Abstract
Aim: Mitochondrial cytochrome c oxidase (COX), the last enzyme of the respiratory chain, catalyzes the reduction of oxygen to water and therefore is essential for cell function and viability. COX is a multimeric complex, whose biogenesis is extensively regulated. One type of control targets cytochrome c oxidase subunit 1 (Cox1), a key COX enzymatic core subunit translated on mitochondrial ribosomes. In Saccharomyces cerevisiae, Cox1 synthesis and COX assembly are coordinated through a negative feedback regulatory loop. This coordination is mediated by Mss51, a heme-sensing COX1 mRNA-specific processing factor and translational activator that is also a Cox1 chaperone. In this study, we investigated whether Mss51 hemylation and Mss51-mediated Cox1 synthesis are both modulated by the reduction–oxidation (redox) environment. Results: We report that Cox1 synthesis is attenuated under oxidative stress conditions and have identified one of the underlying mechanisms. We show that in vitro and in vivo exposure to hydrogen peroxide induces the formation of a disulfide bond in Mss51 involving CPX motif heme-coordinating cysteines. Mss51 oxidation results in a heme ligand switch, thereby lowering heme-binding affinity and promoting its release. We demonstrate that in addition to affecting Mss51-dependent heme sensing, oxidative stress compromises Mss51 roles in COX1 mRNA processing and translation. Innovation: H2O2-induced downregulation of mitochondrial translation has so far not been reported. We show that high H2O2 concentrations induce a global attenuation effect, but milder concentrations specifically affect COX1 mRNA processing and translation in an Mss51-dependent manner. Conclusion: The redox environment modulates Mss51 functions, which are essential for regulation of COX biogenesis and aerobic energy production. Antioxid. Redox Signal. 24, 281–298.
Introduction
In biological systems, reduction–oxidation (redox) reactions are central to most cellular processes, including cellular differentiation, proliferation, and death (15). In eukaryotes, the mitochondrion plays a central role in these processes, a case in point being the redox-driven cellular respiration (30) that takes place in the organelle.
The mitochondrial respiratory chain (MRC) is formed by four enzymatic complexes (complexes I–IV) and two mobile electron carriers (cytochrome c and coenzyme Q) that act in concert to transfer electrons from reducing equivalents to molecular oxygen (O2). In each round of electron transfer through the MRC, one electron is donated to oxygen by COX, the terminal oxidase in the chain. As a by-product of respiration, electrons can escape prematurely, reducing oxygen to superoxide anion radicals (O2−), which are rapidly dismutated to H2O2, a membrane-permeable molecule with a longer half-life than most reactive oxygen species (ROS).
The cellular response to ROS displays hormesis (38). High ROS concentrations induce oxidative stress and can cause significant damage to many cellular components. However, elevated ROS also act as redox signaling molecules in the maintenance of physiological functions. For example, the response to oxidative stress is mediated by dedicated transcription factors whose activities are governed by thiol redox-sensitive mechanisms (10, 22).
In recent years, several reports have described the existence of cysteine thiol-based redox switches in heme-regulated proteins (35). Among them, human heme oxygenase-2 (HO-2) is subjected to reversible thiol/disulfide interconversion, with different heme-binding affinities in the oxidized and the reduced states (35). Proteins that bind heme for regulatory purposes contain heme regulatory motifs (HRMs) that usually consist of conserved Cys-Pro core sequences flanked at the C-terminus by a hydrophobic residue [CPX motifs (46, 59)]. In some cases, the redox switch involves the cysteines in the CPX motifs. For example, the transcription factor Rev-erbβ, a member of the nuclear receptor superfamily, undergoes a heme ligand switch upon oxidation of two CPX motif cysteines, which results in lower affinity for heme (14). We recently reported the presence of similar HRMs in the Saccharomyces cerevisiae mitochondrial translational activator, Mss51 (44).
Innovation.
Reactive oxygen species (ROS) and heme mediate essential cell regulatory processes. A mechanism that combines ROS and heme sensing consists of thiol/disulfide reduction–oxidation (redox) switches regulating the function of hemoproteins. In this study, we identified such a mechanism operating in the translational regulation of yeast mitochondrial cytochrome c oxidase (COX) assembly. The key element is Mss51, a heme-binding COX1 mRNA-specific translational activator that coordinates cytochrome c oxidase subunit 1 (Cox1) synthesis and assembly. We show that redox sensing through heme regulatory cysteines modulates Mss51 functions, resulting in Cox1 synthesis attenuation under oxidative stress conditions. Thus, this study adds new dimensions in understanding how mitochondrial translation is regulated in response to oxidative stress.
Mss51 is specifically involved in the biogenesis of cytochrome c oxidase subunit 1 (Cox1). COX is a multimeric enzyme that contains three catalytic core subunits (Cox1, Cox2, and Cox3) encoded in the mitochondrial DNA and eight nucleus-encoded subunits that act as a protective shield of the core (43). Cox1 contains heme A and copper prosthetic groups, which potentially make this protein highly reactive when the metal groups are not correctly inserted (16). During COX assembly, Cox1 constitutes the seed around which the holocomplex is assembled by incorporation of individual subunits and prebuilt modules containing the core subunits (12, 25, 31). For these reasons, COX assembly is a highly controlled process, with the biogenesis of Cox1 being the most regulated step.
Our group and others have reported that Cox1 is the subject of assembly-dependent translational regulation (4, 12, 29, 32–34, 41, 44). Through a negative feedback regulatory loop, Cox1 synthesis and COX assembly are coordinated to optimize COX biogenesis. This coordination is mediated by Mss51, a COX1 mRNA-specific processing factor and translational activator (9), which also acts as a chaperone to stabilize newly synthesized Cox1 and promote its assembly (4, 32). To prevent excessive Cox1 synthesis when the protein cannot be assembled into COX, Mss51 binds to newly synthesized Cox1 and forms a transient complex with additional COX assembly factors that trap Mss51, thereby limiting its availability to act in Cox1 translation (4, 11, 12, 28, 29, 34). In a recent study, our group uncovered that Mss51 binds heme B and contains two CPX motifs located in its N-terminus, whose integrity is required for Mss51 function as a COX1 mRNA translational activator (44).
Since mitochondria are a major source of ROS, we have now explored whether heme binding to Mss51 and consequently the multiple functions performed by this protein could be affected by redox changes induced by oxidative stress. A combination of in vitro and in vivo experimental strategies allowed us to demonstrate that exposure to hydrogen peroxide induces formation of a disulfide bond in Mss51 involving HRM cysteines. We show that a combination of oxidation-induced impairment of heme-dependent and heme-independent roles of Mss51 contributes to decreased mitochondrial COX1 mRNA maturation and attenuated Cox1 synthesis. We conclude that the redox environment modulates Mss51 functions, which are essential for regulation of COX biogenesis and aerobic energy production.
Results
Oxidative stress specifically attenuates Cox1 synthesis in vivo
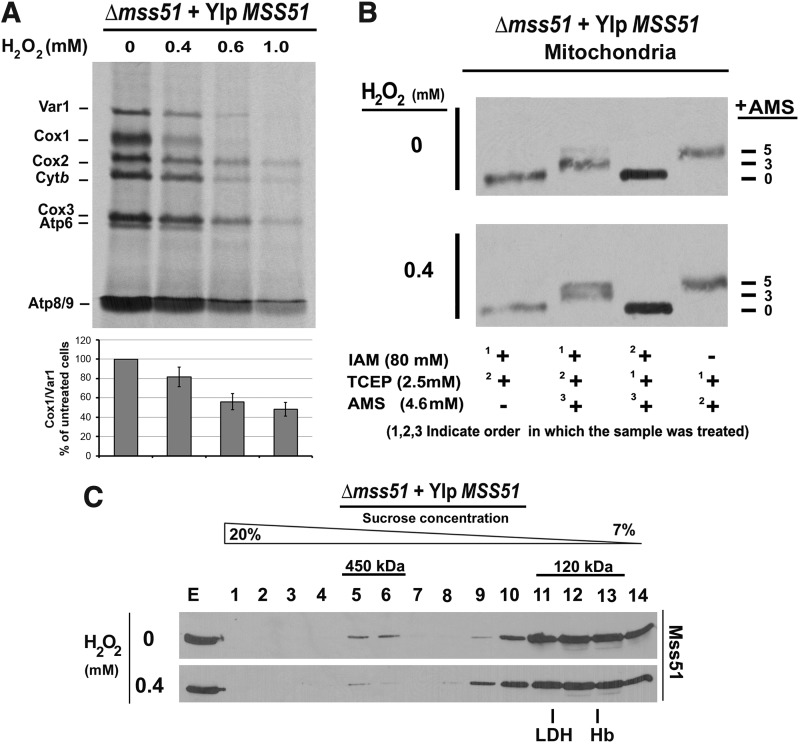
To test the effect of oxidative stress in mitochondrial protein synthesis, we assayed in vivo mitochondrial translation in two S. cerevisiae wild-type (WT) strains, namely W303 and D273, exposed to increasing concentrations of H2O2 at 30°C for 2 h during exponential growth, when the permeability constant for H2O2 is the highest (45). Mild to severe oxidative stress was achieved with H2O2 concentrations in the 0–0.8 mM range (47) at which the cells largely retained their ability to divide. Although severe oxidative stress (0.8 mM H2O2) attenuated overall mitochondrial translation in both strains, mild and medium oxidative stress (up to 0.6 mM H2O2) produced a specific predominant decrease in COX1 mRNA translation, W303 being slightly more affected (Fig. 1A).
FIG. 1.
