Abstract
In this work we address the question whether the enhanced stability of thermophilic proteins has a direct connection with internal hydration. Our model systems are two homologues G-domains of different stability: the mesophilic G domain of the Elongation-Factor thermal unstable protein from E. coli and the hyperthermophilic G domain of the EF-α protein from S. solfataricus. Using molecular dynamics simulation at the microsecond time-scale we show that both proteins host water molecules in internal cavities and that these molecules exchange with the external solution in the nanosecond time scale. The hydration free energy of these sites evaluated via extensive calculations is found to be favourable for both systems, with the hyperthermophilic protein offering a slightly more favourable environment to host water molecules. We estimate that under ambient conditions, the free energy gain due to internal hydration is about 1.3 kcal/mol in favor of the hyperthermophilic variant. However, we also find that at the high working temperature of the hyperthermophile, the cavities are rather dehydrated, meaning that at extreme conditions other molecular factors secure the stability of the protein. Interestingly, we detect a clear correlation between the hydration of internal cavities and the protein conformational landscape. The emerging picture is that internal hydration is an effective observable to probe the conformational landscape of proteins. In the specific context of our investigation, the analysis confirms that the hyperthermophilic G-domain is characterized by multiple states and it has a more flexible structure than its mesophilic homologue.
Keywords: Thermophilic Proteins, Thermal Stability, Protein Hydration, Free-Energy, Molecular-Dynamics
Introduction
Proteins from thermophilic or hyperthermophilic organisms are stable and functional at very high temperatures, up to the boiling point of water. Several molecular factors ensure this extreme stability and their combination results in different thermodynamic strategies for thermal adaptation1-4. With respect to their mesophilic homologues, (hyper)thermophiles are generally enriched in charged amino-acids and cross-linked by a larger number of hydrogen bonds (HB) and salt-bridges that contribute to stabilize the protein fold5-7. They have shorter loops that protect the fold from thermal excitation by reducing the extension of weak spots at the protein surface2-4,8,9. Moreover, an optimized and extended hydrophobic packing in the protein core or at domain interfaces enhances the intramolecular cohesive forces10.
These factors depend, directly or indirectly, on the coupling between the protein and surrounding water. For example it has been demonstrated that the distribution of charged amino-acids at the protein surface and in contact with water can be effectively optimized to increase thermal stability11,12. Moreover, the stabilizing contribution of salt-bridges partially buried in the protein matrix depends on the local polarity5,13,14 while the presence of internal water molecules could alleviate the associated desolvation penalty15,16. In this regard, it was proposed that (hyper)thermophiles could benefit from dense ion-pairing because of a higher internal dielectric constant17. Finally, the presence of water molecules inside internal cavities or superficial pockets also improves the molecular packing and extends the HB connectivity with potential effects on local kinetic stability18. In this regard it is worth noting that many thermophiles are also resistant to high pressure2, in fact internal hydration could reduce the volume of internal voids which are thought to trigger pressure unfolding19 and at the same time make intramolecular HB patterns resistant to temperature and pressure stresses20.
While protein hydration has been the object of intense and debated research21-25 so far only sporadic investigations tried to link the interactions between protein and water to thermal stability, or to extreme adaptation in general26-32. In a previous computational study of two homologous G-domains of different stability, it was observed that the thermal stability of the percolating water HB network enveloping the protein surface correlates to the different stabilities of the two molecules27. Due to the different polarities and morphologies, a strong coupling between the protein surface and water was invoked for the hyperthermophilic domain33,34.
The impact of the surface composition on the organization of the hydration layer was recently highlighted for two orthologous malate dehydrogenase (MDH) proteins, one being halophilic and the other thermophilic and non-halophilic, whose X-ray structures were resolved at high resolution31. At cryogenic temperature, the thermophilic MDH was found to be covered by extended networks of water clusters, mostly in pentagonal configurations, and showed a remarkably high level of hydration. These findings are in line with those reported for the hyperthermophilic b-glicosidase protein35 and question the generality of the conclusion from a previous investigation according to which thermophilic proteins are less hydrated26.
On the other hand, as shown in a more recent work on the homologous G-domains29, the dynamics of individual water molecules in the hydration layer was found to be unaffected by the chemical composition of the proteins, since water relaxation mainly depends on an excluded volume effect36. This result, later verified by other examples25, agrees with the results from NMR experiments32 reporting a similar retardation factor for the hydration water of both an halophilic and a non-halophilic peptide of markedly different amino-acid composition.
Hydration dynamics provides only kinetic insights on the protein/water coupling, however an understanding of water’s contribution to protein stability in extreme conditions needs to be quantified from a thermodynamic point of view. For instance, an enhanced structuring of water at the surface of the protein fold could explain the systematic lower heat capacity of unfolding, ΔCp, measured for (hyper)thermophiles with respect to mesophiles26,37.
Another interesting question concerns the contribution from buried water to the overall stability gap between homologues. A seminal attempt to shed light on such issue was done by Yin, Hummer and Rasaiah30 who used computational means to estimate the stability of water clusters in the nonpolar cavities of the tetrabrachion protein from the hyperthermophilic Staphylothermus marinus, whose optimal growth temperature is 365 K. This coiled coil assembly, made up of four helices, is characterized by a sequence of cavities located along the protein axis and, according to the crystallographic structure resolved at low temperature (T = 100 K), they are filled by water. Yin and coworkers found that at ambient temperature the largest cavity is preferentially hydrated with clusters comprising 4 to 9 molecules, being a necessary threshold to create a convenient environment for the water HB connectivity22,38. By increasing temperature, the cavity dehydrates and the drying process marks the onset of denaturation. For a very similar system, the SNARE protein, the presence of internal water is instead destabilizing39. In the very stable neutral variant (melting temperature Tm = 365 K) the four helices are tightly bound and no buried water is visible in the structure even if long residence molecules were detected by molecular dynamics simulations40. On the contrary, in the less stable protein from S. cerevisiae (Tm = 328 K) three internal cavities are filled by water and when these cavities are removed by punctual mutations, the stability increases as monitored by a shift of Tm by about 6 K39.
Estimating the increase in stability arising from internal water molecules, by performing single point mutations that promote/inhibit water filled cavities is, however, ambiguous. In fact, the different intramolecular interactions, like the HB propensity and the conformational reorganization, caused by the change of the amino-acid also contribute to the overall thermodynamical shift. In some cases, creating a host environment for water is stabilizing, as reported for BPTI41, subtilisin42, lysozyme43 and lipases18, but for other proteins the reverse applies, as reported for iso-1-cytochrome44, protease45 and lysozyme T4 mutants46.
Computer simulations, based on modern all-atom force fields, provide a unique tool to explore the specific role of water inside cavities and pockets22,47-52. Free energy calculations and MD simulations successfully predict the hydration state of pockets of different chemical natures (polar vs nonpolar)22,50,51,53, help to understand the relation between internal hydration and function as in the case of charge separation processes54-57, ligand binding58-60, allostery61,62 and can be used to estimate the contribution to stability50,63.
In this work we continue our investigation of the molecular mechanisms at the origin of extreme thermal stability by comparing the behavior of a representative pair of mesophilic/hyperthermophilic homologues: the G-domains of the Elongation Factor thermo unstable (EF-Tu) protein from the mesophilic Escherichia coli bacterium and EF-1α protein from the hyperthermophilic Sulfolobus solfataricus archaeon. The experimental melting temperatures of the two domains differ by about 40 K64,65 and are qualitatively well reproduced in silico. By using extensive MD simulations at high temperature (T = 360 K) we previously probed the early steps of unfolding of the mesophilic domain. Conversely, in the same timescale and temperature range, the hyperthermophilic protein was found to remain stable9. Interestingly, for the folded states at ambient conditions, we noticed an extended hydration of internal cavities within the proteins. Buried water molecules were suggested to play a major role in regulating stability29. Following this idea, we present here a precise study of both proteins’ internal hydration, and determine the contributions to their different stabilities arising from water molecules buried within the two domains.
Methods
System and MD simulations
The G-domain of the mesophilic EF-Tu (E. coli), PDB code 1EFC66) and hyperthermophilic EF-1α (Sulfolobus solfataricus, PDB code 1SKQ67) proteins were simulated in water solution and at ambient temperature for a time of 0.6 μs for each protein. The mesophilic domain (, 196 a.a.) was solvated with 7440 and the hyperthermophilic domain (, 226 a.a.) with 10673 water molecules. Counter-ions were added to neutralize the systems. In order to explore the behavior at high temperature, multiple simulations were carried out at T=360 K, see9. In particular, for the mesophilic protein, we collected 10 independent runs of length between 400 ns and 1.5 μs, each probing the early steps of the unfolding. For the hyperthermophilic domain, two independent simulations of 1 μs and 0.7 μs were used to probe its stability at high temperature.
