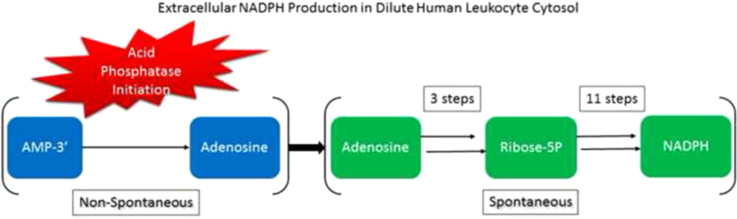
Abstract
Complex metabolism is thought to occur exclusively in the crowded intracellular environment. Here we report that diluted enzymes from lysed human leukocytes produce extracellular energy. Our findings involve two pathways: the purine nucleotide catabolic pathway and the pentose phosphate pathway, which function together to generate energy as NADPH. Glucose6P fuel for NADPH production is generated from structural ribose of purine ribonucleoside monophosphates, ADP, and ADP-ribose. NADPH drives glutathione reductase to reduce an oxidized glutathione disulfide-glutathione redox couple. Acid phosphatases initiate ribose5P salvage from purine ribonucleoside monophosphates, and transaldolase controls the direction of carbon chain flow through the nonoxidative branch of the pentose phosphate pathway. These metabolic control points are regulated by pH. Biologically, this energy conserving metabolism could function in perturbed extracellular spaces.
Abbreviations: AUC, area under the curve; FGT, female genital tract; GC–MS, gas chromatography-mass spectrometry; GSH, reduced glutathione; GSSG, oxidized glutathione; LC–MS, liquid chromatography–mass spectrometry; NET, neutrophil extracellular trap; NO, nitric oxide; NOB-PPP, non-oxidative branch pentose phosphate pathway; OB-PPP, oxidative branch pentose phosphate pathway; Pi, inorganic phosphate; PAP, human prostate acid phosphatase; PNP, purine nucleoside phosphorylase; PNCP, purine nucleotide catabolic pathway; TLS, tumor lysis syndrome; u, units; XOR, xanthine oxidoreductase
Keywords: Extracellular metabolism, Redox regulation, Human leukocytes, Leukocyte and human prostate acid phosphatases, Transaldolase, Glutathione reductase
Graphical abstract
Highlights
-
•
Extracellular metabolism occurs in dilute human leukocyte cytosolic protein solution.
-
•
Extracellular metabolism is activated by acid phosphatases.
-
•
Ribose5P salvage from purine nucleotides and its metabolism produces NADPH.
-
•
Extracellular NADPH generating metabolism influences the redox environment.
-
•
Extracellular metabolism could be a component of inflammation and wound healing.
1. Introduction
The impetus for the experiments reported here occurred while studying the synthesis of nitric oxide (NO) from l-arginine by murine NO synthase in 100,000g supernatants. This reaction is driven by reducing equivalents delivered by NADPH [1]. However, in control experiments, we observed that purine ribonucleotides, which do not carry energy as reducing equivalents, could substitute for NADPH and drive the NO synthase reaction. This result, at that time, did not have a rational explanation. Later, in an attempt to understand this puzzling observation, we used cytosol prepared from human leukocytes to detect NADPH production.
Cytosol, a term often inaccurately employed to describe the aqueous cytoplasm of intact cells, is referred to here as: “that portion of the cell which is found in the supernatant fraction after centrifuging the homogenate, diluted in buffer, at 105,000g for 1 h” [2]. The concentration of cytosolic proteins in experimental reaction mixtures ranged from 75 ng/μl to 200 ng/μl (0.0075% to 0.02% protein solution respectively). This represents cytosolic proteins obtained from 3.38×103 to 9×103 leukocytes which, in cells/μl, correspond to a low to normal adult human peripheral blood leukocyte count [3] based on a calculation of 22.2 pg of cytosolic protein/leukocyte (see Section 2).
Cytosolic proteins contain enzymes that normally function in the crowded aqueous cytoplasm of intact cells (170×103 ng/μl to 350×103 ng/μl protein) [4]. However, after release from human leukocytes and dilution to 75 ng/μl to 200 ng/μl protein, the diluted enzymes generate energy as NADPH that counters oxidative challenge in the form of added GSSG via cytosolic glutathione reductase [5] (Fig. 1, Reaction 14). Coordinated metabolism by released enzymes survives leukocyte homogenization, cytosol preparation and dilution of cytosolic proteins.
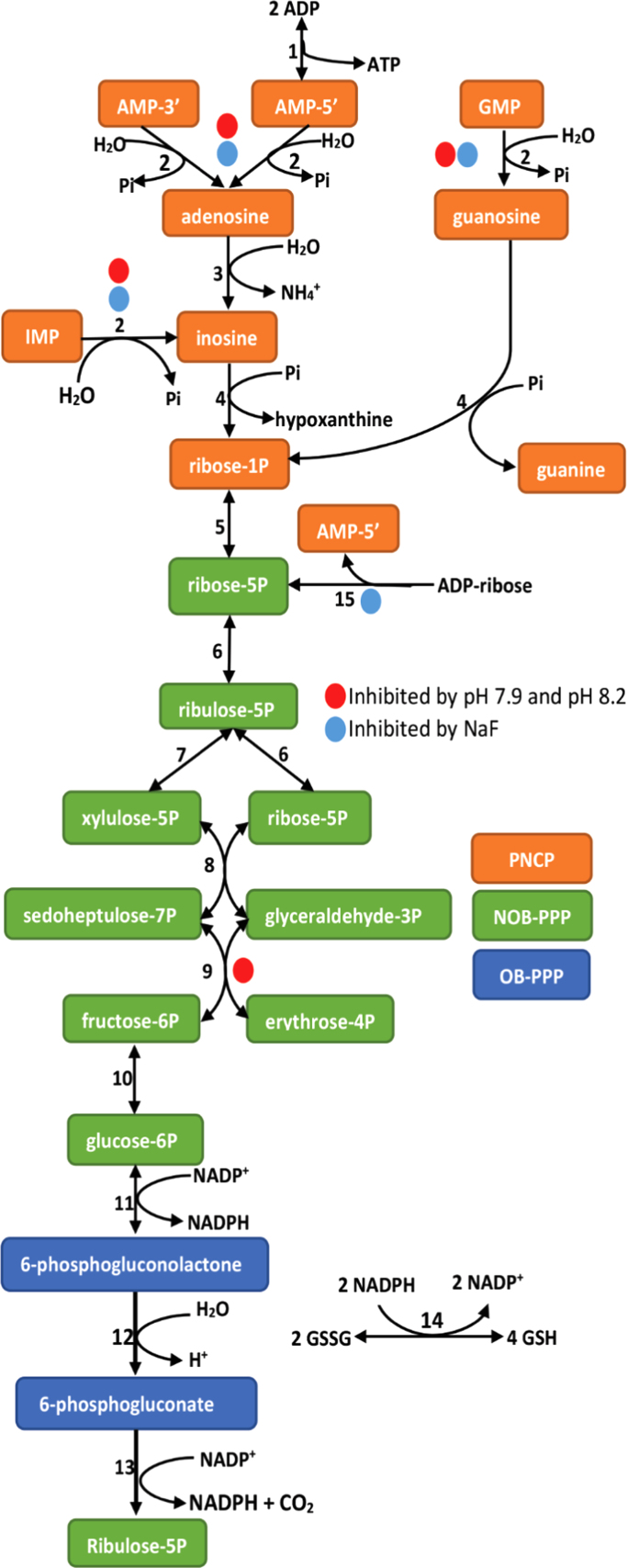
Fig. 1.
Metabolism and NADPH production in dilute cytosol prepared from human leukocytes as measured by reduction of added GSSG to 2GSH by cytosolic glutathione reductase (Reaction 14). Other cytosolic enzymes involved in this energy yielding extracellular metabolism are: 1. adenylate kinase, 2. cytosolic acid phosphatases, 3. adenosine deaminase, 4. purine nucleoside phosphorylase 5. phosphoribomutase, 6. ribulose5P isomerase, 7. ribulose5P epimerase, 8. transketolase, 9. transaldolase, 10. phosphoglucose isomerase, 11. glucose6P dehydrogenase, 12. 6-phosphogluconolactonase, 13. 6-phosphogluconate dehydrogenase, and 15. ADP-ribose pyrophosphatase. Blue coded reactions are inhibited by NaF, and red coded reactions are inhibited by pH 7.9–8.2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Using this experimental system, we observed that ribose5P, a stable structural component of purine ribonucleoside monophosphates, ADP, and ADP-ribose (produced in stressed cells) [6], is salvaged and metabolically converted to glucose6P. Glucose6P is the immediate carbon chain fuel for NADPH production. In effect, this dilute solution metabolism, which generates reducing equivalents carried by NADPH from molecules containing a ribose5P moiety, acts as an extracellular biofuel cell. It is an extracellular energy conserving process that occurs without energy input or a need for oxygen. See Fig. 1 for a summary of the extracellular metabolism discussed.
The enzymatically catalyzed dilute solution sugar phosphatate interconversion reactions reported here are similar to the enzyme free abiotic dilute solution metabolism recently reported to support the theory of a prebiotic origin of metabolism (see Section 4).
2. Materials and methods
2.1. Biochemical reagents
3-(N-Morpholino)propanesulfonic acid, 4-Morpholinepropanesulfonic acid (MOPS), 5,5′-Dithiobis(2-nitrobenzoic acid (DTNB), Adenosine 5′-diphosphoribose sodium salt (ADP-ribose), Dextran from Leuconostoc mesenteroides, d-Glucose 6-phosphate disodium salt hydrate, d-Ribose 5-phosphate disodium salt hydrate, l-Ascorbic acid, l-Glutathione oxidized disodium salt (GSSG), N-[Tris(hydroxymethyl)methyl]glycine (Tricine), Penicillin G potassium salt, Potassium phosphate monobasic anhydrous (KH2PO4), β-Nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt hydrate (NADPH), β-Nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP+), Adenosine 3′-monophosphate (AMP-3′), Adenosine 5′-monophosphate sodium salt (AMP-5′), Adenosine 5′-diphosphate sodium salt (ADP), Adenosine 5′-triphosphate disodium salt (ATP), Ammonium molybdate tetrahydrate, Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA), Guanosine 5′-monophosphate disodium salt hydrate (GMP), Inosine, Inosine 5’-monophosphate disodium salt (IMP), l-Glutathione reduced (GSH), N,N-Bis(2-hydroxyethyl)glycine (Bicine), Phenol red solution 0.5%, Sodium chloride, Sodium fluoride (NaF), Sodium phosphate dibasic (Na2HPO4), Sodium phosphate monobasic monohydrate (NaH2PO4·H2O), were from Sigma-Aldrich, St Louis, MO, USA. Heparin, sodium solution and N-[2-Hydroxyethyl]piperazine-N′-[2-Ethanesulfonic Acid] (HEPES) were from USB, Cleveland, OH, USA. Human prostate acid phosphatase was from Lee Biosolutions Inc., St Louis, MO, USA. Sulfuric acid was from Mallinckrodt Baker, St Louis, MO, USA.
