Abstract
Introduction: Clinical, biodiversity, and environmental biobanks share many data standards, but there is a lack of harmonization on how data are defined and used among biobank fields. This article reports the outcome of an interactive, multidisciplinary session at a meeting of the European, Middle Eastern, and African Society for Biopreservation and Biobanking (ESBB) designed to encourage a ‘learning-from-each-other’ approach to achieve consensus on data needs and data management across biobank communities.
Materials, Methods, and Results: The Enviro-Bio and ESBBperanto Working Groups of the ESBB co-organized an interactive session at the 2013 conference (Verona, Italy), presenting data associated with biobanking processes, using examples from across different fields. One-hundred-sixty (160) diverse biobank participants were provided electronic voting devices with real-time screen display of responses to questions posed during the session. The importance of data standards and robust data management was recognized across the conference cohort, along with the need to raise awareness about these issues within and across different biobank sectors.
Discussion and Conclusion: While interactive sessions require a commitment of time and resources, and must be carefully coordinated for consistency and continuity, they stimulate the audience to be pro-active and direct the course of the session. This effective method was used to gauge opinions about significant topics across different biobanking communities. The votes revealed the need to: (a) educate biobanks in the use of data management tools and standards, and (b) encourage a more cohesive approach for how data and samples are tracked, exchanged, and standardized across biobanking communities. Recommendations for future interactive sessions are presented based on lessons learned.
Introduction
The biobanking landscape comprises a diverse and expanding collection of institutions, researchers, and practitioners who, regardless of their different functions, share a common need for best practices to implement data standards, ethical regulations, and risk management.1,2 These regulatory, ethical, and operational standards must continually evolve to keep biobanks in step with technical and scientific advancements, and the present and future demands of their stakeholders and clients. However, procedures, policies, and standards are designed with limited consideration given to the potential advantage of adapting those created by other thematic biobanks. Encouraging and sustaining cooperation and knowledge-sharing across globally dispersed and diverse biobanks is challenging, and scaling-up interactions is a limiting factor in terms of resources, costs, and coordination.
Data constitute a ‘universal language’ across biobank disciplines as they are the result of sample collection, management, and use. Additionally, genomics research technologies that apply increasingly sensitive biomolecular analyses are rapidly evolving, increasing the intrinsic value of all associated data. In any biobank the value and utility of a biospecimen or biological resource is determined by a) its fitness-for-purpose (assurance that the quality of the biospecimen meets the standard(s) of its end use; and b) the quality of the associated and attributed information (information used to describe, annotate, and authenticate the biospecimen as well as the data that provide a record of the processing and pre-analytical variables to which it has been exposed3). The greater the quality of data linked to a biospecimen, the better a sample will be characterized and qualified for future use. A high-quality sample is worthless if it has not been appropriately annotated or is improperly supported by information that assures its identity and quality, and describes its collection and use.10–16
In addition to preserving biospecimens as physical entities, biobanks need robust formats and standards to store information about their processing such as a) permissions including the ethical and regulatory documentation needed to acquire, transfer, or collect biospecimens; b) data that annotate collection, processing, and pre-analytical variables; c) associated data related to environmental or clinical information which characterize a specimen and validate its use; and d) data that enable standardized access and exchange of information (e.g., genomic sequencing data).
This article reports the outcome of a multidisciplinary, interactive session at the 2013 ESBB meeting in Verona, Italy entitled “The Life in Data,” designed by the Society's ESBBperanto and Enviro-Bio Working Groups. Individuals from across the globe, representing diverse types of bio-repositories and thematic disciplines, who would otherwise have only limited opportunity to interact, were brought together to gauge their level of knowledge and opinions regarding existing data standards, and to provide a forum for knowledge-sharing, debate, and consensus. Using a series of biobank-related questions on specific topics, the interactive session captured a ‘snapshot’ of informed opinion through a real-time survey. Guidelines and a ‘tool’ for planning, developing, and delivering an interactive conference session are presented as outputs of the experiences of the session planners, presenters, and participants. The value of incorporating similar interactive sessions at future conferences is discussed.
Materials, Methods, and Results
The interactive session, which started with a poll of the audience for demographic information, was designed to follow the lifecycle of a biobank sample through the stages of permissions and consent, sample collection, sample classification, research and data standardization. Each of these four stages was illustrated by examples from the environmental, biodiversity, and clinical sectors. Each stage was comprised of three parts—a scene-setting presentation, multiple-choice questions to solicit audience opinions, and projection of results in real-time with open discussion to provide continual interaction of participants. The poll was conducted using Televoter™ technology (Logos AV.COM., s.r.l.) with Showvote™ 2000 software collating and displaying the votes.
