Summary
Correlative and functional studies support the involvement of the RUNX gene family in hematological malignancies. To elucidate the role of epigenetics in RUNX inactivation, we evaluated promoter DNA methylation of RUNX1, 2, and 3 in 23 leukemia cell lines and samples from acute myeloid leukemia (AML), acute lymphocytic leukemia (ALL), and myelodysplatic syndromes (MDS) patients. RUNX1 and RUNX2 gene promoters were mostly unmethylated in cell lines and clinical samples. Hypermethylation of RUNX3 was frequent among cell lines (74%) and highly variable among patient samples, with clear association to cytogenetic status. High frequency of RUNX3 hypermethylation (85% of the 20 studied cases) was found in AML patients with inv(16)(p13.1q22) compared to other AML subtypes (31% of the other 49 cases). RUNX3 hypermethylation was also frequent in ALL (100% of the 6 cases) but low in MDS (21%). In support of a functional role, hypermethylation of RUNX3 was correlated with low levels of protein, and treatment of cell lines with the DNA demethylating agent decitabine resulted in mRNA re-expression. Furthermore, relapse-free survival of non-inv(16)(p13.1q22) AML patients without RUNX3 methylation was significantly better (p=0.016) than that of methylated cases. These results suggest that RUNX3 silencing is an important event in inv(16)(p13.1q22) leukemias.
Keywords: acute myeloid leukemia, RUNX3, DNA methylation, gene expression, inv(16)(p13.1q22)
Introduction
The RUNX family of transcription factors represents the DNA binding α-chain partners of the heterodimeric core binding factor (CBF) complex (Blyth et al, 2005). Each of the RUNX proteins, RUNX1 (AML1), RUNX2, and RUNX3 (AML2) can form heterodimers with CBFβ. In the CBFB-MYH11 subtype of human acute leukemia, the chromosomal translocation resulting in an inversion in chromosome 16 [inv(16)(p13.1q22), referred only as inv(16) in the remaining of this report] encodes a chimeric protein in which CBFβ is fused to smooth muscle myosin heavy chain (MYH11) (Liu et al, 1993). Although the exact mechanism of leukemogenesis by this chimera is unknown, it is thought that CBFB-MYH11 sequesters RUNX1 in the cytoplasm and antagonizes its normal function (Shigesada et al, 2004). Although the role of RUNX1 in hematopoiesis has been previously well-established (Okuda et al, 1996), recent data have indicated that the RUNX3 may also play a key role in the development of human acute leukemias (Cheng et al, 2008).
DNA hypermethylation of the RUNX3 promoter and gene downregulation have been reported for human solid tumors (Kim et al, 2004), including colon (Silva et al, 2013; Subramaniam et al, 2009), bladder (Kandimalla et al, 2013; Kim et al, 2004), stomach (Fan et al, 2011; Imamura et al, 2005), and lung (Yanagawa et al, 2007) cancers. In support of this gene’s function as a tumor suppressor, mice lacking Runx3 develop gastric epithelial hyperplasia and tumors as a result of stimulated proliferation and suppressed apoptosis in epithelial cells (Li et al, 2002). In leukemia, several studies reported downregulation of RUNX3 expression in inv(16) AML. Debernardi et al. characterized 28 patients with AML using gene expression profiling and found that RUNX3 was downregulated in patients with inv(16) AML and overexpressed in patients with acute promyelocytic leukemia (t15;17) (Debernardi et al, 2003). Two later independent studies also using gene expression profiling confirmed the downregulation of RUNX3 in inv(16) AML (Gutierrez et al, 2005; Sun et al, 2007). The role of RUNX3 in hematopoiesis has been characterized in zebrafish models in which Runx3 inhibition led to a decline in the number of mature blood cells (Crosier et al, 2002). Thus, the role of RUNX3 in tumorigenesis, its downregulation in hematopoietic cells, and the finding that RUNX3 is involved in hematopoiesis in animal models all suggest that this gene may contribute to the development of leukemia in humans.
Given the decreased gene expression of RUNX3 and prior data showing no somatic mutations in this gene (Otto et al, 2003), we sought to analyze the promoter methylation status of this gene as well as its two other family members, RUNX1 and RUNX2, in patients with AML. We found that RUNX3 was hypermethylated in the majority of patients with AML inv(16). RUNX1 and RUNX2 hypermethylation were rare, and RUNX3 methylation was low in non-inv(16) AMLs.
Material and Methods
Cell lines and AML patient samples
Eleven human leukemia cell lines of myeloid origin (K562, BV173, HL60, NB4, THP1, U937, ML1, OCI-AML3, HEL, MOLM13, and KBM5R) and 12 of lymphoid origin (MOLT4, Jurkat, Peer, T-ALL1, CEM, J-TAG, BJAB, RS4, ALL1, Raji, REH, and Ramos) were used in this study. All cell lines were obtained from the American Type Culture Collection and were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA, USA) and penicillin–streptomycin (Invitrogen, Carlsbad, CA). Cell suspensions from bone marrow aspirate specimens from patients with AML, MDS, and ALL prior to any therapy were obtained from established tissue blocks at The University of Texas MD Anderson Cancer Center. Peripheral blood samples were obtained from 4 healthy volunteers, and CD34+ cells were obtained from another 4 individuals. All samples from healthy donors were collected using Ficoll-Paque density centrifugation.
DNA extraction and bisulfite modification
DNA was extracted from leukemia cell lines and samples from patients and healthy volunteers using standard phenol-chloroform methods. DNA was subsequently treated with sodium bisulfite as previously described (Estecio et al, 2006). Briefly, 2 µg of genomic DNA from each sample was denatured in 0.2 M NaOH at 37°C for 10 min and then incubated with 3M sodium bisulfite at 50°C for 16 hours. DNA was then purified using the Wizard cleanup system (Promega, Fitchburg, WI), and desulfonated with 0.3 M NaOH at 25°C for 5 min. DNA was precipitated with ammonium acetate and ethanol, washed with 70% ethanol, dried, and resuspended in water.
