Summary
Persistence of latent virus represents a major barrier to eradicate HIV even in the current antiretroviral therapy era. A critical limitation to eliminate these viral reservoirs is the lack of reliable methods to detect, quantify, and characterize cells harboring low levels of virus. However, recent work of several laboratories indicates that PCR and viral amplification based technologies underestimate or overestimate the size of the reservoirs. Thus, new technologies and methodologies to detect, quantify, and characterize these viral reservoirs are necessary to monitor and eradicate HIV. Recent developments in imaging technologies have enabled the development or improvement of detection protocols and have facilitated the identification and quantification of several markers with exquisite resolution. In the context of HIV, we developed new protocols for detection of low amounts of viral proteins. In this chapter, we describe several antibody-based technologies for signal amplification to improve and detect low amounts of HIV proteins in cells, tissues, and other biological samples. The improvement in these techniques is essential to detect viral reservoirs and to design strategies to eliminate them.
Keywords: HIV, reservoirs, eradication, AIDS, QVOA, detection
1. Introduction
HIV has become a chronic disease and despite the reduction in viremia to often to undetectable levels by antiretroviral therapy (ART), treatment is still not curative. A major obstacle to complete HIV eradication is the generation of viral reservoirs that sequester the virus in infected individuals (1–3). The best characterized HIV reservoir is a small population of resting CD4+ memory and naïve T cells (1,2,4), but other reservoirs in macrophages and astrocytes (3–5) also have been described. Currently, identification and quantification of viral reservoirs is mainly performed by PCR or cell reactivation based technologies, but both detection systems have interpretation and technical difficulties, including the need for large amounts of blood, extensive time allocation, high cost, and significant differences in assay sensitivity (6–8).
We have developed several comprehensive, integrated, and highly sensitive assays to analyze viral reservoirs by simultaneously examining integrated HIV DNA (sensitivity equal to one copy of HIV DNA per cell) or HIV mRNAs (sensitivity for few molecules) and viral proteins (sensitivity of few proteins, protocols described below). Because the detection is by imaging techniques, it does not require cell purification or amplification of the HIV components for the identification of a small number of viral reservoirs among millions of uninfected cells. We achieved this sensitivity using highly specific signal amplification systems as well as improved microscopy and optic devices as described recently (9,10). In addition to the HIV products, we are able to detect several cellular/molecular markers to analyze further viral trafficking, cellular activation, compartmentalization, and HIV interacting proteins including histone acetylates, apolipoproteins, and others (up to 5–6 colors). Our approach enables improved techniques of antigen recovery, staining, and confocal analysis resulting in outstanding identification and quantification of viral reservoirs. By using these methods, we are able to analyze millions of cells and focus only on the cells positive for viral HIV DNA/mRNA/protein using confocal microscopy, improved equipment, and imaging software. In this chapter, we will focus on 2 methods of detection of low levels of HIV proteins in cells. These methods can then be combined with assays for detection of HIV DNA and/or mRNA in the same samples, to obtain the most sensitive and reliable detection of viral reservoirs.
Some of the technical improvements described here include: 1) Improved cell and tissue preparation to conserve antigens and nuclei acids during the processing of the sample even in archival materials; 2) The use of larger pieces of tissue or numbers of cells to analyze millions of cells using big pinholes to generate large optical sections to detect any positive signal; 3) The development of novel protocols enables signal amplification for antibodies. 4) The use of state of the art confocal systems and the automation of microscopes allows us to perform fast scanning of large areas in 3 dimensions to identify the few HIV-infected cells by 3D reconstructions and deconvolution; 5) The use of a spectrum detection and unmixing system to detect extremely narrow wavelengths and eliminates auto-fluorescence; 6) Improved detection systems include cameras with recovery of 90% of photons per frame instead of the high resolution cameras for microscopy that only recover approximately 50% of photons and 7) Lastly, improved software and algorithms detect and quantify the signals generated by the different viral components (see details using other latent pathogens in (11,12)).
The combination of all these factors enables us to detect, quantify, and localize specific signals from HIV reservoirs.
2. Materials
2.1 Tissue sections
Any tissue section can be analyzed for viral reservoirs. The important point is preservation and size of the section (10–300 µm) to allow analysis of millions of cells.
Alcohol/Xylenes
Phosphate buffered saline (PBS) and Tris buffered saline (TBS)
Citrate
Fish Gelatin
Horse serum
Sudan Black
Sodium borohydrate
Pontamine sky blue and 6.6’-[(3, 3’-dimethoxy[1,1’-biphenyl]-4,4’-diyl)bis(azo)]bis[4-aminuteso-5-hydroxy-1,3-naphthalenedisulfonic acid], tretrasodium
Toluidine blue
Triton-X
Biotin blocking reagents
Strepavidin conjugated to different fluorochromes or beads
Alexa conjugated secondary antibody- Goat Anti-Rabbit IgG
Prolong Gold anti-fade agent with DAPI
2.2. Leukocytes
Whole blood or leukopacks from HIV infected or uninfected individuals.
