Highlight
An integrated approach of metabolomics and transcriptomics was applied to understand regulatory networks associated with biosynthesis of anthocyanins that are differentially regulated in light-red- and dark-purple-colored potato cultivars.
Key words: Anthocayanin, metabolomics, colored potato, RNA-seq, ultrapressure liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF-MS).
Abstract
To gain insights into the regulatory networks related to anthocyanin biosynthesis and identify key regulatory genes, we performed an integrated analysis of the transcriptome and metabolome in sprouts germinated from three colored potato cultivars: light-red Hongyoung, dark-purple Jayoung, and white Atlantic. We investigated transcriptional and metabolic changes using statistical analyses and gene–metabolite correlation networks. Transcript and metabolite profiles were generated through high-throughput RNA-sequencing data analysis and ultraperformance liquid chromatography quadrupole time-of-flight tandem mass spectrometry, respectively. The identification and quantification of changes in anthocyanin were performed using molecular formula-based mass accuracy and specific features of their MS2 spectra. Correlation tests of anthocyanin contents and transcriptional changes showed 823 strong correlations (correlation coefficient, R 2>0.9) between 22 compounds and 119 transcripts categorized into flavonoid metabolism, hormones, transcriptional regulation, and signaling. The connection network of anthocyanins and genes showed a regulatory system involved in the pigmentation of light-red Hongyoung and dark-purple Jayoung potatoes, suggesting that this systemic approach is powerful for investigations into novel genes that are potential targets for the breeding of new valuable potato cultivars.
Introduction
To date, more than 635 anthocyanins have been identified in numerous fruits, vegetables, and flowers (Wu and Prior, 2005; He and Giusti, 2010; He et al., 2010). The anthocyanin derivatives of delphinidin, petunidin, and malvidin are sources of purple and dark colors, whereas the derivatives of cyanidin and pelargonidin are the main pigments in bright-red-colored fruits (Jaakola, 2013). Although several genes encoding proteins implicated in anthocyanin biosynthesis and regulation have been identified, insights into the regulation of each anthocyanin biosynthesis pathway have remained future objectives. Recently, the integration of large-scale datasets derived from high-throughput functional genomics techniques have been applied successfully to studies on the functions of genes regulating tissue development (Persson et al., 2005), environmental responses (Wang et al., 2006; Cho et al., 2008), and plant metabolism (Hirai et al., 2007; Gutiérrez et al., 2008). In particular, transcript and metabolite datasets have been combined through correlation and clustering analyses and further represented as connection networks between genes and metabolites in many plants (Urbanczyk‐Wochniak et al., 2003; Hoefgen and Nikiforova, 2008; Saito et al., 2008), including Arabidopsis (Hirai et al., 2004, 2007), tomatoes (Alba et al., 2005; Mounet et al., 2009), and potatoes (Stushnoff et al., 2010). The acquisition and integration of an “omics” dataset (i.e. transcriptome, proteome, and metabolome) represents a useful approach for the establishment of a strategy to identify potential genes regulating the determination of pigmentation in potatoes.
Colored potatoes have attracted research interest due to their anthocyanin content with enhanced antioxidant capacities (Truong et al., 2009; Stushnoff et al., 2010; Chong et al., 2013). Solanum tuberosum cvs Hongyoung and Jayoung are pigmented potato cultivars originated from a cross made between the white-colored Atlantic and deep dark-purple-colored AG34314 cultivars through the potato breeding program of the National Institute of Highland Agriculture Research Center in 2003 (Park et al., 2009a, b). Hongyoung has a light-red skin and light-red flesh, while Jayoung has dark-purple skin and dark-purple flesh (see Supplementary Fig. S1 available at JXB online).
Most anthocyanins have been identified by the combined methods of ultraviolet/visible (UV/Vis) spectrometry and mass spectrometry (MS). In positive ionization mode, the [M+] ion and mass fragmentation patterns of anthocyanins are the same as the [M+H]+ ion and fragmentation patterns of flavonol (Wu and Prior, 2005; Sun et al., 2012). Anthocyanins (480–540nm) and non-anthocyanin phenolic compounds (<400nm) have maximum absorbance at different UV/Vis wavelengths. Thus, a liquid chromatography (LC) mass spectrometer equipped with a photodiode array detector is usually used to distinguish anthocyanin and flavonol glycosides (Lin et al., 2011; Sun et al., 2012). A recent study showed that MS data acquired in the negative ionization mode using ultrahigh-performance LC with high-resolution MS provided a series of characteristic ions for anthocyanins (e.g. [M-2H]–, [M-2H+H2O]–, and formic acid adducts) (Sun et al., 2012), suggesting that the integrative analysis of mass ions and mass fragmentations acquired from both the positive and negative modes can distinguish and identify anthocyanin and flavonol glycosides.
In this study, we explored the regulatory networks of anthocyanin biosynthesis in colored potatoes at the level of the transcriptome and metabolome. We focused on the differential expression of anthocyanin metabolites and their regulatory genes in light-red Hongyoung and dark-purple Jayoung potatoes compared with those of a white Atlantic potato cultivar. Connection networks were mapped on the basis of correlation analyses between metabolites and transcripts to highlight the regulatory genes associated with anthocyanin metabolites. Our findings provide new insights into the molecular mechanisms associated with the biosynthesis and regulation of anthocyanin in the pigmentation of potatoes, and highlight the usefulness of an integrated approach for understanding this process.
Materials and methods
Plant material
Medium-sized (80–150g) potato tubers from three different potato cultivars, Hongyoung, Jayoung, and Atlantic, were stored at a low temperature (4 °C) for 4 months after harvesting in the Dae-Gwal-Lyeong area (800 m above sea level), Korea. After storage, sprouts of potato tubers were induced at room temperature for 1 month with scattered light conditions. Whole sprouts were collected, immediately frozen in liquid nitrogen, and then stored at –80 °C prior to metabolite extraction.
Metabolite extraction
Frozen spouts of potatoes (200mg) were ground using a bead beater (4.5ms−1, 25s, three repetitions, MP 24X4, FastPrep-24; MP Biochemicals), suspended in methanol (600 μl) with a 0.125% formic acid solution, kept at 4 °C for 30min, sonicated at 4 °C for 20s (20kHz, 250W, three repetitions, Bioruptor-KRB-01; Bop-Medical Science), and centrifuged at 3000rpm for 15min at 4 °C. Finally, the supernatant solution was centrifuged at 13 000rpm for 10min at 4 °C and used for metabolomics analysis.
Metabolite profiling using ultraperformance LC quadrupole time-of-flight tandem MS (UPLC-Q-TOF-MS)
Metabolite profiling was conducted using a UPLC system (ACQUITY UPLC; Waters, Milford, MA, USA) and hybrid Q-TOF tandem mass spectrometry (Triple-TOF-MS) (Triple TOF 5600 system; AB SCIEX, Concord, ON, Canada). Chromatographic separation was performed on an ACQUITY UPLC BEH C18 column (2.1 mm×100 mm×1.7 μm; Waters) using mobile phase A (0.1% formic acid in deionized water) and mobile phase B (0.1% formic acid in acetonitrile). Mobile phase B was increased linearly from 3% at 0min to 50% at 3min to 70% at 4min to 100% at 10min, and then held at 100% until 10.5min. Finally, solvent B was decreased to 3% at 11min and held at 3% until 12min. The flow rate was maintained at 0.4ml min–1. Mass data acquisition was performed in both positive [electrospray ionization-positive (ESI+)] and negative (ESI–) modes using the following parameters: ion spray voltage of 5.5kV in ESI+ and –4.5kV in ESI–; nebulizer gas (gas 1) of 55 psi; heater gas (gas 2) of 65 psi; curtain gas of 30 psi; turbo spray temperature of 600 °C; and declustering potential of 100V in ESI+ and –100V in ESI–. For TOF MS2 data, information-dependent acquisition was used with the following conditions: survey scans of 250ms; product ion scans of 70ms; high-resolution mode; declustering potential of 90V; collision energy of 35V in ESI+ (–35V in ESI–); and collision energy spread of 15V. The TOF-MS and information-dependent acquisition scan was operated with the mass range of m/z 50–1600. TOF-MS and product ion calibration was performed in both high-sensitivity and high-resolution modes using a calibrant delivery system prior to analysis.
LC-MS data files (Wiff format files) including MS and MS2 spectra data were converted to mzXML files using MSConvert in the Proteowizard software (version 3.04999) (Patti et al., 2012). The converted raw data were further processed using MZmine software (version 2.10) and outputted as a retention time m/z dataset.
Multivariate statistical analysis
The intensities of mass peaks for each sample were sum-normalized and Pareto-scaled using the SIMCA-P+ software package (version 12.0; Umetrics, Umeå, Sweden). Principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) with data from 18 samples (three cultivars×six biological replicates) were performed to observe differences in metabolic composition among the three potato cultivars. The reliability correlation [p(corr)] values of all metabolites from the S-plot of the OPLS-DA were extracted using the first component. We selected metabolites satisfying the following criteria as potential markers: (i) high confidence [|p(corr)|>0.6] in discriminations between Hongyoung and Atlantic, between Jayoung and Atlantic, and between Jayoung and Hongyoung; (ii) mean intensities in one potato cultivar significantly different from those of another cultivar (P<0.05); and (iii) a minimum of a 2-fold change. The P value was calculated using an independent two-sample t-test.
Targeted selection of anthocyanins and their intermediates
Targeted selection of anthocyanins and their intermediates was performed based on follow information: (i) molecular formula and exact mass information of the compounds on phenylprophanoid and flavonoid pathways were established throughout the KEGG (http://www.genome.jp/kegg) and PMN (plant metabolic network, http://www.plantcyc.org) databases and literary references (see Supplementary Fig. S2 and Table S2 available at JXB online) (He et al., 2010; Stushnoff et al., 2010; Jaakola, 2013); (ii) specific daughter ions of anthocyanins through literary references on similar compounds (Sun et al., 2012); and (iii) MS2 spectra of standard compounds and metabolome databases, including METLIN (http://metlin.scripps.edu/) and LIPD MAPS (http://www.lipidmaps.org/). Finally, fragmented or adducted mass features were analyzed for selected ions. The identification of anthocyanin-related compounds is summarized in Supplementary Table S3 available at JXB online.
