Highlight
BGLU6 is a single trait locus that causes natural qualitative variation in the accumulation of flavonol 3-O-gentiobioside 7-O-rhamnoside in Arabidopsis thaliana. BGLU6 encodes a glycoside hydrolase-type flavonol 3-O-glucoside: 6″-O-glucosyltransferase.
Key words: Arabidopsis thaliana, flavonoid, flavonol glucosyltransferase, glycoside hydrolase-type, natural variation, whole genome association mapping.
Abstract
Flavonols are colourless secondary metabolites, primarily regarded as UV-protection pigments that are deposited in plants in their glycosylated forms. The glycosylation of flavonols is mainly catalysed by UDP-sugar-dependent glycosyltransferases (UGTs). Although the structures of flavonol glycosides accumulating in Arabidopsis thaliana are known, many genes involved in the flavonol glycosylation pathway are yet to be discovered. The flavonol glycoside profiles of seedlings from 81 naturally occurring A. thaliana accessions were screened using high performance thin layer chromatography. A qualitative variation in flavonol 3-O-gentiobioside 7-O-rhamnoside (F3GG7R) content was identified. Ler × Col-0 recombinant inbred line mapping and whole genome association mapping led to the identification of a glycoside hydrolase family 1-type gene, At1g60270/BGLU6, that encodes a homolog of acyl-glucose-dependent glucosyltransferases involved in the glycosylation of anthocyanins, possibly localized in the cytoplasm, and that is co-expressed with genes linked to phenylpropanoid biosynthesis. A causal single nucleotide polymorphism introducing a premature stop codon in non-producer accessions was found to be absent in the producers. Several other naturally occurring loss-of-function alleles were also identified. Two independent bglu6 T-DNA insertion mutants from the producer accessions showed loss of F3GG7R. Furthermore, bglu6 mutant lines complemented with the genomic Ler BGLU6 gene confirmed that BGLU6 is essential for production of F3GGR7. We have thus identified an accession-specific gene that causes a qualitative difference in flavonol glycoside accumulation in A. thaliana strains. This gene encodes a flavonol 3-O-glucoside: 6″-O-glucosyltransferase that does not belong to the large canonical family of flavonol glycosyltransferases that use UDP-conjugates as the activated sugar donor substrate.
Introduction
Flavonols are colourless natural pigments that are among the most abundant flavonoids in plants. Usually they accumulate in the form of mono-, di-, or triglycosides. Glycosylation enhances the solubility of flavonols and thus is likely to be critical for their transport to and storage in their final destination in the vacuole or cell wall. There is evidence that their biological roles differ across plant species, given that they have, for example, an important role in the maintenance of fertility in Zea mays and Petunia hybrida (Mo et al., 1992; van der Meer et al., 1992) but not in Arabidopsis thaliana or Eustoma grandiflorum (Burbulis et al., 1996; Nielsen et al., 2002). Because flavonol glycosides absorb UV-B light in the region of 280–320nm, they are regarded as effective UV filters with a major function in tissue protection (Ormrod et al., 1995; Olsson et al., 1998; Solovchenko and Schmitz-Eiberger, 2003). Additionally, flavonols act as UV flower pigments that confer both attractive and defensive functions to insects (Gronquist et al., 2001). They also function as co-pigments, sandwiched between anthocyanin molecules, to modify the colour of flowers and fruits (Goto and Kondo, 1991; Andersen and Jordheim, 2006). Auxin retention and transport, as well as subsequent physiological processes dependent on auxin movement, are also modulated by the accumulation of flavonols (Peer et al., 2011 and references therein; Yin et al., 2014). However, in spite of the obvious importance of flavonols for the plant, the relationship between their structure and function remains unclear (Pollastri and Tattini, 2011 and references therein).
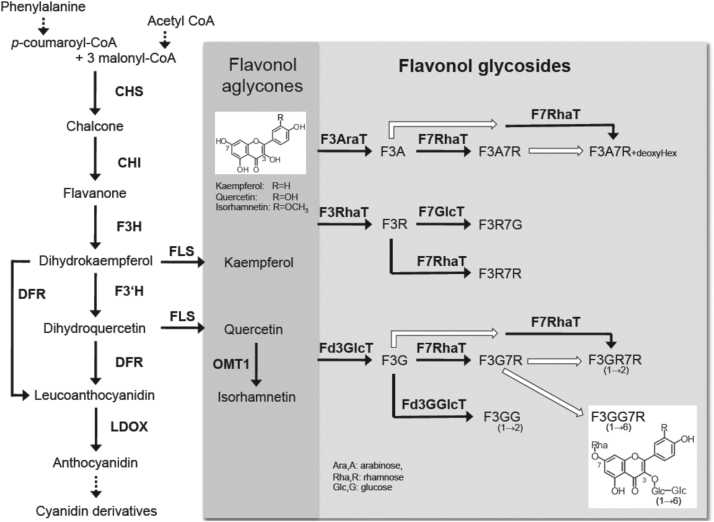
Flavonol synthesis is regulated by a variety of developmental and environmental cues (Winkel-Shirley, 2002). The expression of genes in this pathway is light dependent (Pelletier and Shirley, 1996; Cain et al., 1997; Stracke et al., 2010) and regulated by both light quality and quantity through distinct photoreceptors (Wade et al., 2001; Hemm et al., 2004). The expression of flavonol biosynthesis genes is controlled in organ- and tissue-specific patterns by at least three R2R3-MYB transcriptional activators in A. thaliana that function without a basic helix-loop-helix partner (Stracke et al., 2007; Stracke et al., 2010). Production of flavonol glycoside 1 (PFG1, MYB12), PFG2 (MYB11), and PFG3 (MYB111) activates, in parallel, the biosynthetic enzyme encoding genes CHALCONE SYNTHASE (CHS), CHALCONE ISOMERASE (CHI), FLAVANONE 3-HYDROXYLASE (F3H), and FLAVONOL SYNTHASE 1 (FLS1) (Mehrtens et al., 2005; Stracke et al., 2007). The enzymes encoded by these four genes, together with the enzyme flavonoid 3′-hydroxylase (F3′H), which converts dihydrokaempferol to dihydroquercetin, are required for the formation of the core flavonol aglycone (Fig. 1). The accumulation of distinct flavonol glycosides is mediated by specific glycosyltransferases (GTs) (Winkel-Shirley, 2001).
Fig. 1.
Proposed flavonol biosynthesis and modification pathway in A. thaliana.
The compounds detected in A. thaliana are given. White arrows indicate proposed but unidentified reactions. The metabolite flavonol 3-O-gentiobioside 7-O-rhamnoside (F3GG7R), which is in the focus of this paper, is shown in the white box. CHI, chalcone isomerase; CHS, chalcone synthase; DFR, dihydroflavonol reductase; F3AraT, flavonol 3-O-arabinosyltransferase; F3H, flavanone 3-hydroxylase; F3RhaT, flavonol 3-O-rhamnosyltransferase; F3′H, flavonoid 3′-hydroxylase; F7GlcT, flavonol 7-O-glucosyltransferase; F7RhaT, flavonol 7-O-rhamnosyltransferase; Fd3GGlcT, flavonoid 3-O-glucoside glucosyltransferase; Fd3GlcT, flavonoid 3-O-glucosyltransferase; FLS, flavonol synthase; LDOX, leucoanthocyanidin reductase (anthocyan synthase); OMT1, O-methyltransferase1. Modified according to Saito et al. (2013) and Stracke et al. (2010), and incorporating data from Yonekura-Sakakibara et al. (2014).
Several A. thaliana flavonol GTs have been identified by transcriptome co-expression- and genome-wide similarity-analyses and were functionally characterized, mainly by studying loss-of-function mutants and substrate analyses of recombinant proteins. This lead to the identification of genes encoding a flavonol 3-O-rhamnosyltransferase (F3RhaT, UGT78D1) as well as a flavonol 7-O-glucosyltransferase (F7GlcT, UGT73C6) (Jones et al., 2003), a flavonoid 3-O-glucosyltransferase (F3GlcT, UGT78D2) (Tohge et al., 2005), a flavonol 7-O-rhamnosyltransferase (F7RhaT, UGT89C1), a flavonol 3-O-arabinosyltransferase (F3AraT, UGT78D3) (Yonekura-Sakakibara et al., 2007; Yonekura-Sakakibara et al., 2008), a flavonoid 3-O-glucoside 2″-O-glucosyltransferase (Fd3GGlcT, UGT79B6) (Yonekura-Sakakibara et al., 2014), and a recombinant GT that can utilize flavonol 3-O-glucosides as substrate (UGT79B1) (Yonekura-Sakakibara et al., 2012). Glycosylation of flavonols is catalysed by UDP-sugar-dependent glycosyltransferases (UGTs) that are active in the cytosol (Yonekura-Sakakibara and Hanada, 2011). However, even in the highly studied model plant A. thaliana, we remain some distance from comprehensively defining the complete flavonol glycoside biosynthesis pathway. The known flavonol structures and the likely presence of unidentified minor flavonol glycosides suggest that there are as yet unidentified genes encoding modification enzymes.
