Sentinel surveillance in Minnesota revealed that enterotoxigenic E. coli (ETEC) and non-O157 Shiga toxin-producing E. coli are common enteric pathogens; ETEC was the second leading bacterial pathogen in an urban site; 39% of ETEC were domestically acquired.
Keywords: enterotoxigenic Escherichia coli, pathogenic Escherichia coli, Shiga toxin-producing Escherichia coli
Abstract
Background. Enterotoxigenic Escherichia coli (ETEC) and non-O157 Shiga toxin-producing E. coli (STEC) are not detected by conventional culture methods. The prevalence of ETEC infections in the United States is unknown, and recognized cases are primarily associated with foreign travel. Gaps remain in our understanding of STEC epidemiology.
Methods. Two sentinel surveillance sites were enrolled: an urban health maintenance organization laboratory (Laboratory A) and a rural hospital laboratory (Laboratory B). Residual sorbitol MacConkey (SMAC) plates from stool cultures performed at Laboratory A (1996–2006) and Laboratory B (2000–2008) were collected. Colony sweeps from SMAC plates were tested for genes encoding STEC toxins stx1 and stx2 (1996–2008) and ETEC heat-labile and heat-stable toxins eltB, estA 1, 2 and 3 (2000–2008) by polymerase chain reaction (PCR)-based assays.
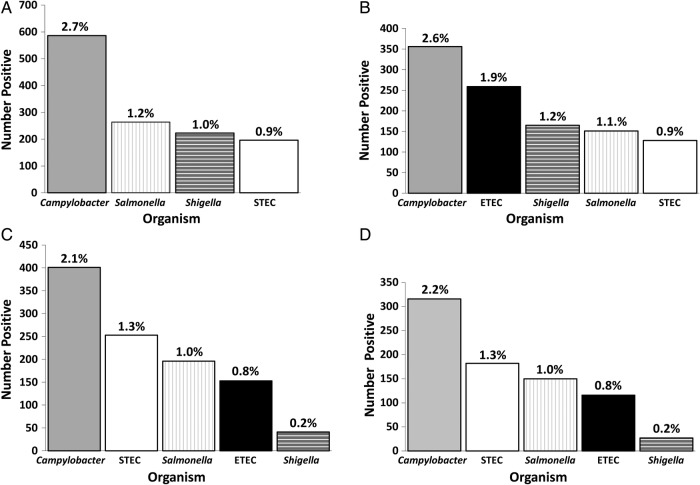
Results. In Laboratory A, a bacterial pathogen was identified in 7.0% of 21 970 specimens. During 1996–2006, Campylobacter was the most common bacterial pathogen (2.7% of cultures), followed by Salmonella (1.2%), Shigella (1.0%), and STEC (0.9%). Among STEC (n = 196), O157 was the most common serogroup (31%). During 2000–2006, ETEC (1.9%) was the second most common bacterial pathogen after Campylobacter (2.6%). In Laboratory B, of 19 293 specimens tested, a bacterial pathogen was identified for 5.5%, including Campylobacter (2.1%), STEC (1.3%), Salmonella (1.0%), and ETEC (0.8%). Among STEC (n = 253), O157 was the leading serogroup (35%). Among ETEC cases, 61% traveled internationally.
Conclusions. Enterotoxigenic E. coli and STEC infections were as common as most other enteric bacterial pathogens, and ETEC may be detected more frequently by culture-independent multiplex PCR diagnostic methods. A high proportion of ETEC cases were domestically acquired.
Enterotoxigenic Escherichia coli (ETEC) is considered to be the leading cause of traveler's diarrhea [1–3]. Although Shiga toxin-producing E. coli (STEC) O157 is a well documented cause of gastroenteritis and hemolytic uremic syndrome (HUS) in the United States, non-O157 STEC also have been recognized as important pathogens, with a wide range of clinical presentations, from mild illness to HUS [4, 5]. Both ETEC and non-O157 STEC have been implicated in outbreaks in the United States [6, 7]. In contrast to STEC O157, ETEC and non-O157 STEC are not detected by conventional stool culture methods in clinical laboratories. Culture-independent diagnostic tests (CIDTs), including enzyme-linked immunoassay tests to detect Shiga toxins, became commercially available to clinical laboratories around 2000. The adoption of these tests has been increasing since [8, 9], leading to a corresponding increase in the recognition of non-O157 STEC as an important cause of illness [10–13]. However, because commercially available assays for ETEC have been lacking, and the adoption of Shiga toxin assays by clinical laboratories has been neither universal nor uniform, the true importance of these types of diarrheagenic E. coli as enteric pathogens in the United States is unknown.
The objective of our study was to use long-term sentinel surveillance to determine the frequency with which ETEC and non-O157 STEC infections occur in a rural and an urban setting in Minnesota, and the relative frequency of these pathogens compared with other bacterial enteric pathogens.
METHODS
Surveillance
Two sentinel sites were enrolled in this study: Laboratory A, a large health maintenance organization laboratory that served the Minneapolis-St. Paul metropolitan area; and Laboratory B, a hospital laboratory that served a small city and surrounding rural area that is rich in animal agriculture, particularly dairy production. The exact population size served by these 2 laboratories is unknown. Every stool submitted for bacterial culture at both laboratories was plated on a sorbitol MacConkey agar (SMAC) plate. Residual SMAC plates from every stool culture were sent to the Minnesota Department of Health (MDH) Public Health Laboratory (PHL) for STEC and ETEC polymerase chain reaction (PCR) testing regardless of the culture results at the clinical laboratory [5]. Only 1 isolate per person was included.
