Abstract
Gli1 is a downstream transcriptional factor of Sonic hedgehog pathway in mammalians, and has been recognized as a proliferative indicator of carcinogenesis. However, its actual role in prognosis among solid malignancies remains unclear. Therefore we performed this meta-analysis aiming to discover the correlation between Gli1 positivity and clinical prognosis in patients suffering from diverse carcinomas. A total of 39 studies containing 4496 cases were selected into our quantitative analysis via electronic database search. Original data of 3-year, 5-year, 10-year overall survival and disease-free survival were extracted and calculated using odds ratio and Mantel-Haenszel model. Subgroup analysis was also conducted to clarify the possible confounding factors. P < 0.05 was considered significant in statistics. Gli1 redundancy was associated with worse 3-year, 5-year, 10-year overall survival and disease-free survival in solid malignancies. Different source regions, sample-size, mean-age and detection approaches had no impact on the negative prognostic effect of Gli1 over-expression. Nevertheless, stratified by cancer type and subcellular localization, cytoplasmic Gli1 expression and Gli1 positivity in intracranial tumors was not correlated to poorer 3-year and 5-year prognosis. The over-expression of Gli1 is a credible indicator of poorer prognosis in most of solid malignancies, irrespective of intracranial tumors.
As a life-threatening disorder and costly burden of healthcare system, cancer has drawn tremendous academic attention to the mechanisms towards its formation and uncontrollable spread. To date, accumulating evidence has disclosed the close kinship between normal embryonic development processes and malignant proliferation, both of which share similar cellular mechanisms and morphological alterations1. Including the conserved Sonic hedgehog (Shh) signaling, aberrant activation of pivotal developmental pathways generally facilitates carcinogenesis through a direct signal transduction in a variety of tumors, implicating a potential therapeutic targeted strategy for future medication2,3.
It is well documented that the Sonic hedgehog signaling initially emerged as an essential pathway involved in tissue growth and patterning during the fetal development4. The pathway has been strictly inhibited in mature tissues to ensure the physiological function and prevent abnormal proliferation. Following studies subsequently confirm its out-of-control plays a crucial role within the tumor origination and metastatic dissemination, identified as a tumor-stimulating pathway in diversified malignancies5. The transduction of this relatively conserved pathway is mediated by interactions between Shh, Patched and Smoothened proteins. As long as ligand Shh binds to trans-membrane protein Patched, the normally activity-inhibited Smoothened protein is then released to activate the translocation of cytoplasmic Gli1 into nucleus. Gli1 acts as a transcriptional factor downstream of the Shh pathway, whose nuclear translocation serves as a hallmark indicator of Shh pathway activation, culminating in up-regulation of target genes especially certain proliferative oncogenes6. Given the tumorigenic feature of Gli1, a specific targeted therapy may be prone to benefit patients with better survival quality and longer lifespan. Nevertheless, the prognostic impact of Gli1 presence in solid malignancies remains in controversy, despite an overwhelming majority of evidence has explored a negative prognostic value of Gli1 over-expression across miscellaneous neoplasms. Failed to draw a similar conclusion, Pizem et al.7 presented with a better survival status of patients under stronger nuclear Gli1 expression, suggesting an astonishing positive prognostic significance in circumstance of Gli1 redundancy. Therefore, in the present study through gathering available evidence, we carried out an exhaustive meta-analysis as well as subgroups analysis to verify the prognostic influence of Gli1 positivity across solid malignancies, aiming to provide more theoretical supports for targeted regimens.
Results
Eligible studies
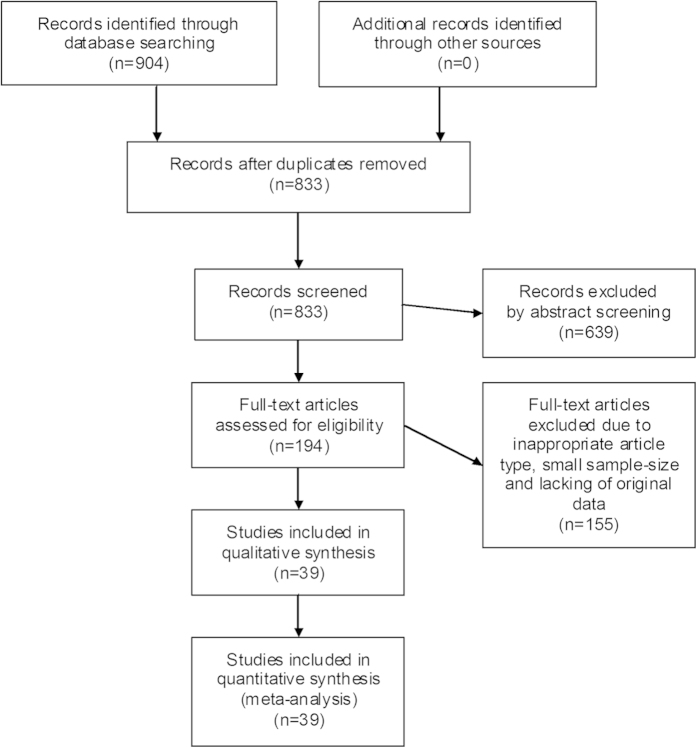
According to the selection criteria, most of the preliminarily included entries were eliminated on account of duplicated data, inappropriate article type or inadequate original information. Eventually, a total of 39 observational studies consisting of 4496 cases were retained for subsequent pooling calculation. Figure 1 concisely displayed the selection workflow of all eligible studies in our meta-analysis.
Figure 1. Selection flow chart of the meta-analysis.
Demographic characteristics of included studies
As for the source regions of included studies, the majority were carried out in China (n = 21), followed by USA (n = 6), Japan (n = 5) and other sporadic nations. None of the eligible entries scored less than six by Newcastle-Ottawa Scale, revealing a high methodological quality across all studies. Studies concerning breast cancer occupied the largest proportion of cancer type among all primary literatures (n = 6), followed by hepatocellular carcinoma (n = 5), esophageal malignancy (n = 4) and remaining types of solid neoplasm. The sample-size ranged from 19 to 339, with a median of 90 patients. A total of 38 studies described the correlation of overall survival and Gli1 expression, while 11 trials reported relationship between disease-free survival and Gli1 presence. Other detailed features were recorded and summarized in Table 1.
