Abstract
There continues to be a significant increase in the number and complexity of hydrophobic nanomaterials that are engineered for a variety of commercial purposes making human exposure a significant health concern. This study uses a combination of biophysical, biochemical and computational methods to probe potential mechanisms for uptake of C60 nanoparticles into various compartments of living immune cells. Cultures of RAW 264.7 immortalized murine macrophage were used as a canonical model of immune-competent cells that are likely to provide the first line of defense following inhalation. Modes of entry studied were endocytosis/pinocytosis and passive permeation of cellular membranes. The evidence suggests marginal uptake of C60 clusters is achieved through endocytosis/pinocytosis, and that passive diffusion into membranes provides a significant source of biologically-available nanomaterial. Computational modeling of both a single molecule and a small cluster of fullerenes predicts that low concentrations of fullerenes enter the membrane individually and produce limited perturbation; however, at higher concentrations the clusters in the membrane causes deformation of the membrane. These findings are bolstered by nuclear magnetic resonance (NMR) of model membranes that reveal deformation of the cell membrane upon exposure to high concentrations of fullerenes. The atomistic and NMR models fail to explain escape of the particle out of biological membranes, but are limited to idealized systems that do not completely recapitulate the complexity of cell membranes. The surprising contribution of passive modes of cellular entry provides new avenues for toxicological research that go beyond the pharmacological inhibition of bulk transport systems such as pinocytosis.
Introduction
Increasingly, nanomaterials such as C60 fullerenes are incorporated into a wide variety of consumer products including sporting goods and batteries and are under review by the U.S. Food and Drug Administration and other agencies for approval in a broad array of drugs, medical devices, and consumer products. In comparison to bulk material of the same chemical composition, the inherent larger surface area per volume and sub-cellular size of nanomaterials provide the significant potential for unanticipated and potentially adverse interactions with biological components.1,2
Icosahedral in shape and with molecular diameter of approximately 1 nm, C60 fullerenes are aromatic structures with π-orbital electrons that are readily donated for radical scavenging at biological pH,1,3 and are known to interact with cells. For example, the chemical attachment of drugs such as Paclitaxel and Doxorubicin to C60 fullerenes has under experimental conditions improved pharmacokinetics and putatively enhanced therapeutic properties.4 Similarly, Doxorubicin bound to fullerenes displays increased solubility when compared to the drug alone.5
Aside from the potential for occupational exposures, fullerenes are produced by a wide array of naturally occurring events such as volcanic eruptions and wildfires and are produced by anthropogenic sources that employ low-temperature combustion of fossil fuels. Release of fullerenes into the atmosphere may result in widespread exposure to humans through the inhalation route.6 Due to their small aerodynamic radii these particles gain access to the alveolar space and resident macrophages on the airway surface. Nanomaterials with an effective aerodynamic radius of 0.1-1 μm are sufficiently small to cross the alveolar epithelium and enter the adjacent bloodstream and circulating monocytes. Larger clusters of nanomaterial (> 1 μM) exit the lung via the mucociliary escalator and may travel from the esophagus to the gut, through which they encounter the enteric immune system.7
Therefore, a greater understanding of interactions between nanomaterials and the various components of the innate immune system will enable the creation of better predictive models of adverse outcomes following exposure. As the barrier between the intracellular and extracellular compartments, the lipid membrane is an important structural component of cells. Investigations into the mechanisms by which nanomaterials interact with the lipid membrane, however, are challenging. Essentially every aspect of the nanomaterial, particularly composition and surface chemistry, affects the way it interacts with the lipid membrane. Size, shape, charge, presence of functional groups on the structure, protein corona formation, and ability to interact with receptors on the cellular surface for endocytosis are factors that determine the entry mechanism and rate of entry of the nanomaterials into the cell.8
Current research into the behavior of nanomaterials in biological systems, especially within the lipid membrane, has been limited by the ability to identify and track the nano-sized aggregates with currently available imaging technology. Due to the extremely small scale of the material and the complexity of living systems, in this paper we report on a three-pronged approach to determine the cellular distribution of C60 fullerenes with a particular focus on their interaction with the lipid membrane. In vitro experiments in RAW 264.7 immortalized mouse macrophages via biochemical/cytochemical techniques and biophysical measurements of model membranes via NMR are cross-referenced with computational models of nano-lipid interactions.
Methods
Physical Characterization of Pristine and Functionalized Fullerenes
Pristine fullerenes have very low solubility in water (< 10-9 mg/L) and in the past they have, with some controversy, been dissolved in solvents such as tetrahydrofuran (THF) or have been replaced by functionalized fullerenes.9 Solvents are not an ideal vehicle for fullerenes due to their inherent toxic potential.10 Functionalization of fullerenes with hydroxyl groups or polyethylene glycol, for example, increases solubility but alters the way the nanomaterial interacts with biological systems.5,9 In this study fullerenes were prepared via the method of Deguchi et al.11 and Spohn et al.12 Specifically, this study combined the solvent exchange method with centrifugal filtration, which has been shown to remove major by-products tetrahydro-2-furanol, γ-butyrolactone, and peroxides identified in samples prepared in THF by solvent exchange.12,13 This two-step method yielded the advantage of producing pristine or Tb@C60 fullerenes in water, without the contamination of THF byproducts that may unnecessarily produce skew the results of biological studies by the formation of chemically-induced radical species.
Full characterization of the Tb@C60 was performed and results were compared with pristine C60 fullerenes to confirm that the Tb@C60 is a proper substitute. We did this comparison with two separate methods: A Malvern Zetasizer was used to determine zeta potential and aggregate size; a Nanosight LM 14C was used to determine and compare the aggregate size of terbium particles and fullerenes. C60 fullerenes prepared on three separate days were analyzed for charge and aggregate size on each instrument.
Cell Culture
RAW 264.7 immortalized mouse macrophages were obtained from American Type Culture Collection. They were maintained in Dulbecco's Modified Eagle Medium with 10% fetal bovine serum and 1% penicillin-streptomycin-glutamate solution. The cells were grown at 37 °C and 5% CO2. Cells were utilized until passage 30. Cell culture used both pristine fullerenes and terbium endohedral fullerenes.