In A. thaliana, a single qualitative trait locus (QTL) on chromosome 1 has been described that corresponds to different glycosylated flavonols (Keurentjes et al., 2006). This QTL was mapped to 88.6 cM using a recombinant inbred line (RIL) population derived from a cross between the accessions Ler and Cvi. RILs carrying the Ler allele at this locus accumulate flavonols containing dihexosyl glycosides, whereas lines carrying the Cvi allele at this position do not. The authors speculate that a GT catalysing the production of flavonol dihexosides seems to be active or present in Ler but not in Cvi, thus affecting the flavonol composition. Alternatively, however, the QTL could encode a regulator of such a structural gene. The same qualitative trait was described by Stracke et al. (2009) between the accessions Col-0 and Nö-0. The flavonol derivatives accumulating in Nö-0 seedlings, but not in Col-0 seedlings, have been structurally identified as kaempferol 3-O-gentiobioside 7-O-rhamnoside (K3GG7R) and quercetin 3-O-gentiobioside 7-O-rhamnoside (Q3GG7R) (Fig. 1), also named kaempferol/quercetin 3-O-beta-[beta-D-glucosyl(1→6)D-glucoside]-7-O-alpha-L- rhamnoside (Stracke et al., 2009), which were first described as accumulating in Arabidopsis ‘Wassilewskija-2’ leaves (Veit and Pauli, 1999).
Here, we describe the mapping of this QTL using Ler × Col-0 RILs and whole genome association mapping. We identify the causal polymorphisms and the protein encoded by the affected locus BETA GLUCOSIDASE 6 (BGLU6). We confirm that BGLU6 is essential for the production of F3GG7R using knockout mutants and their complementation with a genomic fragment. Thus, in this study, we identify an accession-specific gene that leads to a qualitative difference in flavonol glycoside accumulation in A. thaliana, and encodes a newly identified type of flavonol glucosyltransferase that does not belong to the large canonical family of UGTs.
Material and methods
A. thaliana accessions, mapping population, and insertion mutants
Seeds of all used natural accessions of A. thaliana were obtained from the European Arabidopsis Stock Centre (NASC): Nordborg collection (N22660; Supplementary Table S1), Ws-4 (N5390). A RIL population of extensively genotyped lines [100 lines (N1899), 200 lines (N4858)] derived from a cross between the accessions Col-0 and Ler (Lister and Dean, 1993) was used for QTL mapping (http://arabidopsis.info/CollectionInfo?id=45). T-DNA insertion lines for At1g60270/BGLU6 were obtained from INRA Versailles (bglu6-1, line FLAG_338H01 in Ws-4) and NASC (bglu6-2, JIC SM line GT_5_84554 with stock ID N162904 in Ler). Homozygous insertion lines were identified by PCR-based genotyping. In FLAG_338H01, the wild-type allele was detected with primers RS1252 (5′-TAAGACCTCTCGTCATATCCACTCG-3′) and RS1224 (5′-TTGTGTGACGAAGAAACAAAGTATATGG-3′), and the insertion allele with primers RS1233 (5′-GAAGATGTGAA GTTGATGGTGGACA-3′) and RS1237 (5′-CGTGTGCCAGGT GCCCACGGAATAGT-3′). In GT_5_845554, the wild-type allele was detected with primers RS1233 and RS1228 (5′-CTAGGAGTAA GGAGAGAGGTTGCTCTGC-3′), and the insertion allele with primers RS1228 and B006 (5′-ACGGTCGGGAAACTAGCTCTAC-3′). The resulting PCR products were sequenced to determine the exact insertion positions.
Plant growth conditions
Surface-sterilized A. thaliana seeds were sown on 0.5 Murashige and Skoog medium, on 1.5% sucrose, 0.8% agar plates. They were kept for one day at 4°C in the dark, and then transferred to a phytochamber with 16h of light illumination per day at 21°C and 18.5°C at night. Seedlings were harvested on the 6th day after transfer into the light (5-day-old seedlings).
Linkage mapping of the F3GG7R trait
The genotype data of 542 molecular markers distributed across all five chromosomes in the Lister and Dean RIL population (Lister and Dean, 1993) was obtained from the European Arabidopsis Stock Center (NASC, http://arabidopsis.info/CollectionInfo?id=45). CAPS makers were generated from the information obtained from Max-Planck Arabidopsis SNP Consortium database (Schmid et al., 2003; Torjek et al., 2003). Additional markers were generated from the Ler genomic sequence (GenBank AM748036). All additional markers are listed in Supplementary Table S2. New markers and the F3GG7R trait were integrated into the A. thaliana genetic map to identify markers significantly linked (base 10 logarithm of odds of 3 and higher) to the F3GG7R trait using Mapmaker/EXP version 3.0b as described in Lander et al. (1987).
Genome-wide association analysis of F3GG7R
The association mapping was performed using the GWAPP web interface, a web application for genome-wide association mapping in Arabidopsis (http://gwas.gmi.oeaw.ac.at) (Seren et al., 2012). The accelerated mixed-model option with default setting was used to identity associations between the F3GG7R phenotype measured from 81 accessions and about 206 000 single nucleotide polymorphisms (SNPs) available from the web interface.
Flavonol glycoside extraction and detection
Methanolic extracts were produced from 50mg of plant material in 2mL reaction tubes by addition of 0.4mL 80% methanol and about 10–15 zirconia beads of 1mm diameter (Roth). Samples were homogenized in a Precellys 24 homogenizer (Bertin Technologies) at a speed of 6500rpm, two times for 45s. Homogenized samples were incubated for 10min at 70°C and centrifuged for 10min at 15 000g in a standard bench-top centrifuge. Supernatants were vacuum-dried in a SpeedVac at 60°C. The dried pellets were dissolved in 1 µL of 80% methanol mg−1 starting material. HPTLC with subsequent diphenylboric acid-2-aminoethyl ester (DPBA) staining of flavonoids was performed according to Wagner and Bladt (1995). Two microliters of methanolic extracts were spotted on 10cm × 10cm silica-60 HPTLC plates (Merck) used as the stationary phase. Adsorption chromatography was carried out using a system of ethyl acetate, formic acid, acetic acid, and water (100:26:12:12, v/v/v/v) as the mobile phase in a closed glass tank. Separated phenylpropanoid compounds were stained by spraying with a 1% DPBA (w/v) (Neu, 1957) solution in methanol, followed by spraying with 5% methanolic polyethylene glycol 4000 (w/v) solution. The stained chromatograms were examined under UV light (312nm) and photographed.
LC-MS was performed as described by Tohge and Fernie. (2010). All data were processed using Xcalibur 2.1 software (Thermo Fisher Scientific). Peak identification and annotation were performed by a combination approach using standard chemical confirmation (Nakabayashi et al., 2009), MS/MS profiling, retention time profiling, and mutant analysis (Tohge et al., 2007; Yonekura-Sakakibara et al., 2008).
Transcriptome co-expression analysis
The ATTED-II database (Obayashi et al., 2007) (version 7.1 from 17 August 2013; http://atted.jp/) was used to identify BGLU6 co-expressed genes. The strength of gene co-expression is given as Mutual Rank (MR), which is calculated based on the rank of Pearson’s correlation coefficient. Lower MR values indicate stronger gene co-expression.
RNA isolation and cDNA synthesis
RNA was isolated from 50mg plant material using the NucleoSpin® RNA Plant kit (Macherey-Nagel) according to the manufacturer’s instructions. cDNA was generated from 1 µg total RNA as described previously (Stracke et al., 2007).
cDNA cloning
The nucleotide sequence of the full-length coding sequence (CDS) of BGLU6_Ler was obtained by sequencing a BGLU6_Ler entry construct. PCR was performed with Phusion® polymerase (New England BioLabs) using primers containing attB recombination sites, with cDNAs from 5-day-old A. thaliana seedlings as template (primers were RS1253, 5′-attB1-CAATGAAAAAGACTTTTGCTCTGATTACC-3′; and RS1254, 5′-attB2-GGTCGGAGTAAGGAGAGAGGTTGCTCTGC-3′). The resulting amplicon was introduced into the GATEWAY® vector pDONRzeo using BP clonase (Invitrogen).
