Abstract
PcINF1 was previously found to induce pepper defense response by interacting with SRC2-1, but the underlying mechanism remains uninvestigated. Herein, we describe the involvement of SGT1 in the PcINF1/SRC2-1-induced immunity. SGT1 was observed to be up-regulated by Phytophthora capsici inoculation and synergistically transient overexpression of PcINF1/SRC2-1 in pepper plants. SGT1-silencing compromised HR cell death, blocked H2O2 accumulation, and downregulated HR-associated and hormones-dependent marker genes’ expression triggered by PcINF1/SRC2-1 co-overexpression. The interaction between SRC2-1 and SGT1 was found by the yeast two hybrid system and was further confirmed by bimolecular fluorescence complementation and co-immunoprecipitation analyses. The SGT1/SRC2-1 interaction was enhanced by transient overexpression of PcINF1 and Phytophthora capsici inoculation, and SGT1-silencing attenuated PcINF1/SRC2-1 interaction. Additionally, by modulating subcellular localizations of SRC2-1, SGT1, and the interacting complex of SGT1/SRC2-1, it was revealed that exclusive nuclear targeting of the SGT1/SRC2-1 complex blocks immunity triggered by formation of SGT1/SRC2-1, and a translocation of the SGT1/SRC2-1 complex from the plasma membrane and cytoplasm to the nuclei upon the inoculation of P. capsici. Our data demonstrate that the SGT1/SRC2-1 interaction, and its nucleocytoplasmic partitioning, is involved in pepper’s immunity against P. capsici, thus providing a molecular link between Ca2+ signaling associated SRC2-1 and SGT1-mediated defense signaling.
Elicitins, conserved extacellular proteins in Phytophthora and Pythium spp., which might have evolved to the benefit of pathogens, trigger immunity in the host plants such as Nicotiana benthamiana due to the presence of ligands that have been evolved under the selective pressure. Unlike the well-characterized typical PAMPs and effectors, elicitins are rather narrowly conserved and transported into plant cells by receptor-mediated endocytosis1 possibly through a clathrin-mediated way2,3, thereby triggering hypersensitive reaction (HR) cell death in the host plants, which is a hallmark of effector-triggered immunity (ETI)4,5,6,7,8,9. These features suggest their roles as elicitors in the continuum between typical PAMPs and effectors. Upon treatment with elicitins, cellular responses including depolarization of the plasma membrane, potassium and chloride ion efflux, a large calcium ion influx, alkalization of the extracellular medium, and production of either reactive oxygen species or nitric oxide are activated and culminated in the HR and plant immunity10. These processes are believed to be initiated by the recognition and binding of elicitins to their corresponding receptors in plant plasma membranes. Although NgRLK1, a receptor like kinase, and NbLRK1, a lectin-like receptor kinase, both in Nicotiana, have been shown to interact with the elicitins capsicein and INF1, respectively11,12,13. However, the receptors of most elicitins have not been identified. PcINF1, a member of the elicitins from Phytophthora capsici exhibiting a high sequence homology with orthologs in other Phytophthora species, was found to trigger HR cell death in tobacco. Additionally, our previous study further found that PcINF1 triggers HR cell death and defense response in pepper and Nicotiana benthamiana plants by interacting with SRC2-114 which was found to be induced by pathogen inoculation as well as by treatment with abiotic stresses. Noticeably, the SRC2-1 contains a conserved C2 domain, which was originally identified in Ca2+-dependent isoforms of protein kinase C and is present in most Ca2+-binding proteins involved in protein-protein interactions, binding of phospholipids, and targeting of proteins to the membrane in response to Ca2+ signalling15,16,17. Since there is no any kinase domain in SRC2-1 as in NgRLK1, NbLRK1 and other typical PRRs, and C2 domains have been implicated in protein-protein interactions2. SRC2-1 may form complexes with other proteins such as receptor-like kinases to elicit defense response in host plants. However, little is known about the components of these complexes, as well as the molecular events leading to the defense response triggered by PcINF1.
The highly conserved eukaryotic co-chaperone SGT1 (suppressor of the G2 allele of skp1) is an important protein component shared by PTI (PAMP-triggered immunity)3 and ETI (effector-triggered immunity)18,19,20,21,22. When involved in ETI, SGT1 mediates R proteins, such as Rps4, N5, Rpi-blb220, Rx123, R3a24, RPS425, and MLA26, controlling their stability and activity as well as nucleocytoplasmic balance22,27. SGT1 also facilitates the recognition of effectors by R proteins28. However, some R proteins might function independently of SGT129. SGT1 has also been found to participate in the regulation of plant PTI. For example, SGT1 is significantly induced by Xcc-derived flg22, a typical PAMP, as well as the flg22 domain-containing hook-associated protein (Fla) in citrus and in Nicotiana benthamiana30,31. SGT1 functions as an integral component in both ETI and PTI by interacting with other immunity-associated components such as RACK1, Rac1, RAR1, Rboh, HSP90, and HSP7022,32,33,34,35,36,37,38,39 to create multi-protein networks. Recently, it was found that SGT1 might act as a receptor for the type III effector AvrBsT6 by interacting with receptor-like cytoplasmic kinase1 in pepper. How SGT1 mediates the signaling of the network/s, however, it is not fully understood; other interacting partners of SGT1 may exist. Further identification and characterization of binding partners may help to elucidate the mechanisms of plant immunity.
In the present study, SGT1 was found to be essential in the defense response induced by PcINF1 by interacting with SRC2-1. Furthermore, the translocation of the SGT1/SRC2-1 complex from membrane and cytoplasm into nuclei was induced by pathogen inoculation and is required for their roles in immunity. Additionally, a functional association between the two proteins was observed through their synergistic response to pathogen inoculation.
Results
Transcriptional expression of SGT1 in response to Phytophthora capsici inoculation and PcINF1/SRC2-1 transient overexpression
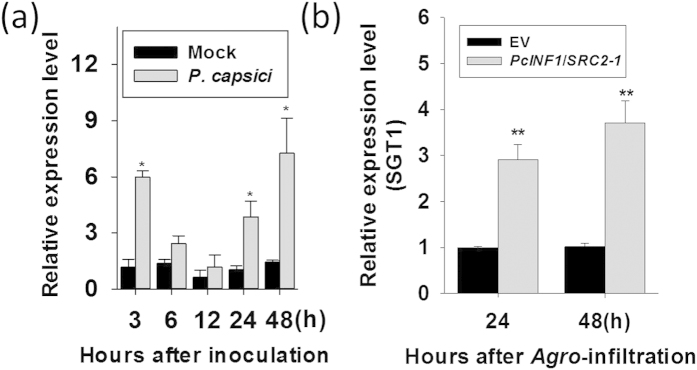
Phytophthora capsici is a causal agent of phytophthora blight of pepper, one of the most important diseases in pepper production, and elicitins are conserved extracellular proteins in P. capsici and other Phytophthora and Pythium spp. triggering immunity in the host plants. Our previous study found that SRC2-1 from pepper plants perceive and interact with PcINF1 and trigger HR cell death and immunity14, but the underlying mechanism remains unknown. To isolate potential SRC2-1-interacting partners, a pepper cDNA library generated from P. capsici-inoculated pepper leaves was previously screened with SRC2-1 as a bait using GAL4-based yeast two-hybrid system, one positive clone acquired was identified to be SGT1. Since SGT1 have been implicated in plant immunity40, the expression of SGT1 was assayed by real-time PCR in P. capsici-inoculated pepper plants to test its potential role in immunity against P. capsici inoculation. The result showed that the expression of SGT1 was enhanced compared to the mock treatment (Fig. 1a). Our previous study found that PcINF1, an elicitin from P. capsici, triggered HR cell death and defense response in pepper plants by interacting with SRC2-114. To test if SGT1 is involved in the immunity mediated by PcINF1/SRC2-1, an Agrobacterium-mediated transient expression system was employed to characterize the effects of PcINF1/SRC2-1 simultaneously transient overexpression on the expression of SGT1. Agrobacterial cells containing 35S::PcINF1/35S::SRC2-1 fusion vector were infiltrated into pepper leaves, and the transcript level of SGT1 was assayed by real-time RT-PCR (Fig. 1b). The result showed that the transcript levels of PcINF1 and SRC2-1 were both significantly increased in the pepper leaves transiently expressing PcINF1 and SRC2-1 (Supplementary Fig. S1), additionally, SGT1 was also upregulated by the PcINF1/SRC2-1 transient overexpression, indicating that SGT1 may be involved in pepper immunity mediated by PcINF1/SRC2-1.
Figure 1. Transcript levels of SGT1 in response to Phytophthora capsici infection and the transient overexpression of PcINF1/SRC2-1 in pepper leaves.
(a) The SGT1 transcript levels detected at different time points in pepper leaves inoculated with P. capsici. The 4-weeks-old pepper plants growing in the greenhouse were used for P. capsici infection. After P. capsici inoculation, the leaves next to the infected leaves of the pepper plants were harvested for RNA extraction at different time points. Pepper leaves treated with MgCl2 was used as a negative control (Mock). (b) The SGT1 transcript levels in pepper leaves transiently overexpressing the PcINF1/SRC2-1 fusion protein. Agrobacterial cells harboring the construct containing the PcINF1/SRC2-1 fusion vector or the empty vector (EV) were infiltrated into the leaves of the healthy pepper plants. 24 h and 48 h after Agro-infiltration, the Agro-infiltrated pepper leaves were collected for total RNA extraction. (a,b) Experiments were repeated with at least four independent biological repetitions. Error bars indicate standard deviation (SD). Asterisks indicate a significant difference using a Fisher’s protected least significant difference (LSD) test. *P < 0.05; **P < 0.01.
