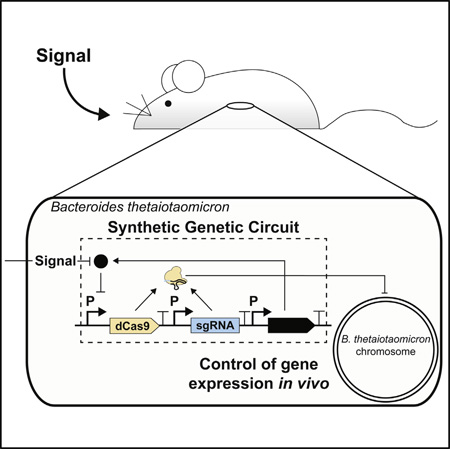
SUMMARY
Engineering commensal organisms for challenging applications, such as modulating the gut ecosystem, is hampered by the lack of genetic parts. Here, we describe promoters, ribosome-binding sites, and inducible systems for use in the commensal bacterium Bacteroides thetaiotaomicron, a prevalent and stable resident of the human gut. We achieve up to 10,000-fold range in constitutive gene expression and 100-fold regulation of gene expression with inducible promoters and use these parts to record DNA-encoded memory in the genome. We use CRISPR interference (CRISPRi) for regulated knockdown of recombinant and endogenous gene expression to alter the metabolic capacity of B. thetaiotaomicron and its resistance to antimicrobial peptides. Finally, we show that inducible CRISPRi and recombinase systems can function in B. thetaiotaomicron colonizing the mouse gut. These results provide a blueprint for engineering new chassis and a resource to engineer Bacteroides for surveillance of or therapeutic delivery to the gut microbiome.
Graphical abstract
INTRODUCTION
Bacteroides species are prominent Gram-negative anaerobic symbionts of the mammalian gut microbiome (Human Microbiome Project Consortium, 2012), comprising 30% of culturable anaerobes in the human gastrointestinal tract (Salyers, 1984). Of the Bacteroides genus, Bacteroides thetaiotaomicron is both prevalent (present in 46% of humans) (Human Microbiome Project Consortium, 2012) and abundant (up to 1010 per g stool) (Salyers, 1984), making it a promising organism for both understanding and manipulating the gut environment. Stable and robust colonization of the densely populated gut environment is facilitated by the metabolic diversity of Bacteroides (Lee et al., 2013). Specifically, B. thetaiotaomicron and its relatives are equipped with an extensive repertoire of saccharolytic enzymes and serve as primary fermenters of host-, diet-, or microbially-derived polysaccharides (Martens et al., 2008). Moreover, members of Bacteroidetes, the phylum to which B. thetaiotaomicron belongs, were among the most stable components of the human gut microbiota in a 5-year longitudinal study (Faith et al., 2013), making them useful candidates for long-term cellular diagnostics and therapeutics in the gastrointestinal tract.
To date, multiple microorganisms have served as chassis for engineered microbial therapies of human disease. Recombinant, attenuated strains of Salmonella spp. (Nemunaitis et al., 2003) and Listeria monocytogenes (Rothman and Paterson, 2013) were used as vectors for anti-cancer therapies in several human trials. Lactococcus lactis has been implemented as a chassis for the production of therapeutic molecules targeting human inflammatory diseases (Steidler et al., 2003). In mouse models, strains of Escherichia coli were engineered to produce molecules that reduce food intake and obesity (Chen et al., 2014). However, compared to E. coli (Kotula et al., 2014) and L. lactis (Steidler et al., 2003), which undergo depletion or clearance within days of administration, Bacteroides populations exhibit low variation in abundance and long-term colonization (Lee et al., 2013).
Genetic parts and circuits enable control over the level and timing of expression of multi-gene traits in response to environmental conditions. Recently, new techniques in DNA construction and high-throughput screening have led to a resurgence in part design, including a revisiting of the organization of the “expression cassette” (Leavitt and Alper, 2015; Nielsen et al., 2013). For model organisms, large libraries and computational models of promoters, ribosome binding sites, and terminators enable fine control of multi-gene systems (Nielsen et al., 2013). Insulators have been designed for integration between parts to ensure that parts can be swapped without impacting function (Brophy and Voigt, 2014; Geyer, 1997; Mutalik et al., 2013). However, a challenge with engineering non-model organisms has been a lack of these tools, which limits the sophistication of the systems that can be constructed (Mutalik et al., 2013).
Few genetic parts and inducible systems are available for B. thetaiotaomicron and its relatives. Previous efforts have co-opted natural glycan-sensing systems (Hamady et al., 2008) or the classic E. coli lac operon (Parker and Smith, 2012) for inducible genetic systems, yielding systems that span a 10- to 50-fold range of expression. Replicative plasmids (Smith et al., 1992) and integrative transposons (Wang et al., 2000) have been built for the introduction of heterologous genes. However, unlike most other prokaryotes, the unique major sigma factor in Bacteroides binds to a −33/−7 consensus sequence (TTTG/TAnnTTTG) (Bayley et al., 2000; Vingadassalom et al., 2005). Moreover, the strength of translation initiation is poorly correlated with the level of ribosome binding site (RBS) complementarity to the 16S rRNA of the host organism (Wegmann et al., 2013). Compared to the E. coli RBS, Bacteroides RBS strength is more sensitive to secondary structures (Accetto and Avguštin, 2011), depleted in GC content (Wegmann et al., 2013), and predicted to rely more heavily on interactions with ribosomal protein S1. These unique promoter and RBS architectures in Bacteroides preclude the direct incorporation of genetic systems developed in other organisms (Smith et al., 1992). A lack of genetic part libraries hinders the introduction of multi-gene pathways, such as those that could produce a metabolic product designed to treat disease.
Multiple cellular states are naturally maintained within Bacteroides populations via reversible recombinases that vary expression of cell-surface polysaccharides (Coyne et al., 2003). Recombinases have been harnessed to build counters and integrated memory-and-logic devices (Bonnet et al., 2013; Friedland et al., 2009; Siuti et al., 2013; Yang et al., 2014). By connecting these switches to environmental sensors, cellular memory can be used to infer exposure to a particular set of conditions. This is useful if direct readout of a reporter from a microenvironment is not possible. For example, an engineered toggle switch enabled E. coli to record exposure to antibiotics in the mouse gut (Kotula et al., 2014). In addition to toggle switches, memory has recently been achieved using dynamic genome editing (Farzadfard and Lu, 2014).
CRISPR-Cas9 technologies have revolutionized genome editing because their ease of reprogramming to target user-defined DNA sequences with a single guide RNA (sgRNA) (Mali et al., 2013). CRISPR interference (CRISPRi) employs a catalytically inactive version of the endonuclease Cas9 (dCas9) to regulate gene expression at target DNA sequences by blocking transcription by RNA polymerase (Qi et al., 2013). The specificity of dCas9 repression is governed by sequence homology and is independent from host machinery (Mali et al., 2013). This has enabled implementation of CRISPRi in both bacterial and eukaryotic systems (Qi et al., 2013), permitting both the construction of synthetic gene circuits as well as the study of natural biological networks (Nielsen and Voigt, 2014).