Table 1. Demographic information of included studies.
| Reference | Country | Cancer type | No. | Mean age | Male/Female | TNM stage | Follow-up (range) months | Gli1(−/+) No. | 3-year OS(−/+)% | 5-year OS(−/+)% | 10-year OS(−/+)% | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Li et al.12 | China | Gallbladder carcinoma | 93 | NA | 39/54 | I–IV | 32(5–66) | 27/66 | 48.1/24.2 | 14.8/7.6 | NA | 7 |
| Xie et al.9 | China | Gallbladder carcinoma | 32 | 67.0 ± 12.0 | 11/21 | I–IV | 20 | 16/16 | 18.8/12.5 | NA | NA | 7 |
| Hong et al.13 | China | Lung cancer | 55 | 61.9 ± 8.1 | 34/21 | I–IV | NA | 31/24 | 74.2/50.0 | 64.5/20.8 | 54.8/12.5 | 8 |
| Ishikawa et al.14 | Japan | Lung cancer | 102 | 64.8 ± 9.8 | 68/34 | II–IV | NA | 87/15 | 59.8/40.0 | 41.4/20.0 | NA | 7 |
| Che et al.15 | China | Liver cancer | 46 | 51.7 ± 11.2 | 40/6 | NA | 30(1–83) | 21/25 | 57.1/24.0 | 47.6/24.0 | NA | 7 |
| Zheng et al.16 | USA | Liver cancer | 139 | NA | NA | NA | NA | 65/74 | 63.1/35.1 | 44.6/13.5 | 27.7/13.5 | 6 |
| Tang et al.17 | China | Liver cancer | 108 | NA | 77/31 | I–IV | 17(2–82) | 63/45 | 39.7/13.3 | 20.6/11.1 | NA | 6 |
| Xu et al.18 | China | Liver cancer | 63 | NA | 49/14 | I–IV | NA | 6/57 | 83.3/35.1 | NA | NA | 7 |
| Zhang et al.19 | China | Liver cancer | 58 | 51.7 | 43/15 | I–IV | 23(3–36) | 31/27 | 41.9/22.2 | NA | NA | 6 |
| Chaudary et al.20 | Canada | Cervical cancer | 85 | 47.0 | All female | I–IV | 72(9–127) | 43/42 | NA | NA | NA | 7 |
| Pressey et al.21 | USA | Rhabdomyosarcoma | 68 | NA | 46/22 | I–IV | 70(0-123) | 26/42 | 84.6/92.9 | 80.8/92.9 | 80.8/92.9 | 7 |
| Yan et al.22 | China | Glioma | 65 | NA | 18/47 | II–IV | >3 years | 18/47 | 77.8/42.6 | 61.1/42.6 | NA | 7 |
| Ding et al.23 | China | Colon cancer | 96 | NA | 60/36 | I–IV | 38(6–60) | 20/76 | 85.0/48.7 | 25.0/6.6 | NA | 7 |
| Xu et al.24 | China | Colon cancer | 228 | NA | 108/120 | I–III | 52(5–109) | 56/172 | 96.4/70.9 | 91.1/65.1 | NA | 7 |
| Liao et al.25 | China | Ovarian cancer | 44 | NA | All female | I–IV | 64(5–111) | 33/11 | 75.8/45.5 | 69.7/18.2 | 66.7/0.0 | 7 |
| McCann et al.26 | USA | Ovarian cancer | 19 | 61.0 ± 13.6 | All female | III–IV | NA | 10/9 | 40.0/11.1 | 40.0/0.0 | 20.0/0.0 | 8 |
| Ciucci et al.27 | Italy | Ovarian cancer | 56 | 54.0 | All female | III–IV | 35(9–127) | 46/10 | 69.6/40.0 | 63.0/30.0 | 52.2/30.0 | 8 |
| He et al.28 | China | Bladder cancer | 118 | 56.0 | 105/13 | I–III | NA | 23/95 | 78.3/61.1 | NA | NA | 7 |
| Sverrisson et al.29 | China | Bladder cancer | 261 | NA | 194/67 | I–IV | 33 | 60/201 | 40.0/17.9 | 33.3/12.4 | NA | 7 |
| Haaf et al.30 | Germany | Breast cancer | 186 | 56.0 | All female | NA | 78(0–148) | 66/120 | 86.4/83.3 | 80.3/66.7 | 77.3/57.5 | 8 |
| Xu et al.31 | USA | Breast cancer | 139 | NA | All female | I–IV | 94 | 94/45 | 81.9/80.0 | 76.6/68.9 | 64.9/60.0 | 7 |
| O’Toole et al.32 | USA | Breast cancer | 267 | 55.0 | All female | NA | NA | 184/83 | 90.8/85.5 | 89.7/71.1 | 79.9/65.1 | 7 |
| Li et al.33 | China | Breast cancer | 284 | 52.0 | All female | I–III | 62(3–83) | 48/236 | 95.8/81.8 | 95.8/77.1 | NA | 7 |
| Ramaswamy et al.34 | USA | Breast cancer | 289 | 54.5 | All female | NA | 96(1–139) | 89/200 | 95.5/88.0 | 89.9/78.0 | 71.9/51.0 | 8 |
| He et al.35 | China | Breast cancer | 290 | NA | All female | NA | NA | 131/159 | 96.9/96.6 | 92.4/82.4 | NA | 6 |
| Souzaki et al.36 | Japan | Neuroblastoma | 92 | NA | 56/36 | I–IV | NA | 30/62 | 80.0/95.2 | 70.0/93.5 | NA | 7 |
| Mori et al.8 | Japan | Esophageal cancer | 104 | 63.0 | 92/12 | I–IV | NA | 52/52 | 71.2/48.1 | 71.2/34.6 | NA | 6 |