The Silencing of SGT1 compromises the immunity triggered by PcINF1/SRC2-1
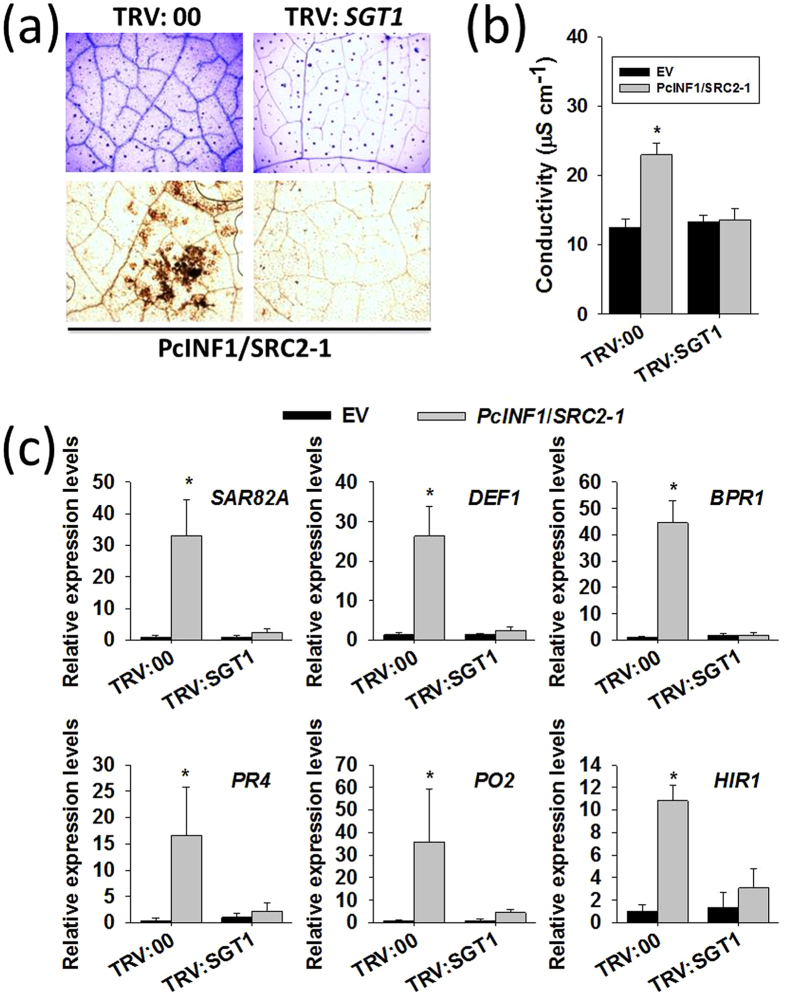
To further confirm the potential role of SGT1 in the defense response triggered by PcINF1/SRC2-1, the silencing of SGT1 was performed by a VIGS (virus induced gene silence), which has been successfully used in Solanaceae, such as pepper, tomato, and tobacco14,41,42,43. To avoid the mistargeting of any potential orthologs of SGT1, fragment of 213 bps in length corresponding to a specific region of the 3′UTRs of SGT1 was chosen to construct VIGS vectors (The specificity of the region was confirmed by BLASTN in pepper genome databases: http://passport.pepper.snu.ac.kr/?t=PGENOME and http://peppersequence.genomics.cn/page/species/index.jsp). The efficiency of SGT1 silencing was examined by the real time RT-PCR and the result showed that the expression of SGT1 was significantly decreased in the TRV:SGT1 pepper plants (Supplementary Fig. S2). In SGT1-silenced pepper plants, the HR cell death manifested with the darker trypan blue staining triggered by the transient co-overexpression of PcINF1/SRC2-1 significantly decreased compared to the control plants (Fig. 2a), and was consistent with an increased ion leakage in the PcINF1/SRC2-1 co-overexpressed and SGT1-silenced pepper plants (Fig. 2b). In addition, the H2O2 accumulation triggered by the co-overexpression of PcINF1/SRC2-1 manifested by the DAB staining (Fig. 2a), which is closely related to HR cell death by previous studies40,44, was also found to be compromised by SGT1 silencing. Furthermore, the SA-dependent SAR82A, BPR1 and PR4, JA-dependent DEF1, HR associated marker gene HIR1 and defense-associated PO2 (triggered by the PcINF1/SRC2-1 co-overexpression) were also consistently compromised by the silencing of SGT1 (Fig. 2c). All these results suggest that SGT1 is involved in pepper immunity mediated by PcINF1/SRC2-1.
Figure 2. Silencing of SGT1 restrained the defense response triggered by PcINF1/SRC2-1 complex in pepper plants.
(a) Trypan blue and DAB staining of SGT1-silenced and unsilenced pepper leaves transiently expressing PcINF1/SRC2-1 vector. Trypan blue and DAB staining were performed 48 h after Agro-infiltration to detect the cell death and H2O2 content. (b) Electrolyte leakage measurement of SGT1-silenced pepper leaves transiently expressing PcINF1/SRC2-1. The transcript level of SGT1 in pepper leaves was first silenced using the VIGS technology. The GV3101 cells harboring the constructs of PcINF1/SRC2-1 or empty vector were infiltrated into the leaves of SGT1-silenced and unsilenced pepper leaves. The leaves were collected 48 h after Agro-infiltration for the electrolyte leakage measurement. (c) Relative expression levels of defense-related marker genes in pepper leaves 48 h after Agro-infiltration with GV3101 cells harboring the PcINF1/SRC2-1 fusion vector, compared to empty vector, which was set to a relative expression level of “1”. CaActin and 18S ribosomal RNA were used to normalize the transcript level. (b,c) Experiments were repeated with at least four independent biological repetitions. Error bars indicate SD. Asterisks indicate a significant difference using an LSD test. *P < 0.05; **P < 0.01.
Simultaneously transient overexpression of SGT1 and SRC2-1 induced heightened cell death and defense responses in pepper leaves
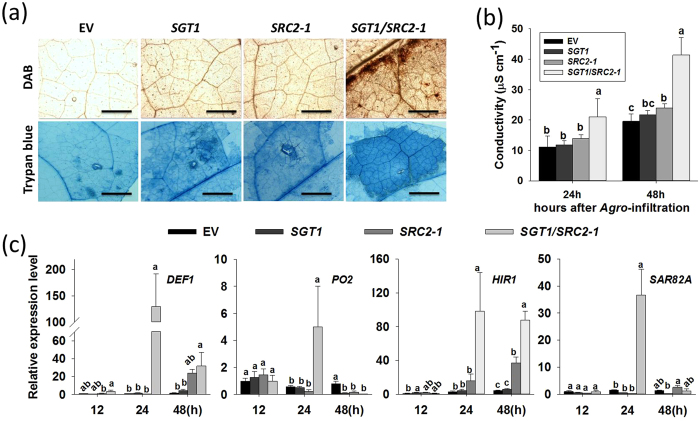
Since both SRC2-1 and SGT1 are involved in the pepper defense response against P. capsici inoculation as well as the simultaneously transient overexpression of PcINF1/ SRC2-114, it appears that these two proteins are functionally related. To test this possibility, the effect of the simultaneously transient co-overexpression of SRC2-1 and SGT1 on the defense response in pepper plants was assayed. Agrobacterial cells containing 35S::SGT1 and 35S::SRC2-1 were infiltrated into pepper leaves individually or simultaneously. Infiltrated pepper leaves were then harvested at either 48 HAI (for trypan blue staining, DAB staining, and conductivity measurements), or at 12, 24, and 48 HAI for total RNA isolation to detect the transcriptional expression of defense-associated marker genes. Our results showed that both SGT1 and SRC2-1 were successfully overexpressed in the pepper leaves infiltrated with agrobacteria cells carrying 35S::SGT1 and 35S::SRC2-1 (Supplementary Fig. S3), besides, transient co-overexpression of SGT1 and SRC2-1 in pepper leaves induced significant HR-like cell death displayed by the trypan blue staining, the accumulation of H2O2, and the ion leakage measurement (Fig. 3a/b). This was coupled with significant up-regulation of HR-marker genes HIR1, SA-dependent SAR82A, JA-dependent DEF1, and active oxygen metabolism-associated PO2. Up-regulation of these genes occurred at 12, 24, or 48 HAI with maximum expression occurring at different time points for each gene (Fig. 3c). Additionally, the heightened cell death and the accumulation of H2O2, as well as the up-regulation of HR-, SA-, and JA-associated marker genes DEF1, PO2, HIR1, and SAR82A, were triggered by the simultaneous co-overexpression of SGT1 and SRC2-1.
Figure 3. Transient co-expression of SGT1 and SRC2-1 induced cell death in pepper leaves.
(a) The DAB and trypan blue staining of the Agro-infiltrated pepper leaves. The DAB and trypan blue staining assay were performed at 48 h after infiltration with GV3101 cells harboring the constructs containing SGT1, SRC2-1, SGT1/SRC2-1, and EV (empty vector), respectively. (b) Electrolyte leakage in Agro-infiltrated pepper leaves at 24 and 48 hours post infiltration. The pepper leaves was infiltrated with GV3101 cells harboring constructs containing SGT1, SRC2-1, SGT1/SRC2-1, and EV (empty vector) grown to an OD595 of 0.4, the infiltrated pepper leaves were collected at different time points for the electrolyte leakage measurement. (c) Quantitative real-time RT-PCR analysis of the transcript levels of stresses-associated marker genes in pepper leaves transiently expressing SGT1 and SRC2-1 at different time points after inoculation with P. capsici. Expression values are normalized by the expression level of CaActin and 18S ribosomal RNA. (b,c) Data represent mean ± SD from three independent biological repetitions. Different letters indicate statistically significant differences using an LSD test (P < 0.05).