Methylation analysis with pyrosequencing
PCR primers for the CpG island region overlapping the proximal promoters (P2) of RUNX1, RUNX2, and RUNX3 were designed using Biotage’s PSQ primer design software (Biotage AB, Uppsala, Sweden). Optimal annealing temperatures for each of these primers were tested using gradient PCR. PCR conditions and primers are presented in Table 1. PCR reactions were performed in a total volume of 20 µl, and the entire volume was used for each pyrosequencing reaction. Briefly, PCR product purification was done with streptavidin-sepharose high-performance beads (GE Healthcare Life Sciences, Piscataway, NJ), and co-denaturation of the biotinylated PCR products and sequencing primer (3.6 pmol/reaction) was conducted following the PSQ96 sample preparation guide. Sequencing was performed on a PSQ HS 96 system with the PSQ HS 96 SNP reagents kit (Biotage AB) according to the manufacturer’s instructions. The degree of methylation was calculated using the PSQ HS 96A 1.2 software.
Table I.
Sequences of primers used in this study.
| Gene | Oligos | Sequence to Analyze |
|---|---|---|
| RUNX1 | F: TGAATGAGAGTGTTTGGAAATGAA | GYGGTYGGYGGTTAATTATAGGTTTTT |
| Size: 123bp | R: Bio-AACTAATACCCAAAAAACCTATAATTAACC | |
| P: TTTTTTAGTTAGTTTGTATAGGG | ||
| RUNX2 | F: GGGGTTGGATTGTTGAATTTATAT | GGGGYGTTAGYGTATTTAGGA |
| Size: 132 bp | R: Bio-CCTAAAAAAACCAAAAACTTAAATTTACA | |
| P: ATAGGATTTTAATTGGGAG | ||
| RUNX3 | F: Bio-TTGTTTAGGGTTTGTGATTTTTGTAATT | ACRACRTCACCAA |
| Size: 113 bp | R: ACCAAACCAAAAAAATAAATAACCTCTTT | |
| P: TTAAAACCACTCCCAAT |
Immunohistochemistry of leukemia patient-derived samples
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded sections of bone marrow biopsy specimens. Sections were de-paraffinized, re-hydrated, and submitted to heat-induced epitope retrieval. Samples were incubated with an anti-RUNX3 rabbit antibody or a negative control rabbit antibody (AML2 a400 affinity-purified antibody kindly provided by Kun-Sang Chang) at a dilution of 1:2000 at room temperature (RT) in a dark humidified chamber for 60 minutes. Detection of the primary antibody was achieved with the EnVision+ system (DakoCytomation, Denmark) containing secondary antibodies conjugated to a horseradish peroxidase complex (HRP). Slides were incubated at RT in a dark humidified chamber for 30 min and were developed with the chromogen 3,3’-diaminobenzidine (DAB)/H2O2 (DakoCytomation). Slides were counterstained with hematoxylin, dehydrated, mounted, and cover-slipped.
Treatment with 5-aza-2’deoxycitidine
Seven leukemia cell lines (Raji, HL60, Molt4, ALL1, Jurkat, RS4, and PEER) were treated with the hypomethylating agent decitabine (5-aza-2'-deoxycytidine) to study the effects of epigenetic modulation. Cells were plated at low density 6 – 8 hrs before treatment with decitabine at concentrations of 1, 3, 5, and 10 µM for 3 days. Control samples were treated with dimethyl sulfoxide (DMSO). Media containing decitabine or the same volume of vehicle were changed every 24 hours. Total cellular RNA was extracted with Trizol (Invitrogen) according to the manufacturer’s protocol.
Analysis of gene expression with real-time PCR
Three µg of total RNA was used for reverse transcription (RT) reactions. RT reactions were performed using the Moloney murine leukemia virus RT enzyme (Invitrogen) according to the manufacturer’s protocol. Quantitative PCR reactions were performed using inventoried TaqMan gene expression assays for RUNX3 and GAPDH and TaqMan universal PCR master mix (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. Experiments were performed in triplicate for each data point. The signal of RUNX3 was normalized to the endogenous reference GAPDH to obtain the relative threshold cycle and then related to the CT of the control to obtain the relative expression level of the target gene.
Statistical analysis
DNA methylation and clinicopathogical values were compared between groups using the non-parametric Mann-Whitney U test. Survival curve comparison was done with the Log-rank (Mantel-Cox) test. Patients that did not respond to therapy were excluded from the relapse fee survival (RFS) analysis. All calculations were done in GraphPad software (GraphPad Software, Inc., La Jolla, CA).