2.2. Source of human leukocytes and human serum
Three healthy male donors ranging in age from 28–75 y.o. provided the peripheral blood leukocytes and the serum used in these experiments. The University of Utah Health Science Center and the Salt Lake City VA Medical Center Institutional Review Boards approved this research. Written informed consent was obtained from each of the participants.
2.3. Isolation of human leukocytes
Human venous blood (180–200 ml total) was collected aseptically into four 50 ml syringes containing heparin at a final concentration of 20 units/ml. The heparinized blood was mixed with an equal volume of 0.9% NaCl solution containing 2% Dextran (MW=400,000–500,000 from Sigma Cat. no. D103) and held at room temperature for 40 min to permit sedimentation of erythrocytes. The leukocyte rich supernatant was transferred to 50 ml tubes and centrifuged 5 min at 700g to sediment leukocytes. The few contaminating erythrocytes were lysed by addition of 5 ml of sterile distilled deionized water to the cell pellet for 20 s, followed immediately by the addition of 45 ml 0.9% NaCl. The leukocytes were washed three more times in 0.9% NaCl by centrifugation at 200g for 5 min. Total number of cells was determined by microscopy using a hemocytometer.
2.4. Preparation of the 100,000g supernatant (cytosol) from human peripheral blood leukocytes
Pelleted cells were re-suspended in a buffer containing H2O+15 mM HEPES+phenol red 8 mg/L, pH 7.4 at 108 cells/ml and disrupted by sonication (Bronson ultrasonic homogenizer). Cell lysis was monitored by light microscopy and the process stopped when >99% of the cells were disrupted. Two other methods of cell disruption were used with identical results (a cell disrupter bomb and a Potter-Elvehjem tissue grinder). The buffer containing the disrupted cells was centrifuged at 100,000g at 4° for 60 min (Beckman L5-5B centrifuge, SW 41 rotor). The 100,000g supernatant was filtered through a 0.22 µm filter, aliquoted, and the protein concentration determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc. Catalog no. 500-0006). The aliquoted 100,000g supernatant protein was frozen at −70° until used. Cytosol remains metabolically active for more than 1 year after preparation when frozen at −70°. Proteinase inhibitors were not added to cell lysates or to cytosol during preparation, storage, or the bioassay. We define the 100,000g supernatant prepared from lysates of human peripheral blood leukocytes as cytosol. The term cytosol was first proposed in 1965 by H.A. Lardy. His definition is quoted by Clegg [2]. Cytosol approximates the aqueous cytoplasm of intact cells.
We calculated the average contribution of cytosolic protein of human peripheral blood leukocytes to be 22.2 pg/cell. Total cellular protein of human peripheral blood neutrophils has been reported to be 50 pg/cell [7]. The aqueous cytoplasm fraction has been measured to be 57% of total human neutrophil cellular protein [8]. From these measurements, cytosolic protein is ~28.5 pg protein/human neutrophil. Lymphocytes are 20–45% of human peripheral blood leukocytes [3]. Circulating lymphocytes are small “resting” non-activated cells with scant aqueous cytoplasm. Our measurement of 22.2 pg cytosolic protein/cell likely approximates that of the heterogenous population of circulating human peripheral blood leukocytes. Example of how we calculated average cytosol protein contribution per leukocyte: 16.67 ml of a cytosolic protein stock solution containing 0.6 mg of cytosolic protein/ml was prepared from 450×106 peripheral blood leukocytes (16.67 ml×0.6 mg/ml cytosolic protein=10 mg total cytosolic protein). 10 mg cytosolic protein=10×109 pg cytosolic protein÷450×106 leukocytes=22.2 pg cytosolic protein/leukocyte.
Human peripheral blood leukocytes should be used as the source of cytosolic protein to reproduce the dilute solution extracellular metabolism reported here. Human DLD-1 colon adenocarcinoma cells and murine EMT-6 mammary adenocarcinoma cells require significantly higher concentrations of cytosolic protein to carry out metabolism that is less efficient (unpublished data).
2.5. Collection and dialysis of human serum
Human serum was obtained by centrifugation (250g) of aseptically collected clotted blood and stored at 4° until use. 10 ml of serum was placed into dialysis tubing (diameter 11.5 mm) with molecular weight cut-off 3500 (Spectrum Laboratories Inc. Cat. no. 132720). The tubing was then immersed in 4 l of normal saline and kept there with agitation for 24 h at 4°. After 24 h, the tubing was transferred to 4 l of fresh normal saline. The process was repeated one more time for total of three changes of dialysate. The serum was removed and filtered through 0.22 µm filter.
2.6. Basic assay for measurement of extracellular metabolism
Experiments were carried out in 96 well microtiter plates with a final volume (which included all additives and reagents) of 80 µl. All reagents and additives used were dissolved in distilled deionized water containing 15 mM HEPES and 8 mg/liter phenol red at pH 7.4. The basic constituents of the 80 μl reaction mixtures were: 2 mM Mg(acetate)2, 50 mM NaCl, 100 units/ml penicillin G, and 8 mg/L phenol red. The choice of organic buffer used was based on the pH poise desired for the experiment being performed: pH 6.9 (15 mM MOPS), pH 7.4 (15 mM HEPES), pH 7.9 (15 mM Tricine), and pH 8.2 (15 mM Bicine). Reaction mixture pH was monitored using an Orion Research Digital pH meter model 611. Concentrations of cytosolic protein, GSSG (always 3 mM), the nicotinamide nucleotide hydride ion carrier (NADP+), and the carbon chain fuel sources for each experiment are indicated in the Figure and/or in the Figure legends of the experiments reported. The microtiter plate was vortexed (to mix all reagents), sealed with pressure film (to prevent evaporation) and incubated for 20 h (unless otherwise indicated) in the dark at 37°. At the end of incubation, aliquots of reaction mixture were removed for measurement of reduced glutathione, determination of inorganic phosphate and/or analysis of metabolites via gas chromatography/mass spectrometry or liquid chromatography/mass spectrometry.
2.7. Reduced glutathione assay
We used a modification of the enzymatic, kinetic glutathione 96 well microtiter plate assay described by Allen et. al. [9] to measure GSH. Procedure for measurement of GSH: To prepare the DTNB reagent add 1 ml of the DTNB stock solution to 14 ml of phosphate buffered saline (PBS). Pipet 125 µl of the DTNB reagent into a microtiter well. Add a 4 µl sample of experimental reaction mixture to the DTNB reagent in a microtiter well and mix. Incubate at room temperature for 5 min. Measure absorbance at 420 nm using a TECAN GENios microplate reader with an appropriate range of standards.
2.8. Inorganic phosphate assay
We used a modification of the inorganic phosphate assay described by Ames [10] which is based on the principle of phosphomolybdate complex reduction by ascorbic acid. This assay was adapted for absorbance readings using small sample volumes from experiments carried out in a microtiter plate. Procedure for measurement of inorganic phosphate: Pipet 75 µl of the ascorbic acid/molybdate reagent into a microtiter well. Add a 32 µl sample from the experimental reaction mixture to the reagent in the microtiter well, mix, and incubate for 1 h at 37°. Measure absorbance at 820 nm using a TECAN GENios microplate reader with an appropriate range of standards. Samples with higher phosphate levels were diluted to measurable concentrations prior to assay.
2.9. Metabolite extraction and analysis by GC–MS and LC–MS
The method of Jiye et. al. [11] was used with slight modification to remove protein by precipitation. In brief, 684 μl of −20°, 90% methanol (aq.) was added to 76 μl of the individual tubes containing the assay to give a final concentration of 80% methanol. The samples were incubated for one hour at −20° followed by centrifugation at 30,000g for 10 min using a rotor chilled to −20°. The supernatant containing the extracted metabolites was then transferred to fresh disposable tubes and completely dried en vacuo.
GC–MS analysis was performed on each metabolite present in three replicate samples with a Waters GCT Premier mass spectrometer fitted with an Agilent 6890 gas chromatograph and a Gerstel MPS2 autosampler. Dried samples were suspended in 40 μl of a 40 mg/ml O-methoxylamine hydrochloride (MOX) in pyridine and incubated for one hour at 30°. 25 μl of this solution was added to autosampler vials. 30 μl of N-methyl-N-trimethylsilyltrifluoracetamide (MSTFA) was added automatically via the autosampler and incubated for 60 min at 37° with shaking. After incubation 3 μl of a fatty acid methyl ester standard solution was added via the autosampler followed by 1 μl of the prepared sample injected into the gas chromatograph inlet in the split mode with the inlet temperature held at 250°. A 10:1 split ratio was used for analysis. The gas chromatograph had an initial temperature of 95° for one minute followed by a 40°/min ramp to 110° and a hold time of 2 min. This was followed by a second 5°/min ramp to 250°, a third ramp to 350°, then a final hold time of 3 min. A 30 m Phenomex ZB5-5 MSi column with a 5 m long guard column was employed for chromatographic separation. Helium was used as the carrier gas at 1 mL/min. GC–MS data was collected using MassLynx 4.1 software (Waters). Metabolites were identified and their peak area was recorded using QuanLynx (Waters). Metabolite identity was established using chemical library developed using pure purchased standards. These data were transferred to an Excel spreadsheet. The total area for all metabolites observed in each sample was summed, each metabolite was divided by this summed total area and multiplied by 100 to produce percent metabolite AUC.
