Highlight
We report two novel reduced-height maize mutants caused by mutations within a putative inositol polyphosphate 5-phosphatase-encoding gene. Auxin-mediated internode elongation probably requires regulation of inositol and/or phospholipid metabolism.
Key words: Brachytic, br3, brevis plant1, bv1, cell elongation, development, growth, internode elongation, plant height.
Abstract
In maize (Zea mays L.), as in other grass species, stem elongation occurs during growth and most noticeably upon the transition to flowering. Genes that reduce stem elongation have been important to reduce stem breakage, or lodging. Stem elongation has been mediated by dwarf and brachytic/brevis plant mutants that affect giberellic acid and auxin pathways, respectively. Maize brevis plant1 (bv1) mutants, first identified over 80 years ago, strongly resemble brachytic2 mutants that have shortened internodes, short internode cells, and are deficient in auxin transport. Here, we characterized two novel bv1 maize mutants. We found that an inositol polyphosphate 5-phosphatase orthologue of the rice gene dwarf50 was the molecular basis for the bv1 phenotype, implicating auxin-mediated inositol polyphosphate and/or phosphoinositide signalling in stem elongation. We suggest that auxin-mediated internode elongation involves processes that also contribute to stem gravitropism. Genes misregulated in bv1 mutants included genes important for cell wall synthesis, transmembrane transport, and cytoskeletal function. Mutant and wild-type plants were indistinguishable early in development, responded similarly to changes in light quality, had unaltered flowering times, and had normal flower development. These attributes suggest that breeding could utilize bv1 alleles to increase crop grain yields.
Introduction
In grasses including Zea mays, plant height at maturity largely results from stem elongation (Moore et al., 1991). This elongation occurs primarily after the shoot apical meristem initiates the production of reproductive tissues in response to developmental and environmental signals (Siemer et al., 1969; Colasanti and Muzynski, 2009). Prior to the reproductive transition, the shoot apex is below or near the surface of the soil. New leaves emerge from the pseudostem, a whorl composed of the leaf sheaths of younger leaves. For example, when eight leaf tips have emerged from the maize whorl, the shoot meristem can be below ground. Cell division and cell expansion within the internodes extend the shoot apex through the whorl. At maturity, the apex terminates with the mature tassel.
Tall crop plants can have poor agronomic performance due to lodging, especially in soils with a high nitrogen content. Several reduced-height mutants have been utilized in cereal breeding. For example, the semi-dwarfing gene sd1 was used in the rice semi-dwarf cultivar IR8, which produced record yields in Asia (Monna et al., 2002; Spielmeyer et al., 2002). Similarly reduced height1 (rht1) in wheat and dwarf-3 in sorghum have increased plant yields by reducing stem lodging (Peng et al., 1999; Khush, 2001; Barrero Farfan et al., 2012). In maize, one class of reduced-height mutants including anther ear1 (an1), dwarf plant1 (d1), dwarf plant2 (d2) dwarf plant3 (d3), dwarf plant5 (d5), dwarf plant8 (D8), and dwarf plant11 (D11) are deficient in gibberellic acid (GA) biosynthesis or signalling. A second class, including brachytic1 (br1), brachytic2 (br2), and brevis plant1 [bv1, formerly termed brachytic3 (br3)] are gibberellin insensitive. GA mutants have stunted growth habits very early in development and the mutations affect organ size and sex determination (Phinney, 1956; Fujioka et al., 1988a, b; Bensen et al., 1995; Spray et al., 1996; Wang et al., 2013). Flowers may be bisexual, thereby affecting seed production. In contrast, the phenotypes of the brachytic/brevis plant mutants are less pleiotropic than those of the anther ear/dwarf mutants. These mutants, known for over half a century, can have leaves similar in size to wild-type leaves and inflorescences with normal, unisexual male and female flowers (Li, 1931; Kempton, 1941; Multani et al., 2003; Pilu et al., 2007), and are thus candidates to reduce the plant height of cultivated maize.
Molecular characterization of reduced-height maize mutants has identified genes involved in GA or auxin biosynthesis and signalling. For example, DWARF1 and DWARF3 genes encode a gibberellin 3-oxidase and a cytochrome P450, respectively, enzymes important for GA biosynthesis, (Winkler and Helentjaris, 1995; Chen et al., 2014). Brachytic2 (Br2) encodes an ATP-binding cassette phosphoglycoprotein (Multani et al., 2003). BR2 facilitates indole-3-acetic acid (IAA) transport across the nodal/intercalary meristem region of the maize stem (Multani et al., 2003; Knöller et al., 2010), analogous to the function of ABCB1 within Arabidopsis thaliana (Noh et al., 2001; Geisler et al., 2005).