Simultaneous silencing of SGT1 and SRC2-1 compromises immunity to a greater extent compared to single gene silencing in pepper plants against P. capsici inoculation
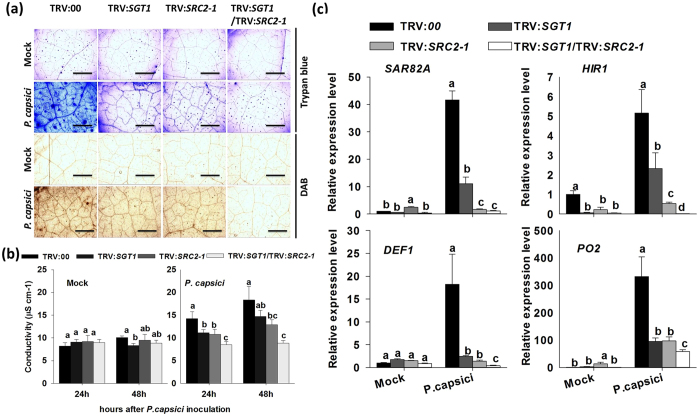
To characterize the functional relationship between SGT1 and SRC2-1 by loss of function by VIGS, GV3101 cells containing the constructs TRV:SGT1 or TRV:SRC2-1 were infiltrated into 4-leaf-age pepper plants individually or simultaneously. The silencing efficiency was examined by qPCR (Supplementary Fig. S2). The results show that SGT1 and SRC2-1 were successfully silenced in the corresponding pepper plants. The silencing of either SGT1 or SRC2-1 conferred decreased HR cell death and H2O2 accumulation, manifested by weaker trypan blue and DAB staining, and also a decrement on electrolyte leakage (Fig. 4a,b). Additionally, the down-regulation of HR cell death and SA- and JA-dependent immunity-associated marker genes was observed (Fig. 4c). However, simultaneous SGT1 and SRC2-1 silencing reduced cell death indicators and the expression of immunity-associated genes (including SA-dependent SAR82A, HR-marker genes HIR1, JA-dependent DEF1, and active oxygen metabolism-associated PO2) to a much lower level compared to that resulting from the individual silencing of either gene alone or the unsilenced control plants.
Figure 4.
Silencing of SGT1, SRC2-1, and SGT1/SRC2-1 compromises the hypersensitive cell death response in pepper leaves inoculated with P. capsici. (a) The trypan blue and DAB staining of SGT1-, SRC2-1-, and SGT1-/SRC2-1- silenced pepper leaves 24 h post inoculation with P. capsici. SGT1 and SRC2-1 were silenced in pepper plants using the virus-induced gene silencing technology. The P. capsici zoospores were dissolved in sterilized ddH2O and infiltrated into the leaves of the pepper plants with a syringe without needle. (b) Electrolyte leakage measurements of the VIGS and control pepper leaves collected at different time points after P. capsici inoculation. The P. capsici inoculated leaves of the VIGS and control pepper plants were harvested at 24 and 48 hpi for electrolyte leakage measurement. (c) Quantitative real-time RT-PCR analysis of the expression of stress-related marker genes in leaves of VIGS and control (TRV:00) pepper plants 24 h after inoculation with P. capsici. Expression values are normalized to the expression level of CaActin and 18S ribosomal RNA. (b,c) Data represents the mean ± SD from three independent biological repetitions. Different letters indicate statistically significant differences as analyzed by an LSD test (P < 0.05). Mock, treated with 10 mM MgCl2.
Analysis of SGT1 and SRC2-1 inter-dependence in immunity induction using VIGS and transient overexpression
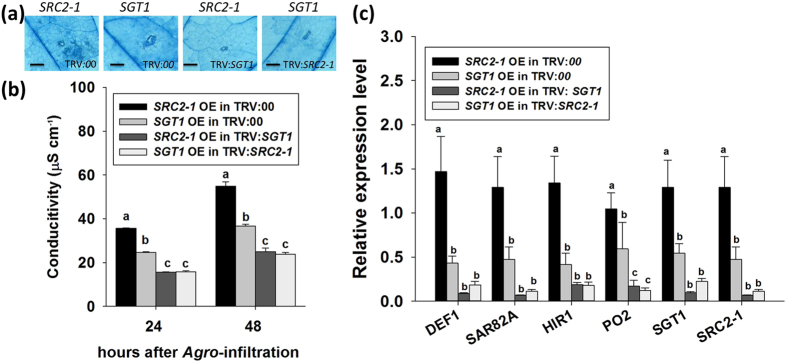
To investigate whether SGT1 and SRC2-1 are interdependent on each other for their roles in pepper immunity, a combination of VIGS and transient overexpression was employed to investigate the function of each protein independently. The results show that in pepper plants with SRC2-1 silenced, but with SGT1 transiently overexpressed, the cell death intensity and electrolyte leakage iron were both lower than that in control plants transiently overexpressing SGT1 (Fig. 5a,b). Additionally, the transcriptional expression of HR-associated HIR1, SA-dependent SAR82A, and JA-dependent DEF1 were significantly lower than that in control plants transiently overexpressing SGT1 (Fig. 5c). Similarly, the cell death intensity and the expression of these immunity-associated genes were also inhibited in the SGT1-silenced pepper plants transiently overexpressing SRC2-1 when compared to the control plants overexpressing SRC2-1 (Fig. 5).
Figure 5. The transcript levels of stress-related marker genes in SGT1- and SRC2-1-silenced pepper leaves transiently expressing either SRC2-1 or SGT1, respectively.
(a) The trypan blue staining of VIGS pepper leaves transiently overexpressing SGT1 or SRC2-1. GV3101 cells harboring the SGT1 and SRC2-1 overexpression vectors were infiltrated into the SRC2-1- and SGT1-silenced pepper leaves, respectively. The trypan blue staining assay was performed at 48 hpi. (b) Electrolyte leakage measurements of SGT1- or SRC2-1 silenced pepper leaves 24 and 48 h post infiltration with GV3101 cells containing SRC2-1 or SGT1 overexpression vectors. (c) Quantitative real-time PCR analysis of the defense response associated marker genes in the VIGS and control pepper leaves transiently overexpressing either SRC2-1 or SGT1. (b,c) Data represents the mean ± SD from three independent biological repetitions. Different letters indicate statistically significant differences as analyzed with an LSD test (P < 0.05).
SGT1 interacts with SRC2-1
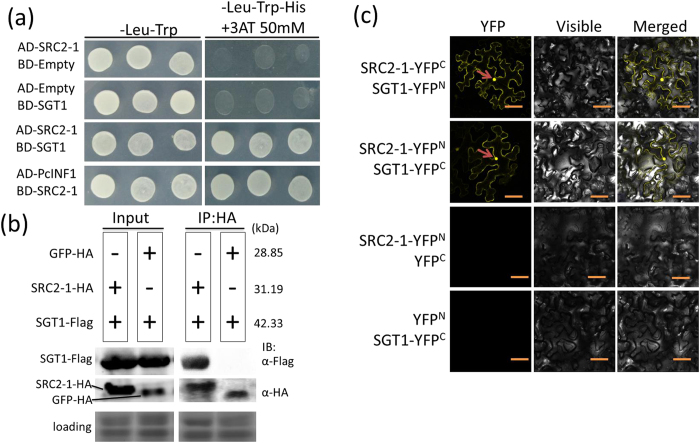
The above data suggests that SGT1 is involved in PcINF1/SRC2-1 mediated pepper immunity, and that SRC2-1 and SGT1 are functionally related with each other. Furthermore, it appears that SRC2-1 and SGT1 may be functionally related in a protein-protein interacting manner. To test this possibility, the interaction between the two proteins was assayed using the yeast two-hybrid (Y2H) system. The Y2H analysis showed that SRC2-1 interacts with SGT1 (Fig. 6a).
Figure 6. The interaction identification between SGT1 and SRC2-1 or PcINF1 by yeast two- hybrid, co-IP and BiFC.
(a) The interaction confirmation between SGT1 and SRC2-1 by yeast two-hybrid. Yeast transformants expressing the corresponding serial vectors were plated on SD-Leu-Trp-His media containing 50 mM 3-amino-1, 2, 4-triazole (3AT), a competitive inhibitor of the His3p enzyme. The PcINF1/SRC2-1 interaction was used as positive control. (b) The SGT1/SRC2-1 interaction confirmation by co-IP. SRC2-1-HA/SGT1-Flag and GFP-HA/SGT1-Flag protein complexes co-expressed in N. benthamiana leaves (OD595 = 0.8). Anti-HA and anti-Flag antibodies were used to detect SRC2-1 (GFP) and SGT1, respectively. The interaction of GFP-HA and SGT1-Flag were used as a negative control. Coomassie brilliant blue staining of the membrane was to show the equal loading. IB: immunoblotting. (c) BiFC assay of the SGT1/SRC2-1 interaction in N. benthamiana leaves infiltrated with Agrobacterium tumefaciens (OD595 = 0.4). The fluorescent signals were detected using a confocal microscope 48 h after infiltration. Bars = 50 μm.
Co-IP assays were used to investigate SGT1/SRC2-1 binding. Co-IP was performed using the same transient co-expression system for leaves of N. benthamiana. This result suggests that SRC2-1 binds to SGT1 (Fig. 6b). The interaction of PcINF1 and SRC2-1 was used as positive control. The negative control experiment, which used GFP-HA and SGT1-Flag, showed no sign of interaction (Fig. 6b).
Additionally, SGT1/SRC2-1 interaction was further confirmed by BiFC analysis. The N- and C-terminal of yellow fluorescent protein (YFP) were fused to SGT1 and SRC2-1 to generate SGT1-YFPN and SRC2-1-YFPC. The reciprocal constructs (SGT1-YFPC and SRC2-1-YFPN) were also generated. All of the destination constructs were introduced into the Agrobacterium strain GV3101 for Agrobacterium-mediated transient overexpression in N. benthamiana leaves. 48 HAI, YFP signal in N. benthamiana leaves was detected using the leica laser confocal microscopy, with SRC2-1-YFPN/YFPC and SGT1-YFPC/YFPN being used as negative controls. Transient co-expression of SGT1-YFPC/SRC2-1-YFPN and SGT1-YFPN/SRC2-1-YFPC in N. benthamiana leaves resulted in the presence of yellow fluorescence in the membrane, cytoplasm, and nuclei, indicating that SGT1 interacts with SRC2 in the membrane, cytoplasm and the nucleus of the plant cells. The YFP signal was not detected in the negative control (Fig. 6c).
