Abstract
Recent genome- and epigenome-wide studies demonstrate that the DNA methylation is controlled in part by genetics, highlighting the importance of integrating genetic and epigenetic data. To better understand molecular mechanisms affecting gene expression, we used the candidate susceptibility gene paraoxonase 1 (PON1) as a model to assess associations of PON1 genetic polymorphisms with DNA methylation and arylesterase activity, a marker of PON1 expression. PON1 has been associated with susceptibility to obesity, cardiovascular disease, and pesticide exposure. In this study, we assessed DNA methylation in 18 CpG sites located along PON1 shores, shelves, and its CpG island in blood specimens collected from newborns and 9-year-old children participating (n = 449) in the CHAMACOS birth cohort study. The promoter polymorphism, PON1-108, was strongly associated with methylation, particularly for CpG sites located near the CpG island (P << 0.0005). Among newborns, these relationships were even more pronounced after adjusting for blood cell composition. We also observed significant decreases in arylesterase activity with increased methylation at the same nine CpG sites at both ages. Using causal mediation analysis, we found statistically significant indirect effects of methylation (β(95% confidence interval): 6.9(1.5, 12.4)) providing evidence that DNA methylation mediates the relationship between PON1-108 genotype and PON1 expression. Our findings show that integration of genetic, epigenetic, and expression data can shed light on the functional mechanisms involving genetic and epigenetic regulation of candidate susceptibility genes like PON1.
Keywords: methylation, arylesterase, paraoxonase, mediation
Background
Epigenetic modifications, including DNA methylation, chromatin modifications, and non-coding RNAs, are heritable and affect gene expression without a change in DNA sequence [1, 2]. Because of their impact on gene expression, there is a growing recognition that epigenetics may play an important role in key biological processes and mechanisms of disease development [3–5]. DNA methylation is an important epigenetic mechanism and is also the most commonly measured epigenetic mark. Available methodologies for the measurement of DNA methylation are relatively high throughput and cost-effective, allowing for its use in population studies. Interest in DNA methylation has grown rapidly in recent years resulting in the publication of numerous studies examining associations of DNA methylation with environmental exposures as well as adverse health outcomes in human populations [6–9].
In genetic epidemiology, concepts of genetic architecture such as the existence of haplotype blocks and linkage disequilibrium (LD) transformed the way in which genetic association studies were designed, analyzed, and interpreted [10]. Epigenome-wide data emerging from recent studies are now providing analogous information on the methylome that can further inform epigenetic analysis for population studies. For instance, relationships of methylation at CpG sites in the same gene are often performed as one CpG site at a time. However, recent studies have shown that methylation at nearby CpG sites can be highly correlated with each other, although over shorter distances than observed with single-nucleotide polymorphisms (SNPs) in the genome [11, 12]. Furthermore, methylation may be partially controlled by genetics [11, 12]. Therefore, to understand the molecular mechanisms explaining how DNA methylation can affect health through gene expression changes, it will be important in population studies to integrate data on DNA methylation with data on genetic polymorphisms and gene expression [13, 14].
Here, we use paraoxonase 1 (PON1) as a model for integrating different types of molecular data that can be used to understand the role of epigenetics on gene expression. PON1 is a multifunctional enzyme involved in oxidant defense [15], and its genetic variants and lower levels have been linked to adverse health outcomes including adverse neurodevelopment, cardiovascular disease, and obesity [16–22]. We previously studied PON1 variability in Mexican-American children and their mothers from the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS) birth cohort and found a much larger than previously known (>100-fold) range of levels and substrate-specific activities defined in part by age and genotype [23, 24]. This broad variability of PON1 within populations suggests differential susceptibility among individuals. PON1 sequencing and haplotype analyses in this cohort have shown that the PON1 promoter polymorphism (PON1-108; rs705379) is the strongest known predictor of PON1 gene expression and protein levels. Even so, it explains less than 25% of the variability of PON1 protein levels; furthermore, incorporation of other genetic variants explained <3% of additional variability. This means that other factors beyond genetics, including epigenetics, may contribute to modulation of PON1 gene expression.
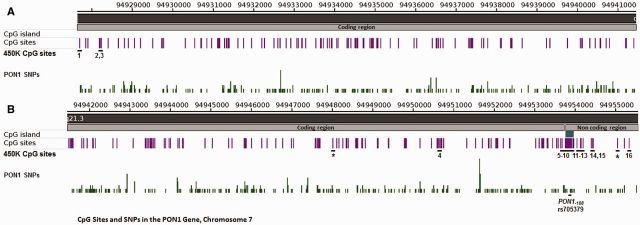
There are a total of 287 CpG sites located in the PON1 gene including one CpG island in the promoter region comprising 19 CpG sites (Fig. 1). Beyond the CpG island, there are an additional 66, 48, and 146 CpG sites within shores, shelves, and open sea regions, respectively. One recent study showed that a PON1 SNP located in a miRNA binding site (miR-616) was associated both with changes in PON1 expression and increased risk of ischemic stroke and carotid atherosclerosis [25]. These data underscore the vital influence of epigenetic marks like miRNA and DNA methylation on PON1 and demonstrate further the clinical significance of PON1 variability [26]. To our knowledge, few studies of PON1 epigenetics in relation to molecular phenotype have been reported [27, 28].
Figure 1.
CpG sites and SNPs in the PON1 gene. This map of PON1 spans chromosome 7 from coordinates 94 927 671 to 94 941 000 in (A) and 94 941 000 to 94 955 500 in (B). It shows all 287 CpG sites and the subset of CpG sites included in the 450K BeadChip assay. There is one CpG island in the non-coding region (B) and the distribution of SNPs across the gene is also shown
The purpose of this study is to determine the relationship of DNA methylation in the PON1 gene with PON1 genetic polymorphisms and with gene expression at the protein level in CHAMACOS children. In addition to expanding molecular characterization of PON1 variability to epigenetics, our data can serve as a model for integrating genetic, epigenetic, and expression data on candidate susceptibility genes.
Results
PON1 CpG Sites
We used the data from a 450K BeadChip array to assess CpG sites located in the PON1 gene in blood specimens collected from 449 children. Samples were assessed at two time points, once at birth (n = 378) and again at the age of 9 years (n = 247). The 450K Bead Chip array included 18 of the 287 PON1 CpG sites, which are described in Table 1 and shown in Fig. 1. The majority of CpG sites interrogated were located in the promoter region and included several sites in the CpG island (n = 4), shores (n = 9), and shelves (n = 1).
Table 1.
