Abstract
In the light of emerging antibiotic resistance mechanisms found in bacteria throughout the world, discovery of drugs that potentiate the effect of currently available antibiotics remains an important aspect of pharmaceutical research in the 21st century. Well-established clinical tests exist to determine synergy in vitro, but these are only optimal for low-throughput experimentation while leaving analysis of results and interpretation of high-throughput microscale assays poorly standardized. Here, we describe a miniaturized broth microdilution checkerboard assay and data analysis method in 384-well plate format that conforms to the Clinical Laboratory and Standards Institute (CLSI) methods. This method has been automated and developed to rapidly determine the synergism of current antibiotics with various beta-lactamase inhibitors emerging from our antimicrobial research efforts. This technique increases test throughput and integrity of results, and saves test compound and labor. We facilitated the interpretation of results with an automated analysis tool allowing us to rapidly qualify inter- and intraplate robustness, determine efficacy of multiple antibiotics at the same time, and standardize the results of synergy interpretation. This procedure should enhance high-throughput antimicrobial drug discovery and supersedes former techniques.
Introduction
The global increase in antibiotic resistance is occurring at an alarming rate such that many of the currently available FDA-approved drugs are no longer effective.1–3 In 2013, the Centers for Disease Control reports that ∼2 million illnesses and ∼23,000 deaths are caused by antibiotic-resistant bacteria annually in the United States.4 The majority of these resistant organisms are well characterized, both phenotypically and genotypically, and whole cell antimicrobial testing is routine in clinics around the world.3 However, it is clear that even when armed with such information physicians are still challenged with new and emerging resistant strains entering their clinical settings. The European Centre for Disease Prevention and Control reports that between 2009 and 2012 resistance to third-generation cephalosporins in Klebsiella pneumonia and Escherichia coli has increased significantly within the EU and European Economic Area.5 This is further complicated by the fact that bacteria acquire resistance through mechanisms such as intra- and interstrain transmission by transposable elements.3,6 Much of the resistance is conferred to the bacteria by beta-lactamases, enzymes that are effective against the cell wall synthesis inhibitors, commonly referred to as beta-lactam molecules.3,7 Current combination therapies do exist such as with amoxicillin and clavulanic acid, which inhibit extended-spectrum beta-lactamases, and, when used properly, render these antibiotics effective again.8 However, this combination (Augmentin; GlaxoSmithKline, Philadelphia, PA) and other novel antibiotic combination therapies, such as NXL104 (AstraZeneca, London, United Kingdom) and ceftazidime (GlaxoSmithKline), are less effective than expected against other strains, in particular, those exhibiting class B metallo-beta-lactamases and class D beta-lactamases, for which there are no clinically relevant inhibitors.9,10
Complicating this issue is the fact that at the present time, there is a lack of consistent, rapid, cost-effective, and noncontroversial testing methods for predicting synergy of current antibiotic combinations with new antimicrobial agents against novel or unknown clinical isolates. Although strip-based Etests (bioMérieux USA, Durham, NC) and various other methods have been established by Clinical Laboratory and Standards Institute (CLSI) for lower throughput testing on agar plates, in tubes, and in 96-well plates, there are currently no agreed-upon testing and analysis designs for use with novel compounds in checkerboard microplate testing.11 Comparison studies have been performed and results may vary widely based on the method of interpretation used.12 Lacking a better method for interpreting results lends itself to controversy and demonstrates the need for a more standardized approach to understanding antibiotic efficacy in combination with current beta-lactamase inhibitors (BLIs).
Currently, many laboratories perform drug discovery-based screens in high-density formats. This allows for rapid production of reliable results. Here, we have miniaturized a standard antimicrobial synergy assay to 384-well plate format (wpf). The automation and instrumentation necessary for this task are fairly common as described in the methods. Implementing synergy assays into 384 wpf results in a major increase in throughput and data acquisition compared to the typical 96 wpf. This results in concomitant difficulty in interpreting results, for which we generated a novel, customizable, in-house analysis tool known as the Synergy RunTool.