RNA sequencing (RNA-seq) data analysis
RNA-seq paired-end libraries were prepared using the Illumina TruSeq RNA Sample Preparation Kit version 2 (Illumina, San Diego, CA, USA). Starting with total RNA isolated using PureLink® Plant RNA Reagent (Life Technologies Korea LLC, Seoul, Korea), mRNA was purified using poly(A) selection or rRNA depletion; next, RNA was chemically fragmented and converted into single-stranded cDNA using random hexamer priming. Next, the second strand was generated to create double-stranded cDNA. Library construction began with the generation of blunt-end cDNA fragments from double-stranded cDNA. Then, an A base was added to the blunt ends to make them ready for the ligation of sequencing adapters. After size selection of the ligation products, the ligated cDNA fragments that contained adapter sequences were enhanced via PCR using adapter-specific primers. The library was quantified with a KAPA library quantification kit (Kapa biosystems KK4854) following the manufacturer’s instructions. Each library was loaded onto the Illumina Hiseq2000 platform and high-throughput sequencing was performed to ensure that each sample met the desired average sequencing depth. Image analysis and base calling were performed using the Illumina pipeline with default settings.
For mRNA sequencing, total RNA (10 μg) was isolated from sprouts of Atlantic, Hongyoung, and Jayoung using a PureLink® RNA Mini kit (Life Technologies Korea LLC) and used to create normalized cDNA and PCR-amplified datasets according to the Illumina RNA-seq protocol; then, the RNA was sequenced by Illumina HiSeq2000 (242M 100bp paired-end reads). Sequence data with base-pair qualities in the upper Q ≥20 were extracted by SolexaQA. Trimming resulted in reads with a mean length of 80.14bp across all samples; a minimum length of 25bp was applied during sequence trimming. The gene annotation used S. tuberosum Group Phureja DM1-3 516R44 (CIP801092) Genome Annotation version 3.4 mapped to the pseudomolecule sequence (PGSC_DM_v3_2.1.10_pseudomolecules.fa) downloaded from Solanaceae Genomics Resource at Michigan State University (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml) (Consortium, 2011)
Transcript profiles and annotation
mRNA libraries generated from each sample were sequenced using Illumina HiSeq2000 (100bp paired ends). Reads for each sequence tag were mapped to the reference with the Bowtie software (Langmead et al., 2009). The number of mapped clean reads for each gene was counted and normalized using the DESeq package in R (Anders and Huber, 2010). Only the genes that mapped with read counts of 100 or above in all experimental samples were retained for further analysis. Fold change and binomial tests were used to identify differentially expressed genes between each sample. The false discovery rate calculated via DESeq was applied to identify the threshold of the P value in binomial tests and analyses.
Gene Ontology (GO) and KEGG pathway functional enrichment analyses were performed via the Gene Ontology Database and DAVID (http://david.abcc.ncifcrf.gov/tools.jsp), respectively (Huang et al., 2008). GO consists of terms that provide a more global representation of gene functions using a controlled vocabulary; DAVID comprises web-accessible programs that provide a comprehensive set of functional annotation tools that can be used by investigators to understand the biological meaning behind a large list of genes. The gene lists generated by annotated TAIR (The Arabidopsis Information Resource) ID of transcripts of up- and down-regulated differentially expressed genes were classified into MapMan BINs using the MapCave tool (http://mapman.gabipd.org/web/guest/mapcave), which is linked with three databases (Arabidopsis thaliana TAIR8, Arabidopsis thaliana TAIR9, and TAIR release 10) (see Supplementary Table S4 available at JXB online).
Integrative analysis of metabolome and transcriptome
Pearson correlation coefficients were calculated for metabolome and transcriptome data integration. For this, the mean of all biological replicates of each cultivar in the metabolome data and the mean value of expression of each transcript in the transcriptome data were calculated. The fold changes in each pigmented potato (Hongyoung and Jayoung) were then calculated in both the metabolome and transcriptome data and compared with the control cultivar (Atlantic). Finally, the coefficients were calculated from log2(fold change) of each metabolite and log2(fold change) of each transcript using the EXCEL program (see Supplementary Table S5 available at JXB online). Correlations corresponding to a coefficient with R 2>0.9 were selected (see Supplementary Table S6 available at JXB online). Metabolome and transcriptome relationships were visualized using Cytoscape (version 2.8.2).
Results
Metabolic differences among the three potato cultivars
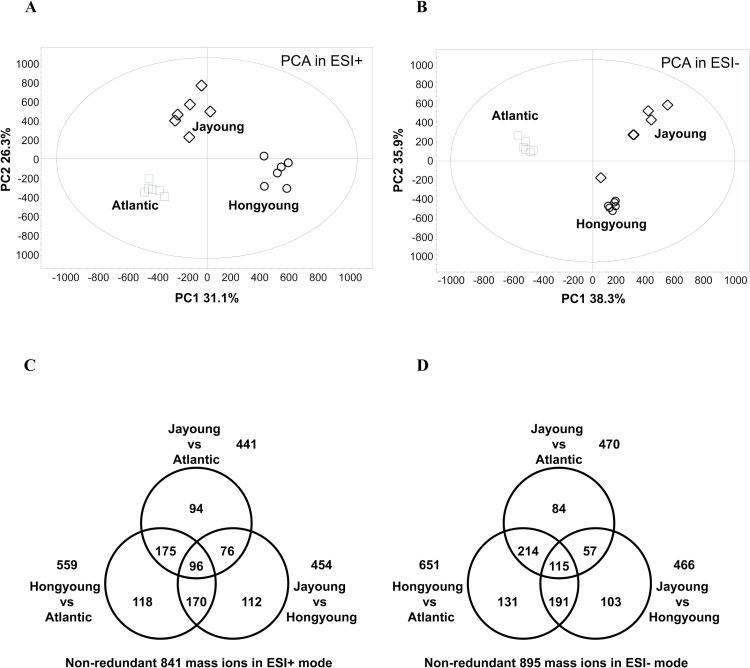
To compare the metabolite composition involved in the pigmentation of the three different potato cultivars, datasets obtained from UPLC-Triple-TOF-MS in the ESI+ (ESI–) mode were subjected to PCA. The results showed that the three potato cultivars were clearly separated in the PC1×PC2 score plots (Fig. 1A, B). Indeed, the first principal component (PC1) in ESI+ mode (31.1% of the total variables) and PC1 and PC2 in ESI– (38.3 and 35.9%, respectively) were clearly separated between Hongyoung and Atlantic. The differences between Jayoung and Atlantic resulted from PC2 (26.3% variables) in ESI+ mode and PC1 (38.3%) in ESI– mode. Furthermore, score plots and S-plots of OPLS-DA were used for modeling the differences between two potato cultivars (see Supplementary Fig. S3 available at JXB online). The selection of variables responsible for the differences was performed through statistical analysis as described in Materials and methods. A total of 556 (651), 441 (470), and 454 (466) mass ions were selected between Hongyoung and Atlantic, between Jayoung and Atlantic, and between Jayoung and Hongyoung in the ESI+ (ESI–) mode, respectively. In total, 841 and 895 mass ions were selected in the ESI+ and ESI– modes, respectively (Fig. 1C, D).
Fig. 1.
PCA score plot of colored potatoes and numbers of potential markers for each. PCA score plots were derived from metabolite ions acquired from ESI+ (A) and ESI–(B) mode. Potential markers were selected by comparing quantitative differences of mass ions in ESI+ (C) and ESI– (D) mode between Hongyoung and Atlantic, between Jayoung and Atlantic, and between Hongyoung and Jayoung.
Differential accumulation of anthocyanin derivatives between Hongyoung and Jayoung
Anthocyanins are glycosides and acylglycosides of anthocyanidin aglycones that are biosynthesized through the flavonoid pathway via the phenylpropanoid pathway (Stushnoff et al., 2010; Jaakola, 2013). Cyanidin (Cy), delphinidin (Dp), pelargonidin (Pg), peonidin (Pn), petunidin (Pt), and malvidin (Mv) are six common anthocyanins that are grouped according to the hydroxyl pattern or methoxy substitutions of B ring (Supplementary Fig. S2). Among them, the anthocyanin derivatives Dp, Pn, and Mv are sources of purple and dark colors, whereas the derivatives of Cy and Pg are the main pigments in bright-red-colored fruits (Jaakola, 2013).
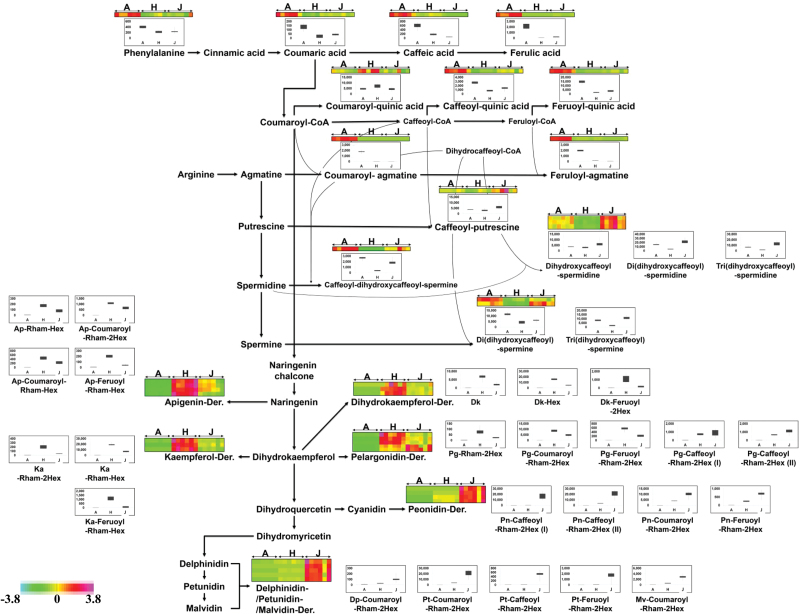
Among the mass ion peaks detected in our metabolomics analysis, anthocyanins and their intermediates were selected using product-ion scanning and precursor-ion scanning based on their molecular formula in MS1 and MS2 spectral data, respectively (see Supplementary Fig. S4 available at JXB online). The selected peaks were identified by interpretation of their MS2 fragment patterns and are summarized in Supplementary Table S3. The identified anthocyanins and their relevant compounds were rearranged to their corresponding positions in an anthocyanin biosynthesis pathway established based on KEGG, PMN, and literature references. Figure 2 shows that the composition of compounds on the anthocyanin biosynthesis pathways were specifically different depending on the potato cultivars (i.e. white Atlantic, light-red Hongyoung, and dark-purple Jayoung). In particular, the compositions of flavonoids downstream of the phenylpropanoid pathway were highly different between Hongyoung and Jayoung, indicating that these metabolites might play a crucial role in determining the pigmentation in potato. Apigenin (Ap), kaempferol (Ka), dihydrokaempferol (Dk), and Pg derivatives were shown to be most abundant in Hongyoung, whereas Pn, Pt, Dp, and Mv derivatives were shown to be the more abundant in Jayoung.
