Abstract
Atherosclerosis and its attendant clinical complications such as myocardial infarction, stroke, and peripheral artery disease, are the leading cause of morbidity and mortality in western societies. In response to biochemical and biomechanical stimuli, atherosclerotic lesion formation occurs from the participation of a range of cell types, inflammatory mediators, and shear stress. Over the past decade, microRNAs have emerged as evolutionarily conserved, non-coding small RNAs that serve as important regulators and “fine-tuners” of a range of pathophysiological cellular effects and molecular signaling pathways involved in atherosclerosis. Accumulating studies reveal the importance of miRNAs in regulating key signaling and lipid homeostasis pathways that alter the balance of atherosclerotic plaque progression and regression. In this review, we highlight current paradigms of microRNA-mediated effects in atherosclerosis progression and regression. We provide an update on the potential use of miRNAs diagnostically for detecting increasing severity of coronary disease and clinical events. Finally, we provide a perspective on therapeutic opportunities and challenges for miRNA delivery in the field.
Keywords: MicroRNAs, atherosclerosis, vascular, lipoprotein
Subject Terms: Atherosclerosis, Mechanisms, Pathophysiology, Vascular Biology
Introduction
Atherosclerosis represents a chronic inflammatory disease of the arterial wall initiated by endothelial injury and subendothelial lipoprotein retention, particularly at sites of disturbed blood flow.1 The pathogenesis of atherosclerotic lesion formation is a multistage process involving both immune and non-immune cellular constituents of the vessel wall. Research over the last 3 decades has uncovered key signaling and molecular regulatory pathways involved in the initiation and progression of atherosclerotic plaques. The recent emergence of microRNAs (miRNAs) as important regulators of pathophysiological processes such as cellular adhesion, proliferation, lipid uptake and efflux, and generation of inflammatory mediators has provided novel molecular insights into their impact on these pathways in atherosclerosis, and identified new therapeutic targets. In addition, the appreciation that miRNAs can be detected extracellularly including in circulating blood raises the potential for their use as biomarkers for diagnosis, prognosis, or in response to cardiovascular therapeutics. This review highlights the role of miRNAs implicated in regulating critical aspects of atherosclerotic lesion formation and regression. We also review the potential use of miRNAs diagnostically for detecting increasing severity of coronary disease and clinical events.
Brief Primer on MicroRNAs
MiRNAs were first identified in C. elegans in 1993, however, their functional roles in human disease were not appreciated for nearly a decade later.2 MiRNAs are evolutionarily conserved, small (average ~18–24 nucleotides), single-stranded noncoding RNAs that regulate gene expression at the post-transcriptional level by typically binding to the 3′-untranslated region (3′-UTR) of specific target mRNA sequences (utilizing a conserved ~7–8 nucleotide seed sequence), thereby leading to reduced protein expression by blocking mRNA translation and/or by promoting mRNA degradation.3–8 It is estimated that >60% of all protein-coding genes are directly regulated by miRNAs.9 Furthermore, a given miRNA may bind to and regulate more than one target, sometimes as part of the same signaling pathway. Conversely, a given mRNA may harbor several distinct miRNA binding sites within its 3′-UTR, adding multiple levels of regulation. As such, miRNAs are “fine-tuners” of gene expression patterns in response to pathophysiological stimuli.
Intergenic miRNAs are located within the genome between gene-coding regions and are transcribed by their own promoters. By contrast, intronic miRNAs are found within intronic sequences of protein-coding genes and often share the same upstream promoters and transcriptional regulation with their host genes. MiRNA biogenesis proceeds in response to a well-defined series of cellular events. 2, 3, 5 MiRNAs are transcribed primarily by RNA polymerase II (more rarely by polymerase III) to generate the primary miRNA transcript, termed pri-miRNAs that harbor a canonical hairpin structure including a 5′ cap and 3′ poly-A tail.4 The pri-miRNAs are converted to ~70 nt-long hairpin precursor miRNAs (pre-miRNAs) by the Drosha/DGCR8 (Di George Syndrome Critical Region Gene 8) complex. Subsequently, the pre-miRNA is exported from the nucleus to the cytoplasm by Exportin 5 where it undergoes further processing by the RNase III enzyme complex Dicer/TRBP (TAR RNA-binding protein) producing a mature miRNA duplex.10 This double-stranded product is composed of the mature miRNA guide strand and the miRNA passenger strand (miRNA-5p and miRNA-3p, respectively). The guide strand is typically incorporated in the RNA-induced silencing complex (RISC) that contains Argonaute2 (Ago2), which facilitates the miRNA binding to the target mRNA leading to mRNA degradation of translation inhibition.11 Accumulating studies indicate that both miRNA-3p and miRNA-5p strands can be loaded into the RISC complex to target mRNA expression. Finally, mature miRNAs (and pre-miRNAs) can be released from the cell and packaged into a range of microvesicles (ie. exosomes, apoptotic bodies, microparticles) that are detectable in peripheral circulation (sometimes in association with RNA-binding proteins such as Ago2) and may also be taken-up within tissues by cell-to-cell communication.
MicroRNA regulation of lipoprotein homeostasis
Cholesterol homeostasis is essential for cellular physiology, and altered levels of cellular or systemic cholesterol are associated with metabolic diseases. In the circulation, cholesterol is carried on lipoproteins, which can both deliver [eg. low density lipoprotein (LDL)] and remove [eg. high density lipoprotein (HDL)] cholesterol from cells and tissues to mediate cholesterol homeostasis. Imbalances that favor the accumulation of cellular cholesterol, such as high levels of LDL cholesterol (LDLc) and/or low levels of HDL cholesterol (HDLc), promote atherosclerosis. The recent discoveries of miRNAs that control LDL and HDL abundance and function have greatly expanded our understanding of the regulatory circuits governing plasma lipoprotein levels.
The liver plays a major role in both the production and clearance of lipoproteins, and a number of hepatic-enriched miRNAs have been identified that functionally regulate lipoprotein metabolism. miR-122 was the first miRNA implicated in lipoprotein metabolism and its expression is highly enriched in the liver12. Loss-of-function experiments in both mice and non-human primates identified miR-122 as a crucial regulator of cholesterol and fatty acid synthesis, and thus lipoprotein homeostasis12, 13. Notably, miR-122 function appears to be broadly required for the expression of hepatocyte-specific genes, rather than the specific targeting of lipid metabolism pathways14. By contrast, miR-223 and miR-27b act as key post-transcriptional regulatory hubs controlling networks of cholesterol and lipoprotein metabolism genes15, 16. miR-223 represses genes involved in cholesterol biosynthesis (HMGS1, SC4MOL) and HDL uptake (SRB1), and Mir223−/− mice display increased HDL-C levels, as well as hepatic and plasma total cholesterol15. miR-27b is a cholesterol-responsive hepatic miRNA that represses a number of targets (PPARG, GPAM, ANGPTL3 and NDST1) involved in lipid metabolism and lipoprotein remodeling16. In addition to these gene network-regulating miRNAs, miR-30c was shown to have a potent effect on the production of apoB-containing lipoproteins (VLDL, LDL). miR-30c targets the microsomal triglyceride transfer protein (MTTP), a protein essential for the lipidation of of nascent apoB, and also reduces de novo lipogenesis by targeting lysophosphatidylglycerol acyltransferase 1 (LPGAT1) 17. Lentiviral overexpression of miR-30c in mice reduces the assembly and secretion of apoB-lipoproteins17, leading to decreased levels of plasma total and LDL cholesterol. Furthermore, studies in apolipoprotein E (apoE)-deficient mice (Apoe−/−) mice showed that miR-30c overexpression mitigated hyperlipidemia and atherosclerosis without inducing steatosis17, an undesirable side effect associated with conventional MTP inhibitors.
MicroRNA regulation of plasma levels of LDLc through targeting of the LDL receptor (LDLR) has also been reported. LDLR expression in the liver promotes the clearance of circulating LDL particles, and is a major determinant of plasma cholesterol levels. Two recent studies identified miR-148a as a negative regulator of LDLR expression and activity, and showed that inhibition of miR-148a in mice could increase the clearance of circulating labeled LDL and decrease plasma LDLc levels18, 19. Notably, single-nucleotide polymorphisms (SNPs) in the promoter region of miR-148A are associated with altered LDLc in humans19, suggesting that altered expression of this miRNA may contribute to dyslipidemias. Indeed, analysis of genome-wide association study (GWAS) data identified 3 other miRNAs predicted to target the LDL-R (miR-128-1, miR-130b and miR-301b) that lie in close proximity to human SNPs associated with abnormal levels of plasma lipids19. Like miR-148a, inhibition of miR-128-1 with locked nucleic acid (LNA) anti-sense oligonucleotides increased hepatic LDLR expression and LDL clearance in mice19. In addition to LDLR, miR-148a and miR-128-1 also target additional genes involved in lipid and energy metabolism (miR-148a: ABCA1, AMPKa1, CPT1a and SIK1; miR-128-1: ABCA1, SIRT1 and IRS1)19, suggesting important roles for these miRNAs in regulating metabolic pathways.
miRNAs have also been identified to act as critical regulators of HDL biogenesis and cholesterol efflux. These pathways control levels of plasma HDLc and the reverse cholesterol transport pathway through which excess cholesterol is removed to the liver for excretion. The ATP-binding cassette transporter ABCA1 plays a central role in these processes by controlling cholesterol efflux across the cell membrane onto lipid-poor apoA120 to mediate both hepatic HDL biogenesis and the removal of excess cholesterol from peripheral cells, particularly Mø in atherosclerotic plaques. A number of miRNAs have been identified that target ABCA1 to reduce cholesterol efflux to apoA1 in vitro, including miR-3321–27, miR-75828, miR-2629, miR-10630, miR-14431, 32, as well as the above mentioned miR-128-118 and miR-148a19. Of these, inhibition of miR-3321–27, miR-14431, 32, miR-128-118 and miR-148a19 has also been tested in vivo and shown to increase plasma levels of HDLc in mice or monkeys. Circulating levels of HDLc are also regulated by hepatic clearance via the scavenger receptor BI (SR-BI), which has been shown to be targeted by miR-22333, miR-455-5p34, miR-9633, miR-18533, and miR-125a34. However, only miR-223 has been manipulated in vivo and shown to impact plasma HDLc levels15.
Epidemiologic studies have shown an atheroprotective role for HDL, which is thought to be associated with its ability to mediate reverse cholesterol transport. Of the miRNAs targeting the HDL/reverse cholesterol transport pathway, the miR-33 family has been the most extensively studied in vivo using pre-clinical animal models. miR-33a and b are intronic miRNAs that are co-expressed with their host genes, SREBF2 and SREBF1, which code for transcription factors that regulate cholesterol and fatty acid synthesis/uptake23–25. Thus, transcription of SREBF2 and SREBF1 also results in expression of miR-33a and b, which cooperate with their host genes to balance cellular lipid levels by repressing genes that oppose SREBP-regulated pathways, such those involved in cholesterol efflux (ABCA1, ABCG1)23–25 and fatty acid oxidation (HADHB, CROT, CPT1A, PRKAA1)21, 28, 35. Inhibition of miR-33 using modified anti-sense or LNA oligonucleotides increased hepatic ABCA1 expression and plasma HDLc levels in both mice and monkeys by 40–50% 23–25, 27, 36, 37, and enhanced cholesterol transport from macrophages to the plasma, liver and feces by up to 80%27. This increase in reverse cholesterol transport is likely the combined effects of derepression of ABCA1 and additional miR-33a/b targets such as Abcb11 and Atp8b1 that promote cholesterol excretion into bile38. Importantly, HDL isolated from monkeys treated from with anti-miR-33 was shown to retain its anti-inflammatory properties, particularly its ability to promote macrophage cholesterol efflux and to protect endothelial cells from cytokine induced inflammation27.