To perform extensive simulations, we employed the NAMD parallel software68. The two proteins were modeled using the chamm2269 force field and water was represented by the charmm-TIP3P model. The system preparation and simulation protocol are detailed in our previous work9,27,29.
Hydration dynamics
The exchange dynamics of water molecules between the protein hydration shell and the bulk solution was analyzed by computing the survival probability function Nw(t). To this aim we first defined the hydration shell by detecting water oxygen atoms within 4.5 Å of any heavy atom of the protein. The survival probability Nw(t)70,71 counts how many water molecules, initially located within the hydration shell, remain in the shell for the subsequent time span t. This is formally defined as:
| (1) |
where the indicator function Pj(tn, t) takes the value 1 if the j-th water is found continously in the hydration shell between times tn and tn + t, or zero otherwise. Nt is the number of simulation time-frames of duration t. By computing Nw(t) we also measured the distribution of the continuous residence times τr of water in the hydration shell. Since we are interested in long time behavior, the analysis was performed on reduced trajectories with a time resolution of 20 ps.
Free energy calculations
Free energy differences were computed to estimate the water contribution to protein stability, measured on a limited number of molecules that hydrate the interior of the proteins. These water molecules were extracted from the ensemble of water molecules that were found within the hydration shell with a residence time τr > 5 ns. The frequency distributions of the contact of these molecules with individual amino acids of the proteins allowed to detect their most probable locations. Two particularly hydrated cavities were found in each protein. For a short stretch of the trajectory (100 ns), water molecules inside these pockets were ranked depending on their cumulative residence time, and among those having the larger total residence times, we selected a small subset for our calculations. For each selected molecule we extracted three representative configurations separated in time by at least 5 ns one from another. In order to characterize the localization and binding dynamics of the water molecules, Figure S1 of the Supplemental Information (SI) reports the time evolution of distances between water oxygens and a reference atom in each pocket.
For each site, a local hydration free energy, or equivalently, a local excess chemical potential for water was calculated. Starting from each representative configuration, an equilibration step was first conducted in the NPT ensemble. During the equilibration, all protein atoms and the water molecule of interest were restrained around their initial positions by a harmonic potential.
Free energy calculations were carried out following the framework used by Olano and Rick50 to study hydration free energy of internal cavities in BPTI and barnase, which we briefly describe below. The excess free energy of hydration is obtained from a virtual thermodynamic cycle and therefore results from two independent components, the free energy required to remove a water molecule from the bulk solution (ΔGwat) and the free energy required to add the water molecule to the protein cavity (ΔGprot). Both were estimated via the free energy perturbation (FEP) method72.
At first a water molecule in the bulk liquid is decoupled from the rest of the system:
| (2) |
In the second step, the non-interacting water (H2Oni), once localized inside the protein’s empty cavity (EC), is coupled to the rest of the system. This step is then further decomposed into a localization process and a coupling process:
| (3) |
| (4) |
where refers to a non-interacting water molecule which is restrained inside the protein cavity. Localization inside the cavity is enforced by a harmonic potential added to the Hamiltonian of the system. This potential confines the motion of the water molecule in its reference site inside the cavity of interest. Since we were interested in calculating the hydration free energy for many sites inside the cavities, we designed an ad hoc strategy to implement the restraint with respect to these positions.
For this purpose, for each internal water we identified the carbon atoms of the protein that, in the equilibrated configuration, lie within 5 Å of the oxygen atom of the water molecule. Among them, all possible combinations of five carbons were considered and the distance d0 between their center of mass and the oxygen of the water molecule was measured. Finally the combination of the five carbons that produced the shortest distance (usually less than 0.1 Å) was selected. The center of mass of the carbons was then defined as the reference site and used to implement the harmonic restraint, with d(t) the time-dependent distance between the water oxygen and the carbons’ center of mass (kharm = 10 kcal/(mol Å2)). The value of the constant was chosen such that it reproduces the fluctuations of the oxygen around the reference site in the unbiased trajectory. This procedure avoids the definition of a static reference point in space and thus the need of fitting the protein structure to a reference configuration during the dynamics50.
For the non-interacting molecule the work of localizing due to Uharm is associated to a free energy contribution:
| (5) |
where kB is the Boltzmann’s constant, T the temperature, and ρ the bulk density of water.
The local binding free energy of a water molecule in a given site was calculated by adding all the components:
| (6) |
In our implementation, the free energy ΔGinter is given by two terms. The first one measures the coupling between the water molecule and the environment in the presence of the harmonic restraint and is calculated using the FEP method as implemented in NAMD, while the second one removes the contribution from the harmonic restraint in the interacting system (see below). A coupling parameter λ taking values in the [0, 1] interval was used to gradually switch off first the electrostatic and then the VdW interactions of the water molecule in the cavity; by this annihilation process we accessed the quantity −ΔGinter. The FEP procedure consisted of 11 windows along the coupling parameter λ. For each value of λ, the system was equilibrated for 500 ps, then statistics were accumulated for 2 ns. The simulations were performed in the NPT ensemble, and by using the same protocol as in our previous work9. In a similar way, we estimated ΔGwat by simulating a system of pure water of 512 molecules. We obtained ΔGwat = 6.2 ± 0.1 kcal/mol at T = 300 K, and ΔGwat = 5.6 ± 0.1 kcal/mol at T = 360 K. While in ref.50 the harmonic restraint is progressively removed along the coupling process so as to disappear completely for a fully interacting water molecule, in our implementation this term was removed only in a separate step and its contribution to ΔGinter was estimated via an independent thermodynamic integration. In this procedure the harmonic constant kharm was progressively scaled to zero (11 steps), and the free energy contribution of the restraint was evaluated by computing ∫ dk⟨∂Uharm/∂kharm⟩k along the path72. The ensemble average ⟨…⟩k was estimated for a given value of the scaled constant kharm = k.
We also point out that during the annihilation process, when all the interactions between the water molecule and the rest of the system are scaled down, other molecules may overlap with the annihilated one, thus occupying the reference site. In order to avoid artifacts arising from such overlaps, we added a harmonic repulsive wall between the tagged water molecule and those nearby. We verified that without this repulsive wall the overlaps occur rarely and only in the last windows of the FEP decoupling procedure; the effect on the estimated free energy is generally comparable to the statistical error. The harmonic restraint as well as the repulsive wall were implemented using the Collective Variables Module available in NAMD73.
Clustering
In order to correlate the protein conformational fluctuations and the change of the internal hydration we have performed a cluster analysis of the trajectories sampled by the two systems at ambient temperature9. For the clustering procedure we used as a collective variable the fraction of native contacts Q defined below.
The number of native contacts for a given Cα site is the number of Cα atoms located within a cut-off distance of 8 Å in the crystallographic structure. Thus, the fraction of native contacts, referring to the whole molecule, is defined as
| (7) |
where NCα is the number of Cα atoms, having native Cα contacts in the reference state and li(t) of them appearing also at time t .
The clustering was done using the well-established leader algorithm74 with the following procedure. Each snapshot was represented by a vector of length equal to the number of Cα atoms (NCα), whose i–th component of the vector is the quantity . Then, the distance between two snapshots was defined as
| (8) |
The output of the clustering was mapped onto a network that was drawn with the use of a forced-based algorithm of the Gephi software75.
Results
The exchange kinetics of internal water molecules
As a preliminary investigation we detected the water molecules that penetrate the internal cavities of the two proteins. In many crystallographic structures the positions of structural water molecules are resolved together with the protein. In the structure of the whole EF-Tu66 and EF-1α67 proteins, a few molecules are visible in the interior of each G-domain. However, these structures were resolved in the presence of substrates which possibly make inaccessible alternative sites. Moreover, according to several computational studies49,51 when the protein is thermalized at ambient conditions locations alternative to the crystallographically determined sites are also sampled.