Fig. 2.
Biosynthetic pathway of anthocyanins. This pathway is constructed based on the KEGG pathway and literary references. Dk, dihydrokaempferol; Ka, kaempferol; Ap, apigenin; Pg, pelargonidin; Pt, petunidin; Pn, peonidin; Dp, delphinidin; Mv, malvidin; Rham, rhamnose; Hex, hexose; Coumaroyl, coumaric acid; Caffeoyl, caffeic acid; Feruoyl, ferulic acid; A, Atlanic potato; H, Hongyoung potato; J, Jayoung potato. Each colored cell represents the normalized intensity of each compound ion according to the color scale (six biological replicates×three cultivars, n=18). Box-and-whisker plots are shown for changes of phenylprophanoids, polyamine conjugates, flavone glycosides, flavonol glycosides, dihydroflavonol glycosides, and anthocyanins in each potato cultivar. Maximum and minimum values of a metabolite among six biological replicates are represented at the upper and lower ends of the whisker, respectively, and their 75th and 25th percentiles are represented at upper and lower ends of the box, respectively.
Differential expression of genes among Atlantic, Hongyoung, and Jayoung
Transcriptome analysis of sprouts showed that 756 and 519 transcripts had at least a 4-fold change in Hongyoung and Jayoung, respectively, compared with Atlantic. There were 482 and 274 up-regulated and down-regulated transcripts in Hongyoung, and 248 and 273 in Jayoung, respectively (Supplementary Fig. S5 available at JXB online). In total, 1044 non-redundant transcripts were collected and classified into 20 groups (main BINs) based on their annotated functions (Table 1). The transcripts were categorized as unknown (37.7%), regulation of protein activity (10.3%), stress (9.6%), other metabolism (7.3%), transcriptional regulation (5.7%), secondary metabolism (5.3%), and signaling (4.0%) (Table 1).
Table 1.
Functional categories of the genes differentially expressed in Hongyoung and Jayoung compared with those of the corresponding control, Atlantic
| BIN Codea | Annotation | Hongyoung | Jayoung | Total number of non-redundant genes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | ||||||||
| Number | % | Number | % | Number | % | Number | % | Number | % | ||
| BIN 1 | Photosynthesis | 1 | 0.2 | 5 | 1.8 | 1 | 0.4 | 1 | 0.4 | 6 | 0.6 |
| BIN 10 | Cell wall | 13 | 2.7 | 8 | 2.9 | 4 | 1.6 | 2 | 0.7 | 23 | 2.2 |
| BIN 11 | Lipid metabolism | 7 | 1.5 | 7 | 2.6 | 4 | 1.6 | 3 | 1.1 | 17 | 1.6 |
| BIN 13 | Amino acid metabolism | 4 | 0.8 | 0 | 0.0 | 1 | 0.4 | 2 | 0.7 | 6 | 0.6 |
| BIN 16 | Secondary metabolism | 29 | 6.0 | 16 | 5.8 | 22 | 8.9 | 7 | 2.6 | 55 (35b) | 5.3 |
| BIN 17 | Hormone | 16 | 3.3 | 6 | 2.2 | 5 | 2.0 | 6 | 2.2 | 30c | 2.9 |
| BIN 20 | Stress | 45 | 9.3 | 21 | 7.7 | 24 | 9.7 | 37 | 13.6 | 100 | 9.6 |
| BIN 21 | Redox | 1 | 0.2 | 1 | 0.4 | 1 | 0.4 | 0 | 0.0 | 3 | 0.3 |
| BIN 27.Ad | RNA processing/transcription/ RNA binding | 5 | 1.0 | 5 | 1.8 | 1 | 0.4 | 6 | 2.2 | 16 | 1.5 |
| BIN 27.3 | Regulation of transcription | 34 | 7.1 | 9 | 3.3 | 14 | 5.7 | 13 | 4.8 | 60c | 5.7 |
| BIN 28 | Chromatin structure/DNA synthesis and repair | 6 | 1.2 | 2 | 0.7 | 5 | 2.0 | 2 | 0.7 | 12 | 1.1 |
| BIN 29 | Protein synthesis/folding | 4 | 0.8 | 2 | 0.7 | 4 | 1.6 | 3 | 1.1 | 11 | 1.1 |
| BIN 30 | Signaling | 15 | 3.1 | 14 | 5.1 | 11 | 4.5 | 11 | 4.0 | 42c | 4.0 |
| BIN 31 | Cell | 5 | 1.0 | 4 | 1.5 | 2 | 0.8 | 3 | 1.1 | 12 | 1.1 |
| BIN 33 | Development | 12 | 2.5 | 4 | 1.5 | 4 | 1.6 | 2 | 0.7 | 18 | 1.7 |
| BIN 34 | Transport | 23 | 4.8 | 7 | 2.6 | 4 | 1.6 | 6 | 2.2 | 36 | 3.4 |
| BIN 35 | Not assigned | 156 | 32.4 | 118 | 43.1 | 85 | 34.4 | 125 | 46.0 | 394 | 37.7 |
| BIN Ae | Carbohydrate metabolism | 10 | 2.1 | 4 | 1.5 | 5 | 2.0 | 3 | 1.1 | 19 | 1.8 |
| BIN Bf | Other metabolism | 52 | 10.8 | 28 | 10.2 | 28 | 11.3 | 24 | 8.8 | 108 | 10.3 |
| BIN Cg | Regulation of protein activity | 44 | 9.1 | 13 | 4.7 | 22 | 8.9 | 16 | 5.9 | 76 | 7.3 |
| Total | 482 | 100.0 | 274 | 100.0 | 247 | 100.0 | 272 | 100.0 | 1044 | 100.0 | |
a BIN codes of genes were produced according to MapMan classification using the MapCave tool (http://mapman.gabipd.org/web/guest/mapcave).
b Represents the number of genes for anthocyanin biosynthesis.
c Represents the number of genes that were selected to analyze the correlation test between gene and metabolite.
d BIN 27.A includes RNA processing (BIN 27.1), transcription (27.2), and RNA binding (27.4).
e BIN A is carbohydrate metabolism-related BINs: major carbohydrates (BIN 2), minor carbohydrates (BIN 3), glycolysis (BIN 4), fermentation (BIN 5), gluconeogenesis/glyoxylate cycle (BIN 6), OPP cycle (BIN 7), TCA/organic acid transformation (BIN 8), and mitochondrial electron transport/ATP synthesis (BIN 9).
f BIN B is other metabolism-related BINs: nitrogen assimilation (BIN 12), S-assimilation (BIN 14), metal handling (BIN 15), cofactor/vitamin synthesis (BIN 18), tetrapyrole synthesis (BIN 19), nucleotide metabolism (BIN 23), biodegradation of xenobiotics (BIN 24), C1-metabolism (BIN 25), and miscellaneous (BIN 26).
g BIN C is protein activity regulation-related BINs including protein targeting (BIN 29.3), protein post-translational modification (BIN 29.4), and protein degradation (BIN 29.5).
