Abstract
Observational epidemiological studies have associated plasma lipid concentrations with risk for coronary heart disease (CHD), but these studies cannot distinguish cause from mere correlation. Human genetic studies, when considered with the results of randomized controlled trials of medications, can potentially shed light on whether lipid biomarkers are causal for diseases. Genetic analyses and randomized trials suggest that low-density lipoprotein (LDL) is causal for CHD whereas high-density lipoprotein (HDL) is not. Surprisingly, human genetic evidence suggests that lipoprotein(a) [Lp(a)] and triglyceride-rich lipoproteins (TRLs) causally contribute to CHD. Gene variants leading to higher levels of plasma apolipoprotein B-containing lipoproteins [LDL, TRLs, and/or Lp(a)] consistently increase risk for CHD. For TRLs, the most compelling evidence revolves around lipoprotein lipase (LPL) and its endogenous facilitator (APOA5) and inhibitory proteins (APOC3, ANGPTL4). Combined, these genetic results anticipate that, beyond LDL, pharmacologic lowering of TRLs and/or Lp(a) will reduce risk for CHD, but this remains to be proven through randomized controlled trials.
Keywords: Atherosclerosis, coronary disease, genetics, genetics, association studies, lipids and lipoproteins
A brief summary of genetic studies of lipid risk factors for atherosclerosis
Atherosclerotic vascular disease, particularly coronary heart disease (CHD) and its complication of myocardial infarction (MI), is the leading cause of death worldwide. Although many environmental factors influence the risk of CHD, genetics play an important role as well. A parental history of premature CHD is associated with a two- to three-fold increase in one’s personal CHD risk.1 Plasma lipoproteins—low-density lipoprotein (LDL), high-density lipoprotein (HDL), triglyceride-rich lipoproteins (TRLs), and lipoprotein(a) [Lp(a)]—have all been found to be associated with CHD risk in observational epidemiological studies.2–4 Whereas LDL (as assessed by its cholesterol content, LDL-C) and TRLs (as assessed by triglycerides) are directly associated with CHD risk, HDL (as assessed by its cholesterol content, HDL-C) is inversely associated with CHD risk. These lipid traits are themselves partly genetically determined, with about one-half of the inter-individual variation in plasma lipid concentrations attributable to genetic variants.5
Over the past few decades, a variety of human genetics approaches have identified genes that are involved in lipoprotein metabolism and, thus, have possible involvement in the pathogenesis of CHD. Traditional linkage analyses and candidate gene sequencing have pinpointed genes responsible for familial hypercholesterolemia (FH) as well as other inherited lipid disorders.6 These genes harbor rare DNA variants—usually deleterious loss-of-function mutations, but occasionally gain-of-function mutations—that so greatly affect protein function that they singlehandedly produce highly aberrant plasma lipid levels that can potentially result in CHD.
Common DNA variants with small or moderate effects on gene function also modulate plasma lipid levels, albeit in a polygenic fashion. Family-based studies are not well suited to identifying these variants; instead, population-based association studies are needed. The primary population-based study design used to date has been the common variant association study (also commonly referred to as a genome-wide association study, GWAS), in which large cohorts of unrelated individuals are genotyped at millions of single nucleotide polymorphisms (SNPs) across the genome. For each SNP, individuals with one genotype at the SNP are compared with individuals with another genotype to assess whether there is a phenotypic difference. Such “lead SNPs” indicate a genomic locus harboring a gene or other functional element that influences the phenotype. With respect to plasma lipid levels, a GWAS with about 100,000 people identified 95 loci with LDL-C, HDL-C, triglycerides, and/or total cholesterol,7 and a subsequent association study with almost 190,000 people increased the yield to 157 loci.8
More recently, next-generation sequencing technologies have enabled the “rare variant association study” (RVAS). As rare variants occur too infrequently to allow association tests of individual variants, RVASs require aggregating rare variants into sets and comparing the aggregate frequency in cases vs. controls or mean quantitative trait values in those who carry a set versus those who do not. RVASs have identified coding variants with large effects on gene function in large cohorts of unrelated individuals, effectively combining the advantages of GWASs and traditional family-based studies. Such RVASs have been able to identify a number of novel genes associated with lipids.9,10
As related below, these discoveries have yielded surprising insights into the causal roles of plasma lipids in the pathogenesis of CHD and into the appropriate use of lipid biomarkers to predict the clinical efficacy of lipid-modifying agents in the reduction of CHD risk.
The Mendelian randomization study design
Starting in the 1960s with the seminal finding in the Framingham Heart Study that plasma total cholesterol concentration was associated with future risk for CHD,11 hundreds of biomarkers have been reported to be associated with CHD risk in observational epidemiological studies. What is unclear is how many of these biomarkers are themselves causal in the pathobiology of CHD, and how many are simply proxies for other causal processes. While any of these biomarkers is potentially useful for cardiovascular risk prediction, only the causal biomarkers represent potential therapeutic targets. The gold standard for proving that a biomarker is causal is a randomized controlled trial (RCT) that demonstrates that an intervention specifically targeting the biomarker reduces the risk of CHD. Such RCTs typically require following thousands or tens of thousands of individuals for several years, making them a time-consuming and costly proposition.
The principles of human genetics offer an alternative study design, called Mendelian randomization, that is akin to an RCT that has already been carried out by nature.12,13 DNA variants can be used as instruments to assess whether a biomarker that has been found to have an epidemiological association with risk for a disease is truly causal for the disease. If (1) a DNA variant is known to directly influence the biomarker level (e.g., a non-coding variant in a promoter or enhancer that alters the expression of the gene that encodes the biomarker) or the activity of a protein that directly influences the biomarker level (e.g., a coding variant that affects the function of an enzyme that metabolizes the biomarker), and (2) the biomarker is truly causal for a disease, then (3) the DNA variant should be associated with disease risk to an extent consistent with the size of the effect of the DNA variant on the biomarker level and, in turn, the size of the effect of the biomarker on the disease process. Assuming that the Mendelian randomization study is adequately powered—which can require hundreds of thousands of individuals if the effect of the DNA variant on the biomarker is small—if the expected association between the DNA variant and disease risk is not apparent, it would argue that the biomarker is not causal for the disease.