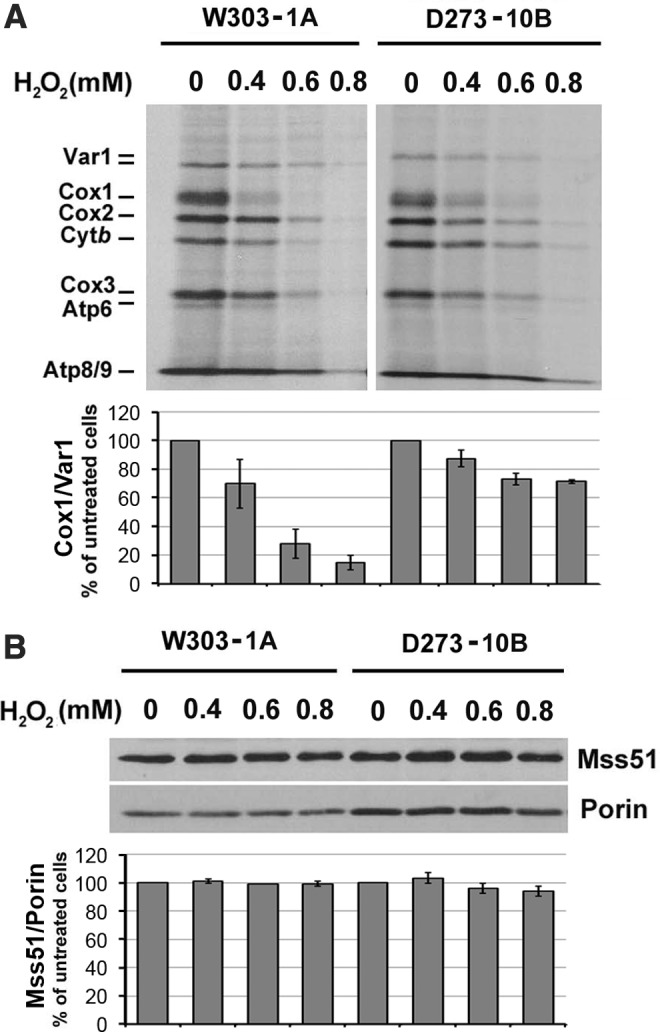
H2O2-induced oxidative stress attenuates Cox1 synthesis in vivo. (A) In vivo mitochondrial protein synthesis (pulses of 15 min) in the indicated strains. Yeast cultures were grown to early exponential phase (OD600 = 1) and subjected to treatment with different concentrations of H2O2 for 2 h at 30°C. (B) Immunoblot analysis performed using whole yeast cell lysates and an anti-Mss51 antibody. Bar graphs represent mean ± SD of percent of untreated control in three independent experiments. Cox1, cytochrome c oxidase subunit 1; OD, optical density; SD, standard deviation.
Cox1 synthesis in S. cerevisiae is under the control of the MSS51 and PET309 gene products that are also involved in maturation of the COX1 mRNAs (9, 24). We hypothesized that defective function of these translation activators could be the reason behind the attenuation of Cox1 synthesis by oxidative stress. Since we have previously shown that Mss51 plays key control roles in these processes, including regulatory heme sensing through CPX motifs (44), we decided to focus on the effect oxidative stress could have on Mss51. The H2O2-induced Cox1 synthesis defect was not, however, the result of Mss51 instability since the treatment did not change the steady-state level of the protein (Fig. 1B). In the following sections, following in vitro and in vivo approaches, we describe the effects of oxidative stress on Mss51 biochemical properties and physiological functions.
Mss51-TF can form a disulfide bond in vitro
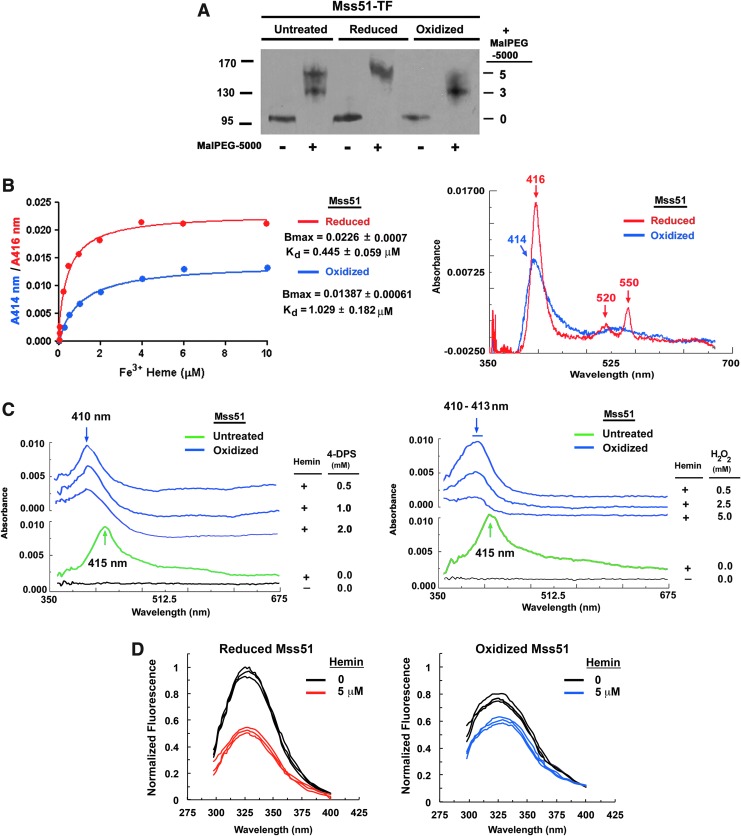
In all S. cerevisiae strains, including W303 and D273, the peripheral inner membrane protein, Mss51, contains four conserved cysteine residues. Two of these cysteines (C85 and C96) are part of CPX motifs located in the hydrophilic N-terminal region of the protein (44) that protrudes into the mitochondrial matrix (57). The other two conserved cysteines (C281 and C362) are located in the C-terminus. In some strains, such as D273, this region contains a fifth cysteine (C310), which is not relevant for protein function, as will be shown later. We have previously expressed in Escherichia coli and purified a recombinant D273 Mss51 protein fused to a trigger factor (TF) N-terminal tag for increased solubility and preceded by a 6xHis-tag for affinity chromatography purification using a cobalt resin (44). Now, we analyzed the redox state of the cysteines in recombinant Mss51 and their capacity to form a disulfide bond in vitro by thiol trapping using Mal-PEG5000 (methoxypolyethylene glycol maleimide), which binds to reduced cysteines, adding 5 kDa per residue. As shown in Figure 2A, as purified, the recombinant Mss51-TF preparation contained two protein fractions, one with a disulfide bond (three PEG) and the other fully reduced (five PEG). Mss51-TF was further reduced with DTT (dithiothreitol) or oxidized with 4-DPS (aldrithiol-4) to confirm the formation of a disulfide upon oxidation that is lost in reducing conditions (Fig. 2A).
FIG. 2.
The redox state of recombinant Mss51-TF determines hemin coordination. (A) Mss51-TF redox state in vitro by thiol trapping. Purified recombinant Mss51-TF aliquots were left untreated, reduced (10 mM dithiothreitol), or oxidized (1 mM 4-DPS) and subsequently exposed to Mal-PEG5000. Proteins were resolved in SDS-PAGE under nonreducing conditions and Mss51 was detected by immunoblotting. (B) Difference spectroscopy titration of hemin binding to reduced or oxidized Mss51-TF performed with increasing concentrations of Fe3+ hemin. The titration curves (left panel) were generated from fits to equation, 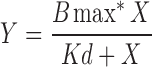 , describing a single binding site using GraphPad Prism software. The right panel displays the overlay of titration curves showing peaks, maxima, after binding 2 μM hemin. (C) Difference absorption spectra of hemin-bound native Mss51-TF treated with increasing concentrations of 4-DPS (left panel) and H2O2 (right panel), as shown. The results were obtained and analyzed as explained in (B). (D) Fluorescence emission spectra of reduced and oxidized Mss51-TF upon binding to hemin. Spectra were recorded at 25°C in 50 mM Tris-HCl pH 8.0 (excitation at 295 nm). The fluorescence maximum of reduced Mss51-TF was arbitrarily set to 1 fluorescence unit. For each condition, plots from three independent samples are presented. 4-DPS, aldrithiol-4; redox, reduction–oxidation; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
, describing a single binding site using GraphPad Prism software. The right panel displays the overlay of titration curves showing peaks, maxima, after binding 2 μM hemin. (C) Difference absorption spectra of hemin-bound native Mss51-TF treated with increasing concentrations of 4-DPS (left panel) and H2O2 (right panel), as shown. The results were obtained and analyzed as explained in (B). (D) Fluorescence emission spectra of reduced and oxidized Mss51-TF upon binding to hemin. Spectra were recorded at 25°C in 50 mM Tris-HCl pH 8.0 (excitation at 295 nm). The fluorescence maximum of reduced Mss51-TF was arbitrarily set to 1 fluorescence unit. For each condition, plots from three independent samples are presented. 4-DPS, aldrithiol-4; redox, reduction–oxidation; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
The binding capacity of recombinant Mss51 to hemin depends on the redox state of the protein
We have previously shown that recombinant Mss51-TF binds hemin-agarose beads with high specificity (44). Now, we aimed to test whether the redox state of Mss51 influences its affinity for heme. The recombinant protein, either reduced with DTT or oxidized with 4-DPS, was dialyzed and both samples were subjected to hemin-binding titrations using a double-beam spectrophotometer.
When reduced Mss51 was titrated with Fe3+ heme, the difference spectra exhibited absorbance peaks in the Soret region at 416 nm and in the α (550 nm) and β (520 nm) bands (Fig. 2B). Although this pattern is characteristic of cytochrome c coordination, in which heme is bound to the protein by one or more common two thioether bonds involving sulfhydryl groups of cysteines, we have previously shown that heme binds to Mss51 tightly, but in a noncovalent manner, since it can be extracted with acidic acetone (44). Reduced Mss51 binds Fe3+ (Fig. 2B) and Fe2+ (not shown) with similar affinities (∼0.445 μM). When oxidized Mss51 was titrated with Fe3+, the spectra changed in several ways, with the Soret peak slightly shifted to 414 nm and significantly weakened (low Bmax). We also observed the substitution of the 4550 and 4520 nm peaks by a diffuse, unresolved broad peak in the 525–540 nm region (Fig. 2B), suggesting a heme ligand switch. The oxidized protein binds Fe3+ hemin with affinity (inverse of Kd) more than two-fold lower (Kd = 1.029 μM) than the reduced protein (Kd = 0.445 μM). These results suggest that a thiol/disulfide redox switch could operate in the regulation of heme binding to Mss51 and the regulation of Mss51 functions.
Subsequently, we tested whether the interaction between Mss51 and hemin is affected by oxidation of hemin-bound Mss51 by two different oxidizing agents, 4-DPS and H2O2. For this experiment, native untreated Mss51-TF was allowed to bind hemin, which was monitored spectrophotometrically, as explained earlier. Treatment with 4-DPS of Mss51 bound to hemin resulted in a shift of the Soret peak from 415 nm (untreated protein peaks between 414 and 416 nm since it is a mixed population of reduced and oxidized protein) to 410 nm (Fig. 2C-left panel). Exposure of hemin-bound Mss51 to a wide range of H2O2 concentrations (from 0.5 to 5 mM) also resulted in a shift of the Soret peak from 415 nm in the native protein to a broad peak of 410–413 nm after oxidation, which was better defined in the mildest conditions (Fig. 2C-right panel). The shifts observed after both treatments are consistent with a change in hemin coordination, a ligand switch, upon Mss51 oxidation. These proposed ligand switches were accompanied by a gradual attenuation of the signal (Fig. 2C). These results support a model depicting Mss51-hemin interactions that are dictated by the redox state of the protein before and after Mss51 is hemylated.