LC–MS analysis was performed on each metabolite present in three replicate samples as previously described [12]. In brief a Phenomenex (Torrence, CA) 3.0 mm×150 mm Gemini-NX C18 (5 μm) column with a Phenomenex Security Guard column filled with the same packing material. The chromatographic system consisted of an integrated Shimadzu HPLC system consisting of two LC-10AD pumps, column oven and a CBM-20A controller. A PE200 autosampler with a cooling unit set to 4° was used for sample handling. A PE Sciex API 365 mass spectrometer modified with an Ionics EP 10+ source was used for analyte detection. A mobile phase consisting of solvent A (water with 15 mM ammonium formate/6.5 mM N-dibutylamine) and solvent B (methanol/6.5 mM N-dibutylamine) was used for elution of samples. The initial condition was 5% B with an initial hold time of 3 min followed by a ramp to 73% B over 21 min. A second ramp to 90% B was employed over the next min with a 1 min hold. The column was brought back to 5% B over two min and re-equilibrated for 9 min. The flow rate was 0.3 mL/min at 24°. Mass spectrometer transition optimization was performed using a syringe pump. For each metabolite optimized it was dissolved in buffer A as a 1 mg/mL solution. Infusion was performed at 20 μl/min while 10% B/90% A buffer was co-infused using the HPLC at 0.3 mL/min. To each sample was added 50 μl of 10 mM K2PO4 pH 7 followed by a brief sonication using a water bath sonicator. Each sample was clarified by 10 min of centrifugation at 20,000g followed by transfer to an autosampler vial. Samples kept at 4° until analysis. After analysis each metabolites peak height was recorded in Excel. The total area for all metabolites observed in each sample was summed, each metabolite was divided by this summed total area and multiplied by 100 to produce percent metabolite AUC.
2.10. Statistical analysis
The data were analyzed using the biostatistical software GraphPad Prism® 4.0. Graphical data models were created using multiple grouping variables and standard two-way ANOVA tests. Analysis of variance was based on 3 or more replicates of each tested variable, from which the Standard Error of the Mean (SEM) and Standard Deviation (SD) were calculated.
3. Results
3.1. Initiation of dilute solution extracellular energy conserving metabolism
The first enzymatic steps in extracellular NADPH generation is the release of purine ribonucleoside monophosphate components of larger molecules. Purine ribonucleoside monophosphates must be salvaged from molecules such as RNA (not evaluated in this study), ADP, and ADP-ribose. Purine ribonucleoside monophosphates are stable molecules unless acted upon by acid phosphatases. The conversion of purine ribonucleoside monophosphates to purine ribonucleosides by acid phophatases with 3′-5′ nucleotidase activity is the enzymatic step that initiates extracellular dilute solution metabolism. Purine ribonucleosides, once released, undergo spontaneous catabolism that generates NADPH in dilute cytosol.
To determine requirements for initiation of catabolism that generates extracellular energy as NADPH, the reduction of added GSSG to 2GSH by cytosolic glutathione reductase was measured. We used NaF to inhibit the production of purine ribonucleosides from larger molecules that are carbon chain fuel sources for NADPH generation as determined by GSH production (Fig. 2A). NaF inhibits lysosomal [13], prostate [14], and macrophage/osteoclast [15] acid phosphatases. NaF also inhibits ADP-ribose pyrophosphatase [16] and prevents the generation of AMP-5′ and ribose5P from ADP-ribose (Fig. 1, Reaction 15).
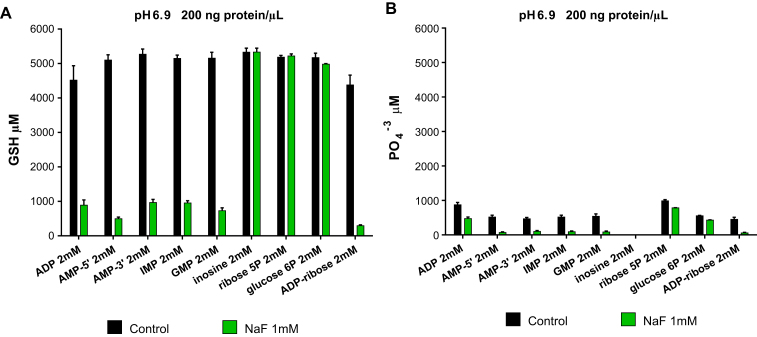
Fig. 2.
Molecules with a structural ribose moiety and sugar phosphates are carbon chain fuel sources for NADPH production in dilute cytosol. (A) Effect of cytosolic acid phosphatases and NaF on catabolism of carbon chain fuels or (B) phosphate ester hydrolysis in the same experiments as (A). NADPH generation was determined by measurement of μM GSH produced from 3mM GSSG added to each experiment. X-axes show carbon chain fuel added to each experiment. X-axes (color coded bars) also show experiments with and without NaF (A,B). Y-axes show μM GSH generated in experiments (A) or μM Pi produced by cytosolic acid phosphatases (B). 200 ng/μl cytosolic protein and NADP+ 20 μM were added to all experiments. Incubations were for 1200 min at pH 6.9 at 37°. Data are the mean (±SEM) of three separately performed experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
NaF decreased NADPH generation as measured by decreased GSH production (Figs. 2A and S1C) and phosphate ester hydrolysis (Figs. 2B and S1D) from purine ribonucleoside monophosphates, ADP, and ADP-ribose. NaF did not inhibit GSH generation when inosine [PNP substrate] [17], ribose5P [NOB-PPP substrate] [18], or glucose6P [OB-PPP substrate] [18] were the carbon chain fuel sources or the carbon chain fuel (Fig. 2A). These results show that cytosolic acid phosphatases initiate catabolism of purine ribonucleoside monophosphates (including AMP-5’ produced by adenylate kinase; Fig. 1, Reaction 1) and that cytosolic ADP-ribose pyrophosphatase initiates catabolism of ADP-ribose. In contrast, inosine (a purine ribonucleoside), ribose5P, and glucose6P, when added to cytosol, are catabolized spontaneously (Fig. 2A).
3.2. Pi requirement for the spontaneous extracellular catabolism of purine ribonucleosides by PNP
The Pi released by 3′-5′ nucleotidase activity of extracellular cytosolic acid phosphatases is co-substrate for PNP, which converts inosine to ribose1P/hypoxanthine and guanosine to ribose1P/guanine (Fig. 1, Reactions 2 and 4). Fig. 2A shows that added inosine is spontaneously catabolized and is an effective carbon chain fuel source. Maximal GSH production occurred even though the reaction mixture Pi concentration was below the limit of detection when the experiment was terminated (Fig. 2B). To decrease the Pi available for the PNP reaction we lowered the reaction mixture cytosolic protein concentration from 200 ng/μl (0.02% solution of cytosolic protein) (Fig. 2) to 75 ng/μl (0.0075% solution of cytosolic protein) (Fig. 3). This also lowered the reaction mixture Pi concentration as determined by a need for supplemental Pi for maximal GSH production (Fig. 3). To determine if co-substrate Pi consumption is a marker of PNP catalytic activity, Pi was added to reaction mixtures (except those labeled controls) in increasing amounts (Fig. 3A–D). Experiments shown in Fig. 3A did not have an added carbon chain fuel source or NADP+. This provided a baseline to compare Pi levels measured in metabolically active reaction mixtures (Fig. 3B–D).
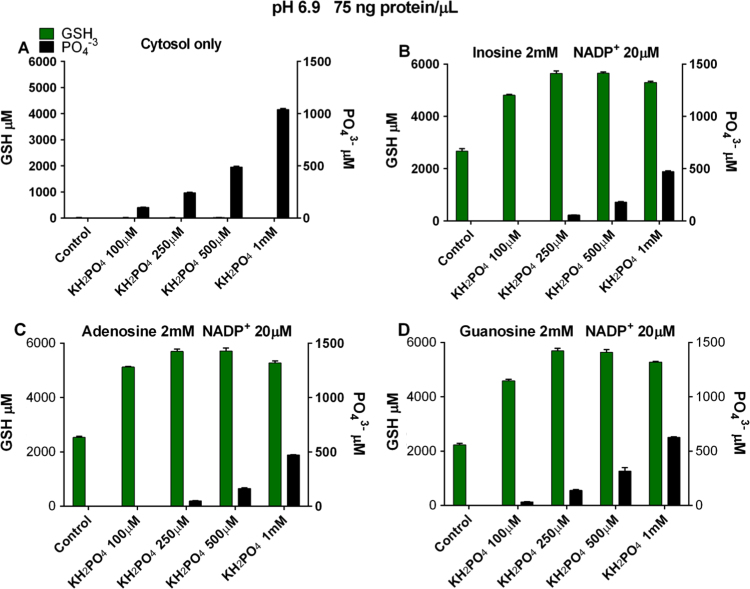
Fig. 3.
Pi requirement for spontaneous extracellular catabolism of purine ribonucleosides. (A) Cytosol only without KH2PO4 (control) or cytosol only with added KH2PO4. (B–D) Cytosol plus an added purine ribonucleoside and NADP+ without KH2PO4 (control) or with added KH2PO4. Presence of a purine ribonucleoside carbon chain fuel source and NADP+ added to individual experiments are indicated above each panel (B–D). NADPH generation was determined by measurement of GSH produced from 3mM GSSG added to all experiments (A–D). X-axes show control experiments (no Pi added) and experiments supplemented with Pi. Y-axes show on the left of panel μM GSH generated and on the right of the panel μM Pi added or consumed (coded bars). 75 ng/μl cytosolic protein added to all experiments (A–D). Incubations were for 1200 min at pH 6.9 at 37°. Data are the mean (±SEM) of three separately performed experiments.
We examined inosine, adenosine (co-substrate for PNP after conversion to inosine by adenosine deaminase, Fig. 1 Reaction 3), and guanosine (co-substrate for PNP) as carbon chain fuel sources. Even though the Pi assay used was not sufficiently sensitive to detect the presence of endogenous cytosolic Pi in control experiments not supplemented with Pi (Fig. 3B–D), sufficient endogenous cytosolic Pi existed to support spontaneous catabolism of purine ribonucleosides and the generation of 2 to 3 mM GSH. In reaction mixtures supplemented with Pi, GSH production increased compared to control experiments without supplemental Pi. Maximal GSH production occurred with 250 μM KH2PO4 added to reaction mixtures with purine ribonucleoside carbon chain fuel sources (Fig. 3B–D). These results show that the extracellular Pi concentration is rate limiting for the spontaneously occurring PNP reaction, and that purine ribonucleoside induced Pi co-substrate consumption is a marker of extracellular PNP catalytic activity.