Multiple studies of miR-33 inhibition or deletion have now been conducted in mouse models of atherosclerosis. Targeted deletion of Mir33 increased plasma levels of HDL and reduced atherosclerotic plaque size in Apoe−/− mice39. Furthermore, treatment of Ldlr−/− mice with established atherosclerosis with a miR-33 inhibitor for 4 weeks, in conjunction with a chow diet, increased plasma levels of HDL-C and markedly regressed atherosclerosis27. Notably, this intervention study was designed to simulate a clinical scenario where plasma LDLc levels were lowered in conjunction with anti-miR-33 treatment. However, two subsequent studies evaluating the effects of miR-33 inhibitors in Ldlr−/− mice on a Western diet showed no effects on HDLc levels, presumably due to decreased hepatic expression of SREBF2/miR-33 under these conditions40, 41. Despite this, Rottlan et al observed reductions in aortic atherosclerotic lesion area after 8 weeks of anti-miR-33 treatment40, suggesting that miR-33 inhibition may have atheroprotective effects beyond raising HDL. This paradox may be explained by a recent report that miR-33 can also instruct macrophage inflammatory polarization by altering the balance of cellular aerobic glycolysis and mitochondrial oxidative phosphorylation42. Targeted deletion or inhibition of miR-33 in macrophages increases oxidative respiration - a metabolic program characteristic of alternatively activated macrophages (AAM) - and induces the expression of genes that define AAM or M2 macrophage polarization. Notably, inhibition of miR-33 also increased macrophage expression of the retinoic acid–producing enzyme Aldh1a2 and retinal dehydrogenase activity both in vitro and in vivo. Retinoic acid can foster the differentiation of atheroprotective regulatory T cells, and consistent with this, Ldlr−/− mice treated with miR-33 inhibitors for 8 weeks on western diet showed the accumulation of M2 macrophages and FoxP3+ T regulatory cells in plaques, and reduced atherosclerosis progression in the absence of changes of plasma HDL42. This study suggests that antagonism of miR-33 is atheroprotective, in part, by promoting M2 macrophage and regulatory T cell polarization to suppress plaque inflammation.
Despite the beneficial effects of miR-33 inhibition on plasma levels of HDLc and atherosclerosis, some studies of genetic deletion43 or antagonism of miR-3344 in conjunction with western diet (WD) feeding have reported an increase in circulating triglycerides and hepatosteatosis. Not all studies have observed these effects of miR-33 inhibitors in mice45, and increases in triglycerides or hepatic lipid levels have not been reported in studies of non-human primates treated with anti-miR33 for up to 16 wks (26, 37). These conflicting findings suggest that more studies of miR-33 are warranted, particularly to understand if effects on triglycerides and hepatosteatosis are species specific or due to potential differences of chemical modifications, delivery, or time of administration of anti-miR-33 oligonucleotides. Nonetheless, these studies highlight the potential utility of miRNA mimics and inhibitors in the treatment of dyslipidemias (Figure 1).
Figure 1. microRNA orchestration of cholesterol homeostasis and macrophage activation in atherosclerosis.
In the liver, miRNAs repress the expression of genes involved in lipoprotein packaging and secretion (eg. miR-30c, miR-27b), uptake (eg. miRNAs targeting LDLR and SRB1), and cholesterol efflux (eg. miRNA targeting ABCA1). miRNA repression of ABCA1 decreases cholesterol efflux to lipid-poor apolipoprotein A-I (apoA-I), and biogenesis of HDL. As the nascent HDL particle mediates free cholesterol (FC) uptake and remodels due to lecithin-cholesterol Acyltransferase (LCAT) conversion of FC to cholesterol ester (CE), secreted microRNAs, such as miR-223, miR-92a and miR-126 are detected on the mature HDL particles. These HDL carried and may mediate extracellular signaling by repressing genes in target tissues and HDL interaction with macrophages and endothelial cells may also result in miRNA exchange (ie. pick-up or delivery via scavenger receptor B1 (SR-B1). MiRNA targeting of SR-B1 and the ABC11 and ATP8B1 transporters reduce selective cholesterol uptake by the liver and excretion, respectively. In macrophages, miRNA targeting of ABCA1 reduces cholesterol efflux and reverse cholesterol transport back to the liver. In addition, miRNAs regulate the polarization of macrophages toward classical M1 (eg-33, miR-155) or alternative M2 (eg. miR-223, miR-27a) inflammatory activation, and also regulate lipoprotein uptake and foam cell formation. (Illustration Credit: Ben Smith).
MicroRNA regulation of endothelial cell inflammation and plaque progression
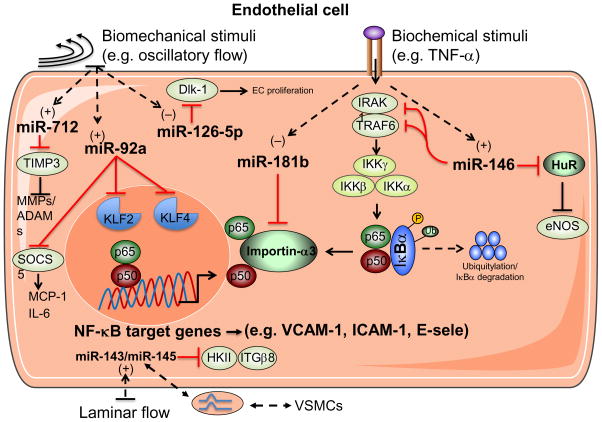
Sustained hyperlipidemia and altered shear stress predispose the vessel wall to atherosclerotic lesion formation. In response to biochemical and biomechanical stimuli, endothelial cells undergo a series of molecular and cellular conformational changes that promote atherogenesis. For example, early induction of adhesion molecule expression such as vascular adhesion molecule (VCAM)-1, intracellular adhesion molecule (ICAM)-1, and E-selectin facilitates leukocyte recruitment to the vessel wall and may be among the earliest hallmarks associated with nascent plaques.46 Several miRNAs have been implicated in atherogenesis by virtue of their ability to directly target the 3′-UTRs of these molecules such as miR-17-3p (targets ICAM-1) and miR-31 (targets E-selectin).47 However, the functional role of both these miRNAs in experimental atherosclerosis remains unknown. Nuclear Factor (NF)-κ B signaling is a major pathway that activates not only these pro-adhesive molecules, but also a range of other pro-inflammatory and pro-thrombotic factors. MiR-181b and miR-146a are two cytokine-responsive miRNAs that regulate distinct components of NF-κB signaling and are atheroprotective 48 (Figure 2).
Figure 2. Endothelial microRNAs regulate vascular inflammation.
In response to biochemical and biomechanical stimuli, microRNAs regulate specific targets in endothelial cells (ECs) that alter the balance of pro- or anti-inflammatory signaling pathways. Biochemical stimuli such as tumor necrosis factor (TNF)-α reduces the expression of miR-181b, while it increases the expression of miR-146. MiR-181b targets importin-α3 in ECs, an effect that prevents cytoplasmic-to-nuclear translocation of nuclear factor κB (NF-κB) family members, p50 and p65, thereby reducing NF-κB-responsive gene expression such as adhesion molecules vascular cell adhesion molecule 1 (VCAM-1), intercellular adhesion molecule 1 (ICAM-1), or E-selectin. Induction of miR-146 targets the 3′-UTR of TNF-receptor-associated factor 6 (TRAF6), IRAK1, to regulate upstream NF-κB signaling and targets HuR which regulates endothelial nitric oxide synthase (eNOS) expression. The expression of miR-92a and miR-712 are increased in response to disturbed flow (d-flow), whereas miR-126-5p expression is reduced. miR-92a targets the transcription factors Kruppel-like factor 2 (KLF2) and 4 (KLF4), and suppressor of cytokine signaling (SOCS5) expression, an effect that decreases anti-inflammatory pathways and increases monocyte chemoattractant protein (MCP)-1 and IL-6 that further promote EC activation. miR-712 suppresses tissue inhibitor of metalloproteinase-3 (TIMP3) thereby increasing the expression of matrix metalloproteinases (MMPs) and A Disintegrin and MMP (ADAMs). In contrast, d-flow reduces expression of miR-126-5p thereby de-repressing its target gene delta-like 1 homolog (Dlk-1), a negative regulator of endothelial cell proliferation. Laminar flow induces the expression of the miR-143/miR-145 cluster, which may be packaged and released extracellularly in microvesicles and taken-up by neighboring vascular smooth muscle cells (VSMCs). Conversely, ECs may enable the passage of miR-143 and miR-145 released from VSMCs through specialized membrane protrusions known as tunneling nanotubes. HKII, hexokinase II. ITGβ8, integrin β 8.
Cytokine-responsive miRNAs that regulate atherosclerosis
MiR-181b
Accumulating studies in mice and human subjects highlight a critical role for miR-181b as an inhibitor of endothelial inflammatory responses by targeting NF-κB signaling in both acute (e.g. sepsis) and chronic (e.g. atherosclerosis) vascular disease states.49, 50 MiR-181b, an intergenic miRNA,51 inhibits NF-κB signaling uniquely in endothelial cells but not leukocytes by targeting importin-α3, a protein important for NF-κB cytoplasmic-nuclear translocation. In leukocytes, the dominant isoform that mediates NF-κB nuclear import is importin-α5, which miR-181b does not target thereby leaving leukocyte NF-κB signaling intact. Nonetheless, upon systemic intravenous delivery, miR-181b decreased endothelial NF-κB activation, an effect that was sufficient to markedly suppress leukocyte recruitment and atherosclerotic lesion formation in atherosclerotic-prone ApoE−/− mice. Importantly, the miR-181b anti-atherosclerotic effects were independent of any changes in lipid profiles. These findings are consistent with endothelial-specific NF-κB (IKKγ) knockout and dominant-negative IκBα transgenic mice that also exhibit protection against atherosclerotic lesion formation.52 A range of pro-inflammatory stimuli (e.g. TNF-α, LPS) reduce miR-181b expression in endothelial cells in vitro. In vivo, miR-181b expression is reduced by 53% in the vascular endothelium and by ~40% in plasma of mice after just 4 weeks of high cholesterol diet in ApoE−/− mice, suggesting that loss of homeostatic control of miR-181b expression may predispose to inflammation in the vessel wall. In line with these observations, miR-181b expression is also reduced in plasma of human subjects with angiographically defined coronary artery disease (CAD) compared to those without CAD.50 Therapeutically, the cell-specific miR-181b inhibitory effects on NF-κB in the vascular endothelium, and not in myeloid cells, may be advantageous in order to maintain optimal protection in response to infectious pathogens. Indeed, miR-181b delivery also inhibits endothelial inflammation and confers protection against sepsis in mice.49 Collectively, these findings provide cogent evidence that miR-181b serves as an important regulator of NF-κB signaling in the vascular endothelium in response to diverse stimuli, and provide new opportunities for anti-inflammatory “replacement” therapy.