Summary of PON1 CpG sites assessed by 450K Beadchip
| CpG site number | Illumina 450K ID | Coordinatesa | Relation to CpG island | Distance from PON1-108 SNP |
|---|---|---|---|---|
| 1 | cg22798737 | 94 927 713 | Open sea | 26 188 |
| 2 | cg01879893 | 94 928 193 | Open sea | 25 709 |
| 3 | cg09416203 | 94 928 249 | Open sea | 25 652 |
| — | cg05433222b | 94 948 020 | Open sea | 5881 |
| 4 | cg24062571 | 94 950 658 | N_Shelf | 3244 |
| 5 | cg07404485 | 94 953 654 | N_Shore | 248 |
| 6 | cg05342682 | 94 953 680 | N_Shore | 221 |
| 7 | cg04155289 | 94 953 771 | Island | 131 |
| 8 | cg19678392 | 94 953 811 | Island | 91 |
| 9 | cg21856205 | 94 953 878 | Island | 24 |
| 10 | cg17330251 | 94 953 956 | Island | 55 |
| 11 | cg01874867 | 94 954 059 | S_Shore | 158 |
| 12 | cg20119798 | 94 954 144 | S_Shore | 243 |
| 13 | cg04871131 | 94 954 202 | S_Shore | 301 |
| 14 | cg23055772 | 94 954 438 | S_Shore | 537 |
| 15 | cg07809369 | 94 954 455 | S_Shore | 554 |
| — | cg17020263b | 94 955 053 | S_Shore | 1152 |
| 16 | cg15887283 | 94 955 348 | S_Shore | 1447 |
aReference hg 19, Chromosome 7.
bThese CpG sites contained SNPs, so their methylation data were not included.
PON1 Methylation
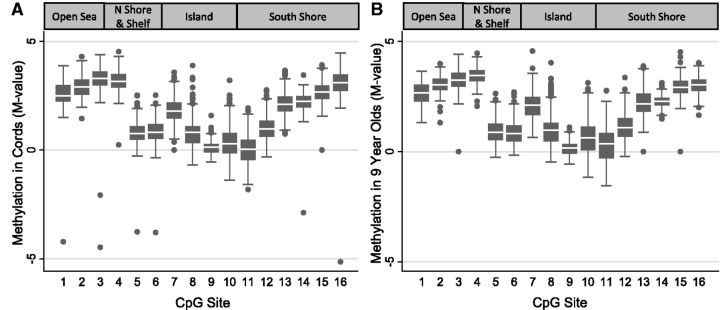
Levels of DNA methylation in newborns and 9-year-old children are shown graphically in Fig. 2. At both ages, average methylation levels (expressed as M values, see Methods) ranged from 2.1 to 3.4 among sites (Sites 1–4 and 14–16) that were further away from the transcription start site (TSS) and were much lower (0.03–2.11) among sites closer in proximity to the TSS (Sites 5–13). At the majority of individual CpG sites, methylation levels were slightly higher at the age of 9 years; these differences by age were no longer statistically significant after adjusting for cell composition in generalized estimating equation models with the exception of one CpG site (Site 4, cg 24062571). For Site 4, we observed slightly higher methylation at the age of 9 years, and this difference persisted after adjusting for cell composition.
Figure 2.
Methylation box plots in newborns (n = 378) and 9-year-old (n = 247) children. DNA methylation was measured using 450K BeadChip. The box plots show methylation levels at 16 CpG sites in the PON1 gene in (A) umbilical cord blood and (B) 9-year-old blood. CpG site numbers correspond to those listed in Table 1
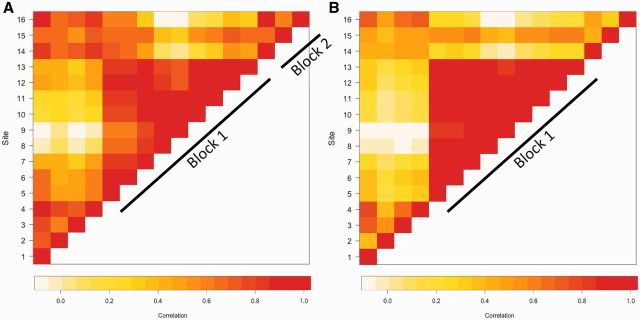
It is well documented in genetic epidemiology that SNPs in close proximity to each other are highly correlated with each other, forming LD blocks that tend to be inherited together [10]. Although less is known about epigenetic architecture, there is a growing understanding that nearby CpG sites may also be correlated with each other [11, 12]. Therefore, we examined correlations among PON1 CpG sites in newborns and in 9-year olds (Fig. 3). Among CpG sites in umbilical cord blood, we identified two methylation blocks (see Methods for definition) containing clusters of highly correlated sites (Fig. 3A). Block 1 contained sites in close proximity to the TSS (Sites 5–13) and spanned 350 base pairs; we will refer to this as the central cluster of PON1 CpG sites or Block 1. Correlation coefficients (r) for CpG site pairs in Block 1 ranged from 0.53 to 0.98. Block 2 (Sites 14–16) spanned 1000 base pairs in length and correlation coefficients ranged from 0.55 to 0.85. Interestingly, sites from Block 2 were also correlated with Sites 1–4 (r ranged from 0.42 to 0.88), and we subsequently refer to these sites (Block 2 and Sites 1–4) as the outer cluster of CpG sites. Among 9-year-old children (Fig. 3B), we observed only one block consisting of Sites 5–13 with correlation coefficients ranging from 0.76 to 0.98. Although sites were again correlated with each other among Sites 14–16, their correlations were not high enough to meet the definition of a methylation block (r2 < 0.4 for most CpG site pairs).
Figure 3.