Materials and Methods
Minimum Inhibitory Concentration and Synergy Protocols
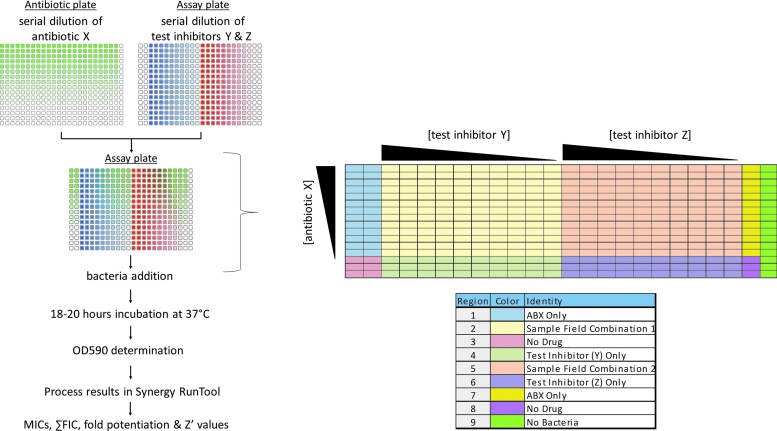
We use a broth microdilution technique with further miniaturization. All additions are automated using a Biomek FX (Beckman Coulter, Inc., Brea, CA) and a Multidrop Dispenser with stackers (Titertek Instruments, Inc., Huntsville, AL). All tests were performed using cation-adjusted Mueller–Hinton broth (CAMHB) (Part No. 297963; Becton, Dickinson and Co., Franklin Lakes, NJ). Enterobacter cloacae BAA-1143, a control strain of bacteria known to produce a high level of AmpC beta-lactamase, was obtained from the ATCC. YMC07/8/B3323 Acinetobacter species transformed with VIM-2 was a generous gift from Kyungwon Lee and Yunsop Chong at the Yonsei University College of Medicine, Seoul, South Korea. Test inhibitor NXL104, a known AmpC inhibitor, was synthesized in-house. Ceftazidime is commercially available (Part No. C3809; Sigma-Aldrich, St. Louis, MO). Imipenem is commercially available (Part No. 1337809; USP, Rockville, MD). We used 384-well plates (Part No. 3701; Corning, Inc., Corning, NY) with a final assay volume of 60 μL, derived from the combination of diluted antibiotic (15 μL), test inhibitor (15 μL), and bacteria (30 μL). As outlined in Table 1 and depicted in Figure 1, CAMHB is dispensed into two separate plates, named the antibiotic plate and the assay plate. The highest concentration of antibiotic is added to the first row of the antibiotic plate using the Beckman Biomek FX, and antibiotic is titrated down the columns using twofold serial dilutions to yield a 13-point curve. Separately, the highest concentration of test inhibitor is added to columns 3 and 13 of the assay plate and diluted across the rows of the plate to yield a 10-point curve. This is followed by a 15 μL transfer of the now diluted antibiotic from the antibiotic plate into the assay plate, making the volume in each well 30 μL. Each well thus contains a unique combination of antibiotic and test inhibitor, which is all tracked and registered through barcode within the Scripps database repository (Assay Explorer; Symyx, Santa Clara, CA). Bacteria, either lab adapted strains obtained from various vendors or clinical isolates obtained from investigators around the world, are diluted from fresh stocks to a working stock concentration of 1 × 106 cfu/mL, and 30 μL of the bacteria is added to the assay plate using a Multidrop Dispenser (Thermo Scientific, Waltham, MA) contained in a BSL-2 enclosure. Each individual well has a final bacterial concentration of 5 × 105 cfu/mL, determined as per CLSI methods.13 The plates are lidded and placed in a humidified chamber within a 37°C incubator for 18–20 h under aerobic conditions. Turbidity results are obtained on the same plate by measuring at OD 590 nm on a Tecan SpectraFluor Plus Reader (Tecan Systems, Inc., San Jose, CA). Plate data are then analyzed using an in-house derived software application called the Synergy RunTool.
Table 1.
Antimicrobial Synergy Assay Protocol
| Step | Plate | Parameter | Value | Description |
|---|---|---|---|---|
| 1 | Antibiotic | Dispense CAMHB | 35 μL | Into each well of rows B–P |
| 2 | Antibiotic | Antibiotic addition; twofold, horizontal serial dilution | 70 μL of highest (antibiotic) | Into each well of row A |
| 3 | Assay | Dispense CAMHB | 15 μL | Into each well of all columns except 3 and 13 |
| 4 | Assay | Test BLI compound addition; twofold, vertical serial dilution | 30 μL of highest (test BLI) | Into each well of columns 3 and 13 |
| 5 | Antibiotic into assay | Transfer antibiotic dilutions into assay plates | 15 μL into each well of the plate | Each well in the assay plate now contains a unique combination of Test BLI and antibiotic |
| 6 | Prepare bacteria to 1E+06 cfu/mL | 10 mL of bacterial solution per (1/2) plate | Bacteria prepared externally in a polypropylene tube | |
| 7 | Assay | Dispense bacteria into plate | 30 μL in each well of columns 1–23 | If testing two different bacteria on the same plate, place bacteria 1 in columns 1–12 and bacteria 2 in columns 13–23 |
| 8 | Assay | Dispense highest (antibiotic) into background control column 24 | 30 μL into each well | Use the same antibiotic solution used in step 2 |
| 9 | Assay | Incubate | 37°C for 18–20 h | Incubate in ample humidification |
| 10 | Assay | Read | OD 600 | Determine OD 600 using Tecan Safire II |
| 11 | Assay | Data analysis | Synergy RunTool | Determination of synergistic effect of antibiotic and test compound |
Step Notes
1. Using the 384-well liquid handle (aka Multidrop) add media to Corning 384-well translucent plates for the purpose of antibiotic dilution.