Here, we have described two novel bv1/br3 mutants. At flowering, mutant plants are one-quarter the height of wild-type plants, and the internodes have shortened, rectangular pith cells. Nonetheless, the plants are indistinguishable from wild-type plants prior to the floral transition. bv1 mutants have the same flowering time and leaf numbers at maturity as wild-type plants and respond in a similar way to changes in red:far-red (R:FR) light ratios. We found that bv1 encodes an inositol polyphosphate 5-phosphatase (5PTase) with WD40 domains. The two mutant genotypes had non-synonymous substitutions within conserved residues of the inositol phosphatase domain, indicating that inositol polyphosphate and/or phosphoinositide signalling is necessary for auxin-mediated cell elongation. Auxin and phosphoinositide/inositol polyphosphate signalling are known components of the stem gravitropic response. Auxin-responsive gene upregulation within the mutant plants suggested the presence but misdistribution of auxin. In addition, genes involved in the production of cell wall and cytoskeleton proteins were misexpressed in the mutant, suggesting perturbation of vesicular transport. Although we did not find evidence of use of bv1 in the past use in plant breeding, we suggest that allelic variation of the gene may contribute to future plant improvement.
Materials and methods
Plant materials
Two maize plants with reduced height and short internodes, similar to br1, br2, and bv1 maize mutants, were identified within two M2 or M3 segregating families originating from the seed of a B73 maternal parent crossed with a B73 paternal parent whose pollen underwent ethyl methanesulphonate (EMS) mutagenesis as described by Till et al. (2004). Seed was provided by Dr Nathan Springer (University of Minnesota, MN, USA). The two mutants were termed 1301 and 1302. Both the maize B73 inbred and a wild-type sibling of 1302, whose progeny did not segregate for the reduced-height phenotype (termed 1302-Sib), were used as wild-type controls.
Red:far-red ratio treatments
Maize plants were grown in the summer of 2011 at the Arkell research station in Guelph, Ontario, Canada, to investigate the effects of neighbouring plants on mutant and wild-type plant heights. Plants were grown in a hydroponic system using 20 l buckets filled with Turface and watered three times daily with a nutrient solution composed of 800g of 28-14-14 NPK fertilizer, 800g of 15-15-30 NPK fertilizer, 400g of NH4NO3, 60g of Micronutrient Mix (Plant Products Co., Canada) and 800g of MgSO4.7H2O dissolved in 20 l of water, and administered in a 100:1 water:solution mix. B73 wild-type and 1302 plants were grown in Turface only (high R:FR) or in Turface covered with grass sod (low R:FR) from germination until anthesis and silking (Rajcan et al., 2004). The sod changed the R:FR ratio to which growing plants were exposed. The ratio was 0.76±0.02 for Turface alone and 0.30±0.02 with sod (Page et al., 2010). The water, nutrient supply, and growing medium of the grass sod were separate from the maize plants. The experiment was a factorial design with two factors: genotype (levels: 1302 and B73) and treatment (levels: sod and no sod) with four replicates for each genotype×treatment combination. Each replicate of the four genotype×treatment combinations had nine buckets, each bucket with two plants. Plant height and number of leaf tips were recorded at 11 time points from 11 to 76 d after planting (leaf tips 3 to 21). The data were analysed using the R aov function, which fits linear models with categorical variables (R Core Team, 2014).
Mapping and allelism tests of reduced-height mutants
Seed of homozygous br1, br2, and bv1/br3 (referred to here as bv1-1) maize mutants was obtained from The Maize Genetics Cooperation Stock Center located at the University of Illinois Urbana/Champaign (stock ids: 113C, 117A, and 506L respectively). We crossed the 1301 and 1302 mutants to inbred lines A619 and Mo17 to generate F2 mapping populations. Mapping populations were grown in Guelph, Ontario, Canada. In the summer of 2009, a small mapping population consisting of 73 F2 plants of a cross between 1302 and A619 was grown. In the summers of 2010 and 2011, 2122 and 1325 plants of the F2 of a cross between 1301 and the inbred Mo17 were grown in the same location. Finally, in 2012, tissue from 491 individuals of the F2 of 1301×Mo17 with the bv1 phenotype were collected. For each population, the heights of individual plants were scored at maturity and classified as tall or short.