Results
RUNX3 is frequently hypermethylated in leukemia cell lines
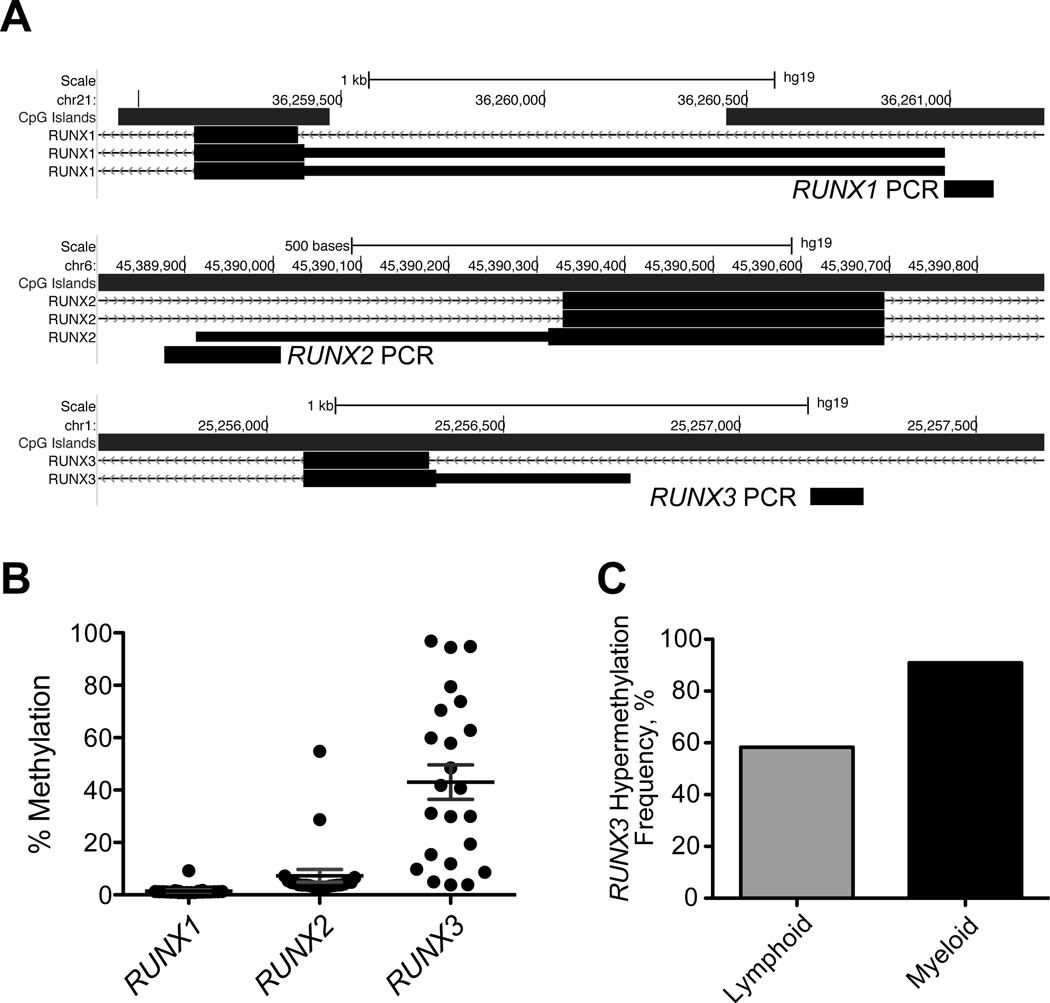
We investigated the DNA methylation status of the RUNX1, RUNX2, and RUNX3 gene promoters (Fig. 1A) using bisulfite PCR followed by pyrosequencing in 23 leukemia cell lines representing AML, CML, and MDS diseases (Table I). Hypermethylation (methylation density equal or above 15%) of RUNX1 and RUNX2 was rare in these cell lines; RUNX1 was not hypermethylated in any of the studied samples, and RUNX2 was hypermethylated in only 2 cell lines (Raji and B-Jab). In contrast, we found that the RUNX3 promoter was hypermethylated in 17 (74%) of the cell lines (Fig. 1B). Interestingly, we observed a trend toward higher frequency of hypermethylation of RUNX3 in cell lines of myeloid (90%) compared to lymphoid (57%) origin (Fig. 1C). The only myeloid cell line with unmethylated RUNX3 was THP1. Whether there was preferential methylation of RUNX3 within disease subgroups could not be determined due the small number of cell lines per disease.
Figure 1.
Gene promoter DNA methylation analysis of RUNX1, RUNX2, and RUNX3 in leukemia cell lines. (A) Schematic representation of bisulfite PCR primers for downstream pyrosequencing analysis. The generated PCR amplicons were within −500bp to +200bp from the transcription start site (TSS) of the alternative promoter (P2) of each gene, and the methylation density of 2 to 3 CpG sites per amplicon was quantified and averaged. (B) DNA methylation density of RUNX1, RUNX2, and RUNX3 in 23 leukemia cell lines. Note that only RUNX3 shows frequent promoter DNA hypermethylation (defined as 15% measured DNA methylation and above). (C) Differential frequency of RUNX3 promoter DNA hypermethylation in lymphoid versus myeloid leukemias.
RUNX3 can be reactivated by treatment with 5-aza-2’-deoxycytidine
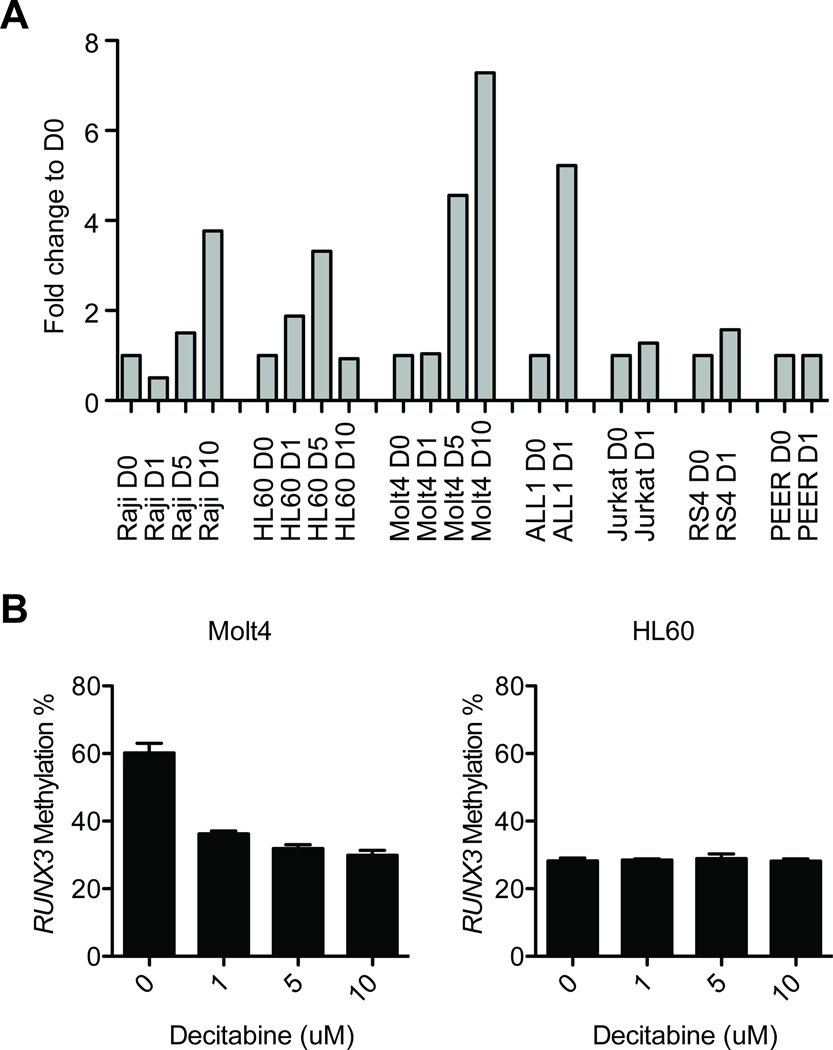
Because demethylating agents like decitabine are efficient in treating leukemias, particularly AML and MDS, we assessed whether treatment with decitabine could elevate the transcription of RUNX3. We first screened 7 leukemia cell lines (Jurkat, RS4, Raji, PEER, ALL1, HL60, and Molt4) for RUNX3 expression using real-time PCR and found that its expression was low to undetectable regardless of RUNX3 methylation status (Fig. 2A). Next, we treated these cells with various doses of decitabine (1 µM, 5 µM, and 10 µM) or only vehicle for 4 days and measured the expression of RUNX3 again. An increase in transcripts (2-fold and above) was seen for each cell line, with the exception of Jurkat and PEER, and an increase in expression 5-fold and higher than baseline was seen for Raji and HL60. Significant decreases in RUNX3 methylation were observed in Molt4 cells after decitabine treatment (decreased from 60% methylation without treatment to 30% after 10 µM decitabine). Despite the increase in gene expression, RUNX3 methylation was unchanged in HL60 (30% methylation before and after exposure to decitabine).