HIV-p24 ELISA (Perkin Elmer, Boston, MA; sensitivity: 12.5 pg/ml) or by COBAS Roche Amplicor v 1.5 (Roche, Germany; sensitivity 20 RNA copies/ml) to detect HIV infection.
Lysis buffer
Ficoll Paque plus
Poly-lysine glass slides
Phorbol myristate acetate (PMA)
Hela cells
Paraformaldehyde (PFA)
3. Methods
3.1 Equipment
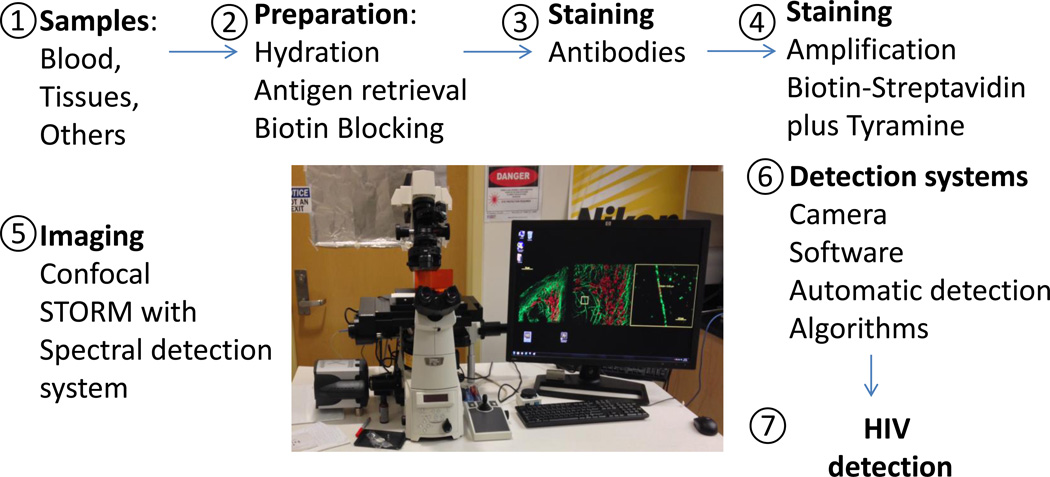
Several types of confocal microscopes can be used depending on the brand. In our case we used an A1 Nikon confocal microscope with spectrum detection and unmixing separation systems. The configuration of the system is described in Figure 1. Using these configurations in addition to better protocols for staining and identification of dim signals, we are able to detect several latent pathogens, including low levels of HIV (9,11).
Figure 1.
Description of the improved sample and microscopy technology to detect low levels of HIV proteins. Every step has been optimized to achieve outstanding resolution. 1 and 2, correspond to improved techniques for tissue preservation. 3 and 4, correspond to the use of new or improved techniques for signal amplification. 5 and 6, correspond to improved imaging and detection systems as well as to software analysis. These improvements result in maximal sensitivity to detect HIV proteins in viral reservoirs as well as in HIV replicating cells.
3.2 Quantification of HIV replication by ELISA or PCR
Viral replication was quantified by measuring HIV-1 p24 concentrations by ELISA using a commercial kit or by PCR.
3.3. Positive and negative controls and sample fixation
ACH2 (a human lymphoid) or OM-10 cells (a monocyte cell line) were used as positive controls because each cell has only 1 integrated copy of HIV-1 DNA and produces significant amounts of viral proteins when stimulated with phorbol myristate acetate (PMA) or TNF-alpha.
Hela cells are used as a negative control representing uninfected cells.
For tissue sections, we use human lymph nodes obtained from individuals with high or undetectable replication, as well as uninfected tissues, as positive and negative controls, respectively.
3.4 Sample preparation
Multiple fixatives can be used for tissues, cells, or fluids including:
70% cold Ethanol (−20 °C for 20 minutes)
Acetone
20 % buffered formalin and subsequent permeabilization using 0.1 % Triton-X for 3 minutes.
Four % paraformaldehyde (PFA) containing 0.1M sodium phosphate buffer, pH 7.4, and subsequent permeabilization using 0.1 % Triton-X for 3 minutes.
3.5 Deparaffining
After fixation, and mounting into paraffin blocks, tissue sections from 10 to 300 µm are deparaffined using Ethanol-Xylene in the following order: Ethanol 30%, 50%, 60%, 70%, 80%, 90% and 100 %, Xylene 1 and 2 (two separate solutions), and then Ethanol 100%, 90%, 80%, 70%, 60%, 50% and 30%, and then PBS for 10 minutes. It is important to include all of the steps to assure the slow and efficient elimination of paraffin. The thickness of the section is also extremely important due to the large numbers and optical sections required for identification of viral reservoirs. Many companies and facilities only prepare sections of 5–10 µm; thus, a special request to the company or training of personnel is required to obtain these types of sections.
3.6 Antigen retrieval
There are several techniques of antigen retrieval depending on the application. For a comprehensive list of antigen retrieval methods visit, www.ihcworld.com/epitope_retrieval.htm. For our applications, we use the boiling citrate buffer method for 15 minutes (pH 6.0) for thicker tissue sections (10 to 300 µm), but we have also obtained good results with microwave-based techniques.