| Yoshikawa et al.37 | Japan | Esophageal cancer | 69 | 60.7 | 58/11 | II–IV | NA | 12/57 | 50.0/0.0 | NA | NA | 7 |
| Zhu et al.38 | China | Esophageal cancer | 100 | NA | 85/15 | I–IV | 23(3–83) | 28/72 | 71.4/51.4 | 50.0/30.6 | NA | 7 |
| Wei et al.39 | China | Esophageal cancer | 35 | 60.0 | 29/6 | I–IV | NA | 10/25 | 60.0/28.0 | 60.0/8.0 | NA | 6 |
| Pizem et al.40 | Slovenia | Medulloblastoma | 65 | 12.4 | 51/14 | NA | 71(2–234) | 25/40 | 60.0/100.0 | 52.0/90.0 | 52.0/80.0 | 8 |
| Buczkowicz et al.41 | Canada | Medulloblastoma | 90 | 6.6 | NA | NA | 80 | 70/20 | 68.6/40.0 | 65.7/30.0 | 60.0/25.0 | 6 |
| Cordeiro et al.42 | Brazil | Medulloblastoma | 40 | NA | 26/14 | NA | NA | 28/12 | 78.6/50.0 | 71.4/41.7 | 64.3/33.3 | 7 |
| Chung et al.43 | USA | Head and neck squamous cell carcinoma | 339 | NA | 263/76 | NA | 96 | 192/147 | 43.2/29.9 | 33.9/17.7 | NA | 7 |
| Saze et al.44 | Japan | Gastric cancer | 41 | 64.8 | 30/11 | I–IV | NA | 19/22 | 57.9/50.0 | 47.4/36.4 | NA | 8 |
| Wang et al.45 | China | Gastric cancer | 121 | 63.0 | 92/29 | I–IV | 30 | 25/96 | 68.8/47.9 | 56.0/26.0 | NA | 8 |
| Jiang et al.46 | China | Pancreatic cancer | 90 | 62.0 | 57/33 | I–IV | 31(0–87) | 35/55 | 57.1/45.5 | 57.1/43.6 | NA | 7 |
| Sheng et al.47 | China | Pancreatic cancer | 57 | NA | 38/19 | I–III | NA | 28/29 | 32.1/10.3 | NA | NA | 6 |
| Liu et al.48 | China | Pancreatic cancer | 62 | NA | 43/19 | I–III | NA | 29/33 | 31.0/9.1 | NA | NA | 7 |
NOS: Newcastle-Ottawa Scale; NA: not available.
Correlation of Gli1 expression with 3-year overall survival and its subgroup analysis
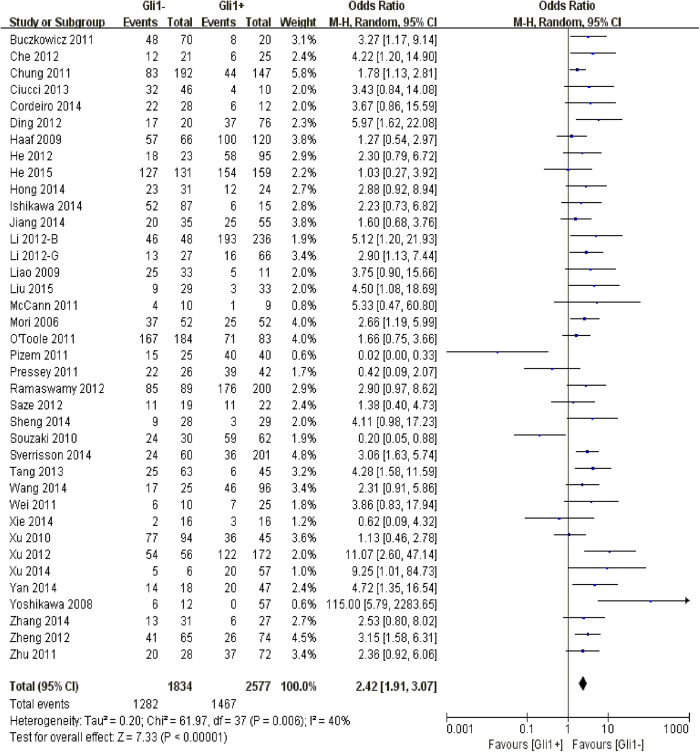
38 observational trials offered original data on 3-year overall survival in terms of different Gli1 expressions. It demonstrated that higher Gli1 activity referred to unfavorable 3-year overall survival (OR: 2.42, 95% CI: [1.91, 3.07], P < 0.00001). A moderate heterogeneity was observed (I2 = 40%) so that a random-effects model was applied for statistical adjustment. In order to explore the possible sources of heterogeneity across studies, we stratified the original articles for subgroup analysis, according to various confounding factors (Fig. 2).
Figure 2. The correlation between Gli1 expression and 3-year overall survival in solid malignancies.
In the subgroup analysis by cancer type, a worse 3-year overall survival of Gli1 positivity was observed in breast cancer (n = 6, OR: 1.63, 95% CI: [1.09, 2.45], P = 0.02, I2 = 1%), cancer of digestive tract (n = 8, OR: 3.51, 95% CI: [1.97, 6.24], P < 0.0001, I2 = 44%), liver cancer (n = 5, OR: 3.54, 95% CI: [2.23, 5.63], P < 0.00001, I2 = 0%), pancreatic cancer (n = 3, OR: 2.47, 95% CI: [1.24, 4.92], P = 0.01, I2 = 7%) and ovarian cancer (n = 3, OR: 3.80, 95% CI: [1.50, 9.61], P = 0.005, I2 = 0%) respectively. Nevertheless, over-expression of Gli1 in intracranial tumors was irrelevant with poorer 3-year prognosis, along with a significant heterogeneity observed (n = 5, OR: 0.99, 95% CI: [0.20, 4.98], P = 0.99, I2 = 84%) (Forest plot was displayed in Supplementary Figure S1).
As for different TNM clinical stages of solid malignancies, a worse 3-year survival was strongly linked to Gli1 positivity in advanced TNM stages (TNM II-IV) (n = 5, OR: 4.51, 95% CI: [1.85, 11.00], P = 0.0009, I2 = 35%) as well as pre-terminal stages (TNM I-III) (n = 5, OR: 4.28, 95% CI: [2.35, 7.79], P < 0.00001, I2 = 0%). Moreover, in studies with coverage to all stages (TNM I-IV), aberrant Gli1 activation likewise suggested a poorer 3-year overall outcome (n = 18, OR: 2.16, 95% CI: [1.55, 3.01], P < 0.00001, I2 = 38%) (Supplementary Figure S2).