Correlation analysis between transcripts and anthocyanin derivatives reveals the differential regulatory network of anthocyanin biosynthesis in Hongyoung and Jayoung
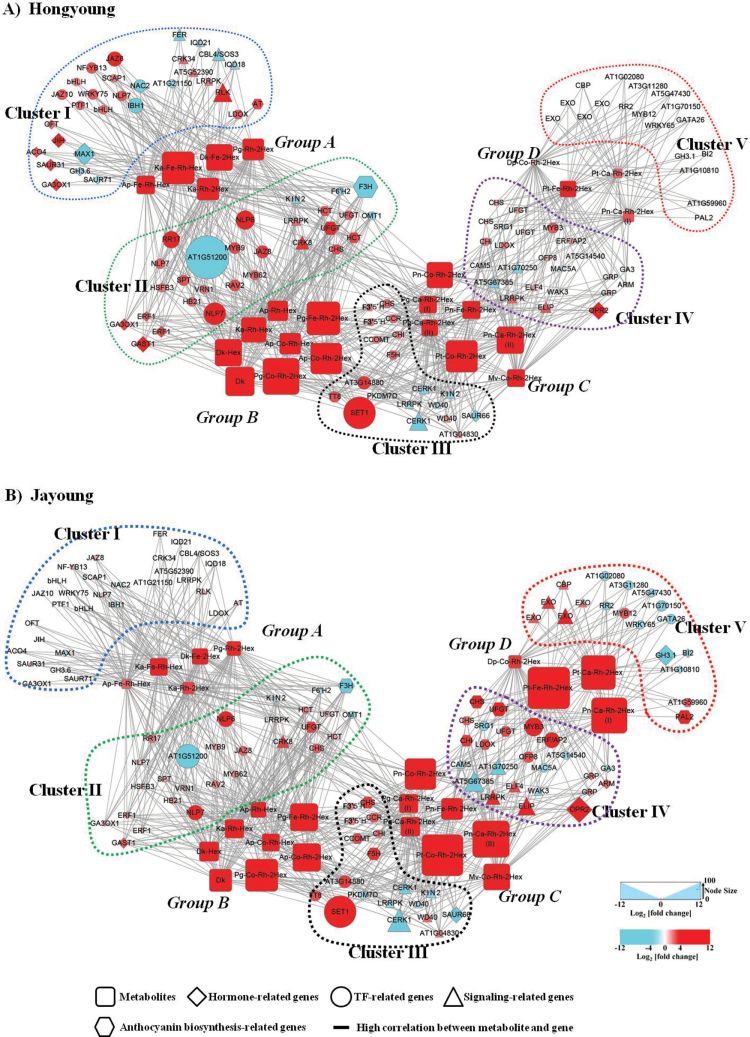
To understand the regulatory network of anthocyanins implicated in the differential distribution of anthocyanin derivatives between Hongyoung and Jayoung, we carried out correlation tests between quantitative changes of metabolites and transcripts in the three different colored potatoes. For this, derivatives of Ka, Dk, Dp, Pg, Pn, Pt, and Mv detected in this study and transcripts categorized into flavonoid metabolism, hormone metabolism, regulation of transcription, and cell signaling were selected from the 1044 genes (Supplementary Fig. S5) differentially expressed in the three potato cultivars. In total, 22 anthocyanin derivatives and 167 transcripts (flavonoid metabolism: 35, hormone metabolism: 30, regulation of transcription: 60, and cell signaling: 42 transcripts) were subjected to Pearson correlation analysis (Supplementary Table S5). The result showed that 119 transcripts had strong correlation coefficient values (R 2>0.9) with 22 metabolites (Supplementary Table S6). Based on the result, interaction networks between the 22 metabolites and 119 transcripts were organized in Hongyoung and Jayoung (Fig. 3 and Supplementary Table S6). The networks showed that the 119 transcripts were grouped into five clusters (I–V) (Table 2) and the 22 metabolites were divided into four groups (A–D) (Table 3). Metabolites in group A containing derivatives of a Pg, two Kas, a Dk, and an Ap were more predominant in Hongyoung compared with Jayoung, and were highly correlated with the transcripts in clusters I and II. On the other hand, group D metabolites including three Pt derivatives and one Dp derivative were more predominant in Jayoung and were showed to be highly connected with the transcripts in clusters IV and V. Group B and C metabolites were highly increased in both Hongyoung and Jayoung compared with Atlantic, although the increase of group C metabolites containing a Pg, three Pt, and an Mv derivative were a little more prominent in Jayoung than Hongyoung. The metabolites in groups B and C were shown to be strongly correlated with the transcripts in cluster III (Table 3 and Fig. 3).
Fig. 3.
Connection network between regulatory genes and anthocyanin-related metabolites. The networks in Hongyoung (A) and Jayoung (B) were visualized with the Cytoscape software (version 2.8.2). Dk, dihydrokaempferol; Ka, kaempferol; Ap, apigenin; Pg, pelargonidin; Pt, petunidin; Pn, peonidin; Dp, delphinidin; Mv, malvidin; Rh, rhamnose; Hex, hexose; Co, coumaric acid; Ca, caffeic acid; Fe, ferulic acid.
Table 2.
Classification of regulatory genes correlated with anthocyanin-related metabolites
| Clustersa | Potato gene | Log2(fold change) | BLASTX TAIR10 Best Hit | BINb | |||
|---|---|---|---|---|---|---|---|
| Hongyoung | Jayoung | AGI | Annotation | Symbol | |||
| Cluster I | PGSC0003DMT400049192 | 2.58 | 0.82 | AT5G05600 | 2-Oxoglutarate (2OG) and Fe(II)- dependent oxygenase superfamily protein, leucoanthocyanidin dioxygenase | LDOX | BIN 16 |
| PGSC0003DMT400069897 | 2.96 | 0.98 | AT2G39980 | HXXXD-type acyl-transferase family protein | AT | BIN 16 | |
| PGSC0003DMT400008366 | 3.07 | 1.24 | AT4G00880 | SAUR-like auxin-responsive protein family | SAUR31 | BIN 17 | |
| PGSC0003DMT400023715 | –2.15 | –0.55 | AT1G56150 | SAUR-like auxin-responsive protein family | SAUR71 | BIN 17 | |
| PGSC0003DMT400028801 | 2.37 | 0.77 | AT2G37980 | O-Fucosyltransferase family protein | OFT | BIN 17 | |
| PGSC0003DMT400036081 | 2.64 | 0.95 | AT1G05010 | Ethylene-forming enzyme, 1-aminocyclopropane-1-carboxylate oxidase 4 | ACO4 | BIN 17 | |
| PGSC0003DMT400040310 | 3.65 | 1.1 | AT1G51760 | Peptidase M20/M25/M40 family protein, IAA- alanine resistant 3, jasmonic acid responsive 3, jasmonoyl-l-isoleucine hydrolase | JIH | BIN 17 | |
| PGSC0003DMT400064343 | –2.27 | –0.6 | AT5G54510 | Auxin-responsive GH3 family protein, IAA- amino acid synthase | GH3.6 | BIN 17 | |
| PGSC0003DMT400074744 | 3.1 | 0.79 | AT1G15550 | Gibberellin 3-oxidase 1 | GA3OX1 | BIN 17 | |
| PGSC0003DMT400095956 | –5.52 | –1.76 | AT2G26170 | Cytochrome P450, family 711, subfamily A, polypeptide 1 | MAX1 | BIN 17 | |
| PGSC0003DMT400014179 | –3.79 | –1.69 | AT2G43060 | ILI1-binding bHLH 1 | IBH1 | BIN 27.3 | |
| PGSC0003DMT400016580 | 2.61 | 0.63 | AT5G13220 | Jasmonate zim domain protein 10 | JAZ10 | BIN 27.3 | |
| PGSC0003DMT400018274 | 2.27 | 0.91 | AT5G51790 | bHLH DNA-binding superfamily protein | bHLH | BIN 27.3 | |
| PGSC0003DMT400022651 | 3.74 | 1.34 | AT1G30135 | Jasmonate zim domain protein 8 | JAZ8 | BIN 27.3 | |
| PGSC0003DMT400035445 | 2.63 | 1.11 | AT4G24020 | NIN-like protein 7 | NLP7 | BIN 27.3 | |
| PGSC0003DMT400041580 | 2.05 | 0.9 | AT5G65590 | Dof-type zinc finger DNA-binding family protein, STOMATAL CARPENTER 1 | SCAP1 | BIN 27.3 | |
| PGSC0003DMT400056352 | 2.66 | 0.84 | AT5G13080 | WRKY DNA-binding protein 75 | WRKY75 | BIN 27.3 | |
| PGSC0003DMT400063260 | 2.76 | 1.22 | AT5G23090 | NUCLEAR FACTOR Y subunit B13 | NF-YB13 | BIN 27.3 | |
| PGSC0003DMT400064428 | –3.15 | –1.25 | AT3G15510 | NAC domain containing protein 2 | NAC2 | BIN 27.3 | |
| PGSC0003DMT400068332 | 2.34 | 0.88 | AT3G02150 | PLASTID TRANSCRIPTION FACTOR 1 | PTF1 | BIN 27.3 | |
| PGSC0003DMT400070825 | –2.18 | –0.87 | AT1G21150 | Mitochondrial transcription termination factor family protein | AT1G21150 | BIN 27.3 | |
| PGSC0003DMT400071861 | 2.07 | 0.64 | AT5G51780 | bHLH DNA-binding superfamily protein | bHLH | BIN 27.3 | |
| PGSC0003DMT400008273 | 2.36 | 0.74 | AT4G11530 | CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE 34 | CRK34 | BIN 30 | |
| PGSC0003DMT400014960 | –3.17 | –1.26 | AT5G24270 | CALCINEURIN B-LIKE PROTEIN 4, SALT OVERLY SENSITIVE 3 | CBL4/SOS3 | BIN 30 | |
| PGSC0003DMT400023467 | –2.02 | –0.65 | AT3G49260 | IQ-DOMAIN 21 | IQD21 | BIN 30 | |
| PGSC0003DMT400027429 | –2.96 | –0.94 | AT3G51550 | Malectin/receptor-like protein kinase family protein, FERONIA | FER | BIN 30 | |
| PGSC0003DMT400029325 | –2.32 | –0.97 | AT1G01110 | IQ-DOMAIN 18 | IQD18 | BIN 30 | |
| PGSC0003DMT400034271 | 4.72 | 1.56 | AT3G22060 | Receptor-like protein kinase-related family protein | RLK | BIN 30 | |
| PGSC0003DMT400037209 | 2.3 | 0.56 | AT5G52390 | PAR1 protein kinase | AT5G52390 | BIN 30 | |
| PGSC0003DMT400046778 | 2.05 | 0.57 | AT4G20140 | Leucine-rich repeat transmembrane protein kinase | LRRPK | BIN 30 | |
| Cluster II | PGSC0003DMT400020466 | 2.57 | 1.25 | AT2G29740 | UDP glucose: flavonoid-3-O-glucosyltransferase | UFGT | BIN 16 |
| PGSC0003DMT400066505 | 3.02 | 1.64 | AT5G67150 | Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase | HCT | BIN 16 | |
| PGSC0003DMT400070444 | –6.26 | –4.13 | AT5G24530 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, flavanone 3-dioxygenase | F3H | BIN 16 | |
| PGSC0003DMT400001145 | 2.74 | 2.03 | AT5G48930 | Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase | HCT | BIN 16 | |
| PGSC0003DMT400030504 | 3.77 | 3.15 | AT5G54010 | UDP-glycosyltransferase superfamily protein | UFGT | BIN 16 | |
| PGSC0003DMT400045239 | –2.38 | –2.02 | AT5G54160 | O-Methyltransferase 1, flavonol 3′-O-methyltrasferase/caffeate O-methyltransferase | OMT1 | BIN 16 | |
