Abstract
Several meningococcal vaccines under development for prevention of serogroup B disease target the factor H-binding protein (FHbp), an immunogenic lipoprotein expressed on the surface of Neisseria meningitidis. Based upon sequence and phylogenetic analyses, FHbp can be classified into 3 protein variants (1, 2 or 3) or 2 subfamilies (A or B). The potential effect of FHbp-containing vaccines on meningococcal carriage is not known. We determined the diversity of FHbp among a population of carriage isolates obtained from Georgia and Maryland high school students in 1998 and 2006–2007. Analysis of the fHbp gene sequence from 408 carriage isolates identified 30 different FHbp protein sequences. The majority of carriage isolates harbored FHbp proteins belonging to variant 2/subfamily A. Association between FHbp proteins and genetic lineage was observed among the carriage isolates. However, split decomposition analysis, together with tests of linkage disequilibrium and pairwise homoplasy suggest recombination at fHbp contribute to allelic diversity. Of note, the FHbp proteins in serogroup B vaccines under development are either absent or not well represented in this carriage population. The FHbp genetic repertoire observed in carriage isolate populations will be useful in understanding the potential impact of FHbp-containing vaccines on meningococcal carriage.
Keywords: Meningococcal vaccine, FHbp, Genetic lineage, Recombination
1. Introduction
Neisseria meningitidis is a major cause of meningitis and meningococcemia world-wide [1]. Based upon the composition of its polysaccharide capsule, most invasive N. meningitidis can be classified into 5 major serogroups – A, B, C, W-135 and Y. Current vaccines licensed in the U.S. target serogroups A, C, W-135 and Y polysaccharide capsules. However, serogroup B polysaccharide-based vaccines have not been feasible since the serogroup B capsule resembles glycosylated proteins expressed in human tissue during fetal development [2,3]. Serogroup B vaccine strategies have therefore focused on antigenic meningococcal outer membrane proteins [4].
Factor H binding protein (FHbp) is a 28 kDa, surface-exposed meningococcal lipoprotein that is capable of inducing high levels of bactericidal antibodies [5,6]. Binding of human factor H by FHbp in the bloodstream of infected individuals permits N. meningitidis to effectively evade host immune responses by preventing complement mediated bacterial cell lysis [7]. Based on sequence diversity, FHbp proteins are classified into either 3 variants (1–3) or 2 subfamilies (A and B) [5,6]. More recently, a modular protein structure has been described for FHbp whereby a combination of 5 distinct variable segments define 6 unique modular groups (I–VI) [8].
N. meningitidis colonizes the nasopharynx of ~10% of the human population. While serogroup C conjugate vaccines have been shown to reduce meningococcal disease incidence in part through indirect effects because of reductions in serogroup C pharyngeal carriage, less is known about the effect of protein-based meningococcal vaccines on pharyngeal carriage [9,10]. The published data are insufficient to draw definitive conclusions about the impact of serogroup B outer membrane protein vaccines on carriage [11–14]. However, high levels of antibody to the surface exposed FHbp may impact carriage and the effect could be substantial given that the presence of FHbp is independent of N. meningitidis serogroup.
Currently, three vaccine strategies employing FHbp are in clinical trials for the prevention of serogroup B meningococcal disease. One strategy utilizes a bivalent vaccine comprised of one FHbp protein from each subfamily (protein IDs 45 [variant 3, subfamily A] and 55 [variant 1, subfamily B]). A second multi-component vaccine targets three outer membrane proteins including a variant 1/subfamily B FHbp antigen (protein ID 1) [15–17]. A third strategy utilizes native outer membrane vesicles from a pathogenic serogroup B strain genetically modified to decrease toxicity and overexpress immunogenic proteins including FHbp protein ID 1 [18]. In the present study, the distribution of FHbp genotypes in carriage isolates among US high school students was determined.
2. Methods
2.1. Isolate collection and data sources
This investigation included 408 isolates obtained from 2 previous meningococcal carriage studies performed in the US – 194 from 1998 and 214 from 2006 to 2007. The first study examined the genetic lineage of 194 carriage isolates from 2730 high school students in 2 Georgia counties in 1998 – one county experiencing a high incidence of meningococcal disease, and the other county for comparison [19,20]. The second carriage study yielded 325 meningococcal isolates from 3 surveillance periods during the 2006–2007 school year from 3314 students in 4 high schools each in Baltimore County, Maryland and Douglas County, Georgia [21]. When multiple isolates were obtained from individual students at different time periods, all isolates with different genotypes (defined by multi-locus sequence typing (MLST) and outer membrane protein genotyping of porA, porB and fetA) were selected for FHbp analysis. For students that had multiple isolates with the same genotype, only the first isolate collected was selected for FHbp analysis. Thus, 111 isolates from 106 students in Maryland and 103 isolates from 83 students in Georgia were selected for evaluation of fHbp genotypes from the 2006 to 2007 study. Serogroup was determined by either slide agglutination or serogroup specific PCR [22]. Isolates that did not agglutinate and did not amplify specific capsular synthesis genes were considered non-groupable, unencapsulated. Isolates that did not agglutinate but amplified a specific capsular synthesis gene were considered groupable, unencapsulated. There were 77 isolates from the Georgia 1998 collection that were non-groupable by slide agglutination and not available for SGS-PCR. To determine if the carriage isolates in this study represented unique fHbp sequences, the neissera.org FHbp database (http://pubmlst.org/neisseria/fHbp/) was searched. In addition, FHbp data from invasive U.S. isolates collected from 2000 to 2007 by the Active Bacterial Core surveillance (ABCs) network were also examined (personal communication, Leonard Mayer) [23,24].
