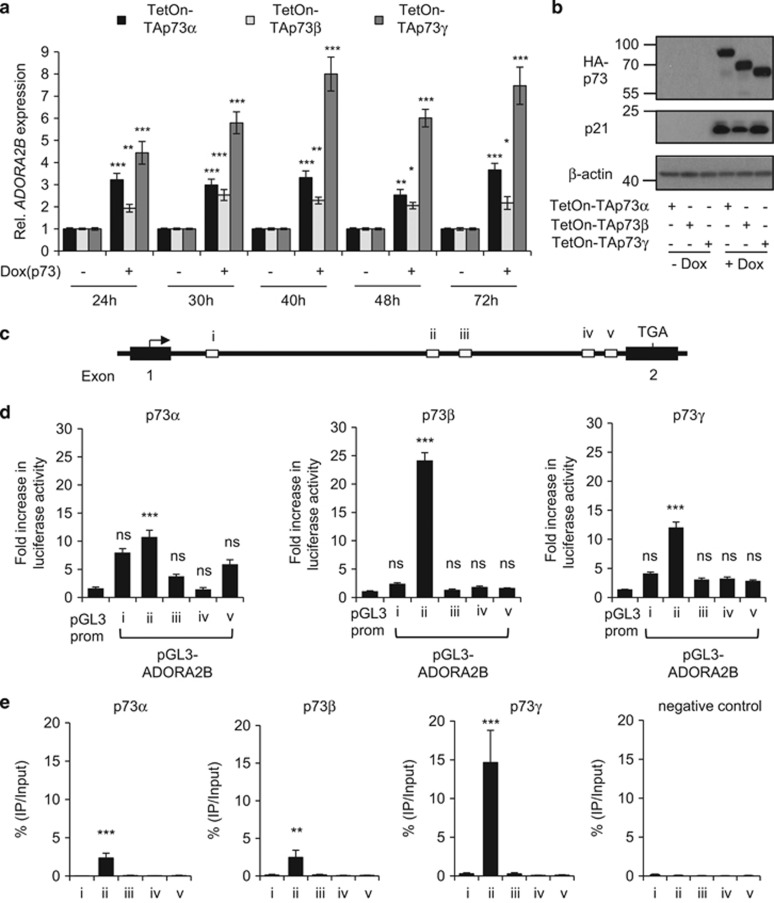
Using previously characterized Saos-2 cells expressing TAp73α, TAp73β or TAp73γ under the control of a tet-inducible promoter,17 we investigated whether these different p73 isoforms could regulate ADORA2B expression. We observed TAp73γ to be a strong inducer (four- to ninefold) of ADORA2B expression. TAp73α and TAp73β could also reproducibly induce ADORA2B expression, albeit to a lesser extent (two- to fourfold) following doxycycline treatment (Figure 1a). The levels of induction of the TAp73 isoforms upon doxycycline treatment are comparable (Figure 1b), indicating that the differential regulation of ADORA2B expression is unlikely due to differences in the TAp73α, TAp73β or TA-p73γ expression levels. Furthermore, induction of each isoform could effectively stimulate p21 expression (Figure 1b), a known target gene of p73,7, 18 indicating that all isoforms are functionally competent. As mentioned earlier, p73 can be transcribed via an alternative internal promoter, giving rise to isoforms lacking the transactivation domain, which are collectively termed ΔNp73. We therefore also examined whether ΔNp73 could regulate ADORA2B expression, and we found that expression of ΔNp73 does not affect ADORA2B levels, as demonstrated in Supplementary Figures S1a and b. We also sought to corroborate the induction of ADORA2B mRNA at the protein level, but we were unable to convincingly detect A2B in our cell systems with sufficient clarity.
Figure 1.
ADORA2B is a p73 target gene. (a) TetOn-TAp73α, -TAp73β or –TAp73γ Saos-2 cells were treated with or without Dox in the presence of zVAD-fmk (50 μM) for 24 h, 30 h, 40 h, 48 h and 72 h before quantification of ADORA2B levels by qRT-PCR (n=3, 9 replicates in total). Statistical analysis was performed by comparing Dox-treated samples to untreated samples for each time. (b) Western blotting analysis of HA-TAp73, p21 and β-actin in TetOn-TAp73α, -TAp73β or –TAp73γ Saos-2 cells treated with or without Dox for 48 h. (c) Previously identified putative p53 binding sites (i-v) within intron 1 of the ADORA2B gene that may be potential p73 binding sites. (d) Saos-2 cells were co-transfected with the TAp73α-, TAp73β- or TAp73γ-expressing construct along with luciferase reporter constructs containing potential p73 binding sites (pGL3-ADORA2B i-v) or vector control (pGL3 prom). Luciferase activity was analysed 24 h after transfection (n=3, 9 replicates in total). The fold increase shown is relative to vector control, which was normalized to 1. (e) Chromatin immunoprecipitation (ChIP) was performed on TetOn-TAp73α, -TAp73β or –TAp73γ Saos-2 cells treated with Dox for 24 h. As a negative control, ChIP was performed on untreated TAp73γ Saos-2 cells. The % input of co-precipitating DNAs were measured by quantitative PCR (n=3, three replicates in total). ns, P>0.05; *P<0.05; **P<0.01; ***P<0.001.
As TAp73 shares a high degree of sequence homology with p53 and can therefore bind to p53-responsive elements,6 we examined whether TAp73 may also transactivate ADORA2B through elements that were previously identified as potential p53-binding sites (Figure 1c).13 Indeed, the co-transfection of TAp73α, TAp73β or TAp73γ with reporter constructs containing the potential p53-binding elements revealed that TAp73 can transactivate ADORA2B (Figure 1d). All three TAp73 isoforms were observed to efficiently transactivate binding site ii. We noted, however, that our luciferase data do not completely parallel our qRT-PCR for ADORA2B induction as shown in Figure 1a. This may be due to the fact that the luciferase assay involves isolated fragments of potential binding sites that are exogenously overexpressed. Therefore, to test more quantitatively whether p73 isoforms bind to this site, we performed chromatin immunoprecipitation (ChIP) assays for the ability of TAp73 isoforms to bind the endogenous elements examined in our luciferase assays. This showed that each isoform associates with binding site ii and in line with our qRT–PCR analysis (Figure 1a), we found that TAp73γ appeared to bind site ii with a greater affinity than TAp73α and TAp73β (Figure 1e). As an indication of specificity, no significant binding was detected using primers that amplify the regions containing the potential binding sites i, iii, iv and v (Figures 1c and e). For comparison, we also examined p53 binding by ChIP, which revealed preferential binding to site ii, indicating that this is the primary site targeted by both p53 and TAp73 (Supplementary Figure 1c).