To determine whether conformational changes occur in Mss51-TF upon reduction/oxidation and/or heme binding, we analyzed the intrinsic fluorescence spectra of the protein in the different conditions. Mss51 has six tryptophan residues and TF contains an additional one. Our analyses revealed that tryptophan fluorescence of Mss51-TF is quenched by 20–25% upon oxidation (Fig. 2D). Heme binding also induces fluorescence quenching by 50% for reduced Mss51-TF and by 20% for the oxidized protein (Fig. 2D). These data indicate that Mss51 undergoes conformational modifications upon changes in hemylation and/or redox state.
Mss51 can be oxidized in a cell-free in organello system upon treatment with hydrogen peroxide
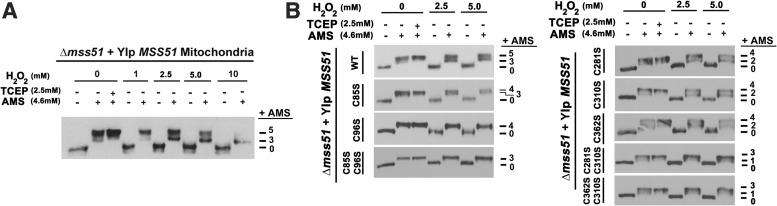
To test whether native mitochondrial Mss51 is susceptible to undergo H2O2–mediated oxidation and to identify the potential cysteines involved, we implemented acute treatments in a cell-free in organello system. For that purpose, mitochondria with an intact outer membrane isolated from a wild-type W303 strain were exposed to increasing high H2O2 concentrations (0–10 mM) during short times to minimize protein degradation. Following treatments, mitochondria were detergent-solubilized and the extracts subjected to thiol trapping using 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid (AMS), which binds to reduced cysteines, adding 0.5 kDa per residue. Despite the risk of forming non-native artifact oxidation events by exposing mitochondrial proteins to atmospheric oxygen during extraction, most of the Mss51 we detected in the nontreated sample was reduced (Fig. 3A). Following H2O2 treatment, Mss51 was shifted from its reduced form to fully oxidized. The amount of the protein becoming oxidized increased with the H2O2 concentration used in the experiment (Fig. 3A).
FIG. 3.
Exposure to H2O2 induces Mss51 oxidation in organello. (A) Mitochondria with intact outer membranes purified in the absence of reducing agents were treated with the indicated H2O2 concentrations and subsequently extracted with 100 mM Hepes pH 7.4 and 0.5% SDS with or without reducing agent, TCEP (tris-[2-carboxyethyl] phosphine, hydrochloride). Oxidation of Mss51 thiols was visualized by addition of AMS and immunoblotting. (B) Oxidation of mutant forms of Mss51-carrying serine substitutions for one or two cysteines. H2O2 treatment was followed by a thiol-trapping assay as in (A).
To identify the cysteines within Mss51 that are affected by H2O2 oxidation, we isolated mitochondria from W303Δmss51 strains carrying integrative constructs expressing D273 Mss51, either wild type or containing substitutions of each cysteine for a serine (Table 1). In organello H2O2 treatment followed by thiol trapping in mitochondrial extracts indicated that the Mss51C96S and Mss51C85SC96S mutant proteins were not oxidized upon treatment with H2O2 (Fig. 3B). These results support a model in which in the wild-type strains, both C85 and C96, are oxidized by H2O2. The fact that C96 can be oxidized in the absence of C85 (showing a shift corresponding to three AMS molecules in Mss51C85S) suggests that C96 can form a more reactive thiol that may react with H2O2, forming a –SOH group. The oxidation of C96 in the absence of C85 affects only one cysteine, which indicates that oxidized C96-SOH cannot react with any of the other cysteines within Mss51. We propose that in the wild-type strain, oxidized C96 could further oxidize C85, forming a disulfide bond, which would affect the way Mss51 coordinates heme binding. Together, these results support the involvement of C85 and C96 in mediating a redox switch regulation of Mss51 functions.
Table 1.
Genotype and Source of Strains Used Trains
| Strain | Genotype | Source |
|---|---|---|
| aW303-1A | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | (48) |
| aW303 Δhem1 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δhem1::LEU2 | (20) |
| aW303 Δmss51 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | (4) |
| aW303 Δcox14 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δcox14::TRP1 | (4) |
| aW303 Δhem1Δcox14 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δhem1::LEU2 Δcox14::TRP1 | (44) |
| aW303 Δmss51+MSS51 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-MSS51 | (44) |
| aW303 Δmss51+mss51C85S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C85S | (44) |
| aW303 Δmss51+mss51C96S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C96S | (44) |
| aW303 Δmss51+mss51C85SC96S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C85SC96S | (44) |
| aW303 Δmss51+mss51F199I | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::Yiplac128-mss51F199I | (44) |
| aW303 Δmss51+mss51C281S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C281S | This work |
| aW303 Δmss51+mss51C310S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C310S | This work |
| aW303 Δmss51+mss51C362S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C362S | This work |
| aW303 Δmss51+mss51C281SC310S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C281SC310S | This work |
| aW303 Δmss51+mss51C362SC310S | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3+LEU2::YIplac128-mss51C362SC310S | This work |
| aW303 Δcox14 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δcox14::TRP1 | (4) |
| aW303Δyta10+ytaE559Q | MATa ade2-1 his3-11,15 trp1-1 leu2,112 YCplac111 (LEU2, CEN): ADH1-YTA10E559Q ura3-52 yta10::URA3 | (1) |
| aW303Δcox14Δyta10+ytaE559Q | MATa ade2-1 his3-11,15 trp1-1 leu2,112 YCplac111 (LEU2, CEN): ADH1-YTA10E559Q ura3-52 yta10::URA3 W303Δcox14::TRP1 | This work |
| αD273-10B/A1 | MATα met6 | (50) |
Hydrogen peroxide-induced oxidative stress specifically attenuates Cox1 synthesis in vivo in an Mss51-dependent manner
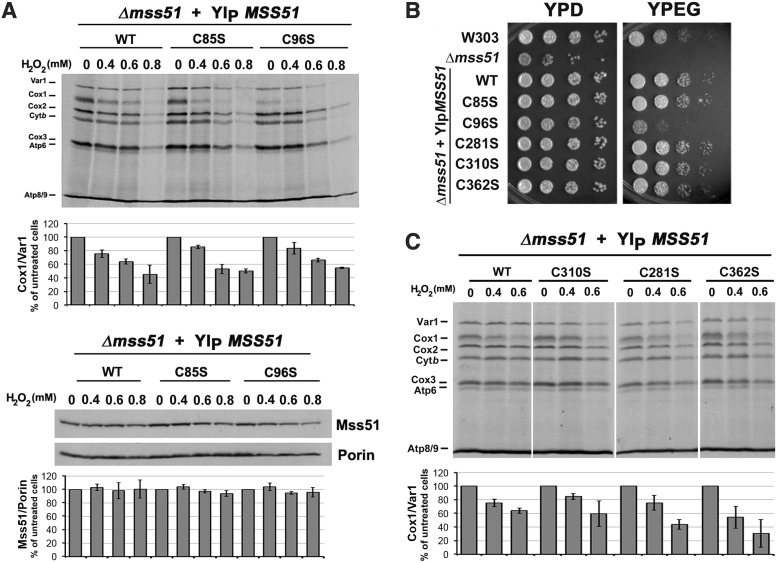
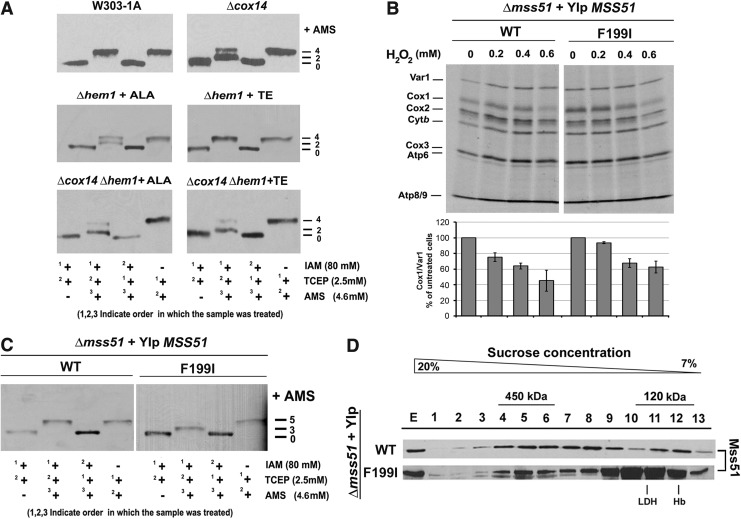
In vivo, wild-type yeast cells treated with 0.2–0.8 mM H2O2 displayed a specific predominant decrease in COX1 mRNA translation (Fig. 1). The effect of H2O2 was further investigated in W303Δmss51 yeast expressing either one copy of the D273 wild-type gene (five cysteine residues) or variants affecting each cysteine in the CPX motifs (mss51C85S and mss51C96S) or the Mss51 C-terminus (mss51C281S, mss51C310S, and mss51C362S). Both CPX mutants synthesized lower levels of Cox1 after treatment (Fig. 4A). Although mss51C96S yeast synthesizes Cox1 very poorly, we were expecting no effect by H2O2 treatment given that Mss51C96C does not undergo oxidation (Fig. 3B). As an explanation, Mss51C96C binds a small amount of heme, probably aberrantly coordinated (44) and perhaps accessible to H2O2. Regarding the mutant strains carrying mss51 substitutions in the C-terminal cysteines, they grew in media containing respiratory substrates (Fig. 4B) and synthesized Cox1 at wild-type levels (Fig. 4C). When these strains were submitted to H2O2 treatment, Cox1 synthesis was as attenuated as in the wild-type strain (Fig. 4C). These results are consistent with the in organello experiments, which showed that Mss51 can be oxidized by H2O2 in the absence of any of the C-terminus cysteines (Fig. 3B).
FIG. 4.
Exposure to H2O2 attenuates Cox1 synthesis in strains expressing mss51 cysteine mutants. (A) In vivo mitochondrial protein synthesis in the indicated strains grown to early exponential phase and subjected to treatment with different concentrations of H2O2 for 2 h at 30°C. The left bottom panel shows an immunoblot analysis performed using whole yeast cell lysates, an anti-Mss51 antibody, and an anti-Porin Ab as a loading control. (B) Growth test using serial dilutions of the indicated strains in complete media containing fermentable (glucose, YPD) or nonfermentable (ethanol-glycerol, YPEG) carbon sources. The plates were incubated at 30°C and the pictures taken after 2 days of growth. (C) In vivo mitochondrial proteins using untreated cultures of the indicated strains. Signals were quantified as in Figure 1.