2. Using the Beckman FX serial dilution technique perform serial twofold dilutions down the rows of the same plate used in step 1.
3. Using the 384-well liquid handle (aka Multidrop) add media to Corning 384-well translucent plates for the purpose of test inhibitor dilution.
4. Using the Beckman FX serial dilution technique perform serial twofold dilutions across the columns of the same plate used in step 3.
5. Transfer samples from the antibiotic source plate prepared in steps 1 and 2 into the assay plate prepared in steps 3 and 4.
6. Fresh bacteria is diluted in CAMHB.
7. Using the Multidrop transfer bacteria into appropriate wells; column 24 being the exception.
8. Prepare column 24 as “No Bacteria” control.
9. Plates are covered and placed at 37°C overnight in a humidified chamber.
10. Read the plates using ABS 590 modality.
11. Use the automated Synergy RunTool to import data, analyze, and archive.
BLI, beta-lactamase inhibitor; CAMHB, cation adjusted Mueller–Hinton broth.
Fig. 1.
Flow diagram of a 384-wpf synergy assay. Separate plates are used to dilute the test inhibitor/s (assay plate) and antibiotics (antibiotic plate), which are then combined using a 384-well transfer device into the assay plate. Test inhibitors (BLIs) are not added to the first two columns of assay plate, just antibiotic and bacteria, allowing for the MIC of antibiotic alone to be measured while also leaving several wells available with no drug serving as positive controls. Bacteria (30 μL) are added to columns 1–23. In column 24, 30 μL of the highest concentration of antibiotic but, no bacteria, is added serving as the negative growth control. A color diagram is included along with reference legend to help identify wells of interest. ABX, antibiotic; BLIs, beta-lactamase inhibitors; MIC, minimum inhibitory concentration; wpf, well plate format.
Synergy RunTool
Minimum inhibitory concentrations (MICs) and synergy results were determined as per CLSI methods by incorporating the Synergy RunTool. The Synergy RunTool facilitates the interpretation of large amounts of data quickly and consistently. A proprietary Excel macro™ spreadsheet was created, in-house, to implement these requirements. A file-cycling system functionality was added to quickly load data files one at a time for viewing at roughly 1-s intervals. For each file loaded, all vital criteria are immediately calculated and a final output is created that easily allows the user to identify compounds of interest. The first step consisted of normalizing the plate to the user-selected high and low controls. In this case, these are bacteria in the presence