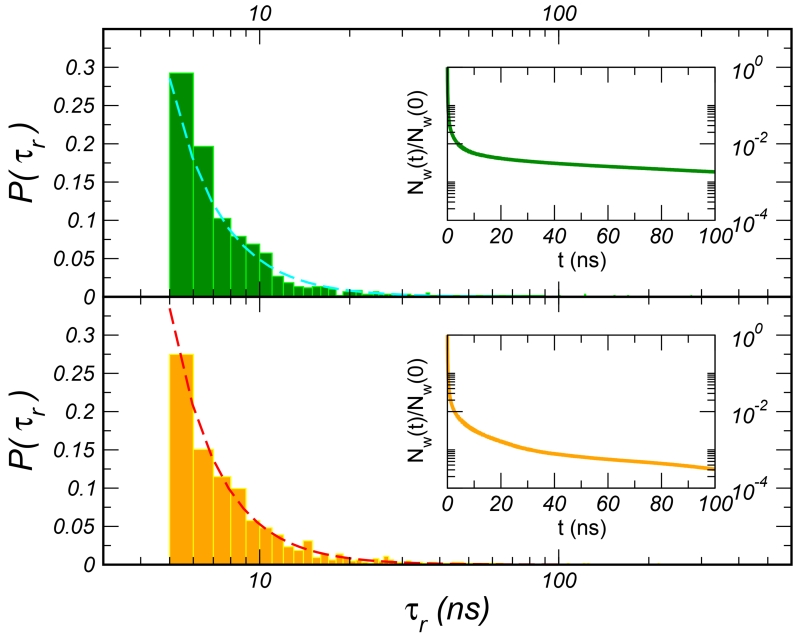
Exchange of these molecules with the external bulk solution occurs, as probed by both NMR23,76 and simulations49,77,78, on the nanosecond, or even longer timescale. In this work, we used this lower bound on the exchange time to fully map the solvent accessibility of the proteins’ interior. As already probed for other globular proteins49,77,78, for these two G-domains, the probability that a water molecule persists in the hydration shell as a function of time, Nw(t), extends with a long tail in the nanoseconds range, as shown in the insets of 1. When performing an exponential fit of this long tail (t> 5 ns) we recover average characteristic times of ⟨τslow⟩ 41 ns and 16 ns for and , respectively. The dynamics concerns about 1% of the water molecules in the shell, the so-called structural water, mainly buried in internal cavities or superficial clefts. The distributions of the residence times τr for this ensemble of molecules is reported in the main plot of 1. The distributions, as commonly observed for other proteins77, decay according to a power law, t−α, with α ~ 2.6. This value is close to what is reported for cytochrome c (2.5)77 and to the assumed distribution of reorientational times to interpret NMR experiments79. In the simulation of the domain, one water molecule gets trapped in the protein matrix for about 80% of the simulation time, ~500 ns, causing the longest value of τslow for this system. In the longest residence time is somewhat shorter, ~250 ns.
Internal cavities sampled by water
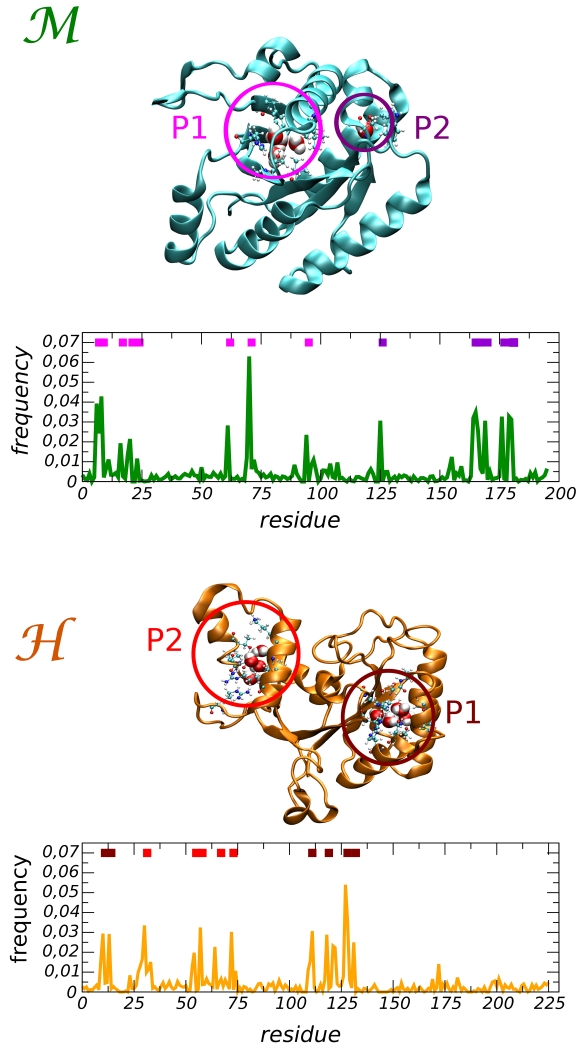
In order to map the internal cavities of the proteins we restricted our attention to those water molecules that have a residence time τr longer than 5 ns. The frequency distribution of their positions along the protein sequence is reported in 2. Two main pockets are found in each system, and their locations are indicated by the colored bars at the top of the x-axis. In the mesophilic domain the largest pocket, marked by the magenta bars, is located in the protein core. There a cluster of molecules is confined by a hydrophobic wall and a polar surface that offers several anchoring sites for hydrogen bonds. The pocket is accessible via a gateway near the P loop, a key region in the GTP binding site. Around this opening two buried water molecules are visible in the X-ray structure, see SI Table S1. The second pocket (violet bars) hosts a single isolated molecule. In our simulation, a single molecule penetrates into this pocket after about 100 ns, and gets trapped for the remainder of the simulation time. The water is caged by three hydrogen bonds formed with the residues S166 (backbone CO and NH groups) and W177 (backbone CO group). Interestingly, the observed binding matches the location of a structural water visible in the X-ray structure (SI Table S1).
In the hyperthermophilic protein, both pockets are filled with clusters of molecules. The first pocket (maroon bars) is found in proximity of the P loop. A portion of the pocket’s surface offers several HB pinning sites, mainly backbone groups, while the other part has a hydrophobic character. In the crystallographic structure three molecules are resolved in this pocket, see Supplemental Information Table S1. The second pocket (red bars) is created by a structural specific motif of the protein: the two small helices α1 [E32-L45] and α2 [E48-E63]. In our previous investigation we have shown that these structures confer stability to this region of the protein and prevent the local unfolding at high temperature that instead occurs in the domain9. The walls of this cavity are cross linked by ionic interactions, i.e. the highly stable ion-pairs R65-D31 and R58-E62, and these charged amino-acids create a highly polar environment. The fluctuations of these links are mediated by the presence of water that can form molecular bridges between the ionic groups (SI Table S3). We stress that in our simulations the ionic groups were all considered charged, however it is widely accepted that it is quite difficult to determine the protonation state of such buried charged groups, that in principle could also be influenced by the presence of water.
Local hydration free energies
We now discuss the contribution to protein stability stemming from these hydrated cavities. From the ensemble of long residence water molecules, we singled out a restricted set of molecules representative of the cavity hydration and with the longest residence times (τr > 15 ns). For practical reasons, this filtering was done considering a short stretch of the trajectory (100 ns). For each one of these molecules three different configurations, A, B, and C, separated from each other by at least 5 ns, were extracted from the trajectory and used to initiate the free energy calculations. This large ensemble allows to estimate the variability induced by the presence of multiple states of the pockets’ hydration, including i) changes in the internal location of the water molecule, ii) changes of the shape of the hosting cavity, and iii) long range effects due to the global fluctuations of the protein. The values of the hydration free energy, , and of the associated statistical error, are reported in Supplementary Tables S4 and S5 for all the pockets and conformers of the and domains, respectively.
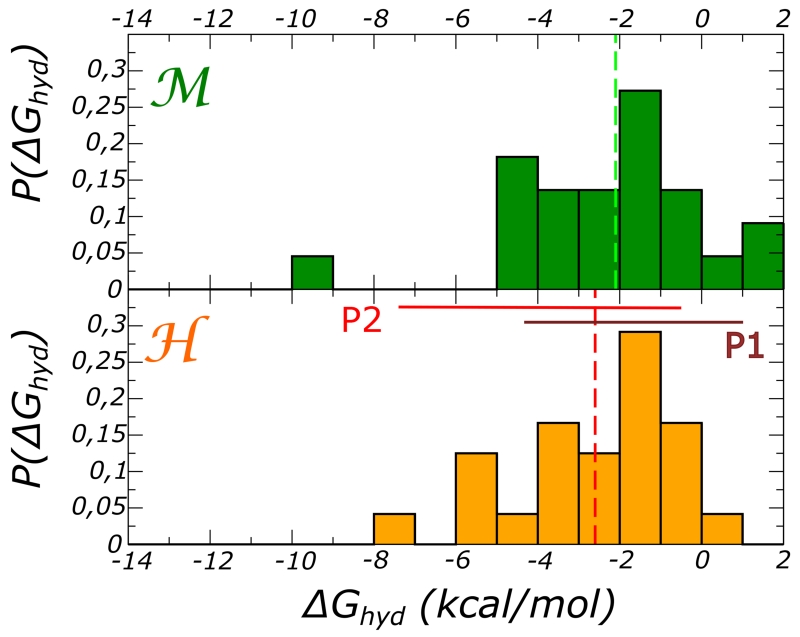
First we note that the hydration free energy is favorable for almost all the selected internal locations. However, we also remark the large variability of the , with a single molecule experiencing different favorable interactions during its confinement in the pocket. This variability can be as high as 4 kcal/mol. In both proteins, some locations provide large contributions to protein stability, for instance −9.0 ± 0.3 () and −7.0 ± 0.3 kcal/mol (). In both these cases, we observe high HB connectivity and strong packing (see data in SI Tables S2 and S3). When focusing on the average value associated to each pocket, we obtain ⟨ΔGi⟩P1 = −1.6 ± 0.3 and ⟨ΔGi⟩P2 = −5.5 ± 0.4 kcal/mol for the domain and ⟨ΔGi⟩P1 = −1.5 ± 0.3 and ⟨ΔGi⟩P2 = −3.6 ± 0.5 kcal/mol for the domain. The values are close to previous estimates of the insertion free energy for water into the cavities of other proteins as the bacteriorhodopsin48, BPTI50, and kinase A80.