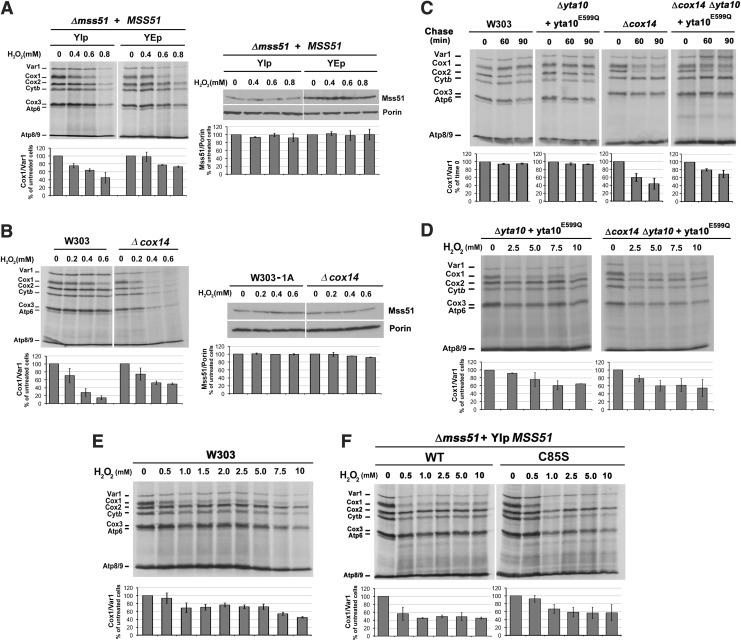
Translational downregulation of Cox1 synthesis is exerted by trapping Mss51 in a high-molecular-weight complex with newly synthesized Cox1, thereby limiting Mss51 availability to promote COX1 mRNA translation (17). The trap can be partially overcome by overexpressing Mss51 (4). In this study, we have observed that the specific Cox1 synthesis decrease induced by H2O2 treatment could be moderately bypassed by overexpression of Mss51 (Fig. 5A). The Mss51 trap is also bypassed in some COX assembly mutants, such as Δcox14 and Δcoa3, in which the trapping complex is destabilized, rendering Mss51 available to promote Cox1 synthesis (4, 11). In these mutants, however, the newly synthesized protein is rapidly degraded (4).
FIG. 5.
Interventions that bypass COX1 mRNA translational downregulation do not bypass the H2O2-induced decrease in Cox1 synthesis. (A–F) In vivo mitochondrial protein synthesis performed as in Figure 4A using the indicated strains. (A, B) The right panel shows an immunoblot analysis of whole yeast cell lysates for Mss51 and Porin as a loading control. (C) The pulse was followed by a chase in the presence of cold methionine and puromycin to inhibit protein synthesis. Signals were quantified as in Figure 1.
We aimed to further test whether some of the oxidation effect on COX1 mRNA translation could be attributed to either degradation of the newly synthesized protein or downregulation of its synthesis by Mss51 trapping. Turnover of mitochondrion-synthesized COX subunits that do not enter the assembly pathway is a function of different proteases, including the m-AAA protease complex Yta10/Yta12 (1), which can be inactivated by an E559Q mutation in the active site of Yta10 (1). The role of Yta10/12 in Cox1 turnover following H2O2 treatment was studied in wild-type and Δcox14 strains harboring the yta10E559Q allele. Treatment of Δcox14 cells with H2O2 resulted in diminished capacity to translate COX1 mRNA, even weaker than in the wild-type strain (Fig. 5B). This phenotype is not contributed by rapid degradation of Cox1 in the absence of Cox14 because substitution of wild-type Yta10 by Yta10E559Q partially restored the detected levels of newly synthesized Cox1 in untreated (Fig. 5C), but not in H2O2-treated, Δcox14 cells (Fig. 5D).
We also tested the effect of H2O2 in Cox1 synthesis using higher concentrations of the oxidant during a shorter time (40 min), which effectively oxidized Mss51 in organello (Fig. 3A, B). Exposure of wild-type W303 and Δmss51 cells expressing wild-type MSS51 or mss51C85S to H2O2 in these conditions produced a specific attenuation of newly synthesized Cox1 (Fig. 5E, F). The maximum effect on Cox1 synthesis was reached with 1 mM H2O2 (Fig. 5E, F), at which Mss51 is partially oxidized (Fig. 3A, B). These results suggest that not all of the Mss51 pool in the cell needs to be oxidized for Cox1 synthesis to be attenuated. Together, the results presented in this section further point to Mss51 as a redox sensor that contributes to control Cox1 synthesis.
Mitochondrial Mss51 can be oxidized in yeast cells upon treatment with hydrogen peroxide
To determine if the H2O2-mediated effect in Cox1 synthesis is contributed by Mss51 oxidation, we designed a strategy to test the redox state of Mss51 in vivo after H2O2 treatment. Wild-type W303 yeast treated with increasing H2O2 concentrations up to 0.8 mM was grown to early exponential phase and used to confirm the in vivo Cox1 synthesis defect (Fig. 6A). To assess the native redox state of Mss51 in its mitochondrial environment, we performed a reverse redox trap assay in mitochondria with intact outer membranes purified from cultures supplemented or not with 0.4 mM H2O2. Mitochondria were first treated under isotonic conditions with an excess of membrane-permeable iodoacetamide (IAM), a compound that covalently binds free thiols. We then followed with denaturation and reduction using SDS (sodium dodecyl sulfate) and TCEP (tris-[2-carboxyethyl] phosphine, hydrochloride), respectively. We subsequently identified the cysteines that had not been bound by IAM previously by incubating the samples with AMS, as explained earlier. We observed the accumulation of Mss51 in two populations, one that corresponds to two cysteines inaccessible by IAM and another with the size of five inaccessible cysteines (Fig. 6B).
FIG. 6.
Mss51 can be oxidized in vivo by H2O2. (A) In vivo mitochondrial protein synthesis performed as in Figure 4A using the indicated strains. (B) Reverse thiol-trapping assay to test the native Mss51 redox state in mitochondria isolated from the cultures used in (A). Numbers 1, 2, and 3 indicate the order in which each sample was treated with the thiol-binding compounds, IAM (iodoacetamide) and AMS (4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid), and the reducing agent, TCEP. Full reduction of Mss51 with TCEP (1) was tested by reducing the protein first, and then adding excess IAM (2), followed by AMS addition (3). Maximum shift of reduced Mss51 was determined by reducing the sample during extraction with TCEP (1), quickly followed by treatment with AMS (2). (C) Sucrose gradient sedimentation analysis of Mss51 high-molecular-mass complexes extracted in native conditions from treated and untreated mitochondria used in (B). The gradients were calibrated with the standards, hemoglobin (Hb, ∼67 kDa) and LDH (∼130 kDa). Fractions were resolved using SDS-PAGE and analyzed by immunoblotting for Mss51. Hb, hemoglobin; LDH, lactic dehydrogenase.
The IAM inaccessibility of two or five cysteines in native Mss51 could be due to interactions of Mss51 with other proteins in complexes, the existence of a disulfide bond, the interaction of Mss51 with heme, or a combination of all these factors. We have previously shown that Mss51 exists predominantly in two high-molecular-weight complexes. In wild-type cells grown to late exponential phase in complete medium containing galactose, the most abundant is a 450 kDa complex, in which Mss51 interacts with newly synthesized Cox1 and other COX assembly factors such as Cox14, Coa3, and mitochondrial Hsp70 Ssc1 (11, 12). Mss51 also forms a 120 kDa heterodimer with Ssc1, which is the source of Mss51 that acts in COX1 mRNA processing and translation (12). Under the culture conditions used here (minimum medium containing galactose, early exponential phase), most of the Mss51 was detected in the 120 kDa complex (Fig. 6C). The H2O2 treatment did not affect Mss51 accumulation in the 120 kDa complex, but the ratio of the 5-Cysteine/2-Cysteine AMS-bound species markedly increased (Fig. 6B). These data further support the concept that oxidation results in the formation of a disulfide bond in Mss51.
The COX1 translational defect upon oxidative stress cannot be solely attributed to loss of heme binding to Mss51
To assess whether heme binding to Mss51 could influence the results of the thiol-trapping experiments, we analyzed the redox state of Mss51 in two strains with disrupted heme biosynthesis, namely W303Δhem1 and W303Δhem1Δcox14. We isolated mitochondria from these strains grown in the presence of either 5-aminolevulinic acid (ALA) or TE (Tween 20-ergosterol). ALA is a heme biosynthesis intermediate that bypasses the need for the HEM1 gene product (5-aminolevulinate synthase). TEs are sources of essential sterol and unsaturated fatty acids whose biosynthesis requires heme and hence allow the mutant cells to grow in heme-deprived media. We have reported that in the Δhem1 strain, most of the Mss51 accumulates in the 450 kDa complex, while in the Δhem1Δcox14 double mutant, most of the Mss51 interacts with Ssc1 in the 120 kDa heterodimer (44). In the wild-type W303 strain grown to late exponential phase in complete media, most of the Mss51 is in the 450 kDa complex, while in the absence of Cox14, this complex is destabilized and most of the Mss51 accumulates in the 120 kDa complex (12).
Reverse thiol-trapping assays indicate that when Mss51 is in the 120 kDa complex, as in the Δcox14 strain, it predominantly has two AMS-bound cysteines (Fig. 7A). Importantly, this pattern remains unchanged when heme is depleted, as in the case of the Δhem1Δcox14 mutant (Fig. 7A). In all, these results support that the two AMS-bound cysteines were not engaged, in vivo, in heme coordination or interactions with other proteins in high-molecular-weight complexes, but rather were oxidized by H2O2.
FIG. 7.
The oxidative stress-induced COX1 mRNA translational defect cannot be solely attributed to loss of heme binding to Mss51. (A) Redox state of Mss51 in mitochondria purified from the indicated strains and analyzed by thiol trapping as in Figure 6B. The Δhem1 mutation was bypassed by growing the cells in media containing 5-aminolevulinic acid (ALA) or expressed by growing cells in media lacking ALA, but supplemented with Tween 20 and ergosterol (TE) for 16 h before mitochondrial purification. (B) In vivo mitochondrial protein synthesis following 35S-methionine incorporation as in Figure 4, using Δmss51 cells expressing from an integrative plasmid either wild-type MSS51 or the point mutant mss51F199I. Signals were quantified as in Figure 1. (C) Native redox state of Mss51 in mitochondria isolated from the indicated strains analyzed by reverse thiol trapping as in Figure 6B. (D) Native distribution of Mss51 in sucrose gradients analyzed as in Figure 6C.
To further test the role of heme binding to Mss51 in the sensitivity of COX1 mRNA translation to H2O2, we used yeast strains expressing either a wild-type D273 MSS51 or a heme-independent variant of MSS51 (mss51F199I) (44). Treatment with H2O2 affected both strains similarly (Fig. 7B). Thiol-trapping experiments showed that Mss51F199I redox state is similar to the redox state of wild-type Mss51 in Δcox14 yeast (Fig. 7C), a strain in which Mss51 also accumulates predominantly in the 120 kDa complex [(12) and Fig. 6D]. Together, the observation that heme coordination by Mss51 can be affected by the redox state of the protein and the fact that bypassing the need for heme binding in the mss51F199I mutant is not sufficient to protect the cells from the effect of H2O2 on Cox1 synthesis suggest that oxidation could additionally affect a COX1 mRNA metabolism step upstream its translation.