Here, we expand the set of genetic tools necessary to precisely and robustly engineer B. thetaiotaomicron for microbiome applications. We report a library of biological parts, comprised of constitutive promoters, inducible promoters, and RBSs that each span output-dynamic ranges of several orders of magnitude (Figure 1A). Constitutive promoters and RBSs were used to characterize the input expression levels required to generate recombinase-based DNA-encoded memory in B. thetaiotaomicron. Externally switchable DNA-based memory devices were then constructed by integrating inducible promoters with recombinases. Additionally, inducible promoters were used to control CRISPRi-based regulation of synthetic and endogenous genes. Finally, we validated the function of these tools in bacteria that have colonized the gut of mice. This includes circuits that respond to stimuli in vivo and either record exposure by altering gene expression or permanently modifying the genome. These devices demonstrate that B. thetaiotaomicron can be used as a platform for predictable gene expression and circuit design for microbiome engineering.
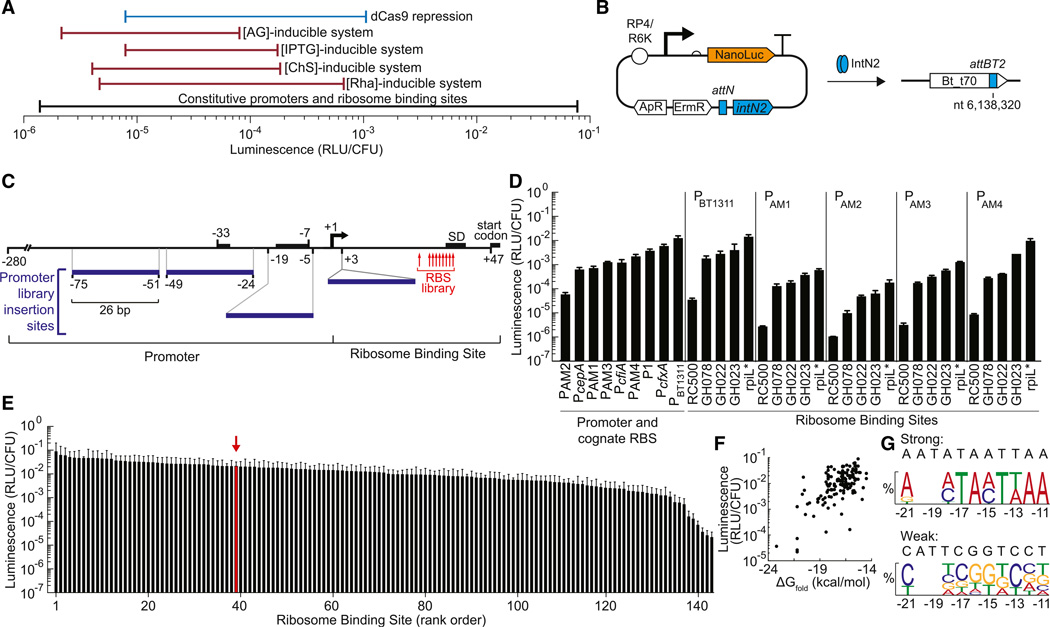
Figure 1. Genetic Parts to Control Expression in B. thetaiotaomicron.
(A) The ranges of gene expression are shown for the different gene regulation systems developed in this manuscript. AG, arabinogalactan; CS, chondroitin sulfate; IPTG, isopropyl beta-D-1-thiogalactopyranoside; Rha, rhamnose; dCas9, catalytically inactive Cas9 endonuclease
(B) Tyrosine integrase IntN2 catalyzes stable integration of pNBU2-based expression constructs into one of two attBT2 sites in the B. thetaiotaomicron genome (Wang et al., 2000). The two attBT2 sites (attBT2-1 at nt 6,217,227 and attBT2-2 at nt 6,138,320) are in the 3′ ends of tRNASer genes (BT_t71 and BT_t70, respectively). ApR, ampicillin resistance cassette; ErmR, erythromycin resistance cassette; RP4, origin of transfer; R6K, origin of replication; NanoLuc, luciferase
(C) Constitutive promoters and ribosome binding sites for the construction of gene expression libraries. The putative −33 and −7 regions of the PBT1311 promoter, the Shine-Delgarno sequence, and the start codon are indicated by black boxes. Numbers below the black boxes represent nucleotide locations relative the PBT1311 transcription start site. The 26 bp sequence introduced in the PAM promoters is shown as blue boxes (see also Figure S1). Numbers at the edges of the blue boxes indicate the PBT1311 nucleotides replaced or the insertion site within the promoter. The location of residues randomized in the rpiL* RBS library are indicated with red arrows (for library A: nt −14, −13, −12; for library B: nt −21, −18, −15; and for library C: nt −17, −16, −11; nt numbering is relative to the translation start site).
(D) Activity was measured for a set of constitutive promoters and their cognate RBSs. For PAM1, PAM2, PAM3, PAM4, the BT1311 RBS was used. Furthermore, PBT1311, PAM1, PAM2, PAM3, and PAM4 were combined with RBSs of varying strengths. Gene expression was measured using a luciferase reporter (NanoLuc) and reported as relative light units/colony forming unit (RLU/CFU).
(E) Three large RBS libraries were constructed and combined with promoter PBT1311. For reference, the parent rpiL* RBS is indicated with a red arrow. The sequences of the RBSs are provided in Table S1. For (D) and (E), error bars represent the SD of three independent biological replicates made on separate days.
(F) The strength of each RBS was compared to the predicted free energy of folding for the mRNA (ΔGfold).
(G) A consensus strong RBS and weak RBS were generated for the rpiL* RBS library using frequency logos that included the 11 strongest and 11 weakest RBSs (residue locations are stated relative to the translation start site). Frequency logos were constructed by comparing the frequency of each nucleotide at each position in that group with the frequency of that nucleotide in that position in the full library. Position −20 and −19 were not randomized and are not shown in the frequency logos.
RESULTS
Landing Pads for Genetic Part and Device Characterization
All genetic parts in this study were characterized using the integration vector pNBU2 to ensure genetic stability of the constructs (Figure 1B). The pNBU2 plasmid encodes the intN2 tyrosine integrase, which mediates sequence-specific recombination between the attN site of pNBU2 and one of two attBT sites located in the 3′ ends of the two tRNASer genes, BT_t70 and BT_t71, on the B. thetaiotaomicron chromosome (Wang et al., 2000). Insertion of the pNBU2 plasmid inactivates the tRNASer gene, and simultaneous insertion into both BT_t70 and BT_t71, is unlikely due to the essentiality of tRNASer.
Expression Control through Promoter and RBS Design
To expand the range of constitutive gene expression that can be implemented in Bacteroides, we constructed and characterized promoter-RBS combinations using the NanoLuc luciferase as a reporter (Figures 1B and 1C). Four promoter variants were constructed based on the constitutive promoter for the B. thetaiotaomicron housekeeping sigma factor BT1311 (PBT1311) (Vingadassalom et al., 2005). Specifically, a 26-bp sequence was substituted or inserted into PBT1311 in regions composing and surrounding the −33 and −7 promoter sequences (Experimental Procedures; Figure S1), regions known to be important for B. thetaiotaomicron promoter activity (Bayley et al., 2000). With NanoLuc as a reporter, the PAM promoters spanned a 20-fold range of expression and had decreased expression levels relative to the PBT1311 parent promoter. For comparison to prior work, we also measured the activities of the previously reported promoter-RBS pairs, PcfxA, PcfiA, P1, and PcepA (Goto et al., 2013; Parker and Smith, 1993; Rogers et al., 1994; Wegmann et al., 2013) (Figure 1D).