Correlation plots of PON1 CpG sites in (A) umbilical cord blood and (B) 9-year-old blood. The colors of the squares represent the correlation between CpG sites (r). Squares are white/yellow if CpG sites are not correlated (r = 0) and red if they are strongly correlated (r ∼ 1). Similar to the concept of LD between SNPs, CpG sites within close proximity of each other tend to be correlated with each other. Here, we identified two methylation blocks (Sites 5–13 and 14–16) where methylation levels were strongly correlated with each at both ages. Block 1 is referred to as the inner cluster of CpG sites, whereas Sites 1–4 and 14–16 are referred to as the outer cluster of CpG sites. CpG site numbers correspond to those listed in Table 1
Relationships of PON1 Genotype with PON1 Methylation
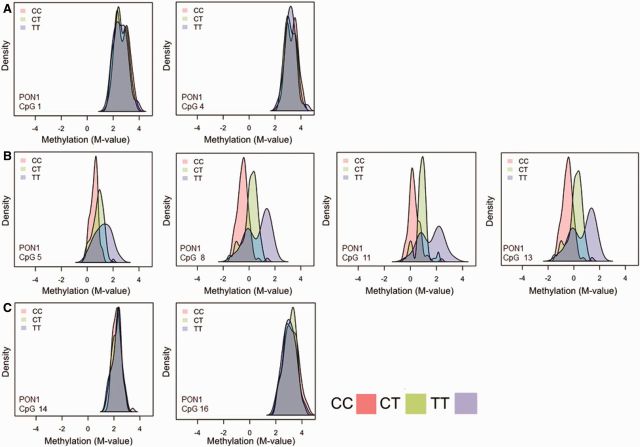
In agreement with other studies [29, 30] we previously found the promoter polymorphism at position -108 (PON1-108) to be the most significant predictor of PON1 expression as measured by arylesterase (AREase) activity in the CHAMACOS cohort [24, 31]. As methylation can also affect gene expression, we examined whether this promoter SNP was associated with methylation at the 16 CpG sites assessed in CHAMACOS newborns and 9-year olds. Figure 3 shows the differences in methylation by PON1-108 genotype. The methylation density plots demonstrate little to no difference in methylation in the outer cluster sites (Fig. 4A and C). However, among sites in methylation Block 1 (Fig. 4B), there is a clear separation by genotype, providing evidence of allele-specific methylation (ASM). Those with the CC genotype, which has been associated with higher levels of gene expression [30], had the lowest levels of methylation, whereas those with the CT genotype had intermediate levels of methylation and those with TT genotypes had the highest levels of methylation. Furthermore, variance was larger within the TT group compared with the CT and CC groups. Similar patterns were observed in 9-year-old children (data not shown).
Figure 4.
Methylation density plots by PON1-108 genotype (CC-pink CT – green TT –purple). Each plot shows density by methylation M-value at eight different CpG sites, from different regions of PON1. Plots in Row A are CpG Sites 1 and 4, respectively. Plots in Row B are from methylation Block 1 (Sites 5, 8, 11, and 13). Plots in Row C show Sites 14 and 16 from methylation Block 2. We include plots from just a few CpG sites that are representative of other CpG sites in the same region. Among CHAMACOS newborns with methylation data (n = 271), 83 were CC, 138 were CT, and 50 were TT. Sites from methylation Block 1 show a distinct separation by PON1-108 genotype
Regression models of PON1-108 genotype with methylation (Table 2) also showed the same trends. We found very strong positive associations of the number of PON1-108T alleles with methylation levels, particularly those in methylation Block 1 (P values ranged from 1.34E-11 to 2.73E-30). For instance, on average, children with the CT genotype had methylation M values that were 0.63 units higher than CC kids at CpG Site 8 located in the CpG island. These associations remain significant even after Bonferonni correction for multiple testing. For Block 1 sites, the variation of methylation explained by PON1-108 genotype as determined by r2 of the regression models ranged from 17 to 39% in cord bloods. As methylation profiles can differ by cell type in blood, we also examined the relationship of PON1-108 genotype adjusting for cell composition in a subset of 82 newborns with differential cell count data (Supplementary Table S1). Results were very similar to unadjusted models; however, the variance of methylation explained by PON1 genotype doubled for Block 1 sites (38–76%). The same but even stronger trends were also seen in 9-year-old children (P values for Block 1 sites ranged from 6.28E-33 to 7.55E-50 and r2 ranged from 0.43 to 0.68); models adjusting for cell composition were very similar to unadjusted models (Table 2 and Supplementary Table S1).
Table 2.
Association of PON1-108 genotypea with DNA methylation in blood in newborns and 9-year olds
| Newborns |
9-Year Olds |
|||||||
|---|---|---|---|---|---|---|---|---|
| CpG Site | CpG site id | Relation to island | N | Beta (95% CI) | P | N | Beta (95% CI) | P |
| 1 | cg22798737 | Open sea | 271 | −0.06(−0.14,0.03) | 0.20 | 218 | 0.00(−0.07,0.06) | 0.97 |
| 2 | cg01879893 | Open sea | 271 | −0.04(−0.12,0.03) | 0.23 | 218 | 0.00(−0.07,0.07) | 0.90 |
| 3 | cg09416203 | Open sea | 271 | −0.08(−0.15, −0.01) | 0.03 | 218 | −0.03(−0.11,0.05) | 0.42 |
| 4 | cg24062571 | N_Shelf | 271 | −0.05(−0.12,0.02) | 0.18 | 218 | −0.01(−0.07,0.06) | 0.86 |
| 5 | cg07404485 | N_Shore | 271 | 0.32(0.24,0.39) | 8.28E-15 | 218 | 0.51(0.45,0.58) | 6.58E-37 |
| 6 | cg05342682 | N_Shore | 271 | 0.32(0.25,0.40) | 5.00E-15 | 218 | 0.52(0.45,0.58) | 5.54E-35 |
| 7 | cg04155289 | Island | 271 | 0.33(0.24,0.42) | 1.34E-11 | 218 | 0.59(0.51,0.67) | 6.28E-33 |
| 8 | cg19678392 | Island | 271 | 0.63(0.54,0.73) | 4.13E-30 | 218 | 0.92(0.82,1.02) | 5.73E-45 |
| 9 | cg21856205 | Island | 271 | 0.26(0.22,0.30) | 2.73E-30 | 218 | 0.35(0.31,0.39) | 4.71E-46 |
| 10 | cg17330251 | Island | 271 | 0.63(0.52,0.74) | 2.15E-25 | 218 | 0.95(0.85,1.04) | 4.56E-48 |
| 11 | cg01874867 | S_Shore | 271 | 0.59(0.49,0.69) | 8.59E-25 | 218 | 0.92(0.82,1.01) | 7.55E-50 |
| 12 | cg20119798 | S_Shore | 271 | 0.38(0.30,0.47) | 2.14E-16 | 218 | 0.69(0.61,0.77) | 3.77E-40 |
| 13 | cg04871131 | S_Shore | 271 | 0.26(0.17,0.34) | 8.04E-09 | 218 | 0.52(0.44,0.61) | 1.55E-24 |
| 14 | cg23055772 | S_Shore | 271 | −0.01(−0.06,0.05) | 0.76 | 218 | 0.00(−0.05,0.05) | 0.86 |
| 15 | cg07809369 | S_Shore | 271 | 0.05(−0.04,0.13) | 0.31 | 218 | 0.20(0.12,0.28) | 1.29E-06 |
| 16 | cg15887283 | S_Shore | 271 | −0.06(−0.14,0.03) | 0.18 | 218 | −0.02(−0.09,0.05) | 0.64 |
CI, confidence interval.
a PON1-108 genotype was coded as 0, 1, and 2 T alleles for CC, CT, and TT children, respectively.