A2B receptor stimulates cell death downstream of p73
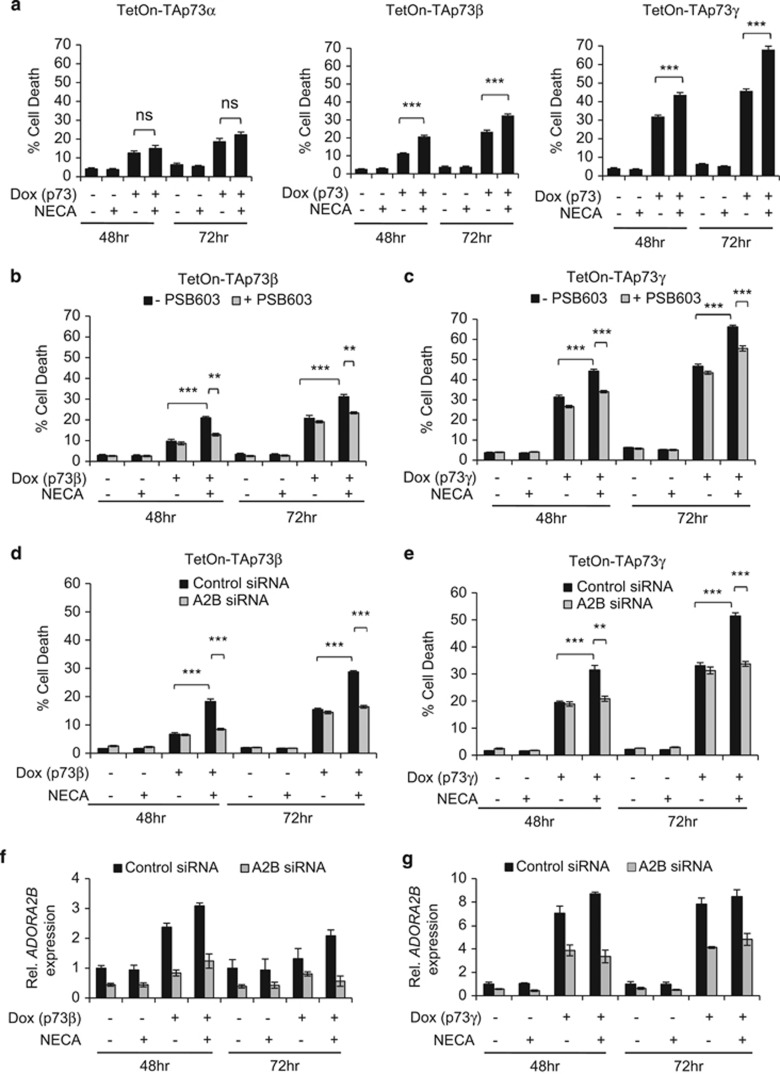
p73 is known to induce programmed cell death by activating the transcription of several p53-responsive pro-apoptotic genes.7, 19, 20 Therefore, we next asked the question whether p73 induction of ADORA2B could contribute to programmed cell death downstream of p73. This was assessed by treating the TetOn-TAp73α, -TAp73β or -TAp73γ Saos-2 cells with dox in combination with the adenosine analogue, adenosine-5'-N-ethylcarboxamide (NECA), which binds to and activates the A2B adenosine receptor. Programmed cell death was assessed using flow cytometry by measuring the percentage of propidium iodide-positive cells with subdiploid (sub-G1) DNA content.21 Upon dox treatment of the TetOn-TAp73α, -TAp73β or -TAp73γ Saos-2 cells, we observed an increase in the proportion of cells displaying a sub-G1 fraction, which is in line with the ability of p73 to stimulate apoptotic cell death (Figure 2a).7, 19 Notably, additional treatment of the cells with NECA resulted in a further increase in this sub-G1 DNA-containing population in TetOn-TAp73β and -TAp73γ, but not -TAp73α Saos-2 cells (Figure 2a). NECA treatment in the absence of Dox did not increase the proportion of apoptotic cells when compared with untreated samples. This indicates that the enhancement in cell death mediated by NECA following TAp73β and TAp73γ activation results from the increased expression of A2B by p73, and not from NECA stimulation of basal levels of A2B.
Figure 2.
A2B stimulates cell death downstream of p73. (a) TetOn-TAp73α, -TAp73β or -TAp73γ Saos-2 cells were treated with or without Dox, and/or NECA for 48 h and 72 h before analysis of cell death by measuring sub-G1 DNA content by flow cytometry (n=3, nine replicates in total). (b) TetOn-TAp73β (n=3, nine replicates in total) or (c) TetOn -TAp73γ (n=3, nine replicates in total) Saos-2 cells were treated with or without Dox, NECA and/or PSB 603 for 48 h and 72 h before analysis of sub-G1 DNA content by flow cytometry. (d, e) TetOn-TAp73β or -TAp73γ Saos-2 cells were transfected with either control or ADORA2B-specific smartpool siRNA before treatment with or without Dox and/or NECA. The effect of ADORA2B knockdown on cell death in TetOn-TAp73β (d) or TetOn-TAp73γ (e) Saos-2 cells were analysed by flow cytometry (n=3, nine replicates in total). (f, g) The efficiency of ADORA2B knockdown in TetOn-TAp73β (f) or TetOn-TAp73γ (g) Saos-2 cells was verified by qRT–PCR. Graph showing ADORA2B knockdown is representative of three separate experiments. ns, P>0.05; **P<0.01; ***P<0.001.
To confirm that the elevated cell death response following TAp73β and TAp73γ stimulation in combination with NECA is due to A2B receptor activation, we employed the use of the A2B-specific antagonist, PSB 60322 and RNAi to knockdown A2B levels. We observed that PSB 603 and A2B knockdown abolishes the enhancement in cell death by NECA in combination with dox (Figures 2b–e), confirming the role of A2B in mediating the cell death response downstream of TAp73β and TAp73γ. However, PSB 603 did not alter the levels of TAp73β and TAp73γ induced by dox treatment (Supplementary Figure 2), discounting the possibility that the inhibition of cell death is attributed to changes in p73 expression. We also tested the effect of NECA treatment on the TetOn-ΔNp73 Saos-2 cells but found that the cells do not die in response to NECA in combination with or without dox treatment (Supplementary Figures S1d). This was expected since ΔNp73 does not upregulate ADORA2B expression, as we described earlier (Supplementary Figure S1a).
p73-A2B receptor mediated cell death is caspase-dependent
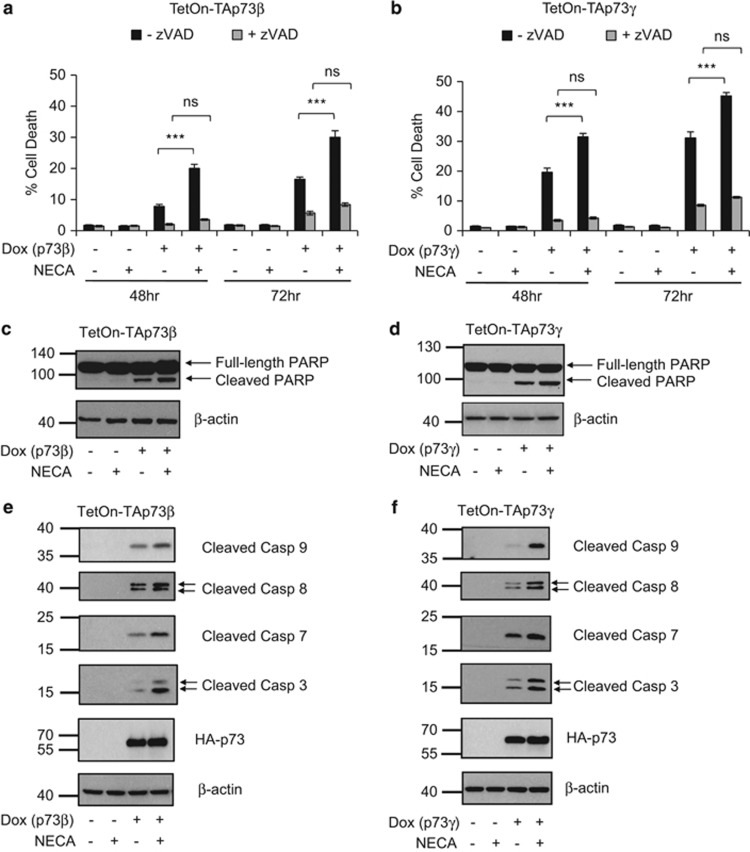
We were next interested to further assess the pathways by which A2B stimulates cell death upon p73 activation. We found that the pre-treatment of cells with the pan-caspase inhibitor, zVAD-fmk, completely blocked the cell death response elicited by A2B signalling downstream of TAp73β and TAp73γ activation (Figure 3a and b). In addition, we found that stimulation of A2B signalling downstream of p73 resulted in enhanced PARP cleavage (Figures 3c and d) and the appearance of cells that were positive for Annexin V and propidium iodide (Supplementary Figure S3). This indicates that A2B likely signals to a caspase-dependent, apoptotic pathway. Furthermore, western blotting analyses revealed that this enhanced cell death response by A2B in combination with TAp73β or TAp73γ activation was accompanied by increased detection of cleaved forms of caspases-9, -8, -7 and -3 (Figures 3e and f).