There were totally two stratified subgroups in terms of mean age among studies. In both groups of mean age >60-year (n = 10, OR: 2.33, 95% CI: [1.52, 3.57], P < 0.0001, I2 = 18%) and mean age <60-year (n = 10, OR: 2.23, 95% CI: [1.35, 3.69], P = 0.002, I2 = 47%), negative Gli1 expression was verified to significantly associate with more favorable 3-year outcome (Supplementary Figure S3).
Subgroups analysis by Gli1 detection methods explored that high Gli1 expression status was identified as a worse prognostic marker of 3-year outcome in mRNA-examined group (n = 8, OR: 2.49, 95% CI: [1.61, 3.84], P < 0.0001, I2 = 7%). Similarly, obvious Gli1 positivity analyzed by immunohistochemistry (IHC) as well predicted an unfavorable 3-year overall survival in solid cancer (n = 30, OR: 2.43, 95% CI: [1.85, 3.21], P < 0.00001, I2 = 47%) (Supplementary Figure S4).
Included studies were divided into three subgroups according to the subcellular localization of Gli1 expression by IHC. Trials based on nuclear staining of Gli1 significantly correlated its over-reactivity with a worse 3-year survival outcome (n = 13, OR: 3.04, 95% CI: [1.82, 5.07], P < 0.0001, I2 = 56%), which made a similar conclusion as group of unspecific expression did (n = 14, OR: 2.27, 95% CI: [1.74, 2.96], P < 0.00001, I2 = 8%). However, the abnormal cytoplasmic existence of Gli1 did not stand for a detrimental tendency on 3-year overall survival through our pooling analysis (n = 3, OR: 1.11, 95% CI: [0.25, 4.83], P = 0.89, I2 = 76%) (Supplementary Figure S5).
Among the subgroups determined by sample size of the studies, elevated Gli1 expression was confirmed to play a worse prognostic role in terms of 3-year overall survival in solid malignancies, regardless of greater amount (>90) (n = 21, OR: 2.27, 95% CI: [1.77, 2.91], P < 0.00001, I2 = 35%) or smaller magnitude (<90) of participants (n = 17, OR: 2.75, 95% CI: [1.63, 4.66], P = 0.0002, I2 = 49%) (Supplementary Figure S6).
Stratified by source regions of the included studies, there was an analogical trend of trials implemented by Asian (n = 26, OR: 2.79, 95% CI: [2.12, 3.69], P < 0.00001, I2 = 30%) or non-Asian investigators (n = 12, OR: 1.83, 95% CI: [1.20, 2.78], P = 0.005, I2 = 50%) that Gli1 over-reactivity was identified as a poorer 3-year prognostic marker in solid tumors (Supplementary Figure S7).
Correlation of Gli1 expression with 5-year overall survival and its subgroup analysis
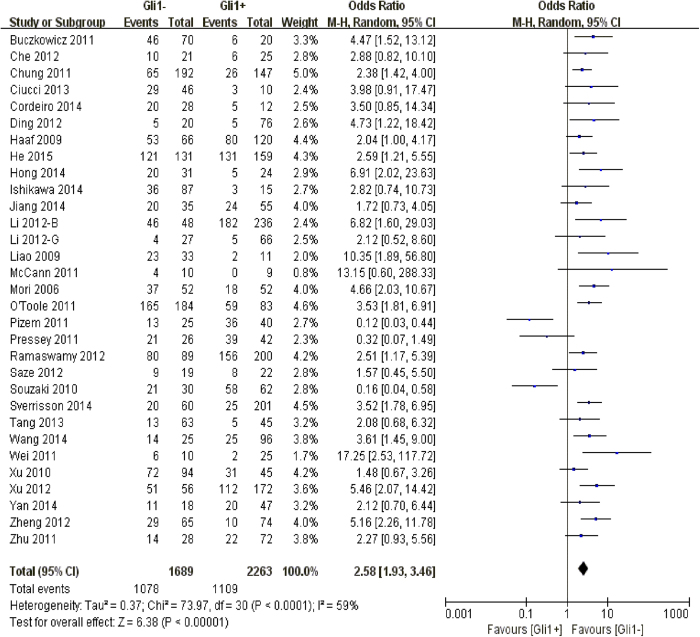
Concerning the 5-year overall survival in solid malignancies, the pooling analysis revealed a negative impact of Gli1 over-expression on clinical prognosis of patients, along with a moderate heterogeneity of undefined source (n = 31, OR: 2.58, 95% CI: [1.93, 3.46], P < 0.00001, I2 = 59%). Thus we performed the following subgroup analysis aiming to clarify the possible confounding elements (Fig. 3).
Figure 3. The correlation between Gli1 expression and 5-year overall survival in solid malignancies.
According to subgroups of different cancer types, Gli1 over-expression in breast cancer (n = 6, OR: 2.52, 95% CI: [1.83, 3.47], P < 0.00001, I2 = 0%), cancer of digestive tract (n = 7, OR: 3.78, 95% CI: [2.50, 5.70], P < 0.00001, I2 = 7%), liver cancer (n = 3, OR: 3.53, 95% CI: [1.96, 6.34], P < 0.0001, I2 = 0%) and ovarian cancer (n = 3, OR: 6.57, 95% CI: [2.30, 18.79], P = 0.0004, I2 = 0%) contributed to a significantly worse 5-year prognosis, except for intracranial tumor, in which Gli1 expression was not associated with the outcome prediction as usual (n = 5, OR: 0.93, 95% CI: [0.21, 4.19], P = 0.92, I2 = 87%) (Supplementary Figure S8).
With respect to subgroups by different TNM stages, our quantitative results suggested that Gli1 abundance in cancer tissues closely correlated to unfavorable 5-year overall survival in all stratified groups, including advanced stages (n = 4, OR: 2.95, 95% CI: [1.44, 6.06], P = 0.003, I2 = 0%), pre-terminal stages (n = 2, OR: 5.85, 95% CI: [2.61, 13.11], P < 0.0001, I2 = 0%) and unselected stages (n = 15, OR: 2.37, 95% CI: [1.46, 3.85], P = 0.0005, I2 = 66%) (Supplementary Figure S9).