| PGSC0003DMT400070465 | –2.14 | –1.68 | AT1G55290 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, Feruloyl CoA ortho-hydroxylase 2 | F6′H2 | BIN 16 | |
| PGSC0003DMT400076179 | 2.83 | 2.63 | AT5G13930 | Chalcone and stilbene synthase family protein | CHS | BIN 16 | |
| PGSC0003DMT400004046 | 4.03 | 2.67 | AT1G75750 | GAST1 protein homolog 1 | GAST1 | BIN 17 | |
| PGSC0003DMT400026627 | 2.9 | 1.38 | AT3G23240 | Ethylene response factor 1 | ERF1 | BIN 17 | |
| PGSC0003DMT400037826 | 2.54 | 1.79 | AT3G23240 | Ethylene response factor 1 | ERF1 | BIN 17 | |
| PGSC0003DMT400086139 | 2.98 | 1.52 | AT1G15550 | Gibberellin 3-oxidase 1 | GA3OX1 | BIN 17 | |
| PGSC0003DMT400002125 | 2.89 | 2.03 | AT1G68840 | Related to ABI3/VP1 2 | RAV2 | BIN 27.3 | |
| PGSC0003DMT400010253 | 2.72 | 1.53 | AT4G36930 | bHLH DNA-binding superfamily protein, SPATULA | SPT | BIN 27.3 | |
| PGSC0003DMT400035433 | 2.54 | 1.22 | AT4G24020 | NIN-like protein 7 | NLP7 | BIN 27.3 | |
| PGSC0003DMT400035439 | 5.61 | 3.83 | AT4G24020 | NIN-like protein 7 | NLP7 | BIN 27.3 | |
| PGSC0003DMT400042922 | 4.27 | 2.1 | AT3G56380 | Response regulator 17 | RR17 | BIN 27.3 | |
| PGSC0003DMT400043852 | 2.19 | 1.33 | AT3G18990 | AP2/B3-like transcriptional factor family protein, reduced vernalization response 1 | VRN1 | BIN 27.3 | |
| PGSC0003DMT400045182 | 2.88 | 1.63 | AT5G16770 | Myb domain protein 9 | MYB9 | BIN 27.3 | |
| PGSC0003DMT400070196 | 2.23 | 1.19 | AT2G41690 | Heat-shock transcription factor B3 | HSFB3 | BIN 27.3 | |
| PGSC0003DMT400081478 | –10.88 | –6.03 | AT1G51200 | A20/AN1-like zinc finger family protein | AT1G51200 | BIN 27.3 | |
| PGSC0003DMT400002991 | 3.21 | 2.41 | AT1G30135 | Jasmonate zim domain protein 8 | JAZ8 | BIN 27.3 | |
| PGSC0003DMT400014383 | 2.44 | 1.8 | AT1G68320 | Myb domain protein 62 | MYB62 | BIN 27.3 | |
| PGSC0003DMT400035440 | 4.76 | 3.94 | AT1G64530 | Plant regulator RWP-RK family protein, NIN-like protein 6 | NLP6 | BIN 27.3 | |
| PGSC0003DMT400045834 | 2.32 | 1.69 | AT2G18550 | Homeobox protein 21 | HB21 | BIN 27.3 | |
| PGSC0003DMT400071481 | 2.29 | 1.25 | AT1G07650 | Leucine-rich repeat transmembrane protein kinase | LRRPK | BIN 30 | |
| PGSC0003DMT400044827 | –2.26 | –1.68 | AT4G16360 | 5′-AMP-activated protein kinase beta-2 subunit protein | KINβ2 | BIN 30 | |
| PGSC0003DMT400053964 | 3.39 | 3.13 | AT4G23160 | Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 | CRK8 | BIN 30 | |
| Cluster III | PGSC0003DMT400001124 | 2.08 | 2.03 | AT5G07990 | Cytochrome P450 superfamily protein, Flavonoid 3′,5′-hydroxylase | F3′5′H | BIN 16 |
| PGSC0003DMT400001125 | 2.08 | 2.08 | AT5G07990 | Cytochrome P450 superfamily protein, Flavonoid 3′,5;-hydroxylase | F3′5′H | BIN 16 | |
| PGSC0003DMT400016512 | 2.22 | 2.94 | AT1G67980 | Caffeoyl-CoA 3-O-methyltransferase | CCOMT | BIN 16 | |
| PGSC0003DMT400025446 | 2.54 | 3.52 | AT4G36220 | Ferulic acid 5-hydroxylase 1 | F5H | BIN 16 | |
| PGSC0003DMT400030428 | 2.17 | 2.26 | AT5G05270 | Chalcone-flavanone isomerase family protein | CHI | BIN 16 | |
| PGSC0003DMT400045220 | 2.41 | 2.74 | AT2G23910 | NAD(P)-binding Rossmann-fold superfamily protein, cinnamoyl-CoA reductase | CCR | BIN 16 | |
| PGSC0003DMT400076178 | 2.4 | 3.29 | AT5G13930 | Chalcone and stilbene synthase family protein | CHS | BIN 16 | |
| PGSC0003DMT400034486 | –2.96 | –4.05 | AT1G29500 | SAUR-like auxin-responsive protein family | SAUR66 | BIN 17 | |
| PGSC0003DMT400010235 | 8.06 | 8.04 | AT2G23380 | SET domain-containing protein, SETDOMAIN 1 | SET1 | BIN 27.3 | |
| PGSC0003DMT400028253 | –1.46 | –2.04 | AT1G08620 | Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein | PKDM7D | BIN 27.3 | |
| PGSC0003DMT400033569 | 2.81 | 2.8 | AT4G09820 | bHLH DNA-binding superfamily protein, TRANSPARENT TESTA 8 | TT8 | BIN 27.3 | |
| PGSC0003DMT400059950 | 3.33 | 3.2 | AT3G14880 | Transcription factor | AT3G14880 | BIN 27.3 | |
| PGSC0003DMT400007965 | 1.73 | 2.05 | AT1G04830 | Ypt/Rab-GAP domain of gyp1p superfamily protein | AT1G04830 | BIN 30 | |
| PGSC0003DMT400018720 | –2.11 | –2.25 | AT1G56120 | Leucine-rich repeat transmembrane protein kinase | LRRPK | BIN 30 | |
| PGSC0003DMT400044824 | –2.36 | –2.47 | AT4G16360 | 5′-AMP-activated protein kinase β2 subunit protein | KINβ2 | BIN 30 | |
| PGSC0003DMT400062862 | 1.94 | 2.36 | AT5G50120 | Transducin/WD40 repeat-like superfamily protein | WD40 | BIN 30 | |
| PGSC0003DMT400073082 | –1.69 | –2.04 | AT5G45760 | Transducin/WD40 repeat-like superfamily protein | WD40 | BIN 30 | |
| PGSC0003DMT400073506 | –2.57 | –3.17 | AT3G21630 | Chitin elicitor receptor kinase 1 | CERK1 | BIN 30 | |
| PGSC0003DMT400073513 | –4.56 | –5.76 | AT3G21630 | Chitin elicitor receptor kinase 1 | CERK1 | BIN 30 | |
| Cluster IV | PGSC0003DMT400018670 | –1.4 | –2.67 | AT1G17020 | Fe(II)/ascorbate oxidase, senescence-related gene 1 | SRG1 | BIN 16 |
| PGSC0003DMT400022254 | 1.61 | 3.66 | AT5G13930 | Chalcone and stilbene synthase family protein | CHS | BIN 16 | |
| PGSC0003DMT400029916 | 1.73 | 3.99 | AT5G54060 | UDP-glucose:flavonoid 3-O-glucosyltransferase | UFGT | BIN 16 | |
| PGSC0003DMT400049165 | 1.32 | 2.44 | AT5G13930 | Chalcone and stilbene synthase family protein | CHS | BIN 16 | |
| PGSC0003DMT400062562 | 1.18 | 2.95 | AT5G54010 | UDP-glycosyltransferase superfamily protein | UFGT | BIN 16 | |
| PGSC0003DMT400030430 | 2.17 | 3.27 | AT5G05270 | Chalcone-flavanone isomerase family protein | CHI | BIN 16 | |
| PGSC0003DMT400058554 | 2.02 | 2.91 | AT4G22880 | Leucoanthocyanidin dioxygenase | LDOX | BIN 16 | |
| PGSC0003DMT400023914 | 1.12 | 2.1 | AT1G22690 | Gibberellin-regulated family protein | GRP | BIN 17 | |
| PGSC0003DMT400030493 | –0.83 | –2.09 | AT5G25900 | GA requiring 3 | GA3 | BIN 17 | |
| PGSC0003DMT400075778 | 1.04 | 2.52 | AT5G37490 | ARM repeat superfamily protein | ARM | BIN 17 | |
| PGSC0003DMT400048325 | 4.37 | 6.46 | AT1G76690 | 12-Oxophytodienoate reductase 2 | OPR2 | BIN 17 | |
| PGSC0003DMT400066182 | 1.32 | 2.05 | AT5G59845 | Gibberellin-regulated family protein | GRP | BIN 17 | |
| PGSC0003DMT400000838 | 1.55 | 2.8 | AT5G19650 | Ovate family protein 8 | OFP8 | BIN 27.3 | |
| PGSC0003DMT400007511 | 1.6 | 3.94 | AT1G19210 | Integrase-type DNA-binding superfamily protein, ERF/AP2 transcription factor family | ERF/AP2 | BIN 27.3 | |
| PGSC0003DMT400021046 | –1.03 | –2.21 | AT1G07360 | CCCH-type zinc fingerfamily protein, MOS4- ASSOCIATED COMPLEX SUBUNIT 5A | MAC5A | BIN 27.3 | |
| PGSC0003DMT400032803 | –0.82 | –2 | AT5G14540 | Protein of unknown function (DUF1421) | AT5G14540 | BIN 27.3 | |
| PGSC0003DMT400078477 | 2.69 | 4.63 | AT1G22640 | Myb domain protein 3 | MYB3 | BIN 27.3 | |
| PGSC0003DMT400003078 | 1.67 | 2.9 | AT2G40080 | EARLY FLOWERING 4 | ELF4 | BIN 30 | |
| PGSC0003DMT400016494 | 2.74 | 4.51 | AT3G22840 | Chlorophyll A-B binding family protein, EARLY LIGHT-INDUCABLE PROTEIN | ELIP | BIN 30 | |
| PGSC0003DMT400044079 | –2.75 | –4.99 | AT5G67385 | Phototropic-responsive NPH3 family protein | AT5G67385 | BIN 30 | |
| PGSC0003DMT400070439 | –1.06 | –2.06 | AT2G27030 | Calmodulin 5 | CAM5 | BIN 30 | |
| PGSC0003DMT400070451 | –1 | –2.22 | AT1G21240 | Wall-associated kinase 3 | WAK3 | BIN 30 | |
| PGSC0003DMT400036503 | –2.67 | –3.87 | AT1G70250 | Putative receptor serine/threonine kinase, protease inhibitor/seed storage/LTP family protein | AT1G70250 | BIN 30 | |