The distribution of the free energies are plotted in 3. The vertical lines indicate the averages, ⟨ΔGi⟩ = −2.1 ± 0.3 kcal/mol and ⟨ΔGi⟩ = −2.6 ± 0.4 kcal/mol for and , respectively. For the domain the distribution is shaped by the contributions from water hydrating the larger pocket (P2), on the contrary for the protein, the distribution reflects the different contribution from the two pockets.
The calculated is the free energy contribution to the protein’s stability from a single buried molecule localized in a specific site i and calculated for different states k of the local environment, including the presence of surrounding water molecules having different residence times. According to the analysis presented in48,53, our values correspond to the insertion free energy of a molecule in the presence of n − 1 others. The ensemble of conformers Ai, Bi, Ci helps us to sample several initial conditions of the hydrated cavities, i.e. different values of n − 1 (see SI Tables S2 and S3).
Global free energy of internal hydration
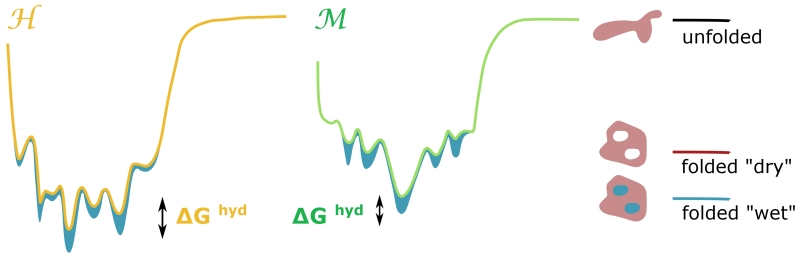
As discussed earlier, both proteins host several long residence water molecules at the same time. Therefore, in order to evaluate the total stabilization due to internal water molecules, we need to estimate their cumulative contribution, as represented schematically in 4. Hence, we determine a global free energy of internal hydration for both proteins based on our local estimates of water binding free energy.
For a given starting configuration k, the calculations described above provide local hydration free energies for each of 8 identified binding sites. Stability of an internal water molecule depends on many degrees of freedom, belonging to the protein as well as neighboring water. Direct calculation of well-converged free energy estimates could therefore be prohibitively long. We choose to obtain each binding free energy by combining data from three independent, local simulations (starting from different configurations). Each simulation is considered to sample one representative region of phase space, hence, their results are combined as if merging samples within the exponential average of FEP:
| (9) |
We assume additivity, meaning that the global free energy of internal hydration is estimated as the sum of local free energies weighted by the occupancy ni of each site:
| (10) |
In turn, the mean occupancy of each site is obtained from the binding free energies as:
| (11) |
C is a factor common to all sites in one protein, and is fitted so that the total predicted occupancy matches the average number of bound water molecules measured in long simulations, in other words:
| (12) |
We find the optimal value of C to be 0.6 for both the mesophile and thermophile.
The term nw can be calculated by scanning the trajectories and counting how many long-residence molecules are present at the same time in the interior of the proteins. Considering the molecules with residence time τr > 15 ns we obtain ⟨nw⟩ = 4.7 for both proteins and for a less strict condition (τr > 5 ns) ⟨nw⟩ = 9.8 and ⟨nw⟩ = 10.4 for the and proteins, respectively.
Within this framework and counting ⟨nw⟩ with the stricter condition (τr > 15 ns), we find ΔGhyd = −15.7 ± 0.5 kcal/mol for the mesophile, and −17.0 ± 0.6 kcal/mol for the hyperthermophile, meaning that the relative stabilization of the protein with respect to by internal hydration is ΔΔGhyd = 1.3 ± 0.5 kcal/mol. By releasing condition (12) on the total occupancy, the relative stabilisation of the protein with respect to is slightly larger, ΔΔGhyd = 2.5 ± 0.9 kcal/mol. The same numerical results are obtained with an alternative formulation of the problem that we report in SI. The obtained value for the relative stability ΔΔGhyd is a small but statistically meaningful number, and its possible effect on protein thermal stability is discussed below.
Unfortunately, for our EF-Tu and −1α G-domains, no thermodynamic values are available for the unfolding free energy ΔGunf(T). The magnitude of the stability gap between homologues, i.e. ΔΔGunf(T) at ambient conditions, can vary substantially depending on the pair. In some cases this gap can be as high as 20 kcal/mol but for many pairs its just on the order of a few kcal/mol81,82. In order to quantify the possible effect of the stability gain due to the internal hydration (ΔΔGhyd) we have considered two model systems. The first one is represented by a pair of homologues close to our proteins, namely the EF-1α from rabbit (mesophile) and from the thermophilic bacterium T. thermophilus83. The second is a pair of mesophilic/hyperthermophilic proteins belonging to CheY family. For these pairs the available thermodynamic data83,84 allow to estimate the unfolding free energy as a function of temperature and hence construct the stability curve, ΔGunf(T). For each protein we upshifted the stability curve by ΔΔGhyd = 1.3 kcal/mol (see above) and as expected the melting temperature of the proteins increases, ΔTm is about 6 K. Depending on the value of the specific heat of unfolding , that controls the curvature of the stability curve, the shift of the melting temperature could be even larger. Thus, even a small contribution as that arising from internal hydration can result in an enhanced thermal stability. The details of this qualitative modelling are provided in SI, see Fig. S.3.
Alternative strategies to estimate the free energy contribution to stability stemming from the hydration of large cavities have been presented before and reporting for non-polar cavities and channels cooperative effects22,48,53. For example in hydrophobic pockets of large volumes favorable hydration is found to take place only above a given threshold: about 4/5 water molecules should be hosted to compensate the loss of HBs due to the cavity nature22. Still, for cavities offering HB acceptor/donor sites, the size and cooperative effects are expected to be less important; for example in the case of a protein kinase, the progressive hydration of an internal cavity is associated to a stable free energy contribution of about −3 to −2 kcal/mol per added molecule80. A detailed analysis of the problem is reserved for future work. Obviously, our conclusions also depend on the reconstructed distributions p(ΔGi), here assumed to be representative of the cavities’ hydration, and on the residence time threshold used to determine the number of buried water molecules, ⟨nw⟩.
While hereby we have considered only the contribution from internal hydration, we stress that a more complete view of the role of hydration on protein thermal stability would require to account for the changes in the solvatation of the external surface as well. This can be only approached using specific modelling techniques, as those explored recently by Harano85 and Ham86.
Hydration and global protein flexibility
The number of internal waters fluctuates over time and the penetration/escape dynamics correlates with the breathing soft-modes of the protein that visits different conformational states78,87. For our proteins, the fluctuations are rather high (~ 40%) and have characteristic oscillation period ranging from tenths to one hundred nanoseconds. As a consequence the internal water contribution to protein stability varies substantially with the proteins’ conformational fluctuations.