The effect of PcINF1 transient overexpression and Phytophthora capsici inoculation on SRC2-1/SGT1 interaction
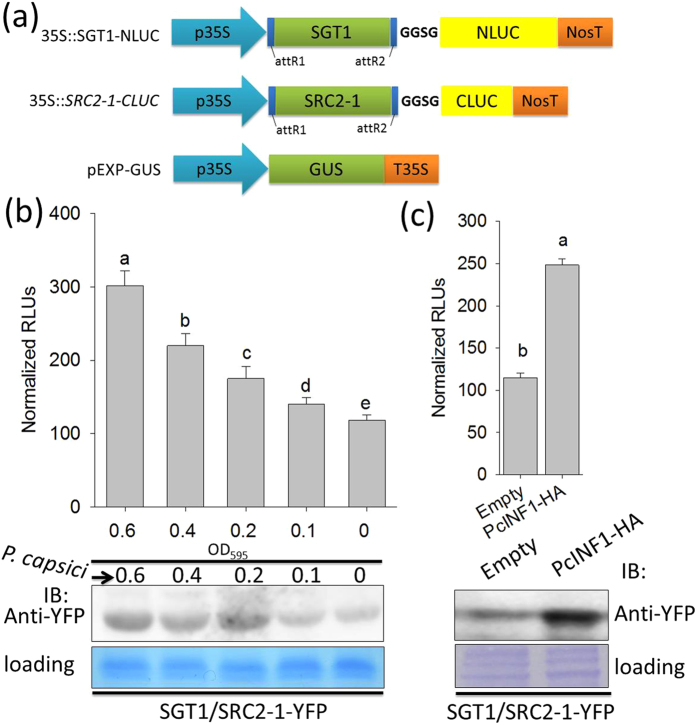
The functional relationship and the direct interaction between SRC2-1 and SGT1 suggest the involvement of a SRC2-1/SGT1 complex in the defense response triggered by PcINF1. To test its possible mode of action, the interaction between SRC2-1 and SGT1 was detected by assays of luciferase complementation imaging (FLuCI)7, immunoblot and fluorescent signals detection under microscope in pepper leaves inoculated with P. capsici. For the luciferase complementation imaging assay, the GV3101 cells carry the split-LUC vector (35S::SGT1-NLUC and 35S::SRC2-1-CLUC) were co-infiltrated into the pepper leaves, followed by inoculated with P. capsici. The result showed that the LUC activities was increased in response to P. capsici inoculation (Fig. 7a/b), which was also confirmed by immunoblot result as well as the result in fluorescent signals detection under microscope (Fig. 7 and Supplementary Fig. S4). Additionally, the interaction intensity was also found to be enhanced with the increase of dosage of inoculated P. capsici (Fig. 7b and Supplementary Fig. S4A). Consistently, the interaction intensity between SRC2-1 and SGT1 was also found to be enhanced by transient overexpression of PcINF1 in pepper plants (Fig. 7c and Supplementary Fig. S4B).
Figure 7. Phytophthora capsici and its elicitin PcINF1 triggered the formation of SGT1/SRC2-1 complex in pepper plants.
(a) Linear structures of the gateway destination vectors of 35::SGT1-NLUC and 35S::SRC2-1-CLUC for studying their protein interaction using the floated luciferase activity measurement. The pEXP-GUS vector was used for GUS activity measurement to normalize the total protein. (b) Analysis of the protein level of SGT1/SRC2-1 complex in pepper leaves infected by P. capsici using western-blot (upper) and luminescence activity measurement assay (bottom). GV3101 cells containing the BiFC vector, SGT1/SRC2-1-YFP, were infiltrated into the leaves of pepper plants (OD595 = 0.3) and the infiltrated plants were growing in the greenhouse for 48 h, followed by infected with different densities of P. capsici zoospores. The total protein of the pathogen-inoculated pepper leaves were extracted for protein levels detection. For the luminescence activity assay, GV3101 cells carrying the split-LUC vectors (35S::SGT1-NLUC and 35S::SRC2-1-CLUC) were vacuum-infiltrated into the leaves of pepper plants and maintained in the greenhouse for 24 h, followed by P. capsici inoculation. The inoculated pepper leaves were harvested for LUC activity measurement at different time points. (c) The protein level detection of SGT1/SRC2-1 complex in pepper leaves transiently overexpressing PcINF1-HA and empty vector. The full length ORF of SGT1 and SRC2-1 were first fused to the N-terminal or C-terminal of YPF, respectively, to generate SGT1-YFPC and SRC2-1-YFPN. Fragments of SGT1-YFPC and SRC2-1-YFPN in satellite vectors were together recombined into the BiFC destination vector PE3519 to generate SGT1/SRC2-1-YFP. Healthy pepper leaves were first Agro-infiltrated with the BiFC vector, SGT1/SRC2-1-YFP, and maintained in the greenhouse for 48 h, followed by Agro-infiltration with agrobacterial cells containing the PcINF1-HA and empty vector (OD595 = 0.3). The anti-GFP antibodies were used to detect the SGT1/SRC2-1 complex. (b,c) The total protein of the treated pepper leaves was extracted and the anti-GFP antibodies were used to detect the SGT1/SRC2-1 complex. Coomassie brilliant blue staining of the membrane was to show the equal loading. Data represent means ± SD for eight independent experiments. Different letters above the bars indicate significantly different means as determined by Fisher’s protected LSD test: lowercase letters, P < 0.05; uppercase letters, P < 0.01.
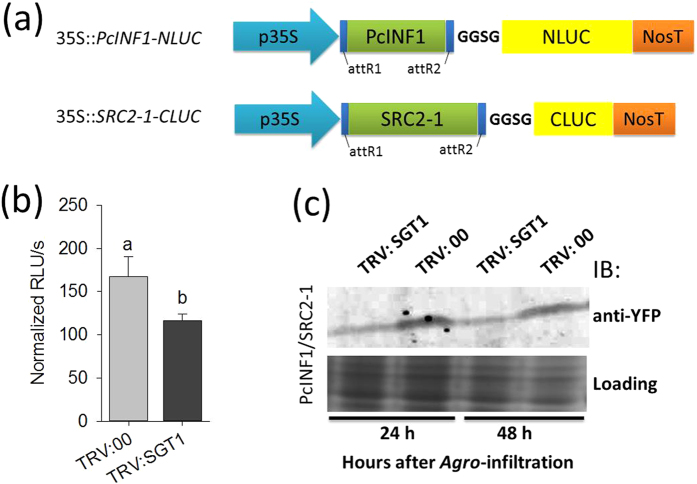
The effect of SGT1-silencing on PcINF1/SRC2-1 interaction
Since the interaction of SRC2-1 with PcINF1 is required in the defense response triggered by PcINF114. To test if PcINF1/SRC2-1 interaction is affected by SGT1, the effect of SGT1 silencing on PcINF1/SRC2-1 formation was assayed by luciferase complementation analysis and co-IP. For the luciferase complementation analysis, the GV3101 cells carrying the split-LUC vectors (35S::PcINF1-NLUC and 35S::SRC2-1-CLUC) were co-infiltrated into the leaves of SGT1-silenced pepper plants and kept in the greenhouse, 24 h after infiltration, the pepper leaves were harvested for LUC activity assay. The result showed that the interaction intensity of SGT1 and SRC2-1 was significantly decreased in the SGT1-silenced pepper leaves compared to the control plants (Fig. 8a/b). For the immunoblot assay, GV3101 cells containing PcINF1/SRC2-1-YFP vector (BiFC vector) were infiltrated into leaves of SGT1-silenced pepper plants generated by VIGS, which were harvested at 48 HAI for protein extraction. The extracted proteins were separated by SDS-PAGE, after which the PcINF1/SRC2-1 complex was detected by immunoblotting with anti-GFP antibody (sigma). This result showed that the interaction between PcINF1 and SRC2-1 was decreased by the silencing of SGT1 (Fig. 8c).
Figure 8. The effect of SGT1-silencing on PcINF1/SRC2-1 interaction.
(a) Linear structure of the gateway destination vectors 35::SGT1-NLUC, and 35S::SRC2-1-CLUC for studying SGT1/SRC2-1 interaction using the floated luciferase activity measurement. Vector pEXP-GUS vector was used for GUS activity measurement to normalize the total protein. (b) The effect of SGT1-silencing by VIGS on the PcINF1/SRC2-1 interaction by luciferase complementation analysis. Luminescence values were measured in the SGT1-silenced and unsilenced pepper leaves transiently expressing 35S::PcINF1-NLUC and 35S::SRC2-1-CLUC 48 h post infiltration. Data represent means ± SD for eight independent experiments. Different letters above the bars indicate significantly different means as determined by Fisher’s protected LSD test: lowercase letters, P < 0.05; uppercase letters, P < 0.01.(c) The effect of SGT1-silencing by VIGS on the PcINF1/SRC2-1 interaction by western-blot analysis. GV3101 cells harboring the PcINF1/SRC2-1 construct were infiltrated into the leaves of SGT1-silenced and unsilenced pepper plants. The protein level of PcINF1/SRC2-1 was detected using the western-blot. Anti-GFP antibodies were used to detect the PcINF/SRC2-1-YFP complex. Coomassie brilliant blue staining of the membrane was to show the equal loading.
Translocation of SGT1/SRC2-1 complex from membrane to nucleus in P. capsici inoculated N. benthamiana leaves
Agrobacterial cells containing constructs of 35S::SGT1-GFP or 35S::SRC2-1-GFP were infiltrated into N. benthamiana leaves, and the subcellular localization of SGT1-GFP and SRC2-1-GFP fused proteins in N. benthamiana leaves was detected using confocal microscopy. Consistent with the previous study14,15, the results show that SRC2-1 was invariably targeted to the membrane of N. benthamiana cells, whereas the fluorescent signal from SGT1-GFP in N. benthamiana leaves was found throughout whole cells, including in the plasma membrane, cytoplasm, and nucleus (Fig. 9). The observed subcellular localization of SGT1 and SRC2-1 suggests the potentiality for interaction. Bimolecular fluorescence complementation (BiFC) analysis of SGT1 and SRC2-1 interaction in N. benthamiana leaves challenged with P. capsici inoculation illustrated dynamic localization of the SGT1/SRC2-1 complex. At 24 hours post inoculation (hpi), YFP signal observation showed that among the 30 cells that displayed YFP signals, 28 cells exhibited YFP localization in plasma membrane or cytoplasm except for the nuclei, while at 48 and 72 hpi, 27 and 28 cells exhibited their YFP signals in the whole cell including the nuclei among the 30 checked cells with YFP signals, respectively (Supplementary Figs 5–6). In addition, we also detected the protein levels of SRC2-1 in the cytoplasm and the nuclei in N. benthamiana leaves with or without co-expression of SGT1, the result showed that SRC2-1 protein was detected only in the cytoplasm when it expressed alone (Supplementary Fig. S7), but was detected in both of the cytoplasm and the nuclei when SGT1 was co-expressed (Supplementary Fig. S7).
