Abstract
Ovarian cancer is a heterogeneous disease of low prevalence, but poor survival. Early diagnosis is critical for survival, but is often challenging because the symptoms of ovarian cancer are subtle and become apparent only during advanced stages of the disease. Therefore, the identification of robust biomarkers of early disease is a clinical priority. Metabolomic profiling is an emerging diagnostic tool enabling the detection of biomarkers reflecting alterations in tumor metabolism, a hallmark of cancer. In this study, we performed metabolomic profiling of serum and tumor tissue from 158 patients with high-grade serous ovarian cancer (HGSOC) and 100 control patients with benign or non-neoplastic lesions. We report metabolites of hydroxybutyric acid (HBA) as novel diagnostic and prognostic biomarkers associated with tumor burden and patient survival. The accumulation of HBA metabolites caused by HGSOC was also associated with reduced expression of succinic semialdehyde dehydrogenase (encoded by ALDH5A1), and with the presence of an epithelial-to-mesenchymal transition (EMT) gene signature, implying a role for these metabolic alterations in cancer cell migration and invasion. In conclusion, our findings represent the first comprehensive metabolomics analysis in HGSOC and propose a new set of metabolites as biomarkers of disease with diagnostic and prognostic capabilities.
Keywords: biomarker, metabolism, ketone body, hydroxybutyric acid, ovarian high-grade serous carcinoma
INTRODUCTION
Ovarian cancer is the eighth most frequent cause of cancer death among women, and associated with poor overall survival. According to the World Health Organization (WHO) statistics worldwide there were estimated to be 239,000 new cases and 152,000 deaths due to ovarian cancer in 2012 (1). High-grade serous carcinoma (HGSOC) is the most common type of ovarian epithelial carcinoma, accounting approximately 70% of the cases (2). Early diagnosis is critical for the survival of the patients, as for the stage I patients the 5-year survival rate is around 90%, whereas for the stage III and IV patients it is only around 20% (3). However, the diagnosis of ovarian cancer is challenging because symptoms often tend to mimic those of other diseases and appear only in advanced stages of cancer.
The main diagnostic methods for ovarian cancer at the moment include pelvic examination, CA-125 blood test and transvaginal ultrasound. CA-125 and HE4 proteins are the only two biomarkers US FDA approved for monitoring disease recurrence or progression, but not for screening purposes (4). CA-125 levels are raised in approximately 90% of patients with advanced epithelial ovarian cancer, but only in 50% of patients with stage I disease (3). Many studies have suggested CA-125 to be a prognostic factor for overall and progression-free survival in ovarian cancer, but also studies showing contradictory results exist (3). Thus, CA-125, the golden standard for ovarian cancer diagnostics, is relatively good in detecting patients with advanced disease, but its expression is elevated in benign conditions, such as endometriosis, and fluctuations associated with menstrual cycle and pregnancy (5). Therefore new biomarkers, either alone or in combination of the aforementioned proteins, are warranted for more specific and sensitive ovarian cancer diagnostics and prognostics.
Several lines indicate that cancer cells undergo profound metabolic reprogramming to sustain increased energy demand for growth and proliferation (6). This metabolic reprogramming leads to the accumulation of small molecule metabolites in the tumor tissue that can also be secreted in the blood stream, becoming potential biomarkers for cancer detection and disease progression. Of note, metabolic differences in the blood of ovarian cancer patients have already been described (7–9), but the clinical utility of these biomarkers has remained limited due to small study populations, lack of validation by other studies or orthogonal methodologies, as well as lack of clinicopathological data to elucidate the most relevant pathways for progression of the disease. To overcome these challenges, we performed global metabolomics profiling in a large and well-characterized cohort of HGSOC patients. This analysis revealed profound deregulation of hydroxybutyric acid (HBA) metabolism in the blood and tumor tissue of these patients, which can be used as diagnostic and prognostic biomarkers of the disease. We also found that these metabolic alterations were associated with the emergence of EMT, a feature of most invasive and aggressive tumor, suggesting a potential role of HBA metabolism in EMT. Finally, we validated the prognostic signature by analyzing independent, publicly available, gene expression data sets.
MATERIALS AND METHODS
Patients and samples
The data from a global metabolomic profiling were obtained from serum samples of 100 subjects without malignant disease (control group) as well as from 158 ovarian high-grade serous carcinoma patient serum samples. Clinicopathological characteristics of the study cohort are summarized in Supplementary Table S1. Before metabolomic analyses, the case and control group samples were combined into one set and randomized. In addition, the data from tumor tissues were obtained for 124 ovarian cancer patients, of which 112 had matching serum samples in the study.
Serum samples were collected from preoperative and pretreated primary ovarian cancer patients as well as from patients without ovarian cancer. All tissue and serum samples were collected at the Tumor Bank Ovarian Cancer (www.toc-network.de) at the Charité Medical University (Berlin, Germany) between 09/2000 and 02/2011. The Ethics Committee approved the use of the samples for the study. The patient’s informed consent was obtained prior sample collection and documentation of clinical and surgical data. A validated documentation system was used to record the surgical data (IMO-intraoperative mapping of Ovarian Cancer (10)). The study population without ovarian cancer consisted of a group of pelvic mass patients with benign tumors, endometriosis, uterus myomatosus, adnexitis and other conditions. Blood was collected using serum tubes containing clot activators (Vacutainer, BD, Medical-Pharmaceutical System, Franklin Lakes, NJ), and clotted for 30 min to 2 h at room temperature. Serum was separated by centrifugation at 1200g for 15 minutes, aliquoted and stored at −80°C until further analysis.
Tumor tissue samples were collected at the time of surgery and immediately frozen in liquid nitrogen within 15 minutes of the removal and then stored at −80 °C. All tissue samples underwent histopathological assessment to verify histological subtype and high tissue quality. Only specimens presenting at least 50% of tumor area were included in the metabolomic analyses. The majority of tissue samples had approximately 80% of tumor area.