3.7 Leukocyte analysis
To analyze a significant number of circulating leukocytes, whole blood, isolated PBMCs, leukopacks or specific populations of cells isolated using magnetics beads, are isolated, pelleted, and subjected to confocal analysis. The pellets can be generated directly on the glass slide or the centrifuged pellet can be fixed and cut with a cryostat. By doing this protocol, we can reduce the size and volume of cells analyzed such that millions of cells can be evaluated with a better chance of detecting viral reservoirs.
The following five points are critical for the detection protocols of low levels of HIV proteins using minimal amplification, because high autofluorescence can result in false positives. Most of these protocols apply to archival tissues.
3.8 Elimination of lipofuscins fluorescence
Natural autofluorescence is due to flavins, porphyrins, and chlorophyll (mostly in plants). The main problem with these compounds present in tissues and cells is that during cutting and solvent treatments, they become redistributed, resulting in background fluorescence. However, new optical configurations to perform unmixing and spectrum detection can significantly reduce this problem. In addition, treatment of the sample with Sudan Back (0.3% in 70% Ethanol) stirred in the dark for 2 h, will reduce significantly the autofluorescence produced by lipofuscins.
3.9 Elimination of elastin and collagen autofluorescence
This artifact is mainly found in blood vessel walls. Elastin contains several potential fluorophores when there is cross-linking of tricarboxylic amino acid with a pyridinium rings (17,18). In small vessels detection of these products is minimal, but in large vessels it is a significant problem. To eliminate autofluorescence from elastin products, incubate your samples in 0.5% pontamine sky blue and 6.6’-[(3, 3’-dimethoxy[1,1’-biphenyl]-4,4’-diyl)bis(azo)]bis[4-aminuteso-5-hydroxy-1,3-naphthalenedisulfonic acid], tretrasodium salt dissolved in 50 mM Tris buffer before mounting the samples. However, the use of both compounds requires extensive calibration, because pontamine sky blue fluoresces in the red channels. However, if the red channel is not to be used, it is an excellent choice.
An alternative solution is 0.1% toluidine blue for 3 minutes before mounting the samples, but this does not work in all vessels and the interpretation of the fluorescence can become complicated.
3.10 Elimination of fixative-induced fluorescence
Aldehydes react with amines and proteins to generate fluorescent products, especially in samples incubated for a long time in fixatives. This problem occurs most often in fixatives such as glutaraldehyde and formaldehyde. For tissue sections 10 to 300 µm, incubate them 5 times for 15 minutes each in a solution of fresh borohydrate (1 mg/ml dissolved in PBS and prepared on ice). After this process, wash in PBS 3 times and discard the leftover sodium borohydrate.
3.11 Bleaching treatment
Currently there is no company that sells appropriate light boxes, but it is relatively easy to construct. To build a specific wavelength light box (like the one used to detect ethidium bromide in agarose gels), fluorescent tubes, especially for blue, green, red, and far red channels can be purchased from several companies. These can be used to “burn” the autofluorescence in the tissue sections before the staining process.
3.12 Endogenous biotin blocking
If the tissue has endogenous biotin activity, biotin blocking is suggested using a Biotin blocking kit. First, incubate the tissue sections or pelleted leukocytes with avidin solution for 10–30 minutes and then wash in TBST 3 times for 5 minutes. Next, incubate with biotin solution for 10–30 minutes and then wash in TBST 3 times for 5 minutes.
3.14 Antibodies and Biotinylation of antibodies
All antibodies for staining of multiple colors will be described below. IgG biotinylation is performed using commercial Biotin labeling kits.
3.15 Data Analysis
Mean differences are tested by non-parametric Kruscal-Wallis analysis. If a significant F-value is obtained, means are compared with the Bofferonni-Dunn multiple comparison test. A value of p<0.05 is considered significant.
3.16 Multiple methods of signal amplification to detect viral reservoirs
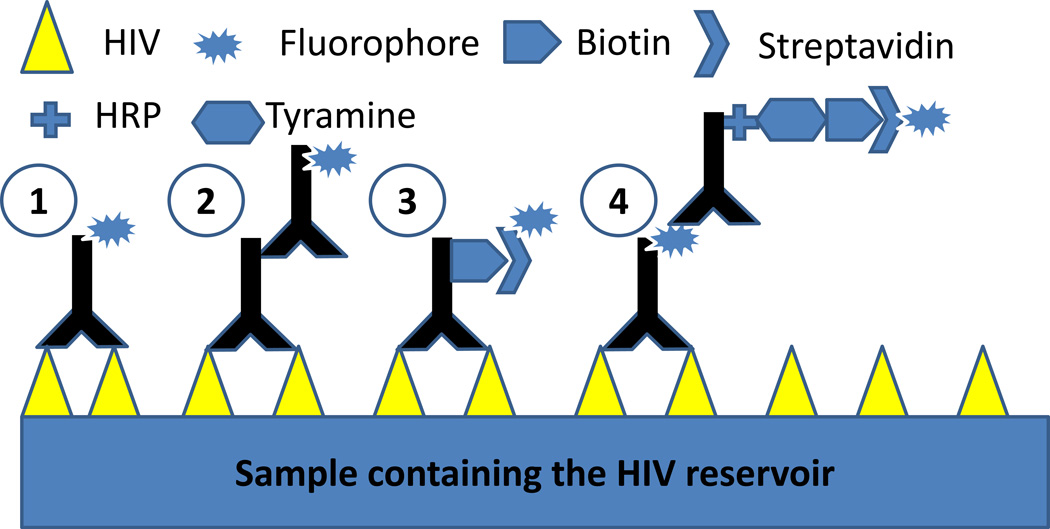
Normally immune detection of proteins requires either a single antibody conjugated to a fluorescent dye or a primary antibody with a secondary antibody that amplifies the fluorescent signal. Most of the times these protocols are inefficient in detecting viral reservoirs or low HIV replication in multiple systems, and only high viral replication can be detected (19,20). As described in figure 2, the combination of better protocols for fixation, antigen recovery, staining, and detection, enables exquisite identification and resolution of HIV proteins, despite minimal to undetectable replication as assayed by ELISA or PCR. A critical component in optimizing these protocols is to determine the expected level of expression of HIV proteins, because most protocols described here are designed to amplify low signals (See Note 1 to decide the best protocol for your application).