Defective Mss51-dependent COX1 mRNA splicing contributes to oxidative stress-induced Cox1 synthesis deficiency
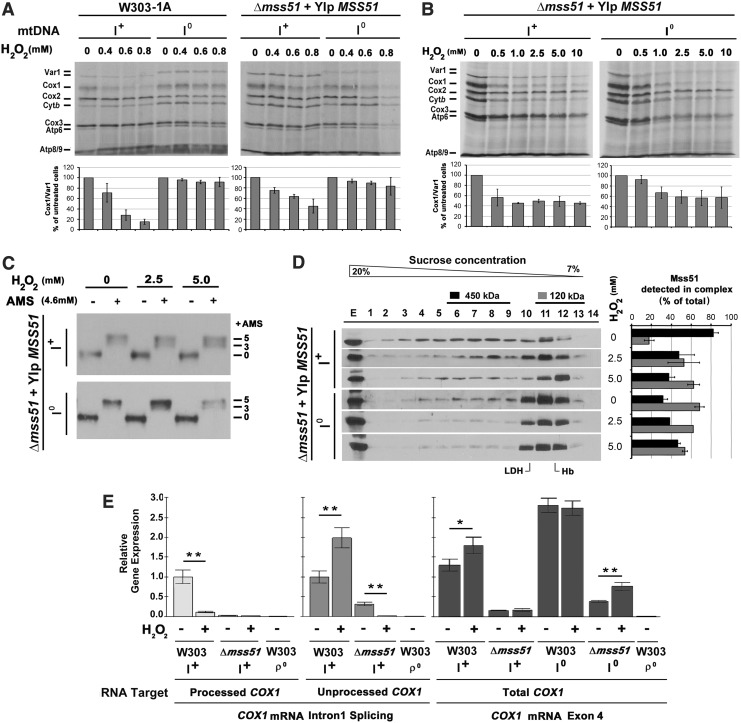
In yeast, the COX1 gene contains introns, which need to be processed to yield the mature mRNA used in translation. Notably, Mss51 was originally shown to be involved in COX1 pre-mRNA splicing (42). For this reason, we tested the effect of H2O2-induced oxidative stress in strains carrying intronless (I0) mtDNA. As shown in Figure 8A and B, when COX1 mRNA splicing was not required, Cox1 synthesis upon H2O2 treatment was less attenuated than in the intron-containing (I+) strains and was fully sensitive only at higher H2O2 concentrations (above 0.5 mM). The milder effect of oxidative stress in the I0 strain is not due to differences in the steady-state level of Mss51, which was as in the intron-containing wild-type strain (not shown).
FIG. 8.
H2O2-induced oxidative stress negatively affects Mss51-dependent processing of intron-containing COX1 mRNA. (A, B) Yeast cultures of intron-containing (I+) and intronless (I0) strains were grown to early exponential phase and treated with the indicated concentrations of H2O2 for 2 h at 30°C. Treatment was followed by in vivo mitochondrial protein synthesis assay as in Figure 3 and signals quantified as in Figure 1. (C) Redox state of Mss51 in organello after treatment with H2O2. Intact mitochondria purified from strains carrying COX1 with (I+) or without introns (I0) were treated under isotonic conditions with the indicated H2O2 concentrations, and then subjected to a thiol-trapping assay as in Figure 3. (D) Native distribution in sucrose gradients of Mss51 from the same strains than in (C) analyzed as in Figure 6C. (E) Steady-state levels of total, processed, and unprocessed COX1 mRNA in the indicated strains measured by quantitative PCR. Levels of total COX1 mRNA were measured using primers within COX1 exon 4. Splicing of COX1 intron 1 was monitored by amplifying COX1 mRNA with primers designed either within intron 1 or at the end of exon 1 and the beginning of exon 2. The rho zero (ρ0) strain, devoid of mitochondrial DNA, was used as a negative control. Expression levels of COX1 mRNA were normalized by actin levels. Error bars represent the mean ± SD of three independent repetitions. *p < 0.05, **p < 0.01.
We assessed the native mass of Mss51 after in organello treatment with high H2O2 concentrations (2.5 and 5 mM) that prevent Cox1 synthesis in strains carrying I0 COX1. For this purpose, we used Δmss51 yeast containing I+ or I0 mtDNA and carrying an integrative plasmid expressing wild-type MSS51 to purify intact mitochondria from mid-exponential phase cultures. In these culture conditions, Mss51 gets distributed in both the 450 and 120 kDa complexes (12). Care was taken to avoid the use of any reducing agent in the purification buffers. After exposing mitochondria carrying I+ or I0 mtDNA to different H2O2 concentrations for 40 min at room temperature (as done for the in organello oxidation assays shown in Fig. 2B), the sample was divided into two aliquots. One aliquot was used for a thiol trap assay, which showed that in both strains, Mss51 was oxidized (Fig. 8C). The second aliquot was extracted with 1% digitonin, as previously established, to analyze Mss51 native size by sucrose gradient sedimentation (12, 44) (Fig. 8D). In the untreated samples, the ratio of Mss51 in the 450 kDa versus the 120 kDa complex was lower in the strain carrying I0 mtDNA, and the exposure to H2O2 lowers the ratio still further. In the intron-containing strain, the H2O2 effect was marked, showing a clear shift in the accumulation of Mss51 from the 450 to the 120 kDa complex. The depletion of the 450 kDa complex in the treated samples is consistent with a decrease of newly synthesized Cox1, which is indispensable to form this complex.
To determine if COX1 mRNA processing could be affected by H2O2-induced oxidative stress, we measured the steady-state levels of total, processed, and unprocessed COX1 message in treated (0.6 mM H2O2 for 2 h, as in Fig. 8A) and untreated wild-type and Δmss51 strains, with or without introns. For this purpose, we used quantitative polymerase chain reaction (q-PCR) with primers specifically designed to amplify either unprocessed (within the intron 1 sequence) or spliced first intron (overlapping exon 1 and exon 2). As a negative control, we included samples from a wild-type strain devoid of mitochondrial DNA (W303 ρ0). The treated intron-containing wild-type sample showed an accumulation of unprocessed COX1 mRNA and a very dramatic decrease of mature message (Fig. 8E). In the absence of Mss51, the amount of processed COX1 is very low with or without oxidative stress, suggesting that Mss51 plays a role in the maturation of at least the first intron within the COX1 mRNA. Accumulation of unprocessed COX1 mRNA in untreated intron-containing Δmss51 yeast was markedly lower than in the untreated wild-type strain, which suggests the message is unstable without Mss51. Upon treatment, the levels of unprocessed COX1 mRNA in the mss51 mutant are drastically decreased, indicating that Mss51 contributes to the stabilization of the unprocessed message during oxidative stress. The total levels of COX1 mRNA in the intron-containing wild-type strain, measured using primers within the COX1 exon 4 sequence, are slightly but significantly increased after H2O2 treatment, while in the wild-type intronless strain, the total levels of COX1 message were unchanged. Importantly, the total levels of COX1 mRNA in the absence of Mss51 are increased after treatment if the transcript does not need to be processed (Fig. 8E), further supporting that Mss51 is necessary for the stability of the unprocessed transcript during oxidative stress.
Altogether, the results presented in this section demonstrate that H2O2-induced oxidative stress negatively impacts not only Mss51-mediated activation of COX1 mRNA translation but also Mss51-dependent COX1 mRNA processing, both of which contribute to the net attenuation of Cox1 synthesis.
Discussion
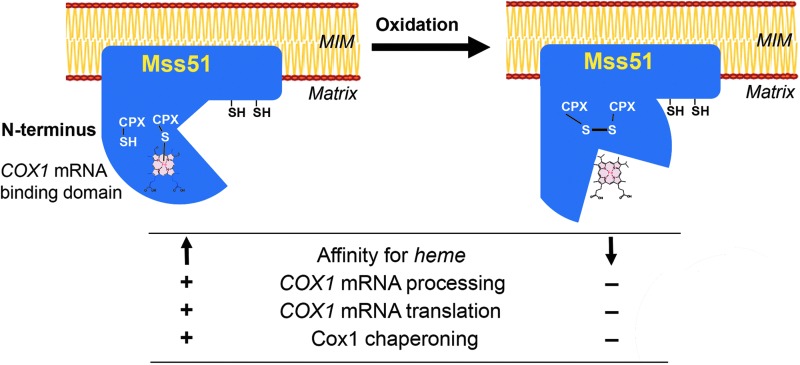
ROS have been identified as mediators of a plethora of critical, physiological cell signaling responses to stress and environmental cues to regulate cell differentiation, propagation, or metabolic adjustments, therefore redox regulation is an area of intense investigation (5, 23, 26, 35, 51, 52, 56). One mechanism by which ROS exert their effects is through the reversible regulation of cysteine thiol-containing enzymes (6, 55). Heme, whose synthesis is stimulated by oxygen, is another important regulatory molecule involved in a variety of biological events related to iron and oxidative metabolism in prokaryotes and eukaryotes by modulating the function of hemoproteins. Heme-regulated proteins contain cysteine-proline-X heme-responsive motifs that are involved in transient heme binding to proteins. We have recently described the presence of two CPX motifs in yeast Mss51, a mitochondrial COX1 mRNA-specific processing factor and translational activator, which also acts as a Cox1 chaperone, forming complexes with COX-specific assembly factors, and the mitochondrial Hsp70 chaperone Ssc1. Heme sensing by Mss51 through its CPX motifs optimizes its functions, thus coordinating synthesis of Cox1 with its assembly into COX, the terminal oxidase of the MRC. In this study, on the basis of six central observations described below, we have concluded that one of the mechanisms by which ROS controls COX biogenesis involves a thiol/disulfide redox switch that disturbs Mss51 functions upon CPX motif cysteine oxidation, thereby limiting COX1 expression (Fig. 9).
FIG. 9.
Effect of H2O2-induced oxidative stress in Mss51 properties and functions. According to our model, oxidative stress induces the formation of a disulfide bond in Mss51, involving the two cysteines in the CPX heme-regulatory motifs, and a change in the conformation of the protein. At least one of the CPX cysteines coordinates heme B in a noncovalent manner. Mss51 oxidation results in heme ligand switch, which lowers heme-binding affinity and induces its release. Oxidation also provokes a change in Mss51 conformation. These changes markedly inhibit Mss51 functions in COX1 mRNA processing and translation and Cox1 assembly chaperoning. In this way, Mss51 acts as a redox sensor to discontinue Cox1 synthesis and COX assembly during potentially harmful oxidative stress conditions.