MiR-146a
MiR-146a is another cytokine-responsive miRNAs that confers atheroprotective properties in the vessel wall. Cytokines such as TNF-α and IL-1β induced expression of miR-146a and miR-146b in a delayed kinetic manner in ECs that coincided with the resolution of inflammatory gene expression.53 MiR-146a overexpression inhibited cytokine responsiveness in ECs suggesting it may participate in a negative feedback mechanism to limit EC inflammatory signaling. Interestingly, miR-146a expression is also increased in human and mouse atherosclerotic plaques.54 Indeed, miR-146a repressed both NF-κB and MAPK signaling pathways by directly targeting HuR, an RNA-binding protein that exerts inhibitory effects on endothelial nitric oxide synthase (eNOS). In addition, miR-146a represses the induction of EC adhesion molecules by targeting upstream adaptor proteins TRAF6 and IRAK1/2.53 In contrast to the more selective inhibitory role of miR-181b on EC NF-κB signaling, miR-146a inhibits NF-κB signaling in both ECs and macrophages.55 Indeed, overexpression of ApoE in ApoE−/− macrophages induced miR-146a expression to reduce macrophage and systemic delivery of miR-146a mimics markedly reduced atherosclerotic lesion progression. The specific cell subsets (e.g. endothelial cells or leukocytes) and mechanisms (e.g. NF-κB-dependent or –independent) by which miR-146a confers these favorable effects in experimental atherosclerosis will require future study. However, given the anti-inflammatory role of miR-146a in regulating a range of immune cells (macrophages, dendritic cells, and T cells,56 it may participate more broadly to limit inflammatory stimuli. Nevertheless, miR-146 is another important cytokine-responsive miRNA that may serve to dampen EC inflammation in a negative feedback manner.
Mechano-sensitive EC miRNAs that regulate atherosclerosis
Atherosclerotic lesions preferentially develop at arterial branch points, bifurcations, and the lesser curvature of the aorta in mice and human subjects.57–59 These findings are explained, in part, by the presence of disturbed flow in these regions which increases endothelial permeability (e.g. by altering the expression and localization of intracellular junctional proteins) and proinflammatory signaling (e.g. NF-κB activation, adhesion molecule expression), thereby promoting leukocyte accumulation and lesion formation.60–63 In contrast, laminar flow confers an anti-inflammatory, anti-adhesive, and anti-thrombotic properties in the vessel wall. A surprisingly large number of miRNAs have been identified as shear stress-responsive by either disturbed flow (d-flow) or laminar flow (L-flow).
miR-92
MiR-92a, a member of the miR-17~92 cluster, is expressed highly in endothelial cells and dynamically regulated by shear stress both in vitro and in vivo.64, 65 Upon exposure of ECs to pulsatile L-flow in vitro, miR-92a expression is reduced, whereas D-flow increases its expression; in addition, miR-92a bound to the 3′-UTR of KLF2, a flow-responsive transcription factor, suggesting a direct role for regulating laminar flow.64 Similarly, in vivo miR-92a expression is highly induced in athero-prone areas of the aortic arch compared athero-resistant regions.66 In vitro studies suggest that miR-92a overexpression suppresses the 3′-UTRs of KLF2 and KLF4. In contrast, miR-92a-mediated inhibition of TNF-α-induced cytokines and leukocyte adhesion can be rescued, in part, by KLF4 siRNAs, indicating partial KLF4 dependency.66 In a separate microarray profiling studies, Loyer et al. identified that upon exposure of ECs to oxLDL and low shear stress, expression of miR-92a and pro-inflammatory markers (MCP-1 and IL-6) are increased in a STAT3-dependent manner.65 Overexpression of miR-92a in ECs reduced expression of KLF2 and KLF4, flow-responsive transcription factors, whereas miR-92a antagonism reduced EC inflammation as reflected by lower phospho-p65 expression. Mechanistically, miR-92a also targets SOCS5 in ECs in the presence of oxLDL and low shear stress conditions.65 While the in vivo role of SOCS5 in atherosclerosis remains unclear, siRNA-mediated knockdown of SOCS5 increased MCP-1 and IL-6 expression in ECs without affecting expression of eNOS, KLF2, or KLF4. In addition, neutralization of miR-92a in LDLR−/− mice decreased EC inflammation and suppressed the progression of atherosclerotic lesion formation. Moreover, mice harboring a genetic deletion of miR-92a in Tie2-expressing endothelial cells were protected from neointimal formation after mechanical arterial injury, attributed, in part, to enhanced re-endothelialization.68 Given the protective role of miR-92a-deficiency in other processes including re-endothelialization after mechanical arterial wire injury67, 68 and angiogenesis after myocardial or peripheral ischemia,69, 70 targeting this miR-92a may hold promise for a range of cardiovascular relevant disease states.
miR-126
miR-126 is among the most abundantly expressed miRNAs in ECs and has been implicated in regulating both inflammation and angiogenesis in a flow-dependent manner. MiR-126 was initially described in vitro to bind to the 3′-UTR of VCAM-1 to limit leukocyte adhesion.71 Subsequent studies revealed that miR-126-deficient mice exhibit impaired vascular integrity and defects in EC proliferation and angiogenesis. 72 Indeed, miR-126 may be induced by KLF2 to control flow-dependent angiogenesis.73 In the context of atherosclerosis, Zernecke A et al. demonstrated that miR-126 was the most abundant miRNA expressed in EC-derived apoptotic bodies where it induced CXCL12 expression by targeting RGS16, a negative regulator of SDF-1/CXCR4 signaling and progenitor cell mobilization.74 Consistent with this pathway, intravenous delivery of endothelial apoptotic bodies mobilized progenitor cells in the circulation, and enhanced their incorporation into aortic plaques, an effect that suppressed atherosclerotic progression in a miR-126-dependent manner. Within the vascular endothelium, Schuber et al. elegantly identified a functional role for the passenger strand, miR-126-5p, as a flow-responsive miRNA that is suppressed by D-flow, an effect that facilitates lesion formation through induction of a negative regulator of EC proliferation termed delta-like 1 homolog (Dlk-1).75 Systemic delivery of miR-126-5p mimics rescued EC proliferation at vulnerable sites and inhibited lesion progression.75 Because the pro-inflammatory transcription factors Ets-1 and Ets-2 also induce miR-126 expression,76 the miR-126 duplex may mechanistically serve to limit pro-inflammatory factors such as TNF-α and Ang-II in a negative feedback manner in the vascular endothelium. However, ECs exposed to L-flow induced KLF2-dependent expression of pri-miR-126 but not miR-126-3p. Moreover, miR-126-5p expression, but not miR-126-3p expression, was lower at predilection sites of ApoE−/− aortas. In addition, delivery of miR-126-5p antagomiRs, but not miR-126-3p antagomiRs, increased lesion formation after EC denudation in ApoE−/− mice, suggesting that miR-126-3p may not be implicated in flow-responsive vascular protection. Finally, atheroprotective L-flow increases the release of miR-126 bound to Ago2 into extracellular microvesicles that serves as a mediator to increase VSMC turnover by targeting genes (e.g. FOXO3, BCL2, and IRS1) implicated in maintaining an atheroprotective VSMC contractile phenotype.77 In this study, miR-126 deficiency inhibited neointimal formation of mouse carotid arteries induced by cessation of blood flow, an effect rescued by miR-126 mimics or by conditioned media from static EC monocultures. These latter findings raise the possibility that despite its atheroprotective in ECs, miR-126 may thin SMCs of the fibrous plaque leading to destabilization. Future long-term studies will be required to assess the relative role of the miR-126-5p/miR-126-3p duplex in response to a range of important biochemical and biomechanical stimuli and in different phases of atherosclerosis.
MiR-712/miR-205
While miRNA biogenesis typically utilizes the canonical Drosha/DGCR8 and Dicer miRNA pathway, accumulating studies also highlight an important role for non-canonical pathways for miRNA maturation.78, 79 miR-712 (human homologue miR-205) was identified as a d-flow responsive miRNA derived from non-canonical pre-ribosomal RNA (using the RN45S gene).80 Functionally, d-flow-induced miR-712 targets TIMP3, an effect that increases MMPs and ADAMs to stimulate pro-inflammatory EC responses. Indeed, neutralization of miR-712 de-represses TIMP3 expression and ameliorates atherosclerotic progression in mice.80 Future studies will be required to determine whether the in vivo neutralization of miR-712 directly affects other relevant cell types, such as immune subsets and vascular smooth muscle cells.
miR-143/miR-145
An emerging paradigm in cell-to-cell communication is the ability of miRNAs to be transferred from one cell to another within tissues. For example, in response to L-flow or KLF2 overexpression in ECs, extracellular microvesicles containing miR-143 and miR-145 are released that confer atheroprotective properties in adjacent VSMCs. Intravenous delivery of these extracellular vesicles also blocked atherosclerotic lesion progression in a miR-143/145-dependent manner.81 Interestingly, miR-143/miR-145, which are expressed higher in VSMCs compared to ECs under basal conditions in vitro, may also participate in VSMC to EC communication via intercellular tubes called tunneling nanotubes (TNTs) in vitro.82 In support of this VSMC-to-EC passage, SMC-specific deletion of the miR-143/miR-145 cluster in mice effectively blocked the induction of miR-143/miR-145 expression in coronary artery endothelial cells in response to trans-aortic constriction-induced pressure overload, an effect that may be mediated by activation of the TGF-β signaling pathway and the miR-143/miR-145 targets hexokinase II and integrin-β8.82 Collectively, while it remains to be determined whether bi-directional extracellular miRNA passage occurs between SMC and EC under atherosclerotic conditions, these findings raise the provocative question of skewed, preferential cell-to-cell “miRNA shifts” that may emerge under atherosclerotic progression or regression, or in response to pharmacologic therapies.
Others: miR-10a, miR-663, miR-155, miR-30-5p
Several other mechano-sensitive microRNAs, such as miR-10a, miR-663, and miR-155, have been identified using a variety of profiling approaches. However, the functional role of these three miRNAs in regulating experimental atherosclerosis in mice has not yet been validated (miR-10a, miR-663) or remains unclear (miR-155). Nonetheless, they may figure prominently at predilection sites in the macrovasculature. For example, miRNA microarray profiling of ECs from the inner aortic arch of pig, an atherosusceptible region, and ECs from the descending thoracic aorta, miR-10a expression was identified as significantly reduced in the atherosusceptible regions.83 Mechanistically, miR-10a targets the 3′ UTRs of MAPK kinase kinase 7 and the β-transducin repeat-containing gene, two key regulators of IκBα degradation. Indeed, miR-10a overexpression inhibits canonical NF-κB signaling in ECs in vitro.83 Therapeutic manipulation of miR-10a in atherosclerotic-prone mice will be required to assess its relative contribution to EC function at predilection sites and lesion pathogenesis. Because miR-10a’s family member, miR-10b, which may also bind to similar consensus sites in target genes, has been implicated in promoting tumor invasion and metastasis, delivery of miR-10 mimetics may require close scrutiny for therapeutic gain. Another miRNA, miR-663, was identified by 2 groups as a D-flow-induced miRNA in ECs.80, 84 Interestingly, human miR-663, like miR-712, may also be derived from the same ribosomal RNA gene, RN45S. Because the XRN1 exonuclease can rapidly degrade the spacer regions from where this rRNA is derived, Son et al examined whether silencing XRN1 may affect miR-663 and miR-712 expression. Indeed, D-flow reduced XRN1 expression in predilection regions in the mouse carotid and aortic arch, and XRN1 deficiency in ECs in vitro significantly increased both miR-663 and miR-712 expression, suggesting another level of regulation and therapeutic modulation of atypical mechano-sensitive miRNAs in the vascular endothelium.80 Finally, miR-155 is increased by L-flow in ECs and in the thoracic aorta compared to the lower aortic arch.75 Mechanistic studies indicate that miR-155 may bind to both anti-inflammatory targets (e.g. eNOS) and pro-inflammatory targets (e.g. MYLK, RhoA, SOCS-1, and Bcl6).75, 85–87 Studies examining the role for miR-155 in experimental atherosclerosis in mice highlight both pro- and anti-atherosclerosis effects depending on the context. For example, LDLR−/− mice harboring bone marrow with miR-155 deficiency exhibited increased atherosclerosis with reduced plaque stability.88 In contrast, miR-155 deficiency in macrophages reduced atherosclerotic plaque size in ApoE−/− mice, an effect thought to be mediated by miR-155’s ability to target BCL6 and, in turn, reduced CCL2-mediated macrophage recruitment.87 Moreover, miR-155 delivery in vivo reduced atherosclerotic lesion formation, an effect that may be mediated by targeting MAP3K10. 89 Because miR-155 also targets myosin light chain kinase (MYLK) in endothelial cells,75 and MYLK-deficiency in ApoE−/− mice reduces atherosclerosis by improving endothelial barrier dysfunction and monocyte migration90, EC-derived miR-155 may be viewed as a potentially protective miRNA in the vessel wall. In line with this hypothesis, miR-155 expression is significantly higher in the thoracic aorta, an area associated with unidirectional shear stress, than in the lower curvature of the aortic arch, a region associated with low shear stress.90 These findings raise nuanced questions for miR-155 in vascular inflammation and future studies will be required to define the EC- and macrophage-specific roles of miR-155 in atherogenesis. Finally, laminar flow shear stress or KLF2 overexpression in ECs induce members of the miR-30-5p family that, in turn, decrease anti-inflammatory markers such as VCAM-1 and ICAM-1 by targeting angiopoietin-2.91 However, a role for miR-30-5p family members in regulating endothelial inflammation and atherosclerosis in mice will require future investigation.