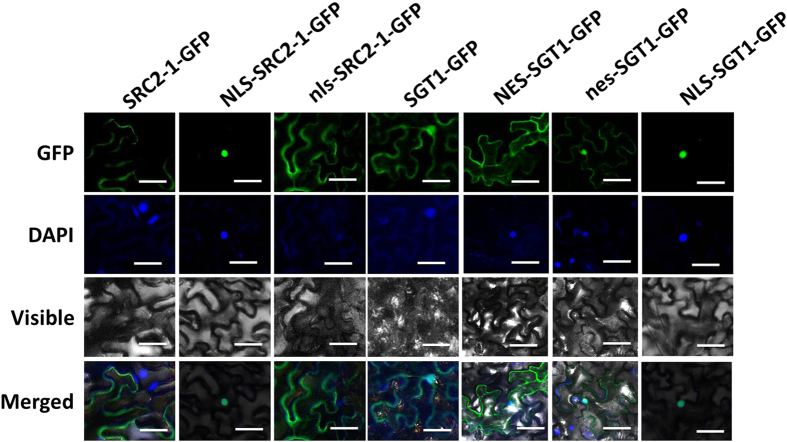
Figure 9. The artificial modulation of subcellular localization of SGT1 and SRC2-1 in cells of N. benthamiana leaves.
The NLS and NES were employed to artificially target SRC2-1 and SGT1 exclusively to the nuclei and cytoplasm, respectively. GV3101 cells containing the SRC2-1-GFP, NLS(nls)-SRC2-1-GFP, SGT1-GFP, NLS-SGT1-GFP, and NES (nes)-SGT1-GFP vectors were infiltrated into the leaves of N. benthamiana plants. 36 h post infiltration, the leaves were harvested and immersed in the DAPI solution and incubated in 37 °C for 1 h and covered with underslip and the fluorescent signals were detected using the Leica confocal laser scanning microscope. Scale bars = 50 μm.
Exclusive nucleus or cytoplasm localization of SGT1/SRC2-1 impaired cell death in pepper plants
To test the possible role of SGT1/SRC2-1 complex translocation from membrane to nucleus in plant immunity, the localization of SGT1/SRC2-1 complex was modified using nuclear localization signal (NLS)45 and nuclear export signal (NES)22. Constructs of SRC2-1 fused to NLS and SGT1 fused to NES, termed 35S::NLS-SRC2-1-GFP and 35S::NES-SGT1-GFP, were generated, and a nonfunctional NES mutant (nes) and a NLS mutant (nls) were also constructed and fused to SGT1 and SRC2-1, respectively, to generate 35S::nes-SGT1-GFP and 35S::nls-SRC2-1-GFP constructs. All of these constructs were transformed into GV3101 cells, and cells containing 35S::NES-SGT1-GFP, 35S::NLS-SRC2-1-GFP, 35S::nes-SGT1-GFP, or 35S::nls-SRC2-1-GFP were infiltrated into leaves of N. benthamiana. GFP signals were then detected with confocal microscopy. The results demonstrated that one NLS tag was sufficient to target SRC2-1 to the nuclei of N. benthamiana leaf cells, and the transient expression of 35S::NES-SGT1-GFP construct exhibited an absence of nuclear GFP fluorescence signals in the N. benthamiana leaf cells, instead, a visible cytoplasmic GFP fluorescence was observed (Fig. 9).
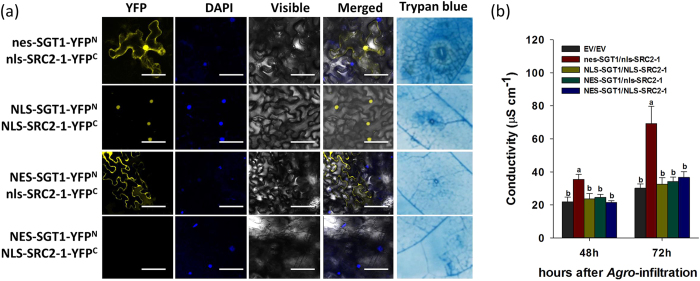
To test the effect of subcellular localization of the SGT1/SRC2-1 complex on pepper immunity, a set of BiFC vectors that include an NLS tag (NLS-SRC2-1-YFPC and nls-SRC2-1-YFPC) and NES tag (NES-SGT1-YFPN and nes-SGT1-YFPN) were constructed. BiFC analysis using different combinations of vectors was performed, and the corresponding induction of immunity associated with these combinations was also assayed. The results show that YFP signals were detected both in the nuclei and cytoplasm of cells of N. benthamiana leaf transiently overexpressing nes-SGT1-YFPN/nls-SRC2-1-YFPC. However, the YFP signals were detected in the nuclei of N. benthamiana leaf cells when NLS-SGT1-YFPN/NLS-SRC2-1-YFPC was simultaneously co-expressed, while the YFP signals were detected in the cytoplasm of N. benthamiana leaf cells when NES-SGT1-YFPN/nls-SRC2-1-YFPC was simultaneously co-expressed. The YFP signal was not detected after co-expression of NES-SGT1-YFPN/NLS-SRC2-1-YFPC (Fig. 10a). Among the vector combinations that produced YFP signals, induced immunity could only be detected (by trypan blue staining and the electrolyte leakage measurement) in leaf cells co-expressing nes-SGT1-YFPN/nls-SRC2-1-YFPC (Fig. 10a/b).
Figure 10. The exclusive nuclear or cytoplasm localization of SGT1/SRC2-1 failed to trigger cell death response in pepper leaves.
(a) BiFC fluorescence and the cell death detection of NLS-(nls-) or NES-(nes-) fused SGT1/SRC2-1 combinations in N. benthamiana leaves 48 h post infiltration. GV3101 harboring the BiFC serial vectors were infiltrated into the leaves of N. benthamiana using a needleless syringe and maintained in the climate chamber, which were harvested and immersed in the DAPI solution and incubated in 37 °C for 1 h before fluorescence detection using the Leica laser scanning microscope. Meanwhile, the infiltrated pepper leaves were also harvested for trypan blue staining. Scale bars = 50 μm. (b) The electrolyte leakage measurement of pepper leaves infiltrated with GV3101 cells containing NLS-(nls-) or NES-(nes-) fused SGT1/SRC2-1 combinations. The infiltrated pepper leaves were harvested for electrolyte ion measurement. Scale bars = 100 μm. Data represents the mean ± SD from three independent biological repetitions. Different letters indicate statistically significant differences as analyzed with an LSD test (P < 0.05).
Discussion
Despite the important roles of SGT1 and SRC2-1 in plant immunity suggested by previous studies14,40, a functional relationship between the two proteins and their potential roles in plant immunity have not yet been established. This study provides evidence of a novel interaction between SGT1 and SRC2-1, and a role for this interaction in pepper immunity induced by PcINF1/SRC2-1.
Using real-time PCR analysis, the response of the transcriptional expression of SGT1 to P. capsici inoculation as well as transient overexpression of PcINF1, an elicitin from P. capsici, was investigated. The qPCR result showed that the mRNA level of SGT1 can be induced in response to the P. capsici inoculation and PcINF1/SRC2-1 co-expression, implying its potential involvement in pepper immunity against P. capsici. This potentiality was further confirmed by loss-of-function assay that employed VIGS. The silencing of SGT1 significantly blocked the cell death and upregulation of immunity associated marker genes in pepper plants challenged with P. capsici or in PcINF1/SRC2-1 transiently overexpressed pepper plants, suggesting the involvement of SGT1 in pepper immunity against P. capsici and in the defense response of pepper triggered by transient overexpression of PcINF1/SRC2-1. As PcINF1 is a member of the conserved elicitin family in P. capsici, the defense response triggered by PcINF1, which is mediated by SGT1, may contribute to the pepper immunity against PTI.
SGT1 was previously found to act as a positive regulator in both ETI18,19,46,47 and PTI pathways9. For example, the overexpression of OsSGT1 in rice significantly increased basal resistance to the virulent bacterial blight Xanthomonas oryzae pv. oryzae PXO999. In addition to its well-established positive roles, SGT1 has also been found to act as a negative regulator of immune receptor accumulation and is thought to assist in maintaining appropriate levels of immune receptor proteins in order to avoid autoimmunity48,49. A recent study found that SGT1, as well as PAR and HSP90, negatively regulates bacterial wilt disease caused by Ralstonia solanacearum in N. benthamiana10. Similarly, SGT1 was found to be induced by the potato virus X TGBp3, and enhanced virus accumulation in N. benthamiana50. These discrepancies might be due to genetic differences both in pathogens and host plants51,52. For example, Arabidopsis SGT1b can antagonize RAR1 to negatively regulate R protein accumulation before infection, and can also function in a RAR1-independent manner to regulate PCD during infection52. Consequently, SGT1 may function differently in plant-pathogen combinations with different effectors and R proteins.
SGT1 is required in PcINF1/SRC2-1 induced pepper immunity by interacting to SRC2-1
SRC2-1, a C2 domain containing protein that was found to act as an interacting partner of PcINF1 in its pepper immunity induction14, was found to synergistically respond to P. capsici inoculation as well as to transient overexpression of PcINF1 with SRC2-1. Also in the present study, the pathogen-responsive W-box and HSE were consistently found to present in the promoters of both SGT1 and SRC2-1 (Supplementary Figs S8/S9 and supplementary Table S3). Given that SGT1 acts as a versatile chaperone, and SRC2-1 contains a C2 domain conferring protein interaction, we speculated that SGT1 and SRC2-1 could potentially be functionally associated in a protein interaction manner. To test this speculation, BiFC experiments were performed and the result potentially demonstrated SGT1 interacts with SRC2-1 which was further confirmed through co-IP experiment, but the potential involvement of other components in the SGT1/SRC2-1 complex cannot be excluded. Additionally, the interaction between PcINF1 and SRC2-1 was found to be decreased by the silencing of SGT1; on the other hand, the transient overexpression of PcINF1 and P. capsici inoculation significantly enhanced the interaction between SGT1 and SRC2-1. All of these results indicated that SGT1 and SRC2-1 are functionally related in the response of pepper to PcINF1 transient overexpression or P. capsici inoculation.