The similarity between a Mendelian randomization study and an RCT arises from Mendel’s first law—the law of segregation—which dictates that each of a parent’s two alleles at the site of a DNA variant has a 50% chance of being passed to a given gamete and thus being transmitted to an offspring. In other words, there is random “assignment” of alleles to offspring. This assignment is unaffected by traditional confounders of observational epidemiology studies, e.g., age, disease status, socioeconomic status, etc. (There is the possibility of confounding via epigenetic phenomena affecting allele transmission, but this is a largely theoretical concern.) The law of segregation renders Mendelian randomization studies fairly impervious to confounding or reverse causation, which is also a major advantage of RCTs.
There are two possible shortcomings to a Mendelian randomization study. First, the study will only be as reliable as the estimates of effect sizes of the DNA variant on the biomarker and the biomarker on disease risk, which are obtained from observational epidemiological studies. If the estimates are unreliable, so too may be the conclusions drawn from a Mendelian randomization study. Second, there is an assumption that the DNA variant only affects disease risk via the biomarker in question, and not through any other mechanisms. If there is pleiotropy, i.e., the DNA variant affects multiple mechanisms, then an observed association between the DNA variant and disease risk may not be due to the biomarker. In this scenario, the biomarker may not itself be causal but instead act as a proxy for another mechanism influenced by the DNA variant.
These issues notwithstanding, Mendelian randomization is potentially a powerful tool to discriminate between causal and non-causal biomarkers for a disease. It is particular useful when considered in combination with RCTs in which pharmacological agents targeting the same biomarkers have been assessed for their effects on disease risk.
Plasma LDL cholesterol as a causal biomarker
There is now ample evidence from human genetics that the plasma LDL-C concentration represents a causal risk factor for CHD. Initial studies of patients with FH identified loss-of-function mutations in the LDLR gene to be linked to very high plasma LDL-C levels and premature CHD, with disease manifesting as early as childhood.14 Biological plausibility emerged from the recognition that the protein product of LDLR, the LDL receptor, resides on the plasma membrane and is responsible for the uptake of LDL particles out of the bloodstream into the cell, within which the LDL particles are degraded. In subsequent studies, mutations in the APOB and PCSK9 genes were identified in FH patients in whom LDLR mutations had been ruled out.15,16 APOB encodes apolipoprotein B (apoB), a key component of LDL particles that is the protein via which the LDL receptor binds to LDL particles and promotes their uptake into cells. Specific APOB mutations result in disruption of the interaction between the LDL receptor and its ligand, apoB, leading to an increased plasma LDL-C concentration and premature CHD. PCSK9 encodes a protein that acts as an antagonist to the LDL receptor by promoting its degradation. Gain-of-function mutations in PCSK9 thus cause FH and premature CHD by inhibiting the removal of LDL particles from the bloodstream. Conversely, loss-of-function mutations in PCSK9 result in increased levels of the LDL receptor on cells and reduction of the blood LDL-C concentration, translating into as much as an 88% reduction of CHD risk.17
These studies of LDLR, APOB, and PCSK9 variants make a strong argument for a casual link between LDL-C and CHD. However, they all impinge on a single biological pathway, cellular LDL particle uptake, that directly modulates the plasma LDL-C concentration in a specific way. What remains to be answered is whether all biological pathways that alter blood LDL-C concentration also alter CHD risk. A partial answer to this question was provided by an analysis of 10 lead SNPs in loci previously identified in GWAS to be primarily associated with LDL-C.18 These SNPs were assessed in a case-control study of about 20,000 individuals with MI and 50,000 control individuals. Nine of the 10 SNPs were found to be associated not only with LDL-C but also with risk of MI in the concordant direction (i.e., the same allele associated with both a decrease in LDL-C and a decrease in MI risk). While the loci included the LDLR, APOB, and PCSK9 genes, they also included a variety of other genes that influence LDL-C through other mechanisms, such as APOE, HMGCR (which encodes the cholesterol synthesis enzyme targeted by the statin drugs), and LPA [which encodes apolipoprotein(a), a component of the LDL-like Lp(a) particle].18–20
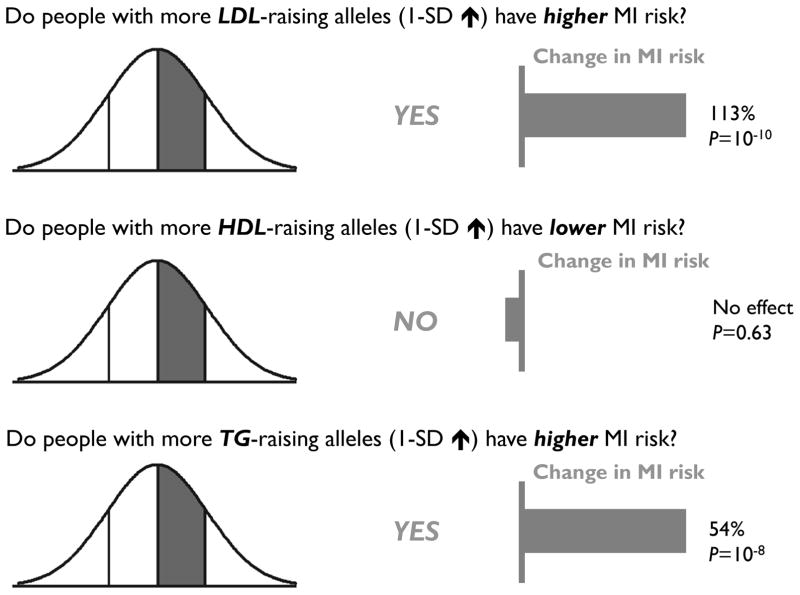
In the same study, a more formal Mendelian randomization analysis was performed for LDL-C in more than 50,000 cases and controls, employing a genetic score comprising 13 SNPs in loci primarily associated with LDL-C.18 Strikingly, whereas a 1-standard deviation (SD) increase in LDL-C (~35 mg/dl increase) was expected to be associated with a 54% increase in MI risk using data from observational epidemiological studies, a 1-SD increase in LDL-C because of genetic score was found to confer a 113% increase in risk (P = 2 × 10−10) (Figure 1). Thus, the analysis suggested that the genetic contribution to the plasma LDL-C level has, if anything, an outsize effect on CHD risk, strongly arguing for a generalized causal relationship between LDL-C and CHD.
Figure 1.
The cumulative effects of genetic variants that raise plasma low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglyceride (TG) levels on the risk of myocardial infarction (MI).