For Laboratory A, STEC testing was performed from 1996 to 2006, and ETEC testing was performed from 2000 to 2006. For Laboratory B, testing both for STEC and ETEC was performed from 2000 to 2008.
During the entire study period, active laboratory-based surveillance for reportable bacterial pathogens was conducted at both laboratories; all Campylobacter, STEC O157, Salmonella, Shigella, Vibrio, and Yersinia isolates cultured at both laboratories were submitted to the MDH PHL for confirmation. During the study period, CIDTs for the detection of Campylobacter and/or STEC were not being used by these sentinel site laboratories.
Laboratory Methods
Nucleic acid extraction was initiated upon receiving a SMAC plate from a sentinel laboratory. Template DNA was prepared from colony sweeps. Six sweeps were made through representative areas of growth, which included all visible colony morphologies, avoiding the primary inoculation area, and mixed using a 1.0 µL disposable loop. One loopful of the mixed sweep material and 200 µL molecular grade water (Sigma) were heated for 15 minutes in boiling water and centrifuged at 16 000 g for 2 minutes. Clear supernatants containing bacterial DNA were withdrawn for PCR analysis.
From January 2000 through July 2005, Shiga toxin genes stx1 and stx2 were detected by PCR using previously described primers and amplification methods [14]. In July 2005, the PCR method of Paton and Paton [15] was implemented to increase Shiga toxin gene detection sensitivity, to detect the intimin-encoding gene eae to identify potential enteropathogenic E. coli, and to detect the alpha-hemolysin-encoding gene hlyA, another marker for STEC.
Specimen sweeps testing positive for stx had up to 24 individual colonies tested for stx by PCR. Shiga toxin gene-positive isolates were identified by standard biochemical methods [16]. Somatic and flagellar antigens were determined using Denka Seiken antisera. If a sample sweep was positive for stx by PCR but a stx-positive colony could not be isolated, the sample was classified as PCR-positive STEC, not isolated. If an individual E. coli colony was Shiga toxin gene-positive and tested negative for O157, it was classified as STEC even if the serogroup could not be determined (e.g., O rough or undetermined).
For ETEC, the plate sweep was tested for estA1, estA2, estA3 (E. coli heat-stable toxin or LT1-encoding genes), and eltB (heat labile or ST1-encoding gene) by Multiplex SYBR Green PCR. Single gene primers and probes were then used to confirm positives by TaqMan PCR. Primer and probe sequences for LT1 and ST1 genes are listed in Table 1. SYBR Green detection assay was 25 µL volume with final concentrations of 1× SYBR Green buffer (SYBR Green Kit, Applied Biosystems), 3.5 mM MgCl2, 1.0 mM dNTPs with dUTP (Applied Biosystems, Foster City, CA), 10 nM fluorescein (Bio-Rad, Hercules, CA), 400 nM estA1 primers, 400 nM estA2,3 primers, 200 nM eltB primers, 0.625 units AmpliTaq Gold (Applied Biosystems), and 1.0 µL sample supernatant DNA. Amplification conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, then 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Samples were positive if the cycle threshold (Ct) value exceeded background level in <40 cycles and the melt curve temperature (Tm) matched those of the controls; estA Tm = 77–77.5°C and eltB Tm = 80.5–81°C. Samples that crossed threshold having incorrect Tm were considered negative. TaqMan probe confirmation has each gene primer and probe PCR assay in 25 µL volume with final concentrations of 1× TaqMan buffer (TaqMan Kit, Applied Biosystems), 5.0 mM MgCl2, 1.0 mM dNTPs with dUTP (Applied Biosystems), 1000 nM specified primer set (estA1, estA2,3, or eltB), 250 nM specific probe (estA1, estA2, estA3, or eltB), 0.625 units Hot Star Taq (QIAGEN), and 2.0 µL sample supernatant DNA. Amplification conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, then 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. A specimen was positive if the Ct value exceeded background level <45 cycles. Specimens positive for SYBR Green but negative on TaqMan assay were subcultured for colony isolation. Up to 24 individual colonies were retested by SYBR Green PCR. Specimens positive only for SYBR Green and without identified positive isolates were classified as negative for ETEC. Isolate identity for selected STEC and ETEC isolates was further confirmed at the Centers for Disease Control and Prevention (Atlanta, GA). Starting in 2005, testing for E. coli virulence factor bfp (bundle-forming pillus encoding gene), indicative of enteropathogenic E. coli (EPEC), was also done by PCR following the methods of Gunzberg [17].
Table 1.