Nucleocytoplasmic distribution of SGT1/SRC2-1 complex is required for its immunity activation
The function of any given protein may be closely associated with its subcellular localization. Similar to the observations made in a previous study15, transiently overexpressed GFP-fused SRC2-1 was exclusively localized to the plasma membrane in N. benthamiana leaf epidermis cells, whereas SGT1 was localized throughout the cells including in the plasma membrane, cytoplasm, and nucleus in the present study. Similar observations of SGT1 were also previously reported in other plant species53. Given that no NLS is found in SGT1, its nuclear targeting is possibly due to the interaction with other nuclear-targeting proteins. Indeed, SGT1 acts as a general adaptor protein in different plant species, interacting with various classes of proteins including R proteins [Prf54 and NLR55], other chaperones [OsCYP256, Hsp9032,57, RAR19, receptor-like cytoplasmic kinase38,40, and type III effector AvrBsT6]. Nucleocytoplasmic distributions have been recently found for TIR-NB-LRRs and CC-NB-LRRs receptors, such as N protein and Rx, which are modulated by SGT1 and are required for full disease resistance22,23,27. Our data showed that this nucleocytoplasmic distribution was also displayed for the SGT1/SRC2-1 interaction complex, as illustrated through its translocation from the membrane and cytoplasm into the nuclei in BiFC assays. By using the NLS and NES-tagged proteins to artificially modify the subcellular localization of SRC2-1 and SGT1, we found the exclusive nuclear or cytoplasmic targeting of SGT1/SRC2-1 complex significantly impaired pepper immunity. This result suggests that the translocation of the SGT1/SRC2-1 complex from the cytoplasm to the nucleus is required for the pepper’s immunity against P. capsici, and this nucleocytoplasmic portioning is potentially mediated by other proteins, such as HSC70 and HSP90, which play roles in nucleocytoplasmic shuttling in several eukaryotic systems58,59,60,61,62, since SGT1 can interact with these proteins by previous studies44,59,63,64. Similarly, the PcINF1/SRC2-1 complex was also found to have a nucleocytoplasmic distribution by our previous study14. It is potential that this nucleocytoplasmic portioning may also be mediated by HSC70 or HSP90. Further experiments are needed to elucidate the potential involvement of HSC70 or HSP90 in SGT1/SRC2-1 nucleocytoplasmic balancing.
Collectively, the data in this study suggest that SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1, and that the nucleocytoplasmic partitioning of the resulting protein complex is required for plant immunity.
Methods
Plant materials and pathogen inoculation
Pepper cultivar (Capsicum annuum L. cv yanshan01) was collected from Yanshan County, Yunnan Province, China. The seeds of N. benthamiana were stored in our lab. The Phytophthora capsici strain used in this study was isolated from Phytophthora blight pepper plants in Huian County, Fujian Province, China. The pepper and N. benthamiana plants were grown as previously described66. For inoculation, P. capsici was cultured in V8 medium (200 g l−1 tomato juice, 3 g l−1 CaCO3) overnight at 28 °C. P. capsici was collected with low speed centrifugation, suspended in sterile ddH2O, and then adjusted to a density of 100 zoospores ml−1. The zoospore suspension of P. capsici was then infiltrated into the lower epidermal of the pepper leaves using a syringe without a needle. The P. capsici-infected pepper plants were then transferred into a climate chamber at 28 °C with 100 mmol photons m−2 s−1, a relative humidity of 70%, and a 16-h light/8-h dark cycle.
Quantitative real-time PCR assay
The pepper leaves collected from treatments were frozen in liquid nitrogen immediately and stored in the −80 °C freezer. The frozen leaf tissues were disrupted in 1.5 ml RNAse-free microcentrifuge tubes using 3 stainless steel beads and the Tissue Lyser II (Qiagen, Dusseldorf, Germany). Trizol (Invitrogen, Carlsbad, USA) was used to extract the total RNA from the disrupted tissues. Both the concentration and the quality of the total RNA were detected using the NanoDrop 2000 (ThermoScientific, Massachusetts, USA). 500 ng of RNA from each sample was used for the first cDNA strand generation using One Step PrimeScriptTM cDNA Synthesis Kit (TaKaRa, Shigo, Japan), which include a genomic DNA digestion step according to the manufacturer’s instructions (TaKaRa). The synthesized cDNA products were diluted ten-fold in sterilized RNAse-free ddH2O for further qPCR analysis. qPCR was performed as a 20-μL reaction (10 μL 2× SYBR Premix Ex-Taq, 0.2 μM forward and reverse primers, and 1 μL cDNA) in a 200-μL PCR tube using SYBR Premix Ex TaqTM (TaKaRa) and a CFXTM Real-Time System with the following program: 95 °C, 30 s; 95 °C, 5 s; 60 °C, 34 s; 95 °C, 15 s, 40 cycles. SYBR green fluorescence was used to detect the amplification of the target genes at each cycle. The Cq (threshold cycle), defined as the real-time PCR cycle at which a statistically significant increase of reporter fluorescence was first detected, was used as a measure for the starting copy numbers of the target gene. Three replicates of each experiment were performed. The data were analyzed by the Livak method70 and expressed as a normalized relative expression level (2−ΔΔCT) of the respective genes. CaActin (GQ339766) and 18S ribosomal RNA (EF564281), which have been confirmed by the published paper66,67, were used to normalize the transcript levels in pepper. The specificities of the qPCR primers were confirmed by the melting curve and the qPCR validation was investigated using the Bio-rad CFX manager 2.0 (Supplementary Table S1). All gene-specific primers are listed in Supplementary Table S1.
Bimolecular fluorescence complementation (BiFC) analysis
To construct vectors for BiFC analysis, the open reading frames (ORFs) of SGT1 and SRC2-1 (flanked by the attB sequence) were amplified with gene-specific primer pairs listed in Supplementary Table S2. Products were introduced into pDONR207 to yield pDONR vectors (named SGT1-207 and SRC2-1-207), and then transferred into the satellite vectors pE3130 and pE3136 to generate SGT1-YFPN and SRC2-1-YFPC. The reciprocal constructs, SGT1-YFPC and SRC2-1-YFPN, were constructed similarly. Fragments of SGT1-YFPN and SRC2-1-YFPC or SGT1-YFPC and SRC2-1-YFPN in satellite vectors were together recombined into the destination vector PE3519, which was then transformed into the Agrobacterium strain GV3101. Agrobacterial cells harboring the BiFC constructs were Agro-infiltrated into the leaves of N. benthamiana plants. Two days after Agro-infiltration, fluorescent signals from N. benthamiana leaves were analyzed using a confocal microscope DM6000 CS (Leica, Solms, Germany). For DAPI staining, N. benthamiana leaves were immersed in DAPI solution (0.1% DAPI and 5% DMSO) and incubated at 37 °C for 1 h before observation.
Generation of Gateway-compatible split-LUC vectors and LUC activity measurement
LUC destination vectors used in this work are purchased using the forms provided at http://www.uni-muenster.de/Biologie.IBBP/agkudla/Plasmids.html. To construct split-LUC vectors, the attB-flanked open reading frames of SGT1 and SRC2-1 without stop codon were amplified with gene-specific primers and the PCR products were introduced into the satellite vector pDONR207. After sequencing confirmation, the SGT1 (PcINF1) and SRC2-1 entry vectors were transformed into the destination vectors (pDEST-GWNLUC and pDEST-GWCLUC) to generate 35S::SGT1-NLUC (35S::PcINF1-NLUC) and 35S::SRC2-1-CLUC, respectively. The yielded SGT1- and SRC2- LUC vectors were transformed into the Agrobacterium stain GV3101. GV3101 cells harboring the various split-LUC constructs were used for infiltration in pepper leaves. GV3101 cells carrying the indicated constructs were centrifuged and resuspended in the infiltration buffer (10 mM MgCl2, 10 mM MES, 150 μM acetosyringone) at an OD595 of 0.4 for LUC activity measurement. Subsequently, the indicated split-LUC constructs and pEXP-GUS were mixed at 1:1 ratio and incubated with gentle shaking at room temperature for 4 hours before infiltration into young but fully expanded leaves of 6–7 week-old pepper plants. After infiltration, the pepper plants were maintained in the greenhouse until LUC activity measurement. Leaf pairs of each pepper plants were infiltrated with the sample in one leaf and the corresponding negative control in the other leaf. Luminescence was quantified in pepper leaves using a floated leaf disc assay15 at appropriate times after infiltration. LUC assay solution (0.1 mM luciferin dissolved in 0.5% v/v DMSO, 10 mM MgCl2 and 10 mM MES/KOH, pH 5.6) was prepared and infiltrated into the agro-infiltrated-pepper leaf before being cut out as leaf discs of 12 mm diameter. The leaf discs were placed in the 96-well microplate with the same LUC assay solution. The light production was measured by a microplate reader (Biotek; http://www.biotek.com). Before LUC activity measurement, the pepper plant were kept in the dark for 10 min to minimize the background fluorescence, and infiltration of LUC assay solution was performed in the low-light condition.
GUS activity measurement
To normalize the LUC values for different expression levels in pepper leaves, GUS activity measurement was performed. Before Agro-infiltration, all the GV3101 cells carrying the indicated constructs were co-infiltrated with a GUS expression vector (pEXP-GUS). To quantitatively measure GUS activity, protein extraction buffer [10% glycerol, 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2% Triton X-100, 10 mM DTT, 1× complete protease inhibitor cocktail (Sigma-Aldrich; http://www.sigmaaldrich.com), and 2% (w/v) polyvinylpolypyrrolidone]16 was used to extract the total protein of infiltrated pepper leave. To measure GUS activity, the rate of p-nitrophenol (γ = 415 nm) release was quantified using a microplate reader (Biotek; http://www.biotek.com).