The most widely used LDL-C-lowering medications are the statins, which have been demonstrated in numerous RCTs in a broad variety of populations to reduce the risk of cardiovascular events.21 In isolation, these RCTs suggest but do not prove that LDL-C is causal for CHD, since statins have well-known pleiotropic effects—besides reducing plasma LDL-C levels, they also reduce plasma C-reactive protein (CRP) levels, which are a marker for inflammation.22 In principle, the beneficial effects of statins could be due to anti-inflammatory effects rather than lipid modification. However, in light of the strong genetic evidence that LDL-C is causal for CHD, it is reasonable to interpret the RCTs as demonstrating that the reduction of LDL-C is a primary mechanism by which statins protect against CHD.
Besides the statins, one of the more commonly used lipid-modifying drugs is ezetimibe, which works to reduce plasma LDL-C levels at least in part by inhibiting the protein product of the NPC1L1 gene.23 NPC1L1 regulates the absorption of dietary and biliary cholesterol in the gastrointestinal tract. Due to negative results in an early RCT assessing the effects of ezetimibe on a non-clinical endpoint, carotid initima-media thickness,24 ezetimibe was felt by many commentators to be an unproven medication despite its unequivocal, safe reduction of plasma LDL-C levels by 15%–20%. Recently, Mendelian randomization studies using variants in or near the NPC1L1 gene found that carriers of the variants not only had lower plasma LDL-C levels but also had decreased CHD risk.25,26 Indeed, the degree of risk reduction exceeded that which would be predicted from observational epidemiological data. Concordant with the genetic data, the Improved Reduction of Outcomes: Vytorin Efficacy International Trial (IMPROVE-IT) ultimately provided RCT data demonstrating that ezetimibe in fact does reduce CHD risk.27
The totality of the existing genetic data and RCT data strongly argue that LDL-C is causal for CHD and, as a corollary, that any therapy that specifically lowers plasma LDL-C levels should also reduce risk for CHD. Alirocumab and evolocumab, which are antibody-based therapeutics that target the PCSK9 protein, substantially reduce LDL-C.28,29 These medicines recently received regulatory approval in spite of the fact that RCT data demonstrating reductions in cardiovascular events were still pending. The genetic evidence undoubtedly provided a strong rationale for approval.
Plasma HDL cholesterol as a non-causal biomarker
There is a strong inverse association of plasma HDL-C concentrations with CHD,2 which for decades lent credence to the notion that pharmacological raising of HDL-C should protect against CHD. Yet recent genetic analyses have in general failed to support a causal role for HDL-C in CHD. As related above, a genetic score comprising LDL-C-associated SNPs was found to have a strong relationship with MI risk. The same study performed a parallel Mendelian randomization study in more than 50,000 cases and controls using a genetic score comprising 14 GWAS SNPs primarily associated with plasma HDL-C levels.18 Whereas a 1-standard deviation (SD) increase in HDL-C (~15 mg/dl) was expected to be associated with a 38% decrease in MI risk using data from observational epidemiological studies, a 1-SD decrease in HDL-C because of genetic score conferred no significant change in MI risk (7% decrease, P = 0.63) (Figure 1).
The same study performed a more focused Mendelian randomization analysis on a coding SNP (Asn396Ser) in the LIPG gene, which encodes endothelial lipase, an enzyme that metabolizes HDL particles but has little effect on plasma LDL-C and triglycerides.18 In order to obtain adequate power for the analysis, the SNP was genotyped in about 20,000 individuals with MI and 95,000 control individuals. Carriers of the LIPG Asn396Ser variant had increased plasma HDL-C levels, on average about 5.5 mg/dL. This degree of increase in HDL-C was expected to be associated with a 13% decrease in MI risk using data from observational epidemiological studies. However, carriers of the LIPG Asn396Ser variant were found to have a negligible change in MI risk (1% decrease, P = 0.85, 95% confidence interval ranging from an 11% increase to a 12% decrease), essentially ruling out an effect of LIPG on the pathogenesis of CHD. In other studies, genetic analyses of both common variants in the ABCA1 gene, which encodes the ATP-binding-cassette transporter A1 involved in reverse cholesterol transport, and rare variants in the same gene that are linked to familial hypoalphalipoproteinemia and Tangier disease, have been unable to demonstrate a relationship between decreased plasma HDL-C levels in affected individuals and increased CHD risk.30
In contrast to LDL-C, the collective genetic data suggest that HDL-C is not causal for CHD risk, at least in a simplistic sense. While the data cannot rule out that there are some biological mechanisms that lead to increased plasma HDL-C levels that also protect against CHD, it seems fair to conclude that not all interventions that raise HDL-C will reduce CHD risk. Further support for this conclusion is provided by RCT data, most notably with the cholesteryl ester transfer protein (CETP) inhibitors, which substantially raise plasma HDL-C levels. Three inhibitors of CETP—torcetrapib, dalcetrapib, and evacetrapib—all raised HDL-C substantially, and each failed to reduce risk for CHD in large-scale RCTs. All three studies—the Investigation of Lipid Level Management to Understand Its Impact in Atherosclerotic Events (ILLUMINATE) trial of torcetrapib, which increased HDL-C by 70%; the dal-OUTCOMES trial of dalcetrapib, which increased HDL-C by 30%; and the Assessment of Clinical Effects of Cholesteryl Ester Transfer Protein Inhibition With Evacetrapib in Patients at a High-Risk for Vascular Outcomes (ACCELERATE) trial of evacetrapib, which was projected to raise HDL-C by more than 90%—were all terminated prematurely due to lack of clinical efficacy.31–33 Indeed, torcetrapib appeared to result in increased cardiovascular events and death, although this has been attributed to off-target, non-lipid-related effects of this particular medication.
Interestingly, CETP has pleiotropic effects on blood lipid levels, and SNPs in or near the CETP gene are not only associated with altered plasma HDL-C levels but also plasma LDL-C in the opposite direction, albeit to a lesser degree. Multiple genetic studies have found that CETP variants are associated with modest changes in CHD risk.18,34,35 Given the pleiotropy at work, it seems likely that the altered CHD risk is due primarily to the effect of CETP on LDL-C rather than HDL-C. This observation leaves open the possibility that the ongoing RCT of a fourth CETP inhibitor—anacetrapib—may find some degree of clinical utility, if the LDL-C-lowering effect is large enough to be beneficial and the HDL-C-raising effect is not harmful.