Subcellular localization studies
The BGLU6_Ler CDS containing GATEWAY® Entry plasmid was recombined with the expression vectors pMDC43 (N-terminal green fluorescent protein (GFP) fusion) and pMDC83 (C-terminal GFP fusion) (Curtis and Grossniklaus, 2003), resulting in constructs encoding GFP fusion proteins whose expression is driven by the cauliflower mosaic virus (CaMV) 35S promoter. Bright Yellow 2 (BY2) cells were treated as described in Haasen et al. (1999). GFP fluorescence was detected after 20h of incubation in the dark using a Leica DM 5500 B microscope (Leica) equipped with a charge-coupled device camera.
Complementation experiments with BGLU6_Ler
A 4050-bp genomic Ler fragment containing the BGLU6 gene, including 1217bp upstream of the translation start and 290bp downstream of the stop codon, was amplified with the primers RS1223 (5′-TCGAATAAACCAAATGAAGAAAGACGTG-3′) and RS1224. A corresponding GATEWAY® Entry clone was produced and the genomic BGLU6 fragment was introduced into the vector pMDC123 (Curtis and Grossniklaus, 2003). The resulting plasmid was transformed into the F3GG7R-negative A. thaliana accession Col-0. Flavonol extracts of T2 seedlings of different transgenic lines were analysed by HPTLC and LC-MS.
Quantitative PCR
Transcript levels of BGLU6 were determined in triplicate by quantitative PCR using the Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen) on a Rotor-Gene 6000 cycler (Corbett Life Science). Gene-specific primers were RS1268 (5′-AGCTGTCTTGGAGTATGTAAAGCA-3′) and RS1269 (5′-ATTTCAGCACACCACCAATGTAAG-3′). As reference gene we used PEX4 (PEROXIN4, At5g25760) determined with the use of the primers RS935 (5′-TTGGACGCTTCAGTCTGTGT-3′) and RS936 (5′-TGAACCCTCTCACATCACCA-3′).
Phylogenetic analysis
BGLU and acyl-glucose-dependent anthocyanin glucosyltransferase (AAGT) protein sequences were aligned by CLUSTALW implemented in MEGA5 (version 5.2.2) (Tamura et al., 2011). A phylogenetic tree was constructed from the aligned protein sequences by MEGA5 using the neighbour-joining method (Saitou and Nei, 1987) with the following parameters: bootstrap method (1000 replicates), Poisson model, uniform rates, and complete deletion.
Results
F3GG7R accumulation in A. thaliana natural variations
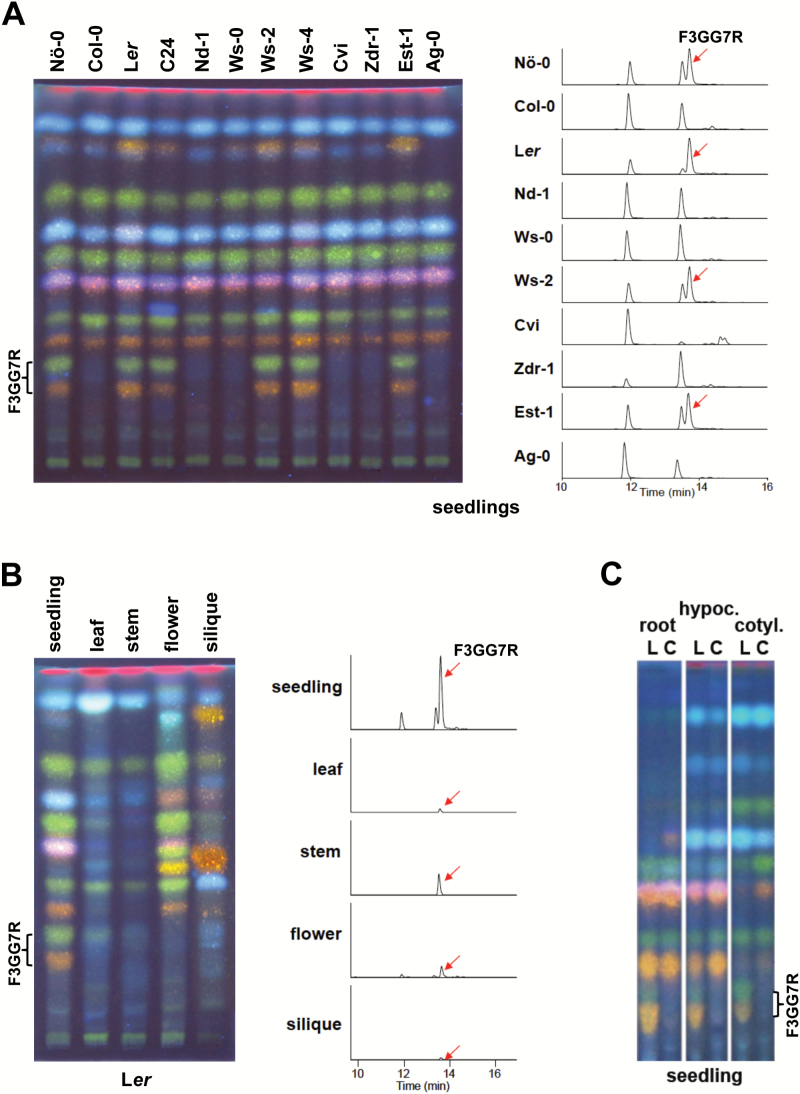
The observation that the secondary metabolites K3GG7R and Q3GG7R are found in A. thaliana seedlings of Ler and Nö-0 accessions, but not in seedlings of Col-0 and Cvi accessions, led us to question whether they accumulate in other natural accessions. For this purpose we used HPTLC to analyse methanolic extracts of plant material for abundant flavonol glycosides. We used a common staining reagent for flavonoid derivatives, DPBA, which visualizes kaempferol (green) and quercetin derivatives (yellow, orange) under UV illumination. Light-grown seedlings of 81 accessions of the Nordborg ecotype collection (N22660) were analysed by HPTLC; 39 accumulated K3GG7R and Q3GG7R (Ler phenotype) while the remainder did not (Col-0 phenotype) (Supplementary Table S1). Representative HPTLC results and the corresponding K3GG7R-specific LC-MS chromatograms from a subset of the Nordborg collection are provided in Fig. 2A. The K3GG7R trait always co-occurred with the Q3GG7R trait (and the isorhamnetin-derivative I3GG7R, as demonstrated by LC-MS analyses Supplementary Table S3), indicating that these traits were most likely influenced by a single locus (from now on called the F3GG7R locus/trait). The F3GG7R trait displayed Mendelian inheritance patterns between Col-0 and Ler, and analysis of F1 seedlings of reciprocal Col-0 × Ler crosses revealed that the Ler allele is dominant (data not shown).
Fig. 2.
Occurrence of the F3GG7R trait. HPTLC (left) and ion-extracted LC-MS chromatograms, m/z 755 (right) of methanolic extracts. In HPTLC, kaempferol derivatives are identified by green fluorescence, while quercetin derivatives appear yellow/orange. (A) Flavonol glycoside accumulation patterns of seedlings from different A. thaliana accessions. (B) Flavonol glycoside accumulation patterns of Ler plant organs. (C) Flavonol glycoside accumulation patterns of Ler (L) and Col-0 (C) seedling organs; hypocotyl (hypoc.) and cotyledon (cotyl.).
F3GG7R accumulation pattern in A. thaliana
Different flavonol derivatives are known to accumulate in distinct organs of A. thaliana plants. Given that the patterns of F3GG7R accumulation might provide information about the expression domain(s) of the causal gene, we performed spatial analysis of F3GG7R accumulation. For this purpose, seedlings and adult plants of the Ler accession were dissected and methanolic extracts were analysed by HPTLC and LC-MS (Fig. 2). Accumulation of F3GG7R compounds was found in all analysed organs of the Ler seedling (cotyledon, hypocotyl, and root; Fig. 2C), and also in adult plant organs at low levels (Fig. 2B).
Mapping of the F3GG7R locus by linkage analysis using RILs
Given the dominant nature of the F3GG7R trait, the location of underlying gene in the A. thaliana genome could be identified by Mendelian mapping. For this purpose, the F3GG7R phenotypes of 95 Ler × Col RIL seedlings from the Lister and Dean mapping population (Lister and Dean, 1993) were evaluated using HPTLC analysis, and the seedlings segregated into 32 lines containing the F3GG7R trait (Ler phenotype) and 63 lines displaying the Col phenotype (Supplementary Table S4). These phenotype scores were used for linkage analysis by comparing the score to the genotype data obtained from 542 polymorphic markers distributed among the five A. thaliana chromosomes (http://arabidopsis.info/CollectionInfo?id=45).