Oligonucleotide Primers and TaqMan Hybridization Probes Used in PCR Assays for Identifying Escherichia coli Heat-Labile (LT1) and Heat-Stable (ST1) Genes Indicative of Enterotoxigenic E. colia
| Geneb,c | Sequence | Product Size |
|---|---|---|
| eltB | 73 bp | |
| eltB-205 | 5′ TAA GAG CGG CGC AAC ATT T 3′ | |
| eltB-277 | 5′ TTC AAT GGC TTT TTT TTG GGA 3′ | |
| eltB probe | 5′ TTG ACT GCC CGG GAC TTC GAC CT 3′ | |
| estA1 | 151 bp | |
| estA1-353 | 5′ AGT CAA CTG AAT CAC TTG ACT CTT CA 3′ | |
| estA1-503 | 5′ CCA GCA CAG GCA GGA TTA CA 3′ | |
| estA1 probe | 5′ AAT CAG AAA ATA TGA ACA ACA CAT TTT ACT GCT GTG AA 3′ | |
| estA2,3 | 139 bp | |
| estA2,3-197 | 5′ CCT TTC GCT CAG GAT GCT AAA C 3′ | |
| estA2,3-335 | 5′ ACA ATT CAC AGC AGT AAT TGC TAC TAT TC 3′ | |
| estA2 probe | 5′ CGA TTC TAG TGT AAT TTT TTC TTT TGA AGA CCC TGC T 3′ | |
| estA3 probe | 5′ AGT AGA GTC TTC AAA AGA AAA AAT CAC ACT AGA ATC A 3′ |
Abbreviations: bp, base pairs; PCR, polymerase chain reaction.
a The SYBR Green and TaqMan primers and probes were developed by Minnesota Department of Health (unpublished data).
b DNA oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
c Hybridization probes (5′ FAM/TAMRA-Q 3′) were synthesized by Operon Biotechnologies, Inc. (Huntsville, AL).
Descriptive and Statistical Analyses
Descriptive analyses were conducted for the entire study period. However, because study time periods differed between the 2 sentinel sites, and ETEC surveillance was initiated in 2000, statistical comparisons of the 2 sentinel sites were restricted to 2000–2006, when surveillance was conducted at both sites. Analyses were conducted using Epi Info 7.1.3.10 (Centers for Disease Control and Prevention, Atlanta, GA).
After the sentinel surveillance study period ended, both laboratories adopted ImmunoCard STAT! EHEC (Meridian Bioscience, Cincinnati, OH) to detect Shiga toxin and sent the enrichment broths to the MDH PHL for confirmation by real-time PCR. Both laboratories still used culture to detect Campylobacter, Salmonella, STEC O157, and Shigella and sent isolates for confirmation at the PHL. To understand how our sentinel surveillance data compared with current surveillance data that include the use of CIDTs for STEC, the frequency of pathogens using sentinel surveillance data was compared to the 2013–2014 surveillance data at each site.
Starting in 2000 until the end of the enrollment period for each site (2006 for site A, and 2008 for site B), ETEC and STEC cases were interviewed about international travel in the 7 days prior to illness onset. The number and proportion of ETEC and STEC cases that reported international travel were described.
RESULTS
Laboratory A (Urban Laboratory)
From Laboratory A, 21 970 SMAC culture plates (cultures) were tested during 1996–2006, with a median of 1997 cultures per year (range, 1589 to 2432) (Table 2, Figure 1). A bacterial pathogen was identified from 1540 (7.0%) cultures overall, with a range of 4.9% to 11% per year. Excluding ETEC, because it was not tested for during the entire study period, Campylobacter was the most common bacterial pathogen isolated, accounting for 586 (2.7%) cultures, followed by Salmonella at 264 (1.2%), Shigella at 223 (1.0%), STEC (O157, non-O157, and not isolated combined) at 196 (0.9%), Vibrio at 6 (0.03%), and Yersinia at 4 (0.02%) (Table 2, Figure 1).
Table 2.
Enteric Bacterial Pathogens Isolated From Patients at an HMO Serving an Urban Geographical Area (Laboratory A), Minnesota, 1996–2006
| 1996 (n = 1905) | 1997 (n = 2048) | 1998 (n = 2243) | 1999 (n = 1972) | 2000 (n = 2432) | 2001 (n = 2114) | 2002 (n = 1997) | 2003 (n = 1667) | 2004 (n = 1589) | 2005 (n = 1722) | 2006 (n = 2281) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) |
| Campylobacter | 52 (2.7) | 70 (3.4) | 74 (3.3) | 34 (1.7) | 79 (3.2) | 60 (2.8) | 58 (2.9) | 39 (2.3) | 34 (2.1) | 45 (2.6) | 41 (1.8) |
| Salmonellaa | 28 (1.5) | 38 (1.9) | 27 (1.2) | 20 (1.0) | 22 (0.9) | 27 (1.3) | 12 (0.6) | 16 (1.0) | 23 (1.4) | 17 (1.0) | 34 (1.5) |
| Shigella | 7 (0.4) | 11 (0.5) | 17 (0.8) | 23 (1.2) | 84 (3.5) | 19 (0.9) | 24 (1.2) | 2 (0.1) | 6 (0.4) | 3 (0.2) | 27 (1.2) |
| Escherichia coli O157:H7 | 8 (0.4) | 8 (0.4) | 3 (0.1) | 9 (0.5) | 10 (0.4) | 7 (0.3) | 2 (0.1) | 1 (0.06) | 0 (0) | 6 (0.3) | 3 (0.1) |