Figure 3.
A2B-induced cell death downstream of p73 is caspase-dependent. (a) TetOn-TAp73β or (b) TetOn-TAp73γ Saos-2 cells were treated with or without Dox, NECA and/or zVAD-fmk (50 μM) for 48 h and 72 h before cell death analysis by measuring sub-G1 DNA content by flow cytometry (n=3, eight replicates in total). (c–f) TetOn-TAp73β or -TAp73γ Saos-2 cells were treated with or without Dox and/or NECA for 48 h (p73β) and 30 h (p73γ), before western blotting detection of (e, f) HA-TAp73, cleaved caspase-9, −8, −7, −3, β-actin and (c, d) PARP. ns, P>0.05; ***P<0.001.
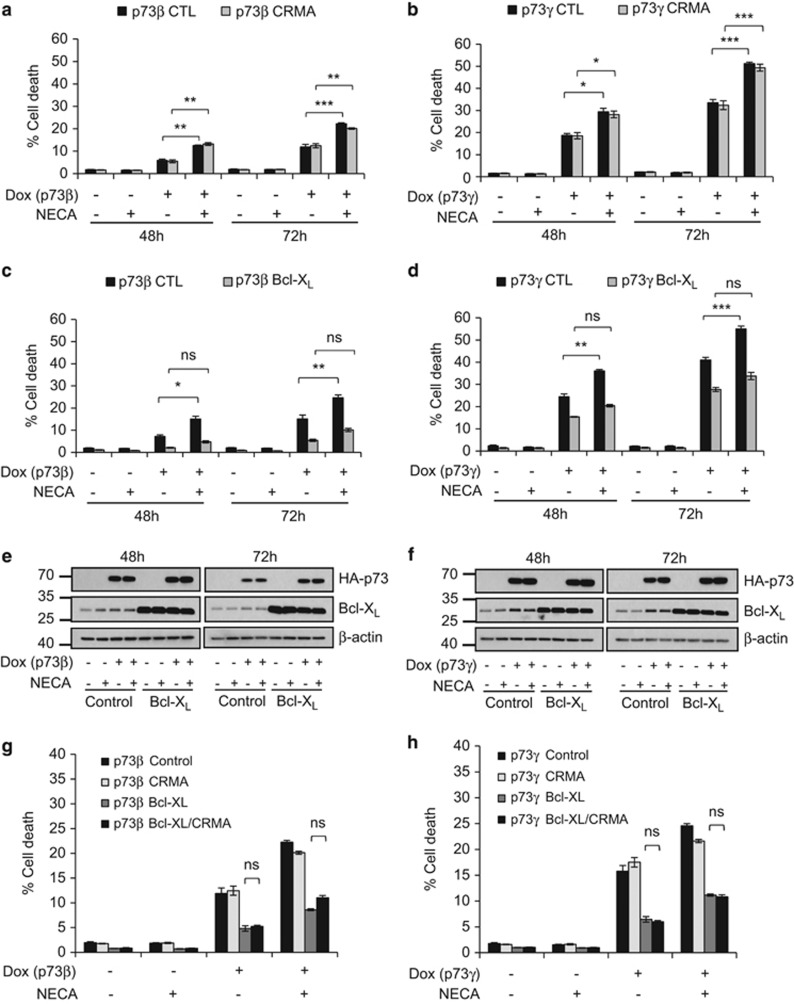
Typically, apoptosis can proceed via two different pathways, that is, the extrinsic/caspase-8-dependent, or the intrinsic-/caspase-9-dependent pathway.23 Crosstalk between the pathways has also been reported.24 In an attempt to elucidate the involvement of either/both of these pathways in mediating the cell death response by A2B and p73, we sought to separately inhibit these pathways and examine the effect on A2B-stimulated death. To attenuate the extrinsic pathway, we ectopically expressed the cowpox viral protein CrmA, which is a potent inhibitor of caspase-8,25, 26 in the TetOn-TAp73β and -TAp73γ Saos-2 cells. We observed that the extent of cell death induced by NECA and TAp73β or TAp73γ was not significantly altered by CrmA (Figures 4a and b). Expression of CrmA, however, effectively attenuated cell death in response to tumour necrosis factor-α (TNF-α) and cycloheximide (Supplementary Figure S4a and b), a combination that triggers caspase-8-dependent apoptosis,27 thereby validating the efficacy of the ectopically expressed viral protein. To attenuate the intrinsic pathway, we overexpressed the pro-survival mitochondrial protein Bcl-XL, which counteracts pro-apoptotic mitochondrial proteins that result in the permeabilization of the mitochondrial outer membrane, release of cytochrome c and activation of caspase-9.28, 29 The overexpression of Bcl-XL was able to prevent the increase in apoptosis caused by A2B stimulation downstream of TAp73β or TAp73γ activation (Figures 4c–f), indicating that A2B signals via the intrinsic apoptosis pathway in this context. In support of this notion, we also observed an enhanced release of cytochrome c upon engagement of A2B signalling downstream of p73 (Supplementary Figure S5). Overexpression of CrmA in combination with Bcl-XL did not further reduce the apoptotic response stimulated by A2B signalling downstream of p73 (Figures 4g and h and Supplementary Figure S4c), indicating that A2B signals primarily to the intrinsic pathway in this context. In this regard, it is also important to note that while cleavage of caspase-8 was observed upon engagement of A2B signalling downstream of p73 (Figures 3e and f), this was markedly reduced in cells expressing Bcl-XL (Supplementary Figure S6). This indicates that caspase-8 cleavage occurs downstream of MOMP as has previously been reported.30
Figure 4.