Gli1 over-expression in different mean-age subgroups similarly imposed a negative effect on 5-year overall survival, especially in the group with older mean-age (Mean-age >60-year: n = 8, OR: 3.54, 95% CI: [2.19, 5.72], P < 0.00001, I2 = 25%) (Mean-age <60-year: n = 8, OR: 2.32, 95% CI: [1.17, 4.58], P = 0.02, I2 = 73%) (Supplementary Figure S10).
Different detection approaches seemed not influence the negative prognostic role of Gli1 redundancy on 5-year overall survival in solid cancer, including PCR-analyzing group (n = 8, OR: 2.85, 95% CI: [1.48, 5.49], P = 0.002, I2 = 48%) as well as immunohistochemistry staining group (n = 23, OR: 2.63, 95% CI: [1.88, 3.69], P < 0.00001, I2 = 63%) (Supplementary Figure S11).
In terms of subcellular localization of Gli1 staining, the excessive expression of Gli1 distributed in nucleus (n = 11, OR: 3.00, 95% CI: [1.61, 5.58], P = 0.0006, I2 = 70%) or unspecific locations (n = 9, OR: 2.61, 95% CI: [2.03, 3.36], P < 0.00001, I2 = 0%) referred to a disappointing 5-year overall survival in a significant way. Nevertheless, a cytoplasmic positivity of Gli1 expression failed to make a similar conclusion via the pooling analysis, displaying a dubious connection with 5-year prognosis (n = 3, OR: 1.34, 95% CI: [0.17, 10.48], P = 0.78, I2 = 88%) (Supplementary Figure S12).
Among the subgroups divided by different amount of sample sizes, no matter with a greater (>90) (n = 20, OR: 2.71, 95% CI: [2.07, 3.53], P < 0.00001, I2 = 44%) or smaller volume (<90) (n = 11, OR: 2.49, 95% CI: [1.04, 5.95], P = 0.04, I2 = 74%) of sample-size, the merged analysis consistently provided an unfavorable effect on 5-year overall survival owing to Gli1 aberrant activation in solid malignancies (Supplementary Figure S13).
Included studies were stratified into Asian group (n = 19, OR: 2.94, 95% CI: [2.06, 4.20], P < 0.00001, I2 = 50%) and non-Asian group (n = 12, OR: 2.10, 95% CI: [1.27, 3.48], P = 0.004, I2 = 70%) on the basis of different source regions. There was a significant association between Gli1 over-expression and poorer 5-year overall survival through our pooling analysis, regardless of the multiple nationality among the studies (Supplementary Figure S14).
Correlation of Gli1 expression with 10-year overall survival and its subgroup analysis
In our pooling analysis, Gli1 positivity was verified to have a strong connection with a worse 10-year prognosis in solid carcinomas, albeit a moderate heterogeneity was observed across studies (n = 13, OR: 2.11, 95% CI: [1.32, 3.39], P = 0.002, I2 = 63%). A subgroup analysis is therefore conducted (Fig. 4).
Figure 4. The correlation between Gli1 expression and 10-year overall survival in solid malignancies.
Studies were stratified according to different types of cancer. Gli1 positivity in both breast cancer (n = 4, OR: 2.10, 95% CI: [1.54, 2.86], P < 0.00001, I2 = 0%) and ovarian cancer (n = 3, OR: 6.28, 95% CI: [1.07, 36.69], P = 0.04, I2 = 38%) acted as an indicator of a worse 10-year overall survival, while its over-expression in medulloblastoma (n = 3, OR: 1.60, 95% CI: [0.25, 10.40], P = 0.62, I2 = 86%) was unable to offer a prognostic prediction as above (Supplementary Figure S15).
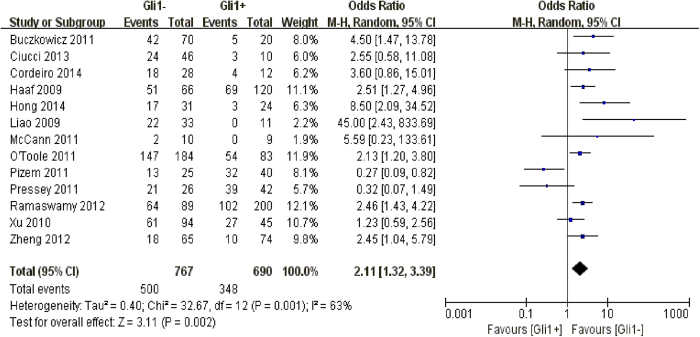
Correlation of Gli1 expression with 3-year and 5-year disease-free survival
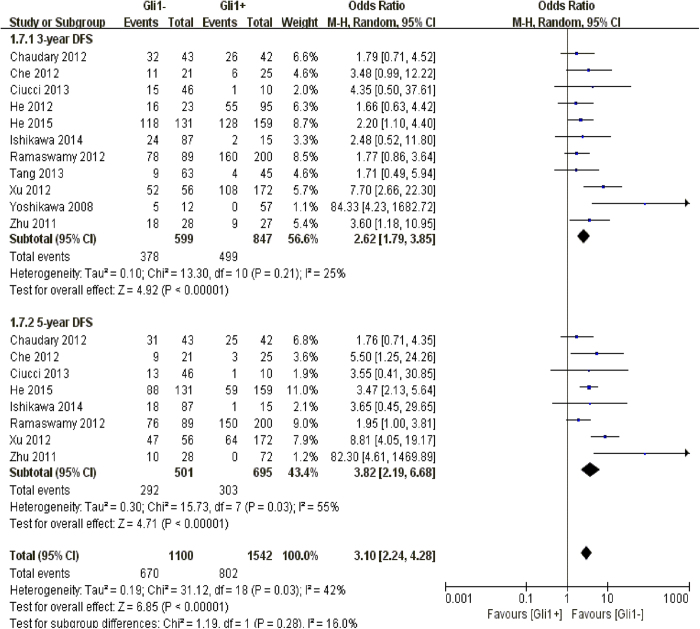
Our quantitative analysis confirmed that high Gli1 expression was shown to be an unfavorable prognostic marker of 3-year (n = 11, OR: 2.62, 95% CI: [1.79, 3.85], P < 0.00001, I2 = 25%) and 5-year disease-free survival (n = 8, OR: 3.82, 95% CI: [2.19, 6.68], P < 0.00001, I2 = 55%) among solid malignancies, along with a moderate heterogeneity existed (Fig. 5).