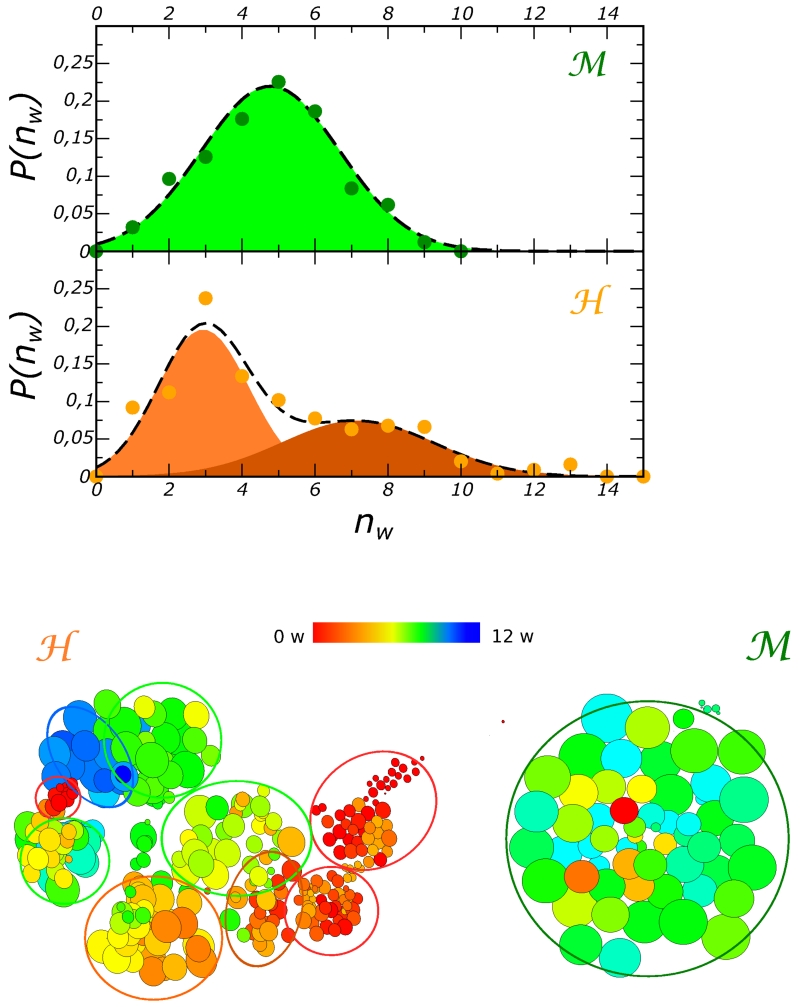
The distribution of the number of long residence water molecule p(nw), is reported in 5 (top panel) for the residence time threshold τr > 15 ns. While for the domain the fluctuations of nw produce an unimodal distribution, for the distribution is bi-modal. This two-state regime relates to the simultaneous hydration dynamics of the two cavities P1 and P2, while in only the hydration of pocket P2 varies substantially in time. Interestingly, we note that despite the qualitative differences, the hydrations dynamics of the internal cavities result in very similar overall compressibilities of the two proteins9, βa ≃ 9 · 10−5 MPa−1. Hence, internal hydration has similar effects on the global flexibility of the two proteins.
In the bottom panel of the figure we provide a more direct mapping of the internal hydration on the conformational landscape, represented as a network of states. In the specific case the network is obtained by clustering the trajectories and using the native contacts order parameter, Q9. The details of the calculation are provided in section Method. The size of each cluster is proportional to its occupation and the color scale reflects the average number of long residence water molecules computed for the configurations in each cluster.
For the domain, two zones of different color appear well separated kinetically; we do also note, however, that some basins that have similar Q values with rather different internal hydration are kinetically connected. For the mesophilic protein, the unimodal distribution of the internal hydration correlates with a compact conformational landscape. The correlation between the fluctuations of the internal hydration and the presence of multiple conformational states measured by conformational order parameters, such as the native contacts Q, supports the notion proposed some years ago76, that the exchange dynamics of internal hydration could be used to probe roughness of the conformational landscape of proteins.
This gives quantitative support to the qualitative sketch of 4, where the free energy landscape of the protein is projected along a generic conformation reaction coordinate. The filled blue zones represent the internal water free energy contribution to stability as it varies along the conformational landscape of the folded state. The thermophile exhibits a broader variety of internal hydration states, some of which confer it additional stability. The tool we have used to investigate the correlation among the dynamics of internal hydration and the protein conformational landscape could result effective to explore the mechanism of unfolding caused by high temperature or pressure and to characterise the kinetic heterogeneity of these processes that have been related to a different transient hydration of internal cavities88.
Stability at high temperature
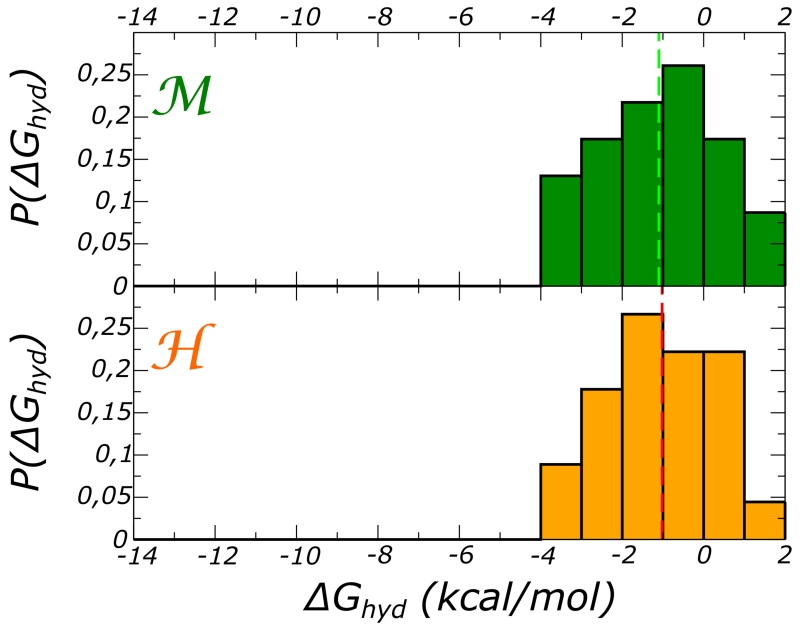
We address here the key issue of high temperature stability. For the same set of water molecules we evaluated the hydration free energy at T = 360 K. The calculations were performed starting from the same protein configurations used at T = 300 K, thus we assumed that the temperature excitation does not distort these representative conformations of the folded state. At high temperature, water binding free energy decreases substantially for most sites, in both proteins; water transfer into some of the protein sites is even unfavorable (6). As a result, there is no significant difference in overall free energy of internal hydration, yielding no clear contribution to the stability gap. The average ⟨ΔGi⟩ is −1.8 and −1.6 kcal/mol, for and . A comparable decrease of the favorable hydration was reported also for nonpolar cavities, and in a less extended temperature window for the favorable hydration of BPTI cavity50.
In the high temperature regime, it is difficult to estimate the overall contribution from internal water to the stability gap between the two proteins because the mesophilic homologue starts to unfold. The early step of unfolding was monitored in a set of independent simulations and occurred within a characteristic time of 100 to 300 ns9. We have estimated the number of long residence waters residing simultaneously in the protein matrix for each of these runs by restraining the calculation to the part of the trajectory before the unfolding event. For the hyperthermophilic domain two simulations of length 0.7 and 1 μs were used. Using a strict threshold (τr > 9.6 ns) we obtain ⟨nw⟩ = 2.6 and ⟨nw⟩ = 2 for and , respectively while for a less strict one (τr > 3.8 ns), the numbers are ⟨nw⟩=3.6 vs ⟨nw⟩ = 4.6 for and , respectively 1. When estimating the ΔGhyd via eq. 10 the contribution of internal hydration to the different stabilities (ΔΔGhyd) can be negligible (0.7 kcal/mol) or even in favour of the mesophile (1.7 kcal/mol) depending on the values of ⟨nw⟩. Therefore our results are not conclusive and not robust for this high temperature regime.
Alternatively, one could investigate the contribution of internal water hydrating the cavity P2 to the kinetic stability of the specific region α1 [E32-L45] and α2 [E48-E63] of the domain. Here, water could link these key secondary structures and indirectly favor the stability of the local ion-pairs. On the other hand, in the mesophilic homologue , the similar region represents the weakest point of the protein structure where unfolding early steps take place9. How transient internal hydration couples to the unfolding of this region and the associated kinetic barriers will be the object of a separate investigation.
Conclusion
In this work we investigated the internal hydration of two homologous proteins and estimated its contribution to the proteins’ different thermal stabilities. Namely, we compared the mesophilic EF-Tu and the hyperthermophilic EF-1α G-domains. Two internal cavities were found to be hydrated in both systems. The location of one of them is common for the two domains with an access near the P-loop at the catalytic site. The other ones are peripheral and domain specific.
In both systems, the per molecule hydration free energy was found to be favorable for all the sites and the protein configurations.
At ambient conditions, the free energy of internal hydration for the hyperthermophilic domain is more favorable than for the mesophilic one by 1.3 kcal/mol, and resulting in a sizeable contribution to the stability gap. At the high working temperature of the hyperthermophile (T = 365 K), the internal hydration free energy is systematically less favorable when compared to ambient condition, giving probably a negligible contribution to the stability gap between the two systems.
We also found a very clear correlation between the conformational fluctuations of the proteins, represented as a connected network of conformational states, and the fluctuations of the number of the long residence waters wetting the interior of the proteins. While the mesophilic protein is characterized by unimodal fluctuations around a single conformational or hydration state, the hyperthermophilic domain visits multiple states for both the protein conformation and the internal hydration. Again, this finding questions the general notion according to which thermophilic proteins are more rigid objects than their mesophilic counterparts.