2.2. Genotyping
The Georgia 1998 and Maryland 2006–2007 carriage isolates were characterized by MLST and OMP genotype profile at the University of Pittsburgh as previously described [25,26]. MLST and OMP genotyping were performed on the Georgia 2006–2007 carriage isolates by the Centers for Disease Control and Prevention using the same methods. All fHbp genotyping was performed at the University of Pittsburgh by PCR amplification and subsequent sequence analysis. Briefly, 1 μl of genomic DNA prepared by boil preparation was PCR amplified with 1.0 μM CDC5UNI (5′-CTATTCTGVGTATGACTAG-3′) and CDC3UNI (5′-GTCCGAACGGTAAATTATYGT-3′) in a final reaction 50 μl reaction volume containing 1X AmpliTaq Gold PCR Buffer (15 mM Tris–HCl, pH 8.0, 50 mM KCl), 2.5 mM MgCl2, 0.2 mM each deoxynucleo-side triphosphate and 1.5 units AmpliTaq Gold DNA polymerase (Applied Biosystems). Cycling conditions were 95 °C for 5 min, followed by 30 cycles of 95 °C for 1 min, 50 °C for 1 min, and 72 °C for 1.5 min with a final extension of 72 °C for 7 min. PCR products were prepared for sequencing by precipitation with 20% polyethylene glycol 8000 (Sigma–Aldrich, St. Louis, MO) in a 2.5 M NaCl solution followed by 2–70% ethanol washes. The cleaned PCR product was resuspended in 30 μl distilled H2O and 1 μl of this product was sequenced with 0.66 pmol of CDC5UNI, CDC3UNI and each of the following primers: 581F, 5′-CCGCCGAACTCAAAGCAGAT-3′; 310R, 5′-TGTAGGCGAACGACGGCGGA-3′; 205R, 5′-CGCCTGACCTTGTCGTTCTT-3′; 540F, 5′-AATCGAACATTTGAAATCGC-3′. Sequencing reactions were performed in 10 μl volumes using Big Dye Terminator v3.1 (Applied Biosystems). Sequencing reactions were cleaned by ethanol precipitation and analyzed on an AB3730 sequence detection system (Applied Biosystems). Resulting sequence files were either assembled into contigs using DNAstarSeqMan software v6.1 (DNAstar, Madison WI) and manually queried against the fHbp sequence typing database at pubmlst.org/neisseria/fhbp/ to obtain alleles or were uploaded to the Meningococcus Genome Informatics Platform (MGIP) website: mgip.biology.gatech.edu for automatic fHbp allele calling [27,28]. In the PubMLST database, unique nucleotide and peptide sequences are arbitrarily assigned allele number in order of discovery [28]. In this manuscript, FHbp protein ID (e.g., FHbp protein ID 25) corresponds to the deduced amino acid sequence peptide ID in the PubMLST database. In addition, FHbp modular groups were assigned according to the PubMLST database. The combination of MLST alleles defined the sequence type (ST). STs were assigned to a clonal complex defined at http://pubmlst.org/neisseria/info/complexes.shtml. The association between FHbp protein sequence and genetic lineage as defined by clonal complex (CC) was determined using the asymmetric Goodman–Kruskal Lambda statistic (λ) [29]. This method calculates the reduction in error for predicting the FHbp protein ID based upon knowledge of the genetic lineage of the isolates.
2.3. Comparative sequence analysis
The software MEGA 4.1 was used to calculate the pairwise distances for fHbp gene sequences using the Kimura 2-parameter model [30]. Estimates of linkage disequilibrium (LD) were performed using two measures: r2 and D′ and the program PERMUTE. The correlation coefficients between physical distance and these LD measures for all fhbp base pairs and the associated p values were calculated using the null hypothesis that in the absence of recombination there should be zero correlation between physical distance and LD measures [31]. The pairwise homoplasy index (PHI) available within SplitsTree 4.11.3 was used to measure the mean incompatibility score of fHbp sequences and SplitsTree 4.11.3 was used to generate a summary network of the relatedness of fHbp sequences for each population [32]. GARD (Genetic Algorithm Recombination Detection) was used to detect discordant phylogenetic signals in alignments of fHbp sequences and provide an estimate of the number and location of break points [33].
3. Results
3.1. Serogroups, capsular genotypes and MLST of carriage population
Capsular phenotype and/or genotype data was available for all 408 carriage isolates (Supplementary Data). Encapsulation was detected by slide agglutination in 35.0% (143/408) of carriage isolates (Table 1). Encapsulated carriage isolates were predominantly from the Georgia 1998 population (117/143, 81.8%) and belonged to serogroup Y (75/117, 64.1%), serogroup B (37/117, 31.6%), serogroup Z (3/117, 2.6%) and serogroup C (2/117, 1.7%) (Table 1). Serogroup B encapsulated carriage isolates were found in both time periods in Georgia but were not observed in the Maryland 2006–2007 collection (Table 1).
Table 1.
Capsular phenotype or genotype of 408 carriage isolates.
| Isolate collection | Encapsulateda (n = 143) |
Unencapsulated (n = 265) |
||||||
|---|---|---|---|---|---|---|---|---|
| SgB | SgC | SgY | SgZ | NGa | Bb | Cb | Yb | |
| GA 1998 | 37 | 2 | 75 | 3 | 77 | NA | NA | NA |
| GA 2007 | 13 | 0 | 4 | 0 | 59 | 17 | 2 | 8 |
| MD 2007 | 0 | 0 | 9 | 0 | 64 | 20 | 3 | 15 |
| Total | 50 | 2 | 88 | 3 | 200 | 37 | 5 | 23 |
Slide agglutination serogroup.
Serogroup specific PCR; NG, non-groupable; NA, not available.
The majority of carriage isolates were unencapsulated by slide agglutination (265/408, 65.0%). Serogroup specific capsular synthesis genes were not detected in the 123 Georgia and Maryland carriage isolates from 2006 to 2007 and SGS-PCR data were not available for the 77 unencapsulated Georgia isolates from 1998 (Table 1). Thus, 75.5% (200/265) of unencapsulated carriage isolates in this study were considered non-groupable. Of the 65 unencapsulated carriage isolates from Georgia and Maryland in 2006–2007 that were groupable by SGS-PCR, 37 (56.9%) were capsular genotype B, 23 (35.4%) were capsular genotype Y and 5 (7.7%) were capsular genotype C (Table 1).
There were 22 clonal complexes represented in the carriage population with the most prevalent types (ST-23, ST-198, ST-41-44) representing 58% of the isolates. Among the 113 ST identified, ST-23 was the most prevalent type representing 25.6% of isolates.
3.2. FHbp proteins among carriage isolates
The gene encoding FHbp was detected in all 408 study isolates (Supplementary Data). A total of 32 unique fHbp gene sequences encoding 30 unique deduced FHbp proteins were identified (Table 2). Of these 30 FHbp protein sequences, 11 belonged to variant 1/subfamily B, 16 belonged to variant 2/subfamily A and 3 belonged to variant 3/subfamily A (Fig. 1). Five FHbp proteins (IDs 4, 13, 19, 21, and 25) comprised the majority of isolates (68%, Table 2). Ten proteins were found in ≤2 isolates. Based upon comparisons with invasive isolates collected during the same time period and same geographic location, there were 12 FHbp proteins identified that were unique to carriage isolates in this study (Table 2 and Fig. 1) (personal communication, Leonard Mayer) [24]. Of the 3 variant 3/subfamily A proteins identified in this study, only FHbp protein ID 70 has been identified in invasive disease [34].
Table 2.