Figure 5. The correlation between Gli1 expression and disease-free survival in solid malignancies.
Sensitivity analysis
Removal of studies featuring intracranial tumors had no substantial impact on the outcomes of 3-year (n = 33, OR: 2.52, 95% CI: [2.12, 3.00], P < 0.00001, I2 = 17%), 5-year (n = 26, OR: 2.95, 95% CI: [2.46, 3.54], P < 0.00001, I2 = 21%) and 10-year overall survival (n = 10, OR: 2.26, 95% CI: [1.47, 3.46], P = 0.0002, I2 = 47%), however, a huge decline on heterogeneity was observed respectively.
Exclusion of studies with mesenchymal origin obtained similar results of 3-year (n = 37, OR: 2.49, 95% CI: [1.97, 3.14], P < 0.00001, I2 = 37%), 5-year (n = 30, OR: 2.71, 95% CI: [2.04, 3.59], P < 0.00001, I2 = 56%) and 10-year overall survival (n = 12, OR: 2.34, 95% CI: [1.49, 3.67], P = 0.0002, I2 = 59%) respectively.
Elimination of studies scoring 6 in NOS assessment was unable to alter the negative prognostic effect of Gli1 positivity in terms of 3-year (n = 30, OR: 2.30, 95% CI: [1.71, 3.08], P < 0.00001, I2 = 49%), 5-year (n = 25, OR: 2.29, 95% CI: [1.63, 3.22], P < 0.00001, I2 = 62%) and 10-year overall survival among solid malignancies (n = 11, OR: 1.95, 95% CI: [1.12, 3.38], P = 0.02, I2 = 67%).
Publication bias
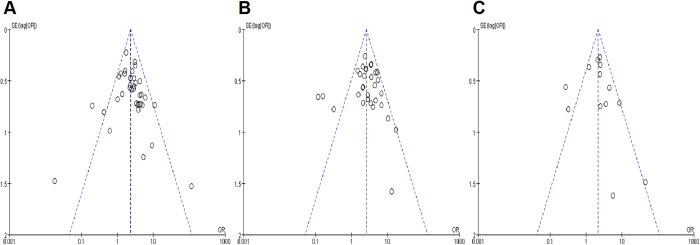
The funnel plots, Egger test and Begg test jointly demonstrated that there was no publication bias concerning 3-year (Egger: P = 0.505; Begg: P = 0.138), 5-year (Egger: P = 0.996; Begg: P = 0.415) and 10-year (Egger: P = 0.653; Begg: P = 0.583) overall survival in our meta-analysis (Fig. 6).
Figure 6. The funnel plots of this meta-analysis.
(A) 3-year overall survival; (B) 5-year overall survival; (C) 10-year overall survival.
Discussion
As the laboratorial evidence has revealed, inactivation of Sonic hedgehog pathway is generally marked by translocation of Gli1 transcriptional factor into the nucleus. The frequently over-expressed Gli1 strongly triggers the carcinogenesis and dissemination in various cancer models. Its oncogenic role takes effects by up-regulation of the downstream target genes, especially including certain detrimental oncoproteins8. The pivotal action of Gli1 inside the molecular carcinogenesis network emphasizes its potential value for targeted treatment modalities. However, from the clinical perspective, a persuasive support of Gli1’s clinical significance is still unavailable, partially due to the uncertainty of the association between Gli1 positivity and prognosis implication. The majority of investigations established potent evidence suggesting an unfavorable impact of Gli1 abnormality on clinical prognosis in a wide spectrum of carcinomas. Nevertheless, on the contrary, several researchers recently highlighted that an obvious advantage on survival duration was obtained in Gli1 over-expression cases, with mechanisms not fully elucidated7. A comprehensive study is therefore in urgent demand.
To our knowledge, the present study is the first and most full-scale meta-analysis systemically exploring the possible prognostic role of Gli1 up-regulation in solid malignancies. On the whole, our quantitative results strongly supported the current mainstream viewpoint that an undesirable impact of Gli1 redundancy was correlated with the 3-year, 5-year, 10-year overall survival and disease-free survival, taking no account of subgroup confounding factors. Additionally, this negative prognostic role was confirmed to be independent of mean-age, source countries, detection measures, sample-size and TNM stages. With respect to subcellular localization, nuclear emergence of Gli1 was identified to tightly associate with awful prognosis while cytoplasmic expression culminated in an absolutely contrary conclusion that there was no obvious connection between Gli1 cytoplasmic expression and worse prognosis in solid malignancies. This inconsistency is probably attributed to the academic consensus that translocation of Gli1 into the nucleus is the hallmark of inactivation of Sonic hedgehog pathway, exclusive of the cytoplasmic positioning pattern9. Moreover, Gli1 lost its prediction efficacy in intracranial tumors by subgroup analysis, displaying an indistinctive prognostic effect on long-term survival. The possible explanation of this exception may be owing to the less possibility of metastasis and cerebrospinal fluid dissemination in medulloblastomas with Gli1 nuclear staining, although the molecular mechanism remains unclear10. Furthermore, Gershon et al. reported a proliferation-inhibitory effect of Gli1 over-expression on neuroblastoma cell lines, which triggered the malignancy towards mature differentiation instead11. In spite of aforementioned plausible reasons, it warrants more investigations to make further explanations to these controversies.
Apart from the inspiring outcomes, limitations still lay in this quantitative meta-analysis. First of all, despite the usage of random-effects model and subgroup analysis, the heterogeneity across studies failed to be eliminated completely, which could result in bias of the outcome in certain extent. Secondly, on account of the lacking of effective data, we merely analyzed the correlation between Gli1 redundancy and prognosis in terms of certain clinical elements. Other parameters that may partially contribute to the heterogeneity were not explored, such as pathological grade and body mass index.