Figure 2.
Schematic of four different methods of immune staining. Method 1 illustrates an antibody directly conjugated to a fluorophore. This technique is commonly used in FACS analysis and also in immunohistochemistry and cytochemistry. Method 2 represents a method providing additional amplification of the signal. Method 3 demonstrates antibodies labeled with biotin and detection is by biotin-streptavidin interactions. Our system, method 4, uses a multistep process that amplifies the number of fluorophores binding to the antigen, resulting in high sensitivity and amplification. Methods 3 and 4 are most appropriate for detection of HIV reservoirs depending on the cells and tissues being analyzed.
3.17 Staining process
As described above and in Figures 1 and 2, fixation, sample preparation, staining, signal amplification, and detection systems are essential for identifying and quantifying low amounts of HIV proteins. Some the improvements of detection and imaging components are recently described in detail (9,10). For viral reservoirs, standard staining using directly conjugated antibodies (Figure 2, method 1) and secondary conjugated antibodies (Figure 2, method 2) are not sufficient to detect low levels of proteins. Thus, the methods using antibody-biotin-streptavidin-fluorophore (Figure 2, method 3) and antibody-fluorophore-antibody to fluorophore conjugated to HRP-tyramine-biotin-streptavidin-fluorophore (Figure 2, method 4) are highly sensitive and adaptable to determine localization, quantification, and trafficking of HIV proteins in cells, tissues, and fluids.
Approach 1
Antibody conjugated to biotin-streptavidin-fluorophore amplification
(See Figure 2, 3rd method):
-
1
Samples are fixed, and prepared as described above (antigen retrieval, and elimination of autofluorescence) according to the sample used, tissue sections or pelleted leukocytes (go to step 4 below).
-
2
If tissue sections are used from paraffin blocks, heat the slides to remove excess paraffin in an oven at 60 °C for 15 minutes and dry at 37 °C overnight.
-
3
Deparaffinize the sections as described above using sequential alcohols and xylenes to eliminate the paraffin slowly.
-
4
Proceed with either antigen retrieval or blocking endogenous biotin as described above, depending upon the tissue or cells being analyzed and the staining process performed. Liver, spleen, and brain are tissues with endogenous biotin.
-
5
Using the set up described in Figure 1, we are able to detect several colors (up to 5–6 colors). We can probe for nucleic acids (DAPI), HIV/SIV proteins (p24, p17, gp120, tat, integrase, or Nef), in combination with different cellular markers including CD4, CD8, GFAP (an astrocyte marker), NeuN or MAP-2 (a neuronal markers) or Iba1 (a microglia/macrophage marker). For pelleted leukocytes, several cellular markers such as CD3, CD4, CD11b, CD11c, CD14, CD16, CD68, and CD163 can also be included in the same staining protocol.
-
6
Tissue sections or pelleted leukocytes are blocked for at least 60 minutes to overnight using blocking solution (0.5M EDTA, 1% horse serum, 1% Ig free BSA, 4% human serum and 1% fish gelatin in PBS).
-
7Samples are incubated overnight in primary antibody at 4 °C. A critical point is to determine how many antibodies can be used concomitantly based on antibody species and isotypes. Several combinations can be used. Some examples are:
- HIV biotinylated antibodies (monoclonal, IgG1)+ CD4 (rabbit antibodies)+ Iba 1 (macrophage marker)+ nuclei acid staining (DAPI).
-
HIV biotinylated antibodies (monoclonal, IgG1) + CD4 (rabbit antibodies)+ Iba 1 (macrophage marker)+ actin staining+ nuclei acid staining (DAPI).A critical point of these experimental approaches is the determination of appropriate negative controls. For the examples described above the following controls are used:
- Purified IgG1 (same concentration as the HIV antibodies)+ rabbit serum or rabbit purified IgG (same concentration as the serum or of the immune IgG) and non-immune goat serum or IgG (same concentration of the Iba-1 IgGs) (See Note 2 to identify the best negative controls for your experiments and antibodies).