Distribution of the 30 FHbp proteins identified in 408 meningococcal carriage isolates from different locations and years.
| FHbp ID | Modular group |
Variant/ subfamily |
Georgia 1998 | Georgia 2006–2007 |
Maryland 2006–2007 |
Total No. (%) | FHbp ID | Modular group |
Variant/ subfamily |
Georgia 1998 |
Georgia 2006–2007 |
Maryland 2006–2007 |
Total No. (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I | 1/B | 0 | 1 | 2 | 3 (<1) | 100 | I | 1/B | 0 | 0 | 2 | 2 (<1) |
| 4 | I | 1/B | 7 | 10 | 26 | 43 (11) | 101 | III | 2/A | 1 | 1 | 1 | 3 (<1) |
| 8 | I | 1/B | 6 | 0 | 0 | 6 (1) | a102 | III | 2/A | 0 | 2 | 5 | 7 (2) |
| 12 | I | 1/B | 4 | 3 | 2 | 9 (2) | a103 | III | 2/A | 0 | 0 | 2 | 2 (<1) |
| 13 | I | 1/B | 7 | 23 | 15 | 45 (11) | 104 | III | 2/A | 0 | 0 | 6 | 6 (2) |
| 14 | I | 1/B | 0 | 1 | 0 | 1 (<1) | a105 | III | 2/A | 0 | 0 | 2 | 2 (<1) |
| 16 | VI | 2/A | 11 | 4 | 6 | 21 (5) | 106 | VI | 2/A | 1 | 1 | 1 | 3 (<1) |
| 19 | VI | 2/A | 7 | 13 | 8 | 28 (7) | a107 | I | 1/B | 0 | 0 | 1 | 1 (<1) |
| 21 | III | 2/A | 15 | 10 | 3 | 28 (7) | a133 | VI | 2/A | 0 | 3 | 0 | 3 (<1) |
| 22 | III | 2/A | 0 | 1 | 1 | 2 (<1) | 205 | III | 2/A | 2 | 0 | 0 | 2 (<1) |
| 23 | III | 2/A | 2 | 3 | 0 | 5 (1) | a208 | I | 1/B | 1 | 0 | 0 | 1 (<1) |
| 24 | III | 2/A | 12 | 6 | 4 | 22 (5) | a271 | V | 3/A | 2 | 0 | 0 | 2 (<1) |
| 25 | III | 2/A | 102 | 12 | 21 | 135 (33) | a272 | I | 1/B | 1 | 0 | 0 | 1 (<1) |
| a70 | V | 3/A | 0 | 1 | 0 | 1 (<1) | a273 | I | 1/B | 1 | 0 | 0 | 1 (<1) |
| a94 | V | 3/A | 11 | 8 | 3 | 22 (5) | a274 | III | 2/A | 1 | 0 | 0 | 1 (<1) |
Proteins unique to carriage isolates in this study.
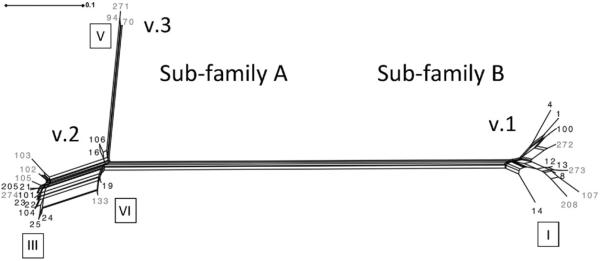
Fig. 1.
SplitsTree phylogram analysis of FHbp proteins from carriage isolates in this study. Variant (v.). Roman numerals in boxes indicate modular groups. The scale represents the number of amino acid substitutions per site.
Among encapsulated carriage isolates 60.8% (87/143) belonged to the ST-23 clonal complex (Table 3), a genetic lineage that is both widely carried and often associated with serogroup Y invasive disease in the U.S. The majority of these isolates (79/87, 90.8%) harbored FHbp ID 25 and were collected from Georgia in 1998 (Table 3A). There were 2 ST-23 clonal complex isolates from Maryland that harbored FHbp protein ID 104 (Table 3A). Both FHbp protein ID 25 and 104 are variant 2/subfamily A proteins. A few serogroup Y, ST-23 clonal complex isolates (6/87, 7.0%) harbored FHbp protein ID 8, a variant 1/subfamily B protein. Among unencapsulated carriage isolates 33.8% (21/65) had the serogroup Y capsule synthesis gene and belonged to the ST-23 clonal complex (Table 3B). While most of these isolates harbored FHbp protein ID 25, there were 4 isolates in this group that carried protein ID 104. Thus, all unencapsulated, capsular genotype Y carriage isolates harbored FHbp variant2/subfamily A proteins (Table 3B).
Table 3.
Distribution of FHbp proteins among encapsulated vs. unencapsulated carriage isolates.
| (A) Encapsulated | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Serogroup | Variant/ subfamily |
FHbp ID | Modular group |
CC | ST | Georgia 1998 | Georgia 2006–2007 | Maryland 2006–2007 | Total |
| Y | 1/B | 8 | I | ST-23 | 1625 | 6 | 0 | 0 | 6 |
| 2/A | 24 | III | ST-167 | 6145 | 0 | 1 | 0 | 1 | |
| 25 | III | ST-23 | 23, 1063, 1625, 2053, 2755, 3801 | 69 | 3 | 7 | 79 | ||
| 104 | III | ST-23 | 183 | 0 | 0 | 2 | 2 | ||
| Subtotal | 75 | 4 | 9 | 88 | |||||
| B | 1/B | 4 | I | ST-198 | 2384 | 2 | 0 | 0 | 2 |
| 12 | I | ST-269 | 2738 | 1 | 0 | 0 | 1 | ||
| 2/A | 16 | VI | None | 2154, 6143 | 1 | 1 | 0 | 2 | |
| ST-35 | 35, 472 | 1 | 1 | 0 | 2 | ||||
| 19 | VI | None | 6127 | 1 | 1 | 0 | 2 | ||
| ST-41/44 | 44, 146, 437, 5110, 6139 | 3 | 8 | 0 | 11 | ||||
| 21 | III | ST-162 | 162, 6133 | 6 | 2 | 0 | 8 | ||
| ST-32 | 32 | 4 | 0 | 0 | 4 | ||||
| ST-41/44 | 2343 | 1 | 0 | 0 | 1 | ||||
| 23 | III | ST-41/44 | 44 | 1 | 0 | 0 | 1 | ||
| 24 | III | None | 955, 2054, 2971 | 3 | 0 | 0 | 3 | ||
| ST-41/44 | 136, 6939 | 6 | 0 | 0 | 6 | ||||
| 25 | III | ST-549 | 5874 | 1 | 0 | 0 | 1 | ||
| 101 | III | ST-32 | 1784 | 1 | 0 | 0 | 1 | ||
| 205 | III | ST-162 | 162 | 2 | 0 | 0 | 2 | ||
| 274 | III | ST-162 | 162 | 1 | 0 | 0 | 1 | ||