Genomic DNA was extracted from leaf tissue samples of F2 plants, using either a GeneEluteTM Plant Genomic DNA Mini prep kit (Sigma, St Louis, MO, USA) or a CTAB-based protocol adapted in-house for 96-well plate extractions. An initial genotyping of 73 individuals was done at the Analytical Genetics Technology Centre of the Princess Margaret Hospital in Toronto, Canada, using the iPLEX Gold Assay (Sequenom, San Diego, CA, USA) using 68 polymorphic single-nucleotide polymorphisms (SNPs) between B73 and A619 over the 10 maize chromosomes (Gore et al., 2009). For fine mapping, the F2 plants grown in years 2010, 2011, and 2012 were genotyped with simple sequence repeat (SSR) markers and SNP markers. SSR markers used included umc1060, umc1966, umc1747, umc1155, umc1822, and bnlg609 from MaizeGDB (http://www.maizegdb.org/ssr.php). PCR for genotyping with SSR markers was performed under the following conditions: 1min of initial denaturation at 95 °C, 35 cycles of 30s denaturation at 95 °C, 30s annealing at 55 °C, and 40s extension at 72°C, followed by 10min of final extension at 72 °C. Additional SNPs (Gore et al., 2009) were assayed using custom allele discrimination TaqMan probes designed with the software PrimerExpress 3.0 (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol.
RNA extraction, pooling, sequencing, and analysis
Plants of genotypes 1301, 1302, 1302-Sib, and B73 were grown following a completely randomized design with four plants per genotype, in a greenhouse in 20 l buckets, using Sunshine Growing Mix #4/LA4 (Sun Gro Horticulture Canada Ltd) as medium and the same nutrients solution as described above. At 14 leaf tips (eight mature leaves with blades fully emerged from whorl with a visible base of an exposed leaf blade, V8), 40 d after planting, approximately 5mm3 samples of stalk tissue from the sixth internode of each plant (counting from the lowest visible internode) were stored in 1.5ml of RNAlater (Qiagen GmbH, Hilden, Germany). At this stage of development for inbred B73, the shoot apical meristem has transitioned into the production of male reproductive structures. Elongation initiates sequentially, with lower internodes initiating growth prior to the upper internodes (Robertson 1994). Internode six elongation has just commenced at the V8 stage. At the time of sampling, the wild-type and mutant plant sixth internodes were each about 1mm in length. Internodes beneath internode 6 were visibly longer in the wild-type plants than in the mutant plants. Samples were kept overnight at 4 °C and then stored at –20 °C. Total RNA of individual samples was isolated using TRIzol (Invitrogen, USA) and purified using a Qiagen RNeasy Mini kit column according to the manufacturer’s instructions. RNA (3 μg) from the four plant samples of each genotype were pooled. cDNA libraries were constructed and sequenced at the Clinical Genomics Center of the Mount Sinai Hospital in Toronto, Ontario, Canada, following the Illumina TruSeq Sample Preparation Guide (Part # 15008136 Rev. A) and using an Illumina Hiseq 2500 system. Paired-end reads of 100bp were aligned with the B73 reference genome (ZmB73 RefGen_v2) using TopHat v.2.0.9 (Schnable et al., 2009; Trapnell et al., 2012). Aligned reads were assembled into putative transcripts with cufflinks (v.2.1.1), and cuffdiff (v.2.1.1) identified significant transcript abundance differences between genotypes (Trapnell et al., 2012). Differences in transcript abundance were considered significant when the Benjamini–Hochberg corrected P values were lower than the default false discovery rate (FDR) of 0.05 (Benjamini and Hochberg, 1995; Trapnell et al., 2012). The number of paired-end reads obtained for 1301, 1302, 1302-Sib, and B73 was 23 993 453, 20 109 205, 23 993 453, and 15 566 118, respectively, with 75.6, 77.9, 75.8, and 81.0% concordant pair alignment rates, respectively. Transcript abundances were quantified and normalized to fragments per kilobase of exon per million fragments mapped (FPKM ).