| PGSC0003DMT400060792 | 2.46 | 3.64 | AT3G47570 | Leucine-rich-repeat protein kinase family protein | LRRPK | BIN 30 | |
| Cluster V | PGSC0003DMT400055529 | 0.24 | 3.92 | AT3G53260 | Phenylalanine ammonia lyase 2 | PAL2 | BIN 16 |
| PGSC0003DMT400058309 | 0.82 | 2.5 | AT1G59960 | NAD(P)-linked oxidoreductase superfamily protein | AT1G59960 | BIN 16 | |
| PGSC0003DMT400016816 | –0.34 | –2.06 | AT1G10810 | NAD(P)-linked oxidoreductase superfamily protein | AT1G10810 | BIN 17 | |
| PGSC0003DMT400037644 | –0.11 | –5.11 | AT2G14960 | Auxin-responsive GH3 family protein | GH3.1 | BIN 17 | |
| PGSC0003DMT400073092 | –0.01 | –2.23 | AT4G18710 | Protein kinase superfamily protein, brassinosteroid-insensitive 2 | BI2 | BIN 17 | |
| PGSC0003DMT400000578 | –0.4 | –2.39 | AT1G02080 | Transcription regulators | AT1G02080 | BIN 27.3 | |
| PGSC0003DMT400023322 | 0.62 | 2.68 | AT2G47460 | Myb domain protein 12 | MYB12 | BIN 27.3 | |
| PGSC0003DMT400027378 | –0.43 | –2.25 | AT1G29280 | WRKY DNA-binding protein 65 | WRKY65 | BIN 27.3 | |
| PGSC0003DMT400048133 | –0.65 | –2.34 | AT3G11280 | Duplicated homeodomain-like superfamily protein | AT3G11280 | BIN 27.3 | |
| PGSC0003DMT400056008 | –0.56 | –2.18 | AT5G47430 | DWNN domain, a CCHC-type zinc finger | AT5G47430 | BIN 27.3 | |
| PGSC0003DMT400058234 | –0.2 | –2.75 | AT1G70150 | Zinc ion binding | AT1G70150 | BIN 27.3 | |
| PGSC0003DMT400066370 | –0.38 | –2.53 | AT4G17570 | GATA transcription factor 26 | GATA26 | BIN 27.3 | |
| PGSC0003DMT400075906 | –0.29 | –2.12 | AT4G16110 | Response regulator 2 | RR2 | BIN 27.3 | |
| PGSC0003DMT400064403 | 0.34 | 2.36 | AT4G27280 | Calcium-binding EF-hand family protein | CBP | BIN 30 | |
| PGSC0003DMT400079206 | 0.11 | 2.6 | AT4G08950 | Phosphate-responsive 1 family protein, EXORDIUM | EXO | BIN 30 | |
| PGSC0003DMT400079207 | 0.96 | 3.97 | AT4G08950 | Phosphate-responsive 1 family protein, EXORDIUM | EXO | BIN 30 | |
| PGSC0003DMT400079208 | 0.41 | 3.26 | AT4G08950 | Phosphate-responsive 1 family protein, EXORDIUM | EXO | BIN 30 | |
| PGSC0003DMT400079209 | 0.4 | 2.11 | AT4G08950 | Phosphate-responsive 1 family protein, EXORDIUM | EXO | BIN 30 | |
AGI, Arabidopsis Genome Initiative Number; TAIR, The Arabidopsis Information Resource.
a Regulatory genes were clustered according to gene and metabolite correlation in Fig. 3, which was calculated using Pearson correlation coefficients (R 2).
b BINs of genes generated according to MapMan classification using the MapCave tool (http://mapman.gabipd.org/web/guest/mapcave)
Table 3.
Classification of anthocyanin metabolites correlated with anthocyanin-related genes
| Log2(fold) ratio of: | |||
|---|---|---|---|
| Hongyoung:Atlantic | Jayoung:Atlantic | Jayoung:Hongyoung | |
| Group A | |||
| Pelargonidin-Rham-2Hex | 5.45 | 3.63 | –1.82 |
| Kaempferol-Rham-2Hex | 5.57 | 3.05 | –2.52 |
| Dihydrokaempferol-Feruloyl-2Hex | 6.68 | 4.06 | –2.62 |
| Apigenin-Feruloyl-Rham-Hex | 4.63 | 2.42 | –2.20 |
| Kaempferol-Feruloyl-Rham-Hex | 8.16 | 4.40 | –3.76 |
| Group B | |||
| Apigenin-Rham-Hex | 5.12 | 3.90 | –1.23 |
| Dihydrokaempferol-Hex | 6.79 | 4.94 | –1.85 |
| Apigenin-Coumaroyl-Rham-2Hex | 7.86 | 7.06 | –0.80 |
| Pelargonidin-Coumaroyl-Rham-2Hex | 9.17 | 8.17 | –0.99 |
| Pelargonidin-Feruloyl-Rham-2Hex | 8.42 | 6.98 | –1.44 |
| Kaempferol-Rham-Hex | 6.35 | 4.77 | –1.58 |
| Apigenin-Coumaroyl-Rham-Hex | 5.39 | 4.43 | –0.96 |
| Dihydrokaempferol | 7.29 | 5.67 | –1.62 |
| Group C | |||
| Pelargonidin-Caffeoyl-Rham-2Hex | 4.16 | 4.87 | 0.71 |
| Petunidin-Coumaroyl-Rham-2Hex | 6.83 | 9.25 | 2.43 |
| Peonidin-Coumaroyl-Rham-2Hex | 6.55 | 7.84 | 1.29 |
| Peonidin-Feruloyl-Rham-2Hex | 4.28 | 5.78 | 1.50 |
| Malvidin-Coumaroyl-Rham-2Hex | 4.44 | 6.66 | 2.22 |
| Group D | |||
| Petunidin-Caffeoyl-Rham-2Hex | 2.37 | 8.61 | 6.24 |
| Petunidin-Feruloyl-Rham-2Hex | >1 | >1 | 6.45 |
| Peonidin-Caffeoyl-Rham-2Hex | 2.90 | 9.30 | 6.39 |
| Delphinidin-Coumaroyl-Rham-2Hex | 1.36 | 3.26 | 1.90 |
Validation of differential gene expression was performed for 14 genes using quantitative PCR (qPCR) with gene-specific primers (see Supplementary Table S1 available at JXB online). Real-time qPCR analysis with RNA isolated from sprouts of Hongyoung, Jayoung, and Atlantic showed that genes in clusters I and II (JAZ8, JAZ10, WRKY75, ERF1, and MYB9) were highly expressed in Hongyoung, whereas MYB3 and UFGT (cluster IV) were highly expressed in Jayoung. The expression levels of most genes in cluster III (CCOMT, two CHI, two CHS, and LODX) were similarly increased in both Hongyoung and Jayoung (Supplementary Fig. S6 available at JXB online).
Transcripts responsible for the differential accumulation of each anthocyanin group
Anthocyanin biosynthesis
The distribution of anthocyanins in the colored potatoes showed that Pg-derivative anthocyanins were abundant in light-red-colored Hongyoung, whereas Pn, Dp, Pt, and Mv derivatives of anthocyanins were enriched in dark-purple Jayoung. Transcripts related to anthocyanin biosynthesis were differentially regulated in each cultivar. Among the 10 genes related to anthocyanin biosynthesis in clusters I and II, homologs for leucoanthocyanidin dioxygenase (LDOX), acyl-transferase, a UDP-glucose:flavonoid O-glycosyltransferase (UFGT: PGSC0003DMT400020466), and hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferases (HCT: PGSC0003DMT400066505) were more strongly up-regulated in light-red Hongyoung compared with Jayoung. In contrast, two homologs from cluster V [phenylalanine ammonia-lyase (PAL2) and NAD(P)-linked oxidoreductase] and six homologs from cluster IV (two UFGTs, two chalcone synthases (CHSs), a chalcone isomerase (CHI) and an ascorbate oxidase) were more strongly up- or down-regulated in dark-purple Jayoung compared with Hongyoung.
Hormones
Of the 12 genes implicated in hormone response in cluster I and four in cluster II (Table 2 and Fig. 3), homologs of a SAUR-like auxin-responsive protein (SAUR71), an IAA-amido synthase (GH3.6), and more axillary branches 1 (MAX1) were significantly down-regulated in Hongyoung compared with Atlantic. However, nine homologs (five in cluster I and four in cluster II) were significantly up-regulated in Hongyoung (Table 2). Hormone-related transcripts of clusters IV and V were highly correlated with group C and D metabolites that were highly increased in Jayoung compared with Hongyoung. The significant down-regulation of four transcripts was observed in Jayoung, including homologs of GA requiring 3 (GA3), GH3.1, brassinosteroid-insensitive 2 (BI2), and an NAD(P)-linked oxidoreductase protein. The four transcripts including gibberellin-regulated family proteins (AT1G22690 and AT5G59845) were up-regulated.
Transcription factors
Transcription factors in clusters I and II were demonstrated to be strongly connected with group A metabolites abundant in Hongyoung. Of these, four transcripts including A20/AN1-like zinc finger family protein were down-regulated, and 21 were up-regulated including NAC2, jasmonate zim domain protein 8 (JAZ8), JAZ10, WRKY75, MYB9, and MYB62 (Table 2 and Fig. 3). In cluster III, which was connected with both group B and C metabolites, a homolog of jumonji (jmj) family protein (PKDM7D) was significantly down-regulated and the genes encoding set domain 1 (SET1), TT8, and a transcription factor were up-regulated in both Hongyoung and Jayoung.