- Several tissues express low levels of endogenous biotin; therefore a control for this expression is required, despite inhibition of biotin binding as described above.
Importantly, negative controls using no primary antibodies or only secondary antibodies are not accurate controls. As described above, by using non-immune IgGs or serum, we consider the possibility of non-specific binding to several proteins such as Fc receptors, especially in immune and inflammated tissues. All cells, tissues, and fluids to some degree have non-specific binding that is necessary to consider, especially in cases of detection of low amounts of proteins, such as found in HIV reservoirs. Thus, specificity of the antibodies must be confirmed by replacing the primary antibody with the appropriate isotype-matched control reagent, anti-IgG1, IgG2A, IgG2B or the IgG fraction of normal rabbit serum depending on the primary antibody being used (see Note 2).
-
8
After incubation with the primary antibodies, at least 5 washes with PBS every 10 minutes are required to eliminate the unbounded antibodies to the antigen.
-
9
To detect antibodies conjugated to biotin, streptavidin conjugated to a fluorophore is necessary. In addition, other streptavidin-conjugated reagents can be used such as beads or gold. Detection of low levels of HIV proteins requires at least 3 hours of incubation.
-
10
After incubation with the secondary antibody or conjugated streptavidin, at least 5 washes with PBS are required to eliminate the unbounded streptavidin or secondary antibodies. Our confocal equipment has unmixing and spectrum detection systems that enable the separation of extremely narrow wavelengths (up to 2.5 nm) to separate multiple colors without overlay (some examples in (9,10))
-
11
After several washes to eliminate unbounded antibodies or dyes, samples are mounted using Prolong Gold anti-fade reagent with DAPI. If beads are used, we suggest using Prolong Diamond anti-fade reagent with DAPI.
-
12
After staining, keep samples in the dark.
-
13
The analysis of thick samples (tissues and pelleted leukocytes) is performed first using a large pinhole just to detect HIV positive signals. After we detect the positive optical section, we perform confocal microscopy in the specific XYZ positive axis with a regular pinhole and good resolution to detect and quantify the localization of viral proteins using 3D reconstructions and deconvolution (see Note 3 for details).
Approach 2
Antibody-fluorophore-antibody to fluorophore conjugated to HRP-tyramine-biotin-streptavidin-fluorophore
(see Figure 2, 4th method (see Note 4 for potential problems):
This method is essentially similar to method 1, but uses additional amplification steps.
Sample fixation, antigen retrieval, biotin blocking, and tissue preparation are similar to what is described for method 1 as well as described in Figure 2.
This method includes antibodies conjugated to a fluorophore to target any HIV protein in a similar manner described above. However, the main difference is that an anti-fluorophore secondary antibody conjugated to HRP (dilutions 1:600 to 1:2000) is used by adding biotinyl-tyramide for 15 minutes in the presence of 0.3 % H2O2 for 20 minutes to amplify the binding of the new fluorophores.
Wash the slides in PBS twice for 10 minutes each.
Incubate in 0.25 mg/ml streptavidin conjugated to any fluorophore for 30 minutes to 3 h in the dark.
Wash three times in PBS every 10 minutes in the dark
Incubate the slides in water for 10 minutes
Mount using prolong with DAPI as described above.
It is important to note that for all of these protocols it is essential to use pure solutions, because any contamination can be amplified and result in false positive signals (See Note 5).
3.18 Improved microscopic analysis to detect low levels of HIV proteins in HIV reservoirs
New confocal systems, including the Nikon A1, have an improved spectral detector and unmixing systems, PMT, and cameras to improve detection and reduce potential cross contamination amount different colors. These systems allow a critical reduction in signal to noise ratio, improving the detection of specific staining. The spectral detector is the mechanism responsible for emitting light through a high-efficiency gate into its individual components, similar to how a prism separates white light into its individual ‘rainbow’ components. The improved spectral detector allows for precise separation of emission wavelengths that are then passed through a 32-photomultiplier tube (PMT) array detector that enables distinction between wavelengths as small as 2.5 nm apart. This precise detection system allows the researcher to separate and analyze specific wavelengths or eliminate autofluorescence. This technology enables the identification of signals that would be impossible to detect with wide-field fluorescence or standard confocal microscopy (for details of these improved technologies see (12,21–23)).