Metabolomic analysis
Tumor samples were prepared by homogenizing 3-4 mg of tissue with Retsch homogenizer (3 min, 20 Hz). The samples were homogenized into 0.9% NaCl, and the volume was adjusted to get tumor tissue concentration of 0.05 mg/μl. 400 μl methanol and 5 μl standard mixture (valine-d8 (37.6 mg/l), heptadecanoic acid (186.5 mg/l), succinic acid-d4 (62.9 mg/l)) were added to 70 μl of the homogenate. For the serum samples 400 μl methanol and 10 μl standard mixture (valine-d8 (37.6 mg/l), heptadecanoic acid (186.5 mg/l), succinic acid-d4 (62.9 mg/l), glutamic acid-d5 (103.5mg/l)) was added to 30 μl of the sample. The samples were vortexed for 2 min. After 30 minutes at room temperature the samples were centrifuged for 5 min at 10000 rpm. 200 μl of the supernatant was moved to a gas chromatography (GC) vial and evaporated to dryness under nitrogen. The samples were derivatized with 25 μl methoxyamine (45°C, 60 min) and 25 μl N-methyltrimethylsilyltrifluoroacetamide (45°C, 60 min) and 50 μl of hexane with retention index compounds and injection standard (4,4′-dibromooctafluorobiphenyl) was added to samples.
For the analysis, a Leco Pegasus 4D GC×GC-TOFMS instrument (Leco Corp., St. Joseph, MI) equipped with a cryogenic modulator was used. The GC part of the instrument was an Agilent 6890 gas chromatograph (Agilent Technologies, Palo Alto, CA), equipped with split/splitless injector. The first-dimension chromatographic column was a 10-m Rxi-5MS capillary column with an internal diameter of 0.18 mm and a stationary-phase film thickness of 0.18 μm, and the second-dimension chromatographic column was a 1.5 m BPX-50 capillary column with an internal diameter of 100 μm and a film thickness of 0.1 μm. A methyl deactivated retention gap (1.5 m × 0.53 mm i.d.) was used in the front of the first column. High-purity helium was used as the carrier gas at a constant pressure mode (40 psig). A 4-s separation time was used in the second dimension. The MS spectra were measured at 45 – 700 atomic mass units with 100 spectra/sec. For the injection, a splitless injection (1.0 μl) at 240 °C was utilized. The temperature program was as follows: the first-dimension column oven ramp began at 50 °C with a 2 min hold after which the temperature was programmed to 240 °C at a rate of 7 °C/min and further to 300 °C at a rate of 25 °C/min and then held at this temperature for 3 min. The second-dimension column temperature was maintained 15 °C higher than the corresponding first-dimension column. The programming rate and hold times were the same for the two columns.
ChromaTOF vendor software (LECO) was used for within-sample data processing, and in-house made software Guineu (11) was used for alignment, normalization and peak matching across samples. The peaks were first filtered based on number of detected peaks in the total profile of all sample runs. The normalization for uncalibrated metabolites was performed by correction for internal standard C17:0. 26 of the metabolites were checked manually in each serum sample and 29 metabolites in each tumor tissue sample for correct integration and identification. Other mass spectra from the GC×GC-TOFMS analysis were searched against NIST05 mass spectral library. After data processing, 1107 metabolite peaks were obtained from serum samples. To remove potential batch effects due to sample preparation or running order, or because of differences in sample storage time at −80 °C, correlation of each metabolite to these variables was inspected by using maximal information coefficient (12) and visual inspection, and filtered out in case correlation was observed. Finally, all the peaks showing zero values in more than 75% of the samples were filtered out. After these filtering steps, 497 metabolites were left for the final analyses. Group information of the samples was not used to guide the data processing or filtering steps at any point. For the tumor tissue samples, only the metabolites aligning to the final serum data were included in the analyses. Supplementary Table S2 provides lower limit of quantification (LLOQ) values for quantified metabolites as well as for each metabolite the number of detected peaks across the whole serum data set.
In some cases GCxGC-TOF-MS data contains several peaks with same identification. This may be due to analytical reasons or e.g. stereoisomeric structures. In cases where the GCxGC-TOF-MS analysis included a reference standard compound, always the peak matching the standard compound was selected as the major peak, otherwise the largest peak was selected to be the main result. The results for all the peaks are shown in Supplementary Table S2.
Nutritional analyses
The following nutritional and lifestyle variables were collected from the patients and investigated for their association with metabolite levels: reported change in food intake, reported feeling of sickness, smoking, BMI, weight change in 3 months, weight change 10 days after operation, daily calorie recall (also divided into protein, fat and carbohydrates) as well as fat percentage (Supplementary Table S4).
Statistical analyses
All statistical analyses were performed using R, version 3.1.2. For comparing mean values between two groups, unpaired t-tests and fold changes were calculated after log2 transformation of the data. In case there were zero values in the data, the data was imputed with a value corresponding to the half of the minimum value of the corresponding molecule across all the samples. Principal component analysis was performed for the full log2-transformed data applying centering and scaling (zero mean, unit variance), and the analysis was blinded to the class information. Analysis of Variance (ANOVA) was used for multiple group comparisons. Correlation analyses were performed either by Pearson or Spearman method, as indicated in each case. Multiple hypothesis correction was evaluated by reporting false discovery rate q-values in addition to p-values. The predictive models for the ovarian cancer diagnosis from serum with metabolites and CA-125 were based on binary logistic regression models. For estimating the performance of the predictive models, random subset cross-validation was applied as follows: the dataset was 1000 times split randomly into training set (comprising 2/3 of the samples), and the constructed model was tested in the validation set (comprising 1/3 of the samples); the ROCR library (13) was used in the analyses.
Survival analysis
Association of the metabolites and genes to survival was investigated by Kaplan-Meier plots with a median split and logrank test (package survival version 2.37-7). In addition, Cox proportional hazards regression test was performed for each metabolite with and without accounting for the residual tumor mass after surgery / age / FIGO stage information. In these models, the metabolite levels were normalized to follow standardized normal distribution.