Our investigation shows that thermal stabilization can be made possible by enhanced internal hydration. However, the thermodynamic gain results from different factors, such as the overall number of structural water and how the local internal polarity is configured. By in silico approaches one can envision a way to enhance thermal stability by using internal hydration and by targeting such factors selectively. One strategy is to tune the internal polarity of a cavity so as to increase the free energy gain. Alternatively, de novo new cavities can be created, possibly by creating pockets near the protein surface, to increase the number of structural waters that contribute to stability. Internal hydration can also be used to kinetically stabilize specific regions of the protein. However, it must be pointed out that the creation of cavities and their hydrated or dry states can result in a complex thermodynamic and kinetic response of the proteins, as pointed out in the context of pressure induced unfolding19,88. Moreover, these changes however must be effective in a broad range of temperatures. A more systematic investigation on the internal and the total hydration of homologues of different protein families is reserved for future work and can benefit from recent methodological development for treating solvation contribution to protein stability85,86.
Supplementary Material
Figure 1.
Frequency distribution of the long residence times τr for the (top) and (bottom) proteins along with the power-law fits (dashed lines). In the figure insets, the fraction of water molecules that continuously stay in the hydration shell as a function of time are plotted.
Figure 2.
Frequency distribution of long residence water molecules in the protein matrix for the (top) and the (bottom) domains. Two pockets are distinguished for each protein and indicated by the colored bars within the graphs. Their location in the 3D protein structure is shown at the top of each graph.
Figure 3.
Frequency distribution of the hydration free energy calculated from representative molecules hydrating the internal cavities of (top) and the (bottom) domains. The calculations were performed at T = 300 K. The vertical lines indicate the average value ⟨ΔGi⟩.
Figure 4.
Schematic view of the contribution from internal water to protein stability (blue zone) as a function of a generic conformational coordinate. The free energy landscape associated with the most stable protein is represented in yellow, while that associated with the less stable protein is represented in green. We assume that the unfolding free energy is decomposed in two terms, one that measures the difference between the unfolded and “dry” folded state (with the internal cavities dehydrated), and another term (blue filled zone) that measures the gain from internal hydration of the cavities. The free energy gap between the two proteins then contains a contribution from the difference in internal hydration free energy, ΔΔGhyd.
Figure 5.
Top panel. Frequency distribution of the number of long-residence water simultaneously located inside the protein matrix, nw. For this plot the residence time threshold is τc > 15 ns. The colored zones represent the area underneath the Gaussian functions fitting the distributions.
Bottom panel. Network representation of the conformational landscapes sampled by the two proteins. The network is obtained by clustering the MD trajectories and using the native contacts order parameter Q. The color map represents the hydration states of internal sites, red indicates fully dehydration, while blue indicates maximal internal hydration.
Figure 6.
Frequency distribution of the hydration free energy at T=360 K calculated for the same set of water molecules used in 3. The top panel refers to ; the bottom one, to the domain.
Acknowledgement
The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013) Grant Agreement no.258748. Part of this work was performed using HPC resources from GENCI [CINES and TGCC] (Grant 2012 c2012086818 and 2013 x201376818) and from CINECA (ISCRA Grant FLOWPROT). We acknowledge the financial support for infrastructures from ANR-11-LABX-0011-01.
Footnotes
The number of long-residence water molecules was estimated by considering the temperature scaling on the time threshold and assuming the escaping process rate-limited by a single barrier. The lower bound at T=360 K is therefore τr > 9.6 ns and the weaker threshold is about 3.8ns.
References
- (1).Nojima H, Ikai A, Oshima T, Noda H. Reversible Thermal Unfolding of Thermostable Phosphoglycerate Kinase. Thermostability Associated With Mean Zero Enthalpy Change. J. Mol. Biol. 1977;116:429–442. doi: 10.1016/0022-2836(77)90078-x. [DOI] [PubMed] [Google Scholar]
- (2).Vieille C, Zeikus GJ. Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability. Microbiol. Mol. Biol. Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kumar S, Nussinov R. How do Thermophilic Proteins Deal with Heat? Cell. Mol. Life Sci. 2001;58:1216–1233. doi: 10.1007/PL00000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sterpone F, Melchionna S. Thermophilic Proteins: Insight and Perspective from In Silico Experiments. Chem. Soc. Rev. 2012;41:1665–1676. doi: 10.1039/c1cs15199a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kumar S, Ma B, Tsai CJ, Nussinov R. Electrostatic Strengths of Salt Bridges in Thermophilic and Mesophilic Glutamate Dehydrogenase Monomers. Proteins. 2000;38:368–83. doi: 10.1002/(sici)1097-0134(20000301)38:4<368::aid-prot3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- (6).Karshiko A, Ladenstein R. Ion Pairs and the Thermotolerance of Proteins from Hyperthermophiles: a ”Traffic Rule” for Hot Roads. Trends Biochem. Sci. 2001;26:550–6. doi: 10.1016/s0968-0004(01)01918-1. [DOI] [PubMed] [Google Scholar]
- (7).Chan C-H, Yu T-H, Wong K-B. Stabilizing Salt-Bridge Enhances Protein Thermostability by Reducing the Heat Capacity Change of Unfolding. PLoS ONE. 2011;6:e21624. doi: 10.1371/journal.pone.0021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Balasco N, Esposito L, Simone AD, Vitagliano L. Role of Loops Connecting Secondary Structure Elements in the Stabilization of Proteins Isolated from Thermophilic Organisms. Protein Sci. 2013;22:1016–1023. doi: 10.1002/pro.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kalimeri M, Rahaman O, Melchionna S, Sterpone F. How Conformational Flexibility Stabilizes the Hyperthermophilic Elongation Factor G-Domain. J. Phys. Chem. B. 2013;117:13775–13785. doi: 10.1021/jp407078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rathi PC, Höffken HW, Gohlke H. Quality Matters: Extension of Clusters of Residues with Good Hydrophobic Contacts Stabilize (Hyper)Thermophilic Proteins. J. Chem. Inf. Model. 2014;54:355–361. doi: 10.1021/ci400568c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sanchez-Ruiz JM, Makhatadze GI. To Charge or not to Charge? Trends Biotechnol. 2001;19:132–5. doi: 10.1016/s0167-7799(00)01548-1. [DOI] [PubMed] [Google Scholar]
- (12).Gribenko AV, Patel MM, Liu J, McCallum SA, Wang C, Makhatadze GI. Rational Stabilization of Enzymes by Computational Redesign of Surface Charge-Charge Interactions. Proc. Natl. Acad. Sci. USA. 2009;106:2601–6. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xiao L, Honig B. Electrostatic Contributions to the Stability of Hyperthermophilic Proteins. J. Mol. Biol. 1999;289:1435–44. doi: 10.1006/jmbi.1999.2810. [DOI] [PubMed] [Google Scholar]
- (14).Gao J, Bosco D, Powers E, Kelly J. Localized Thermodynamic Coupling between Hydrogen Bonding and Microenvironment Polarity Substantially Stabilizes Proteins. Nat. Struct. Mol. Biol. 2009;16:684–691. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fitch C, Karp D, Lee K, Stites W, Lattmanand EE, Garcia-Moreno E. Experimental pKa Values of Buried Residues: Analysis with Continuum Methods and Role of Water Penetration. Biophys. J. 2002;82:3289–3304. doi: 10.1016/s0006-3495(02)75670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Salari R, Chong LT. Effects of High Temperature on Desolvation Costs of Salt Bridges Across Protein Binding Interfaces: Similarities and Differences between Implicit and Explicit Solvent Models. J. Phys. Chem. B. 2012;116:2561–2567. doi: 10.1021/jp210172b. [DOI] [PubMed] [Google Scholar]