A2B engages the intrinsic apoptotic pathway downstream of p73. (a) TetOn-TAp73β or (b) TetOn-TAp73γ Saos-2 cells overexpressing either the control vector or CrmA were treated with or without Dox and/or NECA for 48 h and 72 h before flow cytometry analysis of sub-G1 DNA content (n=3, 9 replicates in total). (c) TetOn-TAp73β (n=3, nine replicates in total) or (d) TetOn -TAp73γ (n=2, six replicates in total) Saos-2 cells overexpressing either the control vector or Bcl-XL were treated with or without Dox and/or NECA for 48 h and 72 h before flow cytometry analysis of sub-G1 DNA content. (e, f) Western blot confirming overexpression of Bcl-XL in TetOn-TAp73β (e) or TetOn-TAp73γ Saos-2 (f) cells. (g) TetOn-TAp73β or (h) TetOn-TAp73γ Saos-2 cells overexpressing control vector, CrmA, Bcl-XL or both CrmA and Bcl-XL were treated with or without Dox and/or NECA for 72 h (p73β) or 48 h (p73γ) before flow cytometry analysis of sub-G1 DNA content (n=2, six replicates in total). ns, P>0.05; *P<0.05; **P<0.01; ***P<0.001.
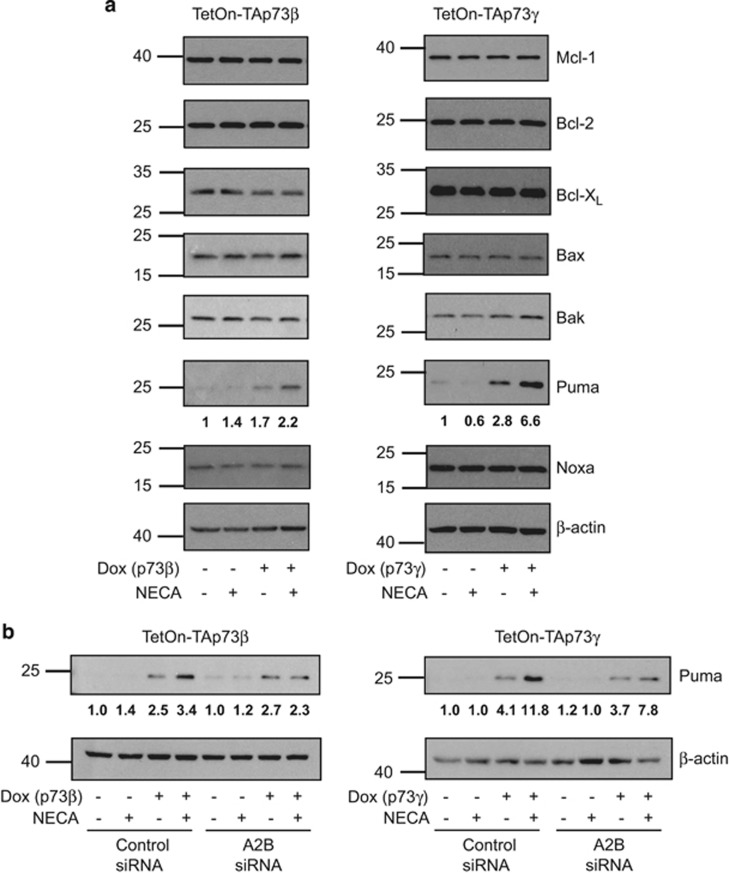
Levels of anti-apoptotic Bcl-2 family proteins are unaltered but Puma is upregulated in response to A2B signalling downstream of p73
As we previously observed A2B signalling to regulate changes in protein levels of Bcl-2, Bcl-XL, Mcl-1 and Puma,13 we decided to examine the levels of these proteins in response to p73 stimulation of A2B signalling. This revealed that while levels of Bcl-2, Bcl-XL and Mcl-1 were unaltered, Puma expression was clearly elevated upon TAp73β or TAp73γ activation in combination with NECA (Figure 5a) and this was found to be A2B dependent (Figure 5b). We also examined levels of pro-apoptotic Bak, Bax and Noxa, but found these proteins to be unaffected (Figure 5a).
Figure 5.
Levels of anti-apoptotic Bcl-2 family proteins are unaltered, but Puma is upregulated in response to A2B signalling downstream of p73. (a) TetOn-TAp73β or -TAp73γ Saos-2 cells were treated with or without Dox and/or NECA for 48 h (p73β) or 30 h (p73γ) before western blotting detection of Mcl-1, Bcl-2, Bcl-XL, Bax, Bak, Puma, Noxa and β-actin. (b) TetOn-TAp73β or -TAp73γ Saos-2 cells transfected with either control or ADORA2B-specific smartpool siRNA were treated with or without Dox and/or NECA for 48 h (p73β) and 30 h (p73γ) before western blotting detection of puma and β-actin. Fold changes in puma levels were quantified by densitometry, normalized to actin and indicated below the relevant western blots.
p73 engages A2B signalling during chemotherapy- and UV-induced cancer cell death
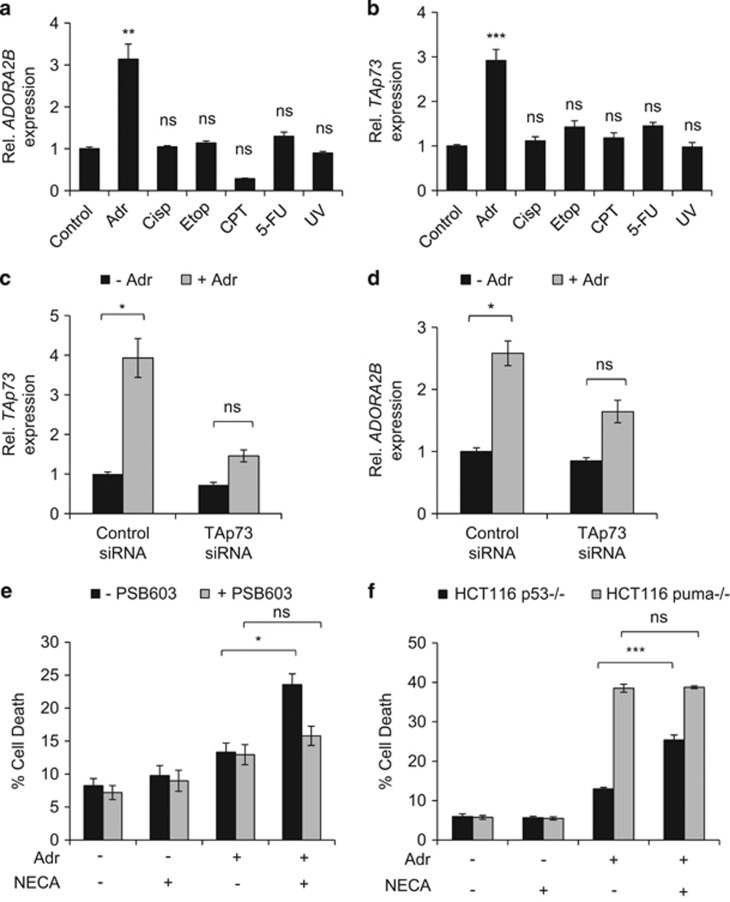
p73, like p53, is known to be activated in response to cellular stress, such as during genotoxic damage.11 We were therefore keen to examine whether p73 activation in response to genotoxic stress could also induce ADORA2B. To test this, we utilized HCT116 cells that are deficient in p53 (HCT116 p53−/−),31 to exclude the involvement in p53 in any ADORA2B induction responses we may observe. We challenged the HCT116 p53−/− cells with a panel of agents known to cause DNA damage and found that adriamycin (doxorubicin) treatment led to an increase in ADORA2B mRNA levels (Figure 6a). This corresponded with an increase in TAp73 expression (Figure 6b). To ascertain whether this increase was dependent on TAp73 activation, we performed RNAi-mediated knockdown of TAp73. This showed that adriamycin causes a significant induction of ADORA2B, which is lost upon knockdown of TAp73 by RNAi (Figures 6c and d).