Four observations identify the Mss51 cysteine residues involved in the regulatory mechanism and offer a mechanistic view. First, reduced recombinant Mss51 binds heme 2.5-fold more tightly than the oxidized protein, which forms a disulfide bond. Second, cellular or mitochondrial oxidation by exogenous H2O2 introduces a disulfide bond in Mss51 involving cysteines, C85 and C96, in the CPX motifs and induces the release of Mss51-bound heme. Third, H2O2-mediated oxidation of hemin-bound recombinant Mss51 results in a change in hemin coordination and subsequent hemin release. Fourth, tryptophan fluorescence quenching experiments indicate that Mss51 undergoes conformational changes upon changes in hemylation and/or redox state.
These four observations suggest the existence of a thiol/disulfide redox regulation of heme binding. Thiol/disulfide redox switch is one mechanism by which oxidative stress is linked to diverse physiological processes. For example, in the case of Rev-erbβ, a heme-binding nuclear hormone receptor that represses a broad spectrum of target genes involved in regulating metabolism and the circadian cycle, a thiol/disulfide redox switch involving the cysteines in two CPX motifs controls heme binding and dissociation (14). In Rev-erbβ, heme iron is hexacoordinated by a CPX motif Cys and His residue. The difference absorption spectra of heme-reconstituted recombinant Rev-erbβ display a profile with Soret peak at 416 nm and α and β peaks at 567 and 537 nm, respectively. Upon oxidation of the sulfhydryl to a disulfide, heme coordination switches and the binding affinity decrease (14). Heme coordination by Mss51 somehow resembles that of Rev-erbβ. The difference spectra revealed a Soret band at 416 nm and in the α and β bands at 550 and 520 nm, respectively. We previously reported that heme B can be extracted from mitochondrial Mss51 using acidic acetone, which indicates noncovalent heme binding to the polypeptide [(44) and Fig. 9]. However, heme binding to recombinant Mss51 was resistant to denaturing conditions such as 2% SDS and 6 M urea (44), which points to a tight interaction between Mss51 and heme as previously reported for other proteins (19). In heme oxygenase-2, the CPX motifs do not bind heme, but instead form a thiol/disulfide redox switch that regulates heme binding to the catalytic domain of the enzyme [reviewed in Refs. (26, 35)].
The fifth and sixth observations explain the physiological consequences of Mss51 oxidation and provide additional mechanistic insight. Fifth, cellular oxidation by exogenous H2O2 markedly attenuates COX1 mRNA processing and Cox1 synthesis in an Mss51-dependent manner. Further exploration of the Mss51 heme and redox dynamics in vivo and in vitro revealed that despite the dissociation of heme from Mss51 during oxidative stress, this event is not the only factor contributing to decreased Cox1 synthesis upon H2O2 treatment. A similar effect was observed in mss51 point mutants that do not bind heme in vivo (44), but can synthesize Cox1 at wild-type levels, such as mss51F199I. This result indicates that the heme-binding independence of Mss51F199I is not sufficient to overcome the negative outcome of Mss51 oxidation in Cox1 synthesis. Mss51 was originally described as a factor required for COX1 mRNA splicing (42). Our data suggest that Mss51 recruitment to the COX1 mRNA 5′ untranslated region (UTR) may facilitate the processing of at least the first intron, a process limited by Mss51 oxidation that does not necessarily require heme binding to Mss51.
Sixth, Mss51 function in translational initiation is also impaired by heme loss upon oxidation. We propose that the negative regulation of COX1 expression triggered by Mss51 oxidation is mediated not only by the loss of heme but also by a conformational change in Mss51 promoted by the oxidation of two cysteines. In organello thiol-trapping experiments showed that the Mss51 cysteines affected by the oxidation stress caused by H2O2 are C85 and C96. We have previously shown that these cysteines present in two CPX motifs within Mss51 N-Terminus are required for Mss51 function in Cox1 synthesis, and in their absence, the amount of heme bound to Mss51 in vivo is barely detectable (44). We propose that an oxidation-induced conformational change in Mss51 affects its ability to promote COX1 mRNA maturation and later steps in translation initiation due to the release of heme. The combination of these two regulatory checkpoints adds significant stringency to the COX1 biogenesis and COX assembly pathway. Additionally, we cannot discard the possibility that oxidative stress could induce changes in additional proteins that could also contribute to the H2O2-mediated COX1 mRNA translational downregulation described here.
Mss51 is part of several high-molecular-mass complexes (4, 12, 25, 29, 34, 44). A point of interest concerns how oxidation affects Mss51 in the several complexes. Because oxidative stress attenuates Cox1 synthesis, Mss51 accumulates mostly in the 120 kDa complex, which is the source of Mss51 for translation activation (12) and presumably also for COX1 mRNA processing. Although Mss51 oxidation could occur also within the multichaperone 450 kDa Cox1 stabilization complex that includes proteins such as Cox14 and Coa3, it is plausible that Mss51 could be more exposed to changes in the redox environment when bound only to the mitochondrial Hsp70 chaperone Ssc1 in the 120 kDa complex.
Heme-mediated regulation often involves the assistance of general chaperones, presumably to assist in changes of hemoprotein conformation upon heme binding and release. For example, in mammals, CPX motifs are present in HRI (hepatic heme-regulated inhibitor-eukaryotic initiation factor 2 alpha (eIF2 alpha) kinase), which associates with Hsp70 and Hsp90 chaperones to coordinate protein synthesis and heme availability in reticulocytes. In the yeast transcriptional activator Hap1 (heme activator protein 1), which mediates oxygen sensing and heme signaling (58), CPX motifs arbitrate heme-dependent formation of complexes with Hsp70 and Hsp90 chaperones that allow Hap1 to bind DNA, thereby activating transcription (18). Future studies are warranted to disclose the role of Ssc1 in heme- and redox-mediated regulation of Mss51 functions.
It remains uncertain whether the regulatory mechanism we have described here for yeast could be conserved in higher eukaryotes. Data present in the literature indicate that in mammalian cells, COX1 synthesis is not specifically blocked by oxidative stress. Both ROS and RNS (reactive nitrogen species) cause damage to mitochondrial components, which results in impaired overall mitochondrial protein synthesis in vascular cells (2). Specifically, treatment with 0.2 mM H2O2 and 0.5 mM peroxynitrite was shown to cause 30% and 70% decrease of overall mitochondrial translation capacity, respectively (2). The regulation of COX1 synthesis and stability differs from yeast to mammals, in part, due to the differences in COX1 mRNA. In mammals, COX1 does not have introns or a long 5′ UTR as it does in yeast. A functional homologue of Mss51 has not been described in mammals, and the single mammalian-specific COX1 mRNA translational activator described so far, TACO1, does not form complexes with newly synthesized COX1 (27, 53). However, COX14 and COA3 are conserved proteins, which interact with and stabilize newly synthesized COX1 in human cells as they do in yeast (7, 27, 54). This has suggested the hypothesis that these proteins could also participate in coupling COX1 synthesis and/or stability with COX assembly. Independently, TACO1, COX14, and COA3 do not contain heme regulatory CPX motifs, and although TACO1 has eight cysteines, there are no data available to allow predictions on whether the protein can form disulfides upon oxidation.
Global downregulation of protein synthesis in response to oxidative stress, as mentioned for mammalian mitochondria, is well described for cytoplasmic translation, where it mostly targets not only translation initiation but also elongation (13). The attenuation of protein synthesis has been proposed to prevent continued gene expression during potentially error-prone conditions as well as allow for the turnover of existing mRNAs and proteins at the same time as gene expression is reprogrammed to deal with the stress (13). In this study, we show that in yeast, high H2O2 concentrations induce a global mitochondrial translation attenuation effect, but milder concentrations specifically affect COX1 mRNA processing and translation, at least in part, in an Mss51-dependent manner. Hence, our results are relevant to the general fields of translation regulation and adaptation to oxidative stress.
In conclusion, the processes identified in this article serve to sense the local submitochondrial redox environment and modulate Mss51 functions accordingly. Physiologically, this is a sophisticated mechanism by which synthesis of the potentially hazardous Cox1 subunit, known to generate pro-oxidant heme A:Cox1 intermediates (16), could be either coordinated with its assembly into COX or attenuated under oxidative stress conditions. Our data indicate that the thiol/disulfide redox switch in Mss51 is one mechanism by which oxidative stress is linked to COX deficiency.
Materials and Methods
Yeast strains and media
All S. cerevisiae strains used are listed in Table 1. Yeast strains were grown in the following standard culture media: YPD (2% glucose, 1% yeast extract, 2% peptone), YPGal (2% galactose, 1% yeast extract, 2% peptone), YPEG (2% ethanol, 3% glycerol, 1% yeast extract, 2% peptone), WO-EG (2% ethanol, 3% glycerol, 0.67% yeast nitrogen base without amino acids), and WO-Gal (2% galactose, 0.67% yeast nitrogen base without amino acids). Strains grown in liquid and solid media were incubated at 30°C unless otherwise indicated.
In vivo mitochondrial protein synthesis
All cultures were grown to early exponential phase in liquid minimum media containing 2% galactose (WO-Gal). Mitochondrial gene products were labeled with [35S]-methionine (7 mCi/mmol, Perkin Elmer) in whole cells at 30°C in the presence of 0.2 mg/ml cycloheximide to inhibit cytoplasmic protein synthesis as reported (3). In pulse/chase experiments, following 15 min pulses, labeling was terminated by addition of excess cold methionine (80 μM) and puromycin (12 μg/ml) and mitochondrial products chased by up to 90 min. Equivalent amounts of total cellular proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to an X-ray film.
Purification of recombinant Mss51-TF
Wild-type or cysteine mutant Mss51 fused to a thrombin-cleavable trigger factor tag (Mss51-TF) was each expressed as an His6-tagged protein from the pColdTF vector (Genescript) in the BL21 Star protein expression strain of E. coli (Invitrogen), extracted, and purified by metal affinity using a Cobalt Sepharose HiTrap HP affinity column as described (44). Briefly, transformed bacteria were grown in M9 minimal media at 37°C to OD600 = 0.8 and gene expression was induced using 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) for 16 h at 25°C. Cells were lysed and homogenized using a French press. Proteins were purified from lysate supernatants by immobilized metal affinity chromatography using Cobalt Sepharose HiTrap HP. The His6-TF-Mss51 proteins were eluted with 500 mM imidazole at 1 ml/min for 20 min. The purity of the purified protein was tested using SDS-PAGE and subsequent gel staining with brilliant Coomassie blue. High-purity Mss51-TF was dialyzed against 50 mM Tris pH 8 for storage and analyses. Dialysis was performed using Spectra/Por membrane tubing (Spectrum Laboratories, California) in 1 L of 50 mM Tris pH 8 at 4°C, with stirring for 8 h. The proteins were stored at −80°C. Approximately 5 mg of purified tagged protein per liter of bacterial culture was obtained.