Audience demographics
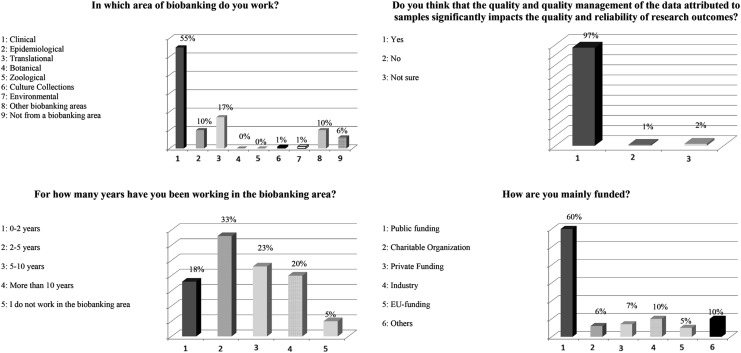
Audience demographics were obtained at the start of the session that involved 160 participants (Fig. 1). The participants represented primarily clinical medicine (55%), epidemiology (10%), and translational research (17%). Participants were funded primarily by public sources (65%) and industry (10%). The amount of time participants had been working in the biobanking field was 0–2 years (18%), 2–5 years (33%), 5–10 years (23%), and >10 years (20%); 5% were not involved in biobanking. The majority (97%) agreed that sample data quality management significantly impacts research quality and reliability.
FIG. 1.
Audience demographics.
Permissions and consent
The first stage was focused on issues related to permissions and consent, a crucial first step in biobanking. The Schistosomiasis Collection at the National History Museum (SCAN)4 was chosen as a case study to focus discussion. Schistosomiasis is a parasitic, neglected tropical disease (NTD) infecting approximately 200 million people globally. Schistosomiasis is caused by blood flukes (trematodes) with a life cycle that includes a snail intermediate host as well as the human or veterinary definitive host. While no human samples are stored at the SCAN repository, samples of parasite larval stages are collected from donors along with associated, relevant demographic data.
The SCAN repository includes both the medical and environmental/biodiversity sectors. SCAN collaborates directly with fieldwork-based projects such as the Schistosomiasis Consortium of Operational Research and Evaluation, which follows the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use Good Clinical Practice (ICH GCP) guidelines and complies with the World Medical Association's Declaration of Helsinki, 1964, amendment 2008.5
The related project, Zanzibar Elimination of Schistosomiasis Transmission,6 was used to describe the permission and consent process. Key issues discussed in this session included consent, open access to data, data protection, harmonization, and benefit sharing.
International legal frameworks for biodiversity biobanks were discussed using the example of the 1993 Convention on Biological Diversity (CBD), which states that biodiversity belongs to its country of origin. The CBD's Nagoya Protocol, which was ratified in 66 states (including the EU) on October 12, 2014,7 addresses access and benefit sharing agreement (ABS) permissions for the utilization of genetic resources. Essential elements of the Nagoya Protocol are prior informed consent, which must be obtained before sample collection, and mutually agreed terms for how benefits are shared. Material Transfer Agreements (MTA) lay out terms of ownership crucial to the consent process, for example, the Global Genome Biodiversity Network (GGBN) agreed to a harmonized set of generic MTAs and Code of Conduct for members.8
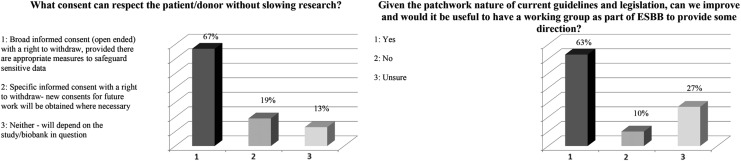
The first poll question asked was “What consent can respect the patient/donor/provider without slowing research?” As shown in Figure 2a, 99% of the audience responded, 67% selecting “Broad informed consent (open ended) with a right to withdraw, provided there are appropriate measures to safeguard sensitive data,” and 19% selecting “Specific informed consent with a right to withdraw—new consents for future work will be obtained where necessary.” One percent of respondents were assigned to faulty electronics or abstained from voting (equivalent to a 1% total error). A majority of 63% agreed on the need for more direction in the form of an ESBB working group (Fig. 2b).
FIG. 2.
Responses to survey questions related to permissions and consent. ESBB, European, Middle Eastern, and African Society for Biopreservation and Biobanking.
Sample collection
The utility of the Sample PREanalytical Code (SPREC, a code for the “management and traceability of biospecimen preanalytical variations”) as a data quality management and standardization tool was considered for clinical, environmental, and biodiversity biobanks.9–12 The point of collection was defined by subject and collector (field personnel, biobank staff, and healthcare professionals). Standard Operating Procedures (SOPs) and well-documented Work Instructions (WIs) were considered essential for sample data quality management, from the point of acquisition, throughout the chain of custody and processing.
As some elements are outside the control of the biobank and cannot be incorporated into the SPREC, good communication between biobank personnel and the collectors was considered vital to assure accurate data collection. This is dependent on well-informed, non-biobank personnel such as nurses, physicians, technical staff, field workers, and expedition teams. Special effort must be taken to include additional instructions that alert collectors (particularly non-biobank staff) to critical factors (e.g., timing, stabilization, temperature, container type). Explaining the significance of SPREC to collectors and biobank staff was considered critical and should involve training in sample and data collection techniques.