Transmission Electron Microscope Imaging
In order to obtain an initial perspective of the interaction between C60 fullerenes and lipids in RAW 264.7 immortalized mouse macrophages, TEM studies were utilized. RAW 264.7 cells were allowed to attach to the culture dish overnight. Subsequently, cells were dosed with 0.1 and 0.36 μg/mL terbium-endohedral C60 fullerenes for 8 hours. They were fixed at 8 hours with glutaraldehyde and sodium phosphate buffer and refrigerated overnight. We took one precautionary step to preclude a likely procedural mishap. Specifically, cell pellets were not post-fixed with osmium tetroxide or counterstained with lead-citrate or uranyl-acetate to prevent the artifact of heavy metal nanometer-sized precipitates. Instead, cell pellets were dehydrated in graded alcohols, cleared, and embedded with epoxy resin for cutting of ultra-thin sections (about 45 nm thickness) in preparation for viewing on a Philips CM-100 transmission electron microscope. (This work was facilitated by the University of Michigan Microscopy and Image Analysis Team.) After the RAW 264.7 cells were dosed and processed as mentioned above. Size-distribution and morphology of C60 clusters was achieved by suspending terbiumendohedral C60 in water, drying on a TEM grid, and imaging on a Phillips CM-100 TEM.
Inhibition of Endocytosis
The role of pinocytosis, endocytosis and phagocytosis in the cellular uptake of C60 was investigated using classical pharmacologic inhibitors (200 μM genistein for 30 minutes, 10 μg/mL chlorpromazine hydrochloride (HCL), or 5 μg/mL cytochalasin A).14–16
Fullerene suspensions were added at a concentration of 4 μg/mL and incubated for 3 hours. Cells were fixed with 4% paraformaldehyde and stained with DAPI and antibodies against fullerenes and clathrin. The fullerene antibody was conjugated to Dylight 594. The clathrin secondary antibody was conjugated directly to FITC. Wide-field microscopy was performed with an Olympus IX-81 Disk Scanning Biological Microscope Imager, with an RGB plugin to quantify the amount of fullerene aggregates in the images. Fullerene aggregates were counted in four images of each inhibitor and four images of control cells exposed to only fullerenes. In each image, seven cells were assessed and the mean amount of red pixels in the images was calculated. Antibodies were obtained from Santa Cruz Biotechnology, Inc., Dallas TX.
Freeze Fracture
To visualize the fullerenes within the lipid membrane, we employed freeze-fracture TEM. RAW 264.7 cells were allowed to attach to culture flasks overnight. Cells were dosed with 0.5 μg/mL terbium-endohedral fullerenes for 8 hours. They were fixed with glutaraldehyde and shipped to NanoAnalytical Laboratory for TEM imaging.
Preparation of Vesicles Containing C60
A lipid precursor was manipulated and exposed to fullerenes as follows: 10 mg of d31-POPC (Avanti Polar Lipids, US) were dissolved in 1 mL of chloroform/methanol (1:1), dried under a stream of nitrogen gas to form a thin lipid film, and then left under high vacuum overnight to remove trace amounts of solvent. The samples were rehydrated with either 30 μL double distilled water, 30 μL of 10.09 μg/mL of C60 in water, or 300 μL of 10.09 μg/mL of C60 in water. All samples were then subjected to five freeze/thaw cycles interspersed with vortexing. The samples were then lyophilized overnight, and then rehydrated with 30 μL of double distilled water and subjected to another five freeze/thaw cycles interspersed with vortexing. The samples were then transferred to a 4 mm, magic-angle spinning rotor for solid-state NMR studies.
Nuclear Magnetic Resonance Spectroscopy
All NMR studies were conducted on a Chemagnetics Infinity 400 MHz spectrometer equipped with a 4 mm, double-resonance, magic-angle spinning probe head. The sample temperature was calibrated using a methanol standard as previously described.17 Static 31P NMR spectra were recorded at 161.1 MHz using a Hahn-Echo pulse sequence with π/2 and π pulses of 5 μs and 10 μs duration, respectively, and an inter-pulse delay of 40 μs. During the echo and acquisition, 60 kHz continuous-wave proton decoupling was applied. Typically 1024 scans were acquired for each spectrum. Static 2H NMR spectra were recorded at 61 MHz using a quadrupolar echo sequence with π/2 pulses of 5 μs and an inter-pulse delay of 50 s. Typically 8192 scans were acquired for each spectrum. The 31P and 2H spectra were referenced to 85% H3PO4 and D2O, respectively. All data processing was performed in matNMR.18 Prior to Fast Fourier Transform, 31P and 2H free induction decays left shifted to the top of the echo with 75 Hz and 250 Hz line broadening added, respectively.
The spectra were Depaked using a weighted Fast Fourier Transform algorithm19 and custom-written routines in Matlab (Mathworks Inc.). The observed quadrupolar splitting's () were used to calculate the order parameters () directly:20–22
where is the static quadrupolar coupling constant (e2qQ/h), which is 167 kHz for a paraffinic C-D bond.23 The order parameter profiles were constructed based on previously published assignments.24 Indeed, to quantify changes along the length of the acyl chains, we constructed order parameter profiles whereby the mobility of a given CD2 group as characterized by the order parameter, SCD, is plotted as a function of its position along the lipid chain.25
Moment analysis was performed using a custom-written routine in Matlab (Mathworks Inc.). The nth moment is calculated as:
where ω is the frequency and S(ω) is the signal intensity at a given frequency.26
Molecular Dynamics Simulations
The NAMD software27 was used to perform MD simulations together with the PLUMED plugin28 for free-energy reconstructions. Atomic interactions were modeled by using CHARMM force field: CHARMM3629 for the lipids, CGenFF30 for fullerenes, and TIP3P31 for water. A time step of 2 femtoseconds (fs) was used to integrate all the forces except for columbic interactions, which were computed every 4 fs with the particle mesh Ewald method32 by using cubic functions on a 0.1 nm grid with a tolerance of 10−6. A cutoff of 1.2 nm was employed for intermolecular forces and the potential was smoothed from 1 nm to the cutoff distance using the X-PLOR33 40 switching function, to avoid truncation effects. All the hydrogen bonds were kept rigid during the simulations with the SHAKE34 algorithm.
A temperature of 310 K was maintained via a Langevin thermostat with a 20 ps−1 dumping coefficient, while the pressure of 1.01325 bar was imposed with the Nosé-Hoover Langevin piston method35,36 with a time constant of 100 fs for the pressure and 50 fs for the barostat temperature. Since the systems are inherently anisotropic, during production runs the dimension of the periodic box normal to the bilayer was allowed to vary independently from the other two axes (NPsT ensemble).
Free-energy landscapes were reconstructed by employing the well-tempered Metadynamics algorithm.37 Gaussians with an initial height of 0.3 kcal/mol were deposited every 0.5 ps, and bias factors between 20 and 35 were used depending on the system.
The initial configuration of lipid bilayers was produced with CharmmGUI38 and then equilibrated for 20-40 ns with a mix of NVT and NPsT simulations. In order to place the C60 cluster in the membrane, a cavity was gradually created in the center of the bilayer. After the cluster was inserted in the cavity, the system was equilibrated again.
