Abstract
Toca 511 is a modified retroviral replicating vector based on Moloney γ-retrovirus with an amphotropic envelope. As an investigational cancer treatment, Toca 511 preferentially infects cancer cells without direct cell lysis and encodes an enhanced yeast cytosine deaminase that converts the antifungal drug 5-fluorocytosine to the anticancer drug, 5-fluorouracil. A panel of established human cancer cell lines, derived from glioblastoma, colon, and breast cancer tissue, was used to evaluate parameters critical for effective anticancer activity. Gene transfer, cytosine deaminase production, conversion of 5-fluorocytosine to 5-fluorouracil, and subsequent cell killing occurred in all lines tested. We observed >50% infection within 25 days in all lines and 5-fluorocytosine LD50 values between 0.02 and 6 μg/ml. Although we did not identify a small number of key criteria, these studies do provide a straightforward approach to rapidly gauge the probability of a Toca 511 and 5-fluorocytosine treatment effect in various cancer indications: a single MTS assay of maximally infected cancer cell lines to determine 5-fluorocytosine LD50. The data suggest that, although there can be variation in susceptibility to Toca 511 and 5-fluorocytosine because of multiple mechanistic factors, this therapy may be applicable to a broad range of cancer types and individuals.
Introduction
The idea of using replication-competent viruses as a direct therapy for the treatment of cancer emerged more than a century ago.1 The γ-retrovirus murine leukemia virus (MLV) is an attractive tool for tumor gene therapy, as its nonlytic life cycle and requirement for host cell division allow for tumor-selective enhanced gene transfer2 and spread throughout the tumor. This MLV-based approach to gene therapy has been used to deliver and express the cytosine deaminase (CD) gene into tumor cells, providing a specific conversion of the well-tolerated antifungal prodrug, 5-fluorocytosine (5-FC), into the potent chemotherapeutic 5-fluorouracil (5-FU), which has been well established in preclinical studies.2–6 Building on this concept, Tocagen's retroviral replicating vector (RRV) named Toca 5117 (vocimagene amiretrorepvec) preferentially infects tumors without immediate cell killing and encodes an optimized yeast cytosine deaminase (yCD) that converts 5-FC into 5-FU within infected tumors.3,4,7,8 We are currently investigating the clinical utility, in recurrent high-grade glioma, of Toca 511 in combination with Toca FC, an investigational orally administered extended-release formulation of 5-FC (NCT01470794, NCT01156584, NCT01985256, and NCT02414165).
CD is lacking or poorly expressed in most human cells but is often expressed in yeast and bacteria as part of the pyrimidine salvage pathway and is responsible for converting cytosine to uracil and ammonia. CD also catalyzes, by means of a deamination step, the conversion of the prodrug 5-FC to the chemotherapeutic drug 5-FU9,10 within cancer cells expressing gene therapy-delivered CD. CD-based prodrug activating gene therapy has been investigated in preclinical animal models and clinical trials for many cancer types, including colon,11,12 liver,13,14 lung,15,16 medulloblastomas,16,17 prostate,10,18 breast,19,20 bladder,21,22 gliomas,3,23,24 head and neck,25,26 sarcomas,27,28 melanoma,11,29 and ovarian13,30 cancers. CD-based cancer gene therapy has proven to be effective in chemotherapy-resistant cancer cell lines,31,32 in combination with chemotherapy,4 and to enhance the effect of radiotherapy.33 To further improve the wild-type CD, we developed a codon-optimized,7 heat-stabilized34 yeast CD (yCD) gene, which resulted in an approximate 3-fold greater enzyme-specific activity compared with the wild-type protein when incorporated into Toca 511.
The primary cytotoxic effects of 5-FU occur through two distinct pathways: inhibition of DNA synthesis by the conversion of 5-FU to fluorodeoxyuridine monophosphate (FdUMP)2,11,35 and perturbation of RNA synthesis by the conversion of 5-FU to 5-fluorouridine triphosphate (5-FUTP)3,16,31,36 through 5-fluorouridine monophosphate (5-FUMP) and 5-fluorouridine diphosphate (5-FUDP) intermediates.7,16,36,37 In addition to direct killing of transduced cells by production of intracellular 5-FU, extracellular 5-FU and related antimetabolites secreted from cancer cells or released during the death of the infected cell result in killing of neighboring cells, a phenomenon known as the “bystander effect.”24,38
In the present study, 9 human tumor cell lines were chosen to represent 3 tumor types: glioblastoma (U-87MG, 8-MG-BA, 42-MG-BA, and T98G), colorectal cancer (COLO 205, HTB-38, and NCI-H508), and breast cancer (AU565 and MB-157). The unique properties of each cell line, such as growth rates, prior treatment with 5-FU, and cell ploidy, are expected to represent some of the diversity of conditions that could influence Toca 511 and Toca FC combination therapy in a clinical setting. A key question in considering this therapeutic strategy for other cancers is their susceptibility toward initial infection with Toca 511, subsequent viral spread throughout the tumor, levels of CD expression, extent of 5-FC to 5-FU conversion, and sensitivity to 5-FU. Therefore, we investigated Toca 511 infection and subsequent 5-FC metabolism in this broad panel of established human cancer cell lines. While other studies39,40 have addressed some of the above-described parameters, this is the first study to integrate all of these parameters with overall transcription patterns by next-generation sequencing, into a single analysis. This integrative approach coupled with additional analyses are complementing clinical endpoints where collectively they hold the potential to reveal predictive biomarkers critical to direct a patient-focused anticancer regimen.41,42 While these experiments were not powered to uncover biomarkers specific to Tocagen's therapy, the results show that Toca 511 combined with Toca FC may be applicable to a broad range of cancer types and individuals.
Materials and Methods
Cell lines and cell culture
U-87MG human glioblastoma (GBM) cells were obtained from the laboratory of Prof. Nori Kasahara, University of California, Los Angeles. T98G human GBM cells (ATCC CRL-1690) and 8-MG-BA and 42-MG-BA human GBM cells were kind gifts from the laboratory of Prof. Walter Gunzburg, Institute of Anatomy, Histology and Embryology, University of Veterinary Medicine, Vienna, Austria. Other cell lines were obtained directly from ATCC (Manassas, VA): NCI-H508 human colorectal metastatic adenocarcinoma cells from a patient treated with 5-FU (ATCC CCL-253); HTB-38 human colorectal primary adenocarcinoma cells (ATCC HTB-38); COLO 205 human colorectal metastatic adenocarcinoma cells from a patient previously treated with 5-FU (ATCC CCL-222); MB-157 human breast medullary carcinoma cells (ATCC CRL-7721); AU565 human breast adenocarcinoma cells from a patient previously treated with 5-FU and isolated from a metastatic pleural effusion (ATCC CRL-2351). U-87MG, T98G, 8-MG-BA, and 42-MG-BA tumor cell lines were cultured in DMEM (Hyclone Lab, Inc., Omaha, NE); MB-157 tumor cell lines were cultured with L-15 Leibovitz (Sigma-Aldrich, St. Louis, MO); HTB-38 tumors were cultured in McCoy's 5A (modified) media (Life Technologies, Grand Island, NY); COLO 205, AU565, and NCI-H508 cancer cells were all cultured with RPM1-1640 media (Sigma-Aldrich). All media additionally contained 10% FBS (Hyclone Lab, Inc.), 1 mM sodium pyruvate (Hyclone Lab, Inc.), and 2 mM glutamax (Life Technologies). All cells were cultured at 37°C in a humidified 5% CO2 incubator, except MB-157, which was cultured at 37°C in a humidified atmospheric (no supplemental CO2) incubator. 5-FC and 5-FU were purchased from Sigma-Aldrich.
Viral transduction and replication kinetics
U-87MG, T98G, 8-MG-BA, 42-MG-BA, HTB-38, and COLO 205 cells were seeded in duplicate at 3 × 105 cells/well and AU565, MB-157, and NCI-H508 cells were seeded at 1 × 105 cells/well in 6-well plates (Corning, Tewksbury, MA) containing 2 ml of complete media. All cell lines were cultured at 37°C in a humidified 5% CO2 incubator, except MB-157, which was cultured at 37°C in a humidified atmospheric (no supplemental CO2) incubator. Twelve to 18 hr postseeding, cells were transduced with Toca 511 or Toca GFP (Toca 511, where yCD is substituted with the enhanced green fluorescent protein [eGFP]) at an multiplicity of infection (MOI) of 0.1 and 10 prepared in complete media in the presence of 4 μg/ml of polybrene (Sigma-Aldrich). Plates were sealed with Breathe-Easy plate sealer (Sigma-Aldrich), centrifuged at 650 × g for 60 min, and returned to a 37°C incubator. The following day the medium was aspirated and replenished with a fresh complete medium. Thereafter, the medium was replenished every two days and/or the cells passaged as needed (if >80% confluency). The cells transduced with Toca GFP were monitored every two or three days by flow cytometry until maximally infected, as determined by maximal percentage of GFP-expressing cells, at which time the appropriate cells were harvested and frozen in 10% DMSO (Sigma-Aldrich)/90% FBS in Nalgene Mr. Frosty Cryo 1°C Freezing Containers. Individual experiments were repeated at least three times.