3.3. Acid phosphatases and pH regulate extracellular metabolism of purine ribonucleoside monophosphates
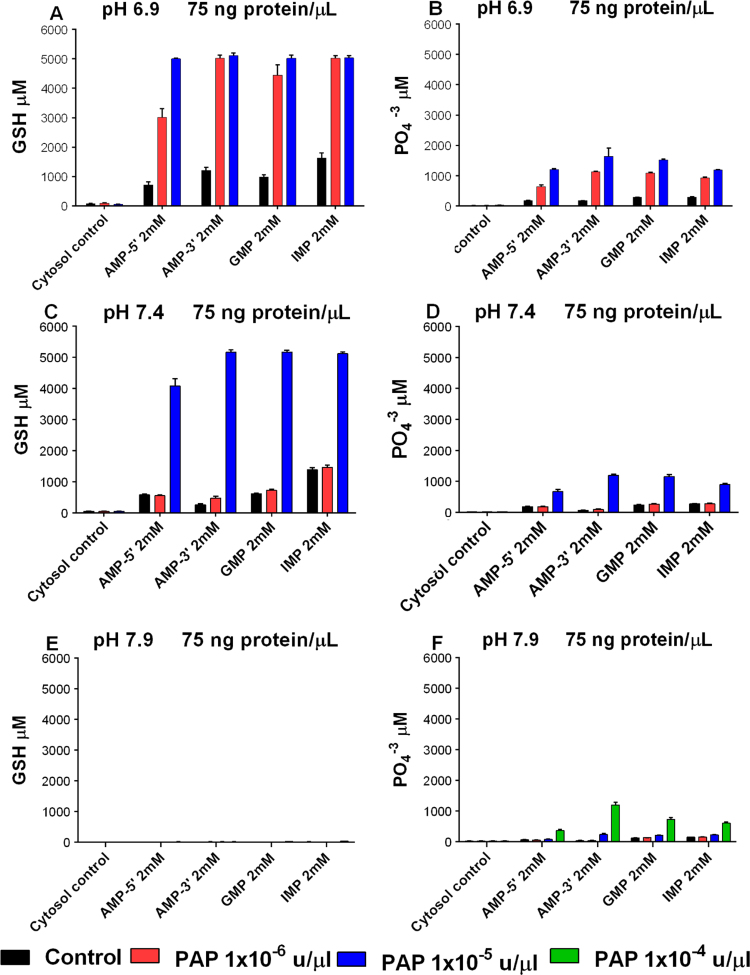
The initiation of extracellular catabolism by acid phosphatases was further evaluated by lowering reaction mixture cytosolic protein concentration to 75 ng/μl (Fig. 4). This reduced endogenous cytosolic acid phosphatase activity for purine ribonucleoside monophosphate carbon chain fuel sources, as measured by GSH production and phosphate ester hydrolysis; compare endogenous cytosolic acid phosphatase activity in experiments illustrated in Fig. 2A and B (200 ng/μl cytosolic protein) to the control experiments (endogenous acid phosphatase activity only) in Fig. 4A and B (75 ng/μl cytosolic protein). In addition we tested the effect of pH and PAP on the initiation of purine ribonucleoside monophosphate catabolism.
Fig. 4.
PAP and pH regulate extracellular catabolism of purine ribonucleoside monophosphates. (A) GSH production without and with added PAP; pH 6.9. (B) Phosphate ester hydrolysis in the same experiments as (A). (C) GSH production without and with added PAP; pH 7.4. (D) Phosphate ester hydrolysis in the same experiments as (C). (E) GSH production without and with added PAP; pH 7.9. (F) Phosphate ester hydrolysis in the same experiments as (E). NADPH generation was determined by measurement of GSH produced from 3 mM GSSG added to each experiment. X-axes show the carbon chain fuel sources added to each experiment. A carbon chain fuel source was not added to cytosol controls. X-axes (color coded bars) also show experiments with and without added PAP. Y-axes show μM GSH generated (A,C,E) or μM Pi produced by cytosolic acid phosphatases or cytosolic acid phosphatases plus added PAP (B,D,F). 75 ng/μl cytosolic protein and 20 μM NADP+ were added to all experiments. Incubations were for 1200 min at the pH indicated at 37°. Data are the mean (±SEM) of three separately performed experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Added PAP increased GSH production and phosphate ester hydrolysis at pH 6.9 and pH 7.4 (Fig. 4A–D). GSH production and phosphate ester hydrolysis initiated by cytosolic acid phosphatases with or without supplemental PAP was inhibited at pH 7.9 (Fig. 4E and F). A higher concentration of PAP, 1×10−4 u/μl, was added to the experiments illustrated in Fig. 4E and F, but this did not reverse the inhibition of GSH production (Fig. 4E). These results show that added PAP supplements the activity of cytosolic acid phosphatases and initiates maximal NADPH generating catabolism of purine ribonucleoside monophosphates at pH 6.9 and pH 7.4. At pH 7.9 the initiation of energy generating metabolism by cytosolic acid phosphatases and by added PAP is totally inhibited, even when phosphate ester hydrolysis occurs after adding 1×10−4 u/μl PAP (Fig.4E and F). This suggests that, in addition to acid phosphatase inhibition, a second site of inhibition exists at pH 7.9.
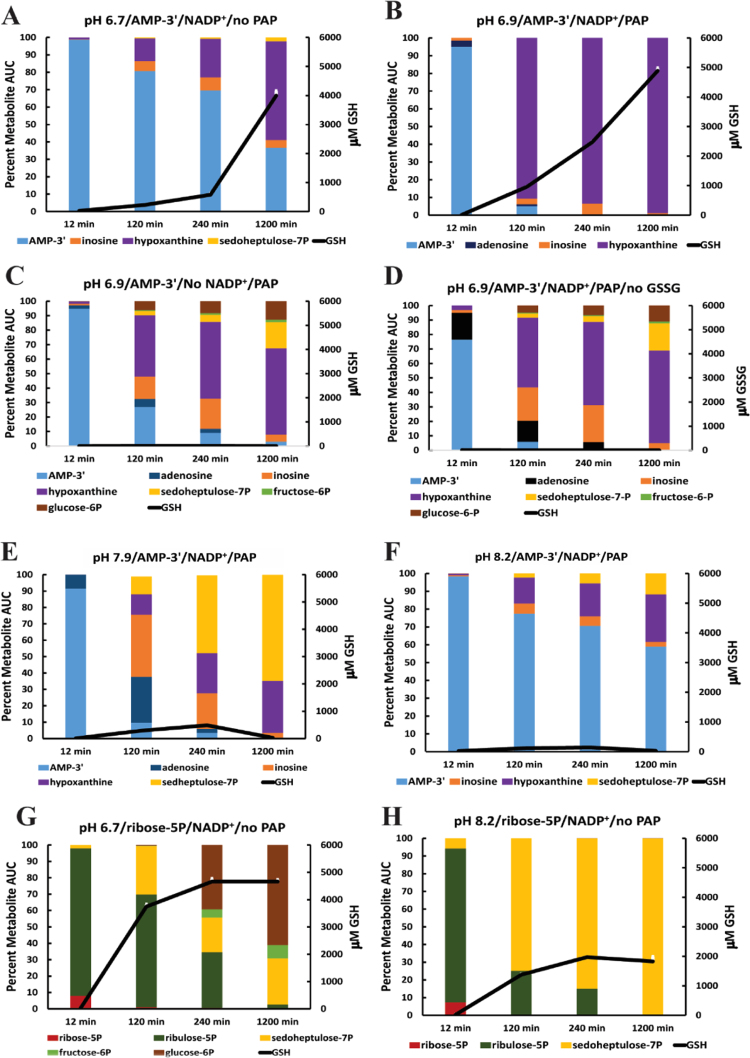
3.4. Analysis of extracellular metabolic flow
GC–MS was used to track metabolites during the catabolism of AMP-3′ by the PNCP/PPP. Results are presented as %AUC of metabolites measured (see Section 2). Triplicate reaction mixtures containing 150 ng/μl cytosolic protein were sampled at 12 min, 120 min, 240 min, and 1200 min. GC–MS analysis and measurement of GSH production was carried-out on each triplicate sample. AMP-3′ catabolism, initiated by cytosolic acid phosphatases at pH 6.7 is shown in Fig. 5A. Significant AMP-3′ remains uncatabolized as determined by the slow decline of AMP-3′ %AUC and slow increase in hypoxanthine %AUC (product of PNP; Fig. 1, Reaction 4) during the 1200 min experiment. However, GSH production (4.5 mM at 1200 min) occurs under conditions of low metabolite flow. To accelerate AMP-3′ catabolism, cytosolic acid phosphatases were supplemented with PAP, 1×10−5 u/μl at pH 6.9 (Fig. 5B). Added PAP caused more rapid metabolite flow as determined by the following: rate of AMP-3′ %AUC decline, inosine and hypoxanthine %AUC increase, appearance of adenosine %AUC, and increased kinetics of GSH production compared to reaction mixtures that were not supplemented with PAP (Fig. 5A vs. B). These experiments show the importance of acid phosphatases in the initiation of extracellular metabolite flow that results in NADPH production.
Fig. 5.
GC–MS analysis of extracellular metabolite flow from AMP-3’ and ribose5P correlated with GSH production. (A) AMP-3′ catabolism initiated by cytosolic acid phosphatases. (B) Effect of added PAP on AMP-3′ catabolism. (C) Effect of not adding NADP+ on AMP-3′ catabolism. (D) Effect of not adding GSSG on AMP-3′ catabolism. (E) Effect of pH 7.9 on AMP-3′ catabolism. (F) Effect of pH 8.2 on AMP-3′ catabolism. (G) Ribose5P catabolism at pH 6.7. (H) Ribose5P catabolism at pH 8.2. The added carbon chain fuel sources were AMP-3′ 2 mM (A–F) and ribose5P 2 mM (G,H). All experiments contained NADP+ 20 μM except (C) and GSSG 3 mM except (D). PAP 1×10−5 u/μl was added to (B–F) but not to (A,G,H). Incubations were at 37°. X-axes show the time of termination for metabolite %AUC and GSH measurements. Note: time not to scale. Y-axes show metabolite %AUC and μM GSH production. GSH production error bars from triplicate samples (mean±SEM) are marked in white. Most fall within the symbol denoting the mean and are not visible.