Survival analysis based on gene expression and copy number was performed for genes of those selected Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that showed significant diagnostic or prognostic metabolomics alterations: fatty acid import and beta oxidation, omega oxidation, ketone body production, pentose phosphate pathway reactions related to 3-erythritol accumulation, leucine degradation reactions related to 3-hydroxyisovaleric acid accumulation, peroxisomal biogenesis factors related to Zellweger syndrome as well as ALDH5A1 gene encoding for SSADH enzyme. TCGA mRNA expression and corresponding clinical information for 489 HGSOC patient samples were obtained from TCGA data portal (14). Copy number alterations for 316 tumors were obtained from cBioPortal (15). Another independent data set and online tool was used to analyze the association of ALDH5A1 to survival in ovarian cancer patients (survival = overall survival, histology = serous carcinoma, follow up threshold = 10 years) (16). Supplementary material for all the TCGA survival analyses are shown in Supplementary Methods and all the R code used to perform these analyses are stored at https://github.com/InesdeSantiago/survivalTCGAOC.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) (17) was used to identify pathways that are correlated with the expression of the ALDH5A1 gene. KEGG pathways from MSigDB curated collection C2 (17) and gene expression signatures of the epithelial-to-mesenchymal transition (EMT) were included in the analysis. EMT signatures were collected, i.e. sets of genes that are known to be upregulated in EMT (EMT+) and downregulated in EMT (EMT−), as described previously (18).
RESULTS
HBAs as serum biomarkers of HGSOC
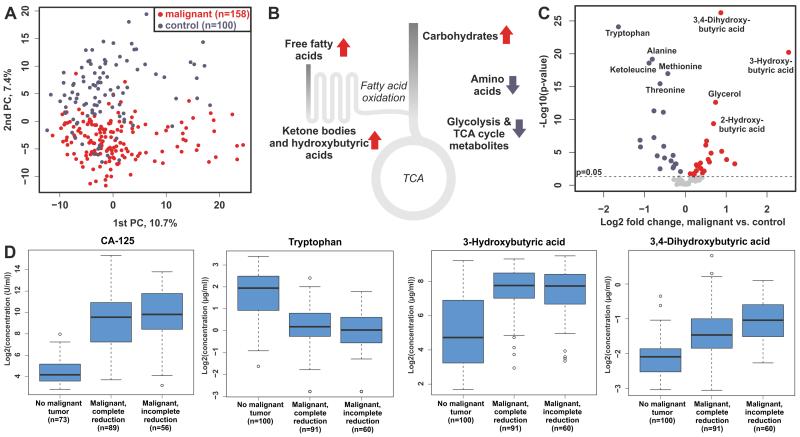
In order to identify metabolic biomarkers of ovarian cancer, we performed global metabolomic profiling of serum samples of 158 ovarian cancer patients and 100 subjects affected by benign ovarian neoplasia and non-neoplastic diseases (Supplementary Table S1). Unsupervised principal component analysis (PCA) revealed that ovarian cancer patients could be separated from the control group (Fig. 1A). Among the metabolites that most significantly differed between the two groups, several amino acids, including tryptophan, alanine, and methionine, were the most depleted (Fig. 1B, C, Supplementary Table S2-3). Interestingly, the depletion of tryptophan in cancer patients is in line with two recent studies (7,19). However, the most significant difference between the cancer patients and control group was observed for 3,4-dihydroxybutyric acid and 3-hydroxybutyric acid, which were found at higher levels in the cancer patients (Fig. 1C, D, Supplementary Table S3). In addition, other HBAs and ketone bodies, sugars, and free fatty acids were accumulated, whilst metabolites related to glycolysis and TCA cycle were found in lower concentrations in the cancer patients. The accumulation of 3-hydroxybutyric acid most likely reflects upregulation of fatty acid oxidation that has been described in ovarian cancer tumors (20) and metastases (21). These results indicate that ovarian cancer exhibits a distinct metabolic signature (Fig. 1A-D) and elevate HBAs appear as key biomarkers for ovarian cancer.
Figure 1. Malignant ovarian tumors cause dramatic metabolic differences in serum.
(A) PCA plot showing the 1st and 2nd principal components and separation of the patients with and without malignant tumors. (B) Summary of the global metabolic differences observed in ovarian cancer patient serum samples. (C) Volcano plot illustrating the most significantly altered metabolites in the serum of patients with malignant tumors. For clarity, only main peaks are shown, and the unidentified compounds and sugars/sugar alcohols are omitted from the figure. (D) Concentration of the most significantly altered metabolites and CA-125 in patients without malignant tumors as well as in patients with malignant tumors that were either completely or incompletely removed during the surgery.
To investigate the diagnostic power of these metabolites, predictive models were constructed and cross-validated, i.e. the study population was split randomly 1000 times into discovery and validation sets, having 2/3 and 1/3 of the samples, respectively. Based on the results, metabolites belonging to the HBA metabolites class showed the highest cross-validated mean AUC value (Supplementary Fig. S1). With the two most significantly altered metabolites, i.e. 3,4-dihydroxybutyric and 3-hydroxybutyric acids, the mean AUC value reached 0.91, and was in the same order as for the clinically used CA-125 (0.96). When the information of these metabolites was combined with CA-125, the mean cross-validated AUC value was 0.98, thus approaching perfect separation of the groups (Supplementary Fig. S2). As a comparison, with 3 random metabolites selected 1000 times the mean AUC was 0.66.
In contrast to CA-125 and 3-hydroxybutyric acid, 3,4-dihydroxybutyric acid, 2,4-dihydroxybutyric acid and 4-hydroxyphenyllactic acid, were statistically significantly associated with the success of tumor reduction during surgery (Table 1), a reflection of tumor spread in the patients. These poorly characterized metabolites showed progressive elevation from patients with benign tumors to patients with incomplete tumor reduction (Fig. 1D), making them better predictors of tumor burden than the clinically used tumor marker CA-125.
Table 1. Diagnostic and prognostic metabolites for ovarian cancer.