Genomic content analysis with propidium iodide
An amount of 5–10 × 106 naïve or maximally infected tumor cells or human peripheral blood mononuclear cells (PBMC) from C.T.L. (Shaker Heights, OH; Cat# CTL-UP1) were fixed with ice-cold 70% ethanol (Sigma-Aldrich) and stored on ice for 2–4 hr before washing the cells with 1 ml of PBS. One milliliter of propidium iodide (PI) solution (20 ml 0.1% Triton X-100 [Sigma-Aldrich], 4 mg DNase-free RNase, 400 μl of 1 mg/ml PI) was used to stain the cells before analysis on the flow cytometer (Canto II; BD, La Jolla, CA). Viable cells were gated through FSC (forward-scattered light) × SSC (side-scattered light) gate and interrogated for the mean fluorescence intensity (MFI) of G1 phase measured in the FL2 channel. The MFI of the known diploid control (PBMC) was used to determine the genomic content of the unknown cancer cells based on the relative shift in MFI. Individual experiments were repeated at least three times.
Proliferation assay
An amount of 1 × 105 naive or maximally infected (Toca 511) cancer cells were labeled in triplicate with 1 ml of 5 mM of carboxyfluorescein succinimidyl ester (CFSE) for 2 min at room temperature according to manufacturer's protocol (Life Technologies). Labeled cancer cells were cultured in 6-well plates as described. Proliferation was measured with flow cytometry as loss of CFSE intensity measured in the FL1 channel after gating viable tumors through an FSC × SSC gate. Cells were analyzed on the flow cytometer at four time points to get an average doubling time throughout the time course. Individual experiments were repeated at least twice.
RNA-sequencing analysis
U-87MG, T98G, 8-MG-BA, 42-MG-BA, NCI-H508, HTB-38, COLO 205, MB-157, and AU565 cell lines were grown to ∼50% confluency in 175 cm2 plates (Corning). Medium was removed and cells were lysed on the plate by addition of 1 ml of Trizol. The Trizol solution was transferred to 1.5 ml microfuge tubes and RNA was extracted by addition of chloroform to 30% final concentration. Tubes were centrifuged at 12,000 × g for 15 min. The aqueous phase was removed and RNA was further purified using Maxwell 16 Total RNA Purification Kit (Promega; Cat.# AS1050). Total RNA was converted into sequencing libraries using the Ovation Human FFPE RNA-Seq kit (Nugen, San Carlos, CA; Cat.# 0340 and 0341). Libraries were sequenced on an Illumina HiSeq 2000 machine generating ∼2 × 107 × 100 base paired-end reads per sample.
Sequence alignment and postalignment processing
Sequence alignment and processing was performed on an Amazon Web Service Instance running Ubuntu 14 Linux environment. Raw sequencing results in the form of FASTQ files were used as input for alignment of the sequencing data to the human ENSEMBL reference RNA database GRCh37. ENSEMBL and human genome (hg19) annotation files were retrieved from the Tophat website, which links to the Illumina iGenome collection. Alignments were performed with Tophat2.43 Tophat2-generated BAM files were converted to CXB files with Cufflinks and used as input into Cuffdiff2 to generate normalized FPKM (Fragments per kb of exon per million reads mapped) values and for differential expression analyses.12,44 HTSeq was used to calculate the number of reads that mapped to each transcript.14,45 SAMseq was used for correlation analyses.15,46 Primary sequencing data files are available via GEO (accession GSE72955). For relative mRNA expression comparisons, gene-centric normalized FPKM values were filtered as follows: mRNAs with average FPKM <1 were removed, remaining mRNAs were log2 (FPKM +1) transformed, and each mRNA was then normalized by subtracting its mean log2 (FPKM +1) value. Clustering was performed with Cluster 3.0 and results were visualized with Java Treeview. Analyses were performed in R (R Core Team, 2013; R Foundation for Statistical Computing, Vienna, Austria).
Cell viability measurement in tissue culture
U-87MG, T98G, 8-MG-BA, 42-MG-BA, NCI-H508, HTB-38, COLO 205, and AU565 cell lines were incubated in triplicate at 37°C in a humidified 5% CO2 incubator (MB-157 was cultured at 37°C in a humidified atmospheric [no supplemental CO2] incubator) with tetrazolium reagent 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS; Promega; Cat# G3581) at 20 μl per 100 μl of media. Thirty-minute time points were chosen for measurements. All samples were assayed on a Tecan Infinite M200 plate reader with absorbance readings taken at 490 nm. Individual experiments were repeated at least twice.
5-FC sensitivity of vector-transduced cell lines in tissue culture
U-87MG, T98G, 8-MG-BA, 42-MG-BA, NCI-H508, HTB-38, COLO 205, MB-157, and AU565 cell lines were maximally infected with Toca 511 vector, expanded, and stored as a bank of frozen cells. Cells were thawed and seeded in triplicate at 1,000 cells/well in 96-well plates (Corning). They were monitored over a six-day period following treatment with decreasing concentrations of the prodrug 5-FC (7-point log-fold serial dilutions were performed from 1290 to 0.00129 μg/ml) and the active drug 5-FU (7-point log-fold dilutions from 1300 to 0.00130 μg/ml). NCI-H508 was supplemented with 1290, 25.8, 0.516, 0.0103, 2.06 × 10−4, 4.13 × 10−6, and 8.26 × 10−8 μg/ml of 5-FC and 1300, 26, 0.52, 0.0104, 2.08 × 10−4, 4.16 × 10−6, and 8.32 × 10−8 μg/ml of 5-FU, respectively. Drug was first added one day after plating and then replenished with the complete medium plus fresh 5-FC or 5-FU every two days for continuous exposure to the fluorinated nucleotides made from these fluorinated pyrimidine bases. Cell growth kinetics were assessed every two days utilizing Promega's CellTiter 96 Aqueous One Solution reagent (MTS) as described above. All time points for each cell line and each concentration were performed in quadruplicate. GraphPad Prism v6 software (La Jolla, Ca) was used to calculate the mean of the time points as well as the LD50 and LD90 of each cell line for nonlinear four-parameter fit of the data points with log inhibitor (response) versus variable slope. The results obtained in this assay are representative of two or more individual experiments ran with most of the tumor cell lines.
Liquid chromatography–mass spectrometry cell preparation
Cell lines were grown to confluency in T-75 cm2 (Corning) tissue culture flasks as described above. At time of harvesting, they were washed with PBS (Hyclone Lab, Inc.) and disassociated with Trypzean Solution (Sigma-Aldrich). U-87MG, T98G, 8-MG-BA, 42-MG-BA, and MB-157 cell lines were seeded in duplicate or triplicate at 2.4 × 104 cells/well; NCI-H508, HTB-38, COLO 205, and AU565 were seeded in duplicate or triplicate at 4 × 104 cells/well to account for slower growth. All 9 cell lines were seeded into 12-well tissue culture plates (Corning) in 1 ml complete growth medium. Plates were seeded in quadruplicate and cells allowed to adhere overnight at 37°C as previously described. Fresh medium supplemented with 20, 66.6, or 200 μg/ml of 5-FC (a negative control without 5-FC was included as a baseline) was replenished 24 hr postseeding at 1 ml/well; cells receiving 5-FU were replenished at 20 and 200 μg/ml. The concentrations of 5-FC added to the tumors were chosen to reflect the approximate range expected to be detected in the blood of patients undergoing current Toca 511-based clinical trial dosing regimens. Incubation at 37°C was continued for an additional 24 and/or 48 hr. At the 24 and 48 hr time points, cells and supernatant were prepared for liquid chromatography–mass spectrometry (LC-MS) analysis and conducted by Southern Research Institute (Birmingham, AL). Medium was transferred from 12-well tissue culture plates into 1.5 ml microfuge tubes at 0.5 ml/tube and stored at −80°C. The cells were rinsed with 1 ml ice-cold PBS and aspirated. An amount of 0.1 ml of ice-cold 60% methanol (Sigma-Aldrich) was added to the cells and the cells were scraped off with a pipet tip. The disassociated cell/methanol suspension was transferred to prechilled 1.5 ml microfuge tubes and stored at −80°C. The controls for the LC-MS experiment included transduced tumors cultured with media alone, nontransduced tumors cultured with 5-FC, individual “spikes” of 5-FC and 5-FU, as well as the associated metabolites FUMP, FdUMP, and FUTP. Individual experiments were repeated twice.