Figure 2.
RUNX3 reactivation after treatment with a demethylating agent. (A) Fold change in RUNX3 expression in 7 leukemia cell lines after treatment with various doses of decitabine (D1=1 µM; D5=5 µM; D10=10 µM) or vehicle (D0) for 3 days. An increase in expression over 2-fold was observed for all cell lines treated with high-dose decitabine (Raji, HL60, and Molt4) and in only ALL1 among the cell lines treated with low-dose decitabine. (B) RUNX3 promoter demethylation after treatment with decitabine.
RUNX3 hypermethylation is associated with cytogenetic status in AML
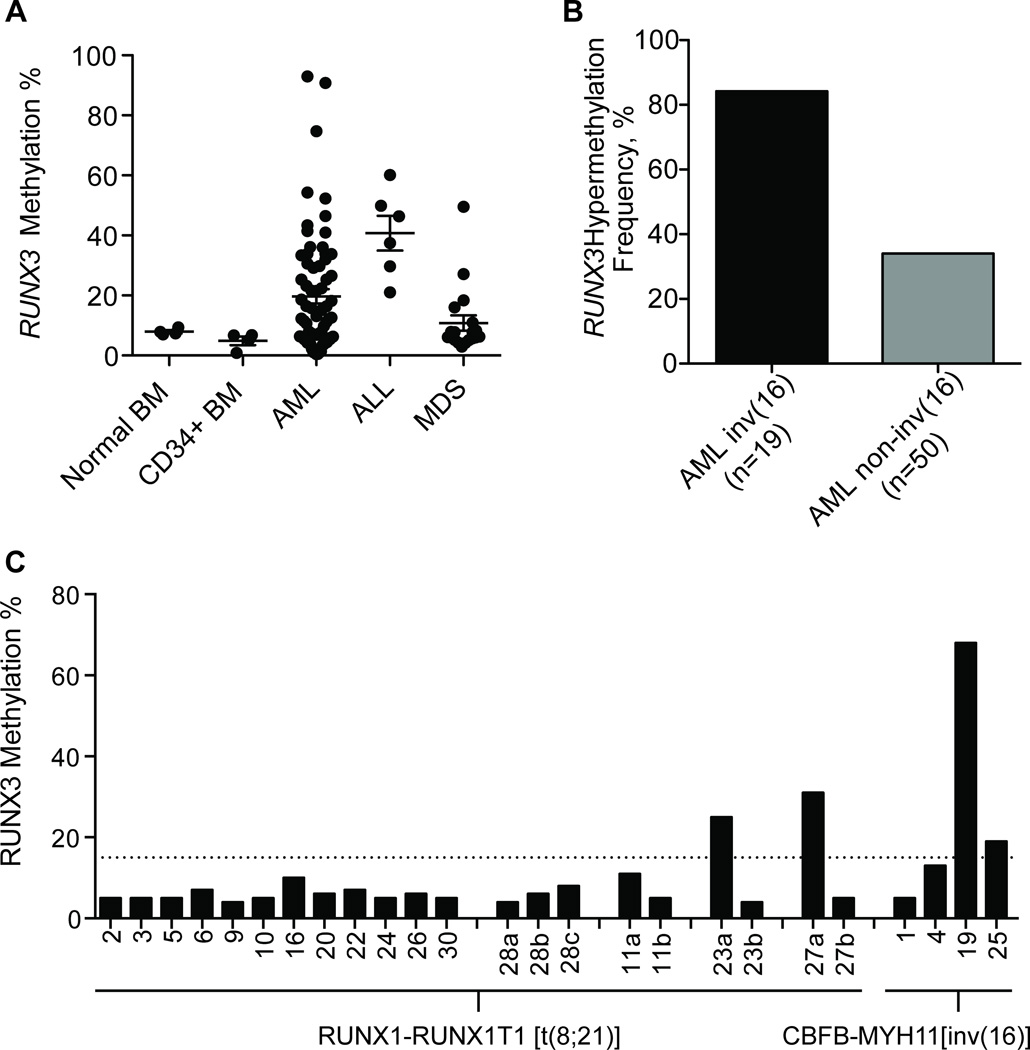
Because of the high frequency of RUNX3 expression in myeloid leukemia cell lines, we studied a larger group of clinical samples composed of AML, ALL, and MDS clinical samples (Table II). In total, 102 samples were evaluated by bisulfite modification followed by pyrosequencing, including 8 control samples (4 whole bone marrow and 4 CD34+ bone marrow cells from healthy donors). RUNX3 promoter methylation was below 15% in normal samples, and hypermethylation was found in 32/69 AML samples (46%), 4/19 MDS samples (21%), and 6/6 ALL samples (100%) (Fig. 3A). Of the 69 AML samples, 20 were classified as inv(16) AML, and 49 were other types of AML. In the AML inv(16) samples, 85% were hypermethylated at the RUNX3 promoter region, whereas only 31% of the other AML subtypes were hypermethylated (Fig. 3B). We also evaluated DNA methylation of RUNX1 and RUNX2 in a subgroup of these samples (66 samples for RUNX1 and 72 for RUNX2) with the same assays and found that, as in cell lines, these genes are almost universally unmethylated; with the exception of a single AML case, all studied samples lacked promoter methylation (data not shown).