Results and Discussion
Regardless of the route of administration, nanomaterials entering the body will interact with cells of the innate immune system. The innate immune system is a nonspecific defense system that uses its component cells (macrophages, dendritic cells, neutrophils, mast cells, and natural killer) to identify, neutralize and dispose of particulate matter that may infect or otherwise damage the various internal compartments of the body. By contrast, the adaptive immune system uses a series of receptors that recognize specific antigens to recognize known threats such as microbial and other particulate stressors.39 Nanomaterials that enter the body via the airway or blood will by virtue of diffusion (e.g., the surface of the lung or in the vascular system) most commonly interact with macrophages in the alveoli or bloodstream. If recognized as foreign by the macrophage, uptake of the nanomaterial by the cell will depend on its physicochemical characteristics such as size, shape, surface charge, and surface functionalization. In general, larger nanoparticles or nanoparticles with a surface charge are more quickly recognized as foreign in comparison to smaller nanoparticles or neutral nanoparticles, respectively.40 In addition to the original composition of the nanomaterial, protein interactions with relatively large (>50 nm hydrodynamic radius) clusters or agglomerates of the nanomaterial that result in stable adsorption of a protein corona alter interactions with the innate immune system. Such larger agglomerates may more freely interact with plasma proteins such as fibrinogen and albumin, further increasing overall size, altering surface charge, reactivity (i.e., passivation of chemical surface of the nanomaterial while ‘opsonizing’ the surface making it biologically recognizable by cell surface receptors), and composition of the agglomerate. Such alteration of the nanomaterial surface may therefore confer on the nanomaterial those biological traits that allow interaction with cell surface receptors.41 If the nanomaterial is recognized as foreign, the macrophage will engulf and attempt to degrade the foreign material using a battery of oxidizing strategies that are effective against the usual microbiological threats to the body.39 However, relatively little is known about the fate and transport of smaller agglomerates or individual nanoparticles (<50nm hydrodynamic diameter) that are unable to have meaningful biochemical/physical interaction with macromolecules and are, themselves, sufficiently hydrophobic to enter the cell.
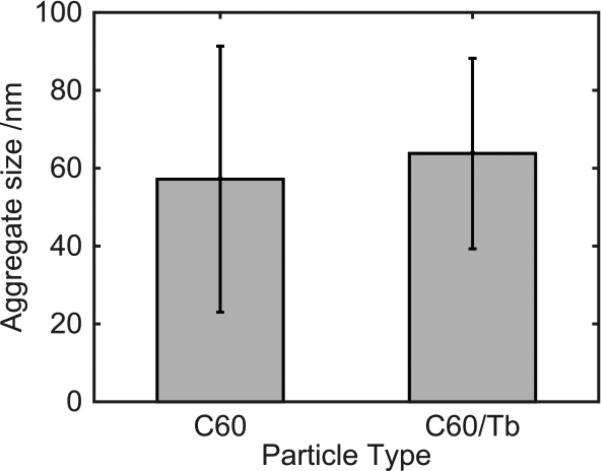
For this multifaceted study, we employed both pristine C60 fullerenes and functionalized fullerenes, of which the latter are required for imaging, particularly transmission electron microscope (TEM). For the functionalized fullerenes, we chose terbium-endohedral fullerenes Tb@C60 since these compounds contain terbium in their core, resulting in an electron-dense spot in the TEM images. We characterized the Tb@C60 nanoparticles with comparison to pristine C60 fullerenes by determining zeta potential and aggregate size. The mean surface charge (zeta potential) of the C60 fullerene was −44.3 mV, and the Tb@C60's was −64.3 mV. The first value is in line with previous observations42,43, while the endohedral fullerene shows an increased propensity to accumulate a negative charge. The most likely explanations for these findings are related to the comparatively big electron cloud surrounding the Tb atom, which has a radius between 0.177 and 0.1 nm (depending on the oxidation state and the definition used). Our analysis does not allow for a clear description of the phenomenon, which may for example be caused by a preferential aggregation of anions on the Tb@C60 surface or by charge transfer (both inter- and intramolecular). However, we don't expect that the increased average negative charge would reflect in a markedly distinct behavior when it comes to interactions with cells or aggregation, as in both cases the values indicate a similarly stable dispersion of nanoparticles. A confirmation of this assumption comes from the comparable size of the aggregates, as measured by the Malvern Zetasizer: C60 and Tb@C60 aggregates had an average hydrodynamic diameter of 91.13 and 99.31nm, respectively (see Figure S1 of the Supplementary Information). The Nanosight LM 14C indicated an average hydrodynamic radius of 55.179 ± 34.16 nm (standard deviation) and 63.765 ± 24.46 nm, respectively (Figure 1). The results show slight differences in measurements by the two instruments. This difference can be attributed to the tendency of the Zetasizer to skew towards larger aggregates. Regardless, the size determinations as well as the zeta potential determination suggest that the aggregates are similar; therefore, Tb@C60 is a reasonable substitution for C60 fullerenes for the purpose of imaging.
Figure 1.
Characterization of C60 Fullerenes and Terbium-Endohedral Fullerenes. Nanosight analysis of C60 fullerenes and Tb@C60 aggregate size: C60 fullerene aggregates were approximately 55.179 nm with a standard deviation of 34.16 nm. Tb@C60 aggregates were approximately 63.765 nm with a standard deviation of 24.46 nm.