| E. coli O157:NM | – | 1 (0.05) | – | – | 1 (0.04) | – | – | – | – | – | – |
| Other STECb | 5 (0.3) | 2 (0.1) | 11 (0.5) | 4 (0.2) | 19 (0.8) | 7 (0.3) | 10 (0.5) | 4 (0.2) | 2 (0.1) | 11 (0.6) | 13 (0.6) |
| Stx-positive; not isolated | 5 (0.3) | 1 (0.05) | 5 (0.2) | 4 (0.2) | 3 (0.1) | 3 (0.1) | 3 (0.2) | 3 (0.2) | 2 (0.1) | 11 (0.6) | 8 (0.4) |
| Enterotoxigenic E. colic | – | – | – | – | 40 (1.6) | 42 (2.0) | 36 (1.8) | 32 (1.9) | 31 (2.0) | 39 (2.3) | 36 (1.6) |
| Aeromonas spp | 1 | – | – | – | – | – | – | – | – | – | – |
| Plesiomonas shigelloides | – | 1 | – | – | – | – | – | – | – | – | – |
| Vibrio hollisae | – | – | 2 | – | – | – | – | – | – | – | – |
| Vibrio parahaemolyticus | – | 1 | – | – | – | – | – | 1 | – | 1 (0.06) | 0 (0.0) |
| Enteropathogenic E. coli | – | – | – | – | – | – | – | – | – | 2 (0.1) | – |
| Yersinia | – | – | – | – | 1 | – | – | – | 1 (0.1) | 1 (0.06) | – |
| No bacterial pathogens | 1799 (94.4) | 1915 (93.5) | 2104 (93.8) | 1978 (95.2) | 2173 (89.4) | 1949 (92.2) | 1852 (92.7) | 1569 (94.1) | 1490 (93.8) | 1586 (92.1) | 2119 (92.9) |
| Total | 1905 (100) | 2048 (100) | 2243 (100) | 1972 (100) | 2432 (100) | 2114 (100) | 1997 (100) | 1667 (100) | 1589 (100) | 1722 (100) | 2281 (100) |
Abbreviations: ETEC, enterotoxigenic E. coli; HMO, health maintenance organization; STEC, Shiga toxin-producing E. coli.
a Salmonella serotypes: Typhimurium, 68 (26%); Enteritidis, 47 (18%); Newport, 16 (6%); Typhimurium var. Copenhagen, 13 (5%); Heidelberg, 13 (5%); other, 107 (41%).
b STEC serogroups: O26, 18 (20%); O103, 18 (20%); O111, 10 (11%); O145, 4 (5%); 20 other serogroups, rough or undetermined, 37 (43%).
c ETEC serogroups: O25, 22 (9%); O6, 18 (7%); O27, 14 (5%); other, 60 (23%); not serotyped, 83 (32%); not isolated, 59 (23%). ETEC testing began in 2000.
Figure 1.
Number and proportion of bacterial pathogens isolated from patients at a health maintenance organization serving an urban geographical area (Laboratory A) and at a laboratory serving a rural agricultural area of Minnesota (Laboratory B). Time frames reported include the entire study period at each laboratory as well as the years when testing was conducted simultaneously at both laboratories. (a) 1996–2006 (n = 21 940 sorbitol MacConkey agar [SMAC] plates tested), Laboratory A. (b) 2000–2006 (n = 13 802 SMAC plates tested), Laboratory A. (c) 2000–2008 (n = 19 293 SMAC plates tested), Laboratory B. (d) 2000–2006 (n = 14 578 SMAC plates tested), Laboratory B. Abbreviations: ETEC, enterotoxigenic E. coli; STEC, Shiga toxin-producing E. coli.
Among the 13 802 cultures tested during 2000–2006, the period when ETEC testing was performed for Laboratory A, Campylobacter remained the most common bacterial pathogen, accounting for 356 (2.6%) cultures. The second most commonly identified pathogen was ETEC, accounting for 256 (1.9%) cultures, followed by Shigella at 165 (1.2%), Salmonella at 151 (1.1%), STEC at 129 (0.9%), Yersinia at 3 (0.02%), Vibrio at 2 (0.01%), and EPEC at 2 (0.01%) (Table 2, Figure 1).
Among the 196 STEC detected over the entire study period, 60 (31%) were O157:H7 or O157:non-motile, 88 (45%) were serogroups other than O157, and 48 (25%) were stx positive but were not isolated (Table 2).
The median annual number of cases for Campylobacter during the study period was 52 compared with a median of 52 during 2013–2014; Salmonella was 23 during the study period vs 25 during 2013–2014; Shigella was 17 vs 8; STEC O157 was 6 vs 5; STEC non-O157 was 7 vs 5; and STEC not isolated was 3 vs 0.
Laboratory B (Rural Laboratory)
For Laboratory B, 19 293 cultures were tested during 2000–2008, with a median of 2044 plates per year (range, 1959 to 2561). A bacterial pathogen was identified in 1069 (5.5%) cultures tested overall, with a range of 4.8% to 7.2% per year. Campylobacter was again the most common bacterial pathogen, accounting for 401 (2.1%) plates tested. Shiga toxin-producing E. coli (O157, non-O157, and not isolated combined) was the second most common bacterial pathogen isolated at 253 (1.3%), followed by Salmonella at 196 (1.0%), ETEC at 153 (0.8%), Shigella at 41 (0.2%), Yersinia at 16 (0.08%), and EPEC at 9 (0.05%) (Table 3, Figure 1).
Table 3.