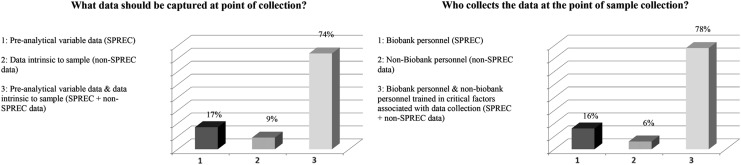
In responses to the question “What data should be captured at the point of collection?” 74% of the audience chose both “pre-analytical variable data and data intrinsic to the sample such as SPREC and non-SPREC related information,” the remainder of the audience chose only one of the two responses (Fig. 3). Thus, the majority vote conveyed a general consensus regarding the need to include both SPREC and associated (non-SPREC) data from the point of collection.
FIG. 3.
Responses to survey questions related to data needed at the point of sample collection. SPREC, Sample PREanalytical Code.
The audience concurred that a clear understanding of the different types of data associated with sample collection is necessary to describe and report sample status. This includes data about collection variables, as well as the associated information that defines the sample; both are vital to sample quality and increase the value of the research and other outcomes that are generated using biobanked samples. For the secondary question, “Who collects the data at the point of sample collection?”, 78% of the audience responded that both biobank staff and importantly non-biobank personnel need to be trained in collecting and recording critical sample acquisition data.
Sample classification
The value of a sample increases with the amount of associated data including site and collection information, information about the sample (e.g., disease definitions), and additional analyses or associated images. The value of a sample as defined by its associated data is clearly distinguishable from the fitness-for-purpose of a sample (quality). However, the associated data may also describe a sample's fitness-for-purpose. This session focused on associated data and the standards available, and used Biospecimen Reporting for Improved Study Quality (BRISQ)13 as an example. The purpose of this standard is to better understand, interpret, compare, and reproduce experimental results that involve human biospecimens. The use of BRISQ to describe research biospecimens in publications has been recommended by the Journal of Pathology,14 Histopathology,15 Nature Publishing Group,16 and Biopreservation and Biobanking.17
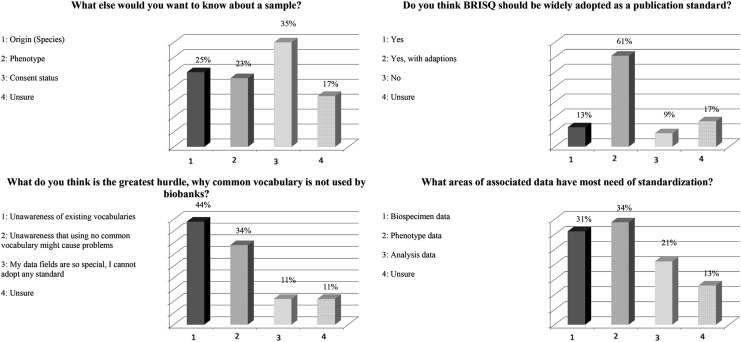
Responses to questions related to sample classification and associated data are shown in Figure 4. Responses to the question “What else do you want to know about a sample?” revealed that data about the origin of a sample was most important for 25% of the audience; 23% wanted to know more about the phenotype, 33% were interested in sample consent status, and 17% were unsure.
FIG. 4.
Responses to survey questions related to sample classification. BRISQ, Bispecimen Repository for Improved Study Quality.
In response to the question “Do you think BRISQ should be widely adopted as a publication standard?” 61% voted yes with adaptions, 13% indicated yes, 9% said no, and 17% were unsure.
Responses to “What do you think is the greatest hurdle for why common ‘vocabulary’ is not used by biobanks?” indicated a lack of awareness of existing vocabularies (44%) and the problems caused by not using controlled vocabularies (34%).
Responses to “What areas of associated data have most need of standardization?” were almost equally distributed between phenotype data (34%), biospecimen data (31%), and analysis data (21%), whereas 13% indicated they were unsure.
Research and data standardization
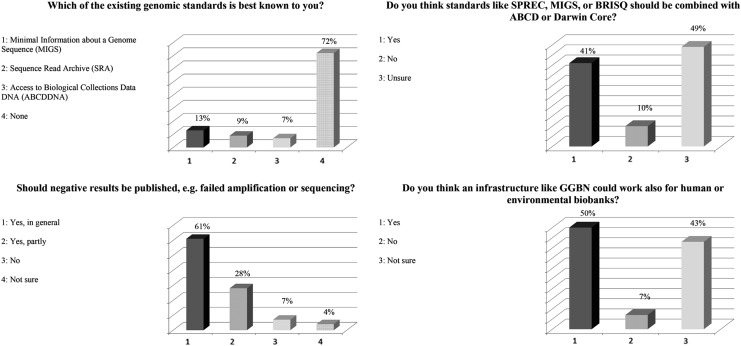
The last stage focused on the use of the sample and data standardization and results are shown in Figure 5. The question, “Which of the existing genomic standards is best known to you?” was posed as a bridge to the previous stage. A majority (72%) of the audience responded ‘none,’ 13% were aware of Minimum Information about a Genome Sequence (MIGS),18 9% knew of the Sequence Read Archive (SRA),19 and 7% knew of Access to Biological Collections Data DNA (ABCDDNA).20
FIG. 5.