The PAM promoters were then combined with RBSs of varying strength to increase the range of expression levels. Previously characterized RBSs GH022, GH023, and GH078 (Wegmann et al., 2013) have an expression range of less than one order of magnitude (Figure 1D). We selected a ribosomal protein RBS (rpiL*) and constructed a weak RBS (RC500) to increase the range of our available RBSs (Figure 1D; Experimental Procedures). This RBS library spanned a >102-fold range when paired with each PAM-derived promoter. When combined, these PAM promoters and RBSs could achieve expression levels over a 104-fold range.
To identify a set of RBSs for fine-tuning gene expression in B. thetaiotaomicron, we generated three randomized rpiL* RBS libraries targeting the most conserved positions of the Bacteroides ribosomal protein RBSs (Wegmann et al., 2013). For each library, we targeted three nucleotides in and around the rpiL* RBS Shine Delgarno sequence. These positions are within or near the RBS region predicted to interact with the ribosomal S1 protein (nt −21 to −11 relative to the start codon of NanoLuc, Figure 1C) (Boni et al., 1991). We achieved 67%–80% coverage of the 64 potential members in each library, resulting in 142 RBS sequences (Figure 1E; Table S1). These RBSs were screened and sequenced and a set of 8 was identified that span 103-fold expression range in approximately even increments (Table S2).
We observed only a weak positive correlation between the minimum free energy of RBS folding (Experimental Procedures) and expression of the NanoLuc reporter (r2 = 0.19) in the rpiL* library (Figure 1F). To visualize the impact of GC content on RBS strength within this library, we generated frequency logos for each nucleotide targeted in the library. The strongest RBSs were GC-depleted relative to the overall library and the weakest RBSs (Figure 1G). Our RBS libraries highlight the distinct AT enrichment of strong Bacteroides RBSs compared to other bacterial species, which results in part failure when constructs are transferred into Bacteroides from other species (Wegmann et al., 2013).
Genetic Sensors and Inducible Systems
To create inducible systems for use in B. thetaiotaomicron, we adapted parts from the large repertoire of systems that govern carbohydrate utilization (Martens et al., 2008). In B. thetaiotaomicron, rhamnose metabolism is mediated by the transcriptional activator RhaR, which activates transcription at the PBT3763 promoter (Patel et al., 2008). To assay the functionality of PBT3763 as an inducible system, we cloned 250 bp of the promoter-RBS region upstream of the start codon of BT3763 into the pNBU2 expression vector to drive expression of NanoLuc. Gene expression was conditional on the concentration of rhamnose and demonstrated a response curve with an output dynamic range of 104-fold (Figure 2A). Fitting the response curve to a Hill function revealed a threshold K of 0.3 mM and a Hill coefficient n = 1.4.
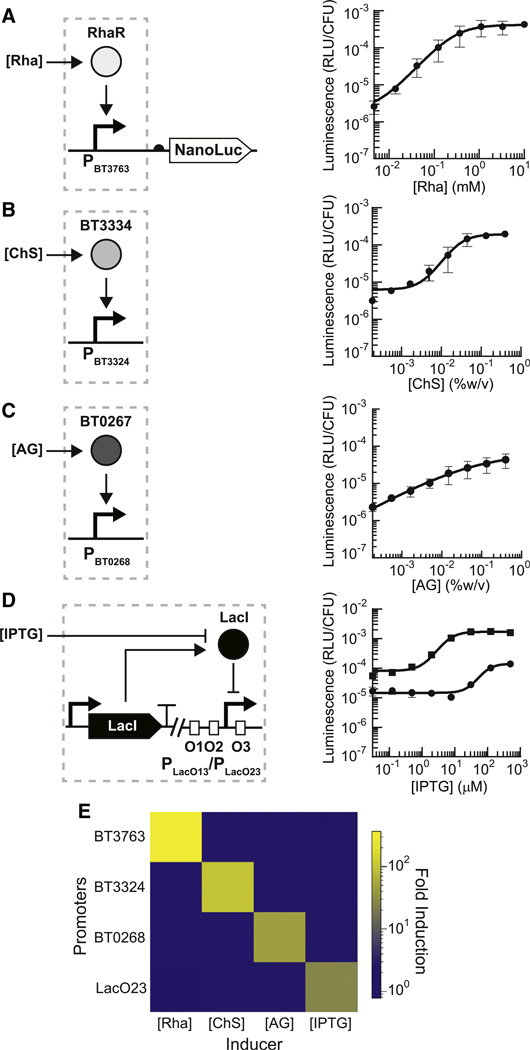
Figure 2. Design and Characterization of Genetic Sensors.
(A–D) Response curves for NanoLuc under the regulated control of the rhamnose (Rha)-inducible promoters (A), chondroitin sulfate (ChS)-inducible promoters (B), arabinogalactan (AG)-inducible promoters (C), or IPTG-inducible promoters (D). LacO1 operator sites were inserted in various regions (O1, O2, O3) of the PcfxA promoter (see also Figure S2). For (D), squares indicate PLacO23 and circles indicate PLacO13. Inducer concentrations were applied as follows: 3-fold serial dilutions starting at 10 mM Rha (A); 3-fold serial dilutions starting at 0.4% for ChS (B) and AG (C); and 4-fold serial dilutions starting at 500 µM for IPTG (D). The leftmost data point in each plot represents the background luminescence in the absence of inducer. Response curves were fit to a Hill function (solid lines).
(E) Orthogonality matrix of sugar-inducible genetic systems incubated with 10 mM rhamnose (Rha), 0.2% chondroitin sulfate (ChS), 0.2% arabinogalactan (AG), or 100 µM IPTG compared to no inducer. Error bars represent the SD of three biological replicates made on different days.
Two-component systems are signal-transduction mechanisms widespread in bacteria for sensing external stimuli. Bacteroides sp. possess a unique variant of these systems, called hybrid two-component systems, that incorporate both the sensor histidine kinase and response regulator of classical two-component systems into a single polypeptide chain (Sonnenburg et al., 2006). Putative hybrid two-component systems, BT3334 and BT0267, were identified in transcriptomic studies to control expression of the chondroitin sulfate (ChS)-inducible PBT3324 promoter and arabinogalactan (AG)-inducible PBT0268 promoter, respectively (Martens et al., 2008, 2011). Chondroitin sulfate induction of PBT3324 and arabinogalactan induction of PBT0268 led to a 60-fold and 29-fold regulation of output gene expression, respectively (Figures 2B and 2C).
Next, we developed an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible system based on the E. coli LacI system. Our design expands upon on a previously developed IPTG-inducible system in Bacteroides (Parker and Smith, 2012) by investigating the position effects of operator sites on gene expression. Pairs of LacO1 operator sites were inserted in the strong PcfxA promoter in three locations (Figures 2 and S2), and the LacI repressor was expressed from the BT1311 promoter. Compared to the unmodified PcfxA promoter, the addition of synthetic operator sites diminished the maximum expression of NanoLuc (Figure S2). This strategy produced two IPTG-inducible promoters that with thresholds at K = 86 µM (PLacO13) and K = 6 µM (PLacO23). The induction of these systems elicited an 8- and 22-fold change in gene expression, respectively (Figure 2D).