In spite of the limitations mentioned above, there are still numerous valuable implications of this comprehensive mete-analysis. Firstly, the formerly recognized oncoprotein Gli1 has been identified as a biomarker for poor prognosis of 3-year, 5-year, 10-year overall survival and disease-free survival in solid malignancies for the first time, irrespective of several exceptions including intracranial tumors. Secondly, nuclear localization instead of cytoplasmic positivity serves as a prognostic predictor in most cancers, which provides a more specific subcellular positioning to guide the clinical evaluations for diagnosis and prognosis.
Methods
Search strategy
We performed a thorough search for available literatures in electronic databases of PubMed and Web of Science until September 2015. The search terms “Gli1 AND (cancer OR neoplasm OR carcinoma OR malignancy)” was applied and we initially identified 904 entries for further examination. Both abstracts and full-texts were elaborately screened to exclude irrelevant articles. Citation lists of retrieved articles were additionally reviewed to guarantee the sensitivity of the search process. Two authors independently carried out this procedure and any discrepancy was resolved by mutual discussion.
Selection criteria
Studies that met the following requirements were considered eligible and selected into our quantitative meta-analysis: 1. English-written articles published from 2000–2015; 2. Studies exploring the correlation between Gli1 expression and prognosis in human solid malignancies; 3. A minimal follow-up duration of 3 years.
Studies were eliminated on the basis of the following criteria: 1. Duplicated or overlapped studies; 2. Studies with a sample-size less than 10 participants; 3. Lack of enough statistical data for further quantification calculation; 4. Review articles or case reports.
All evaluations were separately undertaken by two individuals to warrant the precision of selection process.
Data extraction
By aid of predefined standardized extraction forms, two investigators independently extracted data from each qualified studies, in terms of general information, 3-year survival, 5-year survival, 10-year survival and disease-free survival respectively. The original survival data were obtained from the text, tables or Kaplan-Meier curves for both comparative groups. GetData Graph Digitizer 2.2 helped us to digitize and extract survival information from the Kaplan-Meier curves. A joint decision was offered in the case of any disagreement.
Methodological quality assessment
Given that all of the included studies were observational studies, a Newcastle-Ottawa Scale (NOS) was adopted to assist the quality assessment of each eligible article. The scale was revised with certain adaptive modifications to match the practical needs of the pooled analysis. There were generally three categories contained in the scale, including selection, comparability and outcome, with a maximum score of nine. Studies graded with more than six scores were classified as high quality literatures in methodology. Two reviewers independently conducted the assessment process and a consensus on NOS score of each study had achieved.
Statistical analysis
Review Manager 5.3 was adopted for the quantitative calculation in this meta-analysis. The original data was inserted as dichotomous variables, therefore odds ratio (OR) along with 95% confidential interval (CI) was applied to measure the correlation between Gli1 presence and long-term survival, including the general survival analysis and sub-groups comparison. I2 was designated as an indicator of heterogeneity across studies, with its value below 25% defined as low heterogeneity. In the absence of significant heterogeneity (I2 < 25%), a fixed-effects model was appropriately employed, while a random-effects model took its place in the remaining situations. P < 0.05 certainly signified a statistical significance within the comparisons. Additionally, we implemented a sensitivity analysis examining the consistency of the pooled outcomes. Funnel plots as well as Egger’s test and Begg’s test was used to investigate the internal publication bias across the included studies, based on the calculation by Stata 12.0 software.
Additional Information
How to cite this article: Cheng, J. et al. Prognostic role of Gli1 expression in solid malignancies: a meta-analysis. Sci. Rep. 6, 22184; doi: 10.1038/srep22184 (2016).
Supplementary Material
Acknowledgments
We sincerely appreciate our lab members for providing methodological support. This study has been financially supported by National Natural Science Foundation of China (No. 81372559) and Research Fund of Public Welfare in Health Industry, Health and Family Plan Committee of China (No. 201402015).
Footnotes
Author Contributions J.C. contributed to the design of the study and manuscript writing. J.C., J.G. and P.Y. contributed to the data extraction and analysis process of the study. P.Y. and K.T. contributed to the financial support and revision of the manuscript.
References
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- Omenetti A. & Diehl A. M. The adventures of sonic hedgehog in development and repair. II. Sonic hedgehog and liver development, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol 294, G595–G598 (2008). [DOI] [PubMed] [Google Scholar]
- Hui M. et al. The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy. Breast Cancer Res 15, 203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. & McMahon A. P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- Pasca D. M. M. & Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer 3, 903–911 (2003). [DOI] [PubMed] [Google Scholar]
- Ma X. et al. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas. Carcinogenesis 26, 1698–1705 (2005). [DOI] [PubMed] [Google Scholar]
- Pizem J., Popovic M. & Cor A. Expression of Gli1 and PARP1 in medulloblastoma: an immunohistochemical study of 65 cases. J Neurooncol 103, 459–467 (2011). [DOI] [PubMed] [Google Scholar]
- Mori Y., Okumura T., Tsunoda S., Sakai Y. & Shimada Y. Gli-1 expression is associated with lymph node metastasis and tumor progression in esophageal squamous cell carcinoma. Oncology-Basel 70, 378–389 (2006). [DOI] [PubMed] [Google Scholar]
- Xie F. et al. Aberrant activation of Sonic hedgehog signaling in chronic cholecystitis and gallbladder carcinoma. Hum Pathol 45, 513–521 (2014). [DOI] [PubMed] [Google Scholar]
- Kool M. et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 3, e3088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon T. R. et al. Enteric neural crest differentiation in ganglioneuromas implicates Hedgehog signaling in peripheral neuroblastic tumor pathogenesis. PLoS One 4, e7491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Immunohistochemical evidence of the prognostic value of hedgehog pathway components in primary gallbladder carcinoma. Surg Today 42, 770–775 (2012). [DOI] [PubMed] [Google Scholar]