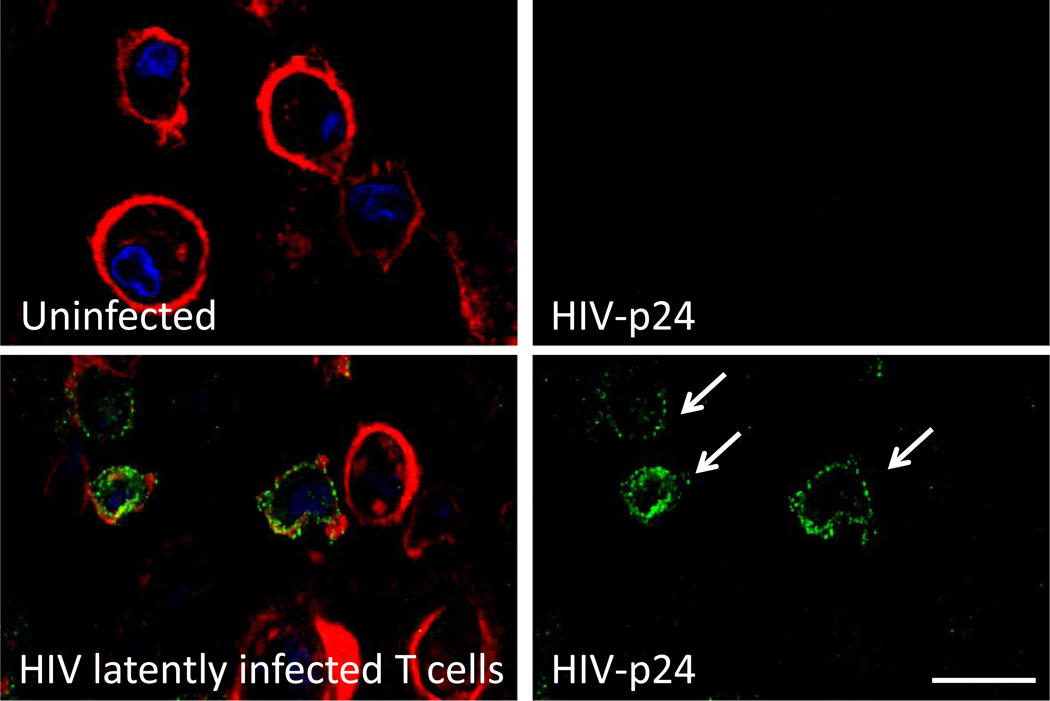
Example 1: As demonstrated in Figure 3, isolated CD4+ T lymphocytes from individuals with undetectable HIV replication as determined by ELISA (<15 pg/ml) and PCR (<20 copies/ml) were negative for HIV-p24 staining using regular immune staining (Figure 4, top panel). Using the staining and microscopy techniques described above, we identified the few latently HIV infected cells, without viral reactivation (Figure 3, lower panel).
Figure 3.
Detection of latent HIV in human CD4+ T lymphocytes using enhanced HIV-p24 staining. Merged images are presented on the right side of the figure. HIV-p24 staining (HIV-p24, green), actin (phalloidin conjugated to Texas red, red) and nuclei (DAPI, blue) detection were performed. Uninfected cells did not show any HIV-p24 staining. Conventional staining without amplification did not result in reliable staining. Enhanced staining using the protocols described above resulted in reliable and reproducible detection of HIV-p24. Bar: 30 µm
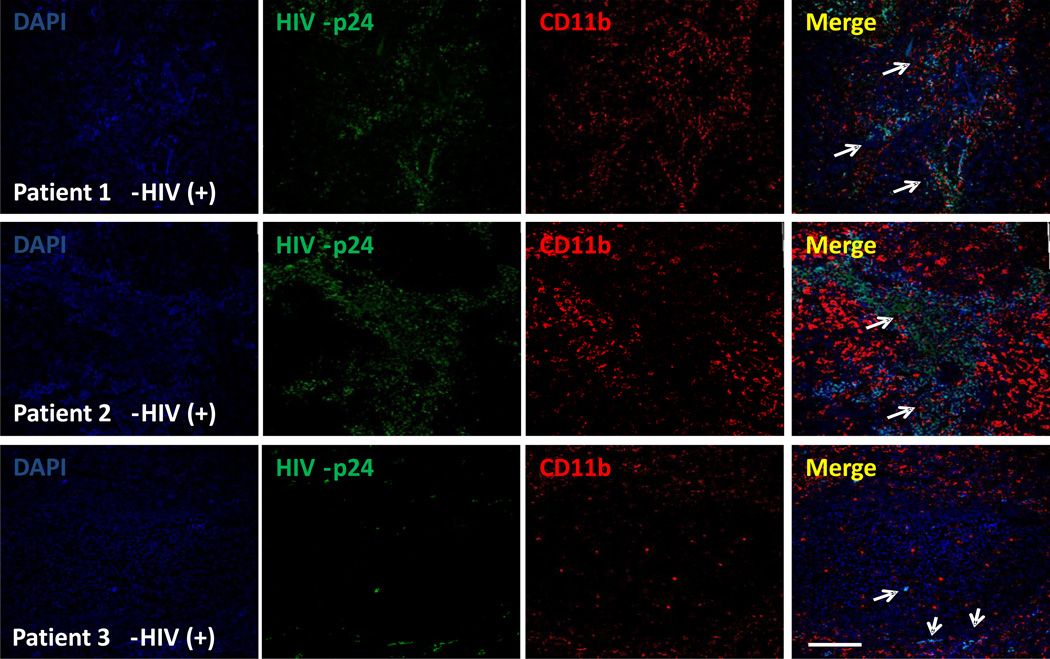
Figure 4.