As the orthogonality of genetic parts is crucial for their simultaneous use, we tested the degree of cross-talk between each inducible system by incubating each engineered strain with the full set of carbohydrate inducers. The inducers themselves bear little structural similarity: rhamnose, a methyl-pentose sugar; ChS, a sulfated glycosaminoglycan composed of chains of acetylgalactosamine and glucuronic acid residues; AG, a polysaccharide composed of arabinose and galactose units; and IPTG, a molecular mimic of allolactose. Functionally, each inducible system was highly orthogonal to each other, with no cross-reactivity observed with any of the combinations (Figure 2E).
Synthetic Genetic Memory
To equip B. thetaiotaomicron with permanent genetic memory, we used serine integrases, which catalyze unidirectional inversion of the DNA sequence between two recognition sequences (Figure 3A) (Grindley et al., 2006). Recently, 11 orthogonal integrases and their recognition sequences were characterized in E. coli (Yang et al., 2014), and a DNA “memory array” composed of a linear concatenation of integrase recognition sequences was used to record the expression of one or multiple integrases in response to a stimulus.
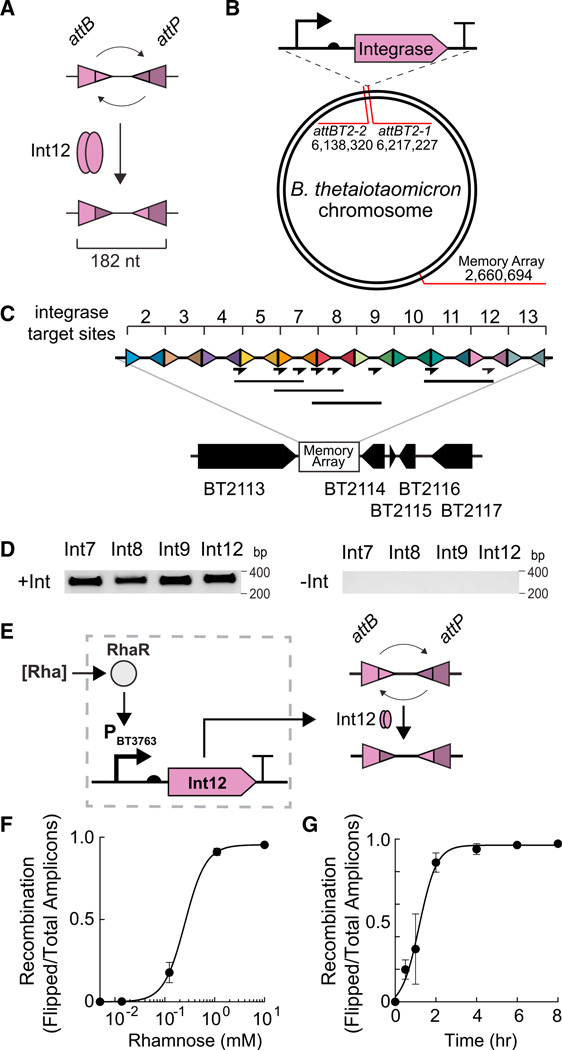
Figure 3. Synthetic Genetic Memory.
(A) Integrases mediate recombination of DNA between integrase binding sites (attB/attP), resulting in the inversion of the intervening spacers.
(B) Schematic of the location of the promoter-RBS-integrase system and the memory array cassettes in the B. thetaiotaomicron chromosome.
(C) Integrase-mediated DNA inversion at each integrase target sequence in the memory array cassette is detected by PCR. Primer pairs (arrows) anneal to the interface of the integrase recognition sites and to the spacer region between recognition sites. PCR amplification occurs only after an inversion event (solid lines below the primer arrows indicate expected amplicons).
(D) Representative PCR products are shown after recombination with integrases Int7, Int8, Int9, or Int12. – indicates no integrase, + indicates the integrase is present. PAM4-rpiL* was used to control expression of each integrase (see also Figure S3.)
(E) Schematic of the rhamnose-inducible recombinase circuit. Transcriptional activator RhaR, produced from the endogenous locus, is activated in the presence of rhamnose causing expression of Int12 from PBT3763. Int12 mediates recombination between the Int12 attB and attP recognition sequences.
(F) Response curve of Int12 memory circuit. Int12 was placed under the control of a subset of PBT3763-rpiL*C51. Inducer concentrations were 9-fold serial dilutions starting at 10 mM rhamnose. The leftmost data point represents the recombination in the absence of inducer. Cells were grown 8 hr at 37°C before harvesting cells and isolating DNA. Absolute quantities of flipped and unflipped memory array in genomic DNA were determined by qPCR using standard curves (Experimental Procedures). The recombination ratio is expressed as the ratio of cells containing a flipped memory array (Flipped) divided by the sum total of cells containing a flipped or unflipped array (Total). Data were fit with a Hill function to guide the eye (see also Figure S3).
(G) Int12-mediated recombination versus time. Cells were induced with 10 mM rhamnose at t = 0. Absolute quantities of flipped and unflipped memory array in genomic DNA were determined by qPCR using standard curves (Experimental Procedures). Recombination ratios were determined as in (F). Data were fit with a sigmoidal dose-response function to guide the eye. For (F) and (G), error bars represent the SD of three biological replicates made on different days. See also Figure S3.
We identified serine integrases that function in B. thetaiotaomicron by cloning the integrases into a strong constitutive expression vectors (PAM4-rpiL*, 1.2 × 10−2 RLU/CFU). To provide a stable, single-copy record of DNA inversion, we incorporated the DNA memory array containing the integrase recognition sequences into the B. thetaiotaomicron chromosome (Figures 3B and 3C; Experimental Procedures). Integrase expression vectors were conjugated into the B. thetaio taomicron memory array strain. Genomic DNA was isolated from transconjugants and analyzed by PCR to detect flipping. Four integrases, Int7, Int8, Int9, and Int12, each catalyzed recombination at the respective recognition sequence in the memory array (Figure 3D), and DNA inversion was not detected in the absence of an integrase (Figure S3A).
To create an inducible memory switch, we cloned Int12 under the control of the rhamnose-inducible promoter with the rpiL*RBS variant C51 (Figures 1E and 3E; Table S1). The Int12 recombinase switch responded to increasing concentrations of rhamnose (Figure 3F) within 2 hr (Figure 3G), with no background detected in the absence of inducer. Notably, expression of Int12 did not impact growth of B. thetaiotaomicron, even when maximally expressed (Figure S3B).