Translated coding sequence similarity searches were performed against the UniProt database (Jain et al., 2009), and alignments were visualized in the software Geneious Basic v.5.6.4 (Meintjes et al., 2012). SNPs relative to the B73 reference genome were identified from aligned RNA sequencing (RNA-Seq) reads using samtools mpileup and filtered with bcftools (Li et al., 2009) for occurrences when the estimated allele frequency of the putative SNP was equal to 1 (AF1=1) and read depth was >8 (DP>8).
The Integrative Genomics Viewer (Robinson et al., 2011) was used to visualize RNA-Seq alignments and manually confirm SNPs. Gene orthologues were identified with information from Gramene (http://www.gramene.org/, release #38, accessed August 2013) and UniProt (http://uniprot.org/, accessed August 2013).
Light microscopy
Transverse and longitudinal sections of fresh tissue were taken from the middle of internodes 1, 2, and 8 of 35-, 63-, and 91-d-old 1302 and B73 plants. Fresh tissue samples were sectioned with a razor blade and stained using 0.05% toluidine blue solution. The height and width of individual cells in longitudinal sections were estimated using ImageJ software.
Results
Genetic and phenotypic characterization of novel bv1 alleles
We identified two reduced-height maize mutants termed 1301 and 1302 in two families segregating for EMS-induced mutations. Self-pollination of both mutants generated populations composed entirely of reduced-height plants. At anthesis in the field, 1301 and 1302 mutants were 48 and 50cm, compared with 191cm for wild-type plants (Figs 1A and 2). The sizes of 1301 and 1302 mutant plant leaves were similar to wild type, and tassels and ears developed normally, although the ears were notably smaller. The ratio of wild-type:reduced-height plants (57:16) within an F2 population (from self-pollinated 1302×A619 hybrids) was consistent with the 3:1 segregation ratio expected for a monogenic recessive allele (χ2 test, P>0.05). Hybrids between 1301 and 1302 mutants failed to complement, suggesting the same gene in both genotypes caused the mutant phenotype. The progeny of crosses between both mutants (1301 and 1302) and br2 mutant was wild type. The progeny of 1302 and bv1-1 (formerly br3) had reduced height (Fig. 1C). We concluded that the reduced-height genotypes were each homozygous for different novel bv1 alleles: bv1-1301 and bv1-1302. The wild-type sibling plant of the bv1-1302 mutant is referred hereafter as Bv1-1302.
Fig. 1.
Maize mutants 1301 and 1302 have reduced height and short internodes compared with wild type plants. (A) An F2 bv1/bv1 plant in the field 63 d after planting from a bv1-1301 mutant×Mo17 cross. (B) Internodes of B73 (left), bv1-1302 mutant (middle), and bv1-1301 mutant (right), 66 d after planting. Leaves were removed. Bar 50cm. (C) F1 progeny of bv1-1 mutant×1302 (left), br2 mutant×1302 (middle), and 1301×1302 (right) 55 d after planting.
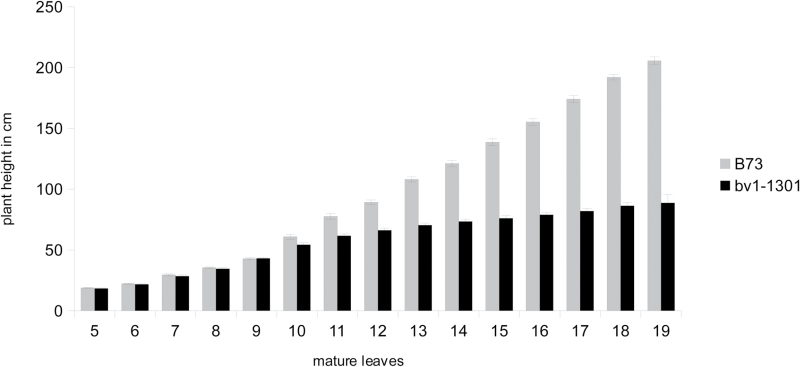
Plant heights of bv1-1301 and bv1-1302 mutants were similar to wild-type heights early in development. In both greenhouse and outdoor bucket experiments, heights of bv1-1301, bv1-1302, and B73 plants were almost identical when a few mature leaves were visible (Fig. 2). In B73, the primary meristem transitioned into an inflorescence meristem when eight mature leaves had emerged from the whorl. The difference in plant height between mutant and wild-type plants was significant when 10 leaves had emerged from the whorl (V10) (Fig. 2).