Figure 6.
p73 upregulates A2B expression in response to chemotherapy. (a, b) p53-null HCT116 cells (HCT116 p53−/−) were treated with or without Adriamycin (0.5 μg/ml), Cisplatin (20μM), Etoposide (20 μM), Camptothecin (2 μM), 5-fluorouracil (5 μM) or UV-irradiation (100J/m2) for 24 h before quantification of ADORA2B (a) and TAp73 (b) mRNA levels by qRT-PCR (n=2–3, 6–9 replicates in total). (c, d) HCT116 p53−/−transfected with either control or TAp73-specific siRNA were treated with Adriamycin (0.5 μg/ml) for 48 h in the presence of zVAD-fmk (50 μM) (to inhibit caspase activation) before quantification of TAp73 (c) and ADORA2B (d) mRNA levels by qRT–PCR (n=3, nine replicates in total). (e) HCT116 p53−/− cells were treated with Adriamycin (0.5 μg/ml), NECA and/or PSB 603 (2 μM) for 48 h before analysis of sub-G1 DNA content by flow cytometry (n=3–5, 9–15 replicates in total). (f) p53-null and puma-null HCT116 cells (HCT116 p53−/−; HCT116 puma−/−) were treated with Adriamycin (0.5 μg/ml) and/or NECA for 48 h before analysis of sub-G1 DNA content by flow cytometry (n=3, eight replicates in total). ns, P>0.05; *P<0.05; **P<0.01; ***P<0.001.
Subsequently, we found NECA treatment to enhance the cell death response triggered by adriamycin in HCT116 p53−/− cells (Figure 6e). This NECA-mediated enhancement of death was found to be A2B-dependent, as the response could be abrogated by PSB 603 treatment (Figure 6e). Previous studies have also reported a role for Puma in p73-induced cell death.19, 32 In this regard, we found that cell death enhancement by NECA did not occur in Puma-null cells indicating that A2B-mediated cell death is also Puma-dependent (Figure 6f). Importantly, NECA treatment on its own (in the absence of adriamycin) does not promote cell death, showing that the p73-A2B pathway is only engaged upon combined treatment with adriamycin and NECA (Figure 6e).
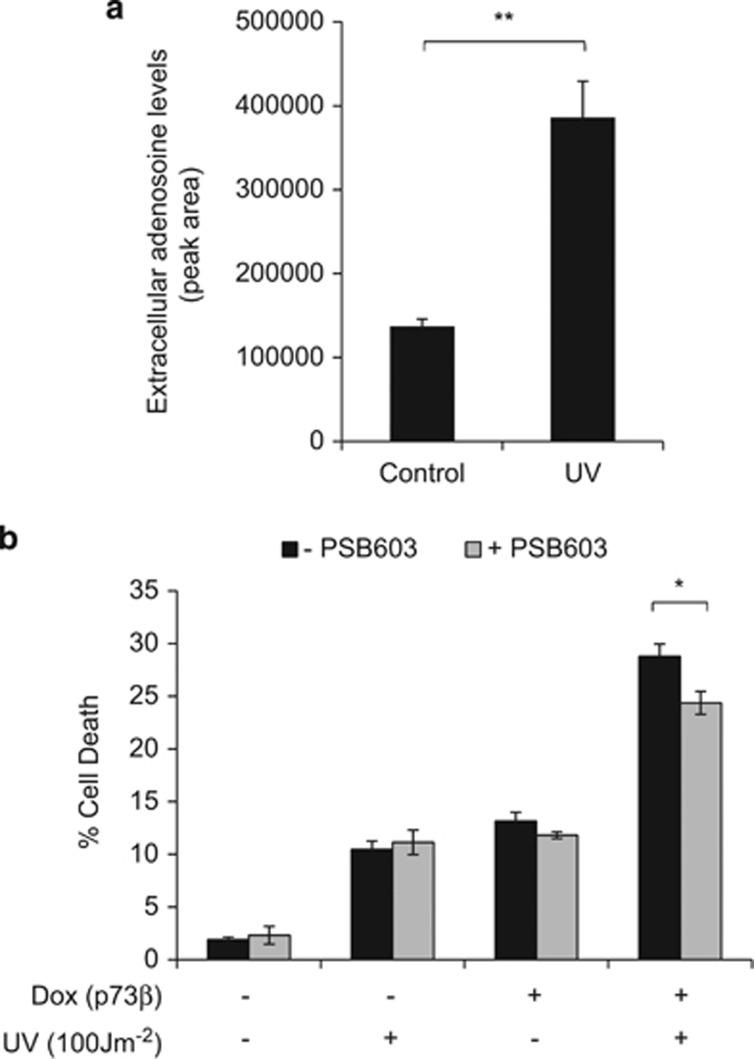
We also observed that exposure of our p73-inducible cells to UV resulted in an accumulation of extracellular adenosine (Figure 7a). Activation of TAp73β in this context resulted in enhanced cell death (Figure 7b). Moreover, treatment with PSB603 resulted in a small, but significant decrease in this cell death response, indicating involvement of A2B in this context (Figure 7b) and highlighting another cell death situation in which the A2B axis can be involved.
Figure 7.
UV irradiation promotes extracellular adenosine accumulation and enhances p73-mediated cell death. (a) Extracellular adenosine detection by LC-MS analysis of conditioned media obtained from TetOn-TAp73β Saos-2 cells treated with or without UV irradiation (100J/m2) for 48 h (n=3). (b) TetOn-TAp73β Saos-2 cells treated with or without Dox, UV irradiation (100J/m2) and/or PSB 603 (1.5 μM) for 48 h before flow cytometry analysis of sub-G1 DNA content (n=3, nine replicates in total). *P<0.05; **P<0.01.
Discussion
Owing to its important role in tumour suppression and its potent cell death-inducing potential, p53 has been highlighted as a credible target for cancer therapy. However, because of the frequent mutation of p53 in human cancer, strategies to activate p53 are limited in their applicability. By contrast, the relatively infrequent inactivation of p73 in cancer makes targeting p73 and its downstream signalling components, a conceivably more promising approach. To this end, it is critical to understand the ways in which p73 can induce programmed cell death. In this study, we described a new signalling pathway that potentiates p73-mediated cancer cell death via activation of the adenosine receptor, A2B. We identified ADORA2B (the gene that encodes A2B) as a new target gene of p73, which can be induced in response to chemotherapy treatment. Strikingly, the combined treatment of chemotherapy with an A2B agonist stimulates more cell death compared with chemotherapy alone. This prominent response occurs in cells deficient in p53 and utilizing this may be a beneficial approach for targeting cancers with deregulated p53 function.
An initial interesting observation from this study is that we only detected induction of p73 and ADORA2B in response to adriamycin. This is particularly intriguing, as other studies have reported p73 induction in response to other drug treatments that we used in our study.33, 34, 35, 36, 37, 38 It may be the case that this is a cell-type-specific effect. However, as we were unable to effectively detect expression of all TA-p73 isoforms at the protein level, it could be that other drugs induce p73 post-transcriptionally in our cells. If this were the case, it still remains that the only drug that induces ADORA2B mRNA in our cells is adriamycin. It is also possible therefore that adriamycin causes post-translational modifications to p73, which facilitate ADORA2B induction, or that adriamycin induces other factors that cooperate with p73 to induce ADORA2B. Whatever the mechanism(s) involved for the selective induction of ADORA2B, it is without question that this is an area worthy of future investigation.