3.5. Effect of blocking reducing equivalent consumption on metabolite profiles
To determine metabolite %AUC patterns when flow of reducing equivalents from the OB-PPP was blocked, the electron carrier NADP+ or the electron acceptor GSSG were not added to reaction mixtures (Fig. 5C and D). In both experiments, metabolites from AMP-3′ flowed through the PNCP and the NOB-PPP, but GSH was not produced (Fig. 5C and D). However, in both cases glucose6P, fructose6P (markers of terminal pathway malfunction), and sedohepulose7P began to accumulate between 12 min and 120 min reaching a maximum during the 240 min to 1200 min interval. This shows that terminal NOB-PPP metabolites accumulate at early time points when the electron carrying cofactor NADP+ for the OB-PPP is not present or when the electron acceptor GSSG was not added to the reaction mixture. The similar metabolite profile of experiments illustrated in Fig. 5C and D reflect controlled redox reactions with no evidence of indiscriminate reducing equivalent flow from the OB-PPP when either NADP+ or GSSG were not added to reaction mixtures.
3.6. Effect of pH 7.9 and pH 8.2 on metabolic flow
GC–MS results, when AMP-3′ catabolism is initiated at pH 7.9 in the presence of 1×10−5 u/μl of added PAP, are shown in Fig. 5E. AMP-3′ hydrolysis occurs but metabolite flow stops at transaldolase (Fig. 1, Reaction 9) as determined by sedoheptulose7P accumulation. Glucose6P and fructose6P are not detected and GSH production is strongly inhibited. Initiation of AMP-3′ catabolism at pH 8.2 in the presence of 1×10−5 u/μl of added PAP is shown in Fig. 5F. There is less AMP-3′ hydrolysis and metabolite flow at pH 8.2 than observed at pH 7.9. This also is indicated by less inosine, hypoxanthine, and sedoheptulose7P accumulation as determined by %AUC; GSH production was strongly inhibited at pH 8.2. Results presented in Fig. 3E and F show that alkaline pH inhibits: [1] acid phosphatase initiation of metabolite flow from AMP-3′ (pH 8.2) and [2] metabolite flow through the NOB-PPP enzyme transaldolase, blocking sugar phosphate interconversion reactions that synthesize glucose6P from sedoheptulose7P and glyceraldehyde3P (pH 7.9 and pH 8.2).
To further evaluate metabolite flow through transaldolase, ribose5P (substrate for the NOB-PPP) was added to cytosol at pH 6.7. This provides an immediate large substrate burden for cytosolic NOB-PPP enzymes (Fig. 5G). At 120 min 3.7 mM GSH was generated and sedoheptulose7P %AUC appeared. By 240 min (GSH 4.7 mM) glucose6P plus fructose6P represented 50% of the metabolite AUC and increased to 70% at 1200 min (GSH remained 4.7 mM) (Fig. 5G). Non-increase of GSH and marked accumulation of glucose6P and fructose6P in this experiment reflects dysfunction of the OB-PPP, ineffective reducing equivalent delivery to glutathione reductase, or impaired enzymatic activity of cytosolic glutathione reductase. The same experiment was carried out at pH 8.2 (Fig. 5H). GSH production reached 2.0 mM at 240 min but was diminished compared to the otherwise identical experiment carried-out at pH 6.7 (Fig. 5G). Sedoheptulose7P %AUC appeared at 12 min, and increased progressively at 120 min, 240 min, and 1200 min, but fructose6P and glucose6P were not detected. This confirms that, at pH 8.2, flow through transaldolase is inhibited when the NOB-PPP is challenged with ribose5P.
Transaldolase is a phosphoprotein [19] that controls the direction of metabolite flow through the NOB-PPP [20]. It has five phosphorylation sites [21] for the ubiquitous and constitutively active protein kinase CK2 [22]. Dephosphorylation of human transaldolase with reagent acid phosphatase results in metabolite flow through the NOB-PPP in the direction of ribose5P to glucose6P [19]. Phosphorylation of transaldolase blocks recycling of ribose5P to glucose6P for oxidation by the OB-PPP [19]. The biochemical phenotype observed in dilute cytosol at pH 7.9 or pH 8.2 (Fig. 5E, F, and H) is the same observed when transaldolase is phosphorylated [19] or deficient [23], [24], [25]. The findings reported in Fig. 5E, F, and H suggest that, in dilute cytosol at alkaline pH, transaldolase becomes phosphorylated by the ubiquitous and constitutively active protein kinase CK2 [22].
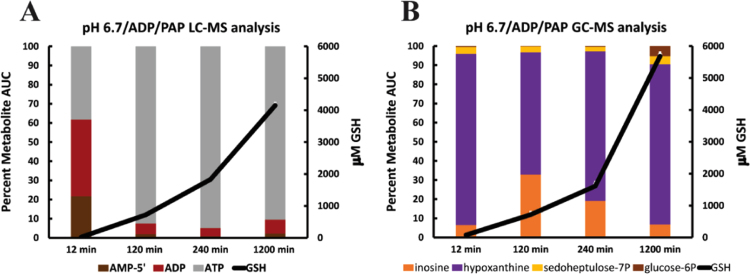
3.7. Metabolite profile of ADP catabolism
The profile of ADP catabolism was examined by LC–MS and GC–MS (Fig. 6A and B) at pH 6.7 in triplicate reaction mixtures. LC–MS showed that AMP-5′ and ATP are generated by cytosolic adenylate kinase [26] (Fig. 1, Reaction 1) from ADP added to cytosol (Fig. 6A). %AUC for ATP is maximal at 120 min and does not diminish during the 1200 min experiment. %AUC of AMP-5′ decreases as GSH production increases (Fig. 6A). Therefore, the action of cytosolic adenylate kinase on ADP supplies AMP-5′ as a carbon chain fuel source for NADPH generation and supplies the reaction mixture with ATP which is not consumed in these experiments. GC–MS metabolite profile and kinetics of GSH production was similar to that observed when AMP-3’ was the carbon chain fuel source (compare Figs. 6B to 5A and B). A small amount of sedoheptulose7P %AUC was measured at all-time points and glucose6P %AUC was observed at the 1200 min. These results show that the action of cytosolic adenylate kinase on ADP supplies the reaction mixture with a carbon chain fuel source for NADPH generation (AMP-5′) and a carrier of phosphoric acid anhydride bond energy (ATP).
Fig. 6.
Cytosolic adenylate kinase initiates ADP catabolism that produces NADPH for GSH production. Each experiment contained 200 ng/μl cytosolic protein, ADP 2mM, GSSG 3mM, NADP+ 5 μM and 1×10−5 u/μl PAP. Incubations were at 37°. (A) LC–MS analysis shows the fate of the ATP and AMP-5′ products of adenylate kinase. (B) GC–MS analysis tracks further catabolism of the AMP-5′ product of the adenylate kinase reaction. X-axes show the time of termination for metabolite and GSH measurements. Note: time not to scale. Y-axes show metabolite %AUC and μM GSH production. Fig. 5 legend describes GSH production.
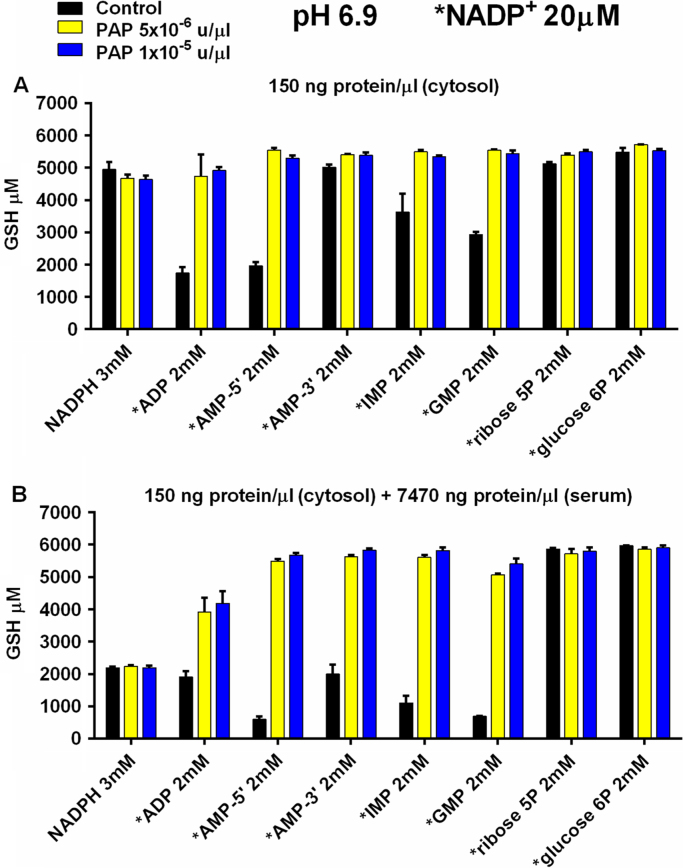
3.8. Effect of dilution of cytosolic proteins with serum proteins on GSH production
NADPH generation by cytosolic proteins diluted with a large excess of dialyzed human serum proteins was evaluated. Experiments with 150 ng/μl cytosolic proteins without added serum proteins (Fig. 7A) were compared to identical experiments with 150 ng/μl cytosolic protein plus 7470 ng/μl of added human serum proteins (Fig. 7B). Cytosolic proteins were 2% of total protein in the experiments reported in Fig. 7B, but were still capable of generating undiminished NADPH from the catabolism of ribose5P, glucose6P and, in the presence of added PAP, from purine ribonucleoside monophosphates. Added reagent NADPH (3 mM) was less effective than metabolically generated NADPH in delivering reducing equivalents to glutathione reductase, as determined by GSH production, in the presence of added serum proteins (Fig. 7B). These experiments show the importance of higher acid phosphatase activity in the presence of a large excess of extraneous serum proteins in extracellular metabolism carried out in dilute cytosol.
Fig. 7.