Reduction and oxidation of recombinant purified Mss51-TF
Purified recombinant Mss51-TF protein (60 ng/μl) was reduced using 10 mM DTT (Sigma-Aldrich) in 50 mM Tris pH 8 at room temperature for 1 h. Purified Mss51-TF (60 ng/μl) was oxidized with 1 mM of Aldrithiol-4 (4-DPS, from Sigma-Aldrich) in 50 mM Tris pH 8 at room temperature for 1 h. Reduced, oxidized, and untreated proteins were dialyzed against 1 L of 50 mM Tris pH 8 for 4 h at 4°C using Spectra/Por membrane tubing (Spectrum Laboratories).
Thiol-trapping analyses of recombinant Mss51-TF and native Mss51
To analyze the redox state of cysteines in recombinant Mss51-TF, untreated, reduced, or oxidized, as explained earlier, 1 μg of purified protein was mixed with 0.5 mM Mal-PEG5000 (Sigma-Aldrich) in 50 mM Tris pH 8 and incubated at room temperature for 30 min. One PEG5000 molecule covalently binds to one reduced cysteine and results in a gel shift increase of 5 kDa. Proteins were resolved in SDS-PAGE under nonreducing conditions, followed by immunoblotting for Mss51.
A reverse thiol trap assay was used to analyze Mss51 redox state in organello. Mitochondria purified from nontreated and 0.4 mM H2O2-treated cultures were first incubated under isotonic conditions with excess of membrane-permeable IAM, a compound that covalently binds free thiols. Mitochondria were then solubilized in the presence of SDS and the reducing agent, TCEP. The cysteines that had not been blocked by IAM were subsequently identified by incubation with AMS, a compound that binds free thiols, adding 0.5 kDa for every molecule bound. Full reduction of Mss51 was tested by reducing the protein first with TCEP, and then adding excess IAM, followed by AMS addition. Maximum shift of reduced Mss51 was determined by reducing the sample with TCEP during extraction, quickly followed by treatment with AMS.
Spectrophotometric properties of hemin-bound recombinant Mss51-TF
Heme titrations were performed by difference spectroscopy at room temperature and aerobically using a double-beam UV-2401PC Shimadzu spectrophotometer as described (14). We used 50 nM of purified wild-type Mss51-TF and hemin in 50 mM Tris buffer pH 8. A baseline was selected by scanning samples from 350 to 675 nm: 50 mM Tris buffer pH 8 in the reference cuvette and 50 mM Tris buffer pH 8 with 50 nM of purified Mss51-TF in the sample cuvette. After the baseline was set, equal concentrations of hemin were added to reference and sample cuvettes. For this assay, we used a fixed low concentration of Mss51 (50 nM) to prevent protein aggregation and increasing concentrations of hemin (250–10.0 μM). We first scanned the Mss51 sample located in the sample cuvette compared with the reference cuvette containing buffer only (50 mM Tris pH 8) in a range wavelength of 350–675 nm. This reading was set as the baseline. Subsequently, increasing concentrations of hemin were added to both the sample and the reference cuvettes, so our readings only reflect the binding of Mss51 with hemin and not the absorbance of the free ligand. To determine the binding parameters, the data obtained from the hemin titrations were plotted and fit to an equation describing a single binding site (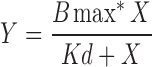 ) using the GraphPad Prism software.
) using the GraphPad Prism software.
Intrinsic tryptophan fluorescence measurements
Tryptophan fluorescence measurements were performed at 25°C on an M5 fluorescence spectrophotometer (Molecular Devices) in stirred 1.0 cm (excitation path) ×0.5 cm (emission path) quartz cuvettes. The excitation wavelength was 295 nm (5 nm bandwidth), and the emission at 300–400 nm was recorded with a bandwidth of 10 nm at a scan speed of 60 nm/min and a step size of 0.2 nm. Samples contained 1 μM Mss51-TF in 1 ml of 50 mM This-HCl pH 8.0. Reduction and oxidation of Mss51-TF were accomplished as described earlier and the samples were dialyzed against the same buffer. The effect of heme binding was tested by addition of 5 μM hemin. All fluorescence spectra were corrected for the buffer (with or without hemin) and the wavelength dependence of the emission monochromator and the photomultiplier.
H2O2 treatment of hemin-bound recombinant Mss51-TF
Recombinant wild-type native Mss51-TF (50 nM in 50 mM Tris pH 8) was allowed to bind 5 mM of hemin. Hemin binding was followed by treatments with increasing concentrations of H2O2 (0–5 mM). Difference absorption spectra were used to monitor hemin binding to Mss51-TF before and immediately after every addition of H2O2.
4-DPS treatment of hemin-bound recombinant Mss51-TF
Recombinant wild-type native Mss51-TF (50 nM in 50 mM Tris pH 8) was allowed to bind 5 mM of hemin. Hemin binding was followed by treatments with increasing concentrations of 4-DPS (0–2 mM). Difference absorption spectra were used to monitor hemin binding to Mss51-TF before and immediately after every addition of 4-DPS.
Purification of mitochondria
Translationally competent mitochondria with an intact outer membrane were purified from yeast cultures grown, unless indicated, in liquid YPGal media (2% galactose, 1% yeast extract, 2% peptone) to mid-exponential phase as reported (12). Briefly, cells were collected by centrifugation at 5K RPM in a Beckman xxx rotor. Cells were then washed once with 100 ml 1.2 M sorbitol/10 g. Washed cells were digested with 0.6 mg/ml of zymolyase (24,600 U/g from Nacalai Tesque) in 1.2 M sorbitol and 20 mM potassium phosphate pH 7.4 for 30 min at 30°C. Spheroplasts were washed once with 1.2 M cold sorbitol. Spheroplasts were pelleted (5K RPM) and resuspended in 10 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 0.6 mM sorbitol. Sheroplasts were mechanically homogenized using a glass/teflon homogenizer. Homogenates were fractionated, keeping the supernatant. Supernatants were further centrifuged at high speed (12K RPM in a Beckman xxx rotor) for 12 min. Mitochondrial pellets were washed and resuspended in 20 mM Hepes and 0.6 mM sorbitol. Under these conditions, the wild-type strain, W303, yields 4–5 g of yeast per liter of culture, which yields 20–25 mg of crude mitochondria. Mitochondria were either used or stored at −80°C.
Distribution of Mss51 in sucrose gradients
Mitochondrial proteins (4 mg) were solubilized in 20 mM Hepes pH 7.4, 0.5 mM PMSF, 1% digitonin, 1.2 mM MgCl2, and 150 mM KCl and the extracts loaded on 7–20% sucrose gradients as reported (12). Gradients were centrifuged for 12 h at 28K RPM in a Beckman xxx rotor at 4°C. After centrifugation, 14 equal fractions were collected from each gradient; the equivalent fractions were pooled and tested for Mss51 distribution by Western blot analyses. Sample fractions were analyzed by SDS-PAGE in 12% acrylamide gels. Proteins were transferred to nitrocellulose membranes and detected using Mss51 antibodies.
H2O2 treatment of yeast cells
Yeast cultures were grown to early exponential phase in minimum galactose media (WO-Gal). For H2O2 treatment, all cultures were adjusted to the same optical density (OD600 = 1). Mild to severe oxidative stress was achieved by supplementing the cultures with increasing H2O2 (Sigma-Aldrich) concentrations in the 0–0.8 mM range (47). Treated and untreated cultures were shaken at 30°C for either 2 h or 40 min, as indicated. Toxicity was assessed by measuring growth arrest and by analyzing effects on mitochondrial protein synthesis.
H2O2 treatment of purified mitochondria and thiol trapping
To identify cysteines susceptible to undergo H2O2 oxidation in mitochondrial Mss51, acute treatments exposing cultures to high H2O2 concentrations during short times were implemented. Mss51 was oxidized in intact mitochondria (cell-free in organello system) maintained in an isotonic buffer (0.6 M sorbitol/100 mM Hepes pH 7.4). One hundred micrograms of purified mitochondria were incubated in the presence of 0–10 mM H2O2 for 40 min, followed by extraction with 100 mM Hepes pH 7.4 and 0.5% SDS with or without 2.5 mM of reducing agent, TCEP, as indicated. Oxidation of Mss51 thiols was visualized by addition of AMS at 4.6 mM.
Reverse transcription–polymerase chain reaction
Yeast cultures were grown to early exponential phase (OD600 = 1–2) in minimum galactose media (WO-Gal). Total RNA was purified using a published acid phenol extraction method (8). Briefly, 40 μg of purified RNA was treated with 3 μl of RNase-free DNAse I at 2000 U/ml (New England Biolabs) in the presence of 15 mM EDTA for 1 h at 37°C. For each reverse transcription–polymerase chain reaction, we used 4 μg of DNase-treated RNA, 4 μM of random hexamers as template (Invitrogen), and 1 mM dNTPs (Invitrogen). This mix was first treated at 65°C for 5 min. To the previous mix, we added 5 mM DTT, 2 U/μl of RNase Out Recombinant Ribonuclease Inhibitor (Invitrogen), and 10 U/μl of Super Script III (Invitrogen). Mixes were incubated at 50°C for 1 h, followed by inactivation for 15 min at 70°C.
Quantitative PCR
To assess the levels of total, processed, and unprocessed COX1 mRNAs by q-PCR, the following primers were designed as reported (49): COX1 (unprocessed): [Forward-Intron1 CAC AAA AGG AAA TAC GAA AAG TGA. Reverse-Intron1 AAA TCT CAT TTT TAT TTG AGT ATT CGG]; COX1 (processed): [Forward-exon 1 GGT ATG GCA GGA ACA GCA AT. Reverse-exon 2 CAG TGA TAA ATT AAG CGA AAC GG]; and COX1 (total) [Forward-exon 4 TGA TCA ATT TTC ATT ACA GCG TT. Reverse-exon 4 GGG TCA CCA CCT CCT GAT AC]. The levels of 21S rRNA and COX2 mRNA were also measured using q-PCR and the following primers: 21S RRNA (Total): [Forward-exon 1 CCG AAA GCA AAC GAT CTA ACT. Reverse-exon 1 GCA AAC CAG ATT TGT CTT TCA C] and COX2 (Total) [Forward CAT GAT TTT GCT ATT CCA AG. Reverse CAT GCT CCA TAG AAG ACA CC]. Expression levels of mitochondrial RNAs were normalized using total mRNA amplifications of the cytoplasmic gene ACT1: [Forward-exon 2 GTG CTG TCT TCC CAT CTA TC. Reverse-exon 2 CTT GGA TTG AGC TTC ATC AC].