Responses to survey questions related to research and data standardization. ABCD, Access to Biological Collection Data; GGBN, Global Genome Diversity Network; MIGS, Minimal Information about a Genome Sequence.
Given the development of genomic technologies and the lack of knowledge of related standards, the focus was on existing standards for genomic data sharing. While still using the initial SCAN case study, the focus was on GGBN as an example of a large-scale, international consortium that is establishing standards for data-sharing and publishing related to genomics data. Published sequence data mostly lack information on the underlying specimens (vouchers).21 Vouchers are biological specimens containing morphological parameters to allow taxonomic verification. They should be deposited in collections (e.g., herbaria or other natural history collections). The GGBN was formed in 2011 with the principal goal of bridging the gap by making high-quality, well-documented vouchered collections that store DNA or tissue samples of biodiversity collections as well as corresponding vouchers across the Tree of Life, available for research through a networked community of biodiversity repositories.
The GGBN Data Portal (http://data.ggbn.org) is based on established systems: GBIF (Global Biodiversity Information Facility),22 BioCASe (Biological Collection Access Service),23 IPT (Integrated Publishing Toolkit),24 and INSDC (International Nucleotide Sequence Data Base Collaboration).25 Each GGBN partner is responsible for its data and samples, and defines conditions for access; scientists can order samples easily via the shared portal. In the case of SCAN, GGBN has to reconcile different sample types and their associations with each other: intermediate host (snail) specimen, snail DNA, parasite specimen, parasite DNA, and relevant sequence data of the various sample types. So far, GGBN is the only platform worldwide to globally aggregate such biodiversity biobank data.
MIGS (Minimum Information about a Genome Sequence)18 is a vocabulary designed by the Genomic Standards Consortium for sharing DNA or RNA sequence information, which provides minimum information about underlying environmental data. Based on MIGS, Darwin Core,26 and ABCDDNA, GGBN has developed the GGBN Data Standard (http://terms.tdwg.org/wiki/GGBN_Data_Standard) to be used with environmental and single-species samples, as well as DNA/RNA and sequence information. Furthermore it can handle SPREC (http://terms.tdwg.org/wiki/ggbn:preparationProcess). When asked “Whether standards like SPREC, BRISQ or MIGS should be combined with ABCD or Darwin Core,” 41% of the audience agreed, 10% chose ‘no’, and 49% were unsure (Fig. 5).
GGBN also promotes publication of negative results (e.g., failed amplification or sequencing); the audience was polled on this issue with the majority (61%) agreeing that negative results should be published, 28% agreeing to partial publication, 10% voting ‘no’, and 7% indicating they were unsure. Finally, the audience was asked if an infrastructure like GGBN could work for human or environmental biobanks: 50% agreed, 43% were unsure, and 7% disagreed (Fig. 5).
Development of the interactive session tool
To provide sufficient time for audience participation, the interactive session was held over 2 hours; the format included four inter-related stages, each organized around a ‘core’ theme or ‘hot topic’; and each stage lasted approximately 10 minutes. At the start of the interactive session, the participants were familiarized with the relevant technology, and the audience demographics were determined. Each stage concluded with one or two multiple-choice questions with clear and understandable options for response in order to understand the baseline comprehension and opinions of the audience and stimulate discussion based on a review of the results.
Participant responses were captured and visualized in real-time, providing data that could be used to stimulate real-time debate. Outcomes were collated and disseminated to the conference participants. All data was made available for further statistical analysis and a more detailed, post-session interpretation.
Post-conference survey
The ESBB's post-conference, online survey carried out after the 2013 conference provided delegate feedback. Twenty-three percent (117) of the total number of conference attendees completed the online survey; 107 answered questions pertaining specifically to the Life in Data Interactive session. Of these, 77 had participated in the conference, representing 18% of the total number of delegates who had actively participated in the session.
A majority (73%) of the respondents were positive about the interactive session, with 25% reportedly very satisfied and 48% satisfied. Twenty-five percent had not attended the session and 2% indicated dissatisfaction. Participants considered the format of the session an excellent way to poll the views of the audience. The recommendation was made that this kind of session should be continued at the annual ESBB meetings, with some refinement to sustain levels of interaction and provide more time for discussion. Furthermore, it was suggested that careful attention be given to the design of questions to ensure that the audience is not being led to a desired response.
Discussion
Audience demographics
While it would have been interesting to know whether abstention from voting was higher for certain questions, these data were unfortunately not available.