Plasma triglyceride-rich lipoproteins as casual
The epidemiological association of plasma triglyceride levels with CHD risk is not as strong as those of LDL-C and HDL-C.3 Nonetheless, genetic evidence is emerging that TRLs as assessed by plasma trigylcerides represent a causal risk factor for CHD. SNPs in at least six genes that modulate plasma triglyceride levels—APOA5, APOC3, ANGPTL4, LPL, APOA4, and TRIB1—have been persuasively linked to CHD.18,36–43 However, Mendelian randomization studies using SNPs associated with plasma triglyceride levels are difficult to interpret due to most of those SNPs having pleiotropic relationships with lipids. Out of 185 lead SNPs for GWAS loci associated with one or more plasmid lipid traits, 94 are associated with triglycerides; of those, just 7 are only associated with triglycerides, with the other 87 also associated with LDL-C and/or HDL-C.44 This is perhaps not surprising, since triglycerides are carried by multiple classes of lipoprotein particles in the blood and, accordingly, the measured plasma triglyceride concentration reflects contributions from multiple physiological processes.
Given this pleiotropy, a statistical framework—termed multivariable Mendelian randomization—to separate the triglyceride-associated effects on CHD risk from the LDL-C- and HDL-C-associated effects on CHD risk was recently developed.44 Confirming previous observations, the isolated LDL-C genetic effect (i.e., contribution of all LDL-C-associated SNPs, after adjustment for the HDL-C and triglyceride effects) confers significantly increased risk of CHD, whereas the isolated HDL-C genetic effect on CHD risk is negligible. In contrast to HDL-C, the isolated triglyceride effect increases the risk of CHD to a comparable degree as the isolated LDL-C effect (Figure 1). This finding suggests that the plasma triglyceride concentration captures risk processes causal for CHD that is independent of the plasma LDL-C concentration.
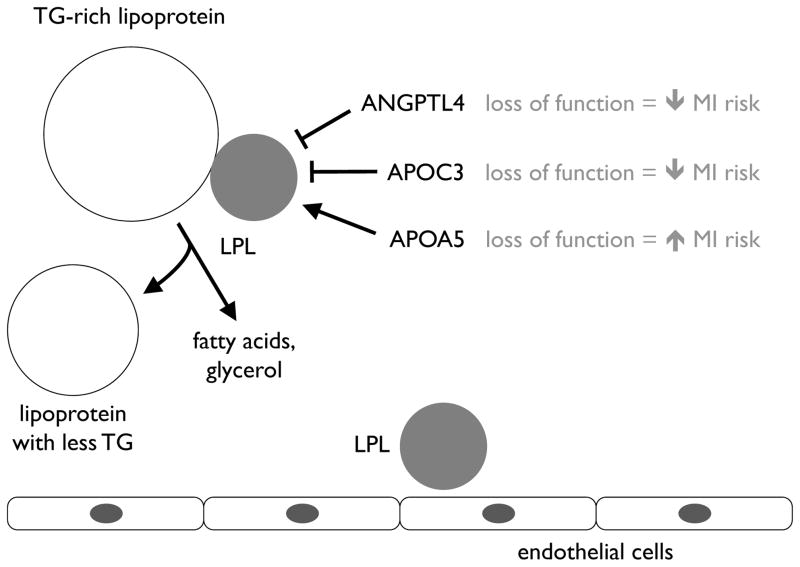
Exactly what risk factor or, potentially, risk factors are embodied in the plasma triglyceride concentration remain to be fully defined, though presumably they involve the metabolism of TRLs that carry triglycerides in the blood. The aforementioned APOA5, APOC3, ANPGTL4, and LPL genes all share the common characteristic that they either encode lipoprotein lipase or encode regulators of lipoprotein lipase, a key enzyme that hydrolyzes triglycerides in various lipoprotein particles (Figure 2). This suggests that lipoprotein lipase is central to a causal pathway for CHD. Another possible casual risk factor may lie in postprandial cholesterol metabolism, specifically the amount of cholesterol in remnant lipoproteins, with which plasma triglyceride levels are strongly correlated. Remnant lipoproteins appear to promote atherosclerosis in much the same way as LDL particles.45
Figure 2.
The causal role of the lipoprotein lipase (LPL) pathway on the risk of myocardial infarction (MI). LPL metabolizes triglyceride (TG)-rich lipoproteins in the bloodstream. ANGPTL4, APOC3, and APOA5 all modulate LPL activity. Variants in all of the genes encoding these proteins influence MI risk.
Of note, RCTs of triglyceride-lowering therapies have yielded ambiguous results, with a trial of gemfibrozil and a trial of fenofibrate resulting in reduced cardiovascular events46,47 (although the latter did not show a difference in the primary endpoint, coronary events) but another trial of fenofibrate and a trial of omega-3 fatty acids showing no such reduction.48,49 This may reflect that the specific mechanism by which plasma triglycerides are lowered, in combination with other factors such as the degree of triglyceride reduction and characteristics of the study population, determines the extent of clinical benefit. Triglyceride-lowering therapies that do so by altering TRLs via the lipoprotein lipase pathway may prove to be particularly efficacious.
Lipoprotein(a) and lipoprotein-associated phospholipase A2 as causal and non-causal biomarkers
Lp(a) is an LDL-like particle that is covalently linked to a protein called apolipoprotein(a), expressed by the LPA gene. The plasma Lp(a) level is notable in that it varies up to 1000-fold among individuals, with the vast majority of the variation determined by genetic variation.50 With plasma Lp(a) being associated with CHD risk,4 a natural question has been whether Lp(a) is a causal risk factor for disease. Mendelian randomization studies using variants in or near the LPA gene have unequivocally demonstrated that genetically elevated Lp(a) results in increased risk of CHD.19,20 Thus, in principle, therapies that specifically reduce plasma Lp(a) concentrations should confer cardiovascular protection.