Table II.
Clinicopathological features of methylated and unmethylated RUNX3 in AML patients.
| Parameter | inv(16) Methylated (n= 13) |
inv(16) Unmethylated (n= 3) |
Non-inv(16) Methylated (n= 16) |
Non-inv(16) Unmethylated (n= 32) |
Mann-Whitney U-Test | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | Mean | SEM | Mean | SEM | inv(16): U vs M |
non-Inv(16): U vs M |
|
| Age, years | 49.77 | 4.21 | 24.33 | 2.40 | 67.38 | 2.21 | 65.25 | 2.80 | 0.015 | 0.844 |
| WBC, K/µL | 35.73 | 8.70 | 41.03 | 25.15 | 11.97 | 6.00 | 20.39 | 4.73 | 0.893 | 0.106 |
| Plt, K/µL | 62.08 | 14.88 | 63.33 | 27.95 | 65.87 | 14.00 | 87.13 | 13.50 | 0.893 | 0.361 |
| Hgb, g/dL | 9.04 | 0.60 | 7.50 | 1.18 | 8.31 | 0.51 | 8.80 | 0.33 | 0.282 | 0.330 |
| Blast, % | 46.08 | 7.67 | 66.33 | 19.74 | 23.27 | 7.61 | 30.06 | 5.83 | 0.252 | 0.328 |
| Mono, % | 25.69 | 5.18 | 18.00 | 18.00 | 9.53 | 3.17 | 9.35 | 2.17 | 0.346 | 0.981 |
| Neut, % | 8.15 | 1.99 | 2.00 | 1.00 | 23.47 | 4.71 | 26.97 | 4.14 | 0.175 | 0.647 |
| Promyelo, % | 0.54 | 0.29 | 0.33 | 0.33 | 0.00 | 0.00 | 0.06 | 0.04 | 1.000 | 0.354 |
| Alb, g/dL | 3.25 | 0.19 | 3.20 | 0.46 | 3.59 | 0.19 | 3.39 | 0.12 | 0.946 | 0.359 |
| LDH, IU/L | 1788.85 | 469.96 | 1273.67 | 386.55 | 1080.13 | 284.84 | 1819.03 | 381.26 | 0.893 | 0.058 |
| Bili, mg/dL | 0.59 | 0.08 | 0.83 | 0.34 | 0.44 | 0.05 | 0.60 | 0.07 | 0.730 | 0.246 |
| Creat, mg/dL | 0.92 | 0.07 | 1.00 | 0.06 | 1.03 | 0.07 | 1.08 | 0.06 | 0.492 | 0.869 |
| UA, mg/dL | 4.48 | 0.38 | 4.80 | 0.60 | 4.67 | 0.45 | 4.87 | 0.42 | 0.563 | 0.879 |
| Fibr, mg/dL | 511.73 | 36.85 | 288.33 | 10.73 | 564.70 | 44.84 | 506.89 | 25.87 | 0.013 | 0.224 |
| BM Bl, % | 55.69 | 5.78 | 67.33 | 8.19 | 49.53 | 6.48 | 52.00 | 4.74 | 0.312 | 0.935 |
| Bm Prog, % | 1.92 | 0.54 | 0.67 | 0.33 | 0.67 | 0.23 | 0.84 | 0.20 | 0.405 | 0.796 |
| BM Mon, % | 6.77 | 1.95 | 4.00 | 2.52 | 2.47 | 1.06 | 3.65 | 1.25 | 0.685 | 0.971 |
| BM Gran, % | 4.54 | 1.57 | 4.67 | 0.88 | 7.27 | 2.39 | 8.26 | 2.09 | 0.408 | 0.860 |
WBC=white blood cell count. Plt=platelet count. Hgb=serum hemoglobin. Blast=bone marrow blasts. Mono=mononuclear cell count. Neut=neutrophil count. Promyelo=promyelocyte count. Alb=serum albumin. LDH=lactate dehydrogenase. Bili=serum bilirubin. Creat=serum creatinine. UA=serum uric acid. Fibr=fibroblast cultures. BM Bl=bone marrow blasts. Bm Prog=bone marrow progenitors. BM Mon=bone marrow monocytes. BM Gran=bone marrow granulocytes. U: unmethylated group. M: methylated group. P values below 0.05 are marked in bold.
Figure 3.
RUNX3 promoter DNA methylation in clinical samples. (A) DNA methylation density of the RUNX3 promoter in 8 control individuals (4 normal bone marrow [BM] and 4 CD34+ BM), 69 acute myeloid leukemia (AML), 6 acute lymphocytic leukemia (ALL), and 19 myelodysplastic syndromes (MDS) patient samples. (B) Differential frequency of RUNX3 promoter DNA hypermethylation in inv(16) versus non-inv(16) AML subtypes. The inv(16) AML group had a 2-fold higher frequency of RUNX3 hypermethylation compared to other AML subtypes. (C) RUNX3 methylation in the RUNX1-RUNX1T1 [t(8;21)] subtype. The letter “a” after the patient number indicates sample collection at admission, and “b” and “c” are post-treatment samples. RUNX3 methylation is infrequent in this group, and it is decreased after treatment. Additional inv(16) samples were evaluated (CBFb-MyH11), and RUNX3 was hypermethylated in half of cases.
A second validation group composed of 16 RUNX1-RUNX1T1 t(8;21) AML samples was evaluated for RUNX3 methylation, and again we observed only rare hypermethylation (2 cases, 12.5%). Four cases had samples collected both at time of admission and at one or two later timepoints after treatment for the disease. In the 2 hypermethylated cases, we observed loss of RUNX3 methylation after treatment. Four new inv(16) AML cases from patients admitted to the hospital after the initial screening for RUNX3 methylation were included with this validation, and in agreement with the previous findings, these showed frequent RUNX3 hypermethylation (2 cases).