The linkage analysis between the F3GG7R segregation data and the genotype data was carried out using MAPMAKER 3.0b (Lander et al., 1993). The F3GG7R locus was mapped on the lower arm of chromosome 1, tightly linked to the restriction fragment length polymorphism (RFLP) marker mi304 (at 86.52 cM), and with a distance of 0 cM to the anchor markers g4026 (87.02 cM, between 22 276 033 and 22 276 876bp) and nga280 (83.83 cM between 20 873 698 and 20 873 802bp). The interval spans a distance of 1.4 Mbp (Supplementary Fig. S1A). To narrow down the area of the F3GG7R locus, CAPS markers between 22.02 Mbp and 23.82 Mbp on chromosome 1 were generated using information gained from the Max-Planck Arabidopsis SNP Consortium database (Schmid et al., 2003; Torjek et al., 2003). However, the mapping interval could not be shortened due to the lack of recombination events in the interval. By screening 200 additional RILs from the Lister and Dean population, we identified three RILs (N4662, N4674, and N4774) that contained recombination events between the markers. Using additional markers, the interval was reduced down to 106 kbp between the markers H98 (22.2 Mbp) and H238 (22.3 Mbp). A Ler BiBAC clone (BAC22K22) (Chang et al., 2003) containing a 211-kb Ler genomic DNA fragment covering the F3GG7R locus was identified and sequenced (GenBank AM748036). Using additional sequence-based markers generated by comparing Ler and Col-0 genome sequences of the region in question, the F3GG7R locus was located between the markers H238 and H139 (22 270 484bp) with an interval of 87kb (Supplementary Fig. S1B, C). Still, this region contains 29 annotated genes (Supplementary Table S5). In addition, there was no indication that this Ler region might include an additional gene that is not present in the corresponding Col-0 region.
According to the Plant Metabolic Network (http://pmn.plantcyc.org/), K3GG7R should be derived from kaempferol 3-O-glucoside 7-O-rhamnoside (K3G7R) and UDP-activated glucose, and the reaction should be catalysed by a UGT family-1 protein for which the corresponding gene has not been identified to date. Consequently, we predicted the candidate gene to encode an enzyme involved in UDP-dependent glucosylation of flavonol glycosides. However, examination of the annotation of the 87-kb genome region did not reveal the presence of a UGT-coding gene (Supplementary Table S5).
Whole genome association mapping of F3GG7R
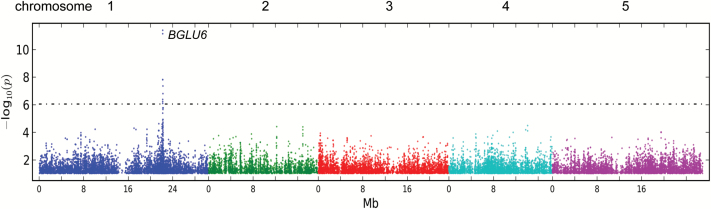
We performed genome-wide association mapping to identify the genes linked to the F3GG7R phenotype. F3GG7R was measured from 81 accessions from the Nordborg collection and used to identify SNPs associated with the accumulation of F3GG7R. We identified eight SNPs that had significant p-values above the cut off of the 5% false discovery rate. All eight SNPs were located between 22 196 382 and 22 267 729bp on pseudochromosome 1. The two highest p-values over -log10(p) = 10 were found on SNPs located at positions 22 222 268bp and 22 222 447bp (Fig. 3) within the gene BGLU6, annotated to encode a glycoside hydrolase family 1 (GH1) protein (Miyahara et al., 2011). An earlier study categorized BGLU6 as a pseudogene because of the lack of key catalytic motifs due to a premature stop codon, which is caused by an SNP located at position 22 222 268bp (Xu et al., 2004). The SNP at position 22 222 447 is located within the ninth intron of BGLU6.
Fig. 3.
Genome-wide association mapping of F3GG7R. Manhattan plots of genome-wide association mapping results for F3GG7R using 81 accessions from the Nordborg collection. The five A. thaliana chromosomes are indicated in different colours. The x-axis indicates the SNP positions in pseudochromosomes. The y-axis indicates the –log10 p-value of association with the SNPs. The grey dotted line displays the p-value cut off of the 5% false discovery rate. (This figure is available in colour at JXB online.)
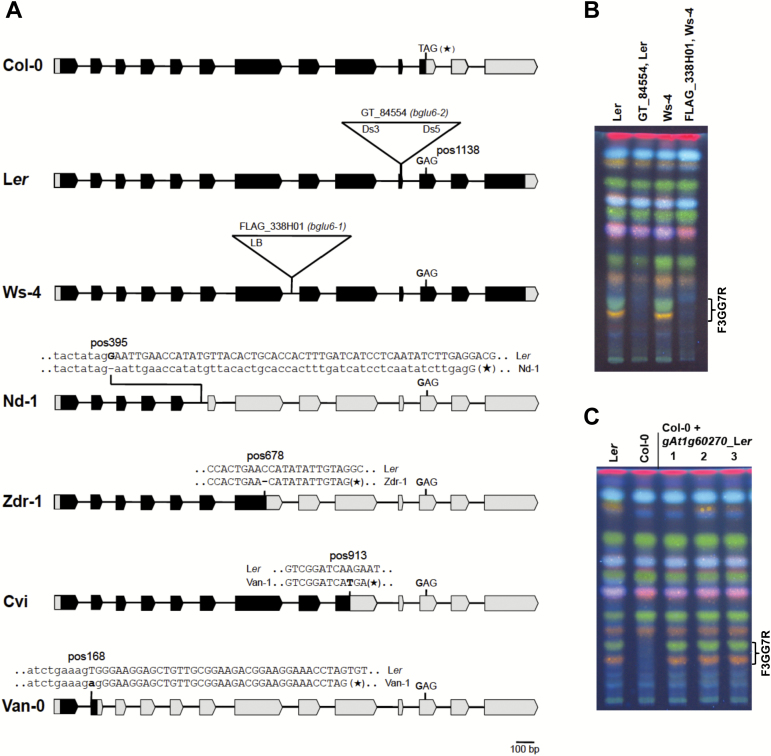
Sequence comparison of the BGLU6 alleles of Col-0 and Ler showed that the SNP at 22 222 268bp of pseudochromosome 1 (corresponding to position 1138bp of the BGLU6 CDS) modifies the stop codon (TAG, present in the Col-0 reference sequence) in the 11th exon into a glutamic acid codon (GAG) (Fig. 4A). Therefore, on the basis of the association data, BGLU6 is the best candidate gene underlying the F3GG7R trait.
Fig. 4.
BGLU6 alleles and their impact on F3GG7R accumulation. (A) BGLU6 alleles. The structure of At1g60270/BGLU6 wild-type and mutant alleles in different A. thaliana accessions and insertion mutants. The exon/intron structure is depicted. Boxed arrows and capital letters indicate exons, while thin black lines and small letters indicate introns. Translated regions are marked in black and untranslated regions in grey. Positions of T-DNA or transposon insertions are given as triangles. SNPs and deletions that are responsible for the loss of function are indicated and their relative position to the start codon (pos) is given. (B, C) Flavonol glycoside patterns, HPTLC of methanolic extracts of seedlings. (B) bglu6 T-DNA insertion mutants. (C) Col-0 complementation lines, carrying a 4.05-kb genomic BGLU6_Ler fragment.
Naturally occurring bglu6 alleles
In addition to the above described bglu6 alleles, which are non-functional because of the SNP at position 22 222 268 (BGLU6 CDS position 1138), further naturally occurring loss-of-function alleles were identified using the 1001 Genomes data and confirmed by Sanger sequencing of amplified genomic and cDNA fragments (Fig. 4A, Supplementary Table S1). A deletion of one nucleotide in the seventh exon (BGLU6 CDS position 678) results in a premature stop codon in the accessions Bor-4, Se-0, Uod-1, and Zdr-1. In Cvi, an SNP in the ninth exon (position 913) alters a glutamic acid codon (GAG) into a premature stop codon (TAG). In two other accessions, splice sites are affected, leading to mis-spliced transcripts. In Nd-1 this is due to a nucleotide deletion at the 5′-splice site of the sixth exon (position 395), and in Van-0 the splice defect is a result of an SNP modifying the 5′-splice site of the second exon (position 168).