Beyond the PON1-108 SNP, we also examined associations of PON1 methylation sites with an additional panel of 40 SNPs and indels distributed throughout the PON1 gene and 4 SNPs located in PON2 and PON3 in a smaller subset of 214 newborns and 157 9-year-old children (Supplementary Tables S2 and S3). As expected, methylation in Block 1 CpG sites were associated with additional SNPs located in the promoter region. These SNPs were all in strong LD with PON1-108. We also observed weaker but significant associations of several other SNPs with methylation of Block 1 CpG sites, including several located further downstream (>2 kb) of PON1 in the PON2 and PON3 genes and two SNPs that were in strong LD with PON155, a common coding SNP.
Relationships of PON1 Methylation with AREase Activity
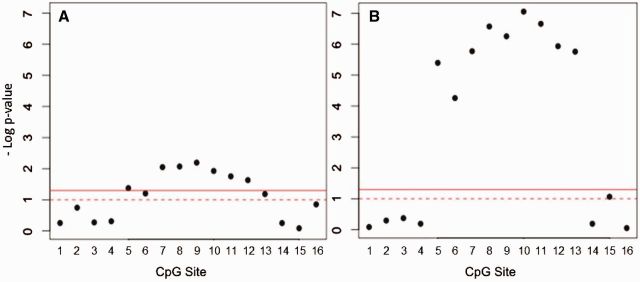
Using AREase as a marker of gene expression at the protein level, we examined associations of PON1 methylation at each of the 16 CpG sites with AREase activity (Fig. 5A, Supplementary Table S4). At both ages, methylation at individual Block 1 CpG sites was significantly associated with AREase activity, and this relationship was noticeably stronger at the age of 9 years (Table 2, Fig. 5B). This may be due to the higher mean, range, and variance of AREase activity observed in 9-year-old children compared with newborns who have very low levels of PON1 protein levels at birth. Effects of cell composition may also contribute to this difference. After adjustment for cell composition in a subset of cord bloods, we observed trends in the same direction but the magnitude of the associations increased and P values were decreased. Results were essentially unchanged in 9-year-old children suggesting that the influence of cell composition on these associations was much stronger in cord blood (Supplementary Table S5).
Figure 5.
Relationship of methylation with AREase activity. The plot shows regression P values for modelling the association of methylation at each individual CpG Site with AREase activity in (A) newborns and (B) 9-year olds. The solid line indicates a P value of 0.05 and the dotted line represents a P value of 0.01
To consider the methylation at multiple correlated CpG sites in one model, we also used PCs analysis. Two PCs explained >95% of the variability of methylation in Block 1, and these PCs were included in the models of AREase activity (Table 3). Both PCs describing cord blood methylation from Block 1 (PC1-1 and PC1-2) were significantly associated with cord AREase activity. In 9-year-old children, only PC1-1 was significantly associated with AREase activity.
Table 3.
PCs regression models of DNA methylation in blood with AREase activity in children
| Newbornsa |
9-Year oldsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| estimate | SE | P | r 2 | estimate | SE | P | r 2 | ||
| Block 1 CpG sites | pc1-1 | 1.31 | 0.62 | 0.035 | 5.84 | 1.09 | 2.78E-07 | ||
| pc1-2 | −8.19 | 2.61 | 0.002 | 0.06 | −8.20 | 6.10 | 0.181 | 0.15 | |
SE, standard error.
aThe newborn model examines association of cord blood methylation with cord AREase activity. The 9-year model examines the association of 9-year-old blood methylation with 9-year AREase activity.
Mediation of PON1-108 Genotype on AREase Activity by PON1 Methylation
Because PON1-108 was the SNP most strongly associated with methylation and expression and prior studies suggest epigenetics can at times mediate effects of genetics on expression, we used the PARAMED module [32, 33] to determine to what extent PON1 methylation mediates the effects of PON1-108 genotype on AREase activity (Table 4). PARAMED relies on causal inference methodologies to estimate the natural direct effect and NIE in the presence of exposure-mediator interactions. We limited this analysis only to the PCs that were significantly associated with AREase activity (PC1-1 and 2 for newborns and PC1-1 for 9-year olds). Among newborns, there was a significant interaction between PON1 genotype and both PCs of Block 1 methylation. Furthermore, the NIE of the first PC of Block 1 methylation (PC1-1) on expression was statistically significant (β(95% confidence interval) = 6.91(1.45,12.37); P = 0.013) providing evidence that methylation in Block 1 mediates the effect of PON1-108 genotype on AREase activity. The NIE for PC1-2 was not statistically significant, suggesting that some portion of the relationship between PON1-108 genotype and AREase is not driven by methylation. Among 9-year olds, we did not observe a significant interaction of genotype and methylation on AREase activity. Additionally, the NIE for PC1-1 did not reach statistical significance. Overall, our mediation analyses suggest that at least in newborns, PON1 methylation in Block 1 mediates the association of PON1-108 genotype on AREase activity.
Table 4.
Mediation analysis: total, direct, and indirect effect
| Newborns |
9-Year olds |
|||||
|---|---|---|---|---|---|---|
| PC1-1 |
PC1-2 |
PC1-1 |
||||
| Estimate (95% CI) | P | Estimate (95% CI) | P | Estimate (95% CI) | P | |
| Total effect | 16.3(−22.1, −10.5) | <0.0005 | −15.6(−21.5, −9.7) | <0.0005 | −30.6(−42.9, −18.3) | <0.0005 |
| Natural direct effect | −23.2(−30.8, −15.6) | <0.0005 | −19.1(−26.3, −11.9) | <0.0005 | −15.6(−35.3,4.19) | 0.123 |
| NIE | 6.9(1.5,12.4) | 0.013 | 3.5(−1.8,8.9) | 0.195 | −15.0(−30.9,0.83) | 0.063 |
CI, confidence interval.
Discussion
In this study, we used PON1 as a model for integration of genetic and epigenetic data and their relationship with gene expression. We expanded our molecular studies of PON1 to DNA methylation at 16 CpG sites in a large cohort of young Mexican-American children from the Salinas Valley, CA. We found evidence of allele-specific methylation between the PON1-108 SNP and methylation sites belonging to methylation Block 1 surrounding the promoter region, demonstrating a genetic contribution to DNA methylation. Furthermore, methylation levels at this Block 1 were strongly associated with AREase activity, a measure of PON1 protein quantity in blood. Mediation analyses provided evidence that methylation can act as a mediator of the PON1-108 SNP on AREase activity
To our knowledge, only one other study has examined associations of PON1 methylation with AREase activity. Similar to our results, de la Iglesia et al. [28] also reported an inverse relationship between methylation at CpG Sites 5–8 and AREase activity in white blood cell specimens collected from 47 obese adults. Our study, relying on a much larger sample size (n = 449) demonstrated that similar relationships also exist at birth and during childhood and found that some of these relationships may be driven by genetics.