RUNX3 protein levels are reduced in AML samples and correlated with promoter hypermethylation
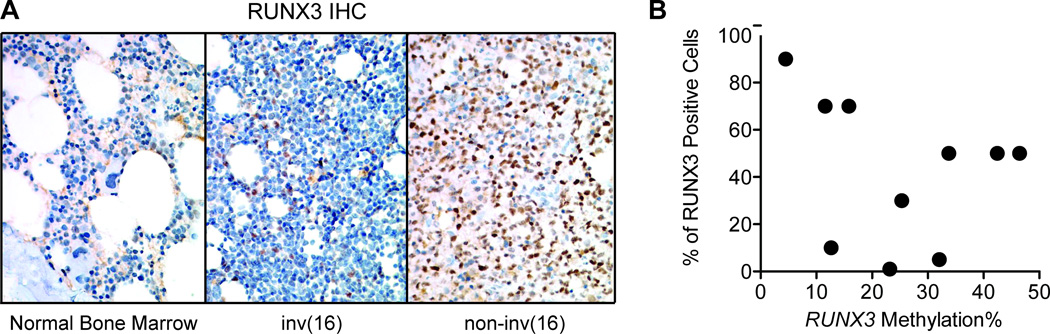
We next sought to verify the expression status of RUNX3 in AML samples. Using immunohistochemistry, we investigated the presence of RUNX3 protein in eight AML samples with variable DNA methylation status and one normal bone marrow sample (Fig. 4A). Except for one case with 90% of cells positive for RUNX3, all tested AML samples showed a decrease in the number of RUNX3-positive cells (ranging from 1% to 70% positive cells). The decrease in RUNX3 positivity was generally correlated with the DNA methylation density in each case (Fig. 4B), although there were one case with low promoter DNA methylation and low RUNX3 protein expression.
Figure 4.
Protein expression of RUNX3 in selected AML samples. (A) Immunohistochemical staining for RUNX3 protein (in brown) in a normal bone marrow sample (left panel), an inv(16) AML case (middle panel), and an non-inv(16) AML case (right panel). (B) Correlation of percent of RUNX3-posivitive cells (y-axis) with DNA methylation (x-axis). The percentage of RUNX3-positive cells decreased with increased promoter DNA methylation.
Patients without RUNX3 methylation have longer relapse-free survival
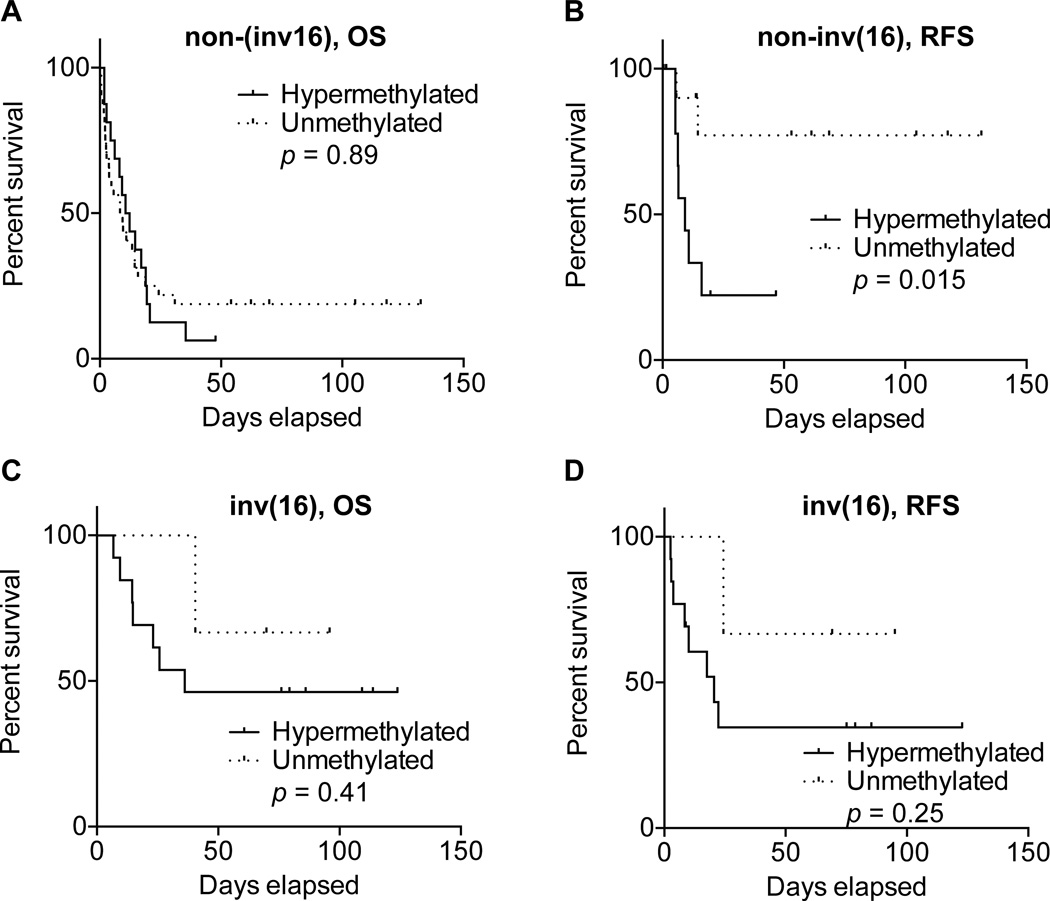
We were able to match clinicopathological information for 16 hypermethylated and 32 non-methylated non-inv(16) AMLs. Using nonparametric tests, we did not observe differences in age or white blood cell and platelet counts (Table II). A significant difference was observed for relapse-free survival (Fig. 5B), with non-methylated RUNX3 cases showing longer relapse-free survival than methylated cases (p=0.015, Log-rank test). It is important to note that in this analysis we excluded 26 patients that did not respond to therapy. However, overall survival did not differ between the groups (Fig. 5A).
Figure 5.
Survival analysis of AML cases categorized by RUNX3 methylation status. (A) Non-inv(16) AMLs have poor overall survival that does not differ between RUNX3 hypermethylated (solid line) and unmethylated groups (dotted line). (B) Relapse-free survival was significantly longer in unmethylated RUNX3 non-inv(16) cases. (C and D) Trend toward longer overall and relapse-free survival in unmethylated RUNX3 inv(16) AML cases but not statistically significant.