The nucleotide variations of BGLU6 CDSs were compared in the 1001 Genomes population, currently comprising 504 accessions. SNP 1138 was the most frequent SNP. Of these accessions, 71% display the Ler allele while only 29% display the Col-0 allele, which causes a premature stop and results in the production of a truncated protein. SNP 1138 has only weak linkage disequilibrium, with three SNPs including silent mutations at positions 819 and 834, and a valine-to-isoleucine exchange of amino acids at position 1177 (Supplementary Table S6). Several SNPs or deletions are unique within the 504 analysed accessions (0.2%); one of them being the already mentioned loss-of-function deletion in Nd-1.
BGLU6 is co-expressed with phenylpropanoid biosynthesis genes
Because the Col-0 and Ler BGLU6 promoter sequences are not significantly different (Supplementary Fig. S2), a similar BGLU6 expression level is assumed in both accessions. We used the ATTED-II database (Obayashi et al., 2007) to identify genes transcriptionally co-expressed with BGLU6, and to estimate potential functional relationships. Within the 100 best correlating co-expressed A. thaliana genes, a striking number of genes related to phenylpropanoid biosynthesis was detected (Supplementary Table S7). These included TRANSPARENT TESTA6 (TT6/F3H, At3g51240), UGT78D2 (At5g17050), UGT89C1 (At1g06000), 4CL3 (At1g65060), UGT75C1 (At4g14090), CHI-like (At5g05270), UF3GT (At5g54060), TT19/GST26 (At5g17220), TT5/CHI (At3g55120), FLS1 (At5g08640), TT3/DFR (At5g42800), BRT1/UGT84A2 (At3g21560), UGT78D1 (At1g30530), and TT4/CHS (At5g13930). Given that phenylpropanoid metabolism is highly regulated at the transcriptional level, this finding strongly supports the involvement of BGLU6 in the flavonoid metabolic pathway.
Identification and analysis of insertional mutants of bglu6
Searching the established insertion mutant databases we identified bglu6 insertion alleles in the F3GG7R-accumulating wild-type accessions Ler and Ws-4. The T-DNA (FLAG_338H01, Ws-4, bglu6-1) and transposon (JIC SM Line GT_5_845554, Ler, bglu6-2) insertions were located in the tenth exon and the seventh intron, respectively (Fig. 4A). Homozygous insertion mutants were identified by PCR and subsequently analysed by HPTLC and LC-MS for F3GG7R accumulation. No insertion mutants displayed accumulation of F3GG7R (Fig. 4B, Supplementary Table S3), indicating two different loss-of-function alleles in different genetic backgrounds. These mutants confirm that a functional BGLU6 is essential for F3GG7R accumulation.
An BGLU6 Ler fragment complements the Col-0 phenotype
A 4050-bp genomic Ler fragment containing the BGLU6 gene, including 1217bp upstream of the translation start and 290bp downstream of the stop codon, was stably introduced in the F3GG7R-negative Col-0 accession via Agrobacterium-mediated transformation. Analysis of T2 seedlings of different transgenic lines indicated the accumulation of F3GG7R (Fig. 4C, Supplementary Table S3), confirming that a functional BGLU6 is required for F3GG7R accumulation.
BGLU6 is predominantly expressed in seedlings
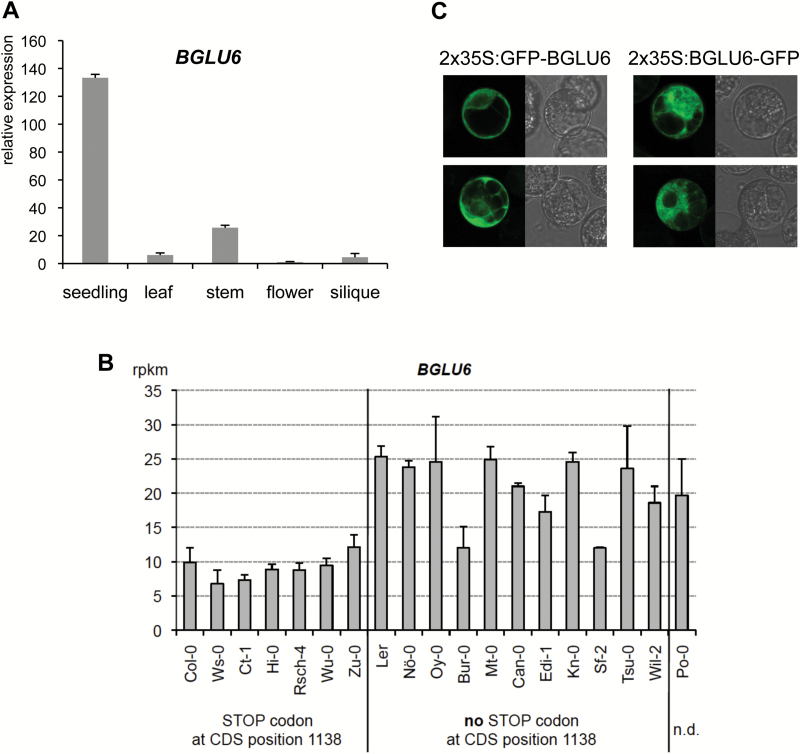
The expression of BGLU6 was monitored in Ler plant organs using quantitative real-time PCR, indicating that BGLU6 is predominantly expressed in seedlings (Fig. 5A). This expression pattern is in good accordance with the high level of F3GG7R found in the seedling (Fig. 2B).
Fig. 5.
BGLU6 is mainly expressed in seedlings and the protein seems to be localized in the cytoplasm. (A) Quantitative PCR data giving relative BGLU6 expression in Ler organs (flower expression level set to 1). (B) Expression of BGLU6 was analysed in seedlings of 19 different A. thaliana accessions using RNA-Seq data from the study of Gan et al. (2011). Expression levels are given in reads per kilobases per million mapped reads (RPKM). n.d., no data available. Different expression levels between the groups with or without a stop codon at CDS position 1138 are significantly correlated (p = 0.000936, Mann–Whitney U-test). (C) Subcellular localization of N- and C-terminal GFP fusion proteins with BGLU6 in transiently transfected BY2 protoplasts. Two representative protoplasts are shown for each construct. Left panels: GFP fluorescence. Right panels: bright field image using Nomarski optics. (This figure is available in colour at JXB online.)
In addition, we analysed the expression of BGLU6 in seedlings of 19 different A. thaliana accessions using RNA-Seq data from the study of Gan et al. (2011). Figure 5B shows that expression levels ranged from 6.8±2.0 to 25.3±1.6 reads per kilobase of exon model per million mapped reads (RPKM). The expression level of BGLU6 alleles without the premature stop codon (at CDS position 1138) was 20.7±5.0 RPKM (n = 11), while it was 9.0±1.8 RPKM (n = 7) for alleles with a premature stop codon. The different expression levels between the two groups were found to be significantly correlated (p = 0.000936, Mann–Whitney U-test). Thus, it seems that the 1138 stop codon may be correlated with a 2.3-times reduction in BGLU6 expression level in seedlings. The reason for this remains unclear.
The BGLU6 protein seems to be localized in the cytoplasm
The software Plant-mPLoc (Chou and Shen, 2010) predicted that the N-terminal amino acid sequences of BGLU6 would contain a putative transit peptide necessary for localization in the vacuole, and PlantLoc (Tang et al., 2013) predicted that BGLU6 would be located in the peroxisome (microbody) or vacuole. To confirm the subcellular localization of BGLU6 protein (encoded by the Ler allele), N- and C-terminal GFP fusion constructs driven by the CaMV 35S promoter were introduced into tobacco BY2 protoplasts. In contrast to the predictions, a clear GFP signal was observed in the cytoplasm of BY2 cells (Fig. 5C), indicating a cytoplasmic localization of BGLU6-GFP. Although it is possible that the distribution of transiently expressed full-length BGLU6-GFP in BY2 cells might differ from that of the native protein, our results suggest that the cytoplasm is a strong candidate location as the final destination of BGLU6.
Discussion
Differences in flavonoid composition between naturally occurring A. thaliana accessions are mainly quantitative rather than qualitative (Routaboul et al., 2012). However, besides the metabolite F3GG7R described in this work, other accession-specific flavonol glycosides have been reported in leaves of some accessions; for example, kaempferol 3-O-rhamnosyl-glucosylglucoside-7-O-rhamnoside in C24 (Stobiecki et al., 2006) and kaempferol-hexosylhexoside-deoxyhexoside in Ms-0 (Tohge and Fernie, 2010). BGLU6 is the first gene to be determined as responsible for the accumulation (or non-accumulation) of an accession-specific flavonol glycoside in A. thaliana. The qualitative difference in flavonoids described in this work is not due to the presence of an additional or missing accession-specific gene, but rather to the loss-of-function of a gene present and transcribed in all analysed accessions. The biological function of F3GG7R is currently unknown; however, the accumulation of F3GGR is clearly expendable in many accessions, which could indicate its function in adaptation to different environmental perturbations such as tolerance of high UV radiation. For instance, glycosylation of flavonols has shown to alter absorbance in the UV-B spectral region and suggested to be instrumental in the protection of plants against UV damage (Smith and Markham, 1998). In this respect we tried to correlate F3GG7R production with the level of UV radiation (UV-index) or the altitude at the collection places of the Nordborg A. thaliana accessions, but found no obvious correlation (Supplementary Fig. S3). Correlations may be confounded by the underlying structure of the Nordborg collection, given that accessions of this collection are not independent populations, but rather are connected through a genetic and geographical population structure (Weigel, 2012).