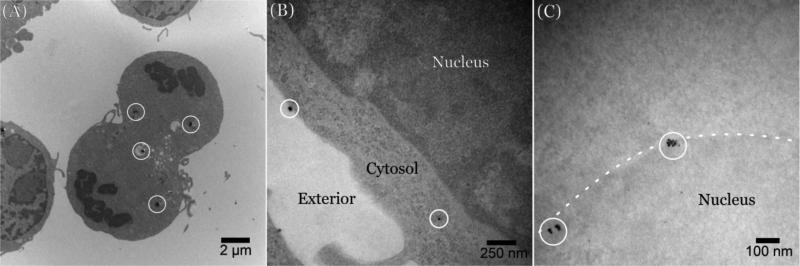
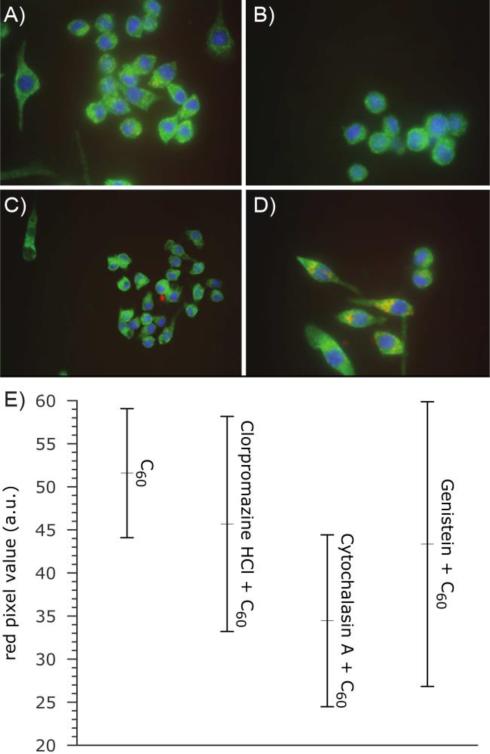
To determine the uptake and localization of fullerenes, we exposed RAW 264.7 immortalized macrophages to Tb@C60. This cellular exposure, at two different concentrations (0.1 μg/mL and 0.36 μg/mL of C60), was followed by TEM analysis of Tb@C60 distribution. Medium power electron microscope images of fixed cells in the absence of lead citrate and uranium acetate stains (which themselves deposit nanometer scale, electron dense crystals) do not provide compelling evidence that bulk transport of C60 nanoparticles is effected through phagocytosis or macropinocytosis (Figure 2). However, several Tb-enhanced clusters of C60 are readily observed either without membranous investment in the cytosol (Figure 2B), in close proximity to, or apparently integrated into cell and nuclear membranes (Figures 2B and 2C). Analysis of epifluorescent images taken from cells preincubated with C60 and detected with a monoclonal antibody raised against pristine C60 enabled detection of cytoplasmic clusters of nanomaterial. Pharmacologic inhibition of endocytosis and macropinocytosis further confirmed minimal involvement of these membrane-mediated mechanisms of cellular uptake (Figure 3). However, spectral analysis of images shows the entry of C60 clusters into the cytoplasm by other mechanisms. While it is possible that individual particles (< 1 nm hydrodynamic diameter) could plausibly pass through channels or pores, it is unlikely that the larger clusters or agglomerates (as large as 80 nm) could take advantage of this pathway. The limitations of epifluorescent microscopy and spectroscopy permit acquisition of signal emitted from both large and small aggregates of C60. However, the main focus of this manuscript is individual, and small clusters of C60 in the range < 5 nm: a size that has been postulated to account for the largest fraction of particles in most preparations of commercially available nanomaterial.44 By contrast, caveolae are estimated to be about 50 nm in diameter45,46 and induce membrane invaginations with a minimal radius of curvature ≥ 90 nm.47 Therefore, it is widely accepted that (at least for caveolin), materials need to be about 100 nm in diameter to trigger this important transport mechanism.
Figure 2.
No contrast transmission electron micrograph of RAW 264.7 immortalized macrophages containing terbium-endohedral C60. (A) High contrast clusters of C60 (inside with circles) are observed in membrane-bound structures and scattered “free” throughout the cytoplasm. (B) Higher power no contrast TEM shows two major clusters of Tb@C60 adjacent to the cell membrane (upper left) and not bounded by lipid membrane in the cytosol. (C) High power examination of the nuclear envelope (dotted line) reveals membrane-associated clusters that are not contained in classical phagocytotic structures.
Figure 3.
Epifluorescent images of immortalized RAW 264.7 macrophages showing lack of role for endocytosis in cellular accumulation of C60. (A) Baseline uptake was established following 3 hrs exposures to C60, and a monoclonal antibody (red) was used to detect and quantify uptake. The effect of classical inhibitors of active membrane-mediated processes for cell entry was determined in the presence of (B) chlorpromazine (clathrin-mediated endocytosis), (C) cytochalasin A (F-actin polymerization) or (D) genistein (caveolin-mediated endocytosis, macropinocytosis and phagocytosis). (E) Statistical analysis of images supports diffusion rather than endo-/phagocytosis as the major route by which small clusters of C60 enter the cell membrane and cytoplasm. The apparent lack of statistically significant diminution in the uptake of C60 induced by the pharmacologic inhibition of clathrin/caveolin-mediated endocytosis suggests that a significant quantity of material enters the cell through other mechanisms.
To further elucidate the observed interaction between the lipids of the cellular membrane and the fullerenes, freeze–fracture TEM was applied to cells dosed with Tb@C60 to visualize the fullerenes within the lipid membrane. Results are reported in the Supplementary Information, Figure S2. The fullerenes from the extracellular side appear to be entering the cytosol. We see aggregates of fullerenes in close proximity to the membrane as well as in the membrane space.
Therefore, the observed proximity of C60 aggregates to lipid membranes prompted investigation of the role of nano-lipid interactions. Using thermodynamically-driven atomistic models of an idealized lipid bilayer and confirmed by NMR analysis, the potential for passive diffusion as a major contributor to cellular C60 loads was examined.
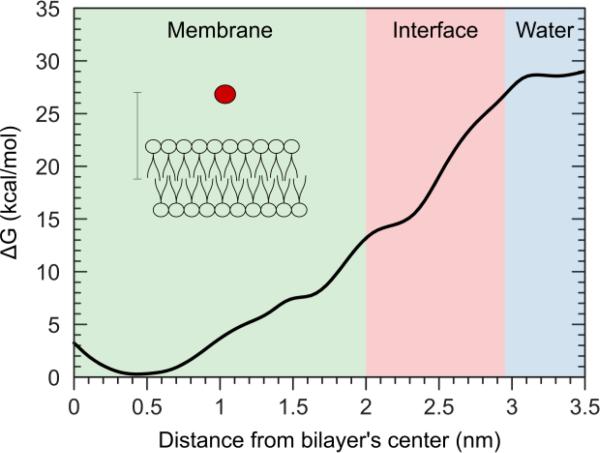
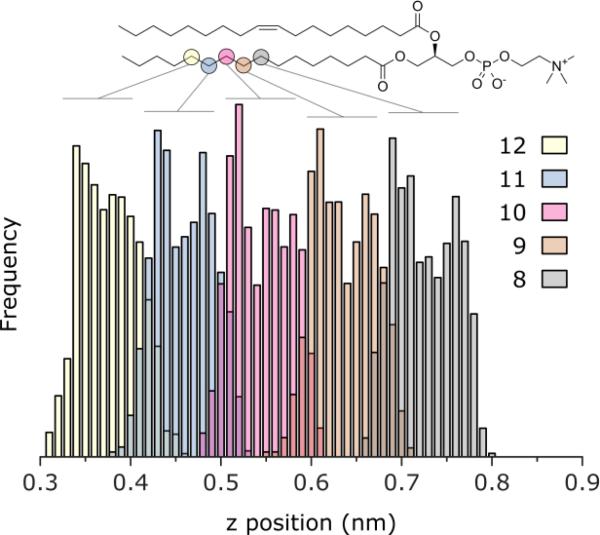
Molecular dynamics (MD) simulations were utilized to gain insights on the interactions of a cellular membrane with nanoparticles. A fully hydrated symmetric lipid bilayer of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) in the presence of a single C60 as well as agglomerates of C60 molecules was analyzed. The interactions between a single C60 molecule and the membrane were evaluated by computing the free energy (FE) as a function of the projection on the bilayer normal of the distance between the C60 molecule and the center of the bilayer. Figure 4 reports the reconstructed FE profile of the permeation of a single C60 in the POPC bilayer. C60 is readily absorbed into the hydrophobic part of the lipid bilayer with a marked increase in the stability of the system (approximately 30 kcal/mol) and with minimal disruption of the hydrophobic environment. The preferred location (minimum of the FE profile) does not correspond to the center of the bilayer, but is shifted from the central plane towards the lipids headgroups by 0.4-0.6 nm. This distance corresponds to the position of the 9th and 10th carbon of the saturated alkyl chains of POPC, as shown by plotting the equilibrium distribution of the atoms’ distance from the bilayer middle plane (Figure 5). The results indicate the permeation process is thermodynamically favored and barrierless.