Moreover, 13 transcripts encoding transcription factors (five in cluster IV and eight in cluster V) were highly correlated with group C and D metabolites abundant in Jayoung (Table 2 and Fig. 4). Of the five transcripts in cluster IV, transcripts for MOS4-associated complex subunit 5A (MAC5A) and a transcription factor were down-regulated and three transcripts for ovate family protein 8 (OFP8), an ERF/AP2 transcription factor, and MYB3 were significantly up-regulated in Jayoung compared with Atlantic. In cluster V, the up-regulation of MYB12 and the down-regulation of seven transcripts including WRKY65, two zinc finger transcription factors, response regulator 2 (RR2), and GATA transcription factor 26 (GATA26) were observed in Jayoung.
Signaling
The expression levels of 30 transcripts encoding proteins implicated in signaling pathways were correlated with flavonoid levels in Hongyoung and/or Jayoung (Table 2 and Fig. 3). Six homologs (four in cluster I and two in cluster II) of genes were significantly up-regulated and five were down regulated both in Hongyoung and Jayoung. The up- or down-regulation of transcripts related to signaling in clusters I and II were more significant in Hongyoung than in Jayoung. Furthermore, the significant down-regulation of genes encoding a phototropic-responsive NPH3 family protein, calmodulin 5 (CAM5), wall-associated kinase 3 (WAK3), and a protease inhibitor/seed storage/LTP family protein in cluster IV were observed in Jayoung. Eight transcripts (three in cluster IV and five in cluster V) were significantly up-regulated, including homologous genes encoding early flowering 4 (ELF4), early light-inducible protein (ELIP), and a leucine-rich repeat protein kinase in cluster IV and a calcium-binding EF-hand family protein (CBP) and exordium (EXO) in cluster V (Table 2 and Fig. 3)
Discussion
In this study, as an effort to elucidate the differential regulation of anthocyanin biosynthesis involved in differential pigmentation of potatoes, a correlation test was performed in light-red-colored Hongyoung and dark-purple Jayoung with 22 anthocyanins and 167 genes categorized to flavonoid metabolism, hormones, transcription factors, and signaling. Of the 167 genes, 119 genes were strongly correlated with the 22 anthocyanins, and the correlation network showed that the genes and metabolites were divided into five clusters (I–V) and four subgroups (A–D) (Fig. 3, Tables 2 and 3). Many of the differentially expressed genes between white and colored potatoes coincided with a recent report performed with potato cultivars “Xin Dang” (white skin and white flesh) and “Hei Meiren” (purple skin and purple flesh) (Liu et al., 2015). In particular, a CHI gene (PGSC0003DMT400030430), two CHSs (PGSC0003DMT400049165 and PGSC0003 DMT400076178), an LDOX (PGSC0003DMT400058554), a flavonoid 3′,5′-hydroxylase (F3′5′H; PGSC0003DMT 400001124), and a basic helix–loop–helix (bHLH) DNA-binding superfamily protein, transparent testa 8 (TT8: PGSC0003DMT400033569) that are increased in the skin and flesh of the purple potato “Hei Meiren” (Liu et al., 2015) were also found to be increased in Hongyoung and Jayoung. However, the relative expression levels of these genes were differential between Hongyoung and Jayoung. Each gene cluster might be functionally connected to the anthocyanin subgroups, regulating the biosynthesis of anthocyanin derivatives that determined the colors of potatoes. Genes in cluster III were shown to be strongly connected with anthocyanins in groups B and C, which were highly accumulated in both light-red Hongyoung and dark-purple Jayoung. The genes in cluster III contained CHS, CHI, and F3′5′H. Expression of these genes was similar in Hongyoung and Jayoung; thus, these genes might be commonly involved in the biosynthesis of anthocyanins in both Hongyoung and Jayoung. Furthermore, genes encoding TT8 and WD40-repeat protein (WD40) in cluster III were strongly up-regulated in both Hongyoung and Jayoung. TT8, a bHLH-type regulation factor, forms a ternary complex with WD40-repeat protein and R2R3–MYB (WD40/bHLH/R2R3–MYB complex); these proteins are involved in the regulation of flavonoid pathways, and specifically in anthocyanin and pro-anthocyanin biosynthesis in Arabidopsis, purple cauliflower, and purple strawberry (Chiu and Li, 2012; Jaakola, 2013; Schaart et al., 2013; Xu et al., 2013). Studies on TT8 promoter activity using WD40 (ttg1), bHLH (tt8, gl3, and egl3), and R2R3–MYB (tt2, myb5, pap1, and pap2) in Arabidopsis mutants showed that the TT8 promoter activity is differentially regulated by various WD40/bHLH/R2R3–MYB complexes (Chiu and Li, 2012; Xu et al., 2013). In our study, the up-regulation of homologous genes encoding TT8 and WD40 in both Hongyoung and Jayoung indicated that the accumulation of anthocyanin in red-light- and dark-purple-colored potato cultivars was commonly regulated by the TT8-mediated pathway.
Of the genes in clusters I and II, we observed that jasmonic acid (JA) signaling-related genes, including JIH, JAZ8, and JAZ10, were up-regulated in light-red Hongyoung compared with dark-purple Jayoung. JA has been known to increase anthocyanin production and to stimulate gene expression of CHS and UFGT. In Arabidopsis, JA activates the degradation of JAZs, a negative regulator of JA, in a SCFCOl1 complex-dependent manner, to abolish the interaction between JAZs and the bHLH/R2R3–MYB complexes, and to stimulate activation of the WD40/bHLH (GL3, EGL3, and TT8)/R2R3–MYB (GL1 and MYB75) complexes, thereby activating the expression of anthocyanin biosynthesis-related genes (Qi et al., 2011, 2013). Of JA and its oxylipin derivatives, JA–Ile (but not JA, methyl jasmonate, or 12-oxo phytodienoic acid) promotes degradation of JAZ by the formation of the SCFCOl1–JAZ complexes (Thines et al., 2007). JIH catalyzes the cleavage of the JA–Ile conjugate, generating 2-hydroxy-JA. NaJIH-suppressed transgenic tobacco plants showed a dramatic increase in JA–Ile levels during herbivore attacks, thereby enhancing their resistance compared with that of wild-type tobacco (Woldemariam et al., 2012). Thus, the up-expression of JIH most likely reduces JA–Ile levels. Taken together, the transcriptional up-regulation of homologous genes encoding JIH, JAZ8, and JAZ10 probably indicates that JAZ degradation-mediated anthocyanin biosynthesis might be inactivated in light-red Hongyoung.
In this study, most auxin-related genes were shown to be negatively correlated with anthocyanin levels. Genes encoding SAUR66, SAUR71, GH3.6, and MAX1 in clusters I and II were negatively correlated with group A or B anthocyanins that are abundant in light-red Hongyoung. Moreover, we observed a negative correlation between the levels of GH3.1 in cluster V and anthocyanins in group D that are enriched in dark-red Jayoung. Auxin-sensitive SAUR66 and SAUR71 genes are responsive to GH3.1 and GH3.6 genes that catalyze the conjugation of amino acids to auxin. MAX1, a member of the CYP711A cytochrome P450 family, has been known to down-regulate genes involved in the flavonoid pathway, including CHS, CHI, F3H, F3′H, FLS, DFR, ANS, and UFGT (Lazar and Goodman, 2006). Liu et al. (2014) showed that auxins regulated the expression levels of anthocyanin biosynthesis genes in red pap1-D Arabidopsis cells, including genes for six transcriptional factors (TTG1, EGL3, MYBL2, TT8, GL3, and PAP1) and four structural genes (PAL1, CHS, DFR, and ANS) (Liu et al., 2014). The results demonstrated the involvement of auxin in anthocyanin biosynthesis. In other words, the anthocyanin biosynthesis in light-red Hongyoung and dark-purple Jayoung might be regulated by the inactivation of negative regulators, including MAX1.
In contrast to the down-regulation of auxin-related genes, genes involved in ethylene were up-regulated in the two pigmented potatoes compared with the white Atlantic, including homologous genes encoding 1-aminocyclopropane-1-carboxylate oxidase 4 (ACO4) in cluster I, an ethylene response factor 1 (ERF1) in cluster II, and an ERF/AP2 transcription factor in cluster IV. ACO4 converts 1-aminocyclopropane-1-carboxylic acid to ethylene, and ERF1 promotes ethylene production via the ethylene signaling cascade. The up-regulation of ACO4 and ERF1 in both Hongyoung and Jayoung indicated that ethylene may be related to the pigmentation of the two pigmented potatoes. In a recent study, exogenous treatment with the ethylene-releasing compound 2-chloroehtylphosphonic acid was reported to result in the accumulation of anthocyanins in grape skins and to stimulate the long-term expression of CHS, F3H, ANS, and UFGT (El‐Kereamy et al., 2003). These results indicated that ethylene was involved in anthocyanin biosynthesis. However, a gene in cluster I encoding NAC2 (also called ANAC092 and ORE1), a positive regulator of ethylene-mediated leaf senescence, was observed to be down-regulated in both Hongyoung and Jayoung compared with Atlantic. Transcriptional expression of NAC2 has been shown to be up-regulated by ethylene insensitive 2 (EIN2), which activates ethylene signaling and induces the expression of senescence-associated genes (Woo et al., 2013). Therefore, the result indicates that anthocyanin biosynthesis in the two pigmented potatoes may not be induced in a senescence-dependent manner activated by the EIN2–NAC2 pathway. Indeed, it has been reported that ethylene suppresses sugar-induced anthocyanin accumulation in Arabidopsis by suppressing the expression of positive regulators of the WD40/bHLH/R2R3–MYB complex and stimulating the expression of the negative R3–MYB regulator MYBL2 (Jeong et al., 2010). Thus, ethylene differentially regulates anthocyanin biosynthesis according to developmental and environmental stimuli. However, studies into their regulation mechanisms are lacking.