Fig. 2.
Plant heights of greenhouse-grown 1301 and B73 plants measured when leaf ligules from five to 19 leaves had emerged (V stage). Error bars represent the standard error (n=16).
Low ratios of red:far-red light and auxin treatment can induce cell expansion (Wolbang et al., 2004), and we investigated whether these factors affected plant height differently within the bv1 mutants compared with wild-type controls. A low R:FR treatment induced similar increases in plant height in bv1-1302 mutants as in wild-type B73 plants. Throughout early development, the plant heights of both mutant and wild-type plants grown in the low R:FR environment were 1–2cm greater than plants not exposed to low R:FR (see Supplementary Fig. S1 at JXB online). In addition, in both wild-type and mutant genotypes, leaf emergence occurred more quickly among the plants not exposed to the low R:FR treatment than in those exposed. Beginning 30 d after planting, at approximately the V4–V5 stage, low R:FR-treated plants had up to one less emerged leaf tip than plants not exposed to low R:FR conditions (see Supplementary Fig. S2 at JXB online). Thus, the low R:FR treatment-induced increase of plant height and decrease of leaf emergence rate were not bv1 dependent. Consistent with the similar response of wild-type and mutant plants to changes in the R:FR ratio, spraying bv1-1301 mutant plants and wild-type plants with IAA did not alter the plant heights of either genotype (data not shown).
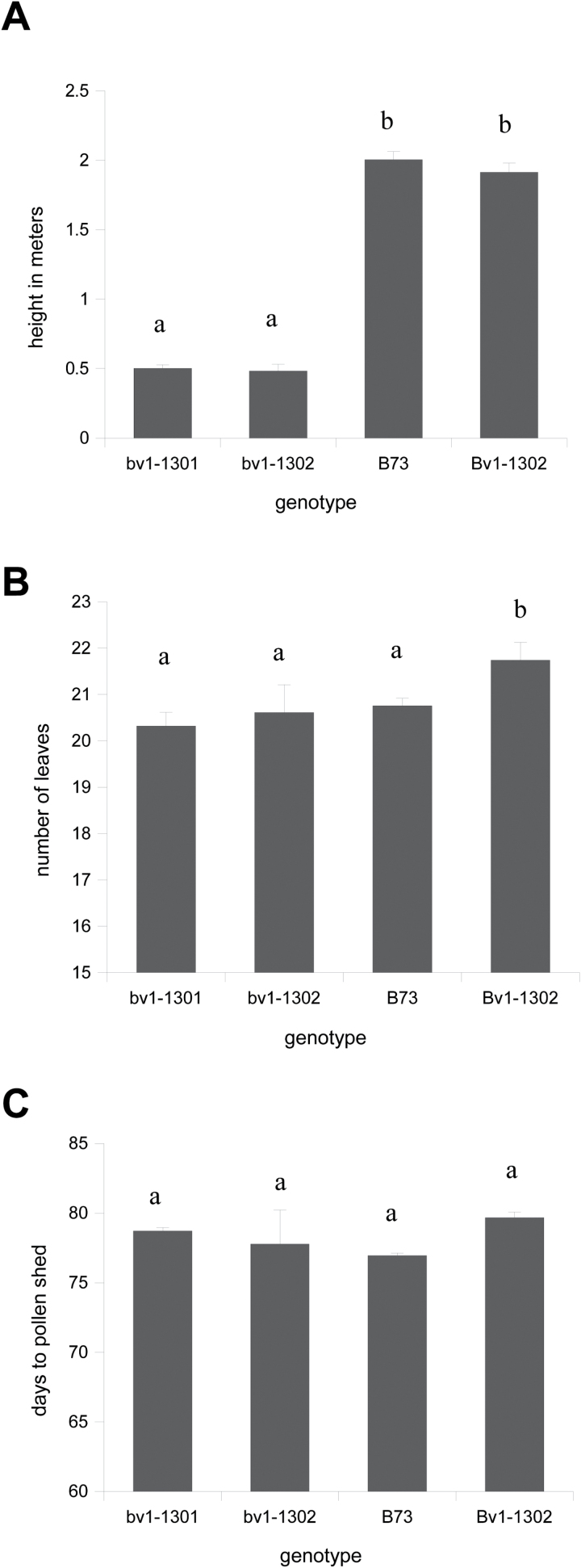
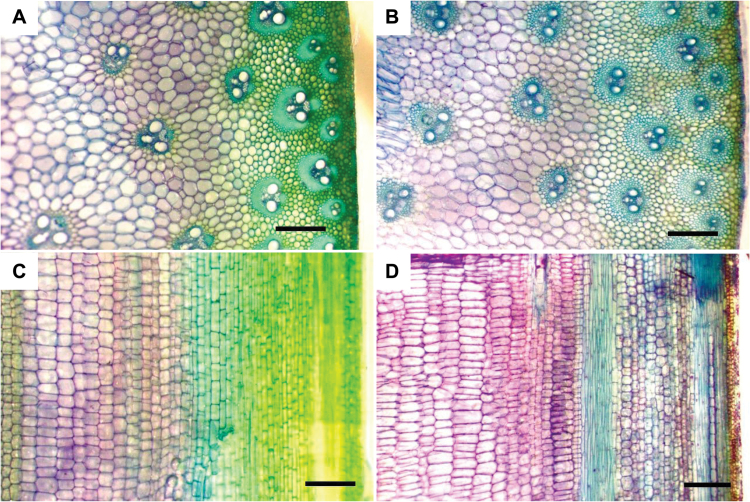
Although both upper and lower internodes of bv1-1301 and bv1-1302 mutants were short relative to wild-type plants, the biggest effect of bv1 was on the upper internodes (Fig. 1A, B and Supplementary Fig. S3 at JXB online). At V14, when the tassel had emerged from the whorl, the maximum internode lengths of bv1-1301 and bv1-1302 mutants were 5.08 and 4.13cm, respectively (Supplementary Fig. S3). In contrast, the maximum B73 internode length was 14.23cm. Lengths of internodes 1 and 2 did not significantly differ (Supplementary Fig. S3). From internodes 3 to 8, the differences between wild-type internodes and mutant internodes increased. Lengths of internodes 8 –14 differed the most between mutant and wild-type plants. The differences varied