Few if any studies that have focused on PON1 have examined associations of genetic polymorphisms with PON1 methylation. However, we were able to extract publicly available data on PON1 from a handful of studies that have taken genome- and epigenome-wide approaches to explore patterns of methylation and relationships with genetics. The same methylation block that we identified in CHAMACOS children (Sites 4–12, Block 1) was also identified as a methylation cluster in two other studies, one a study of whole blood specimens from adults (n = 247) and the other a smaller study of fetal (n = 14) and adult (n = 181) liver samples [11, 34]. In line with our findings, the study of liver specimens demonstrated inverse correlations between PON1 methylation at Sites 4–12 and PON1 gene expression (mRNA) [34] and also observed significant associations of PON1-108 genotype with methylation at the same sites. Our study showed that similar relationships can be observed in children’s blood specimens and when we consider PON1 expression at the protein level (AREase activity)—the final step of the flow of genetic information described in the central dogma of molecular biology.
There is growing interest in the phenomenon of ASM such as that observed between PON1 methylation in the promoter region and PON1-108 genotype. ASM is a hallmark feature of imprinted genes but recent epigenome-wide studies have shown that the most prevalent types of ASM are those where genotype affects DNA methylation in cis among non-imprinted genes [12, 35, 36]. A recent study in blood specimens reported a prevalence of ASM that was cis in nature in 8.1% of heterozygous SNPs and that 22% of the genes exhibiting ASM also displayed allele-specific gene expression, as we observed with PON1 [36]. ASM is a particularly interesting phenomenon because it provides a functional link between genetic polymorphisms and phenotypic differences beyond nonsynonymous SNPs in coding regions [35]. Furthermore, it has been suggested that regions characterized by ASM may be more susceptible to environmental influences [37].
Mechanisms regulating methylation patterns in promoter regions are not well understood although several models through which regulation may occur have recently been proposed [38]. The strong association of the PON1-108 genotype with both methylation and expression is particularly interesting because it serves as an example of genetic regulation of methylation and expression at a transcription factor binding site. Previous studies have demonstrated that this polymorphism is located in a specificity protein 1 (Sp1) binding site [30, 39], which likely activates transcription via methylation. Sp1 binding blocks DNA methyltransferases (DNMTs) from accessing the promoter, leading to lower methylation levels and thus more transcription [38, 40]. The Sp1 binding site at position 108 can be disrupted by the T allele, which in CHAMACOS children was associated both with increased methylation levels and lower expression as measured by AREase activity. Furthermore, results from our mediation analyses suggest that the effect of the PON1-108 polymorphisms on expression is mediated in part by methylation at methylation Block 1.
Although we observed very strong associations between the PON1-108 genotype, DNA methylation, and AREase activity in a large cohort of CHAMACOS children, this study has some limitations. We used AREase activity, a measure of protein levels in blood, as a proxy for gene expression. It is possible, however, that AREase may be more reflective of liver expression as PON1 present in plasma is primarily synthesized in the liver [41]. Relationships observed between blood methylation and AREase activity were strikingly similar to those reported between methylation and mRNA expression in blood and liver samples, possibly because ASM events may not be tissue specific. One study recently observed a concordance of ASM between tissue types and hypothesized that this may be due either to “shared gene regulatory events that occur early in development or an inherent property of DNA sequence that directly affects the propensity of DNA methylation” [36]. Additionally, as the 450 K BeadChip array assays only a select number of CpG sites throughout the methylome, we were only able to interrogate 18 of the 287 CpG sites located in the PON1 gene. Although the coverage of sites in and around the promoter region was quite good, future studies would benefit from a more complete assessment of the PON1 CpG sites across the entire gene using a higher resolution methodology like MethylSeq. Another potential source of bias is related to missingness of biological specimens, which is common in cohort studies utilizing pediatric blood samples. Only 65–71% of participants included in this study had adequate blood volumes for AREase analysis. This could limit generalizability of the relationships that we observed. When we compared children included in our study with all other children in the cohort, however, we did not find that they differed by significant demographic or exposure factors. Finally, our data on differential cell count in cord blood specimens were limited to only a subset of participants. It is still possible that some amount of residual confounding was not accounted for in the models that did not adjust for cell composition. Since adjusting for cell composition did not substantially change results, however, the unmeasured residual confounding is likely small.
Conclusions
Recent genome- and epigenome-wide studies are providing new data showing emerging patterns of a complex interplay between genetics and epigenetics on gene expression that may affect health [11, 12]. Here, we applied some of the new understanding of epigenomic architecture, relationships with genetic polymorphisms, and mediation analysis using PON1 as a model. We found strong evidence of ASM between the promoter polymorphism PON1-108 and methylation in a cluster of CpG sites surrounding the TSS that was also associated with PON1 protein levels. Our results demonstrate that integrating genetic, epigenetic, and expression data can provide evidence of functional mechanisms involving genetic and epigenetic regulation of candidate susceptibility genes.
Methods
Study Subjects
The CHAMACOS study aims to examine the effects of pesticides and other environmental exposures in a population of pregnant women and children living in the agricultural Salinas Valley, CA. Women were eligible for enrollment if they were at least 18 years of age, at less than 20 weeks’ gestation, Spanish or English speaking, eligible for low-income health insurance, receiving prenatal care at one of the local community clinics, and planning to deliver at the public hospital. Six hundred and one pregnant women were enrolled in 1999–2000 and 526 delivered live born singleton newborns [42]. CHAMACOS women were interviewed by bilingual, bicultural interviewers near the end of the first (∼13 weeks’ gestation) and second (∼26 weeks’ gestation) trimesters of pregnancy. Information was obtained on sociodemographic characteristics, mother’s reproductive and medical history, prenatal lifestyle exposures, diet, occupational and residential history, exposures to pesticides and other environmental chemicals, and housing quality.
Methylation of PON1 CpG sites was measured in blood samples collected from children at delivery (umbilical cord blood representing fetal blood) and when they were 9-year old (mean ± SD = 9.3 ± 0.3 years). Our study sample included a total of 449 children who had DNA samples available for PON1 genotyping in addition to methylation analysis at birth and/or at the age of 9 years. Of these children, 176 had samples available at both time points, 202 had samples only at birth, and 71 had samples at the age of 9 years only. Furthermore, among those children with DNA samples available, 247 and 176 had adequate blood volumes available for analysis of arylesterase (AREase) activity at birth and age of 9 years, respectively.