DNA/RNA/protein purification
Genomic DNA (gDNA), RNA, and protein were all extracted from the same cell pellets from each maximally transduced Toca 511-infected tumor cell line according to instructions in the SurePrep RNA/DNA/Protein Purification Kit (Fisher, Pittsburgh, PA). Briefly, transduced cells were harvested as described above and pelleted by centrifugation at 200 × g for 5 min. The supernatants were decanted and cell pellets were washed with PBS and pelleted as above. The cell pellets were mixed with 350 μl of lysis solution, vortexed for 15 sec, mixed with 200 μl of 95% ethanol, vortexed for 10 sec, and applied to the Fisher SurePrep columns. The samples were washed with ethanol; RNA, gDNA, and protein were sequentially purified following ethanol washes after each extraction. RNA concentration was measured using the Nanodrop 8000 (Thermo Scientific, Waltham, MA).
Real-time PCR
TaqMan probe-based real-time PCR (qPCR) was performed as single-targeted 20 μl reactions, prepared in triplicate, on a Bio-Rad (Hercules, CA) CFX96 Real Time System using primers and probes annealing to sequences in the long terminal repeat region of MLV or in the yCD transgene. Primers and probes were designed with PrimerQuest software (Integrated DNA Technologies, San Diego, CA) and synthesized by Integrated DNA Technologies. For qPCR detection of MLV-specific sequences, primers at 300 nM each of MLV-F (5′- AGC CCA CAA CCC CTC ACT C-3′) and MLV-R (5′- TCT CCC GAT CCC GGA CGA-3′), and 100 nM of MLV hydrolysis probe (5′- FAM-CCC CAA ATG AAA GAC CCC CGC TGA CG-3′BHQ_1) were used with iQ PCR Supermix (Bio-Rad). For qPCR detection of yCD-specific sequences, primers at 600 nM each of yCD-F (5′-ATC ATC ATG TAC GGC ATC CCT AG-3′) and yCD-R (5′-TGA ACT GCT TCA TCA GCT TCT TAC-3′), and 100 nM of yCD hydrolysis probe (5′-FAM/TCA TCG TCA ACA ACC ACC ACC TCG T/3′BHQ_1) were used with Bio-Rad's iQ PCR Supermix. Thermal cycling conditions consisted of 95°C for 5 min, followed by three cycles of 95°C for 15 sec and 65°C for 10 sec, followed by 38 cycles of 95°C for 15 sec and 65°C for 30 sec. Absolute quantification and linear regression were performed using a six-log serial dilution standard curve from a proviral-containing plasmid, pAZ3-yCD.7,17 CFX Manager 3.0 software (Bio-Rad) was used to calculate the threshold cycle (Ct) value, defined as the number of cycles at which the fluorescence signal is significantly above the defined threshold, which was inversely correlated to the logarithm of the initial copy number.
Real-time RT-PCR
TaqMan probe-based real-time RT-PCR (RT-qPCR) was performed as multiplexed 20 μl reactions, in triplicate, on a CFX96 Real Time System (Bio-Rad) using primers and probes annealing to sequences in the amphotropic MLV 4070A env, pol, and yCD transgene. Primers and probes were designed with PrimerQuest software (Integrated DNA Technologies) and synthesized by Integrated DNA Technologies. For simultaneous RT-qPCR detection of pol, env, and yCD-specific sequences performed in a single qPCR, primers at 300 nM each of Pol2-F (5′-CAA GGG GCT ACT GGA GGA AAG-3′) and Pol2-R (5′-CAG TCT GGT ACA TGG AGG AAA G-3′); 100 nM of Pol2 hydrolysis probe (5′-HEX/TAT CGC TGG ACC ACG GAT CGC AA/3′BHQ_1); 300 nM each of Env2-F (5′-ACC CTC AAC CTC CCC TAC AAG T-3′) and Env2-R (5′-GTT AAG CGC CTG ATA GGC TC-3′); 100 nM of Env2 hydrolysis probe (5′-TEX615/AGC CAC CCC CAG GAA CTG GAG ATA GA/3′IAbRQSp); 300 nM each of yCD-F (5′-ATC ATC ATG TAC GGC ATC CCT AG-3′) and yCD-R (5′-TGA ACT GCT TCA TCA GCT TCT TAC-3′); and 100 nM of yCD hydrolysis probe (5′-FAM/TCA TCG TCA ACA ACC ACC ACC TCG T/3′BHQ_1) were used with AgPath-ID One-Step RT-PCR Reagent (Life Technologies). Thermal cycling conditions consisted of 46°C for 20 min, followed by 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 55°C for 45 sec. Absolute quantification and linear regression were performed using a seven-log serial dilution RNA standard curve purified from Toca 511 viral vector containing the corresponding targets that underwent RT-qPCR in parallel with the test articles. The Toca 511 standard was originally qualified by comparison with an in vitro synthesized RNA standard quantified by absorbance at 260 nm. CFX Manager 3.0 software (Bio-Rad) was used to calculate the threshold cycle (Ct) value, defined as the number of cycles at which the fluorescence signal is significantly above the defined threshold, which was inversely correlated to the logarithm of the initial copy number.
Western blot
U-87MG, T98G, 8-MG-BA, 42-MG-BA, NCI-H508, HTB-38, COLO 205, MB-157, and AU565 cell lines were grown to confluency in T-75 cm2 tissue culture flasks and harvested for protein purification as described earlier. Lysates were quantified using a BCA Protein Assay (Pierce, Rockford, IL). An amount of 15 μg of total protein was separated by electrophoresis on a 4–12% SDS-polyacrylamide gel at 100 V for 60 min (Bio-Rad; Cat.# 345-0124). The resolved proteins were transferred to a nitrocellulose membrane (Life Technologies) using an iBlot (Life Technologies). Blots were probed overnight at 4°C with a mouse anti-yCD monoclonal antibody (generated for Tocagen at Genscript, NJ, 08854). The next day, blots were rinsed with 1× TBS-tween (0.05%) and probed with antimouse-horseradish peroxidase (HRP) secondary antibody (Southern Biotech, Birmingham, AL) for 1 hr at room temperature. The Western blots were analyzed using the HRP system with chemiluminescence detection (Thermo Scientific). Densitometry of the Western blot was determined using ImageJ software (NIH) and linear regression analysis of the yCD standard was used to quantitate the protein level of yCD in the tumor samples. The results obtained with this Western are representative of two individual experiments ran with most of the tumor cell lines where the protein was isolated directly from a cell pellet and not with SurePrep RNA/DNA/Protein Purification Kit (see above).
Statistical analysis
For MTS assay to assess 5-FC/5-FU sensitivity (LD50 determination), GraphPad Prism v6 software was used to calculate the mean of the time points as well as the LD50 and LD90 of each cell line for nonlinear four-parameter fit of the data points with log inhibitor (response) versus variable slope.
Unless otherwise noted, statistical analyses associated with all correlations between LC-MS, doubling times, infectability, LD50, and next-generation sequencing analysis were performed in R.
Results
Toca 511 infects diverse cancer cell lines
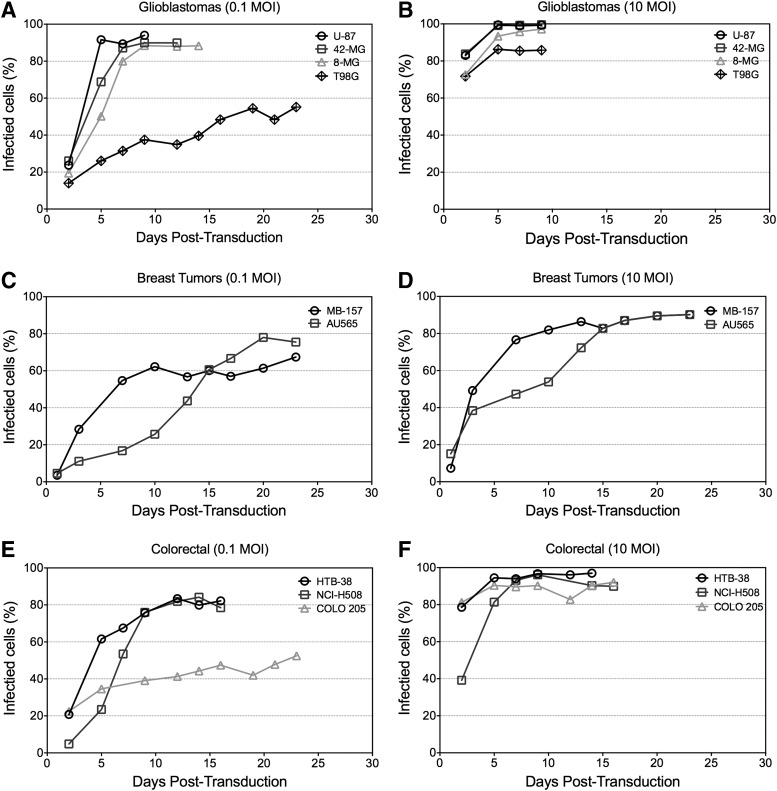
To characterize RRV replication kinetics in potential therapeutic targets, the time taken to reach 50% infection with Toca GFP was measured in multiple cell lines from three different cancer types (Fig. 1 and Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hgtb) at the same MOI. The human GBM cell lines U-87MG and 42-MG-BA supported similar rapid viral replication kinetics. 8-MG-BA had slower viral spread, reaching 50% infection 3 days later than U-87MG at an MOI of 0.1, but at comparable maximal level over time. Less than 60% of T98G cells were transduced beyond 20 days compared with the canonically permissive line U-87MG that achieved 92% transduction in less than 10 days. Colorectal cancer cell lines showed a variation in viral replication kinetics similar to the GBM lines, with HTB-38 and NCI-H508 displaying viral replication kinetics comparable to 8-MG-BA, while COLO 205 was more similar to T98G. Interestingly, the breast cancer cell lines tested showed a comparatively different phenotype from the other two cancer types (GBM and colorectal) with low initial transduction (<20% GFP+) at high MOI, followed subsequently by efficient spread (>80% GFP+). Regardless of replication kinetic differences, the RRVs successfully infected all the cancer cell lines reaching transduction levels that would be expected to be therapeutic.3
Figure 1.