Lipoprotein-associated phospholipase A2 (Lp-PLA2) is an enzyme that is encoded by the PLA2G7 gene and circulates in the plasma, primarily associated with LDL particles. Both Lp-PLA2 mass and activity in the plasma are associated with CHD risk.51 These observations prompted two large RCTs with darapladib, an inhibitor of Lp-PLA2. Both the Stabilization of Atherosclerotic Plaque by Initiation of Darapladib Therapy (STABILITY) trial and The Stabilization Of pLaques usIng Darapladib-Thrombolysis In Myocardial Infarction 52 Trial (SOLID-TIMI 52) found that darapladib did not reduce the risk of CHD,52–53 calling into question whether Lp-PLA2 is a causal risk factor for disease. A subsequently reported Mendelian randomization study using variants in PLA2G7 found no association with CHD risk.54 The consistency of the results of RCTs and Mendelian randomization studies for statins, ezetimibe, and darapladib and their target genes supports the notion that Mendelian randomization studies could potentially be used to prioritize RCTs for those agents most likely to result in the desired clinical outcome.
Implication of lipid and non-lipid causal factors in atherosclerosis
The weight of the genetic evidence suggests that the plasma LDL-C, triglycerides, and Lp(a) concentrations reflect casual risk factors for CHD, whereas the plasma HDL-C concentration does not. This is contrary to the expectations one would have if going purely by observational epidemiological studies, which generally find that HDL-C has the strongest association with CHD. The disparity highlights the need to distinguish between association and causation with respect to biomarkers of disease, with genetic approaches such as Mendelian randomization studies proving to be helpful in this regard.
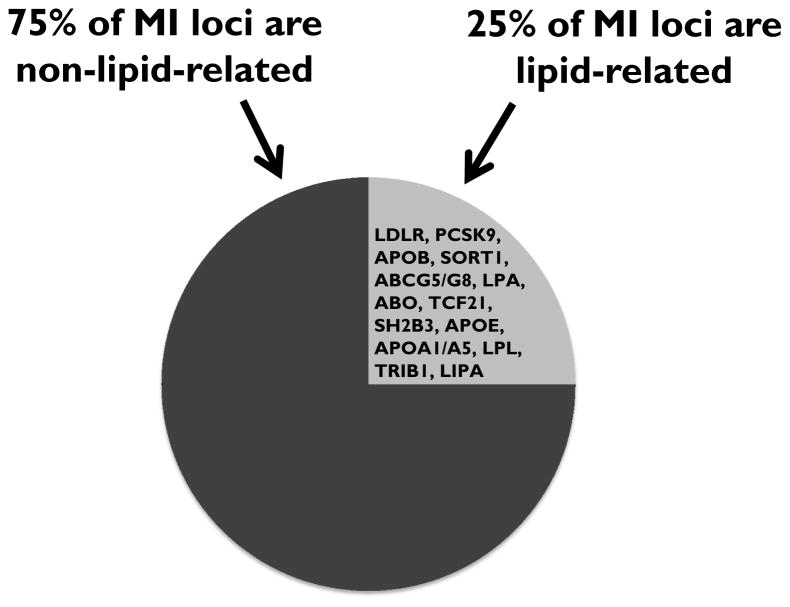
Another notable finding to emerge from human genetic studies is the overall importance of lipid causal factors in atherosclerosis. The largest GWAS reported to date for coronary artery disease identified a total of 55 loci associated with the clinical phenotype.55 Remarkably, 13 of the loci are clearly linked to plasma LDL-C, triglycerides, and/or Lp(a) (Figure 3). The common feature of LDL particles, TRLs, and Lp(a) is that they all carry apoB, reinforcing the notion that apoB-containing particles are generally atherogenic and that approaches that reduce any of these classes of apoB-containing particles may have therapeutic efficacy. Of course, any such approaches will need to be validated with RCTs before clinical use.
Figure 3.
Lipid-associated and non-lipid-associated genes identified in a genome-wide association study (GWAS) on coronary heart disease (CHD).
A much smaller subset of the 55 loci are linked to other recognized risk factors for CHD, including blood pressure and inflammation. The remaining loci appear to contribute to CHD by as of yet unrecognized pathogenetic mechanisms, suggesting that there are quite a few causal risk factors (and possibly therapeutic targets) remaining to be discovered.
Supplementary Material
ABBREVIATIONS
- CHD
coronary heart disease
- MI
myocardial infarction
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- TRL
triglyceride-rich lipoprotein
- Lp(a)
lipoprotein(a)
- FH
familial hypercholesterolemia
- GWAS
genome-wide association study
- SNP
single nucleotide polymorphism
- RVAS
rare variant association study
- RCT
randomized controlled trial
- apoB
apolipoprotein B
- SD
standard deviation
- CRP
C-reactive protein
- CETP
cholesteryl ester transfer protein
- Lp-PLA2
lipoprotein-associated phospholipase A2
Footnotes
DISCLOSURES
None
SOURCES OF FUNDING
None
References
- 1.Lloyd-Jones DM, Nam BH, D’Agostino RB, Sr, Levy D, Murabito JM, Wang TJ, Wilson PW, O’Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 2.Emerging Risk Factors Collaboration. Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, Thompson A, Wood AM, Lewington S, Sattar N, Packard CJ, Collins R, Thompson SG, Danesh J. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarwar N, Danesh J, Eiriksdottir G, Sigurdsson G, Wareham N, Bingham S, Boekholdt SM, Khaw KT, Gudnason V. Triglycerides and the risk of coronary heart disease: 10,158 incident cases among 262,525 participants in 29 Western prospective studies. Circulation. 2007;115:450–458. doi: 10.1161/CIRCULATIONAHA.106.637793. [DOI] [PubMed] [Google Scholar]
- 4.Emerging Risk Factors Collaboration. Erqou S, Kaptoge S, Perry PL, Di Angelantonio E, Thompson A, White IR, Marcovina SM, Collins R, Thompson SG, Danesh J. Lipoprotein(a) concentration and the risk of coronary heart disease, stroke, and nonvascular mortality. JAMA. 2009;302:412–423. doi: 10.1001/jama.2009.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heller DA, de Faire U, Pedersen NL, Dahlén G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- 6.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin SY, Lindström J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin MR, Jula A, Kähönen M, Kaprio J, Kesäniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, März W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njølstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, Isaacs A, Jakobsdottir J, Feitosa MF, Davies G, Huffman JE, Manichaikul A, Davis B, Lohman K, Joon AY, Smith AV, Grove ML, Zanoni P, Redon V, Demissie S, Lawson K, Peters U, Carlson C, Jackson RD, Ryckman KK, Mackey RH, Robinson JG, Siscovick DS, Schreiner PJ, Mychaleckyj JC, Pankow JS, Hofman A, Uitterlinden AG, Harris TB, Taylor KD, Stafford JM, Reynolds LM, Marioni RE, Dehghan A, Franco OH, Patel AP, Lu Y, Hindy G, Gottesman O, Bottinger EP, Melander O, Orho-Melander M, Loos RJ, Duga S, Merlini PA, Farrall M, Goel A, Asselta R, Girelli D, Martinelli N, Shah SH, Kraus WE, Li M, Rader DJ, Reilly MP, McPherson R, Watkins H, Ardissino D, Zhang Q, Wang J, Tsai MY, Taylor HA, Correa A, Griswold ME, Lange LA, Starr JM, Rudan I, Eiriksdottir G, Launer LJ, Ordovas JM, Levy D, Chen YD, Reiner AP, Hayward C, Polasek O, Deary IJ, Borecki IB, Liu Y, Gudnason V, Wilson JG, van Duijn CM, Kooperberg C, Rich SS, Psaty BM, Rotter JI, O’Donnell CJ, Rice K, Boerwinkle E, Kathiresan S, Cupples LA NHLBI GO Exome Sequencing Project. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lange LA, Hu Y, Zhang H, Xue C, Schmidt EM, Tang ZZ, Bizon C, Lange EM, Smith JD, Turner EH, Jun G, Kang HM, Peloso G, Auer P, Li KP, Flannick J, Zhang J, Fuchsberger C, Gaulton K, Lindgren C, Locke A, Manning A, Sim X, Rivas MA, Holmen OL, Gottesman O, Lu Y, Ruderfer D, Stahl EA, Duan Q, Li Y, Durda P, Jiao S, Isaacs A, Hofman A, Bis JC, Correa A, Griswold ME, Jakobsdottir J, Smith AV, Schreiner PJ, Feitosa MF, Zhang Q, Huffman JE, Crosby J, Wassel CL, Do R, Franceschini N, Martin LW, Robinson JG, Assimes TL, Crosslin DR, Rosenthal EA, Tsai M, Rieder MJ, Farlow DN, Folsom AR, Lumley T, Fox ER, Carlson CS, Peters U, Jackson RD, van Duijn CM, Uitterlinden AG, Levy D, Rotter JI, Taylor HA, Gudnason V, Jr, Siscovick DS, Fornage M, Borecki IB, Hayward C, Rudan I, Chen YE, Bottinger EP, Loos RJ, Sætrom P, Hveem K, Boehnke M, Groop L, McCarthy M, Meitinger T, Ballantyne CM, Gabriel SB, O’Donnell CJ, Post WS, North KE, Reiner AP, Boerwinkle E, Psaty BM, Altshuler D, Kathiresan S, Lin DY, Jarvik GP, Cupples LA, Kooperberg C, Wilson JG, Nickerson DA, Abecasis GR, Rich SS, Tracy RP, Willer CJ NHLBI Grand Opportunity Exome Sequencing Project. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94:233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J., 3rd Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 12.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 13.Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5(8):e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehrman MA, Schneider WJ, Südhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci U S A. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derré A, Villéger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 17.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 18.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart A, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett MS, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki ML, Perola M, Havulinna A, de Faire U, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PI, Klungel OH, Maitland-van der Zee AH, Peters BJ, de Boer A, Grobbee DE, Kamphuisen PW, Deneer VH, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WM, Boer JM, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, König IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox KA, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schäfer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O’Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, Bennett D, Silveira A, Malarstig A, Green FR, Lathrop M, Gigante B, Leander K, de Faire U, Seedorf U, Hamsten A, Collins R, Watkins H, Farrall M PROCARDIS Consortium. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 20.Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, Nordestgaard BG. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301:2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- 21.Kapur NK, Musunuru K. Clinical efficacy and safety of statins in managing cardiovascular risk. Vasc Health Risk Manag. 2008;4:341–353. doi: 10.2147/vhrm.s1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridker PM, Cannon CP, Morrow D, Rifai N, Rose LM, McCabe CH, Pfeffer MA, Braunwald E Pravastatin or Atorvastatin Evaluation and Infection Therapy-Thrombolysis in Myocardial Infarction 22 (PROVE IT-TIMI 22) Investigators. C-reactive protein levels and outcomes after statin therapy. N Engl J Med. 2005;352:20–28. doi: 10.1056/NEJMoa042378. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Calvo M, Lisnock J, Bull HG, Hawes BE, Burnett DA, Braun MP, Crona JH, Davis HR, Jr, Dean DC, Detmers PA, Graziano MP, Hughes M, Macintyre DE, Ogawa A, O’neill KA, Iyer SP, Shevell DE, Smith MM, Tang YS, Makarewicz AM, Ujjainwalla F, Altmann SW, Chapman KT, Thornberry NA. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1) Proc Natl Acad Sci U S A. 2005;102:8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastelein JJ, Akdim F, Stroes ES, Zwinderman AH, Bots ML, Stalenhoef AF, Visseren FL, Sijbrands EJ, Trip MD, Stein EA, Gaudet D, Duivenvoorden R, Veltri EP, Marais AD, de Groot E ENHANCE Investigators. Simvastatin with or without ezetimibe in familial hypercholesterolemia. N Engl J Med. 2008;358:1431–1443. doi: 10.1056/NEJMoa0800742. [DOI] [PubMed] [Google Scholar]