| 3/A | 94 | V | ST-198 | 198 | 2 | 0 | 0 | 2 | |
| Subtotal | 37 | 13 | 0 | 50 | |||||
| C | 1/B | 13 | I | ST-60 | 60 | 1 | 0 | 0 | 1 |
| 272 | I | ST-11 | 11 | 1 | 0 | 0 | 1 | ||
| Subtotal | 2 | 0 | 0 | 2 | |||||
| Z | 2/A | 25 | III | ST-103 | 5842, 5989, 6937 | 3 | 0 | 0 | 3 |
| Subtotal | 3 | 0 | 0 | 3 | |||||
| Total | 117 | 17 | 9 | 143 | |||||
| (B) Uneneapsulated | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Capsular genotype |
Variant/ subfamily |
FHbp ID | Modular group |
CC | ST | Georgia 1998 | Georgia 2006–2007 | Maryland 2006–2007 | Total |
| B | 1/B | 1 | I | ST-32 | 32, 33, 749 | 0 | 1 | 2 | 3 |
| 13 | I | ST-60 | 60 | 0 | 0 | 1 | 1 | ||
| 107 | I | ST-269 | 1195 | 0 | 0 | 1 | 1 | ||
| 2/A | 16 | VI | ST-35 | 35, 5914 | 0 | 0 | 2 | 2 | |
| ST-4821 | 6595 | 0 | 0 | 1 | 1 | ||||
| 19 | VI | ST-41/44 | 44, 180, 315, 571, 5919, 6142 | 0 | 4 | 5 | 9 | ||
| 21 | III | ST-162 | 162, 6134, 6593 | 0 | 1 | 2 | 3 | ||
| 22 | III | ST-865 | 865 | 0 | 1 | 0 | 1 | ||
| 23 | III | ST-865 | 865 | 0 | 3 | 0 | 3 | ||
| 24 | III | None | 6131 | 0 | 1 | 0 | 1 | ||
| ST-41/44 | 6135 | 0 | 1 | 0 | 1 | ||||
| 25 | III | None | 5913 | 0 | 0 | 4 | 4 | ||
| 101 | III | None | 3677, 6173 | 0 | 1 | 1 | 2 | ||
| 106 | VI | None | 2048 | 0 | 0 | 1 | 1 | ||
| ST-35 | 6132 | 0 | 1 | 0 | 1 | ||||
| 133 | VI | ST-41/44 | 6130 | 0 | 3 | 0 | 3 | ||
| Subtotal | 0 | 17 | 20 | 37 | |||||
| Y | 2/A | 21 | III | ST-167 | 5918 | 0 | 0 | 1 | 1 |
| 24 | III | ST-167 | 1624 | 0 | 0 | 1 | 1 | ||
| 25 | III | ST-23 | 23, 4245 | 0 | 8 | 9 | 17 | ||
| 104 | III | ST-23 | 183 | 0 | 0 | 4 | 4 | ||
| Subtotal | 0 | 8 | 15 | 23 | |||||
| C | 2/A | 24 | III | ST-35 | 278 | 0 | 1 | 0 | 1 |
| ST-41/44 | 6591 | 0 | 0 | 1 | 1 | ||||
| 105 | III | ST-32 | 32 | 0 | 0 | 2 | 2 | ||
| 3/A | 94 | V | ST-1136 | 6144 | 0 | 1 | 0 | 1 | |
| Subtotal | 0 | 2 | 3 | 5 | |||||
| NG | 1/B | 4 | I | None | 6321 | 0 | 0 | 2 | 2 |
| 12 | I | ST-178 | 178 | 0 | 3 | 2 | 5 | ||
| 13 | I | ST-1157 | 1157, 1649, 6141, 6594 | 0 | 11 | 3 | 14 | ||
| ST-60 | 60, 4146, 6136, 6506 | 0 | 12 | 11 | 23 | ||||
| 14 | I | ST-198 | 198 | 0 | 1 | 0 | 1 | ||
| 100 | I | ST-41/44 | 2578, 5917 | 0 | 0 | 2 | 2 | ||
| 2/A | 16 | VI | ST-35 | 35, 5915, 6140, 6174 | 0 | 2 | 3 | 5 | |
| 19 | VI | ST-41/44 | 2578, 5916 | 0 | 0 | 3 | 3 | ||
| 21 | III | ST-1117 | 1117, 4788, 6137 | 0 | 6 | 0 | 6 | ||
| ST-41/44 | 1127 | 0 | 1 | 0 | 1 | ||||
| 22 | III | ST-60 | 60 | 0 | 0 | 1 | 1 | ||
| 24 | III | ST-167 | 6145 | 0 | 1 | 0 | 1 | ||
| ST-198 | 823 | 0 | 1 | 0 | 1 | ||||
| ST-212 | 5912 | 0 | 0 | 1 | 1 | ||||
| ST-35 | 278 | 0 | 0 | 1 | 1 | ||||
| 25 | III | ST-103 | 103 | 0 | 0 | 1 | 1 | ||
| ST-1157 | 1157 | 0 | 1 | 0 | 1 | ||||
| 102 | III | ST-53 | 53, 6322 | 0 | 2 | 5 | 7 | ||
| 103 | III | ST-53 | 53 | 0 | 0 | 2 | 2 | ||
| 3/A | 70 | V | ST-1136 | 6144 | 0 | 1 | 0 | 1 | |
| 94 | V | ST-1136 | 6144 | 0 | 1 | 0 | 1 | ||
| ST-198 | 39, 198, 6129, 6138, 6146 | 0 | 6 | 3 | 9 | ||||
| Subtotal | 0 | 59 | 64 | 123 | |||||
| NA | 1/B | 4 | I | ST-198 | 823, 2384 | 5 | 0 | 0 | 5 |
| 12 | I | ST-178 | 178, 4210 | 3 | 0 | 0 | 3 | ||
| 13 | I | ST-1157 | 1649, 6803 | 3 | 0 | 0 | 3 | ||
| ST-22 | 1265 | 1 | 0 | 0 | 1 | ||||
| ST-254 | 3808 | 1 | 0 | 0 | 1 | ||||
| ST-60 | 60 | 1 | 0 | 0 | 1 | ||||
| 208 | I | None | 1975 | 1 | 0 | 0 | 1 | ||
| 273 | I | ST-254 | 3808 | 1 | 0 | 0 | 1 | ||
| 2/A | 16 | VI | ST-198 | 198 | 1 | 0 | 0 | 1 | |
| ST-22 | 1265 | 1 | 0 | 0 | 1 | ||||
| ST-35 | 35, 812, 6806, 6941 | 7 | 0 | 0 | 7 | ||||
| 19 | VI | ST-41/44 | 44, 2052, 2056 | 3 | 0 | 0 | 3 | ||
| 21 | III | ST-162 | 162 | 4 | 0 | 0 | 4 | ||
| 23 | III | ST-11 | 2238 | 1 | 0 | 0 | 1 | ||
| 24 | III | None | 955, 2054, 6805 | 3 | 0 | 0 | 3 | ||
| 25 | III | ST-103 | 103, 4399, 5842, 6940 | 7 | 0 | 0 | 7 | ||
| ST-198 | 2047 | 2 | 0 | 0 | 2 | ||||
| ST-23 | 23, 2053, 2755 | 19 | 0 | 0 | 19 | ||||
| ST-549 | 5874 | 1 | 0 | 0 | 1 | ||||
| 106 | VI | None | 2048 | 1 | 0 | 0 | 1 | ||
| 3/A | 94 | V | ST-198 | 198, 3283, 6804, 6938 | 9 | 0 | 0 | 9 | |
| 271 | V | ST-198 | 198 | 2 | 0 | 0 | 2 | ||
| Subtotal | 77 | 0 | 0 | 77 | |||||
| Total | 77 | 86 | 102 | 265 | |||||
CC, clonal complex; ST, sequence type; NG, non-group able; NA, not available.