of dimethyl sulfoxide (DMSO) only and media plus DMSO, that is, no test inhibitor and no bacteria, respectively. The MIC determined by the Synergy RunTool is the lowest concentration of the antimicrobial agent that prevents visible growth of bacteria, which is exactly as defined by CLSI.13 In addition, the Synergy RunTool mathematically determines a unique value to be applied as the MIC cutoff for each compound, which is designated as 3 × standard deviation (SD) + average (Avg) of the “No Bacteria” control. Any well with an OD 590 value greater than the cutoff value has observable growth, whereas those below the cutoff value are translucent and thus determine a unique value for the MIC. The spreadsheet is then able to parse the sample grid to find all relevant MIC values, including the MIC of antibiotic (ABX) alone as well as the MIC of the test inhibitor alone as defined using the following equations:
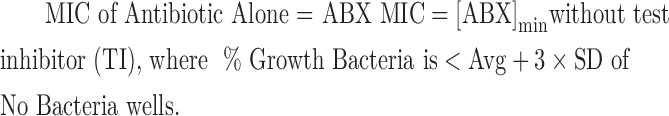 |
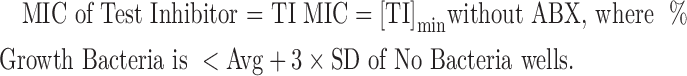 |
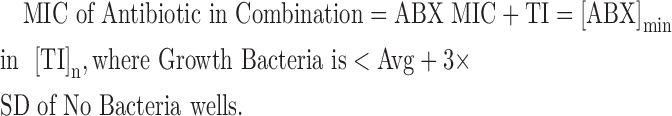 |
 |
A primary metric for determining a successful experiment is whether the measured ABX MIC is within the expected range for the test bacterial strain. An acceptable Z′ > 0.5 is also essential for determining a successful experiment.14 If these two criteria were met, data were processed to determine whether the inhibitors synergize with the antibiotic. Importantly, the Synergy RunTool can also identify the MIC of the combination of test BLI compound and ABX, which is the basis of determining synergy. With these values determined, the Synergy RunTool measures the Total Fractional Inhibitory Concentration (ΣFIC), which is a standard guide for predicting if a compound has a synergistic effect.15 This is calculated with the following equation:
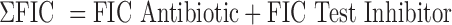 |
where:
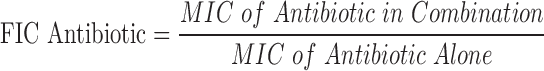 |
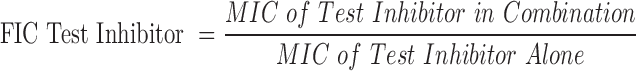 |
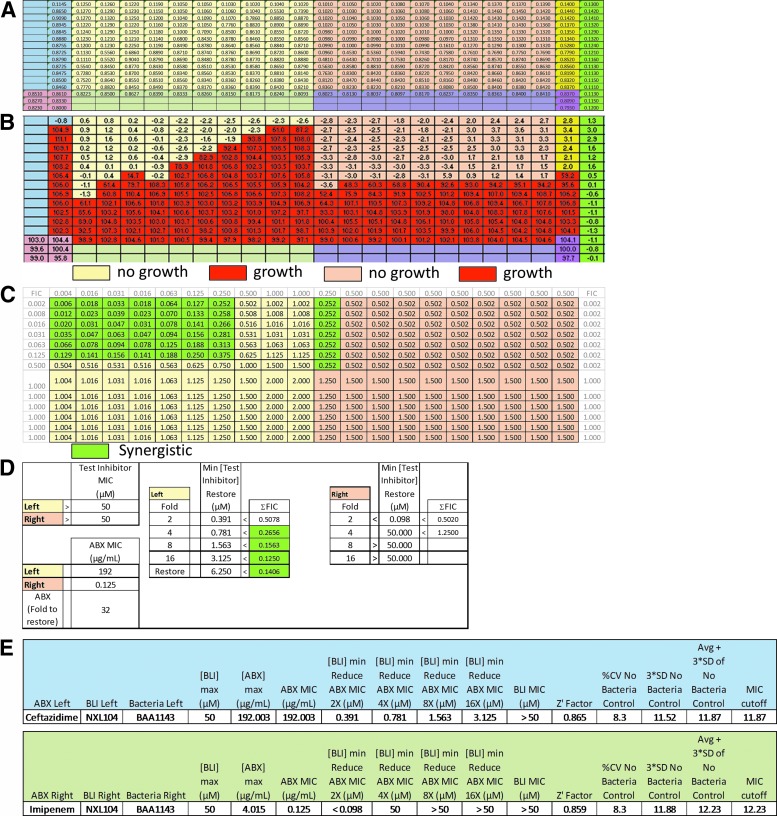
A ΣFIC value is calculated for every well in the plate because every well has a different combination of antibiotic and test inhibitor concentrations. The test inhibitor is considered to have a synergistic effect when ΣFIC ≤0.5 or the value that ultimately equates to the minimum test inhibitor concentration (Min [Test Inhibitor] Restore) necessary to cause a fourfold decrease in ABX MIC of the tested strain. The compound is considered indifferent with the antibiotic when the ΣFIC is >0.5 to <2 and antagonistic when the ΣFIC is ≥2.15 Figure 2 shows a populated RunTool chart that shows the calculated ΣFICs for each well used in a test case.
Fig. 2.