Statistical results and associated metabolic deficiencies/pathways for those metabolites that were significantly altered in ovarian cancer patients and also showed significant result in the log-rank test for overall survival. Results for the CA-125 biomarker are shown as comparison. Statistically significant (p<0.05) results are shown in bold.
| Malignant vs.control group | Tumor reduction | Overall survival | Deficiency, pathway or condition involved | |||||
|---|---|---|---|---|---|---|---|---|
| KM | Cox regression | |||||||
| Metabolite name | p-value | log2 fold | p-value | log2 fold | p-value | p-value | hazard ratio | |
| 3,4-Dihydroxybutyric acid | 5.6E-27 | 0.87 | 0.002 | 0.33 | 0.023 | 0.013 | 1.26 | SSADH deficiency (25) |
| Adipic acid | 1.9E-07 | 0.52 | 0.162 | 0.18 | 0.011 | 0.003 | 1.34 | SSADH deficiency (31), fatty acid omega oxidation, ketosis (40), Zellweger syndromea (37) |
| Erythritol | 2.8E-05 | 0.33 | 6.0E-05 | 0.47 | 0.028 | 0.010 | 1.29 | Deficiencies related to pentose phosphate pathway (41,42) |
| 3-Hydroxyisovaleric acid (β-hydroxy β - methylbutyric acid) | 1.3E-04 | 1.01 | 0.562 | 0.21 | 0.018 | 0.003 | 1.28 | Leucine catabolism (22), ketosis (23) |
| 2,4-Dihydroxybutyric acid | 0.001 | 0.28 | 0.004 | 0.35 | 0.012 | 0.430 | 1.07 | SSADH deficiency (24,25) |
| 4-Hydroxyphenyllactic acid | 0.016 | 0.22 | 0.003 | 0.31 | 4.0E-04 | 4.6E-04 | 1.34 | Zellweger syndromea (38), tyrosinemia (43) |
|
| ||||||||
| CA-125 | 7.2E-52 | 5.01 | 0.112 | 0.68 | 0.513 | 0.729 | 1.04 | - |
Zellweger syndrome is a peroxisomal disorder
Correlation of serum biomarkers with tumor metabolic signatures
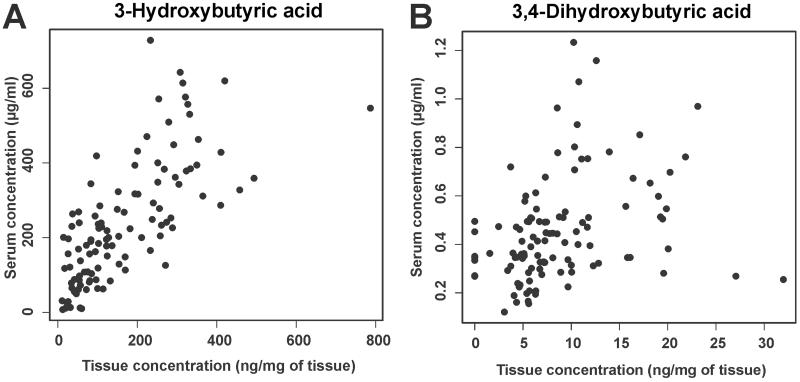
Serum metabolic alterations may be affected by nutritional and health status of the patients. Therefore, we investigated whether physiological and nutritional variables affect the abundance of these diagnostic metabolites. To this aim, nutritional data of 27 patients of the study cohort were obtained and correlated to metabolite levels. BMI was determined at the time of diagnosis and the weight change in 3 months was recorded. 3-hydroxybutyric or 3,4-dihydroxybutyric acids did not show statistically significant association to BMI or weight change, supporting their validity as tumor biomarkers (Supplementary Table S4). These results also suggest that these HBAs may originate from the tumor and do not depend on metabolic processes in other parts of the body. To investigate this hypothesis, tumor tissue samples were analyzed from 112 patients with matching serum sample data. The highest correlations were observed for 3- and 2-hydroxybutyric acids (Fig. 2 and Supplementary Table S5). In addition, concentration of 3,4-dihydroxybutyric acid (Fig. 2) and several molecules of the amino acid and sugar classes showed significant correlations between serum and tissue (Supplementary Table S5). It was also investigated whether tumor samples obtained from different locations showed different metabolic profiles. In general, tumor samples from ovaries, intestine or peritoneum did not show major differences, as only a few metabolites showed statistically significant result (Supplementary Figure S3). In summary, these findings revealed a linear correlation of diagnostically relevant biomarkers between serum and tumor tissue.
Figure 2. Correlation of (A) 3-hydroxybutyric and (B) 3,4-dihydroxybutyric acids in the tumor tissue and serum samples.
Correlation of metabolic signature with cancer patients’ survival
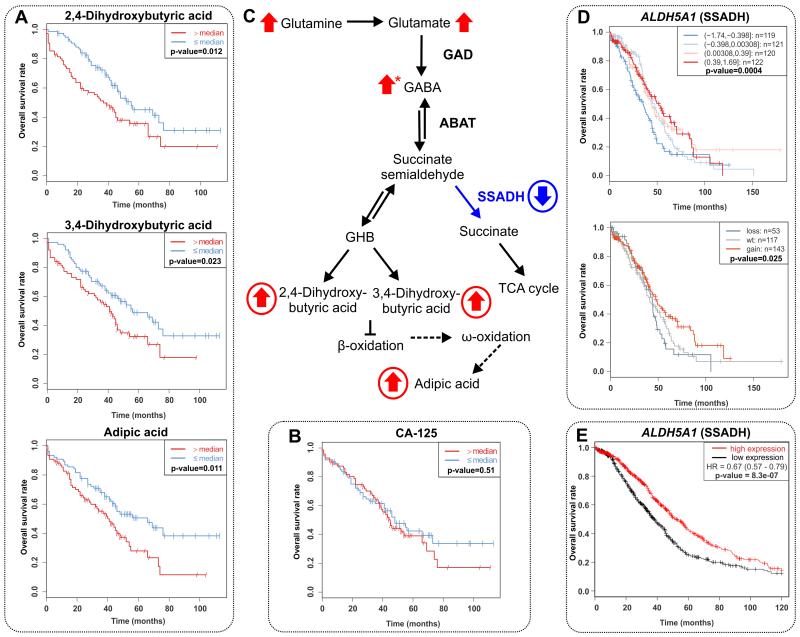
We then investigated whether the observed metabolic features of ovarian cancer could predict cancer progression and survival of the patients. The association with overall survival was investigated by Kaplan-Meier plots with median split together with log-rank test as well as Cox proportional hazards regression models. For comparison, the analyses were performed also for CA-125. Table 1 shows the results for those metabolites with statistically significant result both in the malignant vs. control group comparison as well as in log-rank test for overall survival. In contrast to CA-125, which did not show association with survival, the concentrations of specific metabolites before surgery were found to be strong predictors of the overall survival of the patients and able to predict the survival independently of the success of tumor reduction in the surgery (Fig. 3A, B, Table 1). The most significant result was found for 4-hydroxyphenyllactic acid, which was the only metabolite showing statistically significant result also for progress-free survival (Supplementary Fig. S4). Interestingly, higher levels of 4-hydroxyphenyllactic acid in the serum of ovarian cancer patients was reported recently, although in the previous study the association with survival was not investigated (7). Another significant metabolite was 3-hydroxyisovaleric acid, which showed p-value 0.003 in the cox regression analysis, even after adjusting for tumor reduction and FIGO stage (Supplementary Table S6 and Supplementary Fig. S4). 3-hydroxyisovaleric acid is related to leucine catabolism (22), and especially the reported elevation due to ketogenesis (23) may explain its role as prognostic marker for ovarian carcinomas.