CRISPRi-Mediated Gene Knockdown
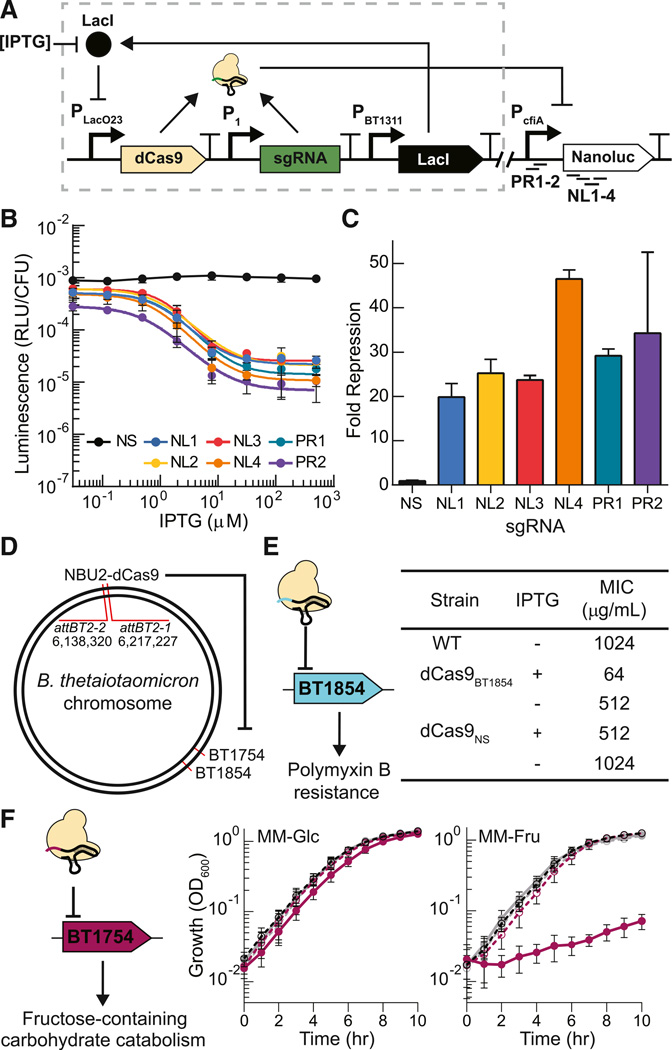
CRISPRi can provide a facile toolbox for constructing synthetic gene circuits and modulating endogenous genes in B. thetaiotaomicron. To demonstrate the use of CRISPRi-mediated gene knockdown for synthetic constructs, production of dCas9 was regulated by the IPTG-inducible PLacO23 system. sgRNAs were constitutively expressed from the P1 promoter and were designed to target the coding sequence of NanoLuc (NL1–4) or the PcfiA promoter (PR1–2) (Figure 4A). A nonsense sgRNA (NS) with no sequence identity to either PcfiA or NanoLuc was used as a negative control. All specifically targeted sgRNAs repressed the expression of NanoLuc (Figure 4B) by 20- to 45-fold with IPTG induction of dCas9 expression (Figure 4C), thus implementing genetic NOT gates in B. thetaiotaomicron. The IPTG-to-NanoLuc response function of sgRNAs targeting the coding sequence or promoter exhibited similar Hill coefficients and lower dissociation constants to the IPTG-to-NanoLuc transfer function of the PLacO23 promoter on its own (n = 1.1–1.4; K = 0.6–1.4 µM IPTG).
Figure 4. CRISPRi-Mediated Repression of Recombinant and Endogenous Genes.
(A) Schematic of dCas9-based repression of NanoLuc. Addition of IPTG induces expression of dCas9, which complexes with constitutively expressed sgRNA targeting the coding sequence of NanoLuc (NL1–4) or the PcfiA promoter (PR1–2). The plasmid backbone separates the NanoLuc cassette and the IPTG-inducible CRISPRi system.
(B) Response curves of dCas9-mediated targeting the coding sequence of NanoLuc (NL1–4), the promoter (PR1–2) or a nonsense sequence (NS). Fourfold serial dilutions of IPTG starting at 500 µM or no inducer were added to cultures. Response curves were fit to a Hill function (solid lines).
(C) Fold repression elicited by various gRNAs in the presence (500 µM) of inducer. Bars are colored to correspond to (B).
(D) Genomic location of endogenous genes targeted using CRISPRi.
(E) Minimum inhibitory concentrations (MICs) of polymyxin B for cells with CRISPRi targeted against BT1854 (dCas9BT1854) compared with wild-type (WT) cells or non-specific control cells (dCas9NS). Reported values are the mode of three independent biological replicates made on three separate days.
(F) CRISPRi was targeted against BT1754 (dCas9BT1754). Growth curves of wild-type (WT) (black), dCas9BT1754 (pink), or dCas9NS (gray) cells in minimal media supplemented with 0.5% glucose (MM-Glc) or 0.5% fructose (MM-Fru) in the presence (full line) or absence (dotted line) of 100 mM IPTG. Error bars represent the SD of three biological replicates made on different days.
To demonstrate the programmable knockdown of endogenous genes in B. thetaiotaomicron, we designed sgRNAs to target mechanisms implicated in the resilience of Bacteroides in the human microbiota. In B. thetaiotaomicron, LpxF, the gene product of BT1854, is required for resistance to inflammation-associated cationic antimicrobial peptides, such as polymyxin B (Cullen et al., 2015). Using the minimum inhibitory concentration (MIC) of polymyxin B as a phenotypic readout, we designed an sgRNA to specifically suppress BT1854 expression. In cells containing the sgRNA targeted against BT1854 (dCas9BT1854), the induction of dCas9 with led to sensitization of the cells to polymyxin B treatment (8- to 16-fold decrease in MIC), while wild-type B. thetaiotaomicron and strains containing dCas9NS demonstrated high levels of polymyxin B resistance in the presence or absence of dCas9 induction with IPTG (Figures 4D and 4E).
Next, we explored dCas9-mediated repression of carbohydrate-utilization pathways in B. thetaiotaomicron, which are important for the bacterium’s ability to successfully and persistently colonize the mammalian gut. The hybrid two-component sensor BT1754 regulates the BT1757-1763/BT1765 fructose-containing polysaccharide utilization locus, and BT1754 is essential for growth on fructose as the sole carbon source (Sonnenburg et al., 2010). We designed a specific sgRNA to repress BT1754 and integrated this system into the B. thetaiotaomicron genome along with an IPTG-inducible dCas9 cassette (dCas9BT1754) (Figures 4D and 4F). Induction of dCas9BT1754 did not affect the growth rate of cells on minimal media (MM)-glucose compared to WT cells and dCas9NS. The generation time G = (log102 × t)/log10(B/B0) ≈ 1 hr (where t is the time interval, and B0 and B are the initial and final concentrations of bacteria, respectively), indicating that neither dCas9 induction nor repression of BT1754 impacts growth on glucose media (Figure 4F). However, induction of dCas9BT1754 drastically decreased the growth rate of the cells in MM-fructose (G = 4.7 hr) while the growth of WT and dCas9NS cells in MM-fructose remained similar (G = 1 hr) to growth in MM-glucose (Figure 4F). Thus, inducible dCas9-mediated repression of endogenous genes can alter both the resistance and metabolic profiles of B. thetaiotaomicron.
Function of Genetic Parts in B. thetaiotaomicron Colonizing the Mouse Gut
We next investigated whether the function of our B. thetaiotaomicron genetic parts and modules can be maintained in the context of a complex microbiota. As wild-type strains of Bacteroides spp. are unable to stably colonize conventional specific-pathogen-free (SPF) mice (Lee et al., 2013; Cullen et al., 2015), we employed an antibiotic regimen that promotes B. thetaiotaomicron colonization without sterilizing the gut microbiota (Figure 5A) (Bloom et al., 2011; Lee et al., 2013). A 10-day treatment of animals with ciprofloxacin and metronidazole prior to bacterial inoculation was sufficient to maintain stable and high levels of colonization for the duration of the experiments (up to 12 days tested) (Figure S4).