Figure 4.
Free energy profile of the permeation of a C60 fullerene into a POPC bilayer as function of the distance on the membrane normal between the fullerene and the bilayer center.
Figure 5.
Distribution of selected carbon atoms of the saturated chain of POPC. Shown as function of the distance on the bilayer normal. Zero corresponds to the center of the bilayer.
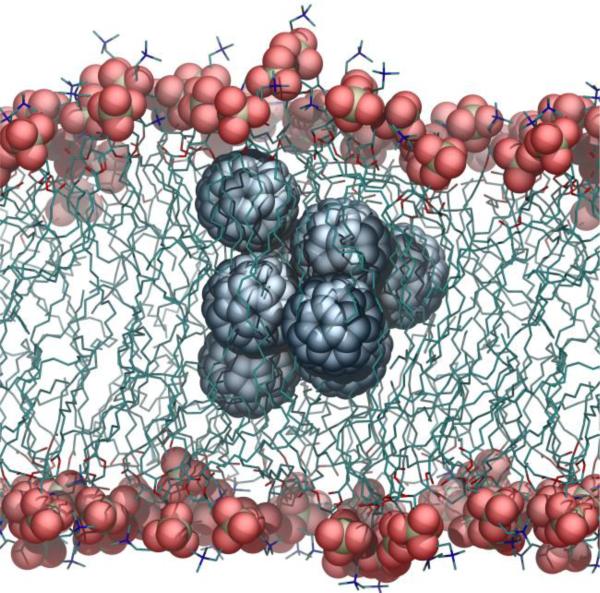
To highlight the differences in interactions between the lipid bilayer with a single or a small cluster of C60 fullerenes, we monitored the properties of a POPC membrane when a cluster of eight fullerenes (approximately 3 nm in diameter) is present in the hydrophobic region. The cluster markedly perturbed not only the acyl chains structure and organization, but also the disposition of the headgroups, causing a localized bulging of the bilayer (Figure 6). This perturbation affects the entire lipid molecules, but is particularly marked in the region that extends from the center of the bilayer to the 9th carbon (about 0.7 nm).
Figure 6.
Small cluster of C60 fullerenes in a POPC lipid bilayer.
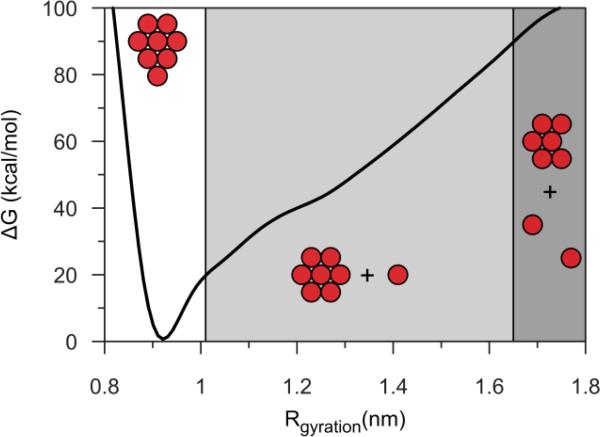
Due to favorable interaction however, fullerene clusters may be short lived in the hydrophobic region of the membrane. Since the dissolution of the C60 clusters could be thermodynamically favored, but slow (relatively to the simulation scale) and therefore hard to observe with an unbiased MD, we employed biased MD to study the behavior of the system during the disaggregation of the C C60 cluster in the membrane. Namely, we explored the changes associated with a change of the cluster radius of gyration. With this approach we are be able to overcome both energetic and kinetic barriers that may affect the solubility of C60 fullerenes. The results show however, that the aggregates are not a metastable state, and disaggregation of the clusters is an extremely unlikely process. As an indicator of adhesion energy, the free-energy profile (Figure 7) shows a clear well around the equilibrium radius (0.915 nm). The energy increases rapidly for small values of the radius of gyration (< 0.9 nm), in agreement with the low compressibility of the cluster; at higher values (about 1.01 nm, which corresponds to approximately 23 kcal/mol) we observe a clear change in slope, which corresponds to the detachment of a single C60 from the cluster. In all cases we see the preferential disaggregation of a C60 situated near the center of the bilayer, likely due to the lower density of that region. At even higher values (approximately 1.85 nm), we observed the detachment of a second C60, but this process requires very high energy (about 90 kcal/mol), making it an extremely unlikely event. Because the detachment of even one or two C60 molecules is a very unfavorable event (23 and 90 kcal/mol, respectively), the disaggregation of a small cluster of C60 in the membrane is a very unlikely occurrence. In conclusion, MD simulations show that particles readily diffuse into biological membranes. A single fullerene causes limited perturbation of the membrane, but clusters of C60 determine bulging and deformation of the membrane. To further analyze the interactions between nanoparticles and lipid bilayers, nuclear magnetic resonance experiments were conducted on vesicles of POPC containing C60.
Figure 7.
Free energy profile of fullerenes with various radii of gyration in a POPC bilayer. A clear well is identified around the equilibrium radius of 0.915 nm. Energy increases rapidly at smaller radii values and shows a change in slope at higher values, corresponding to detachment of a single C60.