- 25.Myocardial Infarction Genetics Consortium Investigators. Stitziel NO, Won HH, Morrison AC, Peloso GM, Do R, Lange LA, Fontanillas P, Gupta N, Duga S, Goel A, Farrall M, Saleheen D, Ferrario P, König I, Asselta R, Merlini PA, Marziliano N, Notarangelo MF, Schick U, Auer P, Assimes TL, Reilly M, Wilensky R, Rader DJ, Hovingh GK, Meitinger T, Kessler T, Kastrati A, Laugwitz KL, Siscovick D, Rotter JI, Hazen SL, Tracy R, Cresci S, Spertus J, Jackson R, Schwartz SM, Natarajan P, Crosby J, Muzny D, Ballantyne C, Rich SS, O’Donnell CJ, Abecasis G, Sunyaev S, Nickerson DA, Buring JE, Ridker PM, Chasman DI, Austin E, Ye Z, Kullo IJ, Weeke PE, Shaffer CM, Bastarache LA, Denny JC, Roden DM, Palmer C, Deloukas P, Lin DY, Tang ZZ, Erdmann J, Schunkert H, Danesh J, Marrugat J, Elosua R, Ardissino D, McPherson R, Watkins H, Reiner AP, Wilson JG, Altshuler D, Gibbs RA, Lander ES, Boerwinkle E, Gabriel S, Kathiresan S. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65:1552–1561. doi: 10.1016/j.jacc.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannon CP, Blazing MA, Giugliano RP, McCagg A, White JA, Theroux P, Darius H, Lewis BS, Ophuis TO, Jukema JW, De Ferrari GM, Ruzyllo W, De Lucca P, Im K, Bohula EA, Reist C, Wiviott SD, Tershakovec AM, Musliner TA, Braunwald E, Califf RM IMPROVE-IT Investigators. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med. 2015;372:2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 28.Sabatine MS, Giugliano RP, Wiviott SD, Raal FJ, Blom DJ, Robinson J, Ballantyne CM, Somaratne R, Legg J, Wasserman SM, Scott R, Koren MJ, Stein EA Open-Label Study of Long-Term Evaluation against LDL Cholesterol (OSLER) Investigators. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- 29.Robinson JG, Farnier M, Krempf M, Bergeron J, Luc G, Averna M, Stroes ES, Langslet G, Raal FJ, El Shahawy M, Koren MJ, Lepor NE, Lorenzato C, Pordy R, Chaudhari U, Kastelein JJ ODYSSEY LONG TERM Investigators. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 30.Frikke-Schmidt R. Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population. Atherosclerosis. 2010;208:305–316. doi: 10.1016/j.atherosclerosis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, Shear CL, Revkin JH, Buhr KA, Fisher MR, Tall AR, Brewer B ILLUMINATE Investigators. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, Leitersdorf E, McMurray JJ, Mundl H, Nicholls SJ, Shah PK, Tardif JC, Wright RS dal-OUTCOMES Investigators. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 33.Eli Lilly and Company. [Accessed October 31, 2015];Lilly to discontinue development of evacetrapib for high-risk atherosclerotic cardiovascular disease. https://investor.lilly.com/releasedetail.cfm?releaseid=936130.
- 34.Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, Keavney B, Ye Z, Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 35.Ridker PM, Paré G, Parker AN, Zee RY, Miletich JP, Chasman DI. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: Genomewide analysis among 18 245 initially healthy women from the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triglyceride Coronary Disease Genetics Consortium and Emerging Risk Factors Collaboration. Sarwar N, Sandhu MS, Ricketts SL, Butterworth AS, Di Angelantonio E, Boekholdt SM, Ouwehand W, Watkins H, Samani NJ, Saleheen D, Lawlor D, Reilly MP, Hingorani AD, Talmud PJ, Danesh J, Rubins HB, Robins SJ, Collins D, Fye CL, Anderson JW, Elam MB, Faas FH, Linares E, Schaefer EJ, Schectman G, Wilt TJ, Wittes J. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 2010;375:1634–1639. doi: 10.1016/S0140-6736(10)60545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jørgensen AB, Frikke-Schmidt R, West AS, Grande P, Nordestgaard BG, Tybjærg-Hansen A. Genetically elevated non-fasting triglycerides and calculated remnant cholesterol as causal risk factors for myocardial infarction. Eur Heart J. 2013;34:1826–1833. doi: 10.1093/eurheartj/ehs431. [DOI] [PubMed] [Google Scholar]
- 38.Do R, Stitziel NO, Won HH, Jørgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, Zuk O, Guella I, Asselta R, Lange LA, Peloso GM, Auer PL, Girelli D, Martinelli N, Farlow DN, DePristo MA, Roberts R, Stewart AF, Saleheen D, Danesh J, Epstein SE, Sivapalaratnam S, Hovingh GK, Kastelein JJ, Samani NJ, Schunkert H, Erdmann J, Shah SH, Kraus WE, Davies R, Nikpay M, Johansen CT, Wang J, Hegele RA, Hechter E, Marz W, Kleber ME, Huang J, Johnson AD, Li M, Burke GL, Gross M, Liu Y, Assimes TL, Heiss G, Lange EM, Folsom AR, Taylor HA, Olivieri O, Hamsten A, Clarke R, Reilly DF, Yin W, Rivas MA, Donnelly P, Rossouw JE, Psaty BM, Herrington DM, Wilson JG, Rich SS, Bamshad MJ, Tracy RP, Cupples LA, Rader DJ, Reilly MP, Spertus JA, Cresci S, Hartiala J, Tang WH, Hazen SL, Allayee H, Reiner AP, Carlson CS, Kooperberg C, Jackson RD, Boerwinkle E, Lander ES, Schwartz SM, Siscovick DS, McPherson R, Tybjaerg-Hansen A, Abecasis GR, Watkins H, Nickerson DA, Ardissino D, Sunyaev SR, O’Donnell CJ, Altshuler D, Gabriel S, Kathiresan S NHLBI Exome Sequencing Project. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, Post W, McLenithan JC, Bielak LF, Peyser PA, Mitchell BD, Miller M, O’Connell JR, Shuldiner AR. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, Lu Y, Tang ZZ, Zhang H, Hindy G, Masca N, Stirrups K, Kanoni S, Do R, Jun G, Hu Y, Kang HM, Xue C, Goel A, Farrall M, Duga S, Merlini PA, Asselta R, Girelli D, Olivieri O, Martinelli N, Yin W, Reilly D, Speliotes E, Fox CS, Hveem K, Holmen OL, Nikpay M, Farlow DN, Assimes TL, Franceschini N, Robinson J, North KE, Martin LW, DePristo M, Gupta N, Escher SA, Jansson JH, Van Zuydam N, Palmer CN, Wareham N, Koch W, Meitinger T, Peters A, Lieb W, Erbel R, Konig IR, Kruppa J, Degenhardt F, Gottesman O, Bottinger EP, O’Donnell CJ, Psaty BM, Ballantyne CM, Abecasis G, Ordovas JM, Melander O, Watkins H, Orho-Melander M, Ardissino D, Loos RJ, McPherson R, Willer CJ, Erdmann J, Hall AS, Samani NJ, Deloukas P, Schunkert H, Wilson JG, Kooperberg C, Rich SS, Tracy RP, Lin DY, Altshuler D, Gabriel S, Nickerson DA, Jarvik GP, Cupples LA, Reiner AP, Boerwinkle E, Kathiresan S. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 42.Folsom AR, Peacock JM, Demerath E, Boerwinkle E. Variation in ANGPTL4 and risk of coronary heart disease: the Atherosclerosis Risk in Communities Study. Metabolism. 2008;57:1591–1596. doi: 10.1016/j.metabol.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varbo A, Benn M, Tybjærg-Hansen A, Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG. Remnant cholesterol as a causal risk factor for ischemic heart disease. J Am Coll Cardiol. 2013;61:427–436. doi: 10.1016/j.jacc.2012.08.1026. [DOI] [PubMed] [Google Scholar]