Enteric Bacterial Pathogens Isolated From Patients at a Hospital Laboratory Serving a Rural Agricultural Area of Minnesota (Laboratory B), 2000–2008
| Pathogen | 2000 (n = 1959) No. (%) | 2001 (n = 1959) No. (%) | 2002 (n = 1987) No. (%) | 2003 (n = 2045) No. (%) | 2004 (n = 2023) No. (%) | 2005 (n = 2044) No. (%) | 2006 (n = 2561) No. (%) | 2007 (n = 2364) No. (%) | 2008 (n = 2351) No. (%) |
|---|---|---|---|---|---|---|---|---|---|
| Campylobacter | 40 (2.0) | 58 (3.0) | 40 (2.0) | 55 (2.7) | 48 (2.4) | 37 (1.8) | 38 (1.5) | 44 (1.9) | 41 (1.7) |
| Salmonellaa | 20 (1.0) | 25 (1.3) | 25 (1.3) | 20 (1.0) | 20 (1.0) | 19 (0.9) | 21 (0.8) | 21 (0.9) | 25 (1.1) |
| Shigella | 6 (0.3) | 5 (0.2) | 0 (0.0) | 8 (0.4) | 2 (0.1) | 4 (0.2) | 2 (0.08) | 2 (0.08) | 12 (0.5) |
| Escherichia coli O157:H7 | 12 (0.6) | 16 (0.8) | 12 (0.6) | 8 (0.4) | 5 (0.2) | 8 (0.4) | 11 (0.4) | 9 (0.4) | 6 (0.3) |
| E. coli O157:motile not H7 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.05) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Other STECb | 5 (0.3) | 12 (0.6) | 7 (0.4) | 5 (0.2) | 9 (0.4) | 9 (0.4) | 16 (0.6) | 21 (0.9) | 25 (1.1) |
| Stx-positive; not isolated | 4 (0.2) | 8 (0.4) | 4 (0.2) | 5 (0.2) | 4 (0.2) | 6 (0.3) | 15 (0.6) | 6 (0.3) | 4 (0.2) |
| Enterotoxigenic E. colic | 11 (0.6) | 16 (0.8) | 17 (0.9) | 14 (0.7) | 16 (0.8) | 25 (1.2) | 17 (0.7) | 26 (1.1) | 11 (0.5) |
| Enteropathogenic E. coli | – | – | – | – | – | 2 (0.1) | 2 (0.08) | 3 (0.1) | 2 (0.09) |
| Yersinia | 1 (0.05) | 1 (0.05) | 1 (0.05) | 1 (0.05) | 1 (0.05) | 4 (0.2) | 1 (0.04) | 4 (0.2) | 2 (0.09) |
| No bacterial pathogens | 1860 (94.9) | 1818 (92.8) | 1881 (94.7) | 1929 (94.3) | 1918 (94.8) | 1929 (94.4) | 2438 (95.2) | 2228 (94.2) | 2223 (94.6) |
| Total | 1959 (100) | 1959 (100) | 1987 (100) | 2045 (100) | 2023 (100) | 2044 (100) | 2561 (100) | 2364 (100) | 2351 (100) |
Abbreviations: ETEC, enterotoxigenic E. coli; STEC, Shiga toxin-producing E. coli.
a Salmonella serotypes: Typhimurium, 39 (20%); Enteritidis, 23 (12%); Newport, 16 (8%); Montevideo, 9 (5%); other, 109 (56%).
b STEC serogroups: O111, 29 (27%); O103, 28 (26%); O26, 23 (21%); O145, 6 (6%); 9 other serogroups, rough, or undetermined, 23 (21%).
c ETEC serogroups: O6, 12 (8%); O169, 8 (5%); other, 66 (43%); not serotyped, 22 (14%); not isolated, 45 (29%).
Among the 253 STEC detected, 88 (35%) were O157:H7 or O157:nonmotile, 109 (43%) were non-O157, and 56 (22%) were stx positive but were not isolated (Table 3).
The median annual number of cases for Campylobacter during the study period was 40 compared to a median of 42 during 2013–2014; Salmonella was 21 during the study period vs 35 during 2013–2014; STEC O157 was 9 vs 8; STEC non-O157 was 9 vs 10; STEC not isolated was 5 vs 1; Shigella was 4 vs 4.
Site Comparisons
To evaluate differences between the urban and rural populations, the proportion of positives for each pathogen from 2000 through 2006 (the time period when surveillance was conducted simultaneously at both sites) was compared. Statistically significant differences were found, with a higher proportion of STEC O157 in the rural site and a higher proportion of ETEC, Shigella, and Campylobacter in the urban site (Table 4). No significant differences in the proportion of Salmonella, non-O157 STEC, or Yersinia were found between the sites.
Table 4.