The Synergy RunTool displaying separate areas of interpretation. (A) The RunTool first imports raw data and displays it overlaid with regions color coded for compound and control allocation. (B) Data are then normalized and MICs determined per the methods described implementing user-defined controls. Bacterial growth versus no growth is automatically assigned the heat map colors depicted biased upon the automatically interpreted MIC values. (C) The same application calculates the ΣFIC using the formula described in the manuscript. The value calculated in each well is used to determine whether the compound is synergistic with the antibiotic assigning a shade of green if it is. A value of ≤0.5 means the compound synergizes with the antibiotic. (D) 384-wpf synergy testing yields two sets of data per plate pertaining to the left and right side of the plate. Results are shown in a color-coded table that quickly allows determination of Test Inhibitor MIC (alone), ABX MIC (alone), synergy shown as the fold potentiation, Min [Test Inhibitor] Restore = the μM concentration required to obtain its designated fold or full restoration of ABX efficacy, and the ΣFIC. Again, a synergistic result is shaded green. In the “Left” result, full restoration of ABX efficacy was achieved. (E) The statistics generated using the Synergy RunTool and also corresponding to the data generated as part of the illustration above. Separate statistics are calculated for each side of the plate. BLI Left is the compound tested in columns 1–12 of the assay plate and BLI Right is the bacteria tested in columns 13–23 of the assay plate. The chart shows values for the maximum BLI (inhibitor) concentration tested, maximum antibiotic (ABX) concentration, and the ABX MIC. The Synergy RunTool also calculates the [BLI]MIN needed to reduce ABX MIC by 2-, 4-, 8-, or 16-fold and the Z′ which determines assay quality as previously discussed. The MIC cutoff is shown and is determined as the sum of 3 × SD plus the average of the no bacteria control, as previously described in the text. All of these data are readily archived in the Scripps database for the ease of access by medicinal chemists. ΣFIC, Total Fractional Inhibitory Concentration.
Describing a test inhibitors' ability to synergize or potentiate the effect of an antibiotic in this manner can be difficult to translate verbally as well as numerically. To facilitate communication of results to investigators and in particular medicinal chemists, we added a feature to help analyze synergy, which we term potentiation. That's the ability of the test inhibitor, which has less effect on its own, to potentiate or allow an antibiotic to be more effective than when not in combination. In this sense, the effect of simultaneous application of antibiotics and test inhibitor is quantified according to the following equation:
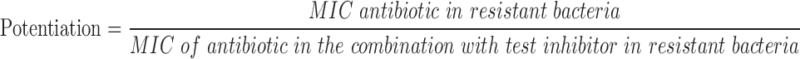 |
By definition, an MIC can vary by twofold day to day but not more. With that and similar to the ΣFICs method, a result of a fourfold increase is the minimum threshold to indicate a synergistic effect. A result of full restoration of antibiotic susceptibility is dependent on the change of the MIC found in resistant versus nonresistant bacteria with the MIC of antibiotic in combination with test inhibitor in resistant bacteria reverting to an MIC found for the antibiotic alone in the nonresistant bacteria.
Results and Discussion
By implementing the 384 wpf, we enable the determination of both MIC and synergy on two bacterial strains and at the same time for two antibiotics and two test inhibitors all on the same microtiter plate. This provides 10 twofold serial dilutions of test inhibitor and 13 for the antibiotic for 130 combinations in one-half of a plate or 260 combinations per plate. This is in addition to determining the MICs and Z′ from controls on the same plate. All are done while conforming to the CLSI-recommended procedures.13 Bacterial strains tested may be laboratory-adapted strains or clinical isolates. The only requirement is that one uses isolates that grow according to CLSI procedures. In addition, the number of serial dilution points for test inhibitor and antibiotic is predicated by the fact that one needs to test and observe at least a fourfold change in MIC in order for synergy to be declared for a test inhibitor and antibiotic combination. In doing so, one can determine synergy with as few as three dilution points, but the number of dilutions to determine restoration or full potentiation may vary from strain to strain and antibiotic tested. As shown in Figure 2, we test and identify the concentration required to restore the antibiotic to full effect against the bacteria, a result that would be missed otherwise. Obviously, this affects the throughput and amount of information able to be obtained on one plate; a situation that is flexible but must be assessed and designed based on user input and their knowledge of the bacteria resistance.