miRNAs regulating leukocyte recruitment and activation in atherosclerosis
Immune responses critically shape atherogenesis. One of the earliest pathogenic events in atherosclerosis is the recruitment of monocytes from the circulation to the artery wall in areas of endothelial dysfunction and lipoprotein retention. Upon differentiation into macrophages, these cells play central roles in the pathophysiology of atherosclerosis by maintaining lipid homeostasis in the vessel wall and secreting inflammation-promoting mediators that act on both immune and non-immune cell types in the artery wall92. Lipoprotein uptake by macrophages in the nascent plaque results in the formation of lipid-laden macrophage foam cells that are hallmarks of atherosclerosis92. For reasons that are poorly understood, these macrophage foam cells persist in the artery wall, setting off a maladaptive immune response that promotes the formation of plaques. These macrophages are a source of inflammatory mediators, including cytokines and chemokines that mediate the recruitment and/or activation of other immune cells, thereby chronically sustaining the inflammation that fuels plaque progression. miRNAs can impact each of these key macrophage processes to influence the progression of atherosclerosis.
Macrophage cholesterol homeostasis is maintained by the balance between cholesterol uptake, endogenous synthesis, esterification/hydrolysis and efflux. A number of miRNAs have been implicated in macrophage cholesterol metabolism metabolism. miR-27a/b, discussed above with regard to lipoprotein homeostasis, can regulate macrophage cholesterol homeostasis by targeting genes involved in cholesterol esterification (ACAT1), uptake (LDL, CD36) and efflux (ABCA1)93. miR-125a-5p and miR-146a decrease lipid uptake and cytokine release in oxLDL-stimulated macrophages, in part by targeting the genes oxysterol binding protein-like 9 and TLR4, respectively94, 95. In addition, miR-155, may also regulate macrophage foam cell formation by targeting the transcriptional repressor HMG box-transcriptional protein 1 (HBP1), which negatively regulates macrophage inhibitory factor (MIF), a protein known to increase the uptake of oxLDL by macrophages96. Furthermore, numerous miRNAs have now been identified that promote foam cell formation by inhibiting macrophage cholesterol efflux via ABCA1, including miR-2629, miR-3323–25, miR-10630, miR-14431, 32, miR-128-118, miR-130b18, miR-148a18, miR-301b18, miR-302a97 and miR-75828. This high degree of miRNA targeting of ABCA1 points to the need for careful fine tuning of macrophage cholesterol efflux to maintain cholesterol homeostasis.
In response to microenvironmental signals, macrophages can initiate different activation programs, including the ‘classical’ pro-inflammatory phenotype (also called M1) and the ‘alternatively’ activated M2 phenotype associated with an anti-inflammatory profile. Atherosclerosis progression is associated with the predominance of an M1 macrophage phenotype in the plaque, whereas plaques undergoing regression are enriched in M2 macrophages92. A growing list of miRNAs is implicated in regulating the balance between the M1 and M2 phenotypes, including miR-let7a98, miR-19a99, miR-21100, miR-27a101, miR-3342, miR-124102, miR-125a103, miR-146a104, miR-155105, miR-214106, miR-223107. However, few of these have been investigated in the context of atherosclerosis. Of note, miR-33, which plays a central role in regulating cholesterol efflux, also regulates macrophage cellular metabolism to alter the cell’s inflammatory phenotype. miR-33 reduces fatty acid oxidation, the metabolic program that fuels M2 macrophages, and promotes aerobic glycolysis, which in turn sustains the inflammatory M1-like macrophage phenotype42. Inhibition of miR-33 metabolically reprograms plaque macrophages to the M2 phenotype involved in resolving inflammation and tissue repair, which in turn promotes the accumulation of atheroprotective T regulatory cells. miR-155, which as described above has a controversial role in atherosclerosis, can also reprogram macrophages from the M2 to M1 phenotype, and thus may play a role in the accumulation of M1 macrophages105. miR-155 expression is significantly higher in CD14+ monocytes from patients with CAD than from healthy controls, and is induced by oxidized forms of LDL in macrophages96. miR-155 acts to repress negative regulators of inflammatory cytokine signaling, such as suppressor of cytokine signaling 1 (SOCS1), Src homology 2 domain-containing inositol-5-phosphatase-1 (SHIP-1), or B cell lymphoma 6 (BCL6), and thereby promoting the release of pro-inflammatory mediators75, 85–87. Conversely, miR-223, which regulates lipid metabolism-related genes in the liver15, can suppress M1 pro-inflammatory pathways and enhance alternative activation, in part by targeting Pknox1107. Finally, miR-27a, which also plays a role in macrophage foam cell formation, can promote markers of M2 macrophages (eg. CD206 and DC-SIGN) and secretion of IL-10101.
Other miRNAs have been implicated in enhancing or reducing macrophage responses to inflammatory stimuli. For example, miR-147 and miR-21 attenuate TLR-associated signaling events in macrophages to limit inflammation108, 109. Likewise, miR-146a/b are induced in macrophages in an NFκB-dependent manner and are involved in inflammation resolution by limiting Toll-like receptor and cytokine signaling. Recently, expression of miR-146a was shown to be induced in macrophages by apolipoprotein E (apoE), a protein with anti-atherosclerotic properties, and to suppress macrophage inflammatory responses in vitro and in vivo55. Furthermore, the transcription factor Krüppel-like factor 2 (KLF2), whose expression in macrophages protects from atherosclerosis, downregulates expression of proatherosclerotic chemokines (eg. CCL2 and CXCL1) by increasing expression of miR-124a and miR-150110. During early atherogenesis, one of the most prominently induced miRNAs in lesional macrophages is miR-342-5p111. This miRNA enhances the production of macrophage inflammatory mediators such as iNOS and IL-6 by suppressing Akt1-mediated suppression of miR-155. Accordingly, inhibition of miR-342-5p in Apoe−/− mice reduces atherosclerosis progression111.
Other immune cells, such as dendritic cells and T cells, also participate in atherogenesis and the regulation of plaque inflammation. By sensing and presenting antigens in the plaque, DCs are positioned at the crossroad of innate and adaptive immune responses. Furthermore T-cell (Th1, Th2, Th17, Treg) and B-cell subpopulations can modulate atherosclerosis development. Thus, miRNAs that regulate immune cell differentiation and/or function would be expected to have wide-ranging effects on plaque evolution. An atlas of miRNA expression patterns in human T and B cell subsets was recently completed112, and their roles in regulating T and B cell differentiation, activation and function were recently reviewed113. However to date, miRNA regulation of these immune cell subsets in atherosclerosis has been underexplored compared to macrophages.
Notably, several miRNAs shown to regulate atherogenic processes in macrophages have also been implicated in dendritic cell function. For example, miR-155 is required for efficient DC maturation by targeting the transcription factor c-Fos114, and its expression increases TLR/IL-1 and type I IFN signaling pathways in human monocyte-derived DCs required to promote antigen-specific T-cell activation115–117. Furthermore, like its role in macrophages, miR-146a reduces TLR signaling and cytokine production in DCs, thereby regulating DC activation118. Moreover, miR-148/152 inhibit the production of pro-inflammatory cytokines (eg. IL-12, IL-6, TNFα) by targeting CaMKIIα, consequently reducing DC-triggered antigen-specific T cell proliferation119. By regulating activation of DCs and their interactions with T and B cells in plaques, these miRNAs are likely to impact the inflammatory milieu of the plaque and atherosclerosis progression.
miRNAs implicated in Vascular Smooth Muscle function and atherosclerosis
VSMCs contribute to the maintenance of vascular wall function by assuming a differentiated, contractile phenotype. In response to vascular injury, VSMCs undergo a switch to a synthetic phenotype, an effect that induces signals that promote migration, proliferation, and inflammation.120 Several miRNAs have been implicated in regulating important VSMC nodal regulators such as the transcription factors (serum-response factor (e.g. SRF/KLF4), co-activators (e.g. myocardin), TGF-β signaling effectors (e.g. Smads), or cytokines/growth factors (e.g. PDGF). While the traditional view of VSMCs in vascular injury states is that they “dedifferentiate” to assume a synthetic, proliferative state,121 recent studies have broadened this concept to suggest that VSMCs may adopt a reprogrammed “transdifferentiated” macrophage phenotype.121, 122 While the role of miRNAs have emerged as important players in the former, their role in VSMC-to-macrophage transdifferentiation remains unexplored. We summarize below major miRNA regulators implicated in VSMC and atherosclerotic lesion formation (Figure 3).
Figure 3. MicroRNA regulation of vascular smooth muscle cell phenotype.
In response to vascular wall injury or atherosclerosis, the expression of the miR-143/miR-145 cluster is markedly reduced in vascular smooth muscle cells (VSMCs). MiR-143 and miR-145 target the transcriptional regulators KLF4, KLF5, myocardin, and ELK-1 important for VSMC phenotypic switching from a contractile, mature, and differentiated cell type to a de-differentiated synthetic, and proliferative cell type. In addition, miR-143/miR-145 target genes important to the regulation of blood pressure such as angiotensin converting enzyme (ACE). In contrast, vascular wall injury increases expression of miR-221 and miR-222, an effect that decreases the expression of the cell cycle regulator c-Kit, p27(Kip1), and p57(Kip2). Induction of miR-21 expression targets phosphatase and tensin homolog (PTEN), thereby increasing the anti-apoptotic regulator B-cell lymphoma 2 (Bcl-2). Microvesicles or exosomes released by neighboring endothelial cells, and carrying microRNAs such as miR-143/miR-145 or miR-126 (bound to Argonaute2 (Ago2)), may be taken up by VSMCs enabling suppression of target genes and altering VSMC functional responses. FOXO3, Forkhead Box O3. IRS1, insulin receptor substrate 1.