Figure 3. Low activity of SSADH enzyme is reflected in the metabolite concentrations and poor overall survival.
Higher concentrations of (A) 2,4-dihydroxybutyric, 3,4-dihydroxybutyric and adipic acids in serum were all associated with poor overall survival of the patients. In contrast, (B) CA-125 did not show association with the survival. (C) Illustration of the pathway and metabolites that are altered due to loss of SSADH activity. Arrow means that the metabolite was upregulated in patients with malignant tumors, and circle that high concentration was associated with worse overall survival. The data for GABA was obtained from (28). The accumulation of adipic acid has been associated with SSADH deficiency, but the mechanism leading to the higher concentration of this metabolite has not been elucidated, and therefore the arrows in this figure refer only to a hypothetical mechanism. (D) Low expression and loss in copy number of ALDH5A1 in tumors were associated with poor survival in TCGA data and. (E) The association of low expression of ALDH5A1 in tumors to poor survival was replicated also in publicly available survival analysis tool of ovarian serous carcinoma patients (n=1137).
Finally, we found that higher concentrations of 3,4-dihydroxybutyric, 2,4-dihydroxybutyric, and adipic acids were associated with worse overall survival of the patients (Fig. 3A, Table 1), which led us to investigate these poorly characterized metabolites in depth. 3,4-dihydroxybutyric and adipic acids showed significant results even when tumor reduction information was incorporated into the model. Addition of patient age into the model raised the p-values for the metabolites, and this observation could be explained by the fact that older patients were associated with poorer survival, together with higher levels of 3,4-dihydroxybutyric acid (Supplementary Fig. S5), potentially linked to extremely low energy intake (Supplementary Fig. S5). Thus, our data indicate an association between 3,4-dihydroxybutyric acid and overall health and nutritional status. However, 3,4-dihydroxybutyric acid showed a statistically significant result and 2,4-dihydroxybutyric a trend with overall survival, even though their concentrations were measured directly from tumor tissue from fewer number of patients (Supplementary Fig. S6). This confirmed that higher concentrations of these metabolites, which are associated with worse survival, originate from altered metabolism of the tumor and not from other physiological processes.
Association of HBA metabolism with low activity of succinic semialdehyde dehydrogenase (ALDH5A1)
We then wanted to investigate the possible mechanisms of accumulation of these dihydroxybutyric and adipic acids in ovarian cancer. Intriguingly, these metabolites accumulate in patients affected by succinic semialdehyde dehydrogenase (SSADH) deficiency (OMIM #271980) (24,25). This syndrome is caused by mutations in the ALDH5A1 gene, which encodes for succinic semialdehyde dehydrogenase, and causes accumulation of gamma-aminobutyric (GABA) and gamma-hydroxybutyric (GHB) acids in the body, leading to various neurological problems in the patients (26). As illustrated in Fig. 3C, 3,4-dihydroxybutyric and 2,4-dihydroxybutyric acids are derived from gamma-hydroxybutyric acid (GHB) (27). Although not identified in our dataset, the dysregulation of SSADH pathway is supported by elevated concentration of GABA in the urine of ovarian cancer patients (28). Furthermore, as mentioned above, only three amino acids were found to be in higher abundance in patients with malignant tumors, two of these (glutamine and glutamate) are directly involved in this pathway and elevation of glycine has been reported in patients with SSADH deficiency (26). Interestingly, the elevation of glutamine, glutamate and GABA have also been reported when comparing the invasive and borderline ovarian tumors (29). Thus, the metabolomics analysis of HGSOC patients revealed a prognostic signature of metabolites related to lowered activity of the SSADH enzyme.
To further validate the metabolomics findings with gene expression data, we analyzed The Cancer Genome Atlas (TCGA) data and selected genes from those pathways that showed significant diagnostic or prognostic metabolomic alterations. Intriguingly, the most significant findings were obtained for the ALDH5A1 gene, as among the selected genes only for ALDH5A1 both low expression and loss in copy number were associated with worse overall survival of the patients (Fig. 3D, Supplementary Table S7). These data (14) show that only three genes (ALDH5A1, PCK2 and GMPR) significantly correlated with patients’ survival across all cohorts (Supplementary Table S8). Thus, the association of decreased expression of ALDH5A1 to worse overall survival was replicated in four different cohorts. These results were also confirmed in another manually curated gene expression database (16) (Fig. 3E). In conclusion, our results indicate that the prognostic metabolomic signature observed in our study is linked to the downregulation of ALDH5A1 and a subsequent lowered activity of the SSADH enzyme.