Effect on NADPH generation of dilution of cytosolic protein with autologous serum protein dialyzed to remove glucose. X-axes (coded bars) show control experiments (PAP not added) and experiments with added PAP. X-axes also show the addition of NADPH or a carbon chain fuel source to experiments. Y-axes show μM GSH generated from 3 mM GSSG. Asterisk marks added 20 μM NADP+. Panel (A) was identical to panel (B) except that 7470 ng/μl autologous dialyzed serum protein was added to each experiment in panel B. Reaction mixtures contained 150 ng/μl cytosolic protein and were incubated for 1200 min at pH 6.9 at 37°. Data represent the mean (±SEM) of 3 separately performed experiments. Cytosolic protein and dialyzed serum were from the same donor.
3.9. Dose response with added NADP+
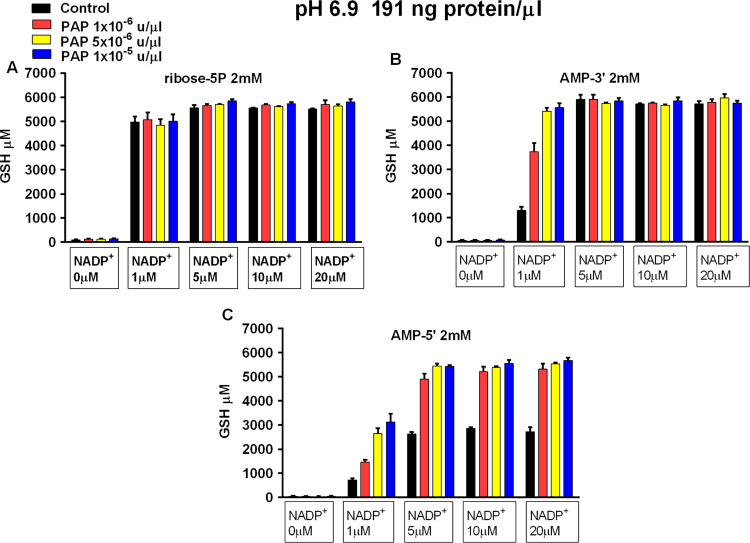
We also determined the lowest concentration of NADP+ that delivers metabolically produced reducing equivalents to glutathione reductase, as measured by GSH production (Fig. 8A–C). AMP-3′ with PAP and ribose5P without PAP support maximal GSH production with 1 μM added NADP+. AMP-5′ with PAP supports maximal GSH production with 5 μM of added NADP+. These results show that NADP+ is effectively cycled as a reducing equivalent conduit between the OB-PPP and glutathione reductase in metabolism carried out in dilute cytosol.
Fig. 8.
Electron carrier requirement in dilute cytosol. Carbon chain fuel sources used in individual experiments are indicated above each panel. X-axes show control experiments and experiments with added PAP (coded bars). X-axes also show the amount of NADP+ added to each experiment. Y-axes show μM GSH generated from 3 mM GSSG. Reaction mixtures contained 191 ng/μl cytosolic protein and were incubated 1200 min at pH 6.9 at 37°. Data represent the mean (±SEM) of 3 separately performed experiments.
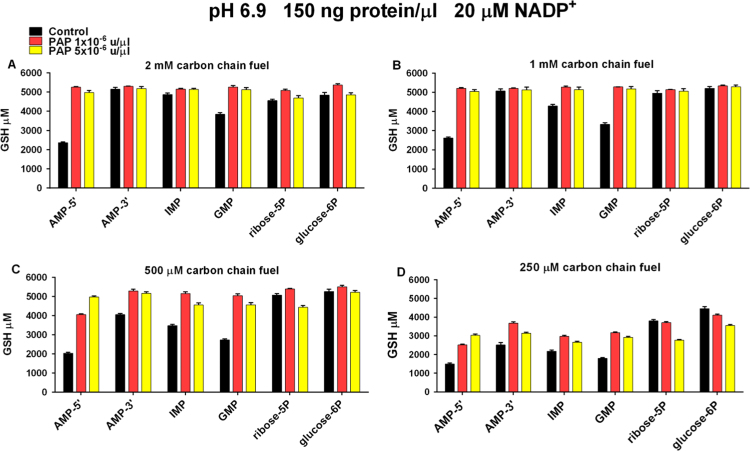
3.10. Dose response with added carbon chain fuel sources
Maximum GSH production from purine nucleoside monophosphates, ribose-5P, and glucose-6P did not decline until the concentration of carbon chain fuels was lowered to 250 μM (Fig. 9A–D). Endogenous cytosolic acid phosphatases produced maximum GSSG reduction from 2 mM AMP-3′ and IMP. Other purine nucleoside monophosphates required PAP for maximal GSH production at 2 mM, 1 mM, and 500 μM carbon chain fuel source concentrations (Fig. 9A–C). PAP increased GSH production from 250 μM purine nucleoside monophosphates but not to maximal levels. Ribose5P and glucose6P maintained maximal GSH production at 2 mM, 1 mM, and 500 μM but GSH production began to decline when their concentrations were lowered to 250 μM (Fig. 9D).
Fig. 9.
Effect of carbon chain fuel source concentration on GSH production by diluted cytosol prepared from lysates of human leukocytes. X-axes show control experiments and experiments with added PAP (coded bars). X-axes also show carbon chain fuel source added to cytosolic protein in each experiment. Y-axes show μM GSH generated from 3 mM added GSSG. The concentration of carbon chain fuel source added to experiments is indicated in the upper portion of each panel. Reaction mixtures contained 150 ng/μl cytosolic protein and were incubated for 1200 min at pH 6.9 at 37°. NADP+ 20 μM was added to all experiments. Data represent the mean (±SEM) of three separately performed experiments.
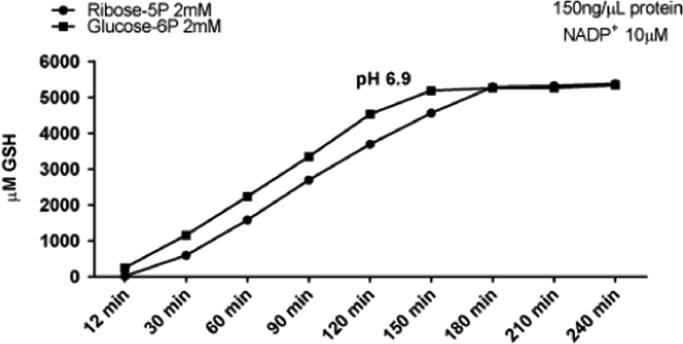
3.11. Kinetics of glucose6P and ribose5P metabolite flow through the PPP
Fig. 10 shows GSH production had linear kinetics between 30 min and 150 min when glucose6P, the carbon chain fuel for the OB-PPP, was added at t=0. At 150 min, 85% of added GSSG was reduced to 2GSH. This redox poise remained constant until the experiment was terminated at 240 min. Enzymes of the NOB-PPP and the OB-PPP are both engaged in metabolism producing NADPH for GSSG reduction when ribose5P is the carbon chain fuel source (Fig. 1). Similar linear kinetics of GSH production were observed between 30 min and 180 min when ribose5P was the carbon chain fuel source added to dilute cytosol at t=0 (Fig. 10). At 180 min, 88% of added GSSG was reduced to 2GSH and the redox poise remained stable until the experiment was terminated. The fact there is only a 30 min difference in maximum GSH production shows efficient function of the NOB-PPP in mediation of the sugar phosphate interconversion reactions that synthesize glucose6P from ribose5P.
Fig. 10.
Comparison of glucose6P and ribose5P as carbon chain fuels by measuring kinetics of GSH production. X-axis shows the time in minutes when GSH measurements were performed. Y-axis shows μM GSH generated from 3 mM added GSSG. Reaction mixtures contained 150 ng/μl cytosolic protein, 10 μM NADP+ and were incubated at pH 6.9 at 37°. Data are the mean (±SEM) of three separately performed experiments. Error bars are not visible and fall within the symbol denoting the mean.
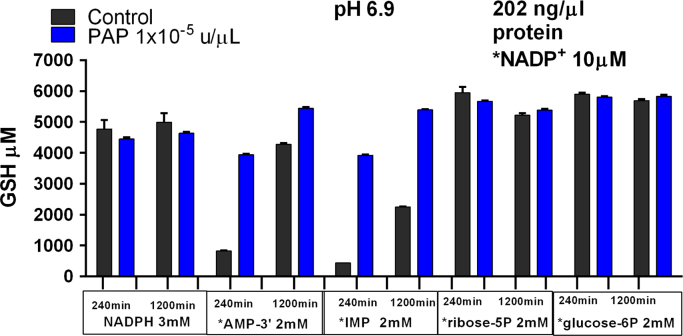
3.12. Kinetics of purine ribonucleoside monophosphate flow through the PNCP and PPP
We observed slower kinetics of metabolite flow through the PNCP and PPP with AMP-3’ as the carbon chain fuel source and a reaction mixture cytosolic protein concentration of 150 ng/μl (Fig. 5A and B). In contrast Fig. 10 shows rapid flow through the PPP of metabolites derived from ribose5P with the same cytosolic protein concentration. Therefore, we increased the reaction mixture cytosolic protein concentration to 202 ng/μl, with and without 1×10−5 u/μl PAP, to further evaluate the different kinetics of PNCP and PPP metabolite flow. AMP-3′ and IMP [acid phosphatase and PNCP substrates] were compared to ribose5P [NOB-PPP substrates], glucose6P [OB-PPP substrate] and reagent NADPH [co-substrate for glutathione reductase] by measuring GSH production at 240 min and 1200 min (Fig. 11). Results showed that GSH production at 240 min was 3936 μM (66% GSSG reduction) for AMP-3′ and 3919 μM (65% GSSG reduction) for IMP. GSH production at 1200 min increased to 5445 μM (91% GSSG reduction) for AMP-3′ and 5394 μM (90% GSSG reduction) for IMP. In the same experiments, maximum GSH production was achieved at 240 min for ribose5P, glucose6P, and reagent NADPH. These results show a more rapid kinetics for AMP-3′ metabolite flow through the PNCP at a cytosolic protein concentration of 202 ng/μl than at 150 ng/μl cytosolic protein. However, these experiments confirm that, under conditions of our experiments, the kinetics of metabolite flow through the PPP is more rapid than through the PNCP.
Fig. 11.