Toca GFP viral replication kinetics in a panel of tumor cell lines. Toca GFP was used to infect selected glioblastomas (A and B), breast carcinomas (C and D), and colorectal adenocarcinomas (E and F) at an MOI of 0.1 or 10 with multiple replicates. To determine the frequency of infected tumors at each MOI, flow cytometry was used to interrogate the number of GFP+ cells among the total tumor population as a function of time. This figure is a representative experiment of two individual experiments. GFP, green fluorescent protein; MOI, multiplicity of infection.
RRVs were capable of infecting the entire panel of human cancer cell lines tested, although with differing rates of infection. Moloney γ-retrovirus virus from which Tocagen's RRVs were developed requires mitosis for productive infection,26,47 and so the rate at which cells divide may influence Toca 511 infectivity. This effect may, in theory, partially explain the differences in viral replication kinetics seen in Fig. 1. We, therefore, measured growth rates of each of the cell lines and compared the results to the infectivity data (1/[time to 50% infection] at an MOI of 0.1, Supplementary Table S1). Results show that, on average, the burden of supporting viral replication retards cell doubling time by 5–15%, yet no meaningful correlation between Toca GFP permissivity and cell doubling time was evident (Pearson r = −0.11). Furthermore, eight of nine cell lines had a doubling time of less than 27 hr, while MB-157 divided every 50 hr, yet still displayed midrange infectivity.
Consistent integration and expression of active CD in cancer cell lines
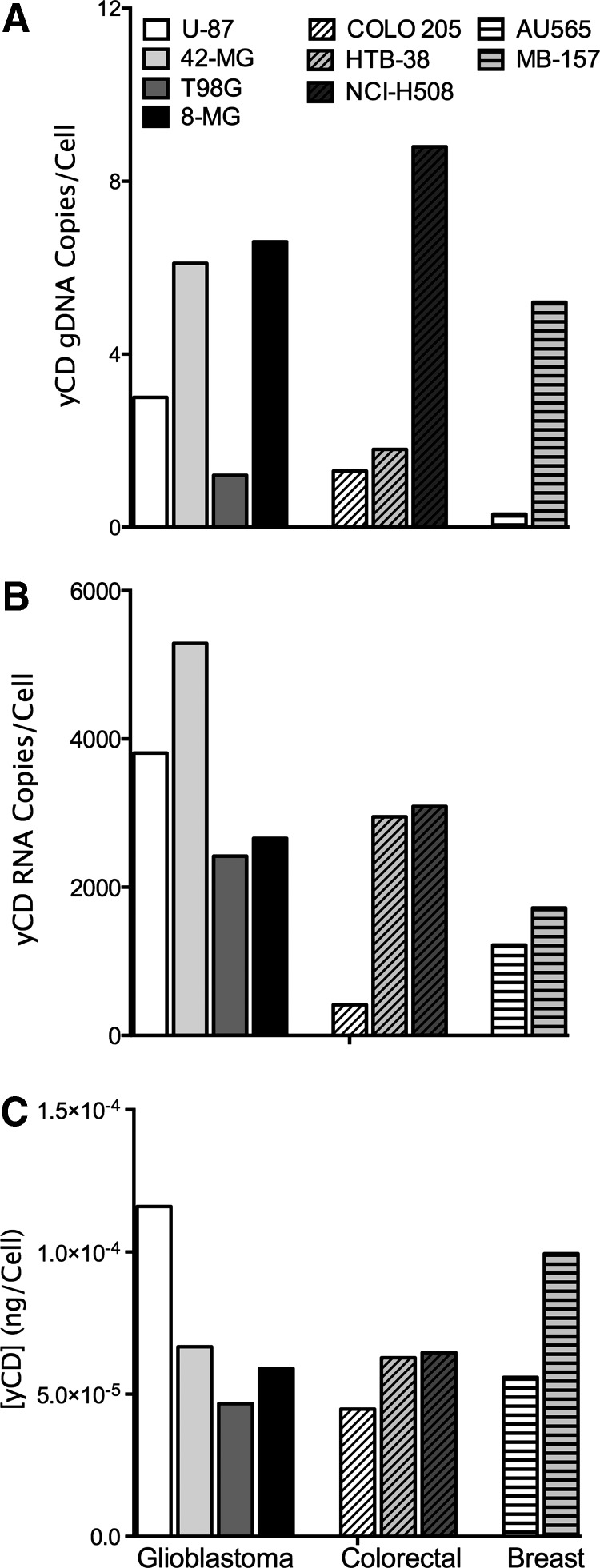
In general, we expect that the level of the yCD protein is dependent on yCD mRNA transcription, mRNA turnover, and translation, which, in turn, is related to the number of yCD DNA transgene integration events. Consequently, all three factors (yCD protein levels, RNA transcription efficiency from the provirus, and number of integrated proviruses [DNA]) have the potential to alter a transduced tumor's sensitivity to 5-FC. Therefore, we quantified the steady-state levels of each of these three related macromolecules in Toca 511-transduced cells. DNA, RNA, and protein were extracted from a single pellet of cells for each line tested and measured (Fig. 2 and Supplementary Table S1). There were 0.3 (AU565) to 8.8 (NC-H508) viral integrations per respective cancer cell genome, with the number of integration events per genome increasing as the rate of infection increased (Pearson r = 0.37 between 1/[days to reach 50% infectivity] and number of integration events per genome). The levels of yCD mRNA correlated with the number of integrations per genome (Pearson r = 0.49) and with protein levels of yCD (Pearson r = 0.48), suggesting a relatively consistent relationship between transgene integration and expression among the different cell lines (Supplementary Fig. S2). Nevertheless, there appeared to be less variation in CD protein levels within cancer cell types than in numbers of integration events and steady-state RNA levels. Levels of the yCD protein in each transduced cancer cell line correlated with the amount of 5-FC converted to 5-FU, which, in turn, influenced tumor killing efficiency. However, the cellular metabolism of 5-FU can also influence cell killing. Therefore, we next investigated the sensitivity of the cells to 5-FC and 5-FU, and the intracellular and extracellular levels of these molecules and downstream metabolites.
Figure 2.
Quantitation of yeast cytosine deaminase (yCD) DNA, RNA, or protein in Toca 511-infected tumors. DNA (A), RNA (B), and protein (C) were all extracted from the same 2 × 106 cells and quantitated with a yCD-specific qPCR, RT-qPCR, or Western blot respectively. DNA and RNA measurements were converted to absolute values using exogenous Toca 511 standard curves, as described in the Materials and Methods. Protein results were normalized and quantitated using densitometry coupled with a yCD protein standard as described in the Materials and Methods. This figure is a representative experiment of two or more individual experiments.
Expression of the CD protein is necessary to sensitize Toca 511-infected cell lines to 5-FC but increased expression does not correlate with increased sensitivity
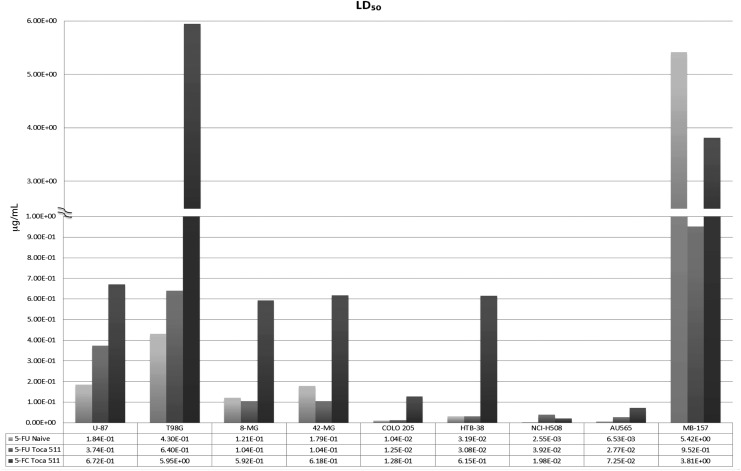
The efficacy of Toca 511 therapy is ultimately tied to the tumor's sensitivity to 5-FU and its cytotoxic metabolites. To assess the tumor's sensitivity to 5-FC or 5-FU, doses of either drug were added to cultures of either naïve or “maximally infected” cell types. Maximal Toca 511 infection was confirmed by qPCR using the ratio of transduced viral envelope and polymerase genes to the approximate total number of genomes available for transduction (Fig. 2 and Supplementary Table S1). The tumors were each cultured for 6 days to determine the LD50 using an MTS assay. Toca 511 infection had little effect on sensitivity to direct addition of 5-FU (Fig. 3). Without expression of yCD, tumors remained insensitive to 5-FC even with the undiluted 5-FC solution, preventing the determination of 5-FC LD50 in naïve tumors (data not shown). If 5-FC enters an infected cell at the same rate and concentration as 5-FU, and yCD converts 5-FC at 100% efficiency, one would expect near identical LD50 for the two fluorinated nucleosides. The sensitivity to 5-FC (LD50) in Toca 511-infected cells varied, ranging from 0.02 (NCI-H508) to 6 μg/ml (T98G), and tracked with cellular 5-FU sensitivity (Pearson r = 0.81, on average 7-fold increase [range 2–20-fold] in 5-FC LD50 vs. 5-FU LD50), which suggests variable but generally efficient import and conversion of 5-FC by yCD. Differences in yCD protein levels, however, did not track with 5-FC sensitivity (Pearson r = −0.14), suggesting that the yCD protein is not the limiting factor for cell killing under these conditions and that downstream 5-FU anabolic or catabolic metabolism may be important in determining the LD50 (Supplementary Fig. S3).