MiR-143/miR-145
The miRNAs miR-143 and miR-145 are co-transcribed as a single primary-miR transcript due to their close proximity and among the highest expressed miRNAs in VSMCs and the medial layer of the vessel wall.123 Indeed, these miRNAs are reduced in the vessel wall in response to vascular injury or the presence of atherosclerosis.124, 125 A series of gain- and loss-of-function studies suggest that miR-143/miR-145 are major regulators of VSMC contractile function. Indeed, miR-143/miR-145-deficient mice exhibit VSMCs with reduced SMC contractile marker expression and function, impaired actin stress fibers and cytoskeletal dynamics, and decreased vessel wall medial thickness.124–126 These miR-143/miR-145-deficient mice also had reduced blood pressure at baseline and in response to vasopressor challenge, an effect that the authors attributed to reduced expression of the target gene angiotensin converting enzyme (ACE). 127 Conversely, delivery of miR-145 in ApoE−/− mice using a lentivirus under the control of the SMC-specific SM22α promoter significantly reduced atherosclerotic plaque size and promoted a more favorable plaque composition with increased fibrous cap area and plaque collagen content, and reduced necrotic core area and macrophage accumulation.128 Consistent with miR-145’s known role in promoting a contractile phenotype in VSMCs, these atherosclerotic stabilizing effects were mechanistically related to reduced KLF4 and increased myocardin expression in the vessel wall of ApoE−/− mice.128 Analogous protective effects of miR-145 have been observed in response to neointimal vascular lesion formation.129 In addition to KLF4 and myocardin, miR-143/miR-145 have been shown to target several other key transcriptional regulators implicated in modulation of VSMC differentiation, such as ELK-1 and KLF5. 124, 125, 130 Because upstream control of miR-143/miR-145 expression is governed in part by serum response factor (SRF), Nkx2.5, and myocardin, miR-143/miR-145 may contribute to ‘fine-tuning’ a SMC-specific transcriptional program important in regulating the contractile VSMC state.
miR-221/miR-222
In contrast to miR-143/miR-145, the miRNAs, miR-221 and miR-222, are increased in response to injury in neointimal lesions.131 In vitro studies have implicated miR-221/miR-222 in regulating PDGF-mediated VSMC proliferation. PDGF induced miR-221/miR-222 in VSMCs, an effect leading to reduced expression of its target genes c-Kit and p27Kip1 and decreased SMC-specific contractile gene expression.132 Using miR-221/miR-222 knockdown approaches, in vivo studies demonstrated that miR-221 and miR-222 deficiency reduced VSMC proliferation and neointimal lesion formation after mechanical injury by targeting p27(Kip1) and p57(Kip2).131 In human atherosclerotic lesions, reduced expression of miR-221 and miR-222 was noted in the shoulder of plaques from patients undergoing carotid endarterectomy (CEA) due to an acute neurological event occurring within 5 days of the CEA. While this study demonstrated a compelling association of reduced miR-221/miR-222 with unstable lesions, it also revealed that the target gene p27(Kip1) was significantly increased. 133 Interestingly, miR-222 expression in isolated human ECs decreased from early lesions to advanced plaques.134 Collectively, these studies provide accumulating evidence that miR-221/miR-222 may play a destabilizing role in atherosclerotic lesions.
MiR-21
While miR-21 has been implicated in promoting VSMC proliferation in response to a range of vascular mechanical injury models, its role remains to be defined in atherosclerotic lesion formation. Neutralization of miR-21 reduced neointimal lesion formation in response to mechanical balloon injury.129 In keeping with its role in regulating VSMC contractile phenotype, miR-21 expression increased significantly in isolated ‘dedifferentiated’ VSMCs compared to mature differentiated VSMCs. Consistent with these findings for miR-21 in rodent vascular injury, delivery of an anti-miR-21-coated stent into balloon-injured human internal mammary arteries using a humanized rat model also revealed protective effects on neointimal lesion formation. 135 Mechanistically, miR-21’s proliferative properties may be due to its ability to target PTEN in VSMCs and indirectly increasing the anti-apoptotic gene Bcl-2.129 Furthermore, miR-21 mediates the induction of the VSMC contractile phenotype by TGF-β and BMP signaling in a unique post-transcriptional processing step implicating that Smad proteins may control DROSHA-regulated maturation of miRNAs. 136 Future studies will be required to assess whether the anti-proliferative effects of miR-21 neutralization also reduce atherosclerotic progression in non-mechanically injury vessels.
MiRNAs as diagnostic markers of atherosclerotic disease severity
Accumulating studies implicate miRNAs as potential diagnostic or prognostic markers in a range of disease states. Because circulating miRNAs can be detected in peripheral blood, saliva, and urine, their expression may be harbingers of various stages of coronary artery disease from subclinical atherosclerotic disease to acute coronary syndromes. Herein, we summarize the profiling of several studies linking specific miRNAs to atherosclerotic disease burden as primarily diagnostic markers. Future studies will be required to further define these miRNAs for their prognostic significance in coronary artery disease (CAD).
Stable CAD
One of the earliest studies that analyzed miRNAs from plasma of eight stable CAD subjects and healthy controls examined candidate miRNAs based on their putative cell-selective expression patterns. In the CAD group, expression of EC-enriched miRNAs (miR-126, miR-17, and miR-92a), VSMC-enriched miRNAs (miR-145), and inflammatory cell enriched miRNAs (miR-155) were all reduced. In contrast, cardiomyocyte-enriched miRNAs (miR-133 and miR-208a) were increased.137 Other groups have identified reduced expression of miRNAs (miR-19a, -484, -155, -222, -145, -29a, -378, -342, -181d, -150, -30e-5p) from whole blood of patients with angiographically defined stable CAD (at least one epicardial vessel with >50% stenosis) compared to healthy subjects.138 Interestingly, when this cassette of 11 miRNAs were compared to another group of subjects with at least 2 cardiac risk factors, but without angiographically defined CAD, there were no differences observed suggesting that this cassette of miRNAs may also be associated with subclinical atherosclerosis.138 Surprisingly, reduced expression of 9 of the miRNA cassette was also associated with reduced expression in CAD or at-risk CAD patients taking ACE inhibitors or ARB antagonists compared to CAD/at-risk CAD patients not taking ACEI/ARBs. No associations were observed in this same cohort for treatment of statins vs no-statins. Another study by Zhu et al identified miR-155 expression in peripheral blood mononuclear cells (PBMCs) or plasma in patients with increasing severity of coronary syndrome and found that miR-155 was lower in patients with unstable angina or acute myocardial infarction than in patients with chest pain syndrome.139 MiR-155 expression was further reduced in patients with 2 or 3 diseased vessels compared to patients with 0 or 1 diseased vessels. However, miR-155 also negatively correlated with a range of traditional cardiac risk factors. In addition, D’Alessandra et al. identified increased expression of miR-337-5p, miR-433, and miR-485-3p out of 178 miRNAs profiled in patients with CAD. Specifically, miR-1, miR-122, miR-126, miR-133a/b, miR-199a, miR-485-3p, and miR-377-5p were increased in subjects with stable angina or unstable angina. Using only miR-1, miR-126, and miR-485-3p, this classified correctly subjects with stable angina compared to controls, whereas miR-1, miR-126, and miR-133a classified patients correctly with unstable angina vs controls in greater than 87% of cases. However, no combination could correctly discriminate between unstable angina and stable angina, suggesting that these markers likely reflect atherosclerotic burden present in both UA and SA patients.140 Moreover, using PBMCs, a higher ratio of miR-135a to miR-147 was used to help discriminate patients with or without CAD.141 In addition, reduced expression of miR-181a in PBMCs of obese patients is associated with prevalence of CAD.142 In contrast, increased expression of miR-146a or miR-146b in patients with CAD was associated with an increased risk of atherosclerosis. Interestingly, the rs2910164 polymorphism in the miR-146a locus is associated with an increased risk of CAD in a Chinese Han population.143 Finally, miR-146a expression was significantly increased in a Japanese cohort of 66 patients with CAD compared to no CAD. Furthermore, 12-months of statin therapy with an ACEI or ARB indicated that miR-146a/b and its regulated targets (IRAK, TRAF6, and TLR4) were significantly decreased.144
Unstable angina
While the diagnosis of acute myocardial infarction can be facilitated using myocardial injury biomarkers such as cardiac troponin I or T, the diagnosis of unstable angina (UA) in patients with normal troponin values can be elusive. Because unstable angina is associated with a higher risk of cardiovascular events, identification of a set of ischemic biomarkers may be useful for diagnosis, risk stratification, and therapeutic decision-making.145 Only a few miRNAs have been uniquely linked to unstable angina compared to stable CAD. However, these findings have not been replicated in larger clinical data sets. For example, expression of miR-134, miR-198, and miR-370 from peripheral blood mononuclear cells was enriched higher in 25 patients with UA compared to 25 with SA. However, these findings were not verified in a separate cohort of 19 UA and 34 SA patients.140 When the comparator group was non-coronary chest pain patients (instead of SA patients), a different cassette of miRNAs (miR-132, miR-150, and miR-186) was found to be increased in UA patients.146 Collectively, these findings suggest that rigorous phenotyping of patients with UA compared to other distinct patient sub-groups (e.g. AMI, SA, and non-coronary chest pain) will be required for appropriate discrimination of this unique and important UA group with elevated cardiac risk.
Acute Myocardial Infarction
While cardiac troponin is used to assist with the diagnosis of acute myocardial infarction (AMI), it may serve as a poor discriminator between AMI subtypes.145 In particular, additional biomarkers are desperately needed to further define Type I MI– ischemic myocardial necrosis due to atherosclerotic-plaque rupture (ACS) vs. Type II – ischemic myocardial not due to ACS, but typically due to supply/demand mismatch.145 Emerging studies indicate that different miRNAs may be released into the peripheral circulation in response to distinct pathophysiological stimuli, suggesting that identification of a cassette of miRNAs may possess improved discriminatory power to diagnose MI subtype or possibly lead to an earlier diagnosis. For example, the cardiac-specific miRNA, miR-208b, is detected in circulation by 3 hrs after AMI and may persist with an elevated expression over 90 days.147, 148 Several other groups validated the potential clinical usefulness of miR-208b as an early biomarker for AMI. Interestingly, miR-208 expression levels were increased in all n=33 patients 4 hrs post-MI compared to 85% for cardiac troponin I. Compared to other cardiac-enriched miRNAs (miR-1, miR-133a, miR-499), use of miR-208b conferred more favorable receiver operating characteristic curve with higher sensitivity and specificity to diagnose AMI compared to non-coronary heart disease patients.149 Similarly, miR-208b and miR-499 were significantly increased in AMI (n=32) compared to controls (n=36) that correlated with plasma cTnT.150 However, in another study that used a larger number of AMI patients (n=224), the diagnostic value for miR-208b (as well as miR-499 and miR-320a) in AMI was significantly lower than that of cardiac troponin I or T.151 Furthermore, while miR-208b and miR-499 expression were increased higher in patients with ST elevation myocardial infarction (STEMI) compared to non-STEMI (NSTEMI) and correlated with hs-cTnT and CK-MB in the plasma 1 hr after onset of chest pain, there diagnostic value was comparable to hs-TnT.152 Nonetheless, in a more focused patient cohort of NSTEMI geriatric subjects (mean age 82.6 years), Olivieri et al. examined plasma levels of miR-1, miR-21, miR-133a, miR-208a, miR-423-5p, and miR-499-5p. Remarkably, miR-499-5p was equivalent to cardiac troponin T in discriminating NSTEMI compared to healthy controls patients or those with acute congestive heart failure (CHF). Furthermore, the diagnostic accuracy of miR-499-5p was higher than either conventional or high sensitivity-troponin T in discriminating NSTEMI and acute CHF (AUC = 0.86 for miR-499-5p vs. cTnT AUC = 0.68 or hs-cTNT AUC=0.70).153 Finally, the bicistronic miRNA cluster of miR-1/miR-133 has also been implicated in AMI,154–156 Despite detection of these 4 miRNAs, miR-208b, miR-499-5p, miR-1, and miR-133a, in the plasma after AMI, these cardiac-enriched miRNAs did not improve diagnostic accuracy of AMI compared to cTnT.157 These findings may reflect the well-established ability of high sensitivity troponins to be released into circulation after generalized myocardial injury.