Comparison of Positive Results in the Urban (Laboratory A) vs Rural (Laboratory B) Laboratories, 2000–2006
| Agent | Urban | Rural | RR (95% CI) | P Value |
|---|---|---|---|---|
| No. (%) | No. (%) | |||
| Campylobacter | 356 (2.6%) | 316 (2.2%) | 1.2 (1.0–1.4) | .02 |
| Enterotoxigenic Escherichia coli | 256 (1.9%) | 116 (0.8%) | 2.3 (1.9–2.9) | <.001 |
| Salmonella | 151 (1.1%) | 150 (1.0%) | 1.1 (0.8–1.3) | NS |
| Shigella spp | 165 (1.2%) | 27 (0.2%) | 6.5 (4.3–10) | <.001 |
| Shigella sonnei | 147 (1.1%) | 23 (0.1%) | 6.8 (4.4–10) | <.001 |
| Shigella flexneri | 13 (0.1%) | 4 (<0.1%) | 3.4 (1.0–11) | .03 |
| Non-O157 Shiga toxin-producing E. coli | 65 (0.5%) | 63 (0.4%) | 1.1 (0.8–1.5) | NS |
| Shiga toxin-producing E. coli O157 | 30 (0.2%) | 73 (0.5%) | 0.4 (0.3–0.7) | <.001 |
| Total Stool Cultures | 13 802 | 14 578 |
Abbreviations: CI, confidence interval; NS, not significant; RR, relative risk.
Six hundred thirty-four ETEC and STEC cases were interviewed about illness and exposures, including 119 STEC and 194 ETEC cases from Laboratory A and 200 STEC and 121 ETEC cases from Laboratory B. Among Laboratory A cases, 125 of 192 (65%) ETEC cases that answered the international travel question reported traveling internationally in the 7 days prior to onset. Travel was evaluated by month of specimen collection and ranged from 3 of 8 (38%) international travelers in May to 20 of 25 (80%) in August. In 3 months (May, September, and November), the proportion of cases that traveled internationally was 50% or lower (38%, 50%, and 40%, respectively). Nineteen of 111 (17%) STEC cases reported traveling internationally; all were non-O157. Among Laboratory B cases, 66 of 119 (55%) ETEC cases reported traveling internationally. By month of specimen collection date, travel ranged from 1 of 5 (20%) international travelers in December to 12 of 15 (80%) in March. In 5 months (February and September through December), the proportion of cases that traveled internationally was 50% or lower (50%, 30%, 42%, 40%, and 20%, respectively). Eleven of 184 (6%) STEC cases reported traveling internationally; 8 were non-O157.
DISCUSSION
This long-term, multisite, sentinel surveillance study in Minnesota is the first to provide extensive data on the frequency of occurrence of ETEC infections compared with other bacterial enteric pathogens. In the urban population, ETEC was the second most common bacterial enteric pathogen behind Campylobacter, and it was more common than other common enteric bacterial pathogens such as Salmonella, Shigella, and STEC. In the rural population, ETEC was the fourth most common bacterial enteric pathogen, but it approached Salmonella in frequency.
The documentation of ETEC as a common cause of gastroenteritis on par with other common enteric bacterial pathogens has important implications for public health. Enterotoxigenic E. coli are not distinguishable from normal flora strains of E. coli by culture, and until recently there has not been a test for ETEC available to clinical laboratories. However, multiplex PCR panels that include ETEC as a target are starting to be adopted by clinical laboratories; it is anticipated that they will become widely used in the near future [18, 19]. As a result, public health agencies that require reporting or submission of clinical materials for ETEC (as is the case in Minnesota) will be confronted with issues related to the following: (1) receiving frequent ETEC reports and interviewing case-patients; (2) confirming, isolating, and serotyping ETEC strains when clinical laboratories submit PCR-positive specimens; and (3) subtyping ETEC isolates for the purpose of more efficient outbreak detection and investigation. The burden of all of these activities will be substantial.
Among the ETEC-positive patients in Minnesota, 61% of those interviewed reported foreign travel. A high percentage was expected, because ETEC is endemic in many developing countries visited by Minnesota travelers and is widely appreciated as the leading cause of traveler's diarrhea [1–3]. However, 39% of ETEC-positive patients in Minnesota who were interviewed did not report foreign travel and thus were presumably domestically acquired. Since a recent history of travel is more likely to lead to testing [20, 21], the proportion of ETEC that are domestically acquired is likely higher than what was found in this study. The sources of these infections were not determined, but produce imported from countries where ETEC is endemic is a likely source. Enterotoxigenic E. coli outbreaks due to imported produce are occasionally identified in Minnesota, other areas of the United States, and other countries [22–25]. These outbreaks are identified because they manifest as groups of ill people associated with discrete events or establishments that are reported to public health; when these groups are interviewed, the symptom and incubation profile that is relatively specific for ETEC becomes apparent, and patients are tested for ETEC. However, the outbreaks of ETEC infections that are identified almost certainly represent the proverbial tip of the iceberg. Because multiplex tests that include ETEC are widely used, pathogen-specific surveillance for ETEC may make it possible to detect ETEC outbreaks associated with produce or other foods distributed through retails settings (ie, grocery stores), in the same way that pathogen-specific surveillance for Salmonella and other pathogens has been so successful in detecting outbreaks associated with imported produce sold at retail [22, 26–28]. Therefore, along with the burden associated with an anticipated increase in ETEC reports comes greater opportunity to detect and control outbreaks.
These findings are also important for clinicians, because they should consider ETEC as a possible etiology of their patients' gastroenteritis and should not simply discount ETEC-positive results if the patient does not have a travel history.