We could not accurately compare clinicopathological features in inv(16) patients because of the low number of non-methylated RUNX3 cases (only 3 with available information versus 13 hypermethylated cases). Nevertheless, we observed a tendency for better both overall survival and relapse-free survival for patients with unmethylated RUNX3 (Figs. 5C and 5D). Other significantly different features, such as age, were unreliable due to the sample size.
Discussion
In this study, we have characterized the methylation status of the RUNX1, RUNX2, and RUNX3 genes in human myeloid leukemias. Our results show that promoter methylation of the RUNX3 gene and downregulation of RUNX3 expression occurs almost universally in inv(16) AMLs, and in cell lines, RUNX3 repression can be reversed by treatment with the hypomethylating agent decitabine. Furthermore, RUNX3 hypermethylation was correlated with shorter relapse-free survival in a limited cohort.
The essential function of RUNX1 in normal hematopoiesis as well as in acute leukemia, specifically the t(8;21) AML M2 subset, has been well characterized (Downing et al, 1993; Lo Coco et al, 1997; Okuda et al, 1996; Peterson and Zhang 2004). RUNX1 knockout is embryonically lethal in mice, and the resultant embryos show an absence of hematopoietic cell development. In contrast, information on RUNX3 function in leukemia is comparatively limited, and several studies of the RUNX3 gene have mostly focused on its role as a tumor suppressor gene in gastric cancer. For example, it has been shown that RUNX3 knockout mice show a phenotype of gastric epithelial hyperplasia (Li et al, 2002).
Some studies suggest that RUNX3 may have a complementary role to RUNX1 in acute myeloid leukemia. Double-knockout Runx1;Runx3 mice present a complex phenotype that includes bone marrow failure and, in a subset of animals, signs of myeloproliferative disease (Wang et al, 2014). RUNX3 also regulates RUNX1 (Brady et al, 2009), which highlights the intricate interplay between RUNX family members.
Studies using cDNA microarray analysis of AML samples have identified RUNX3 downregulation, and in some studies, this was more prominent in the formally called M4Eo subset (Debernardi et al, 2003; Gutierrez et al, 2005). However, Otto et al. previously showed that no somatic mutations could be identified in the RUNX3 gene in AML patients (Otto et al, 2003). We therefore hypothesized that RUNX3 expression in AML may be regulated epigenetically via DNA methylation. Our study showed that the RUNX3 promoter was hypermethylated in specific subsets of AML, specifically inv(16) AML. This finding differs from those of Cheng et al., who found that RUNX3 expression in AML was not mediated by promoter hypermethylation (Cheng et al, 2008). We propose that this difference in results may be due to population characteristics because their study was mostly composed of childhood leukemias and fewer inv(16) cases than we studied.
Scarce information is available regarding the prognostic value of abnormal expression of RUNX3 in leukemia. Lacayo et al. found that RUNX3 overexpression among pediatric patients with AML with mutated FLT3 had an inferior prognosis, but in patients with wild-type FLT3, RUNX3 expression had no predictive value for survival (Lacayo et al, 2004). In contrast, Cheng et al. showed that high RUNX3 expression among childhood AML cases was associated with a shortened event-free survival regardless of their FLT3 status (Cheng et al, 2008). The patient cohort in our study is composed exclusively of adults, and we observe a trend for longer relapse-free survival among the RUNX3 unmethylated subset. However, these data are limited to non-inv(16) patients owing to the rarity of unmethylated RUNX3 cases among the inv(16) subtype. Also, expression data are not available for the group we studied, so direct comparisons to other studies was not possible. It is possible that the trend we observe is not directly related to lack of RUNX3 protein, but rather, that the value of its hypermethylation is a marker of dysregulation of transcriptional programs (given the known interplay between hematopoiesis and RUNX factors) or that RUNX3 is one of several genes concomitantly hypermethylated in leukemia. In respect to the latter, high frequency of methylated genes has been associated with worse survival in leukemia, T-ALL (Roman-Gomez et al, 2005), and MDS (Shen et al, 2010).
In summary, our results suggest that epigenetic dysregulation of RUNX3 is likely an important target in the molecular pathway of leukemogenesis in core binding factor leukemia, and future studies should be dedicated to further characterize the role of RUNX3 in AML.
Acknowledgments
Funding
This work was supported in part by MD Anderson Cancer Center Support Grant P30 CA016672. GGM is also supported by the Edward P. Evans Foundation, the Fundacion Ramon Areces, grant RP100202 from the Cancer Prevention & Research Institute of Texas (CPRIT), and by generous philanthropic contributions to MD Anderson’s MDS/AML Moon Shot Program. MRHE is supported by grant CA153114 from the National Institutes of Health and by MD Anderson’s Center for Cancer Epigenetics.
Footnotes
Competing interest
The authors declare no competing financial interests.
Author contribution
MRHE, SM, CBR, HY, KK and ZF performed the research. MRHE, SM, and GGM designed the research. MRHE, SM, CDD, SAP, and GGM analysed the data. YW, WS, KC, GB, and HK contribute with acquisition of clinical samples and reagents. MRHE, SM, CDD, ZSB and GGM wrote the manuscript.