For both time points (birth and 9 years), children included in the study did not differ from all children in the cohort by other demographic and exposure variables (e.g., poverty level, marriage status, type of work during pregnancy, alcohol and smoking intake during pregnancy, prenatal exposure to DDT/DDE and PBDEs). Study protocols (2010-01-620 and 2010-03-949) were approved by the University of California, Berkeley Committee for Protection of Human Subjects. Written informed consent was obtained from all mothers, and assent was provided by the children at the 9-year assessment.
Blood Collection and Processing
Blood specimens were collected from the umbilical cords of CHAMACOS children after delivery and by venipuncture when children were approximately 9 years old. Heparinized whole blood was collected in BD vacutainers® (Becton, Dickinson and Company, Franklin Lakes, NJ), centrifuged, divided into plasma, buffy coats and red blood cells, and stored at −80°C at the School of Public Health Biorepository, University of California, Berkeley. Stringent conditions in accordance with the Best Practices for Biorepositories were followed [43].
Determination of PON1 Genotypes
DNA was isolated from blood clots as described previously [24]. In total, 39 genetic variants in PON1 and 2 SNPS each in PON2 and PON3 were genotyped. The promoter SNP, PON1-108, was genotyped using a fluorogenic allele-specific assay (Amplifluor, Chemicon, Temecula, CA). It required a two-part nested polymerase chain reaction strategy, in which the region surrounding the SNP was pre-amplified using non-allelic flanking primers and then the amplicon was diluted and used as the template for the Amplifluor assay. The coding polymorphisms, PON1192 and PON155, as well as promoter SNP, PON1-162, were genotyped using the Taqman real-time polymerase chain reaction method. Briefly, primers for the nucleotide sequence flanking the SNP and probes specific for the SNP were custom designed by Applied Biosystems, Inc. (Foster City, CA). The remaining 39 SNPs and indels were genotyped using the multiplex platform iPlex (Sequenom, San Diego, CA). Quality assurance procedures for genotyping of these PON1 SNPs included assessment of randomly distributed blank samples in each plate and duplicates of randomly selected samples with independently isolated DNA from the same subjects. Repeated analysis (4% of samples) in several runs showed a high degree (>99%) of concordance. All discrepancies were resolved with additional genotyping.
Determination of PON1 AREase Activity Levels
AREase activity in plasma samples was measured by determining the rate of phenyl acetate hydrolysis using spectrophotometric methods as described previously [44]. A strong correlation (r > 0.85) between measured PON1 quantity and AREase activity have been demonstrated by ELISA and Western blot-based methods utilizing PON1 antibodies [45, 46], making the AREase assay a reliable measure of PON1 enzyme quantity. All assays were performed in triplicate. Quality assurance included use of internal controls (aliquots of the same sample run on all assay plates), assessment of repeat samples (separate aliquots of the same sample run on different days), and concurrent analyses of specimens from different collections (samples from different time points run on the same plates) [47]. The average coefficient of variation (CV) for repeated samples was 8.5% and the correlation coefficient between repeated runs was 0.94. The average CV for internal controls samples, a measure of inter-assay variability, was 8.7%.
Bisulfite Treatment and Methylation Analyses
DNA was previously isolated from clots using a QIAamp Blood DNA Maxi kit (Qiagen, Inc., Santa Clarita, CA), normalized to 55 µg/ml. Zymo Bisulfite conversion Kits (Zymo Research, Orange, CA) were used to bisulfite convert 1ug aliquots of DNA. Methylation levels at 18 PON1 CpG sites were analyzed as part of a genome-wide methylation assessment using the Illumina Infinium 450k DNA methylation BeadChip. DNA samples were whole-genome amplified, enzymatically fragmented, purified, and applied to the Infinium 450k BeadChips according to the Illumina methylation protocol [48, 49]. BeadChip processing was performed using robotics, and the Illumina Hi-Scan system was used for analysis. Samples included in the analysis had detection P values below 0.01 for 95% of CpG sites. Poor performing CpG sites with P value > 0.01 were excluded. Two PON1 CpG sites mapping to SNPs in the Illumina annotation were also excluded resulting in a total of 16 PON1 CpGs that were included in the subsequent analyses. Raw signal intensities were background corrected and then normalized for color-channel bias using the all sample mean normalization method as described previously by Yousefi et al. [50]. We also applied a second normalization algorithm, beta mixture quantile normalization, to make interpretation between type I and type II probes comparable [51]. Methylation data were expressed as M values, which are calculated as the log2 ratio of the intensities of methylated probe to unmethylated probe [52]. Negative M values therefore indicate that the unmethylated form of a CpG site is more abundant than the methylated form.
Stringent quality control criteria were applied for handling of all samples and DNA methylation data. The quality assurance procedures included use of repeats and internal standards to minimize technical variability. Additionally, samples from both age groups were randomly distributed among different BeadChips to minimize experimental bias and batch effects.
Differential Cell Count
To examine the relationship of blood cell composition with DNA methylation in PON1 CpG sites, we performed differential cell counts in a subset of umbilical cord blood samples (n = 82) as described previously [53]. To prepare heparinized whole blood smears, we used the “gold standard” Wright-Push blood smearing technique [54] followed by staining utilizing a DiffQuik® staining kit. Slides were fixed for 15 minutes at 23°C and were then stained in basophilic dye and eosinophilic dye for 5 seconds each and washed after each staining period. Slides were scored under light microscopy (Zeiss Axioplan) with a magnification lens of 1000x and oil immersion. At least 100 cells were scored for each slide, and a percentage of each cell type (lymphocytes, monocytes, neutrophils, eosinophils, and basophils) was used for data analysis. To ensure consistency and reproducibility of scoring, 100 cells were scored in sets of 3 (3*100 = 300) for a subset of 35 samples. The CV for the repeat scoring in this subset was less than 10%.
Minfi Cell Count Estimation
Whole blood smears were not available for differential cell count in 9-year-old CHAMACOS children. Instead for these children, we used the Bioconductor R package minfi (v1.10.2) [55] to estimate the distribution of six (CD8+ T and CD4+ T lymphocytes, CD56+ natural killer cells, CD19+ B cells, CD14+ monocytes, and granulocytes) different white blood cell types based on their methylation signatures in 450K data. We did not use minfi estimates for cell composition in cord blood samples because as we recently described in Yousefi et al. [53] In Press, proportions of white blood cells in newborns are significantly different from the adult reference samples on which minfi estimates are based [56, 57]. We also found that unlike newborns, minfi estimates of cell composition in older children were much more comparable to those determined by differential cell count. This corresponds with published age-specific reference values [56, 57], which show that compared with adults, blood cell composition in newborns and young children is quite different but begin to resemble adult levels by ages 9–12 years. For comparison of cell type composition in cord bloods to those estimated by minfi in 9-year-old bloods, we use proportions of lymphocytes, granulocytes, and monocytes. For minfi estimates, this required summation of the frequencies for CD8+ T, CD4+ T, natural killer cells, and B cells to calculate the proportion of lymphocytes. For differential cell count, proportions of neutrophils, eosinophils, and basophils were summed to give an estimate of granulocytes.