In contrast to ETEC, our study suggested that EPEC (as defined by the presence of bfp and eae) is not a common pathogen in Minnesota. Our study also provided further data on the frequency of occurrence of STEC infections compared with other bacterial enteric pathogens. In the rural population, STEC was the second most common bacterial enteric pathogen in the rural population, behind Campylobacter. In the urban population, STEC was the fifth most common bacterial enteric pathogen. Culture-independent diagnostic tests for Shiga toxin, which enable the subsequent identification of non-O157 STEC serogroups, have been in use for a number of years, and their use is continuing to increase rapidly [8, 9]. As has been reported in previous studies based on testing of Shiga toxin-positive specimens submitted by clinical laboratories, in our study non-O157 serogroups were detected more commonly than O157. Because of the increasing use of Shiga toxin tests, the same burdens and opportunities for public health as discussed for ETEC are already in play. However, should culture-independent testing for STEC (e.g., immunoassays or multiplex PCRs that detect Shiga toxin or Shiga toxin genes, respectively) become universal, these burdens and opportunities will increase even further. Furthermore, as multiplex PCRs become more widely adopted, many clinical laboratories will likely stop culture for all pathogens, thus delaying the identification of STEC O157 until received at the PHL. This delay could have negative impacts on patient care [29].
Although the order of non-O157 serogroups differed slightly between the urban and rural sites, the top 3 in both sites included serogroups O111, O26, and O103. This result is consistent with previous studies [9–11]. The relative frequency of STEC, including non-O157 cases, compared with other common enteric bacterial pathogens in the study was comparable to that observed in 2013–2014 at both sites despite the differences in identification methods, with 1 minor exception; Salmonella was more common than STEC in the rural site in 2013–2014. This finding supports the hypothesis that increases in the identification of non-O157 STEC in the United States are likely due to detection methods and not an actual increase in incidence [9].
When the urban and rural monitoring sites were compared directly over the same time period, the relative abundance of several pathogens differed between the sites. In the urban population, ETEC and Shigella ranked higher than in the rural population. A higher proportion of STEC O157 in rural areas could be due to a higher likelihood of direct or indirect contact with cattle, cattle run-off, or other agricultural exposures known to be associated with STEC O157 infections [30]. Further study is needed to identify sources for these differences.
This study has some potential limitations. Testing methodology changed over time, and we had no way to verify that all SMAC cultures were in fact submitted. Nonetheless, these data collected over an extended period of time from 2 different sites are a good indication of the impact of the adoption of new CIDTs, including multiplex PCR assays. The identification of STEC other than O157 will continue to increase due to broader adoption of CIDTs. As a result, we will be able to identify a large number of ETEC infections, and we will likely identify a few typical EPEC infections. This strategy will provide new opportunities for outbreak identification and may be useful in better understanding the epidemiology of these pathogens (e.g., proportion of domestically acquired ETEC infections). Unfortunately, the burden on public health laboratories could be quite large. With the adoption of CIDTs, many clinical laboratories will (1) stop bacterial cultures and (2) submit stool or enrichment broths from CIDT-positive clinical tests to public health laboratories for confirmation. In addition, ETEC appears to be as common as other pathogens already under surveillance, and they are burdensome to confirm by culture, which will also add a new burden to the already strained public health laboratories.
CONCLUSIONS
In summary, this study found that when testing all stools submitted to 2 laboratories, the prevalence of ETEC and STEC infections were comparable with other enteric bacterial pathogens. Enterotoxigenic E. coli were the second most common enteric pathogen in the urban laboratory, and STEC were more common in the rural laboratory than the urban one and the second most common enteric pathogen identified in the rural laboratory. Findings of importance for clinicians include that a higher proportion of STEC O157 was found in rural areas, and a high proportion of ETEC (39%) patients did not travel internationally in the 7 days prior to their illness.
Acknowledgments
We thank all the staff at the sentinel sites, Health Partners Central Laboratory, and St. Cloud Hospital laboratory. We also thank the Minnesota Department of Health Public Health Laboratory staff and Acute Disease Investigation and Control Section staff who worked on this project, including C. Taylor, L. Carroll, E. Cebelinski, Y. Xiong, E. Thompson, M. Sullivan, F. Leano, P. Gahr, T. Weber, and Team Diarrhea.