Comparative kinetics of purine ribonucleoside monophosphates and sugar phosphates as carbon chain fuel sources for GSH production. X-axis shows control experiments and experiments with added PAP (coded bars). X-axis also shows the time of termination for GSH measurements and the carbon chain fuel source or reagent NADPH added. Asterisk marks 10 μM added NADP+. Y-axis shows μM GSH generated from 3 mM GSSG. Reaction mixtures contained 202 ng/μl cytosolic protein and were incubated at pH 6.9 at 37°. Data are the mean (±SEM) of three replicate experiments.
3.13. Pyrimidine ribonucleotides are not efficient carbon chain fuel sources
Table S1 compares a series of pyrimidine ribonucleoside monophosphates and diphosphates to ADP and a series of purine ribonucleoside monophosphates as carbon fuel chain sources for the reduction of added GSSG. The results show that pyrimidine ribonucleotides are not efficient carbon chain fuel sources for extracellular NADPH production.
4. Discussion
The in vitro observations reported here must await results of future in vivo experiments to determine their biological significance. However, extracellular metabolism that generates energy as NADPH from purine nucleotide carbon chain fuel sources could have a role at sites of tissue injury and inflammation. The enzymes responsible for extracellular metabolism were present in reaction mixtures containing dilute cytosol (0.0075–0.02% protein) prepared from lysates of human peripheral blood leukocytes. These are cells that rapidly migrate into sites of tissue injury and inflammation. Leukocyte lysis associated with cytosol preparation is the experimental equivalent of cellular necrosis occurring in perturbed tissue. In each case, the integrity of the plasma membrane is lost and intracellular molecules are released into extracellular space. Necrotic cell death, a response to stress by many different cell types, causes inflammation [27]. Tissue perturbations associated with necrosis, and release of intracellular molecules into extracellular space, include microbial invasion, autoimmunity, trauma, chemical toxicity, and ischemia–reperfusion. The degradation of RNA released from necrotic cells produces AMP-3′/AMP-5′ and GMP-3′/GMP-5′. Poly (ADP-ribose) polymerase activation in stressed cells precedes programmed necrosis and produces ADP-ribose polymers [6], [27], [28]. Platelet degranulation during hemostasis releases ADP [3]. All are possible carbon chain fuel sources for extracellular NADPH generation. Extravasation and lysis of erythrocytes in perturbed tissue also could contribute to extracellular NADPH generation by releasing glutathione, NADP+, enzymes and ribose5P containing adenine nucleotides [17], [18], [26], [29]. Catalase is abundant in erythrocytes and is released during lysis [30].
The extracellular NADPH producing metabolism described here could moderate oxidative stress associated with tissue injury and inflammation by regenerating GSH [5] and stabilizing catalase [31]. This would lower extracellular H2O2 toward an optimum concentration for tissue repair [32], [33], [34] and limit damage caused by activated neutrophils, extracellular heme [29] and other pro-oxidant factors.
In this study we used the NADPH dependent glutathione reductase mediated reduction of GSSG to 2 GSH to quantitate NADPH production. This shows that extracellular metabolism can generate GSH for H2O2 removal by glutathione peroxidase. It is of interest that efficient enzymatic removal of H2O2 from perturbed extracellular space either by the glutathione reductase/peroxidase system or by catalase requires a source of extracellular NADPH [5], [31], [35]. Each tetrameric catalase molecule binds four molecules of NADPH. Bound NADPH protects catalase from inactivation by its substrate H2O2 [31]. To maintain maximum catalase activity, a supply of NADPH must be available to replace NADP+ as it is formed [31], [35]. A concentration of NADPH below 0.1 μM is sufficient to maintain catalase in its active form [31]. This suggests that low levels of NADPH production could protect the enzymatic function of catalase in extracellular space. Therefore, a source of NADPH production is necessary for the enzymatic removal of H2O2 from perturbed intracellular or extracellular space.
Another site where extracellular metabolism generating NADPH could occur is the female genital tract (FGT). PAP (Fig. 2C–H) and necrosis can exist in the same proximity in the FGT. Neutrophils, 40–70% of blood leukocytes used for cytosol preparation, migrate into the FGT as a response to insemination [36]. Spermatozoa activate a unique type of programmed necrosis termed NET formation [36], [37]. NETs have antimicrobial activity and are a precedent for continued function of macromolecules extruded from necrotic neutrophils. Neutrophil death associated with NET formation would release enzymes and ribose containing carbon chain fuel sources needed for NADPH generation. The 1% (wt/vol) PAP content of seminal fluid [38] (with no current known function in the fertilization process) could facilitate extracellular energy generating metabolism in the FGT. This could moderate fertilization associated oxidative stress and increase spermatozoa survival.
Certain PPP enzymes have properties relevant to coordinated metabolic function in extracellular space. For example, phosphoglucose isomerase (Fig. 1, Reaction 10) is a multifunctional protein which acts extracellularly as autocrine motility factor and is a marker for certain cancers [39]. Extracellular energy generating metabolism could complement the known extracellular cytokine function of phosphoglucose isomerase by creating a redox environment conducive to cell migration and angiogenesis needed for wound healing or malignant cell spread [29], [32], [33], [34], [39], [40]. It is of interest that phophoglucose isomerase is phosphorylated by protein kinase CK2 [39]. Like transaldolase [19], phosphorylation of phosphoglucose isomerase by protein kinase CK2 inhibits its enzymatic activity [39]. Also transaldolase, glucose-6-phosphate dehydrogenase, and 6-phosphogluconate dehydrogenase form a supramolecular complex in human neutrophils [41]. Supramolecular organization of PPP enzymes, if maintained after cell lysis and cytosol preparation, would facilitate extracellular metabolism. The experiments reported here could provide a model for investigation of supramolecular complex centered metabolism.
There are several examples from the literature relevant to the in vitro extracellular metabolism we observed. The first is the tumor lysis syndrome (TLS) which is a complication of therapy in a small subset of oncology patients [42]. Human tumors with a high proliferation rate, a relatively large mass, and high sensitivity to cytotoxic agents can respond to therapy with rapid cell lysis. This results in extrusion of intracellular contents, including RNA and purine ribonucleotides, into extracellular space. High blood levels of uric acid are produced by extracellular catabolism of extruded RNA and purine ribonucleotides. This is a major complication of the TLS and is associated with renal impairment caused by uric acid crystal nephropathy. The TLS is relevant because it provides in vivo evidence in humans that the PNCP, when coupled to XOR, carries out efficient extracellular metabolism. XOR oxidizes hypoxanthine, a product of PNP (Fig. 1, Reactions 4) to xanthine and then xanthine to uric acid (an antioxidant) [43]. The other products of XOR are either H2O2 or NADH.
The same PNCP enzymes that are active in extracellular space in the TLS also function in dilute cytosol prepared from lysates of human peripheral blood leukocytes in experiments reported here. These in vitro studies show that the other PNP product, ribose1P, becomes the carbon chain fuel source for extracellular NADPH production by the PPP (Fig. 1 Reactions 5 and 6). The experiments demonstrate that, like XOR, the PPP is coupled to the PNCP when carrying out metabolic activity in extracellular space. The metabolic bifurcation created by further metabolism of the PNP products, ribose1P and hypoxanthine, results in the production of an array of redox active molecules that together can modulate the redox environment of perturbed tissue either toward a more reduced or more oxidized redox poise. Proper functioning of this purine ribonucleotide driven extracellular metabolism modulating the redox environment could promote optimal repair of the perturbed tissue. Malfunction could interfere with tissue repair and resolution of inflammation.
The second example relevant to the experiments reported here is a recent study probing the evolutionary origin of core cellular metabolism [44]. The purpose of the study was to provide empirical evidence to support the theory of a prebiotic origin of metabolism, e.g., the metabolism first hypothesis [45]. The experiments demonstrated that abiotic metabolic networks develop under conditions designed to mimic the geochemical and temperature environment of the ancient Archean ocean. Sugar phosphate interconversion reactions similar to those of the NOB-PPP and glycolysis were observed. This enzyme free dilute solution metabolism was facilitated by metals such as ferrous iron that acted as catalysts. The results we report here are similar to these interesting experiments in so far as both involve the same component of core metabolism found in all three domains of life [46] and both produce, in dilute solution, the sugar phosphate intermediates of the NOB-PPP.
In conclusion we show that, in dilute solutions of cytosolic proteins prepared from lysates of human leukocytes, the ribose5P moiety of purine nucleotides is a carbon chain fuel source for NADPH production. The extracellular extraction of ribose5P from purine nucleotides and its conversion to glucose6P requires the combined coordinated action of acid phosphatases, the PNCP, and the NOB-PPP. Glucose6P generated without energy input is then oxidized by the OB-PPP to produce NADPH. The released intracellular contents of necrotic cells and extravasated erythrocytes that undergo lysis contain the purine ribonucleotide substrates and enzymes that carry out this thermodynamically favorable NADPH generating metabolism [17], [18], [26], [29], [30], [35], [41]. If this dilute solution extracellular metabolism can be demonstrated in humans it could modulate the redox environment of perturbed tissue and influence the complex process of repair.
Acknowledgments
We are grateful to R.D. Bloebaum and the Dept. of Veterans Affairs for support. We thank J.P. Kushner, D.L. Granger, J.B. Weinberg, D.A. McClain and J.D. Phillips for critical reading and insightful comments on the manuscript. Special thanks to D.L. Granger for helpful discussions and to J.C. Peterson and F.A. Hibbs for assistance with the manuscript. The authors report no conflict of interest. Partial funding for this work was through NIH Grant 1 P30 DK072437 (J. Cox, Co-I).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2016.02.001.