CD4+ T lymphocytes and a population of macrophages present in human lymph nodes serve as viral reservoirs. Using thicker lymph node tissue sections (200 µm) obtained from HIV infected individuals with undetectable viral replication (for at least one year), tissue staining for Nuclei (DAPI, blue), CD4, Iba-1, or CD11b (red staining), and HIV-p24 viral protein was performed. Using the protocols described above we performed confocal microscopy and subsequent 3D reconstructions. No staining was observed using IgG1 or preimmune sera controls. Quantification of HIV infected cells was performed using the total number of cells (DAPI staining), versus the total number of HIV positive cells. Bar: 40 µm
Example 2: Human lymph nodes were obtained from HIV infected individuals with undetectable viral replication for at least 1 year. Using our signal amplification techniques, we were able to detect HIV proteins (Figure 4) in all cases analyzed (n=9). No signal was detected in uninfected tissues (data not shown). We identified viral reservoirs not only in CD4+ T lymphocytes (data not shown), but also in dendritic cells (CD11b positive cells, Figure 4) as well as in a small population of macrophages (Iba-1 positive cells, data not shown). The tissue distribution of these infected cells was donor dependent. Some donors have diffuse presence of HIV cells while others have well compartmentalized HIV infection.
Most of these techniques also can be combined with detection of HIV DNA and mRNA as described in Note 6. Thus, it is possible to detect HIV DNA or mRNA, viral proteins, and cellular markers at the same time in the same sample.
3.19 Summary of detection properties with different systems and conditions
HIV protein staining
We are able to detect several HIV proteins including HIV-p24, Nef, Vif, and integrase in one latently infected cell among 106 to 108 uninfected cells in blood smears and isolated PBMCs (pelleted preparations) from HIV infected individuals on ART with no detectable viral replication. In tissue sections obtained from HIV infected individuals with no detectable replication at the time of death, we are able to detect 1.6 ±1.2 % of T cells in lymph nodes. In brains obtained from individuals with minimal replication, we detected 5–8% of astrocytes and 3–6 % of microglia/macrophages infected with HIV.
Acknowledgments
This work was supported by the National Institute of Mental Health grants MH075679, MH090958 and MH102113 (to J.W.B.), and MH096625 (to E.A.E.). NIH Centers for AIDS Research (CFAR) Grant AI-051519. We would like to thank the Analytical Imaging Facility at PHRI and Nikon Instruments for microscopy support (http://www.phri.org/facilities/facil_imaging.asp).
Footnotes
The protocols described above are mostly dependent upon the quality of the starting tissue, cell separation, area of the tissue examined, and degree of viral replication. Most of the problems are due to:
Levels of expression and amplification: This is an essential consideration before you start the experiment. How much staining is expected? The amplification system described in Figure 2, method 4, is extremely sensitive; thus, it is not recommended to samples that are expected to have high expression of HIV proteins. In cases for which viral replication is detected by ELISA and PCR, we suggest the use of the technique described in Figure 2, method 2 or 3 instead of method 4. If your detected fluorescence is too strong, leave the stained samples at 4 °C for 1–2 weeks to allow reduction of the signal or repeat the experiment using the same samples using protocol 1 (antibody conjugated to biotin-streptavidin-fluorophore amplification, Figure 2, 3rd method).
Negative controls: Problems with negative control antibodies, endogenous biotin, and Fc receptor expression are also common, resulting in increased background staining. If the background is uniform, changes in gain or unmixing systems in the confocal microscope may be enough to detect specific staining. If the background is non-uniform, repeat the experiment, because uneven staining is not reliable, especially by confocal microscopy. Most of the uneven staining occurs in inflamed, activated tissues that were in formalin for long periods of time. Activated cells expressing high levels of Fc receptors that can bind nonspecifically to the Fc region of the immunoglobulin being used for detection (24).
3D reconstruction: Most of our techniques involve 3D reconstructions and thicker tissue sections. Conventional microscopes are not able to detect specific signals. Analysis by confocal microscopy allows the quantification of thick tissue sections, serial optimal sections, and 3D reconstructions to quantify accurately the total numbers of HIV positive cells. Thus, the use of specific programs to perform 3D reconstruction is required such as Image J from the NIH, or others such as NIS imaging (Nikon, Japan) or Metamorph (Molecular devices, CA) (12,25–28).
Amplification problems: The tyramine amplification method is based on the binding reaction of biotinylated tyramine to phenol derivatives of a protein by peroxidase. This reaction gives nonspecific signals; therefore, it is important to pretreat specimens with methanol containing 0.3% H2O2 to reduce endogenous peroxidase activity.
Purity of the solutions: Due to the high amplification, any cross contamination between samples by using contaminated PBS will generate nonspecific signals. Thus, use different containers for each slide.
Combination of protein and other HIV markers: The described techniques could be combined with detection of HIV DNA and mRNA, resulting in a multicomponent detection system of viral reservoirs. Detection of only viral proteins is not sufficient to demonstrate viral reservoirs, because HIV proteins can be released and taken up by phagocytosis or nonspecific uptake.