Financial support. This work was funded by the Centers for Disease Control and Prevention as part of the Emerging Infections Program, Foodborne Diseases Active Surveillance Network (FoodNet; cooperative agreement U50/CCU511190).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Castelli F, Pezzoli C, Tomasoni L. Epidemiology of travelers’ diarrhea. J Travel Med 2001; 8(suppl 2):S26–30. [DOI] [PubMed] [Google Scholar]
- 2.Steffen R, Hill DR, DuPont HL. Traveler's diarrhea: a clinical review. JAMA 2015; 313:71–80. [DOI] [PubMed] [Google Scholar]
- 3.Kollaritsch H, Paulke-Korinek M, Weidermann U. Traveler's diarrhea. Infect Dis Clin North Am 2012; 26:691–706. [DOI] [PubMed] [Google Scholar]
- 4.Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis 2006; 43:1587–95. [DOI] [PubMed] [Google Scholar]
- 5.Hedican EB, Medus C, Nesser JM et al. Characteristics of O157 versus non-O157 Shiga toxin-producing Escherichia coli infections in Minnesota, 2000–2006. Clin Infect Dis 2009; 49:358–64. [DOI] [PubMed] [Google Scholar]
- 6.Beatty ME, Bopp CA, Wells JG et al. Enterotoxin-producing Escherichia coli O169:H41, United States. Emerg Infect Dis 2004; 10:518–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalton CB, Mintz ED, Wells JG et al. Outbreaks of enterotoxigenic Escherichia coli infection in American adults: a clinical and epidemiologic profile. Epidemiol Infect 1999; 123:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoefer D, Hurd S, Medus C et al. Laboratory practices for the identification of Shiga toxin-producing Escherichia coli in the United States, FoodNet sites, 2007. Foodborne Pathog Dis 2011; 8:555–60. [DOI] [PubMed] [Google Scholar]
- 9.Gould LH, Mody RK, Ong KL et al. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000–2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis 2013; 10:453–60. [DOI] [PubMed] [Google Scholar]
- 10.Hadler JL, Clogher P, Hurd S et al. Ten-year trends and risk factors for non-O157 Shiga toxin-producing Escherichia coli found through Shiga toxin testing, Connecticut, 2000–2009. Clin Infect Dis 2011; 53:269–76. [DOI] [PubMed] [Google Scholar]
- 11.Lathrop S, Edge K, Bareta J. Shiga toxin-producing Escherichia coli, New Mexico, USA, 2004–2007. Emerg Infect Dis 2009; 15:1289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockary VM, Hudson RF, Ball CL. Shiga toxin-producing Escherichia coli, Idaho. Emerg Infect Dis 2007; 13:1262–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning SD, Madera RT, Schneider W et al. Surveillance for Shiga toxin-producing Escherichia coli, Michigan, 2001–2005. Emerg Infect Dis 2007; 13:318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsvik O, Strockbine NA. PCR detection of heat-stable, heat labile, and Shiga-like toxin genes in Escherichia coli. In: Persing D, Smith T, Tenover F et al. eds. Diagnostic Molecular Microbiology. Washington, DC: American Society for Microbiology Press, 1993: pp 271–6. [Google Scholar]
- 15.Paton A, Paton J. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hylA, rfbO111, and rfbO157. J Clin Microbiol 1998; 36:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orskov F, Orskov I. Serotyping of Escherichia coli. In: Bergan T, ed. Methods of Microbiology. Vol. 14 London: Academic Press, 1984: pp 43–112. [Google Scholar]
- 17.Gunzberg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol 1995; 33:1375–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronquist AB, Mody RK, Atkinson R et al. Impacts of culture-independent diagnostic practices on public health surveillance for bacterial enteric pathogens. Clin Infect Dis 2012; 54(suppl 5):S432–9. [DOI] [PubMed] [Google Scholar]
- 19.Khare R, Espy MJ, Cebelinski E et al. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 2014; 52:3667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge VL, Odoi A, Fyfe M et al. Physician diagnostic and reporting practices for gastrointestinal illnesses in three health regions of British Columbia. Can J Public Health 2007; 98:306–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hennessy T, Marcus R, Deneen V et al. Survey of physician diagnostic practices for patients with acute diarrhea: clinical and public health implications. Clin Infect Dis 2004; 38(suppl 3):S203–11. [DOI] [PubMed] [Google Scholar]
- 22.Naimi TS, Wicklund JH, Olsen SJ et al. Concurrent outbreaks of Shigella sonnei and enterotoxigenic Escherichia coli infections associated with parsley: implications for surveillance and control of foodborne illness. J Food Prot 2003; 66:535–41. [DOI] [PubMed] [Google Scholar]
- 23.Yoder JS, Cesario S, Plotkin V et al. Outbreak of enterotoxigenic Escherichia coli infection with an unusually long duration of illness. Clin Infect Dis 2006; 42:1513–7. [DOI] [PubMed] [Google Scholar]
- 24.Devasia RA, Jones TF, Ward J et al. Endemically acquired foodborne outbreak of enterotoxin-producing Escherichia coli serotype O169:H41. Am J Med 2006; 119:168.e7–10. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald E, Møller KE, Wester AL et al. An outbreak of enterotoxigenic Escherichia coli (ETEC) infection in Norway, 2012: a reminder to consider uncommon pathogens in outbreaks involving imported products. Epidemiol Infect 2015; 143:486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barton Behravesh C, Mody RK, Jungk J et al. 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. N Engl J Med 2011; 364:918–27. [DOI] [PubMed] [Google Scholar]
- 27.Collier MG, Khudyakov YE, Selvage D et al. Outbreak of hepatitis A in the USA associated with frozen pomegranate arils imported from Turkey: an epidemiological case study. Lancet Infect Dis 2014; 14:976–81. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention (CDC). Outbreaks of cyclosporiasis--United States, June-August 2013. MMWR Morb Mortal Wkly Rep 2013; 62:862. [PMC free article] [PubMed] [Google Scholar]
- 29.Gould LH, Bopp C, Strockbine N et al. Recommendations for diagnosis of Shiga toxin--producing Escherichia coli infections by clinical laboratories. MMWR Recomm Rep 2009; 58(RR-12):1–14. [PubMed] [Google Scholar]
- 30.Byrne L, Jenkins C, Launders N et al. The epidemiology, microbiology and clinical impact of Shiga toxin-producing Escherichia coli in England, 2009–2012. Epidemiol Infect 2015; 143:3475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]