Representation from biodiversity and environmental biobanks was low compared to human biobanks; while this is typical for ESBB and ISBER meetings it will not be the case for nonclinical conferences. The categorization of biobank areas was made using the type of research primarily supported by the biobank, as described in Watson et al.27 The finding that little is known about data standards is surprising as 86% of the audience had been working in biobanking for >2 years. This is an area of concern as it implies a lack of knowledge about available data standards. Although external biobank stakeholders28 can help ensure biobank sustainability, only 10% of the biobanks represented had funding from the industry sector.
The question on the impact of data quality on research outcomes was designed to get an understanding of the perspective of the audience before the session. As 97% voted that the quality of sample data and the quality of its management had a significant impact on the quality and reliability of research outcomes, it is even more surprising that there was so little awareness of data standards. This is consistent with recent publications that show that research is often not reproducible due to selected reporting.29 More than one-third of irreproducibility in pre-clinical research results is reportedly due to the biological reagents and reference materials used, and about 25% is due to data analysis and reporting issues.30 It would have been valuable to have asked a question on the state of informatics at the organizations represented by the participants, as well as the current method of documenting data (electronic vs. paper). These data could have provided a baseline of understanding upon which to base an education program and would have been related to the outcome of the survey of the ISBER Informatics WG.31
Permissions and consent
The responses to the first question revealed that the majority preferred broad informed consent, indicating concern that a specific consent process would slow research. As biobanking is a partnership between donors and researchers, there is a need to balance and account for the requirements of both groups.32 Responses to the second question indicated dissatisfaction with current regulations and guidelines; a dedicated ESBB working group was suggested to provide support and direction for issues related to permissions and consent such as third party use of material, implications of storing data/samples in public repositories, management and definition of benefit sharing, and balancing obligations with the ability to do research. The Nagoya Protocol will result in additional regulatory burdens with equivalent issues applying to clinical biobanks when balancing patient privacy and the need for research. One concern is the potential for genetic exceptionalism and the impact on data sharing.33,34
Sample collection
The majority of the audience was involved in clinical biobanks where data collection must be integrated with the care of hospital patients involving non-biobank personnel including nurses, clinicians, surgeons, and pathologists. The same is true for biodiversity and environmental sample data collection in remote sites where the logistics of sample movement are difficult and rely on non-biobank personnel.35 While SPREC offers considerable latitude for sample annotation throughout the collection process,10 it does not capture all significant associated data, some of which are available in BRISQ.13
The importance of training collection staff in SPREC and BRISQ tools according to biobank best practices2 was highlighted during the interactive audience debate. SPREC demonstrates how data quality management can be harmonized across the research and biobanking sectors (see Nussbeck et al., 201312), although a comprehensive assessment of the scope and utility of SPREC in relation to other reporting tools is recommended. This will facilitate data management from the point of sample collection throughout the entire biospecimen process and will address data standardization, an important requirement both across and between biobanking communities.36
Sample classification
When polling the participants on the information they want to know about a sample (Fig. 4), the relatively balanced distribution of responses indicated that many may have wanted to select more than one option. Consent status received the most votes (35%) indicating the need for satisfactory permissions not only for use in human biobanks (audience majority), but also for enviro-biobanks with the Nagoya protocol.7,34,37 While a previous survey in 2012 from the ESBBperanto Working Group demonstrated that there is almost no standardization for BRISQ data across tumor biobanks,38 results from our interactive session highlighted a consensus that controlled vocabulary and terminology should be adopted as a standard, but with adaptions according to need (61%). This is, perhaps, an indication of why introducing standards is difficult as many biobanks consider their data requirements to be unique. The lack of awareness of existing standards (44%) and the consequences of not using a common vocabulary (34%) clearly highlight the need for data standardization and use of clear definitions and terminology.36,39
Research and data standardization
Audience responses indicated a knowledge gap regarding genomic standards. Despite the fact that genomic sequencing is at the forefront of clinical and environmental research, 72% of the participants were unaware of the existence of any such standards. In an effort to explain this surprising response, the audience was asked to indicate its involvement in genomic research. The small number of responders underlined a lack of awareness of standards in developing technologies and the difficulty staying abreast of standards emerging from different disciplines, despite their potential multidisciplinary applicability. It would appear that training workshops for data standards and the use of common vocabularies would be useful for future ESBB conferences.9–11,35,39–44
Interactive sessions
Conferences provide timely opportunities for geographically dispersed and operationally diverse communities to come together. Interactive sessions enhance communication between groups that would not usually interact. Greater delegate participation is encouraged through recurrent interaction with session presenters, and as such, the participants have the potential to direct the course of the session. Sessions that focus on biobanking topics of special interest facilitate the sharing and dissemination of information across traditional thematic boundaries, and opinion polls create opportunities to test consensus or divergence about common topics of relevance to biobank communities in real-time.
Delivering interactive sessions at conferences enables delegates to benefit from the presence of stakeholders, vendors, clients, and beneficiaries in attendance, and they have the potential for participation across the wider biobanking community. The inclusion of participatory formats is inexpensive, and a less time-consuming alternative compared to other types of interactions. Importantly, voting provides immediate results and increases the possibility for follow-up questions that can achieve real-time group consensus or stimulate progression that can ultimately influence global biobanking policies.