Atherosclerotic-based biomarkers that may reflect AMI or increased predisposition to plaque rupture or erosion would help discriminate MI subtype. In keeping with this notion, non-cardiomyocyte specific miRNAs may provide further refinement. For example, increased circulating levels of the miR-663 family member, miR-663b (a potential endothelial-enriched and flow-sensitive miRNA implicated in atherosclerosis (see above section on “Mechano-sensitive EC miRNAs that regulate atherosclerosis”) exhibited a high sensitivity (95%), specificity (90%), and accuracy (92.5%) to discriminate AMI (n=20 subjects) compared to controls (n=20 subjects).158 Examination of different subtypes of ischemic injury may also be informative for use of miRNA to assist in diagnostic discrimination of related disease conditions. For instance, plasma levels of miR-21-5p and miR-361-5p were significantly increased in AMI patients (n=17), whereas miR-519e-5p was reduced with comparable diagnostic accuracy to cTnI. In contrast, these three miRNAs were all increased in patients with ischemic stroke (n=9) or pulmonary embolism (n=8), whereas decreased miR-519e-5p was only detected in AMI.159 Collectively, these findings indicate that miRNAs may be useful to discriminate AMI subtype, and raise the possibility of incorporating miRNAs as a tool for linking pathophysiological events, prognosis, and stage-specific therapy.160
Taken together, these findings indicate that miRNAs may be associated with subclinical atherosclerosis. Furthermore, caution should be required for interpretation of similar miRNA profiling studies based upon baseline characteristics of medical treatment.
Therapeutic opportunities and challenges
Nucleic acid-based therapies represent a new frontier in the treatment of human diseases. Several classes of RNA therapeutics are currently under clinical development, including anti-sense oligonucleotides (ASO), small interfering RNA (siRNA) and miRNA mimetics and inhibitors. The field of RNA therapeutics saw a huge leap forward with the recent FDA approval of the first anti-sense oligonucleotide drug, Mipomersen, an antisense oligonucleotide (ASO) drug targeting apoB to treat homozygous familial hypercholesterolemia161–164. This will likely pave the way for other oligonucleotide-based drugs, including miRNA replacement and inhibitor therapies. Notably, pharmacological targeting of miRNAs in disease moves beyond the “one-drug-one-target” mode of treatment offered by siRNAs, and this has both benefits and drawbacks. The ability of miRNAs to target gene networks provides a unique approach for the treatment of disease by modulating gene pathways, but also opens the door to potential unwanted effects on additional target genes. Yet, this type of approach may prove to be more effective at improving complex diseases like atherosclerosis, which involve many pathways and would benefit from a multi-modal therapeutic.
Anti-sense oligonucleotides offer the ability to silence miRNAs to fine tune specific pathways or to reduce miRNAs expression dysregulated by disease. A number of approaches have been used to chemically modify anti-miR oligonucleotides to enhance target affinity, stability and tissue uptake165. These include: (i) locked nucleic acids (LNAs) which have high binding efficiencies and improved stability with the addition of a methylene link between the 2′-oxygen and the 4′-carbon resulting in a “locked” position and reducing the flexibility of the ribose ring166, (ii) ribose 2′-OH group modifications such as 2′methoxyethyl (MOE) and 2′4′-constrained 2′O-ethyl (cEt), and (iii) phosphorothioate (PS) modification. Careful evaluation of these chemical modifications will be required to minimize potential side effects and unanticipated toxicities. For example, phosphorothioate modification may facilitate nucleotide-based drugs to bind and activate platelets eliciting thrombus formation in response to carotid injury, pulmonary thromboembolism, and mesenteric artery injury in mice.167 Unlike their double-stranded counterparts, single stranded anti-miR oligonucleotides can be formulated in saline for subcutaneous or intravenous delivery and do not require lipid-based delivery systems. Upon systemic delivery, these compounds rapidly leave the plasma and are taken up by multiple tissues, most prominently liver, spleen, kidney, adipose tissue and bone marrow168, 169. Once taken up by cells, the anti-miR forms a stable, high-affinity bond with the miRNA reducing the availability of the endogenous miRNA for binding to the 3′UTR of the mRNA target. Preclinical studies in non-human primates using naked anti-miR oligonucleotides have shown promise, particularly in targeting miRNA expression in the liver (eg. miR-122, miR-33)13, 26. Cholesterol analogs have been added to anti-miRs in an attempt increase cellular uptake, and this promotes their incorporation into low and high density lipoproteins (LDL and HDL)170–172. Another approach for miRNA inhibition involves the use of miRNA sponges or decoy transcripts173, which act as competitive inhibitors of the miRNA of interest. miRNA sponges contain multiple binding sites that are complementary to the seed sequence of a miRNA of interest, which “mops up” the miRNA and inhibits its function. miRNA sponges can be delivered using viral vectors, and their expression can be made to be inducible in a specific cell type or developmental stage by using specific promoters. Notably, when vectors containing miRNA sponges were transfected into cells, miRNA target genes were as derepressed as when anti-miR oligonucleotides were used174.
miRNA mimetic therapy using synthetic miRNAs (miRNA mimics) offers the ability to reconstitute a miRNA that is downregulated during disease, or to decrease gene pathways involved in disease pathology. For example, patients with CAD exhibit decreased plasma levels of miR-181b, which is also expressed in ECs,50 and miRNA mimics could be used to restore its expression. The delivery of therapeutic miRNA molecules in vivo faces many of the same challenges as siRNAs due to their double-stranded nature. Drug delivery vehicles such as liposomes, polymeric micelles and lipoprotein-based drug carriers are being developed to deliver these oligonucleotides to cells. Notably, miRNAs were recently shown to be associated with endogenous HDL particles and to deliver their miRNA cargo to other cells175, offering the potential to use HDL infusion as a miRNA delivery vehicle. Some of the current challenges associated with miRNA replacement technology include the ability to target miRNAs to a specific tissue, and the potential requirement of multiple doses of a miRNA mimetic to achieve sustained target repression. Viral vectors can be used as gene delivery carriers for miRNAs, including short hairpin RNAs that can be processed in the target cell into the mature miRNA, a approach that has been used in multiple preclinical studies17, 25, 176. However, viral delivery systems will require careful scrutiny for clinical use.
While miRNA mimics and oligonucleotide inhibitors are taken up more efficiently in the liver, we and others have demonstrated success at penetrating the vascular endothelium of the vessel wall and peripheral blood mononuclear cells49, 50. Because only a minority of miRNAs are tissue-specific, delivery of miRNA mimics or inhibitors in a targeted cell- or tissue-specific rather than a systemic manner may represent a novel opportunity to prevent the development or progression of atherosclerosis. Recent approaches to tailor miRNA delivery to specific tissues include incorporating target sites for tissue-specific miRNAs, such as liver specific miR-122 and miR-192177, which would silence the vector in the liver, but allow it to be functional in other tissues.
A number of miRNA based therapeutics are currently in preclinical development, and two have reached clinical trials. The first is a LNA directed against miR-122 (Miravirsen), which targets hepatitis C virus (HCV) RNA178. Studies in non-human primates showed that miR-122 inhibition resulted in long-lasting suppression of HCV viremia with no evidence of viral resistance or side effects179. In a human phase 2 study Miravirsen demonstrated dose-dependent antiviral activity when given as a 4-week monotherapy that was maintained for more than 4 weeks after the end of therapy180. Notably, HCV RNAs in four of nine patients treated with the highest doses of Miravirsen became undetectable during the study181, demonstrating the potential of anti-miR therapeutics. The second miRNA therapeutic in clinical trial is a double-stranded miRNA mimic of miR-34, which acts as a tumor suppressor by inhibiting multiple oncogenic pathways and stimulating anti-tumor immune responses182. MRX34 is a miR-34 mimic encapsulated in a liposomal nanoparticle formulation that is being tested in patients in a range of advanced solid tumors and hematological malignancies. The results of these first clinical trials of a miRNA mimic and inhibitor are eagerly awaited.
Over the last several decades, significant progress has been achieved to treat atherosclerosis. Most notably, 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase inhibitors (statins) have been widely used to treat patients in the primary and secondary prevention of coronary artery disease. While statins effectively reduce LDL levels and cardiovascular events, a considerable residual burden of coronary artery disease remains even in patients treated with statins. Novel complementary therapeutic approaches will likely be necessary for treating disease states such as atherosclerosis involving complex signaling networks. In this regard, because miRNAs target multiple genes often in the same regulatory network, miRNAs may have tremendous effects on biological pathways, cell function, and homeostasis in the vessel wall, liver, and periphery. Delivery of a cassette of miRNA mimics or inhibitors may thereby offer an attractive therapeutic approach to facilitate “fine-tuning” specific stages of atherosclerotic disease and in the management of its complications.
Supplementary Material
Acknowledgments
Sources of Funding
This work was supported by the National Institutes of Health (HL115141, HL117994, and GM115605 to M.W.F. and HL108182 and HL119047 to KJM), the Arthur K. Watson Charitable Trust (to M.W.F.), and the Dr. Ralph & Marian Falk Medical Research Trust (to M.W.F.).
Non-standard Abbreviations and Acronyms
- AMI
acute myocardial infarction
- ASO
anti-sense oligonucleotides
- Ago2
Argonaute2
- ApoE
apolipoprotein E
- CAD
coronary artery disease
- D-flow
disturbed flow
- eNOS
endothelial nitric oxide synthase
- HDLc
HDL cholesterol
- GWAS
genome-wide association study
- ICAM-1
intracellular adhesion molecule 1
- L-flow
laminar flow
- LDLc
LDL cholesterol
- LDLR
LDL receptor
- miRNA
microRNA
- NF-κB
Nuclear Factor κB
- PBMCs
peripheral blood mononuclear cells
- RISC
RNA-induced silencing complex
- SA
stable angina
- SiRNA
small interfering RNA
- SNPs
single-nucleotide polymorphisms
- SR-BI
scavenger receptor BI
- STEMI
ST elevation myocardial infarction
- TRBP
TAR RNA-binding protein
- UA
unstable angina
- UTR
untranslated region
- VCAM-1
vascular adhesion molecule 1
Footnotes
Disclosures: None.