Standard curves for each primer set were created using dilutions of wild-type untreated cDNAs and their amplification efficiencies used to normalize the data. For q-PCR reactions, we used 96-well PCR plates (BioRad). Twenty nanograms of cDNA in a total of 25 μl was mixed with 12.5 μl of Cyber Green (Quanta) and 0.3 μM of reverse and forward primers. Amplification was followed during 40 cycles at 95°C for 15 min, 95°C for 15 s, 60°C for 30 s, and 70°C for 30 s using a CFX BioRad Connection q-PCR thermocycler. Data were analyzed and plotted using CFX software (BioRad).
Blot signal quantification
For quantification of the signals in mitochondrial protein synthesis and immunoblot experiments, the images were digitized and densitometric analyses performed using the ImageJ software from NIH Image (40). To pool data from independent experiments, values were expressed as percentages of control or untreated samples.
Statistical analysis
All experiments were done at least in triplicate. All data are presented as mean ± standard deviation of absolute values or percent of control. Values were analyzed for statistical significance by Student's t-test. p < 0.05 was considered significant.
Miscellaneous procedures
Standard procedures were used for the preparation and ligation of DNA fragments and for transformation and recovery of plasmid DNA from E. coli (37). Yeast was transformed as described (39). The one-step gene insertion method (36) was used to integrate linear plasmids at the LEU2 locus of yeast nuclear DNA. Site-directed mutagenesis was used to create mutant alleles of MSS51. Protein concentration was measured with the Folin phenol reagent (21). Proteins were separated by SDS-PAGE in the buffer system of Laemmli (17), and membranes with immobilized proteins were treated with antibodies against the appropriate proteins, followed by a second reaction with anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Sigma-Aldrich). The SuperSignal chemiluminescent substrate kit (Pierce) was used for the final detection.
Abbreviations Used
- 4-DPS
aldrithiol-4
- AAA
ATPases associated with diverse cellular activities
- ALA
5-aminolevulinic acid
- AMS
4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid
- Bmax
total density (concentration) of receptors in a sample (heme-binding Mss51 in our case)
- COX
cytochrome c oxidase
- Cox1
cytochrome c oxidase subunit 1
- CPX motif
cysteine-proline-hydrophobic amino acid
- DTT
dithiothreitol
- HAP1
heme activator protein 1
- Hb
hemoglobin
- HEM1
5-aminolevulinate synthase
- HO-2
heme oxygenase-2
- HRI
hepatic heme-regulated inhibitor-eukaryotic initiation factor 2 alpha (eIF2 alpha) kinase
- HRM
heme regulatory motif
- IAM
iodoacetamide
- I+ mtDNA
intron-containing mtDNA
- I0 mtDNA
intronless mtDNA
- Kd
ligand (heme in our case) equilibrium dissociation constant. Affinity of ligand binding is the inverse of Kd
- LDH
lactic dehydrogenase
- m-AAA
mitochondrial matrix AAA proteases
- Mal-PEG5000
methoxypolyethylene glycol maleimide
- MRC
mitochondrial respiratory chain
- mtDNA
mitochondrial DNA
- OD
optical density
- PCR
polymerase chain reaction
- PEG
polyethylene glycol
- PMSF
phenylmethanesulfonyl fluoride
- q-PCR
quantitative PCR
- ρ0
rho zero strain, devoid of mtDNA
- redox
reduction–oxidation
- Rev-erbβ
transcription factor from the nuclear receptor superfamily
- ROS
reactive oxygen species
- SD
standard deviation
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TCEP
tris-[2-carboxyethyl] phosphine, hydrochloride
- TE
Tween 20 plus ergosterol
- TF
trigger factor
- UTR
untranslated region
- WT
wild-type
Acknowledgments
The authors thank Dr. Myriam Bourens for critical reading of the manuscript. A.B. is supported by grants from the National Institutes of Health (NIH), RO1 GM071775, GM105781, and GM112179. I.C.S. is supported by an NIH supplement to RO1 GM071775 to support researchers from minority groups.
Author Contributions
I.C.S. and A.B. conceived and designed the experiments; mostly I.C.S. with some help by A.B. performed the experiments; I.C.S. and A.B. analyzed the data; and I.C.S. and A.B. wrote the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Arlt H, Tauer R, Feldmann H, Neupert W, and Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85: 875–885, 1996 [DOI] [PubMed] [Google Scholar]
- 2.Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA, and Runge MS. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res 86: 960–966, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Barrientos A, Korr D, and Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J 21: 43–52, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrientos A, Zambrano A, and Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J 23: 3472–3482, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourens M, Fontanesi F, Soto IC, Liu J, and Barrientos A. Redox and reactive oxygen species regulation of mitochondrial cytochrome c oxidase biogenesis. Antioxid Redox Signal 19: 1940–1952, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo IA, Ayte J, and Hidalgo E. Reversible thiol oxidation in the H2O2-dependent activation of the transcription factor Pap1. J Cell Sci 126: 2279–2284, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Clemente P, Peralta S, Cruz-Bermudez A, Echevarria L, Fontanesi F, Barrientos A, Fernandez-Moreno MA, and Garesse R. hCOA3 stabilizes cytochrome c oxidase 1 (COX1) and promotes cytochrome c oxidase assembly in human mitochondria. J Biol Chem 288: 8321–8331, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collart MA. and Oliviero S. Preparation of yeast RNA. Curr Protoc Mol Biol Chapter 13: Unit13.12, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Decoster E, Simon M, Hatat D, and Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet 224: 111–118, 1990 [DOI] [PubMed] [Google Scholar]
- 10.Delaunay A, Isnard AD, and Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19: 5157–5166, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontanesi F, Clemente P, and Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem 286: 555–566, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontanesi F, Soto IC, Horn D, and Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol 30: 245–259, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant CM. Regulation of translation by hydrogen peroxide. Antioxid Redox Signal 15: 191–203, 2011 [DOI] [PubMed] [Google Scholar]
- 14.Gupta N. and Ragsdale SW. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erb{beta}. J Biol Chem 286: 4392–4403, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handy DE. and Loscalzo J. Redox regulation of mitochondrial function. Antioxid Redox Signal 16: 1323–1367, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalimonchuk O, Bird A, and Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J Biol Chem 282: 17442–17449, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685, 1970 [DOI] [PubMed] [Google Scholar]
- 18.Lan C, Lee HC, Tang S, and Zhang L. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J Biol Chem 279: 27607–27612, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Lechardeur D, Fernandez A, Robert B, Gaudu P, Trieu-Cuot P, Lamberet G, and Gruss A. The 2-Cys peroxiredoxin alkyl hydroperoxide reductase c binds heme and participates in its intracellular availability in Streptococcus agalactiae. J Biol Chem 285: 16032–16041, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodi T, Fontanesi F, and Guiard B. Co-ordinate regulation of lactate metabolism genes in yeast: the role of the lactate permease gene JEN1. Mol Genet Genomics 266: 838–847, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Lowry OH, Rosebrough NJ, Farr AL, and Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275, 1951 [PubMed] [Google Scholar]
- 22.Ma Q. Role of NRF2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53: 401–426, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailloux RJ, Jin X, and Willmore WG. Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biol 2: 123–139, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manthey GM. and McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J 14: 4031–4043, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McStay GP, Su CH, and Tzagoloff A. Modular assembly of yeast cytochrome oxidase. Mol Biol Cell 24: 440–452, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mense SM. and Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res 16: 681–692, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, and Rehling P. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151: 1528–1541, 2012 [DOI] [PubMed] [Google Scholar]
- 28.Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, and Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol 191: 141–154, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, and Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J 26: 4347–4358, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. 1966. Biochim. Biophys Acta 1807: 1507–1538, 2011 [DOI] [PubMed] [Google Scholar]
- 31.Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, and Van den Bogert C. Assembly of cytochrome-c oxidase in cultured human cells. Eur J Biochem 254: 389–394, 1998 [DOI] [PubMed] [Google Scholar]
- 32.Perez-Martinez X, Broadley SA, and Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J 22: 5951–5961, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Martinez X, Butler CA, Shingu-Vazquez M, and Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol Biol Cell 20: 4371–4380, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, and Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J 26: 4335–4346, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragsdale SW. and Yi L. Thiol/disulfide redox switches in the regulation of heme binding to proteins. Antioxid Redox Signal 14: 1039–1047, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211, 1983 [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, and Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]
- 38.Schieber M. and Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol 24: R453–R462, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiestl RH. and Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346, 1989 [DOI] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, and Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shingu-Vazquez M, Camacho-Villasana Y, Sandoval-Romero L, Butler CA, Fox TD, and Perez-Martinez X. The carboxyl-terminal end of Cox1 is required for feedback-assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J Biol Chem 285: 34382–34389, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon M. and Faye G. Steps in processing of the mitochondrial cytochrome oxidase subunit I pre-mRNA affected by a nuclear mutation in yeast. Proc Natl Acad Sci U S A 81: 8–12, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soto IC, Fontanesi F, Liu J, and Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta 1817: 883–897, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soto IC, Fontanesi F, Myers RS, Hamel P, and Barrientos A. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab 16: 801–813, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sousa-Lopes A, Antunes F, Cyrne L, and Marinho HS. Decreased cellular permeability to H2O2 protects Saccharomyces cerevisiae cells in stationary phase against oxidative stress. FEBS Lett 578: 152–156, 2004 [DOI] [PubMed] [Google Scholar]
- 46.Steiner H, Kispal G, Zollner A, Haid A, Neupert W, and Lill R. Heme binding to a conserved Cys-Pro-Val motif is crucial for the catalytic function of mitochondrial heme lyases. J Biol Chem 271: 32605–32611, 1996 [DOI] [PubMed] [Google Scholar]
- 47.Stone JR. and Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal 8: 243–270, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Thomas BJ. and Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630, 1989 [DOI] [PubMed] [Google Scholar]
- 49.Thornton B. and Basu C. Real-time PCR (qPCR) primer design using free online software. Biochem Mol Biol Educ 39: 145–154, 2011 [DOI] [PubMed] [Google Scholar]
- 50.Tzagoloff A, Akai A, and Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett 65: 391–395, 1976 [DOI] [PubMed] [Google Scholar]
- 51.Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, and Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature 486: 126–129, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayte J, Toledano MB, and Hidalgo E. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Natl Acad Sci U S A 102: 8875–8880, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, Kolesar JE, Lochmuller H, Chevrette M, Kaufman BA, Horvath R, and Shoubridge EA. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat Genet 41: 833–837, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Weraarpachai W, Sasarman F, Nishimura T, Antonicka H, Aure K, Rotig A, Lombes A, and Shoubridge EA. Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am J Hum Genet 90: 142–151, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, and Au SW. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J 22: 127–137, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Panek HR, and O'Brian MR. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol Microbiol 60: 209–218, 2006 [DOI] [PubMed] [Google Scholar]
- 57.Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, and Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol Biol Cell 18: 523–535, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L. and Guarente L. HAP1 is nuclear but is bound to a cellular factor in the absence of heme. J Biol Chem 269: 14643–14647, 1994 [PubMed] [Google Scholar]
- 59.Zhang L. and Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J 14: 313–320, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]