Figure 3.
Sensitivity of parental (naïve) and Toca 511-infected tumor cell lines to 5-FC and 5-FU. Cell viability was determined in vitro by an MTS assay. Cell lines that had been maximally transduced with Toca 511 vector (or nontransduced parental cells prepared in parallel) were seeded, in quadruplicate, at 1000 cells/well in 96-well plates. The LD50 was determined by MTS assay 6 days after treatment with various concentrations of 5-FC or 5-FU. LD50 values in μg/ml for 5-FC or 5-FU are listed along the y axis. Numerical LD50 values in μg/ml are listed in the embedded table for the transduced and naïve tumor cells treated with 5-FC or 5-FU. This figure is a representative experiment of two or more individual experiments. 5-FC, 5-fluorocytosine; 5-FU, 5-fluorouracil.
Import and export of 5-FC and 5-FU is not a rate-limiting step for killing of infected cell lines
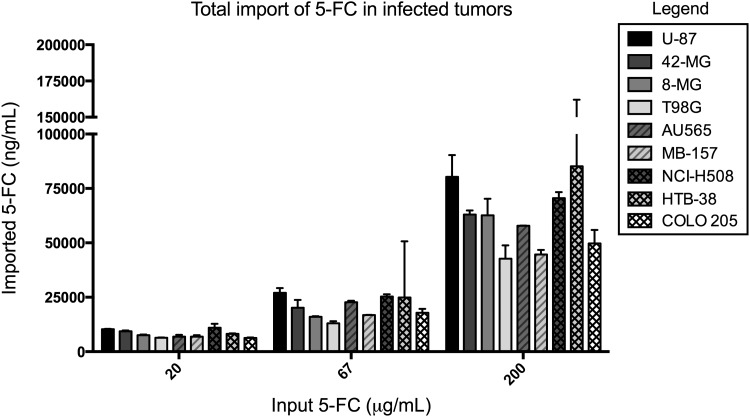
In order to investigate further the factors that influence the response of infected cells to 5-FC treatment, we measured the intracellular (cell extracts) and extracellular (measured from culture supernatant) 5-FC and 5-FU levels after treatment with different concentrations of 5-FC (Figs. 4 and 5). 5-FC is converted by yCD into 5-FU, which, in turn, is metabolized via several different pathways into antimetabolites such as FdUMP (inhibits DNA synthesis), FUMP (inhibits RNA synthesis), and catabolized to nonfunctional fluoro-beta-alanine (FBAL), or exported extracellularly.11,23,29,48 The ability of each transduced cancer cell to import 5-FC and convert it into 5-FU and related antimetabolites is expected to influence the efficacy of Toca 511 therapy. To measure the import and metabolism of fluorinated nucleotides, naïve or maximally infected cell lines were cultured with different concentrations of 5-FC or 5-FU and both intracellular and extracellular 5-FC, 5-FU, FUMP, FUTP, and FdUMP were measured by LC-MS. To determine 5-FC intracellular import at both 24 and 48 hr, 5-FC was added to naïve cancer cells that lack yCD expression and cannot readily convert 5-FC to 5-FU. 5-FC import was similar in most naïve cancer cells regardless of histological origin or growth rate (Supplementary Table S2). In infected cells, 5-FC import, as measured by the sum of intracellular 5-FC and total 5-FU levels, was similar across cell lines with a 2- to 3-fold range per input 5-FC tested (Fig. 4). As expected, 5-FC was similarly imported into both the infected and naive cancer cells.
Figure 4.
Total amount of 5-FC imported into Toca 511-infected tumors. Tumor cells that had been maximally transduced with Toca 511 vector were cultured with different concentrations of 5-FC for 24 hr, and intra- and extracellular 5-FC and 5-FU were measured by liquid chromatography–mass spectrometry (LC-MS). Shown are the 24 hr total intracellular import of 5-FC versus starting 5-FC levels (n = 2, error bars represent standard error of the mean). 5-FC import was determined by the sum of intracellular 5-FC and total (intra- and extracellular) 5-FU levels.
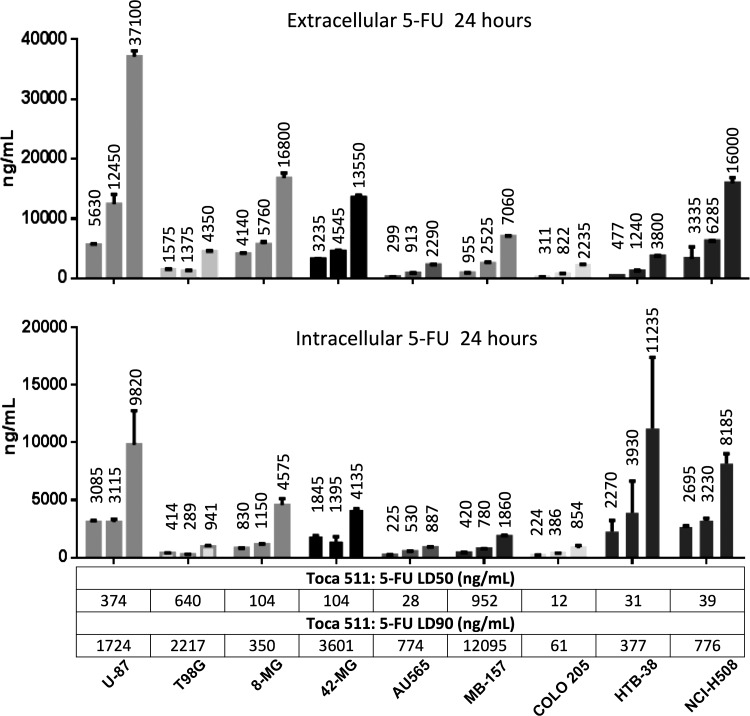
Figure 5.
LC-MS measurement of extra- or intracellular 5-FU converted from 20, 66.6, or 200 μg/ml of 5-FC (respectively) with corresponding 5-FU LD50 and LD90 (determined by MTS assay) for each maximally infected cell line (n = 2; error bars represent standard error of the mean).
To measure the efficiency of 5-FC to 5-FU conversion, 5-FC was added to transduced cancer cells at three different clinically relevant concentrations and 5-FU was quantified by LC-MS 24 and 48 hr later. 5-FC conversion, as measured by total intracellular and extracellular 5-FU, varied by up to 18-fold across the cell lines (Fig. 5).
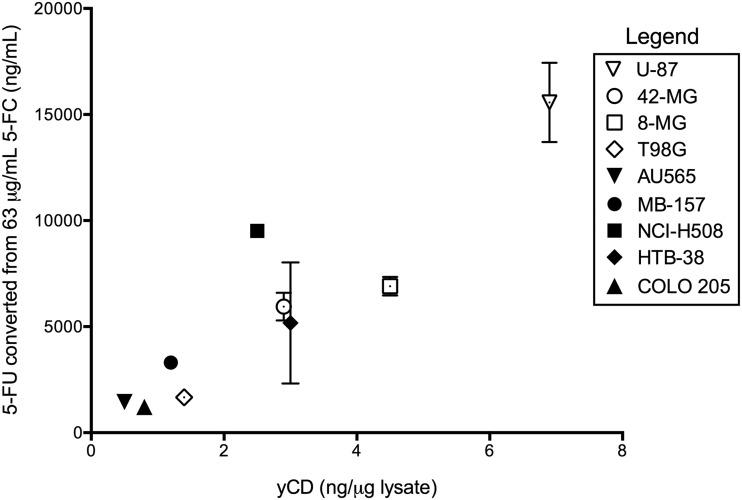
Parenthetically, the Kd for 5-FC of the yCD protein used in this study has been measured as 18 mg/ml (140 mM).34 Consequently, all the 5-FC concentrations used here are well below this Kd. In agreement with the Kd, we have shown that yields of 5-FU continue to increase with increasing 5-FC up to 5.0 mg/ml (38.7 mM) in assays performed using infected U-87 cell extracts (Supplementary Fig. S1). In addition, the LD50 of 5-FU in all cancer cells was reached in the extracellular medium (Fig. 5). The conversion of 5-FC to 5-FU demonstrates a strong correlation to yCD protein levels in the different cell lines over the entire range of 5-FC input (Fig. 6; Pearson r = 0.93–0.94 at 24 h). 5-FU made up the vast majority of total fluorinated metabolites (F-nucs) measured; the other downstream fluorinated metabolites generally contributed less than 10% to the total quantity of measured products. FUTP and FUMP had measurable levels (>5 ng/ml), whereas FdUMP did not (Supplementary Table S2).