Association of loss of SSADH to epithelial-to-mesenchymal transition
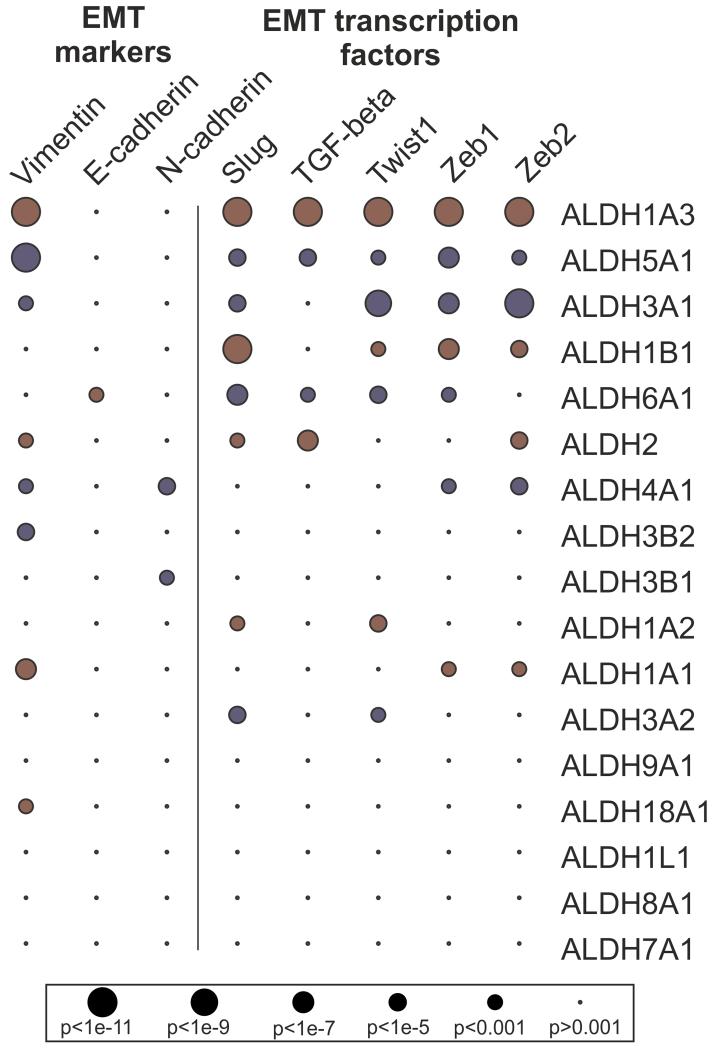
We then investigated whether the loss of SSADH is associated with molecular signatures that could explain its link to poor prognosis in ovarian cancer. To this aim, correlation and gene set enrichment analyses were performed. This analysis revealed that the most significant pathways, negatively correlated with ALDH5A1, were related to cellular adhesion and EMT (Supplementary Table S9). Indeed, low expression of ALDH5A1 was associated with high expression of vimentin and EMT-related transcription factors (Fig. 4). As expected, significant correlation to EMT signature was also found for some other ALDH enzymes (Fig. 4, Supplementary Table S10), including ALDH1A3, which is highly expressed in mesenchymal stem cells (30).
Figure 4. Positive (red) or negative (blue) correlation of ALDH genes to EMT markers or transcription factors.
DISCUSSION
In this study we have performed metabolomics investigation of HGSOC, the most common type of ovarian cancer. Our analysis revealed that HGSOC exhibit a distinct metabolic signature characterized by the accumulation of ketone bodies and HBAs in serum and tumor tissue. Importantly, we have demonstrated that these metabolites can be used for both diagnostic and prognostic purposes, offering valuable clinical opportunities.
The source of the observed accumulation in HBAs is likely to emerge from the deregulation of multiple metabolic pathways. For instance, the increase of 3-hydroxybutyric is due to upregulated fatty acid oxidation, a metabolic feature previously observed in ovarian cancer tumors (20) and metastases (21). The dysregulation of fatty acid metabolism is supported by elevated concentrations of acylcarnitines that have been reported in ovarian cancer patient serum (7), and regions of nuclear magnetic resonance (NMR) spectra including 3-hydroxybutyric acid that have shown signal to separate ovarian cancer patients from controls (8). Previous studies have also reported high AUC values when using principal components or regions of NMR spectra, incorporating also 3-hydroxybutyric acid as predictors of malignancy (8,9). In the present study, we focused our analysis on individual metabolites, which are needed for follow-up studies or when used in clinical practice, confirming that the signal obtained from a small panel of metabolites is sufficient to obtain high cross-validated AUC values. Nevertheless, since the cross-validation may lead to over-optimistic results due to potential selection bias, comparison of novel biomarkers, especially 3,4-dihydroxybutyric acid, against CA-125 has to be validated in independent data sets. Interestingly, the accumulation of 3,4-dihydroxybutyric and 2,4-dihydroxybutyric acid, observed in the metabolic disorder SSADH deficiency (24,25,31), suggested a link between SSADH activity and HGSOC. Indeed, the low mRNA expression of ALDH5A1, the gene encoding for SSADH, was found to be significant prognostic marker in all the investigated data sets. Importantly, our analysis also revealed an unexpected correlation between the loss of the ALDH5A1 and the presence of EMT, a molecular signature associated with invasion and metastasis, another cancer hallmark together with cancer energy metabolism (32). Although we have not provided the molecular link between deregulated metabolism and EMT, recent studies have revealed that decreased activity of the TCA cycle induce EMT. For instance, decreased succinate dehydrogenase activity in ovarian cancer (33), pheochromocytomas and paragangliomas (34); mutations in isocitrate dehydrogenase (35) and loss of citrate synthase in cervical cancer (36) have been linked to EMT. Therefore it is tempting to speculate that lower levels of succinate due to SSADH deficiency may lead to TCA cycle dysfunction and EMT induction. However, it should be noted that the results of the present study are only correlative and more work is required to establish a mechanistic link between SSADH deficiency and EMT.
Among the significant metabolites that were not associated with HBAs or lipid metabolism was 4-hydroxyphenyllactic acid, a product of phenylalanine degradation. Interestingly, a lowered level of amino acids in the cancer patient serum samples was observed, supporting a deregulation of amino acid catabolism in HGSOC. Of note, 4-hydroxyphenyllactic and adipic acids have been reported to elevate in Zellweger syndrome (37,38), which is a peroxisomal disorder caused by mutations in genes participating in peroxisomal biogenesis. Although these genes did not show significant association to survival in the TCGA data set, peroxisomal dysfunction in HGSOC patients cannot be excluded.