Similar to what we previously observed with A2B signalling downstream of p53,13 p73 stimulation of A2B results in a caspase- and Puma-dependent apoptotic cell death. In line with the Puma dependency and similar to what we observed for p53, death from p73 in the presence of adenosine can be repressed by overexpression of Bcl-XL, but not by crmA. This indicates that death proceeds via the mitochondrial pathway without involvement of caspase 8. The engagement of the mitochondrial pathway as a result of p73-mediated stimulation of A2B signalling appears, however, to be different to when A2B signalling is activated by p53. We observed that stimulation of A2B signalling by p73 involves upregulation of Puma, but Bcl-2 and Bcl-XL do not appear to be downregulated upon A2B activation as was the case downstream of p53.13 This difference may suggest that other factors that are activated by p53, but not p73, are responsible for the downregulation of Bcl-2 and Bcl-XL. This may also imply that the downregulation of Bcl-2 and Bcl-XL alone may not be solely responsible for p53-A2B stimulation of cell death, but that it is more the actual balance and interaction between pro- and anti-apoptotic mitochondrial proteins that eventually determines the fate of cells. As a result, this would suggest that activation of A2B signalling by p53 and p73 may result in an additive response through the combined effect on different members of the Bcl-2 family.
In addition to the observed differences in the way p53 and p73 engage adenosine signalling, it was also interesting to see that the cellular response upon stimulation of A2B following activation by p73 was isoform-specific. While all three isoforms of p73 could induce ADORA2B expression, agonist stimulation of A2B only enhances death in response to TAp73β and TAp73γ, but not TAp73α. Previous reports have shown that TAp73β and TAp73γ are, in their own right, much better inducers of cell death than TAp73α.19 This may be due to the fact that different target genes are activated by each of the isoforms due to differences in transcriptional activity as has previously been reported.39, 40 Thus, certain TAp73β- and/or TAp73γ-induced genes may co-operate in cell death induction upon stimulation of A2B signalling. Conversely, it may also be the case that TAp73α, but not TAp73β and TAp73γ, activates gene(s) that promote survival and therefore antagonize cell death responses downstream of A2B. Lastly, of course, it must also be considered that when TAp73α activates A2B this is not part of a cell death response at all. It remains possible that cellular responses other than cell death, which were not examined in this study, could be stimulated by A2B signalling induced in this context. Whatever the reasons for these differences in the response to A2B activation, the differential regulation of A2B function by p73 isoforms is undoubtedly fascinating and warrants further in-depth exploration.
Despite these open questions, the findings from this study certainly corroborate an important conclusion—p73 can engage A2B receptor signalling to trigger cancer cell death in cells exposed to chemotherapeutic drug treatments or agents such as UV, which cause accumulation of extracellular adenosine. As this occurs in the absence of p53, this potentially opens up situations in which modulation of this pathway could be considered as a potential target for cancer therapy. It must be noted, however, that through our analysis of cBioportal for Cancer Genomics (www.cBioportal.org)41, 42 and The Wellcome Trust Sanger Institute's COSMIC (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/)43 databases we observed that allelic loss of ADORA2B has been reported in tumours of lung, ovarian, breast and large intestine among others (Supplementary Figure S7). This indicates that ADORA2B may be a tumour suppressor gene and its connection to cell death pathways downstream of p53 and p73 would certainly support this possibility. If future studies prove this to be the case, then profiling of ADORA2B status may well be prudent in therapeutic situations, which are thought to involve accumulation of extracellular adenosine.
Materials and Methods
Reagents
Adenosine-5'-N-ethylcarboxamide (NECA), Doxycycline (Dox), Adriamycin, Cisplatin, Etoposide, Camptothecin, 5-Fluorouracil, TNF-α and cycloheximide were purchased from Sigma (Dorset, UK). z-Val-Ala-Asp-fluoromethyl ketone (zVADfmk) was purchased from Bachem (Weil am Rhein, Germany). PSB 603 was purchased from Tocris (Bristol, UK).
Cell culture
All cell lines were maintained in DMEM supplemented with 10% FBS and L-glutamine, and cultured in 21% O2 and 5% CO2. Saos-2 and Pheonix Eco were obtained from the ATCC. HCT116 p53−/− and HCT116 puma−/− cells were kind gifts from Bert Vogelstein.31 TetOn-TAp73α Saos-2, TetOn-TAp73β Saos-2, TetOn-TAp73γ Saos-2 and TetOn-ΔNp73 Saos-2 cells were kind gifts from Gerry Melino and previously described.17 All TetOn cells were induced with 1 μg/ml Dox. For all experiments, 20 μM NECA was used. TetOn-TAp73β CrmA Saos-2, TetOn-TAp73γ CrmA Saos-2, TetOn-TAp73β Bcl-XL Saos-2, TetOn-TAp73γ Bcl-XL Saos-2, TetOn-TAp73β CrmA/Bcl-XL Saos-2 and TetOn-TAp73γ CrmA/Bcl-XL Saos-2cells were generated by retroviral infection with pBabe-puro-CrmA or -Bcl-XL (plasmids as previously described13) followed by selection with 1μg/ml puromycin (Sigma) and/or 5μg/ml blasticidin (Life Technologies, Paisley, UK).
RNAi
ADORA2B knockdown was performed using pre-designed siGENOME SMARTpool Human ADORA2B siRNA purchased from Dharmacon (Lafayette, CO, USA). TAp73 isoform-specific knockdown was performed using Silencer Pre-designed TAp73 siRNA (ID: 115665) purchased from Ambion (Life Technologies). siGENOME Non-Targeting siRNA #1 (Dharmacon) and Silencer Negative Control siRNA #1 (Ambion, Life Technologies) were used as control siRNAs, respectively. ADORA2B siRNA oligonucleotides were transfected using Oligofectamine (Invitrogen, Life Technologies) at ~50% confluency according to the manufacturer's instructions for 72 h before treatment. TAp73 siRNA oligonucleotides were transfected using Lipofectamine RNAiMAX (Invitrogen) at ~30% confluency according to the manufacturer's instructions for 72 h before treatment.
Western blotting
Cells were lysed in the buffer containing 50 mM Hepes, 150 mM NaCl, 10 mM EDTA, 100 mM NaF, 10 mM Na4P2O7, 1% Triton X-100, 0.1% SDS and protease inhibitor cocktail (Roche, West Sussex, UK) for 30 min on ice before centrifugation for 15 min at 20 000g. Thirty micrograms of protein was separated by SDS–PAGE and transferred to nitrocellulose membranes using standard techniques. Membranes were probed using standard immuno-blotting techniques with the following antibodies: CrmA (556472, Becton Dickinson, Oxford, UK), Mcl-1 (559027, Becton Dickinson); cleaved caspase-3 (9664, Cell Signalling, Beverly, MA, USA); cleaved caspase-7 (9491, Cell Signalling, Beverly, MA, USA); cleaved caspase-8 (9496, Cell Signalling); cleaved caspase-9 (9501, Cell Signalling); HA-tag (12013819001, Roche); Bcl-XL (AC-0098RUO, Epitomics, Cambridge, UK); Puma (IMG-458-2, Imgenex, Novus Biologicals, Cambridge, UK); Bak (06-536, Millipore, Watford, UK); Noxa (2437, ProSci Inc., Poway, CA, USA); Bax (sc-493, Santa Cruz, Dallas, TX, USA), Bcl-2 (sc-509, Santa Cruz), p21 (sc-397, Santa Cruz); and β-actin (ab8227, Abcam, Cambridge, UK).