Yeast two hybrid assay
The GAL4-based Y2H library screening system was performed according to the manufacturer’s instructions (Invitrogen). The PCR-amplified, attb-containing ORFs of SGT1 and SRC2-1 were introduced into the gateway-compatible vectors pDONR201 and pDONR207, respectively, to generate entry vectors. The entry vectors were then confirmed by sequencing and transformed into the bait vector pDEST32 and the prey vector pDEST22 to generate BD-SGT1 and AD-SRC2-1, respectively. We transformed these constructs into yeast strain MaV203 with positive-control and negative-control vectors and were allowed to grow in the selection media (SD-Leu-Trp-His/+ appropriate concentrations of 3-amino-1, 2, 4-triaole).
Co-immunoprecipitation (Co-IP) assay
To construct vectors for co-IP analysis, SGT1 or SRC2-1 in the pDONR vector was directly introduced into the destination vectors 35S::Flag (pEarleyGate 202) or 35S::6HA (pEarleyGate 201) to generate 35S::SGT1-Flag and 35S::SRC2-1-HA by LR reaction (Invitrogen). Plasmids were transformed into the Agrobacterium strain GV3101, and cells harboring 35S::SGT1-Flag and 35S::SRC2-1-HA were simultaneously infiltrated into leaves of N. benthamiana plants. Leaves were harvested at 48 hours after infiltration (HAI) and total proteins were extracted using protein extraction buffer [10% glycerol, 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2% Triton X-100, 10 mM DTT, 1× complete protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA), and 2% (w/v) polyvinylpolypyrrolidone]69. Extracted proteins were incubated with monoclonal anti-HA magnetic beads (Sigma-Aldrich) at 4 °C overnight. Beads were then collected with a magnet and washed three times with protein extraction buffer. Eluted proteins were immunoblotted using anti-HA-peroxidase antibody or anti-Flag-peroxidase antibody (Sigma-Aldrich).
Agrobacterium-mediated transient overexpression and subcellular localization assay
To construct vectors for transient overexpression of SGT1 and SRC2-1, SGT1 and SRC2-1 from plasmids constructed for BiFC experiments (previously described) were transferred into gateway-compatible destination vectors pK7WG2 and pMDC45 (Invitrogen) using LR reaction to yielded pK7WG2-SGT1, pK7WG2-SRC2-1, pMDC45-SGT1, and pMDC45-SRC2-1. All of these vectors were transformed into the Agrobacterium strain GV3101, and agrobacterial cells containing the appropriate vectors were streaked on solid YEP medium (10 g Bacto peptone, 10 g yeast extract, 5 g NaCl, and 15 g agar in 1 l of water) and grown at 28 °C. Individual clones were selected two days later and further cultivated in YEP liquid medium containing appropriate antibiotics. Cells in liquid medium were collected and then suspended in MMA infiltration buffer (10 mM MgCl2, 10 mM MES, 200 μm acetosyringone). The MMA-suspended cells were infiltrated into the full leaves of pepper or N. benthamiana plants using a syringe without a needle.
For subcellular localization assays, the plasmids described above were used. Green fluorescence in N. benthamiana leaves infiltrated with GV3101 cells containing pMDC45-SGT1 and pMDC45-SRC2-1 was observed using a fluorescence microscope Eclipse Ni-U (Nikon, Tokyo, Japan) at 48 HAI. For visualization of nuclei, leaf samples were stained in DAPI solution and incubated at 37 °C for 1 h before observation.
Nuclear targeting of SRC2-1 and nuclear export of SGT1 using a nuclear localization sequence (NLS) peptide and a nuclear export sequences (NES)
To artificially target SRC2-1 to the nuclei, DNA for a general NLS (5′-GGCCCTAAAAAGAAGCGTAAGGTT-3′)45 was fused to DNA corresponding to the N-terminal of the ORF of SRC2-1. A negative control sequence (nls) was produced by substituting an Asn residue for a Lys (5′- GGCCCTAAAAACAAGCGTAAGGTT-3′)45. To drive nuclear export of SGT1, an NES (CTTGCATTAAAGCTCGCTGGTCTTGATATT) fragment was fused to the N-terminal of the ORF of SGT1. The NLS- and NES-fused ORF of SGT1 and SRC2-1 was first cloned into the pDONR207, sequenced and transferred into the corresponding destination vectors, and eventually introduced into the agrobacterial cells for further study.
The nuclear and cytoplasmic proteins were extracted using the Eukaryotic Nuclear Protein Extraction Kit or Eukaryotic Membrane Protein Extraction Kit, respectively (Thermo scientific).
Virus-induced gene silencing (VIGS) in pepper plants
VIGS technology was used to study the loss-function of SGT1 and SRC2-1. The VIGS assays were performed as described previously14. To re-describe the method briefly, specific 3′ UTR fragments of SGT1 and SRC2-1 around 200 bps in length were amplified and cloned into the vector pTRV2 using the primers listed in Supplementary Table S2. These vectors were then transformed into GV3101 cells, and agrobacterial cells harboring the TRV-derived vectors (TRV1/TRV2-SGT1, TRV1/TRV2-SRC2-1, TRV1/TRV2- SGT1/SRC2-1, and TRV1/empty) were streaked onto solid YEB medium and grown at 28 °C. Individual clones were picked two days later and cultured in liquid YEP medium containing 75 μg ml−1 kanamycin and 75 μg ml−1 rifampicin. GV3101 cells were collected by centrifugation and suspended in MMA infiltration buffer (10 mM MgCl2, 10 mM MES, 200 μM acetosyringone, pH 5.7), and adjusted to an OD595 of 0.6. Suspensions of GV3101 cells containing TRV1/TRV2-SGT1, TRV1/TRV2-SRC2-1, TRV1/TRV2-SGT1/SRC2-1, or TRV1/empty were mixed at the ratio of 1:1 and incubated at room temperature in a shaker at low speed (50 rpm). After gentle shaking for 3 h, the GV3101 cells containing vectors in each combination were co-infiltrated into the cotyledons of 2-week-old pepper seedlings.
Conductivity leakage measurements
The conductivity of leaves was analyzed by measuring ion leakage following the method of Choi et al.70 with slight modification. Six leaf discs (4 cm in diameter) were cut with a hole punch, washed with sterilized ddH2O for three times, and subsequently incubated in 20-ml volume of double-distilled water and maintained in a shaker with gently shaking (at 60 rpm) for 60 min at room temperature. The ion leakage of the bathing solution was measured with a conductivity meter (Mettler Toledo 326 Mettler, Zurich, Switzerland).
DAB and trypan blue staining
DAB (3, 3′-diaminobenzidine) and trypan blue staining were performed after Agro-infiltration using the method70 to visualize H2O2 accumulation and cell death. For DAB staining, the full Agro-infiltrated leaves were immersed in DAB solution (1 mg ml−1 DAB, pH 5.7), protected from light and incubated overnight (for almost 8 h). Leaves were then de-stained with 95% ethanol three times. For trypan blue staining, the full Agro-infiltrated leaves were boiled in a lactophenol-ethanol trypan blue solution (10 ml lactic acid, 10 ml glycerol, 10 g phenol, 30 ml absolute ethanol, and 10 mg of trypan blue dissolved in 10 ml of distilled water) for 5 min and allowed to rest at room temperature overnight. Leaves were de-stained with 2.5 g ml−1 chloral hydrate in distilled water.
Additional Information
How to cite this article: Liu, Z.-q. et al. SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 6, 21651; doi: 10.1038/srep21651 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (C150203, 30971718, and 31372061). The funders had no role in study design, data collection, data analysis, the decision to publish, or the preparation of the manuscript. We would like to thank Prof. Zhengguang Zhang from Nanjing Agriculture University, Nanjing, China for providing the P. capsici strain and the nucleotide sequence of PcINF1. We also would like to thank Prof. Zonghua Wang (Key Laboratory of Bio-pesticide and Chemistry Biology, Ministry of Education, Fujian Agricultural and Forestry University, China) for providing pEarleyGate201, pEarleyGate202, and pEarleyGate203. We also want to thank Mark D. Curtis (University of Zurich) for kindly providing Gateway-destination vectors and Dr. S. P. Dinesh-Kumar (Yale University) for providing pTRV1 and pTRV2.
Footnotes
Author Contributions S.H. and Z.L. designed the experiment(s), Z.L., Y.L., L.S., S.Y., L.S., H.Y., R.W., J.W., Q.T., A.H., M.I.K., J.H., C.L., Y.Z. and W.C. performed the experiment(s), S.H. and Z.L. analyzed the results. All authors reviewed the manuscript.