- 44.Do R, Willer CJ, Schmidt EM, Sengupta S, Gao C, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin SY, Lindström J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stančáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin MR, Jula A, Kähönen M, Kaprio J, Kesäniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, März W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njølstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Altshuler D, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Mohlke KL, Ingelsson E, Abecasis GR, Daly MJ, Neale BM, Kathiresan S. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller M, Stone NJ, Ballantyne C, Bittner V, Criqui MH, Ginsberg HN, Goldberg AC, Howard WJ, Jacobson MS, Kris-Etherton PM, Lennie TA, Levi M, Mazzone T, Pennathur S American Heart Association Clinical Lipidology, Thrombosis, and Prevention Committee of the Council on Nutrition, Physical Activity, and Metabolism; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular Nursing; Council on the Kidney in Cardiovascular Disease. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011;123:2292–2333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 46.Rubins HB, Robins SJ, Collins D, Fye CL, Anderson JW, Elam MB, Faas FH, Linares E, Schaefer EJ, Schectman G, Wilt TJ, Wittes J. Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. Veterans Affairs High-Density Lipoprotein Cholesterol Intervention Trial Study Group. N Engl J Med. 1999;341:410–418. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- 47.Keech A, Simes RJ, Barter P, Best J, Scott R, Taskinen MR, Forder P, Pillai A, Davis T, Glasziou P, Drury P, Kesäniemi YA, Sullivan D, Hunt D, Colman P, d’Emden M, Whiting M, Ehnholm C, Laakso M FIELD study investigators. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366:1849–1861. doi: 10.1016/S0140-6736(05)67667-2. [DOI] [PubMed] [Google Scholar]
- 48.ACCORD Study Group. Ginsberg HN, Elam MB, Lovato LC, Crouse JR, 3rd, Leiter LA, Linz P, Friedewald WT, Buse JB, Gerstein HC, Probstfield J, Grimm RH, Ismail-Beigi F, Bigger JT, Goff DC, Jr, Cushman WC, Simons-Morton DG, Byington RP. Effects of combination lipid therapy in type 2 diabetes mellitus. N Engl J Med. 2010;362:1563–1574. doi: 10.1056/NEJMoa1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ORIGIN Trial Investigators. Bosch J, Gerstein HC, Dagenais GR, Díaz R, Dyal L, Jung H, Maggiono AP, Probstfield J, Ramachandran A, Riddle MC, Rydén LE, Yusuf S. n-3 fatty acids and cardiovascular outcomes in patients with dysglycemia. N Engl J Med. 2012;367:309–318. doi: 10.1056/NEJMoa1203859. [DOI] [PubMed] [Google Scholar]
- 50.Utermann G. The mysteries of lipoprotein(a) Science. 1989;246:904–910. doi: 10.1126/science.2530631. [DOI] [PubMed] [Google Scholar]
- 51.Lp-PLA(2) Studies Collaboration. Thompson A, Gao P, Orfei L, Watson S, Di Angelantonio E, Kaptoge S, Ballantyne C, Cannon CP, Criqui M, Cushman M, Hofman A, Packard C, Thompson SG, Collins R, Danesh J. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet. 2010;375:1536–1544. doi: 10.1016/S0140-6736(10)60319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.STABILITY Investigators. White HD, Held C, Stewart R, Tarka E, Brown R, Davies RY, Budaj A, Harrington RA, Steg PG, Ardissino D, Armstrong PW, Avezum A, Aylward PE, Bryce A, Chen H, Chen MF, Corbalan R, Dalby AJ, Danchin N, De Winter RJ, Denchev S, Diaz R, Elisaf M, Flather MD, Goudev AR, Granger CB, Grinfeld L, Hochman JS, Husted S, Kim HS, Koenig W, Linhart A, Lonn E, López-Sendón J, Manolis AJ, Mohler ER, 3rd, Nicolau JC, Pais P, Parkhomenko A, Pedersen TR, Pella D, Ramos-Corrales MA, Ruda M, Sereg M, Siddique S, Sinnaeve P, Smith P, Sritara P, Swart HP, Sy RG, Teramoto T, Tse HF, Watson D, Weaver WD, Weiss R, Viigimaa M, Vinereanu D, Zhu J, Cannon CP, Wallentin L. Darapladib for preventing ischemic events in stable coronary heart disease. N Engl J Med. 2014;370:1702–1711. doi: 10.1056/NEJMoa1315878. [DOI] [PubMed] [Google Scholar]
- 53.O’Donoghue ML, Braunwald E, White HD, Lukas MA, Tarka E, Steg PG, Hochman JS, Bode C, Maggioni AP, Im K, Shannon JB, Davies RY, Murphy SA, Crugnale SE, Wiviott SD, Bonaca MP, Watson DF, Weaver WD, Serruys PW, Cannon CP, Steen DL SOLID-TIMI 52 Investigators. Effect of darapladib on major coronary events after an acute coronary syndrome: the SOLID-TIMI 52 randomized clinical trial. JAMA. 2014;312:1006–1015. doi: 10.1001/jama.2014.11061. [DOI] [PubMed] [Google Scholar]
- 54.Polfus LM, Gibbs RA, Boerwinkle E. Coronary heart disease and genetic variants with low phospholipase A2 activity. N Engl J Med. 2015;372:295–296. doi: 10.1056/NEJMc1409673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.CARDIoGRAMplusC4D Consortium; Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, König IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikäinen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Müller-Nurasyid M, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schäfer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrières J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kähönen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Trégouët DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvänen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimäki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O’Donnell C, Reilly MP, März W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ Wellcome Trust Case Control Consortium, MuTHER Consortium, DIAGRAM Consortium; CARDIOGENICS Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.