Figure 6.
Correlation between yCD protein levels and conversion of 5-FC to 5-FU in Toca 511-infected tumors. Tumor cells that had been maximally transduced with Toca 511 vector were cultured with different concentrations of 5-FC for 24 hr, and intra- and extracellular 5-FU were measured by LC-MS. 5-FC conversion to 5-FU varied up to 10-fold across cell lines, but strongly correlated with yCD protein levels measured by Western blot (Pearson r = 0.82). Data illustrate the 24 hr time point at 66.6 μg/ml 5-FC, but are representative of all time points and 5-FC concentrations measured.
5-FU is a small, uncharged molecule capable of nonfacilitated diffusion through cellular membranes,23,24,30 yet in certain tissues, its import can be facilitated by equilibrative nucleoside transporters.32,49 To determine the extent of 5-FU uptake, infected tumors were cultured with a high and low dose of 5-FU for 24 or 48 hr before LC-MS quantitation. Levels of 5-FU measured in the supernatant were similar for all tumor cultures at both doses and time points (Supplementary Table S2). 5-FU was equally imported into all tumors reaching 50–100 μg/ml in the high-dose group, values well above the LD90 of all the cell lines (0.06–12.1 μg/ml).
5-FU sensitivity correlates with mRNA expression of nucleotide metabolic factors
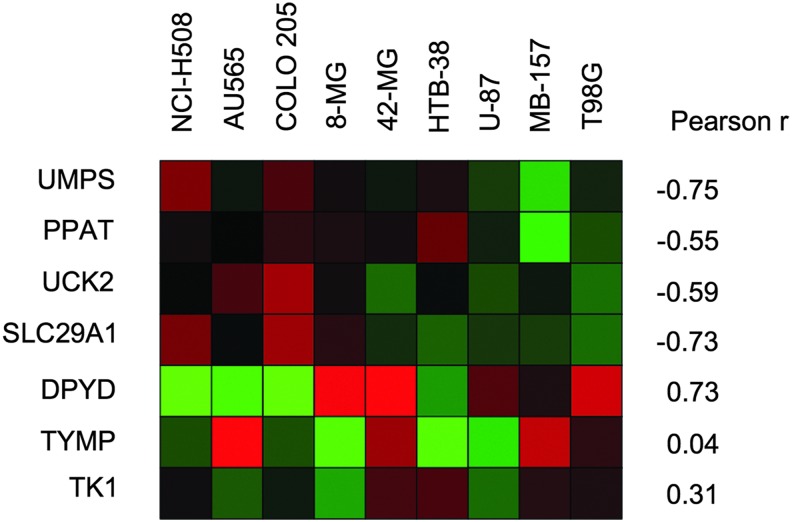
To compare 5-FU and 5-FC sensitivity to expression of mRNAs encoding proteins implicated in 5-FU metabolism, we performed RNA sequencing on all infected cell lines. Among the ∼20 mRNAs known to be involved in 5-FU metabolism, dihydropyrimidine dehydrogenase (DPYD) showed the strongest negative relationship to 5-FU/5-FC sensitivity (Fig. 7; Pearson r = 0.73 for DPYD levels with LD50). DPYD catalyzes the degradation of 5-FU and inactivating mutations in DPYD are associated with severe 5-FU toxicity.50,51 DPYD transcripts were undetected in the three most sensitive human cell lines (HTB-38, NCI H508, and AU656), which were, on average, 10-fold more sensitive to 5-FU/5-FC than the other lines. The relative expression of mRNAs encoding enzymes that convert 5-FU to FUMP positively correlated with 5-FU/5-FC sensitivity (Fig. 7 and Supplementary Table S3); these included uridine monophosphate synthetase (UMPS) and phosphoribosyl pyrophosphate amidotransferase (PPAT), which convert 5-FU to FUMP directly and uridine-cytidine kinase 1 and 2 (UCK1 and UCK2), which convert the intermediate fluorouridine (FUR) to FUMP.9,11 The 5-FC equilibrative nucleoside transporter SLC29A1 also positively correlated with 5-FU/5-FC sensitivity (Fig. 7). In contrast, there was no correlation (Supplementary Table S3) between 5-FU/5-FC sensitivity and the relative expression of mRNAs encoding proteins associated with conversion of 5-FU to FdUMP, including thymidine phosphorylase (TYMP), which converts 5-FU to FUDR, and thymidine kinase 1 (TK1), which converts FUDR to FdUMP.11,48 While low levels of thymidylate synthase (TS) expression can lead to a better response to 5-FU,52 there was no correlation between TS and LD50 in this study.
Figure 7.
Relationship between 5-FC/5-FU sensitivity and relative mRNA expression of 5-FU metabolic genes. Heatmap representation of the relative mRNA expression of seven factors involved in 5-FU nucleotide metabolism across nine human cell lines. The cell lines are ordered from left to right by 5-FC LD50 in Toca 511-infected cells, starting with the lowest LD50. The Pearson correlation compares relative mRNA expression to log10[LD50].
Discussion
Previous literature does not fully address the in vitro parameters of CD gene delivery coupled with 5-FC treatment in the successful killing of multiple cancer cell line types. In addition, many early experiments were performed with bacterial CD, which has a much lower specific activity for 5-FC. The panel of tumors selected for these analyses was varied in their tissue of origin, growth rate, morphological or histological appearance, and clinical exposure to 5-FU before cell line derivation. Within this study, Toca 511 showed a broad range of effective CD transfer across all cancer types tested. While at high MOIs, transduction differences between cell lines were not that marked, at lower MOIs the rate of viral spread separated into three broad groups that were not strongly associated with tumor types from which the cell lines were derived. Regardless of initial MOI, all cell types tested achieved in vitro transduction levels sufficient to kill the cells at clinically achievable levels of 5-FC.
Among the many intrinsic or extrinsic factors that may affect the therapeutic activity of Toca 511, the rate of cell division can be expected to influence both viral replication kinetics and sensitivity to 5-FU.11,24,47,53–55 The doubling time of maximally infected Toca 511 cancer cells was assessed by CFSE dilution at various time points. Regardless of the transduction kinetics measured, the rate of cell division did not alter the susceptibility to 5-FC treatment after maximal yCD gene transfer by Toca 511, suggesting that additional factors beyond cell division are important for susceptibility. It is interesting to note that most infected cell lines had a 5–15% increase in doubling time relative to the naïve cell line, suggesting that the burden of supporting RRV impacts cell proliferation to a small but possibly clinically relevant degree.
Using the time points established in the viral replication kinetic experiments, total DNA, RNA, and protein were all extracted from a single pellet of maximally transduced Toca 511-infected cells, minimizing sampling bias while allowing for a detailed examination of the critical steps after viral transduction. The RRV encoding yCD provided consistent integration of the yCD transgene into the genome of all the cancer cell lines with varying average copy numbers. In all cancer cell lines, with exception of AU565, multiple copies were detected per cell (Supplementary Table S1). The number of yCD transcripts per cell was quantitated with RT-qPCR and revealed substantial expression of both yCD and viral mRNA. Variability between the genomic yCD copy number and expression of mRNA was not unique to the yCD gene as the genomic copy number and expression of other viral mRNA were very similar to that of yCD (Supplementary Table S3). However, translation of yCD transcripts into protein, measured by Western blot, was consistent with the number of transcripts per cell in most cell lines. Even cell lines with relatively low yCD protein expression, such as AU565, could readily convert 5-FC to 5-FU as indicated by MTS and measured in LC-MS endpoints. In all cases, the intracellular concentrations of 5-FU converted from 5-FC were in excess of the LD50. Moreover, T98G and MB-157 converted 5-FC to 5-FU at extracellular concentrations in excess of the LD50 at 5-FC dosing levels expected to be achievable clinically.