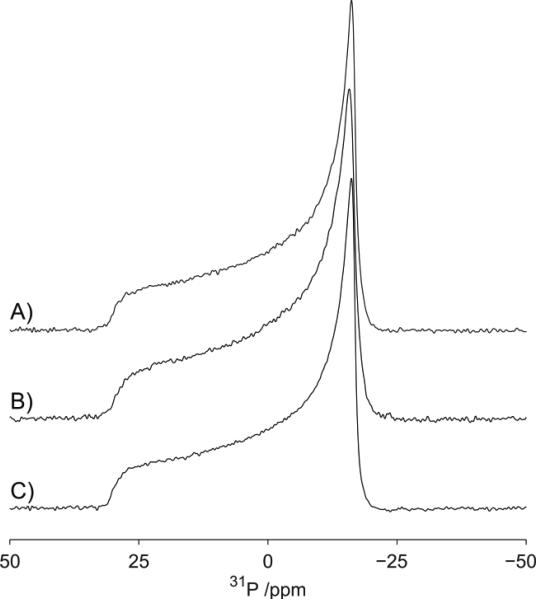
Deuterium and phosphorous NMR spectra were recorded to determine the influence of C60 on the bulk phase behavior of the POPC bilayers irrespective of the location of the C60 within the bilayer as well as their influence on the local structure and dynamics. To assess how C60 perturbs the phase behavior of the lipids48 and assess how its presence alters the structure and dynamics49–53 of the lipid headgroup we have undertaken a 31P NMR study of C60 in lipid vesicles composed of d31-POPC vesicles. The 31P spectra of POPC in absence of C60 is shown in Figure 8A and is consistent with previous studies, with an axially symmetric powder pattern characterized by a chemical shielding anisotropy of approximately −30 ppm. The spectrum is typical for POPC in a liquid crystalline bilayer phase. Because the addition of C60 to a concentration of 10 μg/mL (Figure 8B) does not cause any significant changes to the 31P spectrum of the POPC, we conclude that at these concentrations the C60 has little effect on the structural (geometric) and dynamics properties of the headgroups of the bilayer. The 31P spectra of POPC for increased concentration of C60 (100 μg/mL) are reported in Figure 8C and show a small reduction in the chemical shielding anisotropy, as evidenced by the π/2 edge of the powder pattern shifting from −16.18 to −15.77 ppm. Such a reduction in 31P chemical shielding anisotropy is consistent with small changes in lipid headgroup geometry or dynamics, or both.49–53 Our results therefore, reveal a slight perturbation in head position (geometry) and dynamics at high concentrations. In contrast to studies of surface-modified fullerenes, such as fullerenols, at the concentrations studied, we see no evidence for the disruption of the bilayer such as has been observed for other bilayer systems.54
Figure 8.
Phosphorous spectra of POPC Vesicles by NMR. Prepared in the presence of fullerenes, (A) at 0 μg/mL, (B) 10 μg/mL, (C) and 100 μg/mL. Data acquired at 20 °C.
Furthermore, the observed changes in the distribution of intensity in the 31P spectrum reflect differences in the orientational distribution of the lipids within the sample48,55–57 with an increase in intensity in the low-field region corresponding to an enhanced probability of lipids aligning with their long axis parallel to the magnetic field. On the contrary, typical static 31P NMR spectra of lipid vesicles exhibit a reduced intensity in the low-field region due to the preferred orientation of lipids with their long axes perpendicular to the applied magnetic field, arising from their intrinsic diamagnetic anisotropy. This observation indicates that at concentration of 100 μg/mL the C60 enhances the rigidity of the membrane reducing its ability to deform, in this case in response to the applied magnetic field.
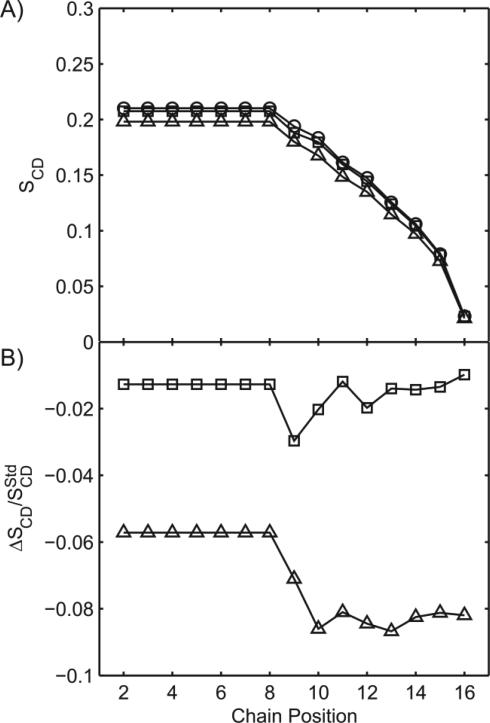
The 2H spectra show that the presence of C60 especially at a concentration of 100 μg/mL also results in an increase in chain mobility, as indicated by the reduction in the width of the quadrupolar splittings and by the order parameter, SCD. The order parameter profiles (Figure 9) obtained for the d31-POPC bilayer in presence of 0, 10 and 100 μg/mL of C60, reveal a small but reproducible concentration-dependent reduction in order parameter. In absence of C60, the observed behavior is consistent with previous research, specifically the order parameter decreases along the length of the acyl chain, indicative of the increased mobility typically found within the center of the lipid bilayer.24 At the lowest C60 concentration (10 μg/mL) the reduction is mostly observed at the 9th and 10th position in the chain. At the highest concentration of C60 (100 μg/mL) a larger reduction in order parameter is observed from the 8th carbon to the methyl end of the acyl chain. Thus, with increasing C60 concentrations, the sn-1 chains become on average more mobile (Figure 9). This is consistent with C60 increasing the disorder and mobility towards the center of the lipid bilayer.
Figure 9.
(A) Order parameter profile for d31-POPC prepared in the presence of fullerenes, at 0 μg/mL (circle), 10 at μg/mL (square), and 100 μg/mL C60 (triangle). (B) Perturbation in order parameter with respect to vesicles in absence of C60. Data acquired at 20 °C.
The agreement between NMR data and simulations suggests that at the lower concentration, the C60 molecules tend to enter the lipid bilayer individually, or possibly as a pair, causing only a limited alteration in the position of aliphatic chains and no changes in the headgroups position. At higher concentration of C60, the agreement between the observed SCD plot and the perturbation of the bilayer computed with MD (Figure 6) suggests the presence of aggregates inside the bilayer.