Statistical Analysis
We used generalized estimating equations to determine the association of age with methylation levels at PON1 CpG, adjusting for batch (coded as categorical variable) and cell composition. For cell composition, we included proportion of monocytes and granulocytes in the model, using the proportion of lymphocytes as baseline.
Correlations of methylation levels at different CpG sites were determined by calculation of Pearson’s correlation coefficients (r). LD methylation blocks were established based on several criteria slightly modified from Shoemaker et al. [12] and Liu et al. [11]: (i) they had to contain at least 3 contiguous CpG sites and (ii) at least 50% of the CpG site pairs had to have methylation levels that were highly correlated with each other (r2 > 0.4).
To determine the relationship between genetic polymorphisms and methylation, we constructed linear regression models where genotype expressed as the number of minor alleles (0, 1, or 2) was the independent variable and the methylation at each CpG site (expressed as M values) was the outcome. Models were also adjusted for batch (coded as a categorical variable). A separate model was run for each individual CpG site. Bonferonni correction was used to account for multiple testing. As there were 16 CpG sites, 3 genetic polymorphisms (PON1-108,PON1192, and PON155), and 2 ages comprising 96 tests, we used an adjusted type I error rate of 0.05/96 = 5.21 E-04. We repeated these models in a subset of newborns (n = 83) who also had differential cell counts available to adjust for cell type distribution. In these models, we included percent lymphocytes, monocytes, basophils, and eosinophils in the models and used the percent neutrophils as the baseline. Similar models were also performed for 9-year olds using minfi estimates of cell composition (n = 218).
We also used linear regression models to examine associations between methylation and AREase activity. We constructed models including methylation at a single CpG site and batch as the independent variables and AREase as the dependent variable. Using Bonferonni correction, we considered P values less than 1.6 E-03 (0.05/32 tests) to be statistically significant. In addition to constructing separate models for each individual CpG site, we also used principal component (PC) analysis to summarize methylation at Block 1 and in the outer cluster of CpG sites (Fig. 2). The PCs explaining ∼95% of the variance of methylation for each group of CpG sites were incorporated as variables in regression models of AREase activity (dependent variable).
Recent studies examining mediation of the relation of genetic polymorphisms on gene expression by epigenetics have employed causal inference methodologies [58–60]. We used the PARAMED module in STATA [32, 33] to determine the extent to which PON1 methylation mediates the effect of PON1-108 genotype on AREase activity. The methodologies applied in this module rely on a counterfactual framework based on parametric regression modelling and can estimate the natural direct effect and natural indirect effect (NIE) in the presence of exposure-mediator interaction. All analyses were performed in STATA (version 12.0; StataCorp, College Station, TX) and R (version 3.1.2; R Development Core Team 2008).
Conflict of interest: None declared.
Supplementary Material
Supplementary Data
Acknowledgements
We are grateful to the laboratory and field staff and participants of the CHAMACOS study for their contributions. We are especially thankful to Dr Lisa Barcellos and Ms Hong Quach for their help with the Illumina 450K methylation assay, and Michael Ha and Girish Motwani who helped with differential cell counts in this study. This work was supported by grants from the National Institute of Environmental Health Science (NIEHS) [PO1 ES009605, R01ES023067] and from the US Environmental Protection Agency (EPA) [RD83451301, R82670901]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIEHS or EPA.
Data availability
Data available in Supplementary Material.
Supplementary data
Supplementary data is available at EnvEpig online.
References
- 1. Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med 2013;34:753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schubeler D. Function and information content of DNA methylation. Nature 2015;517:321–6. [DOI] [PubMed] [Google Scholar]
- 3. Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597–610. [DOI] [PubMed] [Google Scholar]
- 4. Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med 2012;34:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waterland RA. Is epigenetics an important link between early life events and adult disease? Horm Res 2009;71(Suppl 1):13–6. [DOI] [PubMed] [Google Scholar]
- 6. Bailey KA, Wu MC, Ward WO, et al. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol 2013;27:106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Jager PL, Srivastava G, Lunnon K, et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 2014;17:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joubert BR, Haberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 2012;120:1425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Swan EJ, Maxwell AP, McKnight AJ. Distinct methylation patterns in genes that affect mitochondrial function are associated with kidney disease in blood-derived DNA from individuals with type 1 diabetes. Diabet Med 2015;32:1110–1115. [DOI] [PubMed] [Google Scholar]
- 10. Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–9. [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, Li X, Aryee MJ, Ekstrom TJ, et al. GeMes, clusters of DNA methylation under genetic control, can inform genetic and epigenetic analysis of disease. Am J Hum Genet 2014;94: 485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shoemaker R, Deng J, Wang W, et al. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res 2010;20:883–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang YT. Integrative modeling of multiple genomic data from different types of genetic association studies. Biostatistics 2014;15:587–602. [DOI] [PubMed] [Google Scholar]
- 14. Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li HL, Liu DP, Liang CC. Paraoxonase gene polymorphisms, oxidative stress, and diseases. J Mol Med 2003;81:766–79. [DOI] [PubMed] [Google Scholar]
- 16. Harley KG, Huen K, Schall RA, et al. Association of organophosphate pesticide exposure and paraoxonase with birth outcome in Mexican-American women. PLoS One 2011;6: e23923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Infante-Rivard C. Genetic association between single nucleotide polymorphisms in the paraoxonase 1 (PON1) gene and small-for-gestational-age birth in related and unrelated subjects. Am J Epidemiol 2010;171:999–1006. [DOI] [PubMed] [Google Scholar]
- 18. Lawlor DA, Day IN, Gaunt TR, et al. The association of the paraoxonase (PON1) Q192R polymorphism with depression in older women: findings from the British Women's Heart and Health Study. J Epidemiol Community Health 2007;61:85–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leduc V, Poirier J. Polymorphisms at the paraoxonase 1 L55M and Q192R loci affect the pathophysiology of Alzheimer's disease: emphasis on the cholinergic system and beta-amyloid levels. Neurodegener Dis 2008;5:225–7. [DOI] [PubMed] [Google Scholar]