LC-MS analysis revealed that 5-FC was readily imported into all tumors tested at comparable levels. Different concentrations of 5-FC were used to illustrate that increasing concentrations of this prodrug (up to 200 μg/ml) result in increasing concentrations of chemotherapeutic 5-FU. This could be important because the axis of 5-FC-dependent 5-FU sensitivity may be reduced in a clinical setting because of lower levels of transduction, insufficient time for maximal viral spread because of tumor burden, areas of necrosis and scarring, and altered bystander effects related to interstitial pressure dynamics in the tumor. LC-MS analysis confirmed that each of the cancer cell types was able to readily import and export 5-FU, a necessary component for the bystander effect. Consequently, higher dosing levels of 5-FC could be helpful to Toca 511 and Toca FC combination therapy, especially if there is less than 100% infection of cancer cells in vivo.
5-FC sensitivity varied by over 300-fold across cell lines infected with Toca 511, suggesting that variation in sensitivity related to 5-FU production among cancer cell lines may be an important factor in the clinical efficacy of Toca 511 therapy. To address possible factors or pathways that could antagonize or synergize with 5-FC-generated 5-FU sensitivity, RNA-Seq analysis of the tumors was used to compare expression of mRNAs encoding proteins implicated in 5-FU metabolism, to 5-FC sensitivity. This analysis identified several factors whose differential expression may, at least partially, drive differences in 5-FC-generated 5-FU sensitivity. DPYD, which encodes the enzyme responsible for 5-FU degradation, was undetectable in the three most sensitive lines. Severe 5-FU toxicity is observed in cancer patients who have loss-of-function mutations in DPYD, and are undergoing systemic therapy with 5-FU, arguing for a role of this gene in 5-FU pharmacodynamics in vivo.7,51 Conversely, expression of mRNAs encoding key factors in conversion of 5-FU to FUMP (UMPS, PPAT, UCK1, and UCK2) positively correlated with 5-FC sensitivity, as did expression of SLC29A1, which codes for the 5-FC transporter. While thymidylate synthase (TS) expression can be associated with insensitivity to 5-FU,52 we did not see a correlation between TS mRNA expression and LD50 in this study, and this observation is possibly related to the time needed to drive TS upregulation. Expression of gene sets related to apoptosis, nucleotide metabolism, and RNA metabolism also correlated with 5-FU/5-FC sensitivity (Supplementary Table S3).
Human tumors, especially recurrent GBM, are complex mixtures of cells, including malignant cancer cells, inflammatory cells including myeloid-derived suppressor cells (MDSCs) and lymphocytes,56,57 nontransformed cellular matrix, necrotic centers, and acellular fibrotic tracts from previous therapies.58 As noted above, in these clinical circumstances it is unlikely that 100% tumor cell transduction can be achieved, and some form of bystander effect that leads to death of uninfected cells may be necessary for the best clinical outcomes. One form of bystander effect is the 5-FU diffusibility. Another and potentially more powerful form of bystander effect is induction of antitumor immune responses after initial tumor cell killing.3,59 These two may be indirectly related because it seems likely that the more tumor cells that are killed by 5-FU, the greater the inflammation and release of tumor antigens, leading to stronger antitumor immune responses.
In conclusion, the antitumor mechanisms involved in Tocagen's Toca 511 and Toca FC combination therapy are proposed as threefold3: direct killing of the transduced cancer cells through intracellular conversion of 5-FC to 5-FU by CD, a bystander effect through cellular release of 5-FU and uptake by neighboring tumor cells, and induction of a productive antitumor immune response by inflammatory release of tumor antigens. This last may be further potentiated by 5-FU-mediated killing of MDSCs in the tumor, which are reported to be very sensitive to 5-FU.50,60 The experiments discussed in this report demonstrate that 5-FC prodrug and 5-FU were readily imported and released from all cancer cell lines tested. Furthermore, the combination therapy based on Toca 511 and Toca FC administration has broad applicability to infect different cancer types, integrate the transgene, and express amounts of CD needed to convert 5-FC in the tumor to therapeutic levels of 5-FU. Results from these studies suggest a straightforward approach to rapidly gauge the likely magnitude of the effect of the Toca 511 and Toca FC combination therapy in other cancer indications using a single MTS assay of maximally infected cells to determine the LD50 5-FC. Additionally, while current clinical dosing recommendations for antifungal treatment with 5-FC call for levels below 100 μg/ml in plasma,9,61 data from this article suggest that evaluating doses of 5-FC in cancer patients could augment the therapeutic effect. Overall, the data presented here show that it is likely that most cancers are candidates for treatment with Toca 511 and 5-FC.
Supplementary Material
Acknowledgments
All authors are employees of Tocagen Inc. The authors thank Harry E. Gruber, Nicholas A. Boyle, Amanda Omlor, and Alessandro Lobbia (Tocagen, Inc.) for helpful discussions and critical reading of the article. Bernard Ntsikoussalabongui and the Southern Research Institute (Birmingham, AL) are thanked for their services with the LC-MS analysis of fluorinated nucleotides. The authors also thank the ABC2 Foundation (Washington, DC), the National Brain Tumor Society (Watertown, MA), the American Brain Tumor Association (Chicago, IL), the Musella Foundation (Hewlett, NY), and Voices Against Brain Cancer (New York, NY) for financial support.
Author Disclosure
All authors are employees of Tocagen Inc.
References
- 1.Dock G. The influence of complicating diseases upon leukaemia. Am J Med Sci 1904;127:561–592 [Google Scholar]
- 2.Logg CR, Tai CK, Logg A, et al. . A uniquely stable replication-competent retrovirus vector achieves efficient gene delivery in vitro and in solid tumors. Hum Gene Ther 2001;12:921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostertag D, Amundson KK, Lopez Espinoza F, et al. . Brain tumor eradication and prolonged survival from intratumoral conversion of 5-fluorocytosine to 5-fluorouracil using a nonlytic retroviral replicating vector. Neuro-oncology 2012;14:145–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang TT, Hlavaty J, Ostertag D, et al. . Toca 511 gene transfer and 5-fluorocytosine in combination with temozolomide demonstrates synergistic therapeutic efficacy in a temozolomide-sensitive glioblastoma model. Cancer Gene Ther 2013;20:544–551 [DOI] [PubMed] [Google Scholar]
- 5.Wang WJ, Tai CK, Kasahara N. Highly efficient and tumor-restricted gene transfer to malignant gliomas by replication-competent retroviral vectors. Hum Gene Ther 2003;14:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Aguirre ME, Espinoza FL, Martin B, et al. . Abstract 708: Toca 511 gene therapy in combination with 5-fluorocytosine for intratumoral production of 5-fluorouracil in a colon cancer metastasis model. Cancer Res 2014;74:Abstract 708. [Google Scholar]
- 7.Perez OD, Logg CR, Hiraoka K, et al. . Design and selection of Toca 511 for clinical use: Modified retroviral replicating vector with improved stability and gene expression. Mol Ther 2012;20:1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloughesy TF, Kesari S, Kalkanis S, et al. . IT-05 administration of Toca 511 to subjects with recurrent HGG undergoing repeat resection. 2014. In press [Google Scholar]
- 9.Vermes A, Guchelaar HJ, Dankert J. Flucytosine: A review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J Antimicrob Chemother 2000;46:171–179 [DOI] [PubMed] [Google Scholar]
- 10.Lu M, Freytag SO, Stricker H, et al. . Adaptive seamless design for an efficacy trial of replication-competent adenovirus-mediated suicide gene therapy and radiation in newly-diagnosed prostate cancer (ReCAP Trial). Contemp Clin Trials 2011;32:453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat Rev Cancer 2003;3:330–338 [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Li S, Nyati MK, et al. . Regional delivery and selective expression of a high-activity yeast cytosine deaminase in an intrahepatic colon cancer model. Cancer Res 2003;63:658–663 [PubMed] [Google Scholar]
- 13.Gmeiner WH. Novel chemical strategies for thymidylate synthase inhibition. Curr Med Chem 2005;12:191–202 [DOI] [PubMed] [Google Scholar]
- 14.Sia KC, Huynh H, Chinnasamy N, et al. . Suicidal gene therapy in the effective control of primary human hepatocellular carcinoma as monitored by noninvasive bioimaging. Gene Ther 2012;19:532–542 [DOI] [PubMed] [Google Scholar]
- 15.Qiu Y, Peng G-L, Liu Q-C, et al. . Selective killing of lung cancer cells using carcinoembryonic antigen promoter and double suicide genes, thymidine kinase and cytosine deaminase (pCEA-TK/CD). Cancer Lett 2012;316:31–38 [DOI] [PubMed] [Google Scholar]