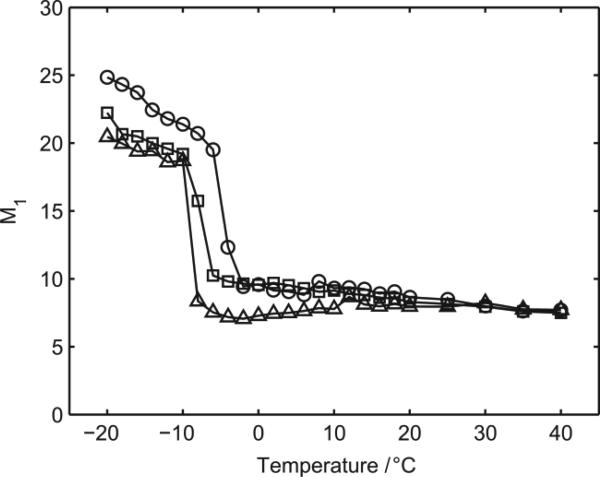
As a consequence of the induced disorder in the lipid chain as function of the concentrations of nanoparticles, we hypothesized a shift in the phase transition temperature toward lower values. This hypothesis was explored with deuterium NMR studies at different temperatures (Figure 10). As the temperature increases, we see a progressive reduction in 1st moment that represents the weighted average deuterium splitting; it provides a convenient unit with which to parameterize the width of the spectra and has been extensively used to characterize lipid phase transitions.22 At −5 °C a rapid reduction (occurring in less than 3 °C) in the 1st moment is observed in POPC bilayers in the absence of the fullerene C60. This rapid reduction arises from the increased chain mobility associated with the cooperative disordering of the lipid chains as the lipids pass through the phase transition between the gel and liquid crystalline phases. The phase transition temperature is similar to previously reported values for d31-POPC.24 Above this temperature, the 1st moment continues to progressively decline as the temperature increases. The addition of the fullerene C60 to the bilayers at concentrations of 10 μg/mL and 100 μg/mL results in a reduction in the phase transition to −8 °C and −9 °C, respectively, indicative of the C60 disrupting the stability of the gel phase. Within the accuracy of the measurements, the temperature range over which the phase transition occurs appears insensitive to the concentration, spanning 3-4 °C in each case, suggesting that the presence of the C60 within the sample has no effect on the cooperative nature of the phase transition. Both above and below the phase transition, the presence of the fullerene C60 within the sample leads to enhanced mobility within the acyl chains as characterized by a lower 1st moment for a given temperature, consistently with our initial hypothesis.
Figure 10.
Temperature dependence of the first moment of deuterium NMR spectra recorded from d31-POPC in the presence of 0 μg/mL (circle), 10 μg/mL (square) and 100 μg/mL C60 (triangle).
Conclusions
The aim of this study was to determine the mechanisms of uptake of C60 fullerenes into compartments of living cells, by using a combination of classical biological, biophysical and computational techniques. Modes of entry studied were endocytosis/pinocytosis and passive permeation of cellular membranes. Cultures of RAW 264.7 immortalized murine macrophage were used as model for immune-competent cells. These cells are well suited to endo-/phagocytosis. Terbium-endohedral C60 fullerenes were employed to provide identification of individual and clusters of nanoparticles in situ. Using transmission electron microscopy, particles were observed in association with or in cell/nuclear membranes as well as unbound by biological membranes in the cytoplasm of immortalized macrophages. Although the cell type chosen for this study is well suited to endo-/phagocytosis, our experiments could not find evidence for a major contribution by this mode of entry.
Since particles were observed in and close to lipid bilayers in vitro, we further analyzed the interactions between nanoparticles and lipid bilayers using molecular dynamics simulations. Computational modeling of C60 interacting with a POPC lipid bilayers, representative of the cellular membrane, show that particles readily diffuse into biological membranes and find a thermodynamically stable equilibrium in an eccentric position within the bilayer. At low concentrations, fullerenes enter the membrane individually and cause limited perturbation; however, at higher concentrations the clusters in the membrane causes deformation of the membrane.
These findings are further supported by NMR experiments of model membranes that reveal deformation of the cell membrane, confirming distortions in membrane morphology that are consistent with the results of atomistic models. However, neither the model nor NMR experiments are able to explain the mechanism of release from lipid bilayers that result in the presence of unbounded particles in the cytoplasm.
Future studies should be aimed at atomistic and molecular mechanisms that take into account integral proteins, the glycocalyx and other complexities found in living cells. It is also important to note that higher concentrations of C60 are required for detection of nanomaterial in nmr studies. One possible confounder might be that accumulation in biological membranes may only occur after saturation of other cellular depots. Additional studies are required to determine whether or not prima faciae accumulation of C60 in the intramembrane space is of primary biological importance.
This study is the first of its kind that suggests a marginal role for endo-/phagocytosis in the cellular uptake of C60. We present new evidence of a major contributor to cellular particle loads through the passive entry and integration of C60 particles into biological membranes. NMR and electron microscopy confirms the ability of C60 to enter and distort membrane morphology their presence in unbound clusters throughout the cell. Moreover, though incomplete, the results of this study provide a plausible mechanism by which particles may integrate into membranes and later escape through as yet unidentified pathways. The biological significance of these findings remains to be determined.
Supplementary Material
ACKNOWLEDGMENT
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number T32ES007062 (KAR), ES R01 088546 (MAP). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
MAP and KAR thank Tessa Adzemovic and Jennifer Fernandez for their technical assistance.
Footnotes
Supplementary Information Available:
Figure S1. Characterization of C60 Fullerenes and Terbium-Endohedral Fullerenes.
Figure S2. Freeze-Fracture Transmission Electron Microscopy on RAW 264.7 Cells.
Figure S3. RGB channels of the epifluorescent images of immortalized RAW 264.7 macrophages shown in Figure 3 of the main text.
Figure S4. Epifluorescent and RGB channels of the control cells with undetectable intracellular fullerene (red channel).