Interestingly, 20% of delegates at the ESBB 2013 session responded to the post-conference online survey. In comparison, 64% of the participants present responded to the questions posed during the session. This highlights a significant increase in response to the real-time surveys in comparison to online surveys, and demonstrates the potential of this mechanism to increase responses to biobanking surveys and questionnaires. Significantly, they empower participants to vote anonymously on important issues and identify gaps that can help to direct the future course of biobank practice.
Recommendations for interactive sessions
The lessons learned from the ESBB's first interactive session have generated the following recommendations. It is expected that these will be refined by the experiences of future interactive session design and delivery teams.
1. Time and resources. The creation of an interactive session requires commitment of time and resources that need to be confirmed before including the session in a program.
2. Presenter coordination. Delivery requires a coordinated and consistent approach by topic presenters. Presenters in interactive sessions must coordinate content development and delivery of their topics to ensure that the ‘storyboard’ of the session is upheld throughout. This is distinct from traditional conference session formats, which comprise presentations developed independently and grouped into a single session.
3. Design and moderation. Careful attention must be paid to the construction and moderation of questions and answers to stimulate delegate participation. It is important that these are designed to be truly representative of all conference participants and are non-biased to avoid leading the audience to particular results.
4. Technology. Technology selected for future interactive sessions should be able to display conclusive summary graphics including absolute numbers of all answers from the session to provide a final correlation of responses from participants. The technology used in the pilot session was limited in this respect.
5. Improving participant interactions. Selected technology should allow questions and answers to be proposed and presented in real-time as part of follow-up to interactive discussions. The ability for participants to interact lends itself to the development of virtual sessions for participants unable to attend the session. Future developments for interactive sessions should permit the session to be completely interactive so that the results of each topic drive the direction of the subsequent one.
Conclusions
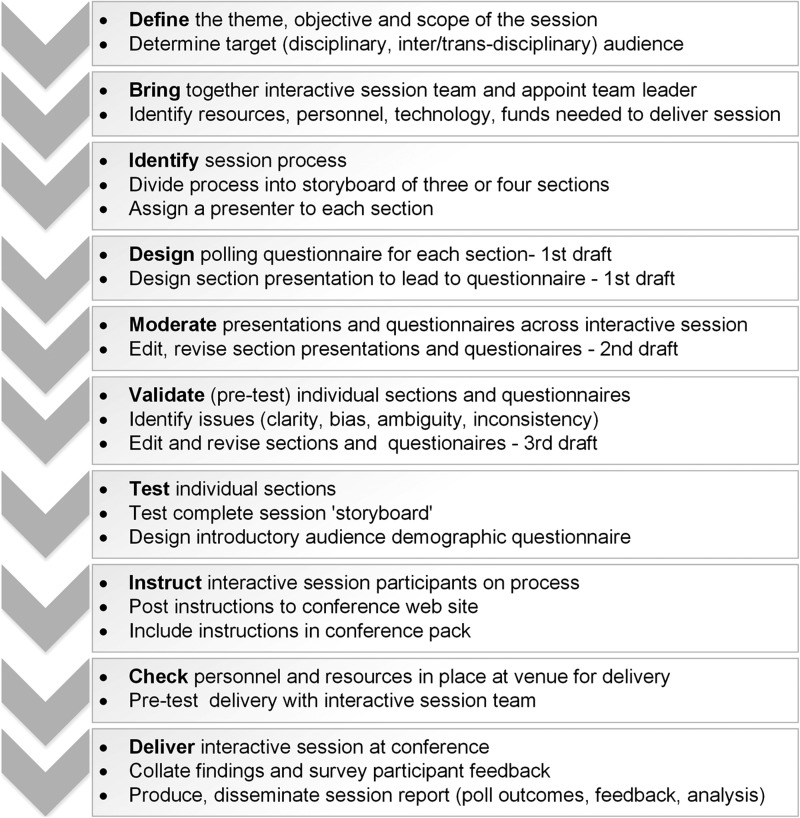
This publication reports on a successful pilot for a new interactive session format that captures data from the biobank community in situ at conferences. Simple guidelines are presented and an interactive session ‘Tool’ (Fig. 6) has been developed to enable the design of future sessions. Lessons learned from this first pilot have highlighted the need to develop communications and information technologies to increase the value of the interactive session by providing total summary graphics and permitting real-time presentation of questions that evolve during the session.
FIG. 6.
A “tool” for designing and delivering an interactive conference session.
This interactive session among biobank representatives highlighted the need for communication and integration between clinical and nonclinical sectors on data standards, terminology, definitions, and vocabularies. The use of data standards is pivotal and some level of harmonization between the standards of clinical and biodiversity biobank data is required. The joint initiative of GGBN and ESBB is a step in this direction.