References
- 1.Hansson GK, Libby P, Tabas I. Inflammation and plaque vulnerability. J Intern Med. 2015;278:483–93. doi: 10.1111/joim.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 8.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–66. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 14.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic acids research. 2008;36:1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA, Sethupathy P, Remaley AT. MicroRNA-223 coordinates cholesterol homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14518–23. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–42. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nature medicine. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y, Fernandez-Hernando C. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nature medicine. 2015;21:1280–9. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nature medicine. 2015;21:1290–7. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell metabolism. 2008;7:365–75. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. The Journal of biological chemistry. 2010;285:33652–61. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17321–6. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12228–32. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–9. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–7. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. The Journal of clinical investigation. 2011;121:2921–31. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2707–14. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS letters. 2012;586:1472–9. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Experimental neurology. 2012;235:476–83. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Aguiar Vallim T, Tarling E, Kim T, Civelek M, Baldan A, Esau C, Edwards P. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circulation research. 2013 doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel AC, Zavadil J, Castrillo A, Jungsu K, Suarez Y, Fernandez-Hernando C. Control of Cholesterol Metabolism and Plasma HDL Levels by miRNA-144. Circulation research. 2013 doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B. MiRNA-185, MiRNA-96 and MiRNA-223 Repress Selective HDL-Cholesterol Uptake through Posttranscriptional Inhibition of Scavenger Receptor Class BI in Hepatic Cells. Molecular and cellular biology. 2013 doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Molecular and cellular biology. 2012;32:5035–45. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Naar AM. MicroRNAs in Metabolism and Metabolic Diseases. Cold Spring Harbor symposia on quantitative biology. 2011;76:225–33. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rayner K, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–409. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Naar AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Science translational medicine. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO molecular medicine. 2012;4:882–95. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. Journal of the American Heart Association. 2012;1:e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice--brief report. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:1973–7. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:455–8. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, Rinehold K, van Solingen C, Fullerton MD, Cecchini K, Rayner KJ, Steinberg GR, Zamore PD, Fisher EA, Loke P, Moore KJ. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. The Journal of clinical investigation. 2015;2015 doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Sowa N, Yahagi N, Shimano H, Matsumura S, Inoue K, Marusawa H, Nakamura T, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nature communications. 2013;4:2883. doi: 10.1038/ncomms3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goedeke L, Salerno A, Ramirez CM, Guo L, Allen RM, Yin X, Langley SR, Esau C, Wanschel A, Fisher EA, Suarez Y, Baldan A, Mayr M, Fernandez-Hernando C. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO molecular medicine. 2014;6:1133–41. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karunakaran D, Richards L, Geoffrion M, Barrette D, Gotfrit RJ, Harper ME, Rayner KJ. Therapeutic Inhibition of miR-33 Promotes Fatty Acid Oxidation but Does Not Ameliorate Metabolic Dysfunction in Diet-Induced Obesity. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:2536–43. doi: 10.1161/ATVBAHA.115.306404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–25. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 47.Suarez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol. 2010;184:21–5. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, Belkin N, Feinberg MW. Endothelial microRNAs and atherosclerosis. Curr Atheroscler Rep. 2013;15:372. doi: 10.1007/s11883-013-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, Hunninghake GM, Vera MP, Blackwell TS, Baron RM, Feinberg MW. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. The Journal of clinical investigation. 2012;122:1973–90. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, He S, Wara AK, Icli B, Shvartz E, Tesmenitsky Y, Belkin N, Li D, Blackwell TS, Sukhova GK, Croce K, Feinberg MW. Systemic delivery of microRNA-181b inhibits nuclear factor-kappaB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circulation research. 2014;114:32–40. doi: 10.1161/CIRCRESAHA.113.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med. 2014;24:105–12. doi: 10.1016/j.tcm.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gareus R, Kotsaki E, Xanthoulea S, van der Made I, Gijbels MJ, Kardakaris R, Polykratis A, Kollias G, de Winther MP, Pasparakis M. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell metabolism. 2008;8:372–83. doi: 10.1016/j.cmet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI, Fish JE. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO molecular medicine. 2013;5:949–66. doi: 10.1002/emmm.201202318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kahonen M, Karhunen PJ, Laaksonen R, Lehtimaki T. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211–7. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 55.Li K, Ching D, Luk FS, Raffai RL. Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-kappaB-driven inflammation and atherosclerosis. Circulation research. 2015;117:e1–e11. doi: 10.1161/CIRCRESAHA.117.305844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saba R, Sorensen DL, Booth SA. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front Immunol. 2014;5:578. doi: 10.3389/fimmu.2014.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies PF, Polacek DC, Handen JS, Helmke BP, DePaola N. A spatial approach to transcriptional profiling: mechanotransduction and the focal origin of atherosclerosis. Trends Biotechnol. 1999;17:347–51. doi: 10.1016/s0167-7799(99)01348-7. [DOI] [PubMed] [Google Scholar]
- 58.Frangos SG, Gahtan V, Sumpio B. Localization of atherosclerosis: role of hemodynamics. Arch Surg. 1999;134:1142–9. doi: 10.1001/archsurg.134.10.1142. [DOI] [PubMed] [Google Scholar]
- 59.Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol Rev. 2011;91:327–87. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiu JJ, Chen CN, Lee PL, Yang CT, Chuang HS, Chien S, Usami S. Analysis of the effect of disturbed flow on monocytic adhesion to endothelial cells. J Biomech. 2003;36:1883–95. doi: 10.1016/s0021-9290(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 61.DePaola N, Davies PF, Pritchard WF, Jr, Florez L, Harbeck N, Polacek DC. Spatial and temporal regulation of gap junction connexin43 in vascular endothelial cells exposed to controlled disturbed flows in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3154–9. doi: 10.1073/pnas.96.6.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miao H, Hu YL, Shiu YT, Yuan S, Zhao Y, Kaunas R, Wang Y, Jin G, Usami S, Chien S. Effects of flow patterns on the localization and expression of VE-cadherin at vascular endothelial cell junctions: in vivo and in vitro investigations. J Vasc Res. 2005;42:77–89. doi: 10.1159/000083094. [DOI] [PubMed] [Google Scholar]
- 63.Yamawaki H, Pan S, Lee RT, Berk BC. Fluid shear stress inhibits vascular inflammation by decreasing thioredoxin-interacting protein in endothelial cells. The Journal of clinical investigation. 2005;115:733–8. doi: 10.1172/JCI200523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Jr, Wang KC, Geary GG, Zhang Y, Wang WC, Huang HD, Zhou J, Li YS, Chien S, Garcia-Cardena G, Shyy JY. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation. 2011;124:633–41. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loyer X, Potteaux S, Vion AC, Guerin CL, Boulkroun S, Rautou PE, Ramkhelawon B, Esposito B, Dalloz M, Paul JL, Julia P, Maccario J, Boulanger CM, Mallat Z, Tedgui A. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circulation research. 2014;114:434–43. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 66.Fang Y, Davies PF. Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:979–87. doi: 10.1161/ATVBAHA.111.244053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iaconetti C, Polimeni A, Sorrentino S, Sabatino J, Pironti G, Esposito G, Curcio A, Indolfi C. Inhibition of miR-92a increases endothelial proliferation and migration in vitro as well as reduces neointimal proliferation in vivo after vascular injury. Basic Res Cardiol. 2012;107:296. doi: 10.1007/s00395-012-0296-y. [DOI] [PubMed] [Google Scholar]
- 68.Daniel JM, Penzkofer D, Teske R, Dutzmann J, Koch A, Bielenberg W, Bonauer A, Boon RA, Fischer A, Bauersachs J, van Rooij E, Dimmeler S, Sedding DG. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovascular research. 2014;103:564–72. doi: 10.1093/cvr/cvu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 70.Hinkel R, Penzkofer D, Zuhlke S, Fischer A, Husada W, Xu QF, Baloch E, van Rooij E, Zeiher AM, Kupatt C, Dimmeler S. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation. 2013;128:1066–75. doi: 10.1161/CIRCULATIONAHA.113.001904. [DOI] [PubMed] [Google Scholar]
- 71.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1516–21. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Developmental cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 75.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nature medicine. 2014;20:368–76. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris TA, Yamakuchi M, Kondo M, Oettgen P, Lowenstein CJ. Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arteriosclerosis, thrombosis, and vascular biology. 2010;30:1990–7. doi: 10.1161/ATVBAHA.110.211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, Chiu JJ, Shyy JY, Chien S. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circulation research. 2013;113:40–51. doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, Baker AH, Woong Seo J, Ferrara KW, Jo H. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nature communications. 2013;4:3000. doi: 10.1038/ncomms4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 82.Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFbeta Triggers miR-143/145 Transfer From Smooth Muscle Cells to Endothelial Cells, Thereby Modulating Vessel Stabilization. Circulation research. 2015;116:1753–64. doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- 83.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13450–5. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol. 2011;300:H1762–9. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun HX, Zeng DY, Li RT, Pang RP, Yang H, Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, Yan GJ, Zhou JG. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension. 2012;60:1407–14. doi: 10.1161/HYPERTENSIONAHA.112.197301. [DOI] [PubMed] [Google Scholar]
- 86.Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, Liao YH. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One. 2012;7:e46082. doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, Weber C, Schober A. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. The Journal of clinical investigation. 2012;122:4190–202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donners MM, Wolfs IM, Stoger LJ, van der Vorst EP, Pottgens CC, Heymans S, Schroen B, Gijbels MJ, de Winther MP. Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice. PLoS One. 2012;7:e35877. doi: 10.1371/journal.pone.0035877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu J, Chen T, Yang L, Li Z, Wong MM, Zheng X, Pan X, Zhang L, Yan H. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One. 2012;7:e46551. doi: 10.1371/journal.pone.0046551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun C, Wu MH, Yuan SY. Nonmuscle myosin light-chain kinase deficiency attenuates atherosclerosis in apolipoprotein E-deficient mice via reduced endothelial barrier dysfunction and monocyte migration. Circulation. 2011;124:48–57. doi: 10.1161/CIRCULATIONAHA.110.988915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Demolli S, Doebele C, Doddaballapur A, Lang V, Fisslthaler B, Chavakis E, Vinciguerra M, Sciacca S, Henschler R, Hecker M, Savant S, Augustin HG, Kaluza D, Dimmeler S, Boon RA. MicroRNA-30 mediates anti-inflammatory effects of shear stress and KLF2 via repression of angiopoietin 2. Journal of molecular and cellular cardiology. 2015;88:111–9. doi: 10.1016/j.yjmcc.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 92.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nature reviews Immunology. 2013;13:709–21. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang M, Wu JF, Chen WJ, Tang SL, Mo ZC, Tang YY, Li Y, Wang JL, Liu XY, Peng J, Chen K, He PP, Lv YC, Ouyang XP, Yao F, Tang DP, Cayabyab FS, Zhang DW, Zheng XL, Tian GP, Tang CK. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis. 2014;234:54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 94.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovascular research. 2009;83:131–9. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 95.Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu J, Sun Z, Shen WF. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS letters. 2011;585:854–60. doi: 10.1016/j.febslet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 96.Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, Zeng A, Zou J, Zhu RF, Han XS, Shen N, Yang HT, Zhao XX, Huang S, Qin YW, Jing Q. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovascular research. 2014;103:100–10. doi: 10.1093/cvr/cvu070. [DOI] [PubMed] [Google Scholar]
- 97.Meiler S, Baumer Y, Toulmin E, Seng K, Boisvert WA. MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:323–31. doi: 10.1161/ATVBAHA.114.304878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, Abraham E, Liu G. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190:6542–9. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, Li N, Gomez-Cabrero A, Reisfeld RA, Xiang R, Luo Y. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33:3014–23. doi: 10.1038/onc.2013.258. [DOI] [PubMed] [Google Scholar]