- 16.Waldorf AR, Polak A. Mechanisms of action of 5-fluorocytosine. Antimicrob Agents Chemother 1983;23:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutova M, Shackleford GM, Khankaldyyan V, et al. . Neural stem cell-mediated CE/CPT-11 enzyme/prodrug therapy in transgenic mouse model of intracerebellar medulloblastoma. Gene Ther 2013;20:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunz C, Focke F, Saito Y, et al. . Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol 2009;7:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moolten FL. Tumor chemosensitivity conferred by inserted herpes thymidine kinase genes: Paradigm for a prospective cancer control strategy. Cancer Res 1986;46:5276–5281 [PubMed] [Google Scholar]
- 20.Yi B-R, Choi KJ, Kim SU, et al. . Therapeutic potential of stem cells expressing suicide genes that selectively target human breast cancer cells: Evidence that they exert tumoricidal effects via tumor tropism (review). Int J Oncol 2012;41:798–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohlhueter RM, McIvor RS, Plagemann PG. Facilitated transport of uracil and 5-fluorouracil, and permeation of orotic acid into cultured mammalian cells. J Cell Physiol 1980;104:309–319 [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi E, Menendez S, Ozu C, et al. . Highly efficient gene delivery for bladder cancers by intravesically administered replication-competent retroviral vectors. Clin Cancer Res 2007;13:4511–4518 [DOI] [PubMed] [Google Scholar]
- 23.Kuriyama S, Masui K, Sakamoto T, et al. . Bystander effect caused by cytosine deaminase gene and 5-fluorocytosine in vitro is substantially mediated by generated 5-fluorouracil. Anticancer Res 1998;18:3399–3406 [PubMed] [Google Scholar]
- 24.Huber BE, Austin EA, Richards CA, et al. . Metabolism of 5-fluorocytosine to 5-fluorouracil in human colorectal tumor cells transduced with the cytosine deaminase gene: Significant antitumor effects when only a small percentage of tumor cells express cytosine deaminase. Proc Natl Acad Sci USA 1994;91:8302–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsujie M, Nakamori S, Nakahira S, et al. . Human equilibrative nucleoside transporter 1, as a predictor of 5-fluorouracil resistance in human pancreatic cancer. Anticancer Res 2007;27:2241–2249 [PubMed] [Google Scholar]
- 26.Hamstra DA, Rice DJ, Fahmy S, et al. . Enzyme/prodrug therapy for head and neck cancer using a catalytically superior cytosine deaminase. Hum Gene Ther 1999;10:1993–2003 [DOI] [PubMed] [Google Scholar]
- 27.Lawrence TS, Rehemtulla A, Ng EY, et al. . Preferential cytotoxicity of cells transduced with cytosine deaminase compared to bystander cells after treatment with 5-flucytosine. Cancer Res 1998;58:2588–2593 [PubMed] [Google Scholar]
- 28.Ramnaraine M, Pan W, Goblirsch M, et al. . Direct and bystander killing of sarcomas by novel cytosine deaminase fusion gene. Cancer Res 2003;63:6847–6854 [PubMed] [Google Scholar]
- 29.Kucerova L, Matuskova M, Pastorakova A, et al. . Cytosine deaminase expressing human mesenchymal stem cells mediated tumour regression in melanoma bearing mice. J Gene Med 2008;10:1071–1082 [DOI] [PubMed] [Google Scholar]
- 30.Chalikonda S, Kivlen MH, O'Malley ME, et al. . Oncolytic virotherapy for ovarian carcinomatosis using a replication-selective vaccinia virus armed with a yeast cytosine deaminase gene. Cancer Gene Ther 2008;15:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman LS. Goodman and Gilman's the Pharmacological Basis of Therapeutics. (McGraw-Hill, New York, NY: ). 1996 [Google Scholar]
- 32.Michaelsen SR, Christensen CL, Sehested M, et al. . Single agent- and combination treatment with two targeted suicide gene therapy systems is effective in chemoresistant small cell lung cancer cells. J Gene Med 2012;14:445–458 [DOI] [PubMed] [Google Scholar]
- 33.Takahashi M, Valdes G, Hiraoka K, et al. . Radiosensitization of gliomas by intracellular generation of 5-fluorouracil potentiates prodrug activator gene therapy with a retroviral replicating vector. Cancer Gene Ther 2014;21:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolworthy TS, Korkegian AM, Willmon CL, et al. . Yeast cytosine deaminase mutants with increased thermostability impart sensitivity to 5-fluorocytosine. J Mol Biol 2008;377:854–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seither RL, Trent DF, Mikulecky DC, et al. . Effect of direct suppression of thymidylate synthase at the 5,10-methylenetetrahydrofolate binding site on the interconversion of tetrahydrofolate cofactors to dihydrofolate by antifolates. Influence of degree of dihydrofolate reductase inhibition. J Biol Chem 1991;266:4112–4118 [PubMed] [Google Scholar]
- 36.Scholer HJ. Flucytosine. In: Antifungal Chemotherapy. Speller DCE, ed. (Wiley, Chichester, UK: ). 1980; pp. 35–106 [Google Scholar]
- 37.Robbins JM, Jolly DJ, Ostertag DG, et al. . Cancer combination therapy and recombinant vectors. US 20140178340 A1 [Google Scholar]
- 38.Chen JK, Hu LJ, Wang D, et al. . Cytosine deaminase/5-fluorocytosine exposure induces bystander and radiosensitization effects in hypoxic glioblastoma cells in vitro. Int J Radiat Oncol Biol Phys 2007;67:1538–1547 [DOI] [PubMed] [Google Scholar]
- 39.Miller CR, Williams CR, Buchsbaum DJ, et al. . Intratumoral 5-fluorouracil produced by cytosine deaminase/5-fluorocytosine gene therapy is effective for experimental human glioblastomas. Cancer Res 2002;62:773–780 [PubMed] [Google Scholar]
- 40.Hlavaty J, Jandl G, Liszt M, et al. . Comparative evaluation of preclinical in vivo models for the assessment of replicating retroviral vectors for the treatment of glioblastoma. J Neurooncol 2010;102:59–69 [DOI] [PubMed] [Google Scholar]
- 41.Neal JW, Gainor JF, Shaw AT. Developing biomarker-specific end points in lung cancer clinical trials. Nat Pub Group 2014;12:135–146 [DOI] [PubMed] [Google Scholar]
- 42.Dietel M, Jöhrens K, Laffert M, et al. . Predictive molecular pathology and its role in targeted cancer therapy: A review focussing on clinical relevance. Cancer Gene Ther 2013;20:211–221 [DOI] [PubMed] [Google Scholar]
- 43.Kim D, Pertea G, Trapnell C, et al. . TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Hendrickson DG, Sauvageau M, et al. . Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Tibshirani R. Finding consistent patterns: A nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res 2013;22:519–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roe T, Reynolds TC, Yu G, et al. . Integration of murine leukemia virus DNA depends on mitosis. EMBO J 1993;12:2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishiyama T, Kawamura Y, Kawamoto K, et al. . Antineoplastic effects in rats of 5-fluorocytosine in combination with cytosine deaminase capsules. Cancer Res 1985;45:1753–1761 [PubMed] [Google Scholar]
- 49.Damaraju VL, Damaraju S, Young JD, et al. . Nucleoside anticancer drugs: The role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene 2003;22:7524–7536 [DOI] [PubMed] [Google Scholar]
- 50.Sun X, Xing L, Deng X, et al. . Hypoxia targeted bifunctional suicide gene expression enhances radiotherapy in vitro and in vivo. Radiother Oncol 2012;105:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Kuilenburg ABP, Meinsma R, Zonnenberg BA, et al. . Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin Cancer Res 2003;9:4363–4367 [PubMed] [Google Scholar]
- 52.Lecomte T. Thymidylate synthase gene polymorphism predicts toxicity in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. Clin Cancer Res 2004;10:5880–5888 [DOI] [PubMed] [Google Scholar]
- 53.Trinh QT, Austin EA, Murray DM, et al. . Enzyme/prodrug gene therapy: Comparison of cytosine deaminase/5-fluorocytosine versus thymidine kinase/ganciclovir enzyme/prodrug systems in a human colorectal carcinoma cell line. Cancer Res 1995;55:4808–4812 [PubMed] [Google Scholar]
- 54.Guo X, Goessl E, Jin G, et al. . Cell cycle perturbation and acquired 5-fluorouracil chemoresistance. Anticancer Res 2008;28:9–14 [PubMed] [Google Scholar]
- 55.Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol 1994;68:510–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowe DB, Bose A, Taylor JL, et al. . Dasatinib promotes the expansion of a therapeutically superior T-cell repertoire in response to dendritic cell vaccination against melanoma. OncoImmunology 2014;3:e27589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker JC, Andersen MH, Schrama D, et al. . Immune-suppressive properties of the tumor microenvironment. Cancer Immunol Immunother 2013;62:1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol Mech Dis 2006;1:97–117 [DOI] [PubMed] [Google Scholar]
- 59.Mullen CA, Coale MM, Lowe R, et al. . Tumors expressing the cytosine deaminase suicide gene can be eliminated in vivo with 5-fluorocytosine and induce protective immunity to wild type tumor. Cancer Res 1994;54:1503–1506 [PubMed] [Google Scholar]
- 60.Alizadeh D, Katsanis E, Larmonier N. Chemotherapeutic targeting of myeloid-derived suppressor cells. OncoImmunology 2014;3:e27359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett JE, Dismukes WE, Duma RJ, et al. . A comparison of amphotericin B alone and combined with flucytosine in the treatment of cryptoccal meningitis. N Engl J Med 1979;301:126–131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.