The interactive session provided a community response that highlighted the need for education to increase the value of the data collected. The development of an associated common, controlled vocabulary accommodating harmonized or equivalent terms would help with data compatibility for wider sample/data querying and exchange. Significant interest was shown for sharing of clinical and nonclinical databases to provide a broader context such as correlating metadata from the human, animal, plant, parasite, pollutant, and environmental sectors to identify patterns of complex conditions. The integration of these biobank data could help answer larger questions about global health for all living organisms.
This exercise also demonstrated a lack of inter-disciplinary knowledge, which could be addressed through more interactive sessions and workshops on this topic, building on the outcomes of this first interactive session.
Acknowledgments
The authors acknowledge the interactive and online survey participants of the ESBB's 2013 annual conference, Verona, Italy; the ESBBperanto and Enviro-Bio Working Groups; ISBER's Biospecimen Science Working Group; and GGBN.
Author Disclosure Statement
No conflicting financial interests exist.
References
- 1.OECD Best Practice Guidelines for Biological Resource Centres. Paris: OECD Publishing; 2012 [Google Scholar]
- 2.2012 Best practices for repositories collection, storage, retrieval, and distribution of biological materials for research international society for biological and environmental repositories (no authors listed). Biopreserv Biobank 2012;10:79–161 [DOI] [PubMed] [Google Scholar]
- 3.Hewitt R, Watson P. Defining biobank. Biopreserv Biobank 2013;11:309–315 [DOI] [PubMed] [Google Scholar]
- 4.Emery AM, Allan FE, Rabone ME, Rollinson D. Schistosomiasis collection at NHM (SCAN). Parasites Vectors. 2012;5:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Medical Association. WMA Declaration of Helsinki—Ethical Principles for Medical Research Involving Human Subjects; 2013. Available from: URL:http://www.wma.net/en/30publications/10policies/b3/ Last accessed 31October2014 [DOI] [PubMed]
- 6.Knopp S, Mohammed KA, Ali SM, et al. . Study and implementation of urogenital schistosomiasis elimination in Zanzibar (Unguja and Pemba islands) using an integrated multidisciplinary approach. BMC Public Health. 2012;12:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Nagoya Protocol on access and benefit sharing of genetic resources. The Nagoya Protocol on access and benefit sharing of genetic resources, analysis and implementation options for developing countries Nijar, Gurdial Singh. Geneva: South Centre [Google Scholar]
- 8.The Global Genome Biodiversity Network (GGBN). GGBN March 2015 Newsletter. Washington, D.C: http://www.ggbn.org/docs/GGBN_March2015_Newsletter_FINAL.pdf Last accessed 6January2016 [Google Scholar]
- 9.Benson EE, Betsou F, Amaral R, Santos LM, Harding K. Standard PREanalytical codes: A new paradigm for environmental biobanking sectors explored in algal culture collections. Biopreserv Biobank 2011;9:399–410 [DOI] [PubMed] [Google Scholar]
- 10.Betsou F, Lehmann S, Ashton G, et al. . Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiol Biomarkers Prevent 2010;19:1004–1011 [DOI] [PubMed] [Google Scholar]
- 11.Lehmann S, Guadagni F, Moore H, et al. . Standard preanalytical coding for biospecimens: Review and implementation of the Sample PREanalytical Code (SPREC). Biopreserv Biobank 2012;10:366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussbeck SY, Benson EE, Betsou F, Guadagni F, Lehmann S, Umbach N. Is there a protocol for using the SPREC? Biopreserv Biobank 2013;11:260–266 [DOI] [PubMed] [Google Scholar]
- 13.Moore HM, Kelly AB, Jewell SD, et al. . Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol 2011;119:92–101 [DOI] [PubMed] [Google Scholar]
- 14.Simeon-Dubach D, Burt AD, Hall PA. Quality really matters: The need to improve specimen quality in biomedical research. J Pathol 2012;228:431–433 [DOI] [PubMed] [Google Scholar]
- 15.Simeon-Dubach D, Burt AD, Hall PA. Quality really matters: The need to improve specimen quality in biomedical research. Histopathology 2012;61:1003–1005 [DOI] [PubMed] [Google Scholar]
- 16.Reducing our irreproducibility. Nature 2013;496:1 [Google Scholar]
- 17.Simeon-Dubach D, Moore HM. BIO comes into the cold to adopt BRISQ. Biopreserv Biobank 2014;12:223–224 [DOI] [PubMed] [Google Scholar]
- 18.Field D, Garrity G, Gray T, et al. . The minimum information about a genome sequence (MIGS) specification. Nature Biotechnol 2008;26:541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database C. The sequence read archive. Nucleic Acids Res 2011;39:D19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holetschek J, Dröge G, Güntsch A, Berendsohn WG. The ABCD of primary biodiversity data access. Plant Biosystems 2012;146:9 [Google Scholar]