- (17).Dominy BN, Minoux H, Brooks CL., 3rd An Electrostatic Basis for the Stability of Thermophilic Proteins. Proteins. 2004;57:128–41. doi: 10.1002/prot.20190. [DOI] [PubMed] [Google Scholar]
- (18).Ahmad S, Kamal MZ, Sankaranarayanan R, Rao NM. Thermostable Bacillus Subtilis Lipases: In Vitro Evolution and Structural Insight. J. Mol. Biol. 2008;381:324–40. doi: 10.1016/j.jmb.2008.05.063. [DOI] [PubMed] [Google Scholar]
- (19).Roche J, Caro JA, Norberto DR, Barthe P, Roumestand C, Schlessman JL, Garcia AE, García-Moreno BE, Royer CA. Cavities Determine the Pressure Unfolding of Proteins. Proc. Natl. Acad. Sci. USA. 2012;109:6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nisius L, Grzesiek S. Key Stabilizing Elements of Protein Structure Identified through Pressure and Temperature Perturbation of its Hydrogen Bond Network. Nat. Chem. 2012;4:711–717. doi: 10.1038/nchem.1396. [DOI] [PubMed] [Google Scholar]
- (21).Levy Y, Onuchic JN. Water Mediation in Protein Folding and Molecular Recognition. Annu. Rev. Biophys. Biomol. Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- (22).Rasaiah JC, Garde S, Hummer G. Water in Nonpolar Confinement: from Nanotubes to Proteins and beyond. Annu. Rev. Phys. Chem. 2008;59:713–40. doi: 10.1146/annurev.physchem.59.032607.093815. [DOI] [PubMed] [Google Scholar]
- (23).Halle B. Protein Hydration Dynamics in Solution: a Critical Survey. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:1207–23. doi: 10.1098/rstb.2004.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ball P. Water as an Active Constituent in Cell Biology. Chem. Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- (25).Fogarty AC, Duboue-Dijon E, Sterpone F, Hynes JT, Laage D. Biomolecular Hydration Dynamics: a Jump Model Perspective. Chem. Soc. Rev. 2013;42:5672–5683. doi: 10.1039/c3cs60091b. [DOI] [PubMed] [Google Scholar]
- (26).Pechkova E, Sivozhelezov V, Nicolini C. Protein Thermal Stability: the Role of Protein Structure and Aqueous Environment. Arch. Biochem. Biophys. 2007;466:40–8. doi: 10.1016/j.abb.2007.07.016. [DOI] [PubMed] [Google Scholar]
- (27).Sterpone F, Bertonati C, Briganti G, Melchionna S. Key Role of Proximal Water in Regulating Thermostable Proteins. J. Phys. Chem. B. 2009;113:131–7. doi: 10.1021/jp805199c. [DOI] [PubMed] [Google Scholar]
- (28).Paredes D, Watters K, Pitman D, Bystro C, Dordick J. Comparative Void-Volume Analysis of Psychrophilic and Mesophilic Enzymes: Structural Bioinformatics of Psychrophilic Enzymes Reveals Sources of Core Flexibility. BMC Struct. Biol. 2011;11:42. doi: 10.1186/1472-6807-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rahaman O, Melchionna S, Laage D, Sterpone F. The Effect of Protein Composition on Hydration Dynamics. Phys. Chem. Chem. Phys. 2013;15:3570–3576. doi: 10.1039/C3CP44582H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yin H, Hummer G, Rasaiah JC. Metastable Water Clusters in the Nonpolar Cavities of the Thermostable Protein Tetrabrachion. J. Am. Chem. Soc. 2007;129:7369–77. doi: 10.1021/ja070456h. [DOI] [PubMed] [Google Scholar]
- (31).Talon R, Coquelle N, Madern D, Girard E. An Experimental Point of View on Hydration/Solvation in Halophilic Proteins. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Qvist J, Ortega G, Tadeo X, Millet O, Halle B. Hydration Dynamics of a Halophilic Protein in Folded and Unfolded States. J. Phys. Chem. B. 2012;116:3436–3444. doi: 10.1021/jp3000569. [DOI] [PubMed] [Google Scholar]
- (33).Melchionna S, Sinibaldi R, Briganti G. Explanation of the Stability of Thermophilic Proteins Based on Unique Micromorphology. Biophys. J. 2006;90:4204–12. doi: 10.1529/biophysj.105.078972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Sterpone F, Bertonati C, Melchionna S. Water Around Thermophilic Proteins: The Role of Charged and Apolar Atoms. J. Phys.: Condens. Matter. 2010;22:284113. doi: 10.1088/0953-8984/22/28/284113. [DOI] [PubMed] [Google Scholar]
- (35).Chi YI, Martinez-Cruz LA, Jancarik J, Swanson RV, Robertson DE, Kim SH. Crystal Structure of the Beta-Glycosidase from the Hyperthermophile Thermosphaera Aggregans: Insights into its Activity and Thermostability. FEBS Lett. 1999;445:375–383. doi: 10.1016/s0014-5793(99)00090-3. [DOI] [PubMed] [Google Scholar]
- (36).Sterpone F, Stirnemann G, Laage D. Magnitude and Molecular Origin of Water Slow-down Next to a Protein. J. Am. Chem. Soc. 2012;134:4116–4119. doi: 10.1021/ja3007897. [DOI] [PubMed] [Google Scholar]
- (37).Ninad VP, Sharp KA. Heat Capacity in Proteins. Annu. Rev. Phys. Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- (38).Liu L, Quillin ML, Matthews BW. Use of Experimental Crystallographic Phases to Examine the Hydration of Polar and Nonpolar Cavities in T4 Lysozyme. Proc. Natl. Acad. Sci. USA. 2008;105:14406–14411. doi: 10.1073/pnas.0806307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Strop P, Kaiser SE, Vrljic M, Brunger AT. The Structure of the Yeast Plasma Membrane SNARE Complex Reveals Destabilizing Water-Filled Cavities. J. Biol. Chem. 2008;283:1113–9. doi: 10.1074/jbc.M707912200. [DOI] [PubMed] [Google Scholar]
- (40).Durrieu M-P, Lavery R, Baaden M. Interactions between Neuronal Fusion Proteins Explored by Molecular Dynamics. Biophys. J. 2008;94:3436–3446. doi: 10.1529/biophysj.107.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Berndt K, Beunink J, Schroder W, Wuthrich K. Designed Replacement of an Internal Hydration Water Molecule in BPTI: Structural and Functional Implications of a Glycine-to-Serine Mutation. Biochemistry. 1993;32:4564–4570. doi: 10.1021/bi00068a012. [DOI] [PubMed] [Google Scholar]
- (42).Pedersen JT, Olsen OH, Betzel C, Eschenburg S, Branner S, Hastrup S. Cavity Mutants of Savinase™: Crystal Structures and Differential Scanning Calorimetry Experiments Give Hints of the Function of the Buried Water Molecules in Subtilisins. J. Mol. Biol. 1994;242:193–202. doi: 10.1006/jmbi.1994.1572. [DOI] [PubMed] [Google Scholar]
- (43).Takano K, Funahashi J, Yamagata Y, Fujii S, Yutani K. Contribution of Water Molecules in the Interior of a Protein to the Conformational Stability. J. Mol. Biol. 1997;274:132–142. doi: 10.1006/jmbi.1997.1365. [DOI] [PubMed] [Google Scholar]
- (44).Hickey DR, Berghuis AM, Lafond G, Jaeger JA, Cardillo TS, McLendon D, Das G, Sherman F, Brayer GD, McLendon G. Enhanced Thermodynamic Stabilities of Yeast Iso-1-Cytochromes C with Amino Acid Replacements at Positions 52 and 102. J. Biol. Chem. 1991;266:11686–11694. [PubMed] [Google Scholar]
- (45).Vriend G, Berendsen H, van der Zee J, van den Burg B, Venema G, Eijsink V. Stabilization of the Neutral Protease of Bacillus Stearothermophilus by Removal of a Buried Water Molecule. Protein Eng. 1991;4:941–945. doi: 10.1093/protein/4.8.941. [DOI] [PubMed] [Google Scholar]
- (46).Xu J, Baase W, Quillin M, Baldwin E, Matthews B. Structural and Thermodynamic Analysis of the Binding of Solvent at Internal Sites in T4 Lysozyme. Protein Sci. 2001;10:1067–1078. doi: 10.1110/ps.02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wade RC, Mazor MH, McCammon JA, Quiocho FA. A Molecular Dynamics Study of Thermodynamic and Structural Aspects of the Hydration of Cavities in Proteins. Biopolymers. 1991;31:919–931. doi: 10.1002/bip.360310802. [DOI] [PubMed] [Google Scholar]
- (48).Roux B, Nina M, Pomes R, Smith J. Thermodynamic Stability of Water Molecules in the Bacteriorhodopsin Proton Channel: a Molecular Dynamics Free Energy Perturbation Study. Biophys. J. 1996;71:670–681. doi: 10.1016/S0006-3495(96)79267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sterpone F, Ceccarelli M, Marchi M. Dynamics of Hydration in Hen Egg White Lysozyme. J. Mol. Biol. 2001;311:409–19. doi: 10.1006/jmbi.2001.4860. [DOI] [PubMed] [Google Scholar]
- (50).Olano LR, Rick SW. Hydration Free Energies and Entropies for Water in Protein Interiors. J. Am. Chem. Soc. 2004;126:7991–8000. doi: 10.1021/ja049701c. [DOI] [PubMed] [Google Scholar]