References
- Tyler B. M. Molecular basis of recognition between phytophthora pathogens and their hosts. Annu Rev Phytopathol 40, 137–67 (2002). [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel. et al. The plant defense elicitor cryptogein stimulates clathrin-mediated endocytosis correlated with reactive oxygen species production in bright yellow-2 tobacco cells. Plant Physiol 146, 1255–66 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam T. et al. Constitutive expression of clathrin hub hinders elicitor-induced clathrin-mediated endocytosis and defense gene expression in plant cells. FEBS Lett 586, 3293–8 (2012). [DOI] [PubMed] [Google Scholar]
- Kamoun S., Klucher K. M., Coffey M. D. & Tyler B. M. A gene encoding a host-specific elicitor protein of Phytophthora parasitica. Mol Plant Microbe Interact 6, 573–81 (1993). [DOI] [PubMed] [Google Scholar]
- Duclos J. et al. Identification of an elicitin gene cluster in Phytophthora cinnamomi. DNA Seq 9, 231–7 (1998). [DOI] [PubMed] [Google Scholar]
- Kamoun S., Hraber P., Sobral B., Nuss D. & Govers F. Initial assessment of gene diversity for the oomycete pathogen Phytophthora infestans based on expressed sequences. Fungal Genet Biol 28, 94–106 (1999). [DOI] [PubMed] [Google Scholar]
- Qutob D., Huitema E., Gijzen M. & Kamoun S. Variation in structure and activity among elicitins from Phytophthora sojae. Mol Plant Pathol 4, 119–24 (2003). [DOI] [PubMed] [Google Scholar]
- Jiang R. H. et al. Elicitin genes in Phytophthora infestans are clustered and interspersed with various transposon-like elements. Mol Genet Genomics 273, 20–32 (2005). [DOI] [PubMed] [Google Scholar]
- Ouyang Z. et al. Elicitin-like proteins Oli-D1 and Oli-D2 from Pythium oligandrum trigger hypersensitive response in Nicotiana benthamiana and induce resistance against Botrytis cinerea in tomato. Mol Plant Pathol 16, 238–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M., Toyoda K., Shiraishi T., Inagaki Y. & Ichinose Y. cDNA cloning and characterization of tobacco ABC transporter: NtPDR1 is a novel elicitor-responsive gene. FEBS Lett 518, 164–8 (2002). [DOI] [PubMed] [Google Scholar]
- Sasabe M. et al. Elicitin-responsive lectin-like receptor kinase genes in BY-2 cells. DNA Seq 18, 152–9 (2007). [DOI] [PubMed] [Google Scholar]
- Kanzaki H. et al. NbLRK1, a lectin-like receptor kinase protein of Nicotiana benthamiana, interacts with Phytophthora infestans INF1 elicitin and mediates INF1-induced cell death. Planta 228, 977–87 (2008). [DOI] [PubMed] [Google Scholar]
- Kim Y. T., Oh J., Kim K. H., Uhm J. Y. & Lee B. M. Isolation and characterization of NgRLK1, a receptor-like kinase of Nicotiana glutinosa that interacts with the elicitin of Phytophthora capsici. Mol Biol Rep 37, 717–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Q. et al. SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J Exp Bot 66, 3683–98 (2015). [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Takata N., Uemura M. et al. Arabidopsis synaptotagmin SYT1, a type I signal-anchor protein, requires tandem C2 domains for delivery to the plasma membrane. J Biol Chem 285, 23165–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. X., Liu H., Knowles S. M. et al. A role for protein kinase casein kinase2 alpha-subunits in the Arabidopsis circadian clock. Plant Physiol 157, 1537–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupwate S. D. & Rajasekharan R. C2 domain is responsible for targeting rice phosphoinositide specific phospholipase C. Plant Mol Biol 78, 247–58 (2012). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. Reactive oxygen species in signalling the transcriptional activation of WIPK expression in tobacco. Plant Cell Environ 37, 1614–25 (2014). [DOI] [PubMed] [Google Scholar]
- Oh S. K., Kwon S. Y. & Choi D. Rpi-blb2-Mediated Hypersensitive Cell Death Caused by Phytophthora infestans AVRblb2 Requires SGT1, but not EDS1, NDR1, Salicylic Acid-, Jasmonic Acid-, or Ethylene-Mediated Signaling. Plant Pathol J 30, 254–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. K., Kim H. & Choi D. Rpi-blb2-mediated late blight resistance in Nicotiana benthamiana requires SGT1 and salicylic acid-mediated signaling but not RAR1 or HSP90. FEBS Lett 588, 1109–15 (2014). [DOI] [PubMed] [Google Scholar]
- Yan Q. et al. GmSGT1 is differently required for soybean Rps genes-mediated and basal resistance to Phytophthora sojae. Plant Cell Rep 33, 1275–88 (2014). [DOI] [PubMed] [Google Scholar]
- Hoser R. et al. Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol 200, 158–71 (2013). [DOI] [PubMed] [Google Scholar]
- Slootweg E. et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22, 4195–215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. I. et al. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J 48, 165–76 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dorey S., Swiderski. & M. Jones J. D. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J 40, 213–24 (2004). [DOI] [PubMed] [Google Scholar]
- Bieri S. et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16, 3480–95 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoser R. et al. Emerging role of SGT1 as a regulator of NB-LRR-receptor nucleocytoplasmic partitioning. Plant Signal Behav 9, e28724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S. et al. Detection of the Virulent Form of AVR3a from Phytophthora infestans following Artificial Evolution of Potato Resistance Gene R3a. PLoS One 9, e110158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyannan S. et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–8 (2013). [DOI] [PubMed] [Google Scholar]
- Ito M., Ohnishi K., Hikichi Y. & Kiba A. Molecular chaperons and co-chaperons, Hsp90, RAR1, and SGT1 negatively regulate bacterial wilt disease caused by Ralstonia solanacearum in Nicotiana benthamiana. Plant Signal Behav 10, e970410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. et al. The destructive citrus pathogen, ‘Candidatus Liberibacter asiaticus’ encodes a functional flagellin characteristic of a pathogen-associated molecular pattern. PLoS One 7, e46447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y. & Shirasu K. The HSP90 complex of plants. Biochim Biophys Acta 1823, 689–97 (2012). [DOI] [PubMed] [Google Scholar]
- Cheng X., Tian C. J., Li A. N. & Qiu J. L. [Advances on molecular mechanisms of plant-pathogen interactions]. Yi Chuan 34, 134–44 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang M., Kadota Y., Prodromou C., Shirasu K. & Pearl L. H. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol Cell 39, 269–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K. & Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem Sci 35, 199–207 (2010). [DOI] [PubMed] [Google Scholar]
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60, 139–64 (2009). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. OsRAR1 and OsSGT1 physically interact and function in rice basal disease resistance. Mol Plant Microbe Interact 21, 294–303 (2008). [DOI] [PubMed] [Google Scholar]
- Nakashima. et al. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20, 2265–79 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai K. K., Li Q. & Liu Y. et al. The MI-1-mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol 144, 312–23 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. H., Kim D. S., Chung E. H. & Hwang B. K. Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation-dependent manner. Plant Physiol 165, 76–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M. & Dinesh-Kumar S. P. Virus-induced gene silencing in tomato. Plant J 31, 777–86 (2002). [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S. P., Anandalakshmi R., Marathe R. & Liu Y. Virus-induced gene silencing. Methods Mol Biol 236, 287–94 (2003). [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M. R., Marathe. & Dinesh-Kumar S. P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30, 415–29 (2002). [DOI] [PubMed] [Google Scholar]
- Noel L. D. et al. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19, 4061–76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel M. R., Corbett A. H. & Hodel A. E. Dissection of a nuclear localization signal. J Biol Chem 276, 1317–25 (2001). [DOI] [PubMed] [Google Scholar]
- Ustun S., Muller P., Palmisano R., Hensel M. & Bornke F. SseF, a type III effector protein from the mammalian pathogen Salmonella enterica, requires resistance-gene-mediated signalling to activate cell death in the model plant Nicotiana benthamiana. New Phytol 194, 1046–60 (2012). [DOI] [PubMed] [Google Scholar]
- Austin M. J. et al. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–80 (2002). [DOI] [PubMed] [Google Scholar]
- Huang S. et al. HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J 79, 427–39 (2014). [DOI] [PubMed] [Google Scholar]
- Song J. T. et al. Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry 69, 1128–34 (2008). [DOI] [PubMed] [Google Scholar]
- Ye C. M., Kelly V., Payton M. & Dickman M. B. & Verchot J. SGT1 is induced by the potato virus X TGBp3 and enhances virus accumulation in Nicotiana benthamiana. Mol Plant 5, 1151–3 (2012). [DOI] [PubMed] [Google Scholar]
- Azevedo C. et al. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–6 (2002). [DOI] [PubMed] [Google Scholar]
- Holt B. F. 3rd., Belkhadir Y. & Dangl J. L. Antagonistic control of disease resistance protein stability in the plant immune system. Science 309, 929–32 (2005). [DOI] [PubMed] [Google Scholar]
- Xing L. et al. The Hv-SGT1 gene from Haynaldia villosa contributes to resistances towards both biotrophic and hemi-biotrophic pathogens in common wheat (Triticum aestivum L.). PLoS One 8, e72571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kud J. et al. SGT1 interacts with the Prf resistance protein and is required for Prf accumulation and Prf-mediated defense signaling. Biochem Biophys Res Commun 431, 501–5 (2013). [DOI] [PubMed] [Google Scholar]
- Bhavsar A. P. et al. The Salmonella type III effector SspH2 specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. PLoS Pathog 9, e1003518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B. et al. OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J 74, 86–97 (2013). [DOI] [PubMed] [Google Scholar]
- Yan J. J., Zhang Y. B. & Ding Y. Binding mechanism between Hsp90 and Sgt1 explored by homology modeling and molecular dynamics simulations in rice. J Mol Model 18, 4665–73 (2012). [DOI] [PubMed] [Google Scholar]
- Kose S., Furuta M. & Koike M. The 70-kD heat shock cognate protein (hsc70) facilitates the nuclear export of the import receptors. J Cell Biol 171, 19–25 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazale A. C. et al. Altered expression of cytosolic/nuclear HSC70-1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. J Exp Bot 60, 2653–64 (2009). [DOI] [PubMed] [Google Scholar]
- Kang K. I. et al. In vivo functional protein-protein interaction: nuclear targeted hsp90 shifts cytoplasmic steroid receptor mutants into the nucleus. Proc Natl Acad Sci USA 91, 340–4 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K., Tsukahara F., Naruse M., Yoshioka T. & Takano K. Regulation of nuclear retention of glucocorticoid receptor by nuclear Hsp90. Mol Cell Endocrinol 213, 131–8 (2004). [DOI] [PubMed] [Google Scholar]
- Echeverria P. C. et al. Nuclear import of the glucocorticoid receptor-hsp90 complex through the nuclear pore complex is mediated by its interaction with Nup62 and importin beta. Mol Cell Biol 29, 4788–97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabka M., Lesniak W., Prus W., Kuznicki J. & Filipek A. Sgt1 has co-chaperone properties and is up-regulated by heat shock. Biochem Biophys Res Commun 370, 179–83 (2008). [DOI] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K. & Shirasu. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100, 11777–82 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F. F. et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ 36, 757–74 (2013). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Kim D. S., Kim N. H. & Hwang B. K. The Capsicum annuum class IV chitinase ChitIV interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses. J Exp Bot 66, 1987–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D. S., Hwang I. S. & Hwang B. K. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D. S. & Hwang B. K. Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23, 823–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D. S., Hwang I. S. & Hwang B. K. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.