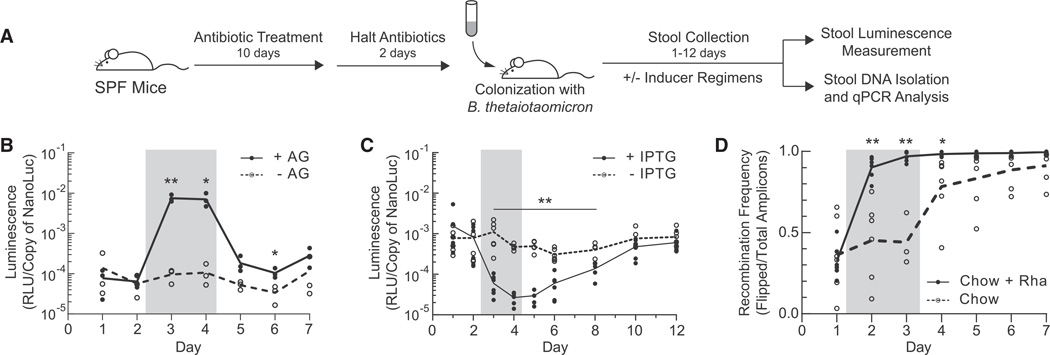
Figure 5. In Vivo Function of Genetic Parts within B. thetaiotaomicron Colonizing the Mouse Gut.
(A) Experimental timeline. Specific-pathogen-free (SPF) Swiss Webster mice were treated for 10 days with ciprofloxacin and metronidazole and gavaged with B. thetaiotaomicron 2 days after cessation of treatment. Bacterial gavage was administered on Day 0 (B–D).
(B and C) Luciferase activity in fecal pellets of mice inoculated with strains possessing the arabinogalactan (AG)-inducible PBT0268 (B) or IPTG-inducible CRISPRi dCas9NL3 (C) systems. Mice were provided drinking water supplemented with 5% arabinogalactan (B) (solid line), or 25 mM IPTG (C) (solid line) for 2 days after stool collection on Day 2 (gray box), or were maintained on normal drinking water throughout the entire experiment (dashed lines). Inducer water was removed on Day 4 after stool collection. Luminescence values were normalized to cell density as determined by qPCR using NanoLuc-specific primers.
(D) SPF mice were colonized with B. thetaiotaomicron containing the rhamnose-inducible integrase construct PBT3763-rpiL*C51-Int12. All mice were exposed to 0.3% rhamnose (w/w) in the plant-based chow. In addition, half of the mice had their drinking water supplemented with 500 mM rhamnose after stool collection on Day 1 (“Chow + Rha,” solid line) while the other half of the mice were maintained on normal drinking water throughout the entire experiment (“Chow,” dashed line). Mice receiving rhamnose-supplemented water on Days 1 and 2 (gray box) were returned to normal water on Day 3 after stool collection. Recombination ratios were determined for fecal DNA as described in Figure 3F. For day 3 “Chow” samples, n = 3. For all other days, n = 6 for both treatment groups. See also Figure S4. For (B)–(D), individual points represent independent biological replicates and the line represents the mean of the group. *p < 0.05, **p < 0.01.
Using this model, we tested the functionality of our inducible systems, CRISPRi, and integrases in vivo. First, SPF mice were colonized with the strain containing the arabinogalactan-inducible PBT0268 promoter-driving expression of NanoLuc (Figure S4A). Within a day of addition of arabinogalactan to the drinking water of the mice, luciferase activity in fecal pellets increased ~75-fold (Figure 5B). Following removal of inducer from the drinking water, luciferase activity in the fecal pellets of mice fed inducer rapidly returned to baseline, demonstrating tight temporal control of gene expression dependent on arabinogalactan.
To investigate whether more complex genetic circuits perform in the context of the mouse microbiome, we evaluated the dCas9NL3 repressor cascade, which is composed of the CRISPRi system as well as the PLacO23 IPTG-inducible promoter, within stably colonized B. thetaiotaomicron. Within 24 hr of adding IPTG to drinking water, CRISPRi elicited an ~20-fold reduction in gene expression compared to the uninduced control (Figure 5C). The fold repression observed in vivo is similar to that measured in vitro. Luciferase activity returned to baseline 6 days following the removal of IPTG from drinking water. Moreover, expression of dCas9 and NanoLuc did not significantly impact in vivo fitness compared to uninduced controls (Figures S4A and S4B).
To test the function of recombinases in vivo, we colonized mice with a B. thetaiotaomicron strain containing the rhamnose-inducible Int12 integrase memory switch (Figure 3E). All mice were fed with plant-based chow that was determined to be composed of 0.3% rhamnose (w/w). In addition, after 1 day of colonization, the drinking water of half of the mice was supplemented with 0.5 M rhamnose to further induce the memory switch. Stool was collected over the course of the experiment, and the absolute number of unflipped (wild-type) and flipped Int12 recognition sequences was determined by qPCR (Figure 5D; Experimental Procedures). A background recombination rate of ~11% per day was detected in mice fed on rhamnose-containing chow but not supplemented with rhamnose in their drinking water (Figure 5D, “Chow”). In mice supplemented with exogenous rhamnose (Figure 5D, “Chow + Rha”), the recombinase switch achieved >90% flipping in % 1 day, a statistically significant increase over mice not supplemented with rhamnose in the water (p < 0.01; Figure 5D).
DISCUSSION
Here, we developed a versatile set of genetic technologies for the manipulation of the abundant gut symbiont B. thetaiotaomicron. Given the distinct transcriptional and translational control mechanisms in B. thetaiotaomicron, we laid the foundation for circuit design in B. thetaiotaomicron by identifying promoters and RBSs for this organism through literature searches and library construction. The resulting toolbox expanded upon the number and expression range of genetic parts available for Bacteroidetes (previous range: 102) (Accetto and Avguštin, 2011; Hamady et al., 2008; Parker and Smith, 2012; Wegmann et al., 2013) and achieved ranges of expression similar to those of libraries characterized for other gut-associated bacteria, including E. coli (range: 104–105) (Cox et al., 2007; Kosuri et al., 2013; Salis et al., 2009) and lactic acid bacteria (range: 103) (Jensen and Hammer, 1998; Kleerebezem et al., 1997). Using a similar approach, we characterized four orthogonal-inducible expression systems. This library of B. thetaiotaomicron promoters and RBSs enabled us to import host-independent machinery, dCas9 and serine integrases, to implement more complex circuits. We envision that similar approaches, coupled with continuing advances in DNA sequencing and synthesis technologies, will enable the rapid and large-scale porting of genetic parts and devices from model organisms into new chassis that can address real-world applications. Lastly, we demonstrated the functionality of these genetic parts in the context of the mouse microbiome. Future work will focus on scaling the complexity of genetic circuits that can be implemented in commensal bacteria and pursuing long-term and quantitative control of microbial and mammalian host biology in vivo.
For microbiome engineering applications, the ability to precisely modulate gene expression in commensal organisms should enable functional studies of the microbiome, non-invasive monitoring of in vivo environments, and long-term targeted therapeutics. For example, our constitutive and inducible systems, integrases, and CRISPRi regulators could be integrated for higher-order computation in B. thetaiotaomicron, such as digital logic, memory, and analog circuits. These engineered commensals could be used to map the dose-dependent and temporal effects of specific surface polysaccharides (Liu et al., 2008) or heterologous pathways (Claesen and Fischbach, 2015) on colonization and maintenance of the gut microbiota and on host health. Higher-order combinations of inducible promoters linked with integrases could achieve Boolean logic with embedded cellular memory (Siuti et al., 2013), enabling surveillance of the gut environment. Furthermore, environmental sensing with analog circuits (Daniel et al., 2013) coupled with precision expression control of heterologous pathways in B. thetaiotaomicron could be exploited for on-demand, localized, and rheostatic delivery of therapeutic molecules. We have also shown that the CRISPRi system can be used to dynamically manipulate bacterial processes in B. thetaiotaomicron by targeting endogenous genes. dCas9-mediated repression could be induced in a commensal library of B. thetaiotaomicron harboring distinct guide RNAs to identify genes required for B. thetaiotaomicron maintenance (Sonnenburg et al., 2010) or interspecies interactions (Mahowald et al., 2009). With these genetic resources, we envision that B. thetaiotaomicron will be a useful platform for cellular sensing, computation, and actuation at the host-microbe interface in the gut.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
B. thetaiotaomicron VPI-5482 (ATCC 29148) (GenBank: AE015928.1) was used for all studies except for the integrase-based memory units. B. thetaiotaomicron VPI-5482 Δtdk was used for the insertion of the memory array and the characterization of integrases.