REFERENCES
- Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nature Reviews Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Brady G, Whiteman HJ, Spender LC, Farrell PJ. Downregulation of RUNX1 by RUNX3 requires the RUNX3 VWRPY sequence and is essential for Epstein-Barr virus-driven B-cell proliferation. Journal of Virology. 2009;83:6909–6916. doi: 10.1128/JVI.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CK, Li L, Cheng SH, Lau KM, Chan NP, Wong RS, Shing MM, Li CK, Ng MH. Transcriptional repression of the RUNX3/AML2 gene by the t(8;21) and inv(16) fusion proteins in acute myeloid leukemia. Blood. 2008;112:3391–3402. doi: 10.1182/blood-2008-02-137083. [DOI] [PubMed] [Google Scholar]
- Crosier PS, Kalev-Zylinska ML, Hall CJ, Flores MV, Horsfield JA, Crosier KE. Pathways in blood and vessel development revealed through zebrafish genetics. The International Journal of Developmental Biology. 2002;46:493–502. [PubMed] [Google Scholar]
- Debernardi S, Lillington DM, Chaplin T, Tomlinson S, Amess J, Rohatiner A, Lister TA, Young BD. Genome-wide analysis of acute myeloid leukemia with normal karyotype reveals a unique pattern of homeobox gene expression distinct from those with translocation-mediated fusion events. Genes, Chromosomes & Cancer. 2003;37:149–158. doi: 10.1002/gcc.10198. [DOI] [PubMed] [Google Scholar]
- Downing JR, Head DR, Curcio-Brint AM, Hulshof MG, Motroni TA, Raimondi SC, Carroll AJ, Drabkin HA, Willman C, Theil KS, Civin CI, Erickson P. An AML1/ETO fusion transcript is consistently detected by RNA-based polymerase chain reaction in acute myelogenous leukemia containing the (8;21)(q22;q22) translocation. Blood. 1993;81:2860–2865. [PubMed] [Google Scholar]
- Estecio MR, Youssef EM, Rahal P, Fukuyama EE, Gois-Filho JF, Maniglia JV, Goloni-Bertollo EM, Issa JP, Tajara EH. LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene. 2006;25:5018–5026. doi: 10.1038/sj.onc.1209509. [DOI] [PubMed] [Google Scholar]
- Fan XY, Hu XL, Han TM, Wang NN, Zhu YM, Hu W, Ma ZH, Zhang CJ, Xu X, Ye ZY, Han CM, Pan WS. Association between RUNX3 promoter methylation and gastric cancer: a meta-analysis. BMC Gastroenterology. 2011;11:92. doi: 10.1186/1471-230X-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez NC, Lopez-Perez R, Hernandez JM, Isidro I, Gonzalez B, Delgado M, Ferminan E, Garcia JL, Vazquez L, Gonzalez M, San Miguel JF. Gene expression profile reveals deregulation of genes with relevant functions in the different subclasses of acute myeloid leukemia. Leukemia. 2005;19:402–409. doi: 10.1038/sj.leu.2403625. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Hibi K, Koike M, Fujiwara M, Kodera Y, Ito K, Nakao A. RUNX3 promoter region is specifically methylated in poorly-differentiated colorectal cancer. Anticancer Research. 2005;25:2627–2630. [PubMed] [Google Scholar]
- Kandimalla R, van Tilborg AA, Zwarthoff EC. DNA methylation-based biomarkers in bladder cancer. Nature Reviews Urology. 2013;10:327–335. doi: 10.1038/nrurol.2013.89. [DOI] [PubMed] [Google Scholar]
- Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW, Bang YJ, Kang GH. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Laboratory Investigation. 2004;84:479–484. doi: 10.1038/labinvest.3700060. [DOI] [PubMed] [Google Scholar]
- Lacayo NJ, Meshinchi S, Kinnunen P, Yu R, Wang Y, Stuber CM, Douglas L, Wahab R, Becton DL, Weinstein H, Chang MN, Willman CL, Radich JP, Tibshirani R, Ravindranath Y, Sikic BI, Dahl GV. Gene expression profiles at diagnosis in de novo childhood AML patients identify FLT3 mutations with good clinical outcomes. Blood. 2004;104:2646–2654. doi: 10.1182/blood-2003-12-4449. [DOI] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- Lo Coco F, Pisegna S, Diverio D. The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias. Haematologica. 1997;82:364–370. [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Otto F, Stock M, Fliegauf M, Fenaux P, Preudhomme C, Lubbert M. Absence of somatic mutations within the Runt domain of AML2/RUNX3 in acute myeloid leukaemia. Leukemia. 2003;17:1677–1678. doi: 10.1038/sj.leu.2403007. [DOI] [PubMed] [Google Scholar]
- Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Prosper F, Heiniger A, Torres A. Lack of CpG island methylator phenotype defines a clinical subtype of T-cell acute lymphoblastic leukemia associated with good prognosis. Journal of Clinical Oncology. 2005;23:7043–7049. doi: 10.1200/JCO.2005.01.4944. [DOI] [PubMed] [Google Scholar]
- Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X, Berry D, Ahmed S, Zhu W, Pierce S, Kondo Y, Oki Y, Jelinek J, Saba H, Estey E, Issa JP. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. Journal of Clinical Oncology. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigesada K, van de Sluis B, Liu PP. Mechanism of leukemogenesis by the inv(16) chimeric gene CBFB/PEBP2B-MHY11. Oncogene. 2004;23:4297–4307. doi: 10.1038/sj.onc.1207748. [DOI] [PubMed] [Google Scholar]
- Silva TD, Vidigal VM, Felipe AV, JM DEL, Neto RA, Saad SS, Forones NM. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncology Letters. 2013;6:1687–1692. doi: 10.3892/ol.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam MM, Chan JY, Soong R, Ito K, Yeoh KG, Wong R, Guenther T, Will O, Chen CL, Kumarasinghe MP, Ito Y, Salto-Tellez M. RUNX3 inactivation in colorectal polyps arising through different pathways of colonic carcinogenesis. The American Journal of Gastroenterology. 2009;104:426–436. doi: 10.1038/ajg.2008.141. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang W, Ramdas L, Stivers DN, Jones DM, Kantarjian HM, Estey EH, Vadhan-Raj S, Medeiros LJ, Bueso-Ramos CE. Comparative analysis of genes regulated in acute myelomonocytic leukemia with and without inv(16)(p13q22) using microarray techniques, real-time PCR, immunohistochemistry, and flow cytometry immunophenotyping. Modern Pathology. 2007;20:811–820. doi: 10.1038/modpathol.3800829. [DOI] [PubMed] [Google Scholar]
- Wang CQ, Krishnan V, Tay LS, Chin DW, Koh CP, Chooi JY, Nah GS, Du L, Jacob B, Yamashita N, Lai SK, Tan TZ, Mori S, Tanuichi I, Tergaonkar V, Ito Y, Osato M. Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Reports. 2014;8:767–782. doi: 10.1016/j.celrep.2014.06.046. [DOI] [PubMed] [Google Scholar]
- Yanagawa N, Tamura G, Oizumi H, Kanauchi N, Endoh M, Sadahiro M, Motoyama T. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]