Flow cytometry
For sub-G1 analysis, total population of cells, including floating and adherent cells, were stained with propidium iodide (PI) and processed for flow cytometric analysis as described previously.44 Acquired events were analysed in FL-2 channel with the CellQuest software (Becton Dickinson). The percentage of cells with sub-G1 DNA content was taken as a measure of the extent of apoptosis in the cell population at the indicated times. For Annexin V and PI co-staining, cells were co-stained with Annexin V-Alexa 488 and PI according to the manufacturer's instructions (Life Technologies). Fluorescence outputs for Annexin V and PI were detected in FL-1 and FL-3 channels, respectively.
Quantitative reverse-transcription polymerase chain reaction
qRT–PCR was undertaken as previously described.45 All samples were normalized to 18S rRNA using primers previously described.46 Primers for ADORA2B are validated QuantiTect primers from Qiagen (Manchester, UK). Primers for TAp73 are forward 5′-CAGACAGCACCTACTTCGACCTT-3′ and reverse 5′-CCGCCCACCACCTCATTA-3′, as previously described.47 qRT-PCR amplification was performed for 40 cycles at 94 °C for 10 s, 55 °C for 30 s, and 72 °C for 30 s with initial activation cycle at 95 °C for 15 min and final extension cycle at 72 °C 10 min.
Luciferase reporter assays
A total of 1.5 × 105 wild-type Saos-2 cells were plated per well in six-well dishes 24 h before co-transfection of luciferase reporter constructs containing potential p53 binding sites (as previously described13) with either control vector or TAp73α-, TAp73β- or Tap73γ-expressing constructs by CaPO4. Twenty-four hours after transfection, cells were lysed in 200 μl of 1 × Luciferase Cell Culture Lysis Reagent (Promega, Southampton, UK) and assayed for luciferase activity according to the manufacturer's instructions using the Veritas Microplate Luminometer.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as described by Nelson et al.48 with some modifications. In brief, cell cross-linking was done by adding 37% formaldehyde to a final concentration of 1% for 8 min at RT, followed by the addition of glycine to a final concentration of 125 mM. Cells were lysed in 1 ml ChIP buffer (150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.5% NP-40, 50 mM Tris–HCl (pH 7.5)) containing 1.5 × protease and phosphatase inhibitor cocktail (Thermo, Renfrew, UK) by passing through a 26-G needle. Nuclei were spun and lysed in 0.5 ml ChIP buffer supplemented with 0.5% SDS and 1 mM sodium butyrate. The lysate was sonicated five times for 30 s with 30 s intervals in the Diagenode Bioruptor (Diagenode, Seraing, Belgium) set to maximum power. Seventy-five microlitres of extract was saved for Input. One hundred and fifty microlitres of extract was diluted five times with ChIP buffer (with no SDS) and used for IP. For each IP 3 μg of antibody was added. The antibody used for p73 ChIPs were rabbit Anti-HA (A190-108A, Bethyl Laboratories, Montgomery, TX, USA) and normal rabbit IgG (X0903, Dako, Glostrup, Denmark) as a negative control; for p53 ChIP, Anti-p53 (DO-1, BD Pharmingen) and mouse Anti-HA (05–904, Upstate, Millipore, Watford, UK) as a negative control. Following overnight incubation at 4 °C with rotation, 50 μl of Protein G magnetic beads (Dynal Life Technologies, Paisley, UK) was added and further incubated for 6 h. Beads were washed once with ChIP buffer, once with high salt buffer (500 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.5% NP-40, 50 mM Tris–HCl (pH 7.5)), once with LiCl buffer (250 mM LiCl, 1 mM EDTA, 0.5% Na-deoxycholate, 0.5% NP-40, 10 mM Tris–HCl (pH 7.5)), one more time with ChIP buffer and once with final wash buffer (0.1% NP-40, 10 mM Tris–HCl (pH 7.5)).
Real-time PCR quantification of immunoprecipitated DNA fragments
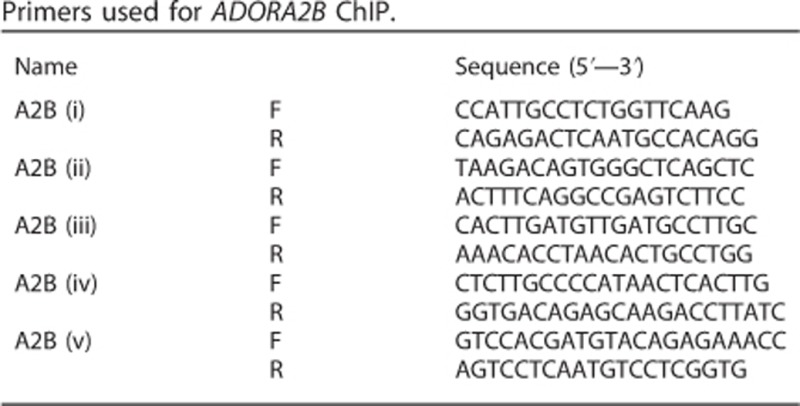
For ChIP experiments, the input and immunoprecipitated samples were assayed by quantitative PCR. PCR reactions contained 2 μl of DNA template and 300 nM of each primer. Quantitative PCR was performed on a BioRad (Hemel Hempstead, UK) CFX 96 units using a 3-min incubation at 95 °C, followed by 40 cycles of 20 s at 95 °C, 20 s at 57 °C (58 °C for region (i) primers) and 20 s at 72 °C (with a plate read after each cycle). The primer sequences used are described in the table below. Data were processed in BioRad CFX Manager 3.0. Occupancy values are expressed as a corrected immunoprecipitation efficiency (% IP/Input [target antibody]−(% IP/Input [control antibody]).
Cytochrome c release assay
The assay for measuring cytochrome c release was adapted from previously described method by Waterhouse and Trapani.49 In brief, adherent and non-adherent cells were collected by trypsinization and collected with 50 μg/ml digitonin (in PBS containing 100 mM potassium chloride and 1 mM EDTA) for 10 min on ice. Cells were subsequently washed with ice-cold PBS, fixed in paraformaldehyde (4% in PBS) on ice for 20 min and incubated in blocking buffer (2% BSA in PBS) for 30 min at room temperature. Cells were then incubated with 1:200 anti-cytochrome c antibody (Clone 6H2.B4, Biolegend, San Diego, CA, USA) overnight at 4 °C. The next day, cells were washed with PBS and incubated with 1:200 anti-mouse-Alexa 488 antibody (A11001, Life technologies) for 1 h at room temperature. Finally, cells were washed with PBS before flow cytometry analysis in FL-1 channel.
Adenosine measurement by liquid chromatography-mass spectrometry
Measurement of extracellular adenosine in conditioned media of UV-treated cells were performed as previously described.13 In brief, the conditioned media was combined with acetonitrile (1:3 volume) and left to shake for 10 min at 4 °C to allow adenosine extraction. Any insoluble material was pelleted and the supernatant was collected for subsequent LC-MS analysis. The remaining cells on the dishes were trypsinized, counted and the adenosine measurements were normalized to cell number.