References
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nature medicine. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner JP, Tawadros R, Pomerantz RJ. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS. 1996;10:573–585. doi: 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Mahlknecht U, Deng C, Lu MC, Greenough TC, Sullivan JL, O'Brien WA, Herbein G. Resistance to apoptosis in HIV-infected CD4+ T lymphocytes is mediated by macrophages: role for Nef and immune activation in viral persistence. Journal of immunology. 2000;165:6437–6446. doi: 10.4049/jimmunol.165.11.6437. [DOI] [PubMed] [Google Scholar]
- 5.Coleman CM, Wu L. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology. 2009;6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durand CM, Blankson JN, Siliciano RF. Developing strategies for HIV-1 eradication. Trends Immunol. 2012;33:554–562. doi: 10.1016/j.it.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson S, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spina CA, et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rella CE, Ruel N, Eugenin EA. Development of imaging techniques to study the pathogenesis of biosafety level 2/3 infectious agents. Pathog Dis. 2014;72:167–173. doi: 10.1111/2049-632X.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subbian S, Eugenin E, Kaplan G. Detection of Mycobacterium tuberculosis in Latently Infected Lungs by Immunohistochemistry and Confocal Microscopy. J Med Microbiol. 2014;63:1432–1435. doi: 10.1099/jmm.0.081091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subbian S, Eugenin E, Kaplan G. Detection of Mycobacterium tuberculosis in latently infected lungs by immunohistochemistry and confocal microscopy. J Med Microbiol. 2014;63:1432–1435. doi: 10.1099/jmm.0.081091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A. 1989;86:2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrer T, Jassoy C, Harrer E, Johnson RP, Walker BD. Induction of HIV-1 replication in a chronically infected T-cell line by cytotoxic T lymphocytes. J Acquir Immune Defic Syndr. 1993;6:865–871. [PubMed] [Google Scholar]
- 15.Butera ST, Roberts BD, Leung K, Nabel GJ, Folks TM. Tumor necrosis factor receptor expression and signal transduction in HIV-1-infected cells. AIDS. 1993;7:911–918. doi: 10.1097/00002030-199307000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto M, Makino M, Kitajima I, Maruyama I, Baba M. HIV-1-infected myelomonocytic cells are resistant to Fas-mediated apoptosis: effect of tumor necrosis factor-alpha on their Fas expression and apoptosis. Med Microbiol Immunol. 1997;186:11–17. doi: 10.1007/s004300050040. [DOI] [PubMed] [Google Scholar]
- 17.Deyl Z, Horakova M, Vancikova O. Changes in pyridinoline content of elastin during ontogeny. Mech Ageing Dev. 1981;17:321–325. doi: 10.1016/0047-6374(81)90050-6. [DOI] [PubMed] [Google Scholar]
- 18.Deyl Z, Macek K, Adam M, Vancikova O. Studies on the chemical nature of elastin fluorescence. Biochim Biophys Acta. 1980;625:248–254. doi: 10.1016/0005-2795(80)90288-3. [DOI] [PubMed] [Google Scholar]
- 19.Orellana JA, Saez JC, Bennett MV, Berman JW, Morgello S, Eugenin EA. HIV increases the release of dickkopf-1 protein from human astrocytes by a Cx43 hemichannel-dependent mechanism. J Neurochem. 2014;128:752–763. doi: 10.1111/jnc.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazleton JE, Berman JW, Eugenin EA. Purinergic receptors are required for HIV-1 infection of primary human macrophages. J Immunol. 2012;188:4488–4495. doi: 10.4049/jimmunol.1102482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murooka TT, Mempel TR. Intravital microscopy in BLT-humanized mice to study cellular dynamics in HIV infection. J Infect Dis. 2013;208(Suppl 2):S137–S144. doi: 10.1093/infdis/jit447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paddock SW, Eliceiri KW. Laser scanning confocal microscopy: history, applications, and related optical sectioning techniques. Methods Mol Biol. 2014;1075:9–47. doi: 10.1007/978-1-60761-847-8_2. [DOI] [PubMed] [Google Scholar]
- 23.Shaner NC. Fluorescent proteins for quantitative microscopy: important properties and practical evaluation. Methods Cell Biol. 2014;123:95–111. doi: 10.1016/B978-0-12-420138-5.00006-9. [DOI] [PubMed] [Google Scholar]
- 24.Buchwalow I, Samoilova V, Boecker W, Tiemann M. Non-specific binding of antibodies in immunohistochemistry: fallacies and facts. Sci Rep. 2011;1:28. doi: 10.1038/srep00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amat F, Lemon W, Mossing DP, McDole K, Wan Y, Branson K, Myers EW, Keller PJ. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat Methods. 2014;11:951–958. doi: 10.1038/nmeth.3036. [DOI] [PubMed] [Google Scholar]
- 26.Ellefsen KL, Settle B, Parker I, Smith IF. An algorithm for automated detection, localization and measurement of local calcium signals from camera-based imaging. Cell Calcium. 2014;56:147–156. doi: 10.1016/j.ceca.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maska M, et al. A benchmark for comparison of cell tracking algorithms. Bioinformatics. 2014;30:1609–1617. doi: 10.1093/bioinformatics/btu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song W, Liu W, Niu X, Wang Q, Sun L, Liu M, Fan Y. Three-dimensional morphometric comparison of normal and apoptotic endothelial cells based on laser scanning confocal microscopy observation. Microsc Res Tech. 2013;76:1154–1162. doi: 10.1002/jemt.22279. [DOI] [PubMed] [Google Scholar]