Synergy testing requires the ability to discern the effects of, in this case, two drugs in combination versus alone. Previous methods of MIC measurement (single drug testing) and synergy testing included low-throughput testing procedures utilizing test tubes and 96 wpf.16–18 These methods are more time, labor, and resource intensive and lack uniformity in analysis. The miniaturization to 384 wpf facilitates faster data collection while still conforming to CLSI standards. Traditional tube synergy testing requires a minimum of 36 tubes to test 25 combinations in a 1 mL volume to achieve reliable interpretation of synergy.13,19 A 96-wpf synergy assay achieves 77 combinations per plate in 0.2 mL volume/well.13 In contrast to both methods, the 384-wpf synergy test is able to obtain 260 test inhibitor combinations per plate using 0.06 mL/well. Both tube and 96-wpf formats dictate lower throughput and less quantitative interpretation, whereas our method allows for the least amount of materials, the smallest assay volume, and a rapid quantitative assessment. The Synergy RunTool generates all the following data in a matter of seconds (Fig. 2): (1) raw data import; (2) SD; (3) median and average value determination of all sample and control regions; (4) normalization of all wells to user selected high and low controls; (5) Z′; (6) signal-to-noise ratio; (7) %CV of the “No Bacteria” control (Percent Coefficient of Variation derived by [SD/Avg] ×100); (8) MIC cutoff based on the “No Bacteria” control (Avg +3 × SD); (9) MIC for every concentration of test inhibitor (n = 10) and ABX (n = 13); (10) ΣFIC of every possible combination of ABX and test inhibitor; (11) resistant versus nonresistant fold difference for ABX and test inhibitor MICs; (12) minimum test inhibitor and ABX concentrations necessary for 2×, 4×, 8×, and 16× fold increase in MIC versus control; and (13) minimum BLI and ABX concentrations necessary to restore resistant bacteria MICs to nonresistant bacteria MICs. From these statistics, we found the following to be the most important: Z′ determination using “No Drug” and “No Bacteria” (assay quality, Z′ ≥ 0.5); ABX MIC-fold difference between nonresistant and resistant bacterial strains (assay quality, Fold ≥8); Avg +3 × SD of “No Bacteria” region (determines MIC cutoff through mathematical interpretation confirming no-visible growth); ΣFIC at [BLI]MIN needed to restore the ABX MIC to its full potential as determined in nonresistant bacteria. These values are essential as they create a standard, noncontroversial method of interpreting quality of synergy assays and results of assays.
To compare our methods with current research, the compound NXL104, a novel but well-known B-lactamase inhibitor, was tested in synergy with ceftazidime, a third generation cephalosporin as shown in Figure 2. In this figure, the test inhibitor (BLI) is NXL104 for both the left and right side of the plate, the antibiotic tested in columns 1–12 is ceftazidime, and the antibiotic tested in columns 13–23 is imipenem. ABX MIC is measured in columns 2 and 23. The Synergy RunTool estimated an MIC of 192 μg/mL (i.e., the concentration of antibiotic in row A is 192 μg/mL) for ceftazidime against ATCC BAA1143; E. cloacae positive for AmpC resistance.20 In column 3, where the concentration of test compound is highest, the MIC of ceftazidime drops to 0.75 μg/mL (twofold titrations down the plate). Livermore et al. synergy tested Enterobacter spp. with AmpC resistance and found that 80% of the isolates had an MIC of 256 μg/mL.17 CLSI predicts the actual MIC for dilution test methods to be within a twofold range of that measurement (128–256 μg/mL). Therefore, the Synergy RunTool's calculation of 192 μg/mL is within the expected CLSI range.13 Livermore et al. also measured the MIC for the combination of NXL104/ceftazidime when NXL104 remained constant at 4 μg/mL; it ranged from 0.25 to 1 μg/mL.17 When tested using our 384-well methods and analyzed with the Synergy RunTool, the highest concentration tested of NXL104 was 50 μM or 1.44 μg/mL. At that concentration, the MIC of ceftazidime was 0.75 μg/mL, also within the range measured and suggested by Livermore et al. We also show that imipenem, as tested on the right half of the plate in Figure 2, is an antibiotic already known to be potent against AmpC-producing strains such as BAA-1143, and as such demonstrated the expected result in terms of potency and synergy with NXL104.21 In this case, it also serves as a reference standard against BAA-1143 bacteria and hence a baseline result for comparison of NXL104's ability to fully restore (potentiate) ceftazidime's efficacy. As shown with data from the experiment above, the Synergy RunTool generates a visual “heat” map (Fig. 2B), demonstrating that as test inhibitor concentration decreased along with a concomitant decrease in the ABX MIC, one creates a pattern of steps. The statistical information generated, as seen in Figure 2E, includes the MIC of antibiotic as well as the concentration of test inhibitor required to decrease, potentiate, and the MIC of antibiotic by 2-, 4-, 8-, and 16-fold. Taken together, we show NXL104 in combination with ceftazidime to demonstrate the ability of our method to determine the fold reduction in MIC, which can also be taken as the amount of potentiation a test inhibitor confers to an antibiotic to restore its function against the bacteria being tested.
Compared to existing literature, our miniaturized broth microdilution checkerboard assay produces accepted results, whereas, importantly, the Synergy RunTool accurately interprets those results.