Statistical tests
Data are presented as mean±s.e. The statistical significance between treatments was determined by Student's two-tailed t-test when comparing two groups and one-way analysis of variance (ANOVA) when comparing multiple groups. P<0.05 was considered statistically significant. In the Figures, P-values are denoted as follows—ns, P>0.05; *P<0.05; **P<0.01; ***P<0.001. All data were analysed with GraphPad Prism.
Acknowledgments
We thank members of the Tumour Cell Death laboratory for critical reading of the manuscript. Work in the Tumour Cell Death Laboratory is supported by Worldwide Cancer Research and Cancer Research UK.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- 1Crighton D, Ryan KM. Splicing DNA-damage responses to tumour cell death. Biochim Biophys Acta 2004; 1705: 3–15. [DOI] [PubMed] [Google Scholar]
- 2Hollstein M, Hergenhahn M, Yang Q, Bartsch H, Wang ZQ, Hainaut P. New approaches to understanding p53 gene tumor mutation spectra. Mut Res 1999; 431: 199–209. [DOI] [PubMed] [Google Scholar]
- 3Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408: 307–310. [DOI] [PubMed] [Google Scholar]
- 4Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Gene Dev 2008; 22: 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Stiewe T, Zimmermann S, Frilling A, Esche H, Putzer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res 2002; 62: 3598–3602. [PubMed] [Google Scholar]
- 6Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci 2000; 113: 1661–1670. [DOI] [PubMed] [Google Scholar]
- 7Jost CA, Marin MC, Kaelin WG Jr. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature 1997; 389: 191–194. [DOI] [PubMed] [Google Scholar]
- 8Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2002; 2: 605–615. [DOI] [PubMed] [Google Scholar]
- 9Oswald C, Stiewe T. In good times and bad: p73 in cancer. Cell Cycle 2008; 7: 1726–1731. [DOI] [PubMed] [Google Scholar]
- 10Bell HS, Dufes C, O'Prey J, Crighton D, Bergamaschi D, Lu X et al. A p53-derived apoptotic peptide derepresses p73 to cause tumor regression in vivo. J Clin Invest 2007; 117: 1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG Jr. Chemosensitivity linked to p73 function. Cancer Cell 2003; 3: 403–410. [DOI] [PubMed] [Google Scholar]
- 12Fredholm BB, IJzerman AP, Jacobson KA, Klotz KN, Linden J. International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev 2001; 53: 527–552. [PMC free article] [PubMed] [Google Scholar]
- 13Long JS, Crighton D, O'Prey J, Mackay G, Zheng L, Palmer TM et al. Extracellular adenosine sensing-A metabolic cell death priming mechanism downstream of p53. Mol Cell 2013; 50: 394–406. [DOI] [PubMed] [Google Scholar]
- 14Blay J, White TD, Hoskin DW. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Res 1997; 57: 2602–2605. [PubMed] [Google Scholar]
- 15Ohta A, Gorelik E, Prasad SJ, Ronchese F, Lukashev D, Wong MK et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc Natl Acad Sci USA 2006; 103: 13132–13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Raskovalova T, Huang X, Sitkovsky M, Zacharia LC, Jackson EK, Gorelik E. Gs protein-coupled adenosine receptor signaling and lytic function of activated NK cells. J Immunol 2005; 175: 4383–4391. [DOI] [PubMed] [Google Scholar]
- 17Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 2000; 19: 4283–4289. [DOI] [PubMed] [Google Scholar]
- 18Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997; 90: 809–819. [DOI] [PubMed] [Google Scholar]
- 19Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem 2004; 279: 8076–8083. [DOI] [PubMed] [Google Scholar]
- 20Martin AG, Trama J, Crighton D, Ryan KM, Fearnhead HO. Activation of p73 and induction of Noxa by DNA damage requires NF-kappa B. Aging 2009; 1: 335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Pellicciari C, Manfredi AA, Bottone MG, Schaack V, Barni S. A single-step staining procedure for the detection and sorting of unfixed apoptotic thymocytes. Eur J Histochem 1993; 37: 381–390. [PubMed] [Google Scholar]
- 22Borrmann T, Hinz S, Bertarelli DC, Li W, Florin NC, Scheiff AB et al. 1-alkyl-8-(piperazine-1-sulfonyl)phenylxanthines: development and characterization of adenosine A2B receptor antagonists and a new radioligand with subnanomolar affinity and subtype specificity. J Med Chem 2009; 52: 3994–4006. [DOI] [PubMed] [Google Scholar]
- 23Long JS, Ryan KM. New frontiers in promoting tumour cell death: targeting apoptosis, necroptosis and autophagy. Oncogene 2012; 31: 5045–5060. [DOI] [PubMed] [Google Scholar]
- 24Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J 1998; 17: 1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Muzio M, Salvesen GS, Dixit VM. FLICE induced apoptosis in a cell-free system. Cleavage of caspase zymogens. J Biol Chem 1997; 272: 2952–2956. [DOI] [PubMed] [Google Scholar]
- 26Zhou Q, Snipas S, Orth K, Muzio M, Dixit VM, Salvesen GS. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J Biol Chem 1997; 272: 7797–7800. [DOI] [PubMed] [Google Scholar]
- 27Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703. [DOI] [PubMed] [Google Scholar]
- 28Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol 2008; 18: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 2010; 11: 621–632. [DOI] [PubMed] [Google Scholar]
- 30Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD et al. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 1999; 144: 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998; 282: 1497–1501. [DOI] [PubMed] [Google Scholar]
- 32Fricker M, Papadia S, Hardingham GE, Tolkovsky AM. Implication of TAp73 in the p53-independent pathway of Puma induction and Puma-dependent apoptosis in primary cortical neurons. J Neurochem 2010; 114: 772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Lunghi P, Costanzo A, Levrero M, Bonati A. Treatment with arsenic trioxide (ATO) and MEK1 inhibitor activates the p73-p53AIP1 apoptotic pathway in leukemia cells. Blood 2004; 104: 519–525. [DOI] [PubMed] [Google Scholar]
- 34Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol cell 2008; 32: 803–814. [DOI] [PubMed] [Google Scholar]
- 35Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 2007; 117: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG Jr, Levrero M et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999; 399: 806–809. [DOI] [PubMed] [Google Scholar]
- 37Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 1999; 399: 814–817. [DOI] [PubMed] [Google Scholar]
- 38Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 1999; 399: 809–813. [DOI] [PubMed] [Google Scholar]
- 39Sauer M, Bretz AC, Beinoraviciute-Kellner R, Beitzinger M, Burek C, Rosenwald A et al. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res 2008; 36: 1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Klanrit P, Flinterman MB, Odell EW, Melino G, Killick R, Norris JS et al. Specific isoforms of p73 control the induction of cell death induced by the viral proteins, E1A or apoptin. Cell Cycle 2008; 7: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011; 39: D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126: 121–134. [DOI] [PubMed] [Google Scholar]
- 45O'Prey J, Skommer J, Wilkinson S, Ryan KM. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell Cycle 2009; 8: 2260–2265. [DOI] [PubMed] [Google Scholar]
- 46Wilkinson S, O'Prey J, Fricker M, Ryan KM. Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity. Gene Dev 2009; 23: 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA 2009; 106: 797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 2006; 1: 179–185. [DOI] [PubMed] [Google Scholar]
- 49Waterhouse NJ, Trapani JA. A new quantitative assay for cytochrome c release in apoptotic cells. Cell Death Differ 2003; 10: 853–855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.