- Hong Z. et al. Activation of hedgehog signaling pathway in human non-small cell lung cancers. Pathol Oncol Res 20, 917–922 (2014). [DOI] [PubMed] [Google Scholar]
- Ishikawa M. et al. Expression of the GLI family genes is associated with tumor progression in advanced lung adenocarcinoma. World J Surg Oncol 12, 253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che L., Yuan Y. H., Jia J. & Ren J. Activation of sonic hedgehog signaling pathway is an independent potential prognosis predictor in human hepatocellular carcinoma patients. Chin J Cancer Res 24, 323–331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. et al. The transcription factor GLI1 mediates TGFbeta1 driven EMT in hepatocellular carcinoma via a SNAI1-dependent mechanism. PLoS One 7, e49581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. et al. The prognostic significance and therapeutic potential of hedgehog signaling in intrahepatic cholangiocellular carcinoma. Clin Cancer Res 19, 2014–2024 (2013). [DOI] [PubMed] [Google Scholar]
- Xu Q. et al. The transcriptional activity of Gli1 is negatively regulated by AMPK through Hedgehog partial agonism in hepatocellular carcinoma. Int J Mol Med 34, 733–741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Evaluation of Jagged2 and Gli1 expression and their correlation with prognosis in human hepatocellular carcinoma. Mol Med Rep 10, 749–754 (2014). [DOI] [PubMed] [Google Scholar]
- Chaudary N. et al. Hedgehog pathway signaling in cervical carcinoma and outcome after chemoradiation. Cancer 118, 3105–3115 (2012). [DOI] [PubMed] [Google Scholar]
- Pressey J. G., Anderson J. R., Crossman D. K., Lynch J. C. & Barr F. G. Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children’s Oncology Group. Pediatr Blood Cancer 57, 930–938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G. N. et al. Endothelial cells promote stem-like phenotype of glioma cells through activating the Hedgehog pathway. J Pathol 234, 11–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. L., Zhou Y., Xiang L., Ji Z. P. & Luo Z. H. Expression of glioma-associated oncogene homolog 1 is associated with invasion and postoperative liver metastasis in colon cancer. Int J Med Sci 9, 334–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Li X., Liu T., Leng A. & Zhang G. Prognostic value of hedgehog signaling pathway in patients with colon cancer. Med Oncol 29, 1010–1016 (2012). [DOI] [PubMed] [Google Scholar]
- Liao X. et al. Aberrant activation of hedgehog signaling pathway in ovarian cancers: effect on prognosis, cell invasion and differentiation. Carcinogenesis 30, 131–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann C. K. et al. Inhibition of Hedgehog signaling antagonizes serous ovarian cancer growth in a primary xenograft model. PLoS One 6, e28077 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci A. et al. Expression of the glioma-associated oncogene homolog 1 (gli1) in advanced serous ovarian cancer is associated with unfavorable overall survival. PLoS One 8, e60145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. C. et al. Expression of hedgehog pathway components is associated with bladder cancer progression and clinical outcome. Pathol Oncol Res 18, 349–355 (2012). [DOI] [PubMed] [Google Scholar]
- Sverrisson E. F. et al. Clinicopathological correlates of Gli1 expression in a population-based cohort of patients with newly diagnosed bladder cancer. Urol Oncol 32, 539–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten H. A. et al. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. Bmc Cancer 9, 298 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. Gli1 promotes cell survival and is predictive of a poor outcome in ERalpha-negative breast cancer. Breast Cancer Res Treat 123, 59–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole S. A. et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res 71, 4002–4014 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y. H. et al. Overexpression of Gli1 in cancer interstitial tissues predicts early relapse after radical operation of breast cancer. Chin J Cancer Res 24, 263–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy B. et al. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res 72, 5048–5059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. et al. The Hedgehog signalling pathway mediates drug response of MCF-7 mammosphere cells in breast cancer patients. Clin Sci (Lond) 129, 809–822 (2015). [DOI] [PubMed] [Google Scholar]
- Souzaki R. et al. Hedgehog signaling pathway in neuroblastoma differentiation. J Pediatr Surg 45, 2299–2304 (2010). [DOI] [PubMed] [Google Scholar]
- Yoshikawa R. et al. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy. Br J Cancer 98, 1670–1674 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W. et al. Correlation of hedgehog signal activation with chemoradiotherapy sensitivity and survival in esophageal squamous cell carcinomas. Jpn J Clin Oncol 41, 386–393 (2011). [DOI] [PubMed] [Google Scholar]
- Wei L. & Xu Z. Cross-signaling among phosphinositide-3 kinase, mitogen-activated protein kinase and sonic hedgehog pathways exists in esophageal cancer. Int J Cancer 129, 275–284 (2011). [DOI] [PubMed] [Google Scholar]
- Pizem J., Popovic M. & Cor A. Expression of Gli1 and PARP1 in medulloblastoma: an immunohistochemical study of 65 cases. J Neurooncol 103, 459–467 (2011). [DOI] [PubMed] [Google Scholar]
- Buczkowicz P., Ma J. & Hawkins C. GLI2 is a potential therapeutic target in pediatric medulloblastoma. J Neuropathol Exp Neurol 70, 430–437 (2011). [DOI] [PubMed] [Google Scholar]
- Cordeiro B. M. et al. SHH, WNT, and NOTCH pathways in medulloblastoma: when cancer stem cells maintain self-renewal and differentiation properties. Childs Nerv Syst 30, 1165–1172 (2014). [DOI] [PubMed] [Google Scholar]
- Chung C. H. et al. Glioma-associated oncogene family zinc finger 1 expression and metastasis in patients with head and neck squamous cell carcinoma treated with radiation therapy (RTOG 9003). J Clin Oncol 29, 1326–1334 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze Z. et al. Activation of the sonic hedgehog pathway and its prognostic impact in patients with gastric cancer. Dig Surg 29, 115–123 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Z. S. et al. Significance and prognostic value of Gli-1 and Snail/E-cadherin expression in progressive gastric cancer. Tumour Biol 35, 1357–1363 (2014). [DOI] [PubMed] [Google Scholar]
- Jiang H. et al. Expression of Gli1 and Wnt2B correlates with progression and clinical outcome of pancreatic cancer. Int J Clin Exp Pathol 7, 4531–4538 (2014). [PMC free article] [PubMed] [Google Scholar]
- Sheng W. et al. The clinicopathological significance and relationship of Gli1, MDM2 and p53 expression in resectable pancreatic cancer. Histopathology 64, 523–535 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Q. et al. Gli1 promotes transforming growth factor-beta1- and epidermal growth factor-induced epithelial to mesenchymal transition in pancreatic cancer cells. Surgery 158, 211–224 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.