The majority of serogroup B encapsulated carriage isolates belonged to clonal complexes that are commonly carried including ST-41/44, ST-162, ST-198 and ST-35 (Table 3A). While ST-41/44 clonal complex is often associated with invasive disease, serogroup B carriage isolates in this clonal complex were comprised of sequence types (STs) commonly carried. There were 5 serogroup B encapsulated isolates that belonged to ST-32, a clonal complex that is also often associated with invasive disease. These isolates harbored FHbp protein IDs 21 and 101 (Table 3A). The majority of FHbp proteins associated with the serogroup B encapsulated carriage isolates (47/50, 94%) belonged to the variant 2/subfamily A group with protein IDs 19, 21 and 24 predominant (Table 3A). There were 37 unencapsulated, carriage isolates that amplified serogroup B capsular synthesis gene. These isolates demonstrated a similar clonal complex distribution as the encapsulated serogroup B carriage isolates with the addition of clonal complex ST-60, ST-4821 and ST-865 (Table 3B). ST-60 and ST-4821 are associated with carriage and invasive disease and ST-865 clonal complex is associated with invasive disease in Taiwan and South Africa (neisseria.org). Similar to the encapsulated serogroup B carriage isolates, the FHbp proteins from the majority of unencapsulated, capsular genotype B carriage isolates (32/37, 86.5%) belonged to the variant 2/subfamily A group (Table 3B).
There were 3 serogroup Z encapsulated carriage isolates that belonged to the ST-103 clonal complex. These isolates like the serogroup Y, ST-23 isolates, harbored FHbp protein ID 25 (Table 3A). The 2 serogroup C encapsulated isolates belonged to the ST-60 and ST-11 clonal complexes and harbored FHbp protein IDs 13 and 272, respectively (Table 3A). Both proteins belong to the variant 1/subfamily B group. FHbp protein ID 272 is unique to this carriage population. Among unencapsulated isolates, there were 5 with capsular genotype C that belonged to ST-32, ST-35, ST 41/44 and ST-1136 clonal complexes (Table 3B). The ST-35 and ST41/44 isolates harbored FHbp protein ID 24 while ST-1136 and ST-32 isolates harbored FHbp protein IDs 94 and 105, respectively. Both of these proteins are unique to this carriage population.
Of the 200 unencapsulated carriage isolates, 123 (61.5%) were non-groupable by SGS-PCR. The majority of these (68.3%) belonged to 3 predominant clonal complexes, ST198, ST1157 and ST-60 with FHbp protein IDs 4, 13 and 94 prevalent (Table 3C). The remaining 77 unencapsulated isolates from the Georgia 1998 collection were unavailable for SGS-PCR testing. ST-198 and ST-23 clonal complexes were prevalent with FHbp protein ID 4, 13, 16, 25 and 94 represented predominantly.
3.3. Association of genetic lineage with FHbp
There was a strong association between clonal complex and FHbp (Fig. 2). The lambda value of 0.9118 for reduction in error of predicting the FHbp protein ID given the ST clonal complex indicates a strong, although incomplete association. Differences in FHbp distribution over time and location could be explained in part by fluctuations in the prevalence of particular clonal complexes. The prevalence of carriage isolates belonging to clonal complexes ST-23 and ST-103 was higher in Georgia in 1998 than in either Georgia or Maryland in 2006–2007. These clonal complexes are associated with FHbp protein ID 25. The carriage rates of clonal complex ST-23 and ST-103 carriage contributed to the high proportion of variant 2/subfamily A in Georgia in 1998 (Table 4A and Fig. 2). In comparison, carriage rates of these clonal complexes in Georgia and Maryland in 2006–2007 were lower and the prevalence of variant 2/subfamily A during this time period was also lower (Table 4A and Fig. 2).
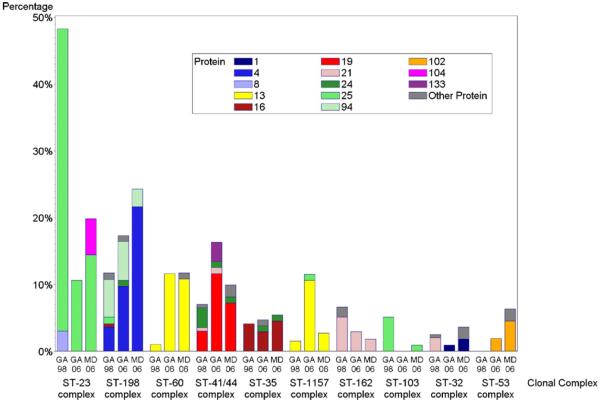
Fig. 2.
Distribution of FHbp proteins in carriage isolates by MLST-defined clonal complex – color coded by FHbp protein.
Table 4.
Distribution of FHbp variant/subfamilies and modular groups from meningococcal isolates collected in Georgia in 1998 (A) and Maryland and Georgia in 2006–2007 (B).
| (A) Variant/subfamily | Modular Group | Georgia 1998 |
|---|---|---|
| 1/B | I | 27(13.9%) |
| IV | 0 | |
| 2/A | III | 135(69.6%) |
| VI | 19(9.8%) | |
| 3/A | II | 0 |
| V | 13(6.7%) | |
| Total isolates | 194 |
| (B) Variant/subfamily | Modular group | Georgia 2006–2007 | Maryland 2006–2007 |
|---|---|---|---|
| 1/B | I | 38(36.9%) | 48(43.2%) |
| IV | 0 | 0 | |
| 2/A | III | 35(34.0%) | 45(40.5%) |
| VI | 21(20.4%) | 15(13.5%) | |
| 3/A | II | 0 | 0 |
| V | 9(8.7%) | 3(2.7%) | |
| Total isolates | 103 | 111 |
The association of FHbp with clonal complex was also evident with variant 1/subfamily B proteins which were more highly represented in Georgia and Maryland in 2006–2007 than in Georgia in 1998 (Table 4B). The increase in the prevalence of FHbp protein ID 4 and 13 in Maryland and Georgia during this time period coincided with an increase in clonal complexes ST-198, ST-1157 and ST-60 carriage (Tables 3B and 4B, Fig. 2 and Supplementary Data).