From the flavonol glycoside composition in seedlings from the F3GG7R-negative accession Col-0 (Saito et al., 2013), it seems likely that flavonol 3-O-glucoside 7-O-rhamnoside (F3G7R) is the substrate of BGLU6, which presumably adds a second glucose residue to the present glucose at the 3 position, forming flavonol 3-O-glucosyl-glucoside-7-O-rhamnoside (Fig. 1). Because these flavonol derivatives are known to have a 1→6 glycosidic linkage between the two glucose residues (Veit and Pauli, 1999; Stracke et al., 2009; Saito et al., 2013), we categorize BGLU6 as a flavonol 3-O-glucoside: 6″-O-glucosyltransferase. Interestingly, as indicated from the flavonol profile analyses of the bglu6-1 mutant and BGLU6 complementation lines (Supplementary Table S3), it seems that the level of the supposed substrate F3G7R is independent of the activity of BGLU6. This may hint to unknown mechanisms that cap the accumulation of flavonol glycosides.
There are some other GTs known to catalyse glycosylation at a sugar moiety attached to flavonol aglycones (FGGTs). For example, UGT707B1 from Crocus sativa (saffron) is involved in the formation of kaempferol- and quercetin 3-O-glucosyl-(1→2)-glucoside (flavonol 3-O-sophorose) (Trapero et al., 2012) and UGT3 from Catharanthus roseus (Madagascar periwinkle) catalyses the 1→6-glucosylation of flavonol and flavone glucosides, forming 3-O-gentiobioside, gentiotrioside, and gentiotetroside in a sequential manner (Masada et al., 2009). Very recently, A. thaliana UGT79B6 was identified as flavonol 3-O-glucoside: 2″-O-glucosyltransferase, needed for the formation of flavonol 3-O-glucosyl-(1→2)-glucoside in pollen (Yonekura-Sakakibara et al., 2014). All three enzymes add a second glucose residue to a flavonol derivative bearing a glucose at the 3 position, forming flavonol 3-O-diglucosides with a 1→2 or 1→6 interglycosidic linkage and use UDP-glucose as the activated sugar donor substrate. Similar to CaUGT3, BGLU6 forms a 1→6 interglycosidic linkage, but by contrast shows structural similarity to GH1 group proteins. Because BGLU6 does not belong to the large family of GTs that utilize UDP-conjugates as the activated sugar donor substrate, it represents an atypical type of flavonol glucosyltransferase.
Until recently it had been assumed that glycosylation of flavonoids was catalysed solely by UGTs. In 2010, a novel glucosylation mechanism was reported in flowers of Dianthus caryophyllus (carnation) and Delphinium grandiflorum, using crude protein extract from plants and purified native and recombinant proteins. The authors found that acyl-glucose-dependent anthocyanin glucosyltransferases (DcAA5GT and DgAA7GT), previously annotated as putative BGLUs, catalysed the formation of anthocyanin 3,5-diglucosides and anthocyanin 3,7-diglucosides using anthocyanin 3-O-glucoside and acyl-glucose as substrates (Matsuba et al., 2010). A similar reaction was reported in A. thaliana (Miyahara et al., 2013), assigning AAGT activity to the GH1 group protein BGLU10. For AtBGLU10, the energy-rich compound sinapate glucose was identified as the substrate of the glycosylation reaction that leads to the addition of glucose to an acyl moiety (4-coumarate) of an anthocyanin modified with sugars and organic acids.
Based on the sequence similarity of BGLU6 to AtBGLU10 and other AAGTs (Supplementary Fig. S4), we hypothesize that BGLU6 most likely could utilize acyl-glucose derivatives (e.g. glucose esters of sinapic or benzoic acid) rather than UDP sugars as the sugar donors.
In order to test this hypothesis, we tried to examine BGLU6 activity in crude protein extracts prepared from A. thaliana Ler seedlings and to analyse its dependence of flavonoid acceptors and glucose donors. In addition, we tried to characterize BGLU6 activity using an in vitro assay with recombinant BGLU6. Unfortunately, both approaches failed. Despite intensive efforts, we could not detect flavonol 3-O-glucoside: 6″-O-glucosyltransferase activity in Ler seedling protein extracts and we were unable to achieve high enough yields of active recombinant protein (from expression in Escherichia coli and Pichia pastoris) to characterize it biochemically. Thus, our functional characterization of the BGLU6 enzyme was limited to that afforded by the expression of this enzyme in a F3GG7R-non-producing A. thaliana background (complementation lines, Fig. 4C, Supplementary Table S3), which at least should be sufficient to give a classification as flavonol 3-O-glucoside: 6″-O-glucosyltransferase for the enzyme’s activity. The question of what the appropriate flavonoid acceptor(s) or glucose donor(s) are remains open.
The amino acid sequences of known GH1-type glucosyltransferases (DcAA5GT, DgAA7GT, AaAA7GT, AtBGLU10) contain the classic highly conserved peptide motifs TFNEP and I/VTENG from GH1 enzymes such as TF/INEA/P and I/VH/L/QENG. The conserved glutamate residues of these key catalytic motifs are not important for distinguishing whether the enzymes are glucosyltransferases or glucosylhydrolases (Matsuba et al., 2010). In BGLU6, the second motif is in accordance with those found in AAGTs; the first motif (TINEG) does not fit, because it has a glycine at the end (Supplementary Fig. S4). Whether this position is related to different substrate specificities (anthocyanins or flavonols) remains to be determined.
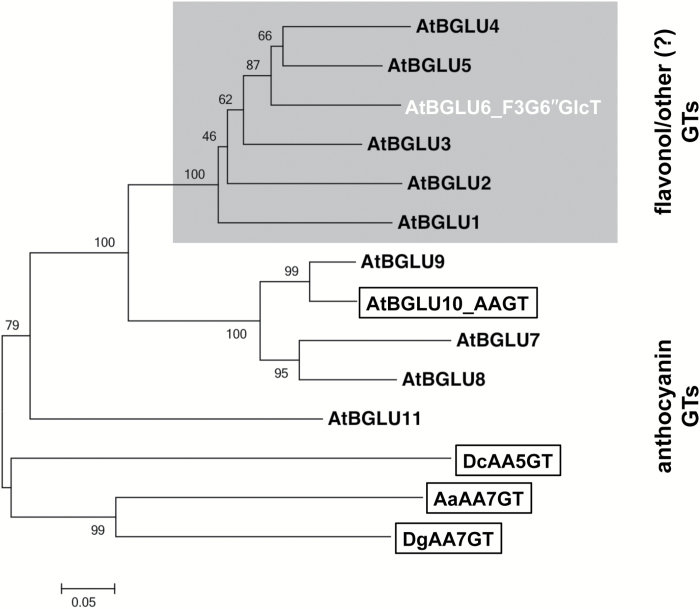
A phylogenetic analysis of known AAGTs and 11 homologous BGLU proteins from A. thaliana, as described by Miyahara et al. (2011), indicated a subgroup separated from all proven AAGTs (Fig. 6). Members of this group, comprising AtBGLU1-5 and AtBGLU6, potentially act on substrates other than anthocyanins, as shown for AtBGLU6 in this study. Further work on these AtBGLUs is required to prove this hypothesis.
Fig. 6.
Unrooted molecular phylogenetic neighbour-joining tree of known AAGTs and homologous BGLU proteins from A. thaliana. The phylogenetic tree was constructed as described in ‘Methods’. The percentages of replicate trees in which the associated proteins clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Bar = 0.05 amino acid substitutions per site. Known AAGTs are shown boxed and BGLU6 is in white. AtBGLUs showing homology to AAGTs were extracted from Miyahara et al. (2011). Abbreviations for species: Aa, Agapanthus africanus; At, Arabidopsis thaliana; Dc, Dianthus caryophyllus; Dg, Delphinium grandiflorum.