- (51).Damjanovi A, Schlessman J, Fitch C, Garcia A, Garcia-Moreno E. Role of Flexibility and Polarity as Determinants of the Hydration of Internal Cavities and Pockets in Proteins. Biophys. J. 2007;93:2791–2804. doi: 10.1529/biophysj.107.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Chakrabarty S, Warshel A. Capturing the Energetics of Water Insertion in Biological Systems: The Water Flooding Approach. Proteins. 2013;81:93–106. doi: 10.1002/prot.24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Oikawa M, Yonetani Y. Molecular Dynamics Free Energy Calculations to Assess the Possibility of Water Existence in Protein Nonpolar Cavities. Biophys. J. 2010;98:2974–2983. doi: 10.1016/j.bpj.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Baudry J, Tajkhorshid E, Molnar F, Phillips J, Schulten K. Molecular Dynamics Study of Bacteriorhodopsin and the Purple Membrane. J. Phys. Chem. B. 2001;105:905–918. [Google Scholar]
- (55).Tashiro M, Stuchebrukhov AA. Thermodynamic Properties of Internal Water Molecules in the Hydrophobic Cavity around the Catalytic Center of Cytochrome c Oxidase. J. Phys. Chem. B. 2005;109:1015–1022. doi: 10.1021/jp0462456. [DOI] [PubMed] [Google Scholar]
- (56).Pisliakov AV, Sharma PK, Chu ZT, Haranczyk M, Warshel A. Electrostatic Basis for the Unidirectionality of the Primary Proton Transfer in Cytochrome C Oxidase. Proc. Natl. Acad. Sci. USA. 2008;105:7726–7731. doi: 10.1073/pnas.0800580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Lee HJ, Svahn E, Swanson JMJ, Lepp H, Voth GA, Brzezinski P, Gennis RB. Intricate Role of Water in Proton Transport through Cytochrome c Oxidase. J. Am. Chem. Soc. 2010;132:16225–16239. doi: 10.1021/ja107244g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Helms V, Wade R. Thermodynamics of Water Mediating Protein-Ligand Interactions in Cytochrome P450cam: a Molecular Dynamics Study. Biophys. J. 1995;69:810–824. doi: 10.1016/S0006-3495(95)79955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Li Z, Lazaridis T. Thermodynamic Contributions of the Ordered Water Molecule in HIV-1 Protease. J. Am. Chem. Soc. 2003;125:6636–6637. doi: 10.1021/ja0299203. [DOI] [PubMed] [Google Scholar]
- (60).Baron R, Setny P, McCammon JA. Water in Cavity-Ligand Recognition. J. Am. Chem. Soc. 2010;132:12091–12097. doi: 10.1021/ja1050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Szep S, Park S, Boder ET, Van Duyne GD, Saven JG. Structural Coupling between FKBP12 and Buried Water. Proteins. 2009;74:603–611. doi: 10.1002/prot.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Prakash P, Sayyed-Ahmad A, Gorfe AA. The Role of Conserved Waters in Conformational Transitions of Q61H K-ras. PLoS Comput. Biol. 2012;8:e1002394. doi: 10.1371/journal.pcbi.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).De Simone A, Dodson GG, Verma CS, Zagari A, Fraternali F. Prion and Water: Tight and Dynamical Hydration Sites Have a Key Role in Structural Stability. Proc. Natl. Acad. Sci. USA. 2005;102:7535–7540. doi: 10.1073/pnas.0501748102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Jensen M, Cool R, Mortensen K, Clark B, Parmeggiani A. Structure-function Relationships of Elongation Factor Tu. Isolation and Activity of the Guanine-nucleotide-binding Gomain. Eur. J. Biochm. 1989;182:247–255. doi: 10.1111/j.1432-1033.1989.tb14824.x. [DOI] [PubMed] [Google Scholar]
- (65).Bocchini V, Adinolfi BS, Arcari P, Arcucci A, Russo AD, Vendittis ED, Ianniciello G, M. Masullo M, Raimo G. Protein Engineering on Enzymes of the Peptide Elongation Cycle in Sulfolobus Solfataricus. Biochimie. 1998;80:895–898. doi: 10.1016/s0300-9084(00)88885-9. [DOI] [PubMed] [Google Scholar]
- (66).Song H, Parsons MR, Rowsell S, Leonard G, Phillips SE. Crystal Structure of Intact Elongation Factor EF-Tu from Escherichia coli in GDP Conformation at 2.05 Å Resolution. J. Mol. Biol. 1999;285:1245–1256. doi: 10.1006/jmbi.1998.2387. [DOI] [PubMed] [Google Scholar]
- (67).Vitagliano L, Ruggiero A, Masullo M, Cantiello P, Arcari P, Zagari A. The Crystal Structure of Sulfolobus Solfataricus Elongation Factor 1a in Complex with Magnesium and GDP. Biochemistry. 2004;43:6630–6636. doi: 10.1021/bi0363331. [DOI] [PubMed] [Google Scholar]
- (68).Phillips JC, Braunand R, Wei W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable Molecular Dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).MacKerell AD, Feig M, Brooks CL., III Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- (70).Impey RW, Madden PA, McDonald IR. Hydration and Mobility of Ions in Solution. J. Phys. Chem. 1983;87:5071–5083. [Google Scholar]
- (71).Marchi M, Sterpone F, Ceccarelli M. Water rotational relaxation and diffusion in hydrated lysozyme. J. Am. Chem. Soc. 2002;124:6787–91. doi: 10.1021/ja025905m. [DOI] [PubMed] [Google Scholar]
- (72).Tuckerman M. Statistical Mechanics. Theory and Molecular Simulation. Oxford University Press; 2010. [Google Scholar]
- (73).Fiorin G, Klein ML, Hénin J. Using Collective Variables to Drive Molecular Dynamics Simulations. Mol. Phys. 2013;111:3345–3362. [Google Scholar]
- (74).Hartigan J. Clustering Algorithms. Wiley; New York: 1975. [Google Scholar]
- (75).Bastian M, Heymann S, Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks. 2009. [Google Scholar]
- (76).Denisov VP, Peters J, Hörlein HD, Halle B. Using Buried Water Molecules to Explore the Energy Landscape of Proteins. Nat. Struct. Mol. Biol. 1996;3:505–509. doi: 10.1038/nsb0696-505. [DOI] [PubMed] [Google Scholar]
- (77).Garcia AE, Hummer G. Water Penetration and Escape in Proteins. Proteins. 2000;38:261–72. [PubMed] [Google Scholar]
- (78).Persson F, Halle B. Transient Access to the Protein Interior: Simulation versus NMR. J. Am. Chem. Soc. 2013;135:8735–8748. doi: 10.1021/ja403405d. [DOI] [PubMed] [Google Scholar]
- (79).Mattea C, Qvist J, Halle B. Dynamics at the Protein-Water Interface from 17O Spin Relaxation in Deeply Supercooled Solutions. Biophys. J. 2008;95:2951–63. doi: 10.1529/biophysj.108.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Knight JDR, Hamelberg D, McCammon JA, Kothary R. The Role of Conserved Water Molecules in the Catalytic Domain of Protein Kinases. Proteins. 2009;76:527–535. doi: 10.1002/prot.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Razvi A, Scholtz JM. Lessons in Stability from Thermophilic Proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Liu C-C, LiCata VJ. The stability of Taq DNA Polymerase Results from a Reduced Entropic Folding Penalty; Identification of other Thermophilic Proteins with Similar Folding Thermodynamics. Proteins. 2013 doi: 10.1002/prot.24458. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- (83).Budkevich TV, Timchenko AA, Tiktopulo EI, Negrutskii BS, Shalak VF, Petrushenko ZM, Aksenov VL, Willumeit R, Kohlbrecher J, Serdyuk IN, et al. Extended Conformation of Mammalian Translation Elongation Factor 1A in Solution. Biochemistry. 2002;41:15342–15349. doi: 10.1021/bi026495h. [DOI] [PubMed] [Google Scholar]
- (84).Deutschman WA, Dahlquist FW. Thermodynamic Basis for the Increased Thermostability of CheY from the Hyperthermophile Thermotoga Maritima. Biochemistry. 2001;40 doi: 10.1021/bi010665t. [DOI] [PubMed] [Google Scholar]
- (85).Maruyama Y, Harano Y. Does Water Drive Protein Folding? Chem. Phys. Lett. 2013;581:85–90. [Google Scholar]
- (86).Chong S-H, Ham S. Protein Folding Thermodynamics: A New Computational Approach. J. Phys. Chem. B. 2014;118:5017–5025. doi: 10.1021/jp500269m. [DOI] [PubMed] [Google Scholar]
- (87).Pizzitutti F, Marchi M, Sterpone F, Rossky PJ. How Protein Surfaces Induce Anomalous Dynamics of Hydration Water. J. Phys. Chem. B. 2007;111:7584–90. doi: 10.1021/jp0717185. [DOI] [PubMed] [Google Scholar]
- (88).Roche J, Dellarole M, Caro JA, Norberto DR, Garcia AE, Garcia-Moreno BE, Roumestand C, Royer CA. Effect of Internal Cavities on Folding Rates and Routes Revealed by Real-Time Pressure-Jump NMR Spectroscopy. J. Am. Chem. Soc. 2013;135:14610–14618. doi: 10.1021/ja406682e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.