3.4. Recombination at fHbp
Despite the strong association between clonal complex and FHbp, there were examples of clonal complexes with diverse FHbp allelic repertoires, indicating that clonal complex cannot be used reliably to predict the FHbp protein ID. Carriage isolates belonging to the ST-23 clonal complex, most commonly associated with serogroup Y disease, encoded FHbp protein ID 8, 25 and 104. FHbp protein ID 25 and 104 belong to variant 2/subfamily A and are closely related, whereas FHbp protein ID 8 is a variant 1/subfamily B member and distantly related to FHbp protein ID 25 (Fig. 1). Carriage isolates belonging to ST-198 clonal complex were primarily associated with FHbp protein ID 4 and 94 which belong to different subfamilies (Fig. 1). Multiple FHbp protein IDs (19, 21, 23, 24, 100 and 133) were associated with the ST-41/44 clonal complex. All of these FHbp belong to variant 2/subfamily A with the exception of FHbp protein ID 100 which belongs to variant 1/subfamily B. Moreover, the genetic distance between FHbp subfamily A and B observed in the SplitsTree analysis suggests that FHbp diversity within genetic lineages is due to horizontal gene transfer (Fig. 1).
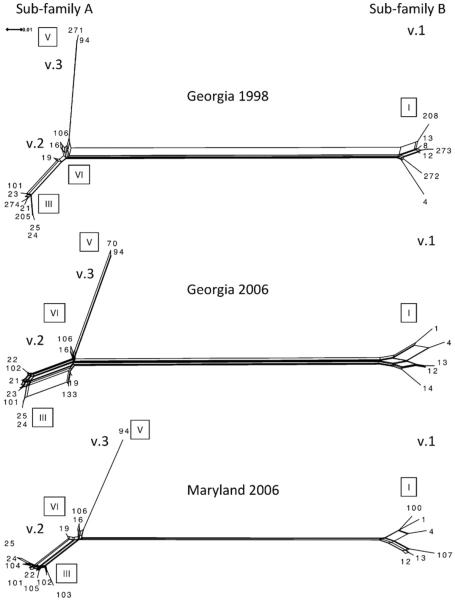
In fact, recombination at fHbp was evident among all 3 carriage populations. The correlation coefficients between physical distance and LD measures (r2 and D′) for all fHbp base pairs generated negative values for all 3 carriage populations (Table 5). These negative values indicate that LD decreases as physical distance increases, an observation consistent with intragenic recombination. In addition, pairwise homoplasy analysis showed statistically substantial evidence for recombination (p < 0.001) in all 3 populations (Table 5). SplitsTree analysis was performed to examine the phylogenetic relationships among the FHbp protein sequences from the different carriage isolate populations. This analysis revealed 3 distinct FHbp variants (Figs. 1 and 3). The network structure of the variant 1/subfamily B sequences was similar among the 3 different carriage populations. However, temporal differences in the networks were evident among the variant 2/subfamily A FHbp protein sequences (Fig. 3). These genetic changes contributed to the diversity of FHbp in the populations and among clonal complexes. For instance, in Georgia 1998, FHbp protein ID 205 and 274 contributed to clonal complex ST-162 allelic diversity in serogroup B carriage (Figs. 2 and 3). While FHbp protein ID 133 contributed to the diversity observed within clonal complex ST41/44 serogroup B isolates in GA 2006 (Figs. 2 and 3) and FHbp protein ID 104 contributes to clonal complex ST-23 serogroup Y diversity in Maryland 2006 (Figs. 2 and 3). These data highlight the importance of recombination as a mechanism to generate FHbp diversity within clonal complexes.
Table 5.
Correlation values between linkage disequilibrium measures (r2 and D') to physical distance (base pairs) and pairwise homoplasy indices (PHI) for fHbp sequences among 3 carriage populations. p values in parentheses.
| Georgia 1998 | Georgia 2006–2007 | Maryland 2006–2007 | |
|---|---|---|---|
| r 2 | −0.371521 (0.001) | −0.369781 (0.001) | −0.397309 (0.001) |
| D' | −0.105531 (0.001) | −0.0497506 (0.001) | −0.0882642 (0.001) |
| PHI | 0.04 (<0.001) | 0.06 (<0.001) | 0.08 (<0.001) |
Fig. 3.
Split decomposition analysis of FHbp protein sequences from 3 carriage isolate populations. Roman numerals in boxes indicated modular groups. Variant 1, 2 and 3 are v.1, v.2 and v.3, respectively. The scale represents the number of amino acid substitutions per site.
GARD analysis identified strong evidence for recombination at nucleotide sites: 291, 471–480 and 582–586 of the fHbp sequence alignments. These sites correspond to variable segment boundaries that define FHbp modular groups and support the hypothesis that recombination around variable segments results in FHbp diversity [8]. As evidence, natural chimeras of the 2 types of variable segments defining modular groups III and VI are apparent in the SplitsTree analysis for the variant 2/subfamily A FHbp (Fig. 3). Interestingly, the variant 1/subfamily B and variant 3/subfamily A FHbp are represented by single modular groups (I and V, respectively, Table 4 and Fig. 3). No natural chimeras (modular group IV) were evident among the variant 1/subfamily B carriage isolates (Table 4). And only chimeric modular group V was detected among carriage isolates bearing variant 3/subfamily A FHbp (Table 4 and Fig. 3). These data support the limited diversity of FHbp variant 1/subfamily B FHbp and variant 3/subfamily A observed among the carriage isolates by SplitsTree analysis.
3.5. FHbp vaccine candidates among carriage isolates
Of the 3 potential FHbp vaccine candidates – protein IDs 1, 45 and 55, there were only 3 isolates among the entire collection, with protein ID 1. These isolates were unencapsulated, serogroup B by SGS-PCR and belonged to the ST-32 clonal complex (Table 3B). There were no isolates with FHbp protein ID 45 or 55
4. Discussion
As highlighted by a decade of serogroup C conjugate vaccine use in the United Kingdom [31], understanding the potential role of new meningococcal vaccines on transmission dynamics of N. meningitidis is important for predicting vaccine effectiveness and introduction strategies. The effects of FHbp-based vaccines on meningococcal carriage are unknown. This study describes the prevalence, distribution and diversity of FHbp genotypes among a recent US carriage population. All carriage isolates amplified FHbp gene sequences. Overall, variant 2/subfamily A FHbp proteins were predominant among carriage isolates from Georgia and Maryland in 1998 and 2006–2007. While the majority of the carriage isolates in this study were unencapsulated, variant 2/subfamily A FHbp was common among encapsulated isolates. Serogroup B genetic lineages commonly associated with invasive disease including ST-32, ST-35, ST-162 and ST-269 were represented in this carriage population and the majority of these isolates harbored FHbp variant 2/subfamily A proteins. This finding is in contrast to investigations of invasive serogroup B disease isolates where the majority of FHbp belong to variant 1/subfamily B [16,34,35]. A recent report has shown increased FHbp subfamily A relative to subfamily B in serogroup B invasive disease from South Africa and more subfamily B in non-serogroup B invasive isolates [36]. All encapsulated carriage isolates in the present study with the exception of 2 serogroup C isolates and 6 serogroup Y isolates, harbored subfamily A FHbp. Most of these carriage isolates were collected from Georgia in 1998 when meningococcal carriage rates were high compared to the 2006–2007 time period.