Appendix A. Supplementary material
Supplementary material
References
- 1.Marletta M.A., Yoon P.S., Iyengar R., Leaf C.D., Wishnok J.S. Macrophage oxidation of l-arginine to nitrite and nitrate: Nitric oxide is and intermediate. Biochemistry. 1988;27:8706–8711. doi: 10.1021/bi00424a003. [DOI] [PubMed] [Google Scholar]
- 2.Clegg J.S. Properties and metabolism of the aqueous cytoplasm and its boundaries. Am. J. Physiol. 1984;246:R133–R151. doi: 10.1152/ajpregu.1984.246.2.R133. [DOI] [PubMed] [Google Scholar]
- 3.Greer J.P., Foerster J., Rodgers G.M., Paraskevas F., Glader B., Arber D.A., Means R.T., Jr. Wintrobe’s Clinical Hematology. 12th edition. Lippincott Williams and Wilkins; Philadelphia, PA: 2009. [Google Scholar]
- 4.Fulton A.B. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 5.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Rad. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 6.Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C., Hurn P.D., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginis I., Zaner K., Wang J.S., Pavlotsky N., Tauber A.I. Comparison of actin changes and calcium metabolism in plastic-and fibronectin-adherent human neutrophils. J. Immunol. 1992;149:1388–1394. [PubMed] [Google Scholar]
- 8.Tomazella G.G., da Silva I., Laure H.J., Rosa J.C., Chammas R., Wilker H.G., de Souza G.A., Greene L.J. Proteomic analysis of total cellular proteins of human neutrophils. Proteome Sci. 2009;7:32. doi: 10.1186/1477-5956-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen S., Shea J.M., Felmet T., Gadra J., Dehn P.F. A kinetic microassay for glutathione in cells plated on 96-well microtiter plates. Methods Cell Sci. 2000;22:305–312. doi: 10.1023/a:1017585308255. [DOI] [PubMed] [Google Scholar]
- 10.Ames B. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 11.Jiye A., Trygg J., Gullberg J., Johansson A.I., Jonsson P., Antti H., Marklund S.L., Moritz T. Extraction and GC/MS analysis of the human blood plasma metabolome. Anal. Chem. 2005;77:8086–8094. doi: 10.1021/ac051211v. [DOI] [PubMed] [Google Scholar]
- 12.Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.C., Cox J.E., Cardon C.M., Van Vranken J.G., Dephoure. N., Redin C., Boudina S., Gygi S.P., Brivet M., Thummel C.S., Rutter J.A. Mitochondrial pyruvate carrier required for pyruvate uptake in yeast, drosophila and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podhajcer O.L., Filmus J.E., Mordoh J. Characterization of lysosomal acid phosphatase from normal and malignant mammary tissue. Clin. Chem. 1986;32:279–282. [PubMed] [Google Scholar]
- 14.Vihko P. Characterization of the principal human prostatic acid phosphatase isoenzyme, purified by affinity chromatography and isoelectric focusing. Part ll. Clin. Chem. 1978;24:1783–1787. [PubMed] [Google Scholar]
- 15.Halleen J.M., Kaija H., Stepan J.J., Vihko P., Väänänen H.K. Studies on the protein tyrosine phosphatase activity of tartrate-resistant acid phosphatase. Arch. Biochem. Biophys. 1998;352:97–102. doi: 10.1006/abbi.1998.0600. [DOI] [PubMed] [Google Scholar]
- 16.Lin S., Gasmi L., Xie Y., Ying K., Gu S., Wang Z., Jin H., Chao Y., Wu C., Zhou Z., Tang R., Mao Y., McLennan A.G. Cloning, expression and characterization of a human Nudix hydrolase specific for adenosine 5′-diphosphoribose (ADP-ribose) Biochem. Biophys. Acta. 2002;1594:127–135. doi: 10.1016/s0167-4838(01)00296-5. [DOI] [PubMed] [Google Scholar]
- 17.Chu S.Y., Cashion P., Jiang M. Purine nucleoside phosphorylase in erythrocytes: determination of optimum reaction conditions. Clin. Biochem. 1989;22:3–9. doi: 10.1016/s0009-9120(89)80062-1. [DOI] [PubMed] [Google Scholar]
- 18.Lionetti F.J. Pentose phosphate pathway in human erythrocytes. In: Yoshikawa H., Rapaport S.M., editors. Cellular and Molecular Biology of Erythrocytes. Univ. Park Press; Baltimore: 1974. pp. 143–166. [Google Scholar]
- 19.Lachaise F., Martin G., Drougard C., Perl A., Vuillaume M., Wegnez M., Sarasin A., Daya-Grosjean L. Relationship between posttranslational modification of transaldolase and catalase deficiency in uv-sensitive repair-deficient xeroderma pigmentosum fibroblasts and SV40-transformed human cells. Free Radic. Biol. Med. 2001;30:1365–1373. doi: 10.1016/s0891-5849(01)00532-9. [DOI] [PubMed] [Google Scholar]
- 20.Wood T. The forward and reverse reactions of transaldolase. FEBS Lett. 1972;25:153–155. doi: 10.1016/0014-5793(72)80474-5. [DOI] [PubMed] [Google Scholar]
- 21.Banki K., Halladay D., Perl A. Cloning and expression of the human gene for transaldolase. A novel highly repetitive element constitutes an integral part of the coding sequence. J. Biol. Chem. 1994;269:2847–2851. [PubMed] [Google Scholar]
- 22.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 23.Verhoeven N.M., Huck J.H.J., Roos B., Struys E.A., Salomons G.S., Douwes A.C., van der Knaap M.S., Jakobs C. Transaldolase deficiency: liver cirrhosis associated with a new inborn error in the pentose phosphate pathway. Am. J. Hum. Genet. 2001;68:1086–1092. doi: 10.1086/320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wamelink M.M., Smith D.E., Jansen E.E., Verhoeven N.M., Struys E.A., Jakobs C. Detection of transaldolase deficiency by quantification of novel seven-carbon chain carbohydrate biomarkers in urine. J. Inherit. Metab. Dis. 2007;30:735–742. doi: 10.1007/s10545-007-0590-2. [DOI] [PubMed] [Google Scholar]
- 25.Hanczko R., Fernandez D.R., Doherty E., Qian Y., Vas G., Niland B., Telarico T., Garba A., Banerjee S., Middleton F.A., Barrett D., Barcza M., Banki K., Landas S.K., Perl A. Prevention of hepatocarcinogenesis and increased susceptibility to acetaminophen-induced liver failure in transaldolase-deficient mice by N-acetylcysteine. J. Clin. Investig. 2009;119:1546–1557. doi: 10.1172/JCI35722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerletti P., Bucci E. Adenylate kinase of mammalian erythrocytes. Biochim. Biophys. Acta. 1960;38:45–56. doi: 10.1016/0006-3002(60)91194-x. [DOI] [PubMed] [Google Scholar]
- 27.Zong W.X., Thompson C.B. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 28.Hitomi J., Christofferson D.E., Ng A., Yao J., Degterev A., Xavier R.J., Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway by a genome wide siRNA screen. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagener F.A.D.T.G., van Beurden H.E., von den Hoff J.W., Adema G.J., Figdor C.G. The heme-heme oxygenase system: a molecular switch in wound healing. Blood. 2003;102:521–528. doi: 10.1182/blood-2002-07-2248. [DOI] [PubMed] [Google Scholar]
- 30.Agar N.S., Sadrzadeh S., M. H., Hallaway P.E., Eaton J.W. Erythocyte catalase: A somatic oxidant defense? J. Clin. Investig. 1986;77:319–321. doi: 10.1172/JCI112294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkman H.N., Galiano S., Gaetani G.F. The function of catalase-bound NADPH. J. Biol. Chem. 1987;262:660–666. [PubMed] [Google Scholar]
- 32.Roy S., Khanna S., Nallu K., Hunt T.K., Sen C.K. Dermal wound healing is subject to redox control. Mol. Ther. 2006;13:211–220. doi: 10.1016/j.ymthe.2005.07.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schäfer M., Werner S. Oxidative stress in normal and impaired wound repair. Pharmcol. Res. 2008;58:165–171. doi: 10.1016/j.phrs.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Huang S.S., Zheng R.L. Biphasic regulation of angiogenesis by reactive oxygen species. Pharmazie. 2006;61:223–229. [PubMed] [Google Scholar]
- 35.Scott M.D., Wagner T.C., Chiu D.T. Decreased catalase activity is the underlying mechanism of oxidant susceptibility in glucose-6-phosphate dehydrogenase deficient erythrocytes. Biochim. Biophys. Acta. 1993;1181(2):163–168. doi: 10.1016/0925-4439(93)90106-b. [DOI] [PubMed] [Google Scholar]
- 36.Alghamdi A.S., Foster D.N. Seminal DNase frees spermatozoa entangled in neutrophil extracellular traps. Biol. Reprod. 2005;73:1174–1181. doi: 10.1095/biolreprod.105.045666. [DOI] [PubMed] [Google Scholar]
- 37.Fuchs T.A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rönnberg l, Vihko P., Sajanti E., Vihko R. Clomiphene citrate administration to normogonadotropic subfertile men: Blood hormone changes and activation of acid phosphatase in seminal fluid. Int. J. Androl. 1981;4:372–378. doi: 10.1111/j.1365-2605.1981.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 39.Yangawa T., Funasaka T., Tsutsumi S., Raz T., Tanaka N., Raz A. Differential regulation of phosphoglucose isomerase/autocrine motility factor activities by protein kinase CK2 phosphorylation. J. Biol. Chem. 2005;280:10419–10426. doi: 10.1074/jbc.M409457200. [DOI] [PubMed] [Google Scholar]
- 40.Terada L.S. Specificity in reactive oxidant signaling: Think globally, act locally. J. Cell Biol. 2006;174:615–623. doi: 10.1083/jcb.200605036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J.B., Espinoza J., Romero R., Petty H.R. Transaldolase is part of a supramolecular complex containing glucose-6-phosphate dehydrogenase in human neutrophils that undergoes retrograde trafficking during pregnancy. Metabol. Clin. Exp. 2005;54:1027–1033. doi: 10.1016/j.metabol.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Wilson F.P., Berns J.S. Onco-nephrology: Tumor lysis syndrome. Clin. J. Am. Soc. Nephrol. 2012;7:1730–1739. doi: 10.2215/CJN.03150312. [DOI] [PubMed] [Google Scholar]
- 43.Harrison R. Structure and function of xanthine oxidoreductase: Where are we now? Free Rad. Biol. Med. 2002;33:774–797. doi: 10.1016/s0891-5849(02)00956-5. [DOI] [PubMed] [Google Scholar]
- 44.Keller M.A., Turchyn A.V., Ralser M. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol. Syst. Biol. 2014;10:725. doi: 10.1002/msb.20145228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luisi P.L. Prebiotic metabolic networks? Mol. Syst. Biol. 2014;10:729. doi: 10.1002/msb.20145351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong H., Tombor B., Albert R., Oltval Z.N., Barabasi A.L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material