Most of the patients in our study population were affected by late-stage ovarian cancer and we had only limited number of patients with early-stage disease. Although CA-125, the golden standard biomarker for ovarian cancer, showed high specificity and sensitivity, as expected, the utility of HBAs as disease biomarkers is underlined by their prognostic power. Furthermore, some of these metabolites can be found accumulated in blood and acetone, which is usually elevated along other ketone bodies, can also be detected in breath (39) suggesting that these metabolites can be detected by minimally invasive techniques. Combining ketone body measurements with CA-125 determination could be used as a powerful diagnostic tool for HGSOC. However, it is worth noting that the level of ketone bodies are affected by co-morbidities, as for instance diabetes and starvation elevate their levels. In the present study, the samples were not taken under fasting conditions, which may cause variability in the nutritional status and affect the results, but importantly, there was no systematic bias in the sample collection between the patients with malignant and benign disease. Nevertheless, as future development of our work is to test the validity of these metabolic biomarkers in a clinical setting, especially in patients with early-stage disease, which still lack appropriate diagnostic tools. Also, the value of these biomarkers in tracking response to therapy should be assessed. Appropriate longitudinal studies are required to fully understand the potential of these markers in clinical use.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Marko Sysi-Aho for statistical advice, Niina Lietzén, Ismo Mattila, Sandra Castillo and Tuulikki Seppänen-Laakso for their contribution to metabolomics analyses, and all the former VTT’s Quantitative Biology and Bioinformatics group members for fruitful discussions.
Funding: This work was supported by the Academy of Finland post-doctoral fellow grants: MH decision 138201 and PG 265966. TA was supported by Academy of Finland (grants 272437, 269862, 279163, 292611) and Cancer Society of Finland. EIA is participant in the Charité Clinical Scientist Program funded by the Charité Universitätsmedizin Berlin and the Berlin Institute of Health. F. Markowetz and I. de Santiago would like to acknowledge the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. Parts of this work were funded by CRUK core grant C14303/A17197 and A19274.
ABBREVIATIONS
- 3,4-DHBA
3,4-dihydroxybutyric acid
- 3-HBA
3-hydroxybutyric acid
- ALDH5A1
aldehyde dehydrogenase 5 family, member A1
- CA-125
cancer antigen 125
- GCxGC-TOFMS
two-dimensional gas-chromatography mass spectrometry
- EMT
epithelial-to-mesenchymal-transition
- HBA
hydroxybutyric acid
- SSADH
succinate semialdehyde dehydrogenase
- TCGA
The Cancer Genome Atlas
Footnotes
Conflict of interest statement: VTT Technical Research Centre of Finland (MH) has filed a patent application, which is partly based on the results of this manuscript. The authors declare no other conflicts of interest.
REFERENCES
- 1.Stewart BW, Wild CP. World Cancer Report 2014. World Heal. Organ.; Geneva: 2014. [Google Scholar]
- 2.Prat J. New insights into ovarian cancer pathology. Ann Oncol. 2012;23 doi: 10.1093/annonc/mds300. [DOI] [PubMed] [Google Scholar]
- 3.Gupta D, Lis CG. Role of CA125 in predicting ovarian cancer survival - a review of the epidemiological literature. J Ovarian Res. 2009;2:13. doi: 10.1186/1757-2215-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen L, Cardenas-Goicoechea SJ, Gordon P, Curtin C, Momeni M, Chuang L, et al. Biomarkers for early detection of ovarian cancer. Womens Health (Lond Engl) 2013;9:171–85. doi: 10.2217/whe.13.2. quiz 186–7. [DOI] [PubMed] [Google Scholar]
- 5.Sarojini S, Tamir A, Lim H, Li S, Zhang S, Goy A, et al. Early detection biomarkers for ovarian cancer. J Oncol. 2012;2012:709049. doi: 10.1155/2012/709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sciacovelli M, Gaude E, Hilvo M, Frezza C. The metabolic alterations of cancer cells. Methods Enzymol. 2014;542:1–23. doi: 10.1016/B978-0-12-416618-9.00001-7. Academic Press Inc. [DOI] [PubMed] [Google Scholar]
- 7.Ke C, Hou Y, Zhang H, Fan L, Ge T, Guo B, et al. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int J Cancer. 2015;136:516–26. doi: 10.1002/ijc.29010. [DOI] [PubMed] [Google Scholar]
- 8.Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, et al. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer. 2005;113:782–8. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- 9.Garcia E, Andrews C, Hua J, Kim HL, Sukumaran DK, Szyperski T, et al. Diagnosis of early stage ovarian cancer by 1H NMR metabonomics of serum explored by use of a microflow NMR probe. J Proteome Res. 2011;10:1765–71. doi: 10.1021/pr101050d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sehouli J, Könsgen D, Mustea A, Oskay-Ozcelik G, Katsares I, Weidemann H, et al. [“IMO”--intraoperative mapping of ovarian cancer] Zentralbl Gynakol. 2003;125:129–35. doi: 10.1055/s-2003-41864. [DOI] [PubMed] [Google Scholar]
- 11.Castillo S, Mattila I, Miettinen J, Oresic M, Hyotylainen T. Data Analysis Tool for Comprehensive Two-Dimensional Gas Chromatography/Time-of-Flight Mass Spectrometry. Anal Chem. 2011;83:3058–67. doi: 10.1021/ac103308x. [DOI] [PubMed] [Google Scholar]
- 12.Reshef DN, Reshef Y a., Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, et al. Detecting Novel Associations in Large Data Sets. Science (80- ) 2011;334:1518–24. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21:3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 14.Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genomewide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-tomesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Wu X, Yin M, Fan L, Zhang H, Zhao F, et al. Clin Chim Acta. Vol. 413. Elsevier B.V.; 2012. Discrimination between malignant and benign ovarian tumors by plasma metabolomic profiling using ultra performance liquid chromatography/mass spectrometry; pp. 861–8. [DOI] [PubMed] [Google Scholar]