The effect of FHbp vaccination on carriage cannot be easily predicted since little is known about mucosal immune responses to protein-based vaccines. In addition, both the expression and immunologic cross-reactivity of protein antigens must be considered as variations in both can have implications for vaccine coverage [37]. Recent clinical trials indicate that FHbp-based vaccines induce serum bactericidal responses in humans [18,38–40]. However, questions remain about long term immunity and the ability of FHbp vaccine candidates to provide cross protective immunity to a meningococcal population with a diverse FHbp repertoire. Limited cross-protection among variant 1/subfamily B FHbp antigens in a set of isogenic meningococcal strains that constitutively express different variant 1/subfamily B FHbp was observed in a recent study and the presence of additional antigens in the vaccine was necessary to provide broad protective immunity [41]. Based upon these results, vaccine coverage for this carriage population would be dependent upon immunity generated by additional antigens in the multi-component vaccine. Studies investigating the diversity of these additional vaccine antigens (NadA and NHBA) are ongoing and will provide further information regarding potential coverage. In addition, since expression of FHbp correlates with serum bactericidal activity, quantification of FHbp expression among these carriage isolates will also be important [42,43].
The SplitsTree analysis of FHbp amino acid sequences demonstrates that similar to invasive isolates, the diversity of FHbp among carriage isolates is influenced by genetic recombination. Substantial recombination at fHbp was evident in all 3 carriage populations investigated and most likely contributed to the allelic variation observed among some clonal complexes. In general, however, genetic lineage of carriage isolates was associated with particular FHbp protein IDs. Recently, a study of invasive serogroup B isolates found multiple FHbp protein IDs among clonal complexes [16]. Similarly, genetic lineages associated with serogroup B carriage isolates in this study (particularly ST-41/44 clonal complex) tended to show the most FHbp diversity.
In summary, the diversity of FHbp in meningococcal carriage isolates collected from Georgia and Maryland high school students from 2 time periods was primarily due to the prevalence of particular clonal complexes within the populations. Recombination at fHbp was evident and contributed to the antigenic diversity within some clonal complexes. Further investigations of meningococcal carriage populations from different locations and time periods are necessary to gain a better understanding of FHbp diversity and the potential impact of FHbp-based vaccines on meningococcal transmission and carriage.
Supplementary Material
Acknowledgements
This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley and Man-Suen Chan located at the University of Oxford [44]. The development of this site has been funded by the Wellcome Trust and European Union. We also thank Dan Granoff and Maria Brooks for helpful discussions, and Scott Kellerman for his assistance with data from the 1998 Georgia carriage study and David Blythe and Pat Ryan for their support of this study.
Funding This study was supported in part by the Centers for Diseases Control and Prevention. Sanofi Pasteur supported the 2006–2007 meningococcal carriage study through a grant to the CDC Foundation.
Dr. Harrison receives funding from the Centers for Disease Control and Prevention and the National Institute of Allergy and Infectious Diseases. He receives research support and lecture fees from Sanofi Pasteur; lecture fees from Novartis Vaccines; and has served as a consultant to GlaxoSmithKline, Novartis Vaccines, Sanofi Pasteur, and Pfizer. Ms. Shutt and Dr. Marsh receive research support from Sanofi Pasteur. Dr. Stephens has research funding from the National Institute of Allergy and Infectious Diseases, the Department of Veterans Affairs and the Georgia Research Alliance.
Footnotes
Financial disclosures Other authors: no disclosures.
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vaccine.2011.06.025.
References
- [1].Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009 Jun;27(Suppl. 2):B51–63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- [2].Finne J, Bitter-Suermann D, Goridis C, Finne U. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J Immunol. 1987 Jun;138(12):4402–7. [PubMed] [Google Scholar]
- [3].Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983 Aug;2(8346):355–7. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- [4].Granoff DM. Review of meningococcal group B vaccines. Clin Infect Dis. 2010 Mar;50(Suppl. 2):S54–65. doi: 10.1086/648966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004 Apr;72(4):2088–100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003 Mar;197(6):789–99. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schneider MC, Exley RM, Chan H, Feavers I, Kang YH, Sim RB, et al. Functional significance of factor H binding to Neisseria meningitidis. J Immunol. 2006 Jun;176(12):7566–75. doi: 10.4049/jimmunol.176.12.7566. [DOI] [PubMed] [Google Scholar]
- [8].Beernink PT, Granoff DM. The modular architecture of meningococcal factor H-binding protein. Microbiology. 2009 Sep;155(Pt 9):2873–83. doi: 10.1099/mic.0.029876-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Russell JE, Jolley KA, Feavers IM, Maiden MC, Suker J. PorA variable regions of Neisseria meningitidis. Emerg Infect Dis. 2004 Apr;10(4):674–8. doi: 10.3201/eid1004.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Trotter CL, Maiden MC. Meningococcal vaccines and herd immunity: lessons learned from serogroup C conjugate vaccination programs. Expert Rev Vaccines. 2009 Jul;8(7):851–61. doi: 10.1586/erv.09.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bjune G. “Herd immunity” and the meningococcal vaccine trial in Norway. Lancet. 1992 Aug;340(8814):315. doi: 10.1016/0140-6736(92)92411-8. [DOI] [PubMed] [Google Scholar]
- [12].Boslego J, Garcia J, Cruz C, Zollinger W, Brandt B, Ruiz S, et al. Efficacy, safety, and immunogenicity of a meningococcal group B (15:P1.3) outer membrane protein vaccine in Iquique, Chile, Chilean National Committee for Meningococcal Disease. Vaccine. 1995 Jun;13(9):821–9. doi: 10.1016/0264-410x(94)00037-n. [DOI] [PubMed] [Google Scholar]
- [13].Holmes JD, Martin D, Ramsay C, Ypma E, Oster P. Combined administration of serogroup B meningococcal vaccine and conjugated serogroup C meningococcal vaccine is safe and immunogenic in college students. Epidemiol Infect. 2008 Jun;136(6):790–9. doi: 10.1017/S0950268807009211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Perkins BA, Jonsdottir K, Briem H, Griffiths E, Plikaytis BD, Hoiby EA, et al. Immunogenicity of two efficacious outer membrane protein-based serogroup B meningococcal vaccines among young adults in Iceland. J Infect Dis. 1998 Mar;177(3):683–91. doi: 10.1086/514232. [DOI] [PubMed] [Google Scholar]