Genetic Parts and Plasmids
Genetic parts and plasmids used in this study are listed in Tables S3 and S4 and will be publically available and deposited in Addgene upon publication. Single guide RNAs (sgRNA) used in this study are listed in Table S5. pNBU2 constructs were conjugated into B. thetaiotaomicron from E. coli. The DNA memory array cassette (Yang et al., 2014) was introduced into the chromosome of a B. thetaiotaomicron Δtdk strain by allelic exchange (Supplemental Experimental Procedures).
Construction of Promoter and RBS Libraries
Promoters and RBS libraries were constructed by PCR using primers with specific or degenerate sequences (Supplemental Experimental Procedures).
Luciferase Assay
NanoLuc luciferase assay was performed with cell lysates or fecal suspensions according to the manufacturer’s suggestions (Supplemental Experimental Procedures). Luciferase activities were measured with an integration time of 1 s at a gain setting of 100 in a BioTek Synergy H1 Hybrid Reader.
Construction and Analysis of the rpiL* RBS Library
The rpiL* RBS was mutagenized by amplifying the plasmid pAT593 (encoding PBT1311-rpiL*-NanoLuc in a pNBU2 vector) with primer oAT617 in combination with primer oAT614, primer oAT615, or primer oAT616 (Table S6), which were each synthesized with degenerate nucleotides at three positions (43 = 64 potential members in each library). The resulting fragments were digested with BbsI, ligated with T4 ligase, and digested with DpnI, yielding three scarless PBT1311-rpiL* RBS libraries.
Growth and Induction
For inducible promoter and dCas9 assays, overnight cultures were diluted 1:100 in fresh tryptone yeast extract glucose medium (TYG) supplemented with inducer. The inducers used were IPTG (Goldman), L-rhamnose (Sigma), chondroitin sulfate A from bovine trachea (Sigma), (+)-arabinogalactan from larch wood (TCI), and fucose (Sigma). Cultures were grown anaerobically at 37°C to an optical density (OD600) of ~0.4–0.8 (~5 hr) before assaying luciferase activity.
In Vitro Genetic Memory Assays
To identify integrases that function in B. thetaiotaomicron, pNBU2-based plasmids containing PAM4-rpiL*RBS-integrase combinations were conjugated into B. thetaiotaomicron Δtdk harboring a chromosomal copy of the memory array. Cells were plated on BHIS+Gm+Erm to select for strains that contained the promoter-RBS-integrase plasmids. DNA was isolated from transconjugants were grown were assayed by PCR for inversion of spacer sequences in the memory array cassette as described (Yang et al., 2014). Control primers specific for unflipped (wild-type) memory array were used to detect the wild-type orientation of the spacer sequences (Table S6).
To characterize the function of integrase Int12 in B. thetaiotaomicron, a pNBU2-based plasmid containing PBT3763-rpiL*C51RBS-Int12 was constructed and conjugated into B. thetaiotaomicron memory array strain described above. For the recombinase response curve, an overnight culture grown in TYG was diluted (1:100) into TYG or TYG supplemented with rhamnose and grown for 8 hr at 37°C. The final OD600 of each culture was measured using a Cary 50 UV-Vis spectrophotometer (Varion), and 500 µl of each cell culture was harvested by centrifugation, flash frozen, and stored at −80°C until DNA was extracted. For the recombinase time course assay, cells were grown in TYG supplemented with 10 mM rhamnose for 8 hr at 37°C, harvested periodically, and stored as described above. DNA was isolated using the DNeasy Blood and Tissue Kit (QIAGEN). Levels of integrase-mediated recombination in vitro were quantified using qPCR with specific to the wild-type (unflipped) (oAT836/837) or recombined (flipped) (oAT836/838) DNA at the target sequence. For absolute quantitation, we used a standard curve generated with purified DNA standards. Flipping ratio was calculated as (copies of flipped DNA)/(copies of flipped DNA + copies of unflipped DNA).
Polymyxin B Susceptibility
Minimum inhibitory concentrations (MICs) were determined by broth microdilution using TYG broth according to modified Clinical and Laboratory Standards Institute (CLSI) guidelines (Supplemental Experimental Procedures).
Fructose/Glucose Growth Assay
Overnight cultures were grown in TYG supplemented with or without 100 µM of IPTG. The following morning, samples were diluted 1:100 in MM supplemented with either 0.5% glucose or 0.5% fructose as the sole carbon source and with or without 100 µM IPTG. Cultures were incubated anaerobically at 37 °C and 200 µl samples were withdrawn from the cultures every hour for 10 hr to monitor growth (OD600) in a BioTek Synergy H1 Hybrid Reader.
Animals
All animal study protocols were approved by the MIT Animal Care and Use Committee. Specific-pathogen-free (SPF) female Swiss Webster mice (5–8 weeks) were purchased from Charles River and were housed and handled in non-sterile conditions. Mice were fed irradiated mouse chow (PicoLab Rodent Diet 20, LabDiet) and provided autoclaved or sterile filtered water for the duration of the experiment. Prior to bacterial gavage, mice were administered ciprofloxacin HCl (0.625 g/l), metronidazole (1 g/l) in sugar-sweetened drinking water for 7 days and were subsequently treated with metronidazole (100 mg/kg) by oral gavage every 24 hr and ciprofloxacin (0.625 g/l) dissolved in drinking water for 3 days. Animals were transferred to a clean cage and provided fresh medicated water on the fifth day of the antibiotic regimen. Two days after the cessation of antibiotic treatment, animals were inoculated with engineered strains of B. thetaiotaomicron (~5 × 108 CFU/mouse) by oral gavage. Mice belonging to the same treatment group were cohoused for the duration of the experiment. Fecal pellets were collected daily for luciferase and qPCR analysis (described below).
Enumeration of B. thetaiotaomicron Strains In Vivo
Strains were enumerated by qPCR with strain-specific primers (Table S6) targeting the NanoLuc (mmD662/663) or flipped (oAT836/838)/unflipped (oAT836/837) memory array alleles in total fecal DNA. For absolute quantitation, we used a standard curve generated with purified DNA standards.
Function of Inducible Promoters and CRISPRi In Vivo
After collection of stool 2 days post-bacterial gavage, mice were provided drinking water supplemented with 25 mM IPTG, 5% arabinogalactan, or kept on sterile water as controls. Normal drinking water was returned to animals 4 days post-bacterial gavage. For the arabinogalactan experiment, animals were transferred to a clean cage following removal of supplemented drinking water. For the CRISPRi experiment, animals were transferred to a clean cage 7 days post-bacterial gavage. Luciferase values were measured daily and normalized to the cell density by determining the copy number of NanoLuc by qPCR using primers mmD662/663.