- 100.Wang Z, Brandt S, Medeiros A, Wang S, Wu H, Dent A, Serezani CH. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS One. 2015;10:e0115855. doi: 10.1371/journal.pone.0115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saha B, Bruneau JC, Kodys K, Szabo G. Alcohol-induced miR-27a regulates differentiation and M2 macrophage polarization of normal human monocytes. J Immunol. 2015;194:3079–87. doi: 10.4049/jimmunol.1402190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Veremeyko T, Siddiqui S, Sotnikov I, Yung A, Ponomarev ED. IL-4/IL-13-dependent and independent expression of miR-124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS One. 2013;8:e81774. doi: 10.1371/journal.pone.0081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Banerjee S, Cui H, Xie N, Tan Z, Yang S, Icyuz M, Thannickal VJ, Abraham E, Liu G. miR-125a-5p regulates differential activation of macrophages and inflammation. The Journal of biological chemistry. 2013;288:35428–36. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vergadi E, Vaporidi K, Theodorakis EE, Doxaki C, Lagoudaki E, Ieronymaki E, Alexaki VI, Helms M, Kondili E, Soennichsen B, Stathopoulos EN, Margioris AN, Georgopoulos D, Tsatsanis C. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J Immunol. 2014;192:394–406. doi: 10.4049/jimmunol.1300959. [DOI] [PubMed] [Google Scholar]
- 105.Cai X, Yin Y, Li N, Zhu D, Zhang J, Zhang CY, Zen K. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. Journal of molecular cell biology. 2012;4:341–3. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 106.Lu S, Gao Y, Huang X, Wang X. Cantharidin exerts anti-hepatocellular carcinoma by miR-214 modulating macrophage polarization. International journal of biological sciences. 2014;10:415–25. doi: 10.7150/ijbs.8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 108.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15819–24. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, Johnson DS, Chen Y, O’Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nature immunology. 2010;11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 110.Manoharan P, Basford JE, Pilcher-Roberts R, Neumann J, Hui DY, Lingrel JB. Reduced levels of microRNAs miR-124a and miR-150 are associated with increased proinflammatory mediator expression in Kruppel-like factor 2 (KLF2)-deficient macrophages. The Journal of biological chemistry. 2014;289:31638–46. doi: 10.1074/jbc.M114.579763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, Heyll K, Corbalan-Campos J, Hartmann P, Thiemann A, Weber C, Schober A. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–19. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 112.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, Maglie S, Marabita F, Mascheroni D, Parente V, Comelli M, Trabucchi E, De Francesco R, Geginat J, Abrignani S, Pagani M. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nature immunology. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 113.Zernecke A. MicroRNAs in the regulation of immune cell functions--implications for atherosclerotic vascular disease. Thrombosis and haemostasis. 2012;107:626–33. doi: 10.1160/TH11-08-0603. [DOI] [PubMed] [Google Scholar]
- 114.Dunand-Sauthier I, Santiago-Raber ML, Capponi L, Vejnar CE, Schaad O, Irla M, Seguin-Estevez Q, Descombes P, Zdobnov EM, Acha-Orbea H, Reith W. Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood. 2011;117:4490–500. doi: 10.1182/blood-2010-09-308064. [DOI] [PubMed] [Google Scholar]
- 115.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226–33. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 116.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2735–40. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jurkin J, Schichl YM, Koeffel R, Bauer T, Richter S, Konradi S, Gesslbauer B, Strobl H. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol. 2010;184:4955–65. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 119.Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J Immunol. 2010;185:7244–51. doi: 10.4049/jimmunol.1001573. [DOI] [PubMed] [Google Scholar]
- 120.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2008;28:812–9. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nature medicine. 2015;21:628–37. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circulation research. 2014;115:662–7. doi: 10.1161/CIRCRESAHA.115.304634. [DOI] [PubMed] [Google Scholar]
- 123.Rangrez AY, Massy ZA, Metzinger-Le Meuth V, Metzinger L. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet. 2011;4:197–205. doi: 10.1161/CIRCGENETICS.110.958702. [DOI] [PubMed] [Google Scholar]
- 124.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–78. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–8. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. The Journal of clinical investigation. 2009;119:2634–47. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lovren F, Pan Y, Quan A, Singh KK, Shukla PC, Gupta N, Steer BM, Ingram AJ, Gupta M, Al-Omran M, Teoh H, Marsden PA, Verma S. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation. 2012;126:S81–90. doi: 10.1161/CIRCULATIONAHA.111.084186. [DOI] [PubMed] [Google Scholar]
- 129.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circulation research. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 130.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circulation research. 2009;105:158–66. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. Journal of molecular and cellular cardiology. 2012;52:245–55. doi: 10.1016/j.yjmcc.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. The Journal of biological chemistry. 2009;284:3728–38. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bazan HA, Hatfield SA, O’Malley CB, Brooks AJ, Lightell D, Jr, Woods TC. Acute Loss of miR-221 and miR-222 in the Atherosclerotic Plaque Shoulder Accompanies Plaque Rupture. Stroke. 2015;46:3285–7. doi: 10.1161/STROKEAHA.115.010567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arteriosclerosis, thrombosis, and vascular biology. 2010;30:1562–8. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 135.Wang D, Deuse T, Stubbendorff M, Chernogubova E, Erben RG, Eken SM, Jin H, Li Y, Busch A, Heeger CH, Behnisch B, Reichenspurner H, Robbins RC, Spin JM, Tsao PS, Schrepfer S, Maegdefessel L. Local MicroRNA Modulation Using a Novel Anti-miR-21-Eluting Stent Effectively Prevents Experimental In-Stent Restenosis. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:1945–53. doi: 10.1161/ATVBAHA.115.305597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating microRNAs in patients with coronary artery disease. Circulation research. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 138.Weber M, Baker MB, Patel RS, Quyyumi AA, Bao G, Searles CD. MicroRNA Expression Profile in CAD Patients and the Impact of ACEI/ARB. Cardiol Res Pract. 2011;2011:532915. doi: 10.4061/2011/532915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhu GF, Yang LX, Guo RW, Liu H, Shi YK, Ye JS, Yang ZH. microRNA-155 is inversely associated with severity of coronary stenotic lesions calculated by the Gensini score. Coron Artery Dis. 2014;25:304–10. doi: 10.1097/MCA.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 140.D’Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, Rubino M, Marenzi G, Colombo GI, Achilli F, Maggiolini S, Capogrossi MC, Pompilio G. Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS One. 2013;8:e80345. doi: 10.1371/journal.pone.0080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, van Berkel TJ, Biessen EA. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010;394:792–7. doi: 10.1016/j.bbrc.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 142.Hulsmans M, Sinnaeve P, Van der Schueren B, Mathieu C, Janssens S, Holvoet P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J Clin Endocrinol Metab. 2012;97:E1213–8. doi: 10.1210/jc.2012-1008. [DOI] [PubMed] [Google Scholar]
- 143.Xiong XD, Cho M, Cai XP, Cheng J, Jing X, Cen JM, Liu X, Yang XL, Suh Y. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat Res. 2014;761:15–20. doi: 10.1016/j.mrfmmm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 144.Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci (Lond) 2010;119:395–405. doi: 10.1042/CS20100003. [DOI] [PubMed] [Google Scholar]
- 145.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD, Katus HA, Lindahl B, Morrow DA, Clemmensen PM, Johanson P, Hod H, Underwood R, Bax JJ, Bonow RO, Pinto F, Gibbons RJ, Fox KA, Atar D, Newby LK, Galvani M, Hamm CW, Uretsky BF, Steg PG, Wijns W, Bassand JP, Menasche P, Ravkilde J, Ohman EM, Antman EM, Wallentin LC, Armstrong PW, Simoons ML, Januzzi JL, Nieminen MS, Gheorghiade M, Filippatos G, Luepker RV, Fortmann SP, Rosamond WD, Levy D, Wood D, Smith SC, Hu D, Lopez-Sendon JL, Robertson RM, Weaver D, Tendera M, Bove AA, Parkhomenko AN, Vasilieva EJ, Mendis S Joint ESCAAHAWHFTFftUDoMI. Third universal definition of myocardial infarction. Circulation. 2012;126:2020–35. doi: 10.1161/CIR.0b013e31826e1058. [DOI] [PubMed] [Google Scholar]
- 146.Zeller T, Keller T, Ojeda F, Reichlin T, Twerenbold R, Tzikas S, Wild PS, Reiter M, Czyz E, Lackner KJ, Munzel T, Mueller C, Blankenberg S. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J. 2014;35:2106–14. doi: 10.1093/eurheartj/ehu151. [DOI] [PubMed] [Google Scholar]
- 147.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet. 2011;4:614–9. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 150.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 151.Devaux Y, Mueller M, Haaf P, Goretti E, Twerenbold R, Zangrando J, Vausort M, Reichlin T, Wildi K, Moehring B, Wagner DR, Mueller C. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med. 2015;277:260–71. doi: 10.1111/joim.12183. [DOI] [PubMed] [Google Scholar]
- 152.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, Corsten MF, Schroen B, Lair ML, Heymans S, Wagner DR. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem. 2012;58:559–67. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 153.Olivieri F, Antonicelli R, Lorenzi M, D’Alessandra Y, Lazzarini R, Santini G, Spazzafumo L, Lisa R, La Sala L, Galeazzi R, Recchioni R, Testa R, Pompilio G, Capogrossi MC, Procopio AD. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. 2013;167:531–6. doi: 10.1016/j.ijcard.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 154.Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, Kempf T, Wollert KC, Thum T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. Journal of molecular and cellular cardiology. 2011;51:872–5. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 155.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, Dong X, Qin S, Zhang C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, Li K, Yu B, Li Z, Wang R, Wang L, Li Q, Wang N, Shan H, Li Z, Yang B. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 157.Li YQ, Zhang MF, Wen HY, Hu CL, Liu R, Wei HY, Ai CM, Wang G, Liao XX, Li X. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics (Sao Paulo) 2013;68:75–80. doi: 10.6061/clinics/2013(01)OA12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Meder B, Keller A, Vogel B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J, Leidinger P, Meese E, Katus HA, Rottbauer W. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol. 2011;106:13–23. doi: 10.1007/s00395-010-0123-2. [DOI] [PubMed] [Google Scholar]
- 159.Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, Chen C, Wang DW. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One. 2014;9:e105734. doi: 10.1371/journal.pone.0105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Feinberg MW. MicroRNAs as pathophysiological targets: An emerging nexus for personalized medicine in heart failure? Trends Cardiovasc Med. 2015 doi: 10.1016/j.tcm.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thomas GS, Cromwell WC, Ali S, Chin W, Flaim JD, Davidson M. Mipomersen, an Apolipoprotein B Synthesis Inhibitor, Reduces Atherogenic Lipoproteins in Patients with Severe Hypercholesterolemia at High Cardiovascular Risk: A Randomized, Double-Blind, Placebo-Controlled Trial. Journal of the American College of Cardiology. 2013 doi: 10.1016/j.jacc.2013.07.081. [DOI] [PubMed] [Google Scholar]
- 162.Mearns BM. Dyslipidemia: Phase III trial of mipomersen in heterozygous FH. Nature reviews Cardiology. 2012;9:674. doi: 10.1038/nrcardio.2012.151. [DOI] [PubMed] [Google Scholar]
- 163.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, Tribble DL, Flaim JD, Crooke ST. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 164.Akdim F, Stroes ES, Sijbrands EJ, Tribble DL, Trip MD, Jukema JW, Flaim JD, Su J, Yu R, Baker BF, Wedel MK, Kastelein JJ. Efficacy and safety of mipomersen, an antisense inhibitor of apolipoprotein B, in hypercholesterolemic subjects receiving stable statin therapy. Journal of the American College of Cardiology. 2010;55:1611–8. doi: 10.1016/j.jacc.2009.11.069. [DOI] [PubMed] [Google Scholar]
- 165.Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104–10. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Grunweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? Bio Drugs. 2007;21:235–43. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 167.Flierl U, Nero TL, Lim B, Arthur JF, Yao Y, Jung SM, Gitz E, Pollitt AY, Zaldivia MT, Jandrot-Perrus M, Schafer A, Nieswandt B, Andrews RK, Parker MW, Gardiner EE, Peter K. Phosphorothioate backbone modifications of nucleotide-based drugs are potent platelet activators. J Exp Med. 2015;212:129–37. doi: 10.1084/jem.20140391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–72. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–6. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 171.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DW, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipid-like materials for low-dose, in vivo gene silencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1864–9. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 173.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen BJ, van den Berg A. Generation of miRNA sponge constructs. Methods. 2012;58:113–7. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 175.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–6. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 177.Geisler A, Jungmann A, Kurreck J, Poller W, Katus HA, Vetter R, Fechner H, Muller OJ. microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 2011;18:199–209. doi: 10.1038/gt.2010.141. [DOI] [PubMed] [Google Scholar]
- 178.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 179.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV Infection by Targeting MicroRNA. N Engl J Med. 2013 doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 181.Janssen HL, Reesink HW, Zeuzem S, Lawitz E, Rodriguez-Torres M, Chen A, Davis C, King B, Levin AA, Hodges MR. A randomized, double-blind, placebo (plb) controlled safety and anti-viral proof of concept study of miravirsen (MIR), an oligonucleotide targeting miR-122, in treatment naíve patients with genotype 1 (gt1) chronic HCV infection. The 62nd annual meeting of the American Association for the Study of Liver Diseases; The Liver Meeting; San Francisco, CA. 2011. [Google Scholar]
- 182.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.