- [15].Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci USA. 2006 Jul;103(29):10834–9. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Murphy E, Andrew L, Lee KL, Dilts DA, Nunez L, Fink PS, et al. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis. 2009 Aug;200(3):379–89. doi: 10.1086/600141. [DOI] [PubMed] [Google Scholar]
- [17].Seib KL, Oriente F, Adu-Bobie J, Montanari P, Ferlicca F, Giuliani MM, et al. Influence of serogroup B meningococcal vaccine antigens on growth and survival of the meningococcus in vitro and in ex vivo and in vivo models of infection. Vaccine. 2010 Mar;28(12):2416–27. doi: 10.1016/j.vaccine.2009.12.082. [DOI] [PubMed] [Google Scholar]
- [18].Keiser PB, Biggs-Cicatelli S, Moran EE, Schmiel DH, Pinto VB, Burden RE, et al. A phase 1 study of a meningococcal native outer membrane vesicle vaccine made from a group B strain with deleted lpxL1 and synX, over-expressed factor H binding protein, two PorAs and stabilized OpcA expression. Vaccine. 2011 Feb;29(7):1413–20. doi: 10.1016/j.vaccine.2010.12.039. [DOI] [PubMed] [Google Scholar]
- [19].Dolan-Livengood JM, Miller YK, Martin LE, Urwin R, Stephens DS. Genetic basis for nongroupable Neisseria meningitidis. J Infect Dis. 2003 May;187(10):1616–28. doi: 10.1086/374740. [DOI] [PubMed] [Google Scholar]
- [20].Kellerman SE, McCombs K, Ray M, Baughman W, Reeves MW, Popovic T, et al. Genotype-specific carriage of Neisseria meningitidis in Georgia counties with hyper- and hyposporadic rates of meningococcal disease. J Infect Dis. 2002 Jul;186(1):40–8. doi: 10.1086/341067. [DOI] [PubMed] [Google Scholar]
- [21].Clark T, Stern E, Pondo T, Arnold K, Harrison L, Vello M, et al. The effect of quadrivalent (A, C, Y,W-135) meningococcal conjugate vaccine on serogroup-specific carriage of Neisseria meningitidis. 16th International pathogenic neissera conference.2008. [Google Scholar]
- [22].Mothershed EA, Sacchi CT, Whitney AM, Barnett GA, Ajello GW, Schmink S, et al. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol. 2004 Jan;42(1):320–8. doi: 10.1128/JCM.42.1.320-328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schuchat A, Hilger T, Zell E, Farley MM, Reingold A, Harrison L, et al. Active bacterial core surveillance of the emerging infections program network. Emerg Infect Dis. 2001 Jan-Feb;7(1):92–9. doi: 10.3201/eid0701.010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang X, Cohn A, Comanducci M, Andrew L, Zhao X, Macneil JR, et al. Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine. 2011 May; doi: 10.1016/j.vaccine.2011.04.092. [DOI] [PubMed] [Google Scholar]
- [25].Harrison LH, Jolley KA, Shutt KA, Marsh JW, O'Leary M, Sanza LT, et al. Antigenic shift and increased incidence of meningococcal disease. J Infect Dis. 2006 May;193(9):1266–74. doi: 10.1086/501371. [DOI] [PubMed] [Google Scholar]
- [26].Harrison LH, Shutt KA, Schmink SE, Marsh JW, Harcourt BH, Wang X, et al. Population structure and capsular switching of invasive Neisseria meningitidis isolates in the pre-meningococcal conjugate vaccine era—United States, 2000–2005. J Infect Dis. 2010 Apr;201(8):1208–24. doi: 10.1086/651505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Katz LS, Bolen CR, Harcourt BH, Schmink S, Wang X, Kislyuk A, et al. Meningococcus genome informatics platform: a system for analyzing multilocus sequence typing data. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W606–11. doi: 10.1093/nar/gkp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Brehony C, Wilson DJ, Maiden MC. Variation of the factor H-binding protein of Neisseria meningitidis. Microbiology. 2009 Dec;155(Pt 12):4155–69. doi: 10.1099/mic.0.027995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Goodman LA, Kruskal WH. Measures of association for cross classifications. J Am Stat Assoc. 1954;49:732–64. [Google Scholar]
- [30].Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007 Aug;24(8):1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- [31].Wilson DJ, McVean G. Estimating diversifying selection and functional constraint in the presence of recombination. Genetics. 2006 Mar;172(3):1411–25. doi: 10.1534/genetics.105.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006 Feb;23(2):254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- [33].Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006 Oct;23(10):1891–901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- [34].Lucidarme J, Comanducci M, Findlow J, Gray SJ, Kaczmarski EB, Guiver M, et al. Characterization of fHbp, nhba (gna2132), nadA, porA, sequence type (ST), and genomic presence of IS1301 in group B meningococcal ST269 clonal complex isolates from England and Wales. J Clin Microbiol. 2009 Nov;47(11):3577–85. doi: 10.1128/JCM.00936-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bambini S, Muzzi A, Olcen P, Rappuoli R, Pizza M, Comanducci M. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine. 2009 May;27(21):2794–803. doi: 10.1016/j.vaccine.2009.02.098. [DOI] [PubMed] [Google Scholar]
- [36].Mothibeli KM, du Plessis M, von Gottberg A, Murphy E, Hoiseth SK, Zlotnick G, et al. Distribution of factor H binding protein beyond serogroup B: variation among five serogroups of invasive Neisseria meningitidis in South Africa. Vaccine. 2011 Mar;29(11):2187–92. doi: 10.1016/j.vaccine.2010.11.072. [DOI] [PubMed] [Google Scholar]
- [37].Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch C, et al. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci USA. 2010 Nov;107(45):19490–5. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dull PM, Pizza M, Toneatto D, DeTora L, Ypma E, Kleinschmidt A, et al. Early clinical development of a novel, multicomponent meningococcal serogroup B vaccine (4CMenB). International Pathogenic Neisseria Conference.2010. [Google Scholar]
- [39].Findlow J, Borrow R, Snape MD, Dawson T, Holland A, John TM, et al. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant Meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin Infect Dis. 2010 Nov;51(10):1127–37. doi: 10.1086/656741. [DOI] [PubMed] [Google Scholar]
- [40].Richmond P, Marshall H, Sheldon E, Jiang Q, Anderson A, Jansen KU, et al. Safety & immunogenicity of serogroup B Neisseria meningitidis (MnB) rLP2086 vaccine in adults and adolescent submects: overview of 3 clinical trials. International pathogenic neisseria conference.2010. [Google Scholar]
- [41].Brunelli B, Del Tordello E, Palumbo E, Biolchi A, Bambini S, Comanducci M, et al. Influence of sequence variability on bactericidal activity sera induced by Factor H binding protein variant 1.1. Vaccine. 2011 Jan;29(5):1072–81. doi: 10.1016/j.vaccine.2010.11.064. [DOI] [PubMed] [Google Scholar]
- [42].Jiang HQ, Hoiseth SK, Harris SL, McNeil LK, Zhu D, Tan C, et al. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine. 2010 Aug;28(37):6086–93. doi: 10.1016/j.vaccine.2010.06.083. [DOI] [PubMed] [Google Scholar]
- [43].Pajon R, Beernink PT, Harrison LH, Granoff DM. Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates. Vaccine. 2010 Feb;28(9):2122–9. doi: 10.1016/j.vaccine.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jolley KA. Internet-based sequence-typing databases for bacterial molecular epidemiology. Methods Mol Biol. 2009;551:305–12. doi: 10.1007/978-1-60327-999-4_21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.