BGLU6 co-expression analysis using the ATTED-II database returned several genes of flavonoid biosynthesis, including genes encoding (i) the enzymes building the flavonol aglycone, like CHS, CHI, F3H, and FLS1; and (ii) the UGTs acting on flavonol substrates like UGT78D2/flavonoid 3-O-glucosyltransferase, UGT89C1/flavonol 7-O-rhamnosyltransferase, and UGT78D1/flavonol 3-O-rhamnosyltransferase. In an alternative approach, BGLU6 was identified in a study by Miyahara et al. (2011) as the third ranked co-expressed gene (by Pearson’s correlation coefficient) using a set of genes that encode enzymes involved in anthocyanin modification as baits, namely UGT78D2 (At5g17050), UGT75C1 (At4g14090), and At5MAT (At3g29590). These findings clearly support the functional inclusion of BGLU6 within the flavonoid biosynthesis pathway.
Studies by Meissner et al. (2008) indicated that the four A. thaliana UGT84A genes (UGT84A1–A4) encode GTs converting hydroxycinnamates to 1-O-glucose esters. UGT84A expression is induced by UV-B radiation and transgenic A. thaliana lines overexpressing single UGT84A genes show increased levels of sinapoyl glucose in seeds and seedlings. UGT84A2, which is co-expressed with BGLU6 (Supplementary Table S7), was identified as the main contributor to the production of 1-O-sinapoyl glucose (Yonekura-Sakakibara et al., 2012). Under the assumption that the energy-rich, cytosol-located compound sinapoyl glucose (Strack and Sharma, 1985) is a substrate of BGLU6, as it is for other known BGLU-like acyl-glucose-dependent glucosyltransferases (AGTs) (Matsuba et al., 2010; Miyahara et al., 2012; Miyahara et al., 2013), a combined activation of genes forming flavonol glycosides and sinapate glucose would be anticipated to accumulate corresponding substrates. In this respect, it would be interesting to analyse the F3GG7R level in a ugt84a2 loss-of-function mutant. Unfortunately, to our knowledge, such a mutant allele (natural or artificially inserted) does not exist in a F3GG7R-producing accession. Further analyses will be required to validate the connection between BGLU6 and UGT84A genes.
Because co-expression reflects similar mRNA-level regulation, it is likely that the transcription factors regulating the expression of genes encoding the biosynthesis enzymes also contribute to the establishment of proper conditions (e.g. pH) and compartments (e.g. vacuole, cell wall) for the accumulation of the modified flavonols. The R2R3-MYB transcriptional regulators MYB12, MYB11, and MYB111 (PFG1–3) have been identified as flavonol branch-specific activators, which activate the transcription of biosynthetic enzymes of the general phenylpropanoid pathway (4CL) and the flavonol pathway [CHS, CHI, F3H, FLS, several UGTs (Mehrtens et al., 2005; Stracke et al., 2007; Nakabayashi et al., 2014)]. Specifically, the flavonol-3-O-rhamnosyltransferase gene UGT78D1 and the flavonol 7-O-rhamnosyltransferase gene UGT89C1, both known to be targets of the PFGs (Stracke et al., 2007), are co-expressed with BGLU6 (Supplementary Table S7), indicating a possible PFG-dependent concerted expression. In this context we wanted to know if BGLU6 is a target gene of the PFGs. Using the NewPLACE database (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?sid=&lang=en&pj=640&action=page&page=newplace), predictive MYB binding sites were found in the 1.2kb BGLU6 promoter region of 10 natural A. thaliana accessions (Supplementary Fig. S2), including proBGLU6_Ler, which was found to be sufficient for functional complementation (Fig. 4C). To analyse if the three PFGs have the capability to directly bind and activate the BGLU6 promoter, we used transient co-transfection of A. thaliana At7 protoplast cells with 35S-driven PFG1-3 as the effector and a 1.2-kb BGLU6_Ler promoter–GUS fusion as the reporter. None of the tested R2R3-MYBs were able to activate the BGLU6 promoter (Supplementary Fig. S5). This result hints that the BGLU6 promoter itself is not a target of the PFG R2R3-MYBs, like the promoters of the flavonol 3-O-arabinosyltransferase UGT78D3 (Yonekura-Sakakibara et al., 2008) and the flavonol 7-O-glucosyltransferase UGT73C6 (Jones et al., 2003), which were not activated in this assay (Stracke et al., 2010). It at least indicates that BGLU6 regulation is different from CHS regulation. However, a potential activation of these genes in planta cannot be formally excluded owing to the possible limitations of the transient co-transfection assay system using hypocotyl-derived At7 protoplasts (Trezzini et al., 1993) or because of missing, unknown co-factors.
We identified BGLU6 as a novel type of flavonol glucosyltransferase not belonging to the large family of GTs that utilize UDP-conjugates as the activated sugar donor substrate. A phylogenetic analysis of all currently known AAGTs and the homologous BGLU proteins from A. thaliana revealed two different subgroups (Fig. 6), one containing all AAGTs and a second containing AtBGLU1 to AtBGLU6. Further analyses of AtBGLUs will answer whether this separation is due to different flavonoid substrate specificities.
Conclusion
In this study we describe a single trait locus causing natural qualitative variation in the accumulation of F3GG7R in A. thaliana. This locus was mapped using RILs and whole genome association mapping and found to encode a flavonol 3-O-glucoside: 6″-O-glucosyltransferase, named BGLU6. Several natural and artificial loss-of-function alleles have been identified, and successful complementation experiments confirmed that functional BGLU6 is required for F3GG7R accumulation. BGLU6 was found to be expressed predominantly in the seedling, with the cytoplasm indicated as a candidate for the subcellular localization of the encoded protein. The BGLU6 protein does not belong to the large canonical family of UGT-type flavonol GTs, but represents a GH1-type GT, which have been heretofore only known to act on anthocyanin substrates.
Accession numbers
At1g60270/BGLU6_Ler gene, cDNA, and protein sequence can be found in the GenBank (NCBI/EMBL/DDBJ) databases under accession number KM047910. The Ler genomic sequence of the F3GG7R mapped interval can be found in GenBank under accession number AM748036.
Supplementary material
Supplementary material is available at JXB online.
Table S1: Listing of SNPs and deletions (Del) in the At1g60270 gene of A. thaliana natural accessions from the Nordborg collection, correlating with the presence or the absence of F3GG7R accumulation.
Table S2: PCR-based markers. PCR primer sets and restriction enzymes used for genotyping of the RILs are listed.
Table S3: LC-MS derived quantitative flavonol profiles of A. thaliana accessions, BGLU6 T-DNA insertion lines, and BGLU6 complementation lines.
Table S4: F3GG7R chemotype of Lister and Dean Ler × Col RILs.
Table S5: List of annotated genes on Col-0 chromosome 1 between the flanking markers H139 and H238.
Table S6: Sequence polymorphisms and their linkage disequilibrium in BGLU6/At1g60270.
Table S7: List of 100 genes showing strongest co-expression with BGLU6.
Fig. S1: Fine-mapping of the F3GG7R locus on chromosome 1.
Fig. S2: BGLU6 promoter, alignment, and putative MYB binding sites.
Fig. S3: Correlation analyses of F3GG7R production with environmental factors at accession collection places.
Fig. S4: Multiple alignment of GH1-type flavonoid glucosyltransferases.
Fig. S5: Co-transfection analysis of target gene specificities in A. thaliana At7 protoplasts.
Acknowledgements
We thank Melanie Kuhlmann for excellent technical help and Thorsten Seidel for assistance with confocal microscopy. We acknowledge the financial support of the German Research Foundation (DFG) and the Open Access Publication Fund of Bielefeld University for the article processing charge.