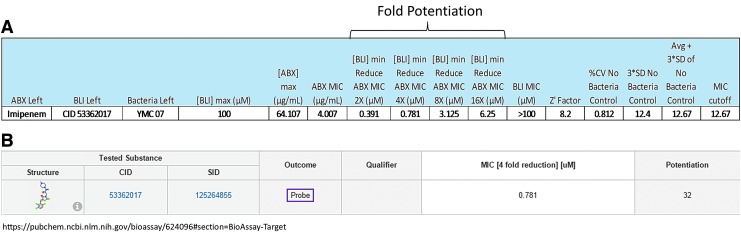
In addition to validating our method using the known synergistic pair described above, we provide further substantiation of our methods ability to determine synergy with different strains of bacteria and various combinations of test inhibitor and imipenem. This work was done in support of medicinal chemistry to determine the potential of test inhibitors to restore antibiotic efficacy against metallo beta-lactamases strains, which ultimately contributed to the identification of two molecular probes, ML121 and ML302.22,23 We tested various lab-adapted and clinical isolates of bacteria from around the world versus numerous test inhibitors (138) in combination with imipenem. In particular, we tested IMP1 and VIM2 metallo-beta-lactamase-expressing strains, including E. coli, Acinetobacter baumannii, and Pseudomonas aeruginosa. The fold potentiation of test inhibitor was different for each test strain and was reported accordingly. The fold restoration can be predicated using nonmetallo-enzyme transformed or wild-type bacteria of the same species, which are known to be sensitive to imipenem. This was done, in particular, for the lab-adapted and transformed strains; e.g., BL21 E. coli (wild type) versus BL21 VIM2 E. coli. Figure 3 represents an example of the outcome found when ML302 (CID 53362017) was tested in combination with imipenem to determine its effect on the clinical isolate YMC07/8/B3323; an Acinetobacter species transformed with VIM-2. A 16-fold potentiation of imipenem's effect was found at 6.25 μM test concentration of ML302. Ultimately, ML302 was able to achieve a 32-fold potentiation of the effect of imipenem in this strain; correlating to a 32-fold reduction in its MIC (Fig. 3B). All synergy data and further information regarding technique and results can be found on the publically available PubChem BioAssay web site (www.ncbi.nlm.nih.gov/pcassay) using the following record identifiers; AID 463099, 504620, 624080, 624082, 624095, and 624096.
Fig. 3.
(A) The statistics generated using the Synergy RunTool corresponding to the data generated as part of the discovery of probe ML302 (CID53362017). The bacteria tested was clinical isolate YMC07/8/B3323, an Acinetobacter spp. expressing VIM2. Fold reduction of the MIC of imipenem by ML302 is shown against this strain as 2×, 4×, 8×, and 16× fold. (B) These data were then archived in the Scripps database and also published to the PubChem web site (https://pubchem.ncbi.nlm.nih.gov/bioassay/624096#section=BioAssay-Target), which is available to the public. Note the maximum potentiation is listed for this and all other compounds published to the link above.
Notably, the methods described here utilize more than one plate to process the synergy assay, which does require cost and time. The automated equipment we used may not be widely available. However, the gain in efficiency when investigating large numbers of test inhibitors over time, such as that required for medicinal chemistry efforts should provide the impetus to move in this direction. In addition, acoustic transfer devices should handle this type of compound combination procedure and may limit the number of plates and manipulations with tips and other plastic ware. However, this too requires more expensive equipment and skilled engineers and in some cases very specialized source plates.
This methodology has been successfully applied to over 138 separate experiments in our lab, many times with multiple bacteria and test inhibitor/antibiotic combinations, equating to 465 synergy test results. We have successfully moved away from lower throughput methods, thus allowing more compounds to be tested simultaneously using reduced resources while generating a larger matrix of data. The creation of the Synergy RunTool easily handles this increased volume of data, which allows for standardization of results' dissemination, aiding communication between chemists and microbiologists in a manner that ensures results are properly archived, accurate, and reproducible. However, future testing using acoustic transfer devices will be done to compare and contrast the outcomes not only for antibiotic combinations but also for drug–drug combination and repurposing studies.
Abbreviations Used
- %CV
percent coefficient of variation
- [BLI]MIN
minimum BLI compound concentration
- ABX
Antibiotic
- Avg
Average
- BLI(s)
beta-lactamase inhibitor(s)
- CLSI
Clinical and Laboratory Standards Institute
- DMSO
dimethyl sulfoxide
- MIC
minimum inhibitory concentration
- SD
standard deviation
- TI
test inhibitor
- wpf
well plate format
- ΣFIC
Total Fractional Inhibitory Concentration
Acknowledgments
The National Institutes of Health (U54MH074404) supported this work. We thank Peter Hodder for his guidance during this effort. Pierre Baillargeon and Lina DeLuca (Lead Identification Division, Translational Research Institute, Scripps Florida) are thanked for their assistance with compound management. We thank Yang-Bo Feng for the synthesis of NXL104. Kyungwon Lee and Yunsop Chong are thanked for their donation of YMC07/8/B3323.