REFERENCES
- 1.Aschberger K, Johnston HJ, Stone V, Aitken RJ, Tran CL, Hankin SM, Peters SAK, Christensen FM. Regul. Toxicol. Pharmacol. 2010;58:455–473. doi: 10.1016/j.yrtph.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen GD, Roursgaard M, Jensen KA, Poulsen SS, Larsen ST. Basic Clin. Pharmacol. Toxicol. 2008;103:197–208. doi: 10.1111/j.1742-7843.2008.00266.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnston HJ, Hutchison GR, Christensen FM, Aschberger K, Stone V. Toxicol. Sci. 2010;114:162–182. doi: 10.1093/toxsci/kfp265. [DOI] [PubMed] [Google Scholar]
- 4.Zakharian TY, Seryshev A, Sitharaman B, Gilbert BE, Knight V, Wilson LJ. J. Am. Chem. Soc. 2005;127:12508–12509. doi: 10.1021/ja0546525. [DOI] [PubMed] [Google Scholar]
- 5.Montellano A, Ros TD, Bianco A, Prato M. Nanoscale. 2011;3:4035–4041. doi: 10.1039/c1nr10783f. [DOI] [PubMed] [Google Scholar]
- 6.Chang R, Violi A. J Phys Chem B. 2006;110:5073–5083. doi: 10.1021/jp0565148. [DOI] [PubMed] [Google Scholar]
- 7.Oberdörster G, Oberdörster E, Oberdörster J. Environ. Health Perspect. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 9.Bosi S, Da Ros T, Spalluto G, Prato M. Eur. J. Med. Chem. 2003;38:913–923. doi: 10.1016/j.ejmech.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Markovic Z, Todorovic-Markovic B, Kleut D, Nikolic N, Vranjes-Djuric S, Misirkic M, Vucicevic L, Janjetovic K, Isakovic A, Harhaji L, Babic-Stojic B, Dramicanin M, Trajkovic V. Biomaterials. 2007;28:5437–5448. doi: 10.1016/j.biomaterials.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Deguchi S, Alargova RG, Tsujii K. Langmuir. 2001;17:6013–6017. [Google Scholar]
- 12.Spohn P, Hirsch C, Hasler F, Bruinink A, Krug HF, Wick P. Environ. Pollut. 2009;157:1134–1139. doi: 10.1016/j.envpol.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, Cho M, Fortner JD, Lee J, Huang C-H, Hughes JB, Kim J-H. Environ. Sci. Technol. 2009;43:108–113. doi: 10.1021/es8019066. [DOI] [PubMed] [Google Scholar]
- 14.dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. PLoS One. 2011;6:e24438. doi: 10.1371/journal.pone.0024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nabi IR, Le PU. J. Cell Biol. 2003;161:673–677. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohdanowicz M, Grinstein S. Physiol. Rev. 2013;93:69–106. doi: 10.1152/physrev.00002.2012. [DOI] [PubMed] [Google Scholar]
- 17.Braun S, Kalinowski H-O, Berger S. 150 and more basic NMR experiments: a practical course. expanded ed. Vol. 2. Wiley-VCH; Weinheim: 1998. [Google Scholar]
- 18.van Beek JD. J. Magn. Reson. 2007;187:19–26. doi: 10.1016/j.jmr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 19.McCabe MA, Wassail SR. Solid State Nucl. Magn. Reson. 1997;10:53–61. doi: 10.1016/s0926-2040(97)00024-6. [DOI] [PubMed] [Google Scholar]
- 20.Davis JH. Biochim. Biophys. Acta BBA - Rev. Biomembr. 1983;737:117–171. doi: 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- 21.Salmon A, Dodd SW, Williams GD, Beach JM, Brown MF. J. Am. Chem. Soc. 1987;109:2600–2609. [Google Scholar]
- 22.Davis JH. Biophys. J. 1979;27:339–358. doi: 10.1016/S0006-3495(79)85222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnett LJ, Muller BH. J. Chem. Phys. 1971;55:5829–5831. [Google Scholar]
- 24.Seelig J, Seelig A. Q. Rev. Biophys. 1980;13:19–61. doi: 10.1017/s0033583500000305. [DOI] [PubMed] [Google Scholar]
- 25.Nezil FA, Bloom M. Biophys. J. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slichter CP. Principles of Magnetic Resonance. Vol. 1. Springer; Berlin Heidelberg: 1990. Berlin, Heidelberg. [Google Scholar]
- 27.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonomi M, Branduardi D, Bussi G, Camilloni C, Provasi D, Raiteri P, Donadio D, Marinelli F, Pietrucci F, Broglia RA, Parrinello M. Comput. Phys. Commun. 2009;180:1961–1972. [Google Scholar]
- 29.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, M. AD., Jr J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price DJ, Brooks CL. J. Chem. Phys. 2004;121:10096. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 32.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 33.Brünger AT. X-PLOR: Version 3.1 : a System for X-ray Crystallography and NMR. Yale University Press; 1992. [Google Scholar]
- 34.Ryckaert J-P, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 35.Feller SE, Zhang Y, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 36.Martyna GJ, Tobias DJ, Klein ML. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 37.Barducci A, Bussi G, Parrinello M. Phys. Rev. Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 38.Jo S, Kim T, Iyer VG, Im W. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 39.Luo Y-H, Chang LW, Lin P, Luo Y-H, Chang LW, Lin P. BioMed Res. Int. BioMed Res. Int. 2015;2015:e143720. 2015. [Google Scholar]
- 40.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Mol. Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. J. Am. Chem. Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 42.Brant J, Lecoanet H, Hotze M, Wiesner M. Environ. Sci. Technol. 2005;39:6343–6351. doi: 10.1021/es050090d. [DOI] [PubMed] [Google Scholar]
- 43.Hyung H, Kim J-H. Water Res. 2009;43:2463–2470. doi: 10.1016/j.watres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Colvin VL. Nat. Biotechnol. 2003;21:1166–1170. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- 45.Marsh M, McMahon HT. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- 46.Parton RG, Simons K. Nat. Rev. Mol. Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 47.Sens P, Turner MS. Biophys. J. 2004;86:2049–2057. doi: 10.1016/S0006-3495(04)74266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seelig J. Biochim. Biophys. Acta-Biomembr. 1978;515:105–140. doi: 10.1016/0304-4157(78)90001-1. [DOI] [PubMed] [Google Scholar]
- 49.Seelig J. Q. Rev. Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 50.Lindström F, Williamson PTF, Gröbner G. J. Am. Chem. Soc. 2005;127:6610–6616. doi: 10.1021/ja042325b. [DOI] [PubMed] [Google Scholar]
- 51.Semchyschyn DJ, Macdonald PM. Magn. Reson. Chem. 2004;42:89–104. doi: 10.1002/mrc.1333. [DOI] [PubMed] [Google Scholar]
- 52.Bechinger B, Seelig J. Biol. Chem. Hoppe. Seyler. 1990;371:758–758. [Google Scholar]
- 53.Doux JPF, Hall BA, Killian JA. Biophys. J. 2012;103:1245–1253. doi: 10.1016/j.bpj.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brisebois PP, Arnold AA, Chabre YM, Roy R, Marcotte I. Eur. Biophys. J. 2012;41:535–544. doi: 10.1007/s00249-012-0809-5. [DOI] [PubMed] [Google Scholar]
- 55.Pott T, Dufourc EJ. Biophys. J. 1995;68:965–977. doi: 10.1016/S0006-3495(95)80272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picard F, Paquet M-J, Levesque J, Bélanger A, Auger M. Biophys. J. 1999;77:888–902. doi: 10.1016/S0006-3495(99)76940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashworth Briggs EL, Gomes RGB, Elhussein M, Collier W, Stuart Findlow I, Khalid S, McCormick CJ, Williamson PTF. Biochim. Biophys. Acta - Biomembr. 2015;1848:1671–1677. doi: 10.1016/j.bbamem.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.