References
- Andersen ØM, Jordheim M. 2006. The anthocyanins. In: Andersen ØM, Markham KR. eds. Flavonoids: chemistry, biochemistry and application . Boca Raton, Florida, USA: CRC Press; pp. 471–553. [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. 1996. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. The Plant Cell 8, 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain CC, Saslowsky DE, Walker RA, Shirley BW. 1997. Expression of chalcone synthase and chalcone isomerase proteins in Arabidopsis seedlings. Plant Molecular Biology 35, 377–381. [DOI] [PubMed] [Google Scholar]
- Chang YL, Henriquez X, Preuss D, Copenhaver GP, Zhang HB. 2003. A plant-transformation-competent BIBAC library from the Arabidopsis thaliana Landsberg ecotype for functional and comparative genomics. Theoretical and Applied Genetics 106, 269–276. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. 2010. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5, e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana . Nature 477, 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Kondo T. 1991. Structure and molecular stacking of anthocyanins - flower color variation. Angewandte Chemie International Edition 30, 17–33. [Google Scholar]
- Gronquist M, Bezzerides A, Attygalle A, Meinwald J, Eisner M, Eisner T. 2001. Attractive and defensive functions of the ultraviolet pigments of a flower (Hypericum calycinum). Proceedings of the National Academy of Sciences of the United States of America 98, 13745–13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T. 1999. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana . The Plant Journal 20, 695–705. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C. 2004. Light induces phenylpropanoid metabolism in Arabidopsis roots. The Plant Journal 38, 765–778. [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J, Schaffner AR, Saito K. 2003. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana . Journal of Biological Chemistry 278, 43910–43918. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, De Vos RCH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M. 2006. The genetics of plant metabolism. Nature Genetics 38, 842–849. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Day MJ, Lincoln SE, Newberg L. 1993. Mapmarker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genetics 121, 174–181. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana . The Plant Journal 4, 745–750. [DOI] [PubMed] [Google Scholar]
- Masada S, Terasaka K, Oguchi Y, Okazaki S, Mizushima T, Mizukami H. 2009. Functional and structural characterization of a flavonoid glucoside 1,6-glucosyltransferase from Catharanthus roseus . Plant & Cell Physiology 50, 1401–1415. [DOI] [PubMed] [Google Scholar]
- Matsuba Y, Sasaki N, Tera M, Okamura M, Abe Y, Okamoto E, Nakamura H, Funabashi H, Takatsu M, Saito M, et al. 2010. A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the petals of carnation and delphinium. The Plant Cell 22, 3374–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. 2005. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology 138, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner D, Albert A, Bottcher C, Strack D, Milkowski C. 2008. The role of UDP-glucose:hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana . Planta 228, 663–674. [DOI] [PubMed] [Google Scholar]
- Miyahara T, Matsuba Y, Ozeki Y, Sasaki N. 2011. Identification of genes in Arabidopsis thaliana with homology to a novel acyl-glucose dependent glucosyltransferase of carnations. Plant Biotechnology 28, 311–315. [Google Scholar]
- Miyahara T, Sakiyama R, Ozeki Y, Sasaki N. 2013. Acyl-glucose-dependent glucosyltransferase catalyzes the final step of anthocyanin formation in Arabidopsis. Journal of Plant Physiology 170, 619–624. [DOI] [PubMed] [Google Scholar]
- Miyahara T, Takahashi M, Ozeki Y, Sasaki N. 2012. Isolation of an acyl-glucose-dependent anthocyanin 7-O-glucosyltransferase from the monocot Agapanthus africanus . Journal of Plant Physiology 169, 1321–1326. [DOI] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. 1992. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proceedings of the National Academy of Sciences of the United States of America 89, 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Kusano M, Kobayashi M, Tohge T, Yonekura-Sakakibara K, Kogure N, Yamazaki M, Kitajima M, Saito K, Takayama H. 2009. Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana . Phytochemistry 70, 1017–1029. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Mori T, Saito K. 2014. Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana . Plant Signaling & Behavior 9, e29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu R. 1957. Chelate von Diarylborsäuren mit aliphatischen Oxyalkylaminen als Reagenzien für den Nachweis von Oxyphenyl-benzo-γ-pyronen. Die Naturwissenschaften 44, 181–182. [Google Scholar]
- Nielsen K, Deroles SC, Markham KR, Bradley MJ, Podivinsky E, Manson D. 2002. Antisense flavonol synthase alters copigmentation and flower color in lisianthus. Molecular Breeding 9, 217–229. [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. 2007. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Research 35, D863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson LC, Veit M, Weissenböck G, Bornman JF. 1998. Differential flavonoid response to enhanced UV-B radiation in Brassica napus . Phytochemistry 49, 1021–1028. [Google Scholar]
- Ormrod DP, Landry LG, Conklin PL. 1995. Short-term UV-B radiation and ozone exposure effects on aromatic secondary metabolite accumulation and shoot growth of flavonoid-deficient Arabidopsis mutants. Physiologia Plantarum 93, 602–610. [Google Scholar]
- Peer WA, Blakeslee JJ, Yang H, Murphy AS. 2011. Seven things we think we know about auxin transport. Molecular Plant 4, 487–504. [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Shirley BW. 1996. Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiology 111, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri S, Tattini M. 2011. Flavonols: old compounds for old roles. Annals of Botany 108, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul JM, Dubos C, Beck G, Marquis C, Bidzinski P, Loudet O, Lepiniec L. 2012. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. Journal of Experimental Botany 63, 3749–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. 2013. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiology and Biochemistry 72, 21–34. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B. 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana . Genome Research 13, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü, Vilhjálmsson BJ, Horton MW, Meng D, Forai P, Huang YS, Long Q, Segura V, Nordborg M. 2012. GWAPP: a web application for genome-wide association mapping in Arabidopsis. The Plant Cell 24, 4793–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Markham KR. 1998. Tautomerism of flavonol glucosides: relevance to plant UV protection and flower colour. Journal of Photochemistry and Photobiology A: Chemistry 118, 99–105. [Google Scholar]
- Solovchenko A, Schmitz-Eiberger M. 2003. Significance of skin flavonoids for UV-B-protection in apple fruits. Journal of Experimental Botany 54, 1977–1984. [DOI] [PubMed] [Google Scholar]
- Stobiecki M, Skirycz A, Kerhoas L, Kachlicki P, Muth D, Einhorn J, Mueller-Roeber B. 2006. Profiling of phenolic glycosidic conjugates in leaves of Arabidopsis thaliana using LC/MS. Metabolomics 2, 197–219. [Google Scholar]
- Strack D, Sharma V. 1985. Vacuolar localization of the enzymatic synthesis of hydroxycinnamic acid esters of malic acid in protoplasts from Raphanus sativus leaves. Physiologia Plantarum 65, 45–50. [Google Scholar]
- Stracke R, De Vos RCH, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B. 2009. Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229, 427–445. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal 50, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B. 2010. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytologist 188, 985–1000. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Li T, Cong P, Xiong W, Wang Z, Sun J. 2013. PlantLoc: an accurate web server for predicting plant protein subcellular localization by substantiality motif. Nucleic Acids Research 41, W441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. 2010. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nature Protocols 5, 1210–1227. [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. 2005. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. The Plant Journal 42, 218–235. [DOI] [PubMed] [Google Scholar]
- Tohge T, Yonekura-Sakakibara K, Niida R, Watanabe-Takahashi A, Saito K. 2007. Phytochemical genomics in Arabidopsis thaliana: a case study for functional identification of flavonoid biosynthesis genes. Pure and Applied Chemistry 79, 811–823. [Google Scholar]
- Torjek O, Berger D, Meyer RC, Mussig C, Schmid KJ, Rosleff Sorensen T, Weisshaar B, Mitchell-Olds T, Altmann T. 2003. Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. The Plant Journal 36, 122–140. [DOI] [PubMed] [Google Scholar]
- Trapero A, Ahrazem O, Rubio-Moraga A, Jimeno ML, Gomez MD, Gomez-Gomez L. 2012. Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus . Plant Physiology 159, 1335–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trezzini GF, Horrichs A, Somssich IE. 1993. Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Molecular Biology 21, 385–389. [DOI] [PubMed] [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JNM, Stuitje AR. 1992. Antisense inhibition of flavonoid biosynthesis in Petunia anthers results in male sterility. The Plant Cell 4, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Pauli G. 1999. Major flavonoids from Arabidopsis thaliana leaves. Journal of Natural Products 62, 1301–1303. [DOI] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI. 2001. Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. The Plant Journal 25, 675–685. [DOI] [PubMed] [Google Scholar]
- Wagner H, Bladt S. 1995. Plant drug analysis: a thin layer chromatography atlas . Berlin: Springer. [Google Scholar]
- Weigel D. 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiology 158, 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2002. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biology 5, 218–223. [DOI] [PubMed] [Google Scholar]
- Xu Z, Escamilla-Trevino L, Zeng L, Lalgondar M, Bevan D, Winkel B, Mohamed A, Cheng CL, Shih MC, Poulton J, et al. 2004. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Molecular Biology 55, 343–367. [DOI] [PubMed] [Google Scholar]
- Yin R, Han K, Heller W, Albert A, Dobrev PI, Zažímalová E, Schäffner AR. 2014. Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytologist 201, 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, et al. 2012. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana . The Plant Journal 69, 154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. 2011. An evolutionary view of functional diversity in family 1 glycosyltransferases. The Plant Journal 66, 182–193. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Nakabayashi R, Sugawara S, Tohge T, Ito T, Koyanagi M, Kitajima M, Takayama H, Saito K. 2014. A flavonoid 3-O-glucoside: 2”-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana . The Plant Journal 79, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. 2008. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. The Plant Cell 20, 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. 2007. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. Journal of Biological Chemistry 282, 14932–14941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.