Quantification of Memory Unit Recombination In Vivo
After collection of stool on Day 1, mice were provided with drinking water supplemented with 0.5 M rhamnose or were maintained on sterile water as controls. Mice were transferred to clean cages and returned to normal drinking water 3 days post-bacterial gavage. Levels of integrase-mediated recombination in vivo were determined by analyzing fecal DNA (1 µl) by qPCR with primers as described for the in vitro analysis. Flipping ratio was calculated as (copies of flipped DNA)/(copies of flipped DNA + copies of unflipped DNA).
Data Analysis, Statistics, and Computational Methods
All data were analyzed using GraphPad Prism version 6.05 (GraphPad Software, http://www.graphpad.com/). Error bars represent the SD of three experiments carried out on different days. Fold repression was calculated by dividing the luminescence of uninduced samples (RLU/CFU) by the luminescence of samples treated with maximum inducer concentration. All response curves were fit to a Hill function: Y = (BmaxXn)/(Kn+Xn) + C, where X is the input, Y is the output, Bmax is the maximum luminescence, n is the Hill coefficient, K is the threshold, and C is baseline luminescence. RBS frequency logos were generated for the region of the rpiL* RBS targeted by mutagenesis by comparing the frequency of each nucleotide at each position within the subpopulation (strongest 7% of RBSs and weakest 7% of RBSs) to the frequency of that nucleotide at that position in the full library.
Supplementary Material
Highlights.
We develop sets of genetic parts for a human commensal bacterium
Promoter and RBS libraries control gene expression over a 10,000-fold dynamic range
Orthogonal, inducible sensors enable synthetic genetic memory and CRISPRi
Genetic circuits respond to stimuli in a complex mouse gut microbiota
ACKNOWLEDGMENTS
We would like to thank N. Shoemaker (University of Illinois at Urbana-Champagne) for helpful advice in culturing B. thetaiotaomicron;M. Fischbach (University of California San Francisco) for pNBU2, pExchange, and B. thetaiotaomicron Δtdk; Robert Citorik (MIT) for technical assistance and helpful discussions; and Jon Kotula (Synlogic Inc.) for advice on mouse experiments. This work was funded by the National Science Foundation (EEC-0540879 grant to the Synthetic Biology Research Center); NIH P50GM098792, 1DP2OD008435, 1R01EB017755, and GM095765; the Defense Advanced Research Projects Agency (DARPA CLIO N66001-12-C-4016); the Defense Threat Reduction Agency (HDTRA1-14-1-0007); the Office of Naval Research (N00014-13-1-0424); and the MIT Center for Microbiome Informatics and Technology. M.M. is a Howard Hughes Medical Institute International Student Research fellow and a recipient of the Qualcomm Innovation Fellowship. M.M., A.C.T., C.A.V., and T.K.L. have filed a provisional application with the US Patent and Trademark Office on this work.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2015.06.001.
AUTHOR CONTRIBUTIONS
M.M. and A.C.T. designed and performed experiments. M.M., A.C.T., C.A.V., and T.K.L. conceived this study, analyzed the data, discussed results, and wrote the manuscript.
REFERENCES
- Accetto T, Avguštin G. Inability of Prevotella bryantii to form a functional Shine-Dalgarno interaction reflects unique evolution of ribosome binding sites in Bacteroidetes. PLoS ONE. 2011;6:e22914. doi: 10.1371/journal.pone.0022914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley DP, Rocha ER, Smith CJ. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 2000;193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- Brophy JAN, Voigt CA. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Guo L, Zhang Y, Walzem RL, Pendergast JS, Printz RL, Morris LC, Matafonova E, Stien X, Kang L, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J. Clin. Invest. 2014;124:3391–3406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesen J, Fischbach MA. Synthetic microbes as drug delivery systems. ACS Synth. Biol. 2015;4:358–364. doi: 10.1021/sb500258b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. USA. 2003;100:10446–10451. doi: 10.1073/pnas.1832655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–623. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Lu TK. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science. 2014;346:1256272. doi: 10.1126/science.1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Goto T, Tanaka K, Minh Tran C, Watanabe K. Complete sequence of pBFUK1, a carbapenemase-harboring mobilizable plasmid from Bacteroides fragilis, and distribution of pBFUK1-like plasmids among carbapenem-resistant B. fragilis clinical isolates. J. Antibiot. 2013;66:239–242. doi: 10.1038/ja.2012.109. [DOI] [PubMed] [Google Scholar]
- Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Hamady ZZR, Farrar MD, Whitehead TR, Holland KT, Lodge JPA, Carding SR. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology. 2008;154:3165–3174. doi: 10.1099/mic.0.2008/019109-0. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, Silver PA. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. USA. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt JM, Alper HS. Advances and current limitations in transcript-level control of gene expression. Curr. Opin. Biotechnol. 2015;34C:98–104. doi: 10.1016/j.copbio.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai Q-A, Tran AB, Paull M, Keasling JD, Arkin AP, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013;10:354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- Nemunaitis J, Cunningham C, Senzer N, Kuhn J, Cramm J, Litz C, Cavagnolo R, Cahill A, Clairmont C, Sznol M. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther. 2003;10:737–744. doi: 10.1038/sj.cgt.7700634. [DOI] [PubMed] [Google Scholar]
- Nielsen AAK, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AAK, Segall-Shapiro TH, Voigt CA. Advances in genetic circuit design: novel biochemistries, deep part mining, and precision gene expression. Curr. Opin. Chem. Biol. 2013;17:878–892. doi: 10.1016/j.cbpa.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Parker AC, Smith CJ. Genetic and biochemical analysis of a novel Ambler class A beta-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 1993;37:1028–1036. doi: 10.1128/aac.37.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AC, Smith CJ. Development of an IPTG inducible expression vector adapted for Bacteroides fragilis. Plasmid. 2012;68:86–92. doi: 10.1016/j.plasmid.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel EH, Paul LV, Patrick S, Abratt VR. Rhamnose catabolism in Bacteroides thetaiotaomicron is controlled by the positive transcriptional regulator RhaR. Res. Microbiol. 2008;159:678–684. doi: 10.1016/j.resmic.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Bennett TK, Payne CM, Smith CJ. Insertional activation of cepA leads to high-level β-lactamase expression in Bacteroides fragilis clinical isolates. J. Bacteriol. 1994;176:4376–4384. doi: 10.1128/jb.176.14.4376-4384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J, Paterson Y. Live-attenuated Listeria-based immunotherapy. Expert Rev. Vaccines. 2013;12:493–504. doi: 10.1586/erv.13.34. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 1984;38:293–313. doi: 10.1146/annurev.mi.38.100184.001453. [DOI] [PubMed] [Google Scholar]
- Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013;31:448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Natl. Acad. Sci. USA. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, Cox E, Remon JP, Remaut E. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 2003;21:785–789. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. An unusual primary sigma factor in the Bacteroidetes phylum. Mol. Microbiol. 2005;56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Shoemaker NB, Wang G-R, Salyers AA. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 2000;182:3559–3571. doi: 10.1128/jb.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U, Horn N, Carding SR. Defining the bacteroides ribosomal binding site. Appl. Environ. Microbiol. 2013;79:1980–1989. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Nielsen AAK, Fernandez-Rodriguez J, McClune CJ, Laub MT, Lu TK, Voigt CA. Permanent genetic memory with >1-byte capacity. Nat. Methods. 2014;11:1261–1266. doi: 10.1038/nmeth.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.