- 21.Droege G, Barker K, Astrin JJ, et al. . The Global Genome Biodiversity Network (GGBN) Data Portal. Nucleic Acids Res 2014;42:D607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen AM. GBIF strategic plan 2012–2016: Seizing the future. Cph: Global Biodiversity Inform Facility 2011; 2011 [Google Scholar]
- 23.Güntsch A, Berendsohn WG, Mergen P. The BioCASE Project—A Biological Collections Access Service for Europe. Ferrantia 2007;51:103–108. 2007. [Google Scholar]
- 24.Robertson T, Doring M, Guralnick R, et al. . The GBIF integrated publishing toolkit: Facilitating the efficient publishing of biodiversity data on the internet. PloS One. 2014;9:e102623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karsch-Mizrachi I, Nakamura Y, Cochrane G, International Nucleotide Sequence Database C. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res 2012;40:D33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wieczorek J, Bloom D, Guralnick R, et al. . Darwin Core: An evolving community-developed biodiversity data standard. PloS One 2012;7:e29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson PH, Barnes RO. A proposed schema for classifying human research biobanks. Biopreserv Biobank 2011;9:327–333 [DOI] [PubMed] [Google Scholar]
- 28.Gemeinholzer B, Droge G, Zetzsche H, et al. . The DNA bank network: The start from a German initiative. Biopreserv Biobank 2011;9:51–55 [DOI] [PubMed] [Google Scholar]
- 29.Open Science C. PSYCHOLOGY. Estimating the reproducibility of psychological science. Science 2015;349:aac4716. [DOI] [PubMed] [Google Scholar]
- 30.Freedman LP, Cockburn IM, Simcoe TS. The economics of reproducibility in preclinical research. PLoS Biol 2015;13:e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fearn P, Michels C, Meagher K, Cada M. 2012 International Society for Biological and Environmental Repositories Informatics Working Group: Survey results and conclusions. Biopreserv Biobank 2013;11:64–66 [DOI] [PubMed] [Google Scholar]
- 32.Simeon-Dubach D, Watson P. Biobanking 3.0: Evidence based and customer focused biobanking. Clin Biochem 2014;47:9. [DOI] [PubMed] [Google Scholar]
- 33.Global Genome Biodiversity Network Standard Material Transfer Agreements (MTAs). Global Genome Biodiversity Network http://ggbn.org/docs/ABS_Guidance/GGBN MTA_June_2015-Final.pdf Last accessed 6January2016
- 34.Lyal CHC. Can we keep it? Managing the impact of the Nagoya protocol on insect collections and research. Antenna. 2014;38:3 [Google Scholar]
- 35.Harding K, Benson EE, Nunes E, et al. . Can biospecimen science expedite the ex situ conservation of plants in megadiverse countries? A focus on the flora of brazil. Crit Rev Plant Sci 2013;11:6 [Google Scholar]
- 36.MacKenzie-Dodds J, Clarke A, Lermen D, et al. . Recent initiatives in biodiversity biobanking: Summary of presentations from the ESBB 2012 Conference. Biopreserv Biobank 2013;11:182–188 [DOI] [PubMed] [Google Scholar]
- 37.OECD Global Science Forum, Second Activity on Policy Issues Related to Scientific Research Collections, Washington, DC, July 17–19, 2008, Final Report on Findings and Recommendations. OECD Global Science Forum. 2008;http://www.oecd.org/science/sci-tech/42237442.pdf Last accessed 6January2016. [Google Scholar]
- 38.Joint Conference of the European, Middle Eastern & African Society for Biopreservation & Biobanking (ESBB) and the Spanish National Biobank Network Granada, Spain November 7–9, 2012: ES-12. ESBBperanto Working Group: Survey on Metadata Standards in Tumor Biobanks. Biopreserv Biobank 2012;10:A45 [Google Scholar]
- 39.Benson E, Betson F, Fuller BJ, Harding K, Kofanova O. Translating cryobiology principles into trans-disciplinary storage guidelines for biorepositories and biobanks: A concept paper. Cryo Lett 2013;34:277–312 [PubMed] [Google Scholar]
- 40.Genebank Standards. Food and Agriculture Organization of the United Nations, Rome, International Plant Genetic Resources Institute, Rome. 1994; ftp://ftp.fao.org/docrep/fao/meeting/015/aj680e.pdf Last accessed 6January2016
- 41.Benson EE. Cryopreservation of phytodiversity: A critical appraisal of theory & practice. Crit Rev Plant Sci 2008;27:79 [Google Scholar]
- 42.Day JG, Benson EE, Harding K, et al. . Cryopreservation and conservation of microalgae: The development of a Pan-European scientific and biotechnological resource (the COBRA project). Cryo Lett 2005;26:231–238 [PubMed] [Google Scholar]
- 43.Pugh RS, Becker PR, Porter BJ, Ellisor MB, Moors AJ, Wise SA. Design and applications of the National Institute of Standards and Technology's (NIST's) environmental specimen banking programs. Cell Preserv Technol 2008;6:14 [Google Scholar]
- 44.Stacey GN. Fundamental Issues for Cell Line Banks in Biotechnology and Regulatory Affairs: Life in the Frozen State: Eds Fuller B, Lane N, Benson EE. CRC Press: 2004:16 [Google Scholar]