In addition to the above genes, Fig. 3 showed that a large number of genes were connected with diverse anthocyanins. Quantitative changes in the secondary metabolites in groups A and B, which were most abundant in the light-red Hongyoung, showed negative or positive correlations with transcriptional changes in genes in clusters I and II. Moreover, the anthocyanin contents in groups C and D, which were most abundant in the dark-purple Jayoung, were negatively or positively correlated with the expression levels of genes in clusters IV and V (Table 2 and Fig. 3). In contrast, the expression level of two F3′5′Hs was not significantly different between Hongyoung and Jayoung (Table 2), while the Dp and Pt derivatives of anthocyanin were highly increased in Jayoung (Table 3). As the biosynthesis of blue Dp-type anthocyanins are known to be driven by the activity of F3′5′H (Ishiguro et al., 2012), there might be other genes responsible for the Dp-based anthocyanin biosynthesis in Jayoung. The more significant up-regulation of LODX, UFGT, CHS, and CHI in cluster IV and PAL2 and NAD(P)-linked oxidoreductase (AT1G59960) in cluster V might have role in the more significant accumulation of Dp and Pt derivatives. LODX converts the colorless leucoanthocyanidins into the colored anthocyanidins, which are inherently unstable under physiological conditions (Lo Piero, 2015). The addition of a glucose moiety in the 3-OH positions of anthocyanidins by UFGT increases the hydrophilicity and stability of anthocyanidins, conveying the flux of flavonoid intermediates towards the synthesis of anthocyanins (Lo Piero, 2015). Putting these together, with the significant increase of early-step genes including PAL2, CHS, and CHI in clusters IV and V, it can be postulated that that UFGT and LODX might have role in driving the flux and accumulation of Dp-type anthocyanins in Jayoung.
In conclusion, we explored the regulatory network connected to anthocyanin biosynthesis using integrated analysis of the metabolome and transcriptome in sprouts of three different colored potatoes: light-red Hongyoung, dark-purple Jayoung, and white Atlantic. Correlation analysis between metabolites and regulatory genes identified the regulatory genes associated with anthocyanin metabolites and provided new insight into the regulatory mechanism underlying the biosynthesis of anthocyanin accumulation in colored potatoes. Moreover, a connection network between changes in transcriptional expression and metabolite levels according to the pigmentation was obtained. The dataset could be harnessed by researchers to utilize genetic approaches to clarify the mechanism of anthocyanin regulation.
Supplementary Data
Supplementary data are available at JXB online.
Table S1. Primer list used in qPCR analysis.
Table S2. Exact mass of aglycones, sugars, and acylated groups found in anthocyanins and flavonoid glycosides.
Table S3. Identification of anthocyanin biosynthesis-related compounds with MS/MS spectra obtained in ESI+ and ESI− modes.
Table S4. List of 167 genes categorized to hormones, signaling, transcriptional regulation, and flavonoid metabolism.
Table S5. The correlation matrix of metabolites (anthocyanins) and gene expression levels.
Table S6. Interaction value between 22 metabolites and 119 genes that has a strong correlation coefficient (R 2>0.9).
Fig. S1. Tubers and sprouts of Solanum tuberosum cvs Atlantic, Hongyoung, and Jayoung.
Fig. S2. Structure and molecular formulae of anthocyanidins.
Fig. S3. Score plots and S-plots of orthogonal partial least-squares discriminant analysis (OPLS-DA) in positive (A) and negative (B) modes.
Fig. S4. Identification of anthocyanin derivatives using MS2 fragmentation.
Fig. S5. The number of differently expressed genes among Hongyoung, Atlantic, and Jayoung.
Fig. S6. Quantitative real-time RT-PCR (qPCR) analysis of genes involved in anthocyanin biosynthetic pathway and putative transcriptional regulators according to different color potato cultivars.
Acknowledgement
This work was supported by a grant from KBSI (T34616) to MHN and also supported by a grant from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisherie s(IPET) through the Golden Seed Project funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA), Republic of Korea (no. 213001-04-3-SB530).
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. 2005. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell Online 17, 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu LW, Li L. 2012. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 236, 1153–64. [DOI] [PubMed] [Google Scholar]
- Cho K, Shibato J, Agrawal GK, Jung Y-H, Kubo A, Jwa N-S, Tamogami S, Satoh K, Kikuchi S, Higashi T. 2008. Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. Journal of Proteome Research 7, 2980–2998. [DOI] [PubMed] [Google Scholar]
- Chong ESL, McGhie TK, Heyes JA, Stowell KM. 2013. Metabolite profiling and quantification of phytochemicals in potato extracts using ultra‐high‐performance liquid chromatography–mass spectrometry. Journal of the Science of Food and Agriculture 93, 3801–3808. [DOI] [PubMed] [Google Scholar]
- Consortium PGS. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. [DOI] [PubMed] [Google Scholar]
- El‐Kereamy A, Chervin C, Roustan JP, Cheynier V, Souquet JM, Moutounet M, Raynal J, Ford C, Latché A, Pech JC. 2003. Exogenous ethylene stimulates the long‐term expression of genes related to anthocyanin biosynthesis in grape berries. Physiologia Plantarum 119, 175–182. [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR. 2008. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1 . Proceedings of the National Academy of Sciences, USA 105, 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Mu L, Yan G-L, Liang N-N, Pan Q-H, Wang J, Reeves MJ, Duan C-Q. 2010. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 15, 9057–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Giusti MM. 2010. Anthocyanins: natural colorants with health-promoting properties. Annual Review of Food Science and Technology 1, 163–187. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K. 2007. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences, USA 104, 6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. 2004. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 101, 10205–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefgen R, Nikiforova VJ. 2008. Metabolomics integrated with transcriptomics: assessing systems response to sulfur‐deficiency stress. Physiologia Plantarum 132, 190–198. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Taniguchi M, Tanaka Y. 2012. Functional analysis of Antirrhinum kelloggii flavonoid 3'-hydroxylase and flavonoid 3',5'-hydroxylase genes; critical role in flower color and evolution in the genus Antirrhinum. Journal of Plant Research 125, 451–456. [DOI] [PubMed] [Google Scholar]
- Jaakola L. 2013. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends in Plant Science 18, 477–483. [DOI] [PubMed] [Google Scholar]
- Jeong S-W, Das PK, Jeoung SC, Song J-Y, Lee HK, Kim Y-K, Kim WJ, Park YI, Yoo S-D, Choi S-B. 2010. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiology 154, 1514–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM. 2006. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proceedings of the National academy of Sciences, USA 103, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L-Z, Sun J, Chen P, Harnly JA. 2011. LC-PDA-ESI/MSn identification of new anthocyanins in purple Bordeaux radish (Raphanus sativus L. variety). Journal of Agricultural and Food Chemistry 59, 6616–6627. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lin-Wang K, Deng C, et al. 2015. Comparative transcriptome analysis of white and purple potato to identify genes involved in anthocyanin biosynthesis. PLoS One 10, e0129148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Shi M-Z, Xie D-Y. 2014. Regulation of anthocyanin biosynthesis in Arabidopsis thaliana red pap1-D cells metabolically programmed by auxins. Planta 239, 765–781. [DOI] [PubMed] [Google Scholar]
- Lo Piero AR. 2015. The state of the art in biosynthesis of anthocyanins and its regulation in pigmented sweet oranges [(Citrus sinensis) L. Osbeck]. Journal of Agricultural and Food Chemistry 63, 4031–4041. [DOI] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M. 2009. Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiology 149, 1505–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Cho J, Cho H, Yi J, Seo H, Choung M. 2009a A new potato cultivar “Hongyoung”, with red skin and flesh color, and high concentrations of anthocyanins. Korean Journal of Breeding Science 41, 502–506. [Google Scholar]
- Park Y, Cho J, Cho H, Yi J, Seo H, Chung M. 2009b A new potato cultivar “Jayoung”, with high concentration of anthocyanin. Korean Journal of Breeding Science 41, 51–55. [Google Scholar]
- Patti GJ, Tautenhahn R, Siuzdak G. 2012. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nature Protocols 7, 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. 2005. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National academy of Sciences, USA 102, 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. 2011. The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana . The Plant Cell Online 23, 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Xie D. 2013. Modified bimolecular fluorescence complementation assay to study the inhibition of transcription complex formation by JAZ proteins. Methods in Molecular Biology 1011, 187–197. [DOI] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. 2008. Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends in Plant Science 13, 36–43. [DOI] [PubMed] [Google Scholar]
- Schaart JG, Dubos C, Romero De La Fuente I, Houwelingen AM, Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG. 2013. Identification and characterization of MYB–bHLH–WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria× ananassa) fruits. New Phytologist 197, 454–467. [DOI] [PubMed] [Google Scholar]
- Stushnoff C, Ducreux LJ, Hancock RD, Hedley PE, Holm DG, McDougall GJ, McNicol JW, Morris J, Morris WL, Sungurtas JA. 2010. Flavonoid profiling and transcriptome analysis reveals new gene–metabolite correlations in tubers of Solanum tuberosum L. Journal of Experimental Botany 61, 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Lin LZ, Chen P. 2012. Study of the mass spectrometric behaviors of anthocyanins in negative ionization mode and its applications for characterization of anthocyanins and non-anthocyanin polyphenols. Rapid Communications in Mass Spectrometry 26, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Truong V-D, Deighton N, Thompson RT, McFeeters RF, Dean LO, Pecota KV, Yencho GC. 2009. Characterization of anthocyanins and anthocyanidins in purple-fleshed sweetpotatoes by HPLC-DAD/ESI-MS/MS. Journal of Agricultural and Food Chemistry 58, 404–410. [DOI] [PubMed] [Google Scholar]
- Urbanczyk‐Wochniak E, Luedemann A, Kopka J, Selbig J, Roessner‐Tunali U, Willmitzer L, Fernie AR. 2003. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Reports 4, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. 2006. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Onkokesung N, Baldwin IT, Galis I. 2012. Jasmonoyl‐l‐isoleucine hydrolase 1 (JIH1) regulates jasmonoyl‐l‐isoleucine levels and attenuates plant defenses against herbivores. The Plant Journal 72, 758–767. [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Nam HG, Lim PO. 2013. Plant leaf senescence and death–regulation by multiple layers of control and implications for aging in general. Journal of Cell Science 126, 4823–4833. [DOI] [PubMed] [Google Scholar]
- Wu X, Prior RL. 2005. Systematic identification and characterization of anthocyanins by HPLC-ESI-MS/MS in common foods in the United States: fruits and berries. Journal of Agricultural and Food Chemistry 53, 2589–2599. [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Gourrierec J, Harscoët E, Berger A, Jauvion V, Scagnelli A, Berger N, Bidzinski P, Kelemen Z. 2013. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytologist 198, 59–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.