- 20. Ryckman KK, Morken NH, White MJ, et al. Maternal and fetal genetic associations of PTGER3 and PON1 with preterm birth. PLoS One 2010;5:e9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eskenazi B, Huen K, Marks A, et al. PON1 and neurodevelopment in children from the CHAMACOS study exposed to organophosphate pesticides in utero. Environ Health Perspect 2010;118:1775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eskenazi B, Kogut K, Huen K, et al. Organophosphate pesticide exposure, PON1, and neurodevelopment in school-age children from the CHAMACOS study. Environ Res 2014;134C: 149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Furlong CE, Holland N, Richter RJ, et al. PON1 status of farmworker mothers and children as a predictor of organophosphate sensitivity. Pharmacogenet Genomics 2006;16: 183–90. [DOI] [PubMed] [Google Scholar]
- 24. Holland N, Furlong C, Bastaki M, et al. Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environ Health Perspect 2006;114:985–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu ME, Liao YC, Lin RT, et al. A functional polymorphism of PON1 interferes with microRNA binding to increase the risk of ischemic stroke and carotid atherosclerosis. Atherosclerosis 2013;228:161–7. [DOI] [PubMed] [Google Scholar]
- 26. Holland N, Lizarraga D, Huen K. Recent progress in the genetics and epigenetics of paraoxonase: why it is relevant to children's environmental health. Curr Opin Pediatr 2015;27: 240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schrader C, Rimbach G. Determinants of paraoxonase 1 status: genes, drugs and nutrition. Curr Med Chem 2011;18: 5624–43. [DOI] [PubMed] [Google Scholar]
- 28. de la Iglesia RMM, Sánchez-Muniz FJ, Zulet MA, et al. Arylesterase activity is associated with antioxidant intake and paraoxonase-1 (pon1) gene methylation in metabolic syndrome patients following an energy restricted diet. EXCLI J 2014;13:416–26. [PMC free article] [PubMed] [Google Scholar]
- 29. Carlson CS, Heagerty PJ, Hatsukami TS, et al. TagSNP analyses of the PON gene cluster: effects on PON1 activity, LDL oxidative susceptibility, and vascular disease. J Lipid Res 2006;47: 1014–24. [DOI] [PubMed] [Google Scholar]
- 30. Deakin S, Leviev I, Brulhart-Meynet MC, et al. Paraoxonase-1 promoter haplotypes and serum paraoxonase: a predominant role for polymorphic position - 107, implicating the Sp1 transcription factor. Biochem J 2003;372:643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huen K, Barcellos L, Beckman K, et al. Effects of PON polymorphisms and haplotypes on molecular phenotype in Mexican-American mothers and children. Environ Mol Mutagen 2011;52:105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley R, Liu H, Dunn G, et al. PARAMED: stata module to perform causal mediation analysis using parametric regression models 2013. Available: https://ideas.repec.org/c/boc/bocode/s457581.html (4 March 2015, date last accessed). [Google Scholar]
- 33. Valeri L, Vanderweele TJ. Mediation analysis allowing for exposure-mediator interactions and causal interpretation: theoretical assumptions and implementation with SAS and SPSS macros. Psychol Methods 2013;18:137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonder MJ, Kasela S, Kals M, et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics 2014;15:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schalkwyk LC, Meaburn EL, Smith R, et al. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet 2010;86:196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gertz J, Varley KE, Reddy TE, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet 2011;7: e1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics 2010;5:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem 2013;288:34287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Osaki F, Ikeda Y, Suehiro T, et al. Roles of Sp1 and protein kinase C in regulation of human serum paraoxonase 1 (PON1) gene transcription in HepG2 cells. Atherosclerosis 2004;176: 279–287. [DOI] [PubMed] [Google Scholar]
- 40. Holler M, Westin G, Jiricny J, et al. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev 1988;2:1127–35. [DOI] [PubMed] [Google Scholar]
- 41. Costa LG, Giordano G, Furlong CE. Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: the hunt goes on. Biochem Pharmacol 2011;81: 337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eskenazi B, Bradman A, Gladstone E, et al. CHAMACOS, a longitudinal birth cohort study: lessons from the fields. J Childrens Health 2003;1:3–27. [Google Scholar]
- 43. ISBER. 2012 best practices for repositories: collection, storage, retrieval, and distribution of biological materials for research. Biopreserv Biobank 2012;10:79–161. [DOI] [PubMed] [Google Scholar]
- 44. Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics 1999;9:745–53. [PubMed] [Google Scholar]
- 45. Connelly PW, Maguire GF, Picardo CM, et al. Development of an immunoblot assay with infrared fluorescence to quantify paraoxonase 1 in serum and plasma. J Lipid Res 2008;49: 245–50. [DOI] [PubMed] [Google Scholar]
- 46. Kujiraoka T, Oka T, Ishihara M, et al. A sandwich enzyme-linked immunosorbent assay for human serum paraoxonase concentration. J Lipid Res 2000;41:1358–63. [PubMed] [Google Scholar]
- 47. Huen K, Richter R, Furlong C, et al. Validation of PON1 enzyme activity assays for longitudinal studies. Clin Chim Acta 2009;402:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288–95. [DOI] [PubMed] [Google Scholar]
- 49. Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011;6:692–702. [DOI] [PubMed] [Google Scholar]
- 50. Yousefi P, Huen K, Schall RA, et al. Considerations for normalization of DNA methylation data by Illumina 450K BeadChip assay in population studies. Epigenetics 2013;8: 1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013;29:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yousefi P, Huen K, Quach H, Motwani G, Hubbard A, Eskenazi B, Holland N. Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environ Mol Mutagen. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Houwen B. The differential cell count. Lab Hematol 2001;7: 89–100. [Google Scholar]
- 55. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dallman P. Blood and blood-forming tissues. In: Rudolph A. (ed.), Paediatrics. New York: Appleton-Century-Crofts, 1977, 1109–12. [Google Scholar]
- 57. Nathan D, Oski F, Orkin S. Nathan and Oski's Hematology of Infancy and Childhood, Vol. 2, 6 edn Philadelphia, PA: Saunders, 2003. [Google Scholar]
- 58. Huang YT. Integrative modeling of multi-platform genomic data under the framework of mediation analysis. Stat Med 2015;34:162–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koestler DC, Chalise P, Cicek MS, et al. Integrative genomic analysis identifies epigenetic marks that mediate genetic risk for epithelial ovarian cancer. BMC Med Genomics 2014;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Y, Aryee MJ, Padyukov L, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 2013;31: 142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Data Availability Statement
Data available in Supplementary Material.