- 20.Fong MY, McDunn J, Kakar SS. Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One. 2011;6:e19963. doi: 10.1371/journal.pone.0019963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieman KM, Kenny H a, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobs C, Sweetman L, Nyhan WL, Packman S. Stable isotope dilution analysis of 3-hydroxyisovaleric acid in amniotic fluid: contribution to the prenatal diagnosis of inherited disorders of leucine catabolism. J Inherit Metab Dis. 1984;7:15–20. doi: 10.1007/BF01805614. [DOI] [PubMed] [Google Scholar]
- 23.Yu WM, Kuhara T, Inoue Y, Matsumoto I, Iwasaki R, Morimoto S. Increased urinary excretion of beta-hydroxyisovaleric acid in ketotic and non-ketotic type II diabetes mellitus. Clin Chim Acta. 1990;188:161–8. doi: 10.1016/0009-8981(90)90160-t. [DOI] [PubMed] [Google Scholar]
- 24.Brown GK, Cromby CH, Manning NJ, Pollitt RJ. Urinary organic acids in succinic semialdehyde dehydrogenase deficiency: evidence of alpha-oxidation of 4-hydroxybutyric acid, interaction of succinic semialdehyde with pyruvate dehydrogenase and possible secondary inhibition of mitochondrial beta-oxidati. J Inherit Metab Dis. 1987;10:367–75. doi: 10.1007/BF01799979. [DOI] [PubMed] [Google Scholar]
- 25.Shinka T, Inoue Y, Ohse M, Ito A, Ohfu M, Hirose S, et al. Rapid and sensitive detection of urinary 4-hydroxybutyric acid and its related compounds by gas chromatography–mass spectrometry in a patient with succinic semialdehyde dehydrogenase deficiency. J Chromatogr B. 2002;776:57–63. doi: 10.1016/s1570-0232(02)00126-5. [DOI] [PubMed] [Google Scholar]
- 26.Pearl PL, Dorsey AM, Barrios ES, Gibson KM. Succinic Semialdehyde Dehydrogenase Deficiency. University of Washington; Seattle: 2013. [PubMed] [Google Scholar]
- 27.Zhang GF, Sadhukhan S, Ibarra RA, Lauden SM, Chuang CY, Sushailo S, et al. Metabolism of gamma-hydroxybutyrate in perfused rat livers. Biochem J. 2012;444:333–41. doi: 10.1042/BJ20112046. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson-Guthrie CS, Guthrie GD, Sutton GP, Baenziger JC. Urine GABA levels in ovarian cancer patients: Elevated GABA in malignancy. Cancer Lett. 2001;162:27–30. doi: 10.1016/s0304-3835(00)00620-0. [DOI] [PubMed] [Google Scholar]
- 29.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 30.Mao P, Joshi K, Li J, Kim S-H, Li P, Santana-Santos L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–9. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishiguro Y, Kajita M, Aoshima T, Watanabe K, Kimura M, Yamaguchi S. The first case of 4-hydroxybutyric aciduria in Japan. Brain Dev. 2001;23:128–30. doi: 10.1016/s0387-7604(01)00181-4. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Aspuria PP, Lunt SY, Väremo L, Vergnes L, Gozo M, Beach J a, et al. Succinate dehydrogenase inhibition leads to epithelial-mesenchymal transition and reprogrammed carbon metabolism. Cancer Metab. 2014;2:1–15. doi: 10.1186/2049-3002-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loriot C, Burnichon N, Gadessaud N, Vescovo L, Amar L, Libé R, et al. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. J Clin Endocrinol Metab. 2012;97:E954–62. doi: 10.1210/jc.2011-3437. [DOI] [PubMed] [Google Scholar]
- 35.Grassian AR, Lin F, Barrett R, Liu Y, Jiang W, Korpal M, et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT) J Biol Chem. 2012;287:42180–94. doi: 10.1074/jbc.M112.417832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C-C, Cheng T-L, Tsai W-H, Tsai H-J, Hu K-H, Chang H-C, et al. Loss of the respiratory enzyme citrate synthase directly links the Warburg effect to tumor malignancy. Sci Rep. 2012;2:785. doi: 10.1038/srep00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Björkhem I, Blomstrand S, Hågå P, Kase BF, Palonek E, Pedersen JI, et al. Urinary excretion of dicarboxylic acids from patients with the Zellweger syndrome. Importance of peroxisomes in beta-oxidation of dicarboxylic acids. Biochim Biophys Acta. 1984;795:15–9. doi: 10.1016/0005-2760(84)90099-7. [DOI] [PubMed] [Google Scholar]
- 38.Mayatepek E, Seppel CK, Hoffmann GF. Increased urinary excretion of dicarboxylic acids and 4-hydroxyphenyllactic acid in patients with Zellweger syndrome. Eur J Pediatr. 1995;154:755–6. doi: 10.1007/BF02276727. [DOI] [PubMed] [Google Scholar]
- 39.Musa-Veloso K, Likhodii SS, Cunnane SC. Breath acetone is a reliable indicator of ketosis in adults consuming ketogenic meals. Am J Clin Nutr. 2002;76:65–70. doi: 10.1093/ajcn/76.1.65. [DOI] [PubMed] [Google Scholar]
- 40.Mortensen PB. Urinary excretion of C4--C10-dicarboxylic acids and antiketogenic properties of adipic acid in ketogenic-stimulated rats due to diabetes, long-chain and short-chain monocarboxylic acids. Biochim Biophys Acta. 1981;664:335–48. doi: 10.1016/0005-2760(81)90056-4. [DOI] [PubMed] [Google Scholar]
- 41.Kardon T, Stroobant V, Veiga-da-Cunha M, Schaftingen E Van. Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase deficiency. FEBS Lett. 2008;582:3330–4. doi: 10.1016/j.febslet.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Valayannopoulos V, Verhoeven NM, Mention K, Salomons GS, Sommelet D, Gonzales M, et al. Transaldolase deficiency: a new cause of hydrops fetalis and neonatal multi-organ disease. J Pediatr. 2006;149:713–7. doi: 10.1016/j.jpeds.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Inborn Errors of Metabolism: From Neonatal Screening to Metabolic Pathways. New York: Oxford University Press; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.