Disclosure Statement
No competing financial interests exist.
References
- 1.Bush K: Antibacterial drug discovery in the 21st century. Clin Microbiol Infect. 2004;10 Suppl 4:10–17 [DOI] [PubMed] [Google Scholar]
- 2.Blizzard TA, Chen H, Kim S, et al. : Side chain SAR of bicyclic beta-lactamase inhibitors (BLIs). 1. Discovery of a class C BLI for combination with imipinem. Bioorg Med Chem Lett 2010;20:918–921 [DOI] [PubMed] [Google Scholar]
- 3.Walsh TR, Toleman MA, Poirel L, Nordmann P: Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev 2005;18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC: Antibiotic resistance threats in the United States, 2013. In: Services USDoHaH (ed.), pp. 1–114. CDC, Atlanta, Georgia, 2013 [Google Scholar]
- 5.European Centre for Disease Prevention and Control: Summary of the Latest Data on Antibiotic Resistance in the European Union, pp. 1–10, Stockholm, 2013 [Google Scholar]
- 6.Normark BH, Normark S: Evolution and spread of antibiotic resistance. J Intern Med 2002;252:91–106 [DOI] [PubMed] [Google Scholar]
- 7.Bebrone C: Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 2007;74:1686–1701 [DOI] [PubMed] [Google Scholar]
- 8.Paterson DL, Bonomo RA: Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 2005;18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore DM, Mushtaq S, Warner M, Miossec C, Woodford N: NXL104 combinations versus Enterobacteriaceae with CTX-M extended-spectrum beta-lactamases and carbapenemases. J Antimicrob Chemother 2008;62:1053–1056 [DOI] [PubMed] [Google Scholar]
- 10.Miossec C: NXL104 β-lactamase inhibitor. Paper presented at the Challenge of Antibacterial Drug Development, San Diego, CA, 2007 [Google Scholar]
- 11.Jorgensen JH, Ferraro MJ: Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 2009;49:1749–1755 [DOI] [PubMed] [Google Scholar]
- 12.Bonapace CR, Bosso JA, Friedrich LV, White RL: Comparison of methods of interpretation of checkerboard synergy testing. Diagn Microbiol Infect Dis 2002;44:363–366 [DOI] [PubMed] [Google Scholar]
- 13.CLSI: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 7th ed., Clinical Laboratory Standards Institute, Wayne, PA, 2006 [Google Scholar]
- 14.Zhang JH, Chung TD, Oldenburg KR: A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999;4:67–73 [DOI] [PubMed] [Google Scholar]
- 15.Orhan G, Bayram A, Zer Y, Balci I: Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J Clin Microbiol 2005;43:140–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody J: Synergism Testing: Broth Microdilution Checkerboard and Broth Macrodilution Methods. 3rd ed. ASM Press, Washington, DC, 2010 [Google Scholar]
- 17.Livermore DM, Mushtaq S, Warner M, et al. : Activities of NXL104 combinations with ceftazidime and aztreonam against carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 2011;55:390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonapace CR, White RL, Friedrich LV, Bosso JA: Evaluation of antibiotic synergy against Acinetobacter baumannii: a comparison with Etest, time-kill, and checkerboard methods. Diagn Microbiol Infect Dis 2000;38:43–50 [DOI] [PubMed] [Google Scholar]
- 19.Minond D, Saldanha SA, Subramaniam P, et al. : Inhibitors of VIM-2 by screening pharmacologically active and click-chemistry compound libraries. Bioorg Med Chem 2009;17:5027–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gootz TD, Sanders CC, Goering RV: Resistance to cefamandole: derepression of beta-lactamases by cefoxitin and mutation in Enterobacter cloacae. J Infect Dis 1982;146:34–42 [DOI] [PubMed] [Google Scholar]
- 21.Jacoby GA: AmpC beta-lactamases. Clin Microbiol Rev 2009;22:161–182, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minond D, Saldanha SA, Spicer T, et al. : HTS Assay for Discovery of Novel Metallo-Beta-lactamase (MBL) Inhibitors. Probe Reports from the NIH Molecular Libraries Program, Bethesda, MD, 2010 [PubMed] [Google Scholar]
- 23.Spicer T, Minond D, Enogieru I, et al. : ML302, a Novel Beta-Lactamase (BLA) Inhibitor. Probe Reports from the NIH Molecular Libraries Program, Bethesda, MD, 2010 [PubMed] [Google Scholar]