from 9.2 to 11.9cm for bv1-1301 mutants and from 9.9 to 12.0cm for bv1-1302 mutants (Supplementary Fig. S3). These short internodes resulted in a severely reduced plant height at maturity (Fig. 3A). Among plants harvested after the tassel had fully emerged from the whorl, internode 8 parenchyma cell architectures in transverse sections were similar between mutant and B73 plants (Fig. 4A, B). We also noted no differences in cell morphology when comparing transverse and longitudinal sections from internodes 1 and 2 (data not shown). However, in longitudinal sections of internode 8, mutant plant parenchyma cells were short and rectangular relative wild-type cells (Fig. 4C, D).
Fig. 3.
Mean plant height (A), number of leaves (B), and days to pollen shed since planting (C) of field-grown 1301, 1302, Bv1-1302, and the B73 inbred, measured at anthesis. Three rows of 16 plants were grown per genotype in a completely randomized design. Bars with different letters were significantly different (P=0.05; n=3). Error bars represent the standard error.
Fig. 4.
Transverse and longitudinal stalk sections of B73 (A, C) and bv1-1302 mutant (B, D) internode 8. Cells of bv1-1302 parenchyma cells had an average height of 66.2 vs 89.5 μm for B73 cells (standard error 3.9 and 3.4 μm, respectively, n=30). Both parenchyma and vascular cells are visible in the four panels. Cells of bv1-1302 were wider than B73 cells (178.9 vs 125.9 μm, respectively; standard error 5.6 and 4.9, respectively, n=30). Samples were taken 91 d after planting after the tassel had fully emerged from the whorl. Bars, 400 μm.
Flowering time is often highly positively correlated with plant height and leaf number (e.g. Khanal et al., 2011). However, at maturity, bv1-1301, bv1-1302, and B73 all had similar leaf numbers and flowering times (P>0.05) (Fig. 3B). Bv1-1302 developed one more leaf (P<0.05) than the other genotypes with no significant difference in days to anthesis (Fig. 3C)). The number of leaves between B73 and mutant plants did not significantly differ at any sampled developmental stage (Supplementary Fig. S2). These results showed that stem elongation can be independent of plant developmental timing.
bv1-1301, bv1-1302, and bv1 have mutations within the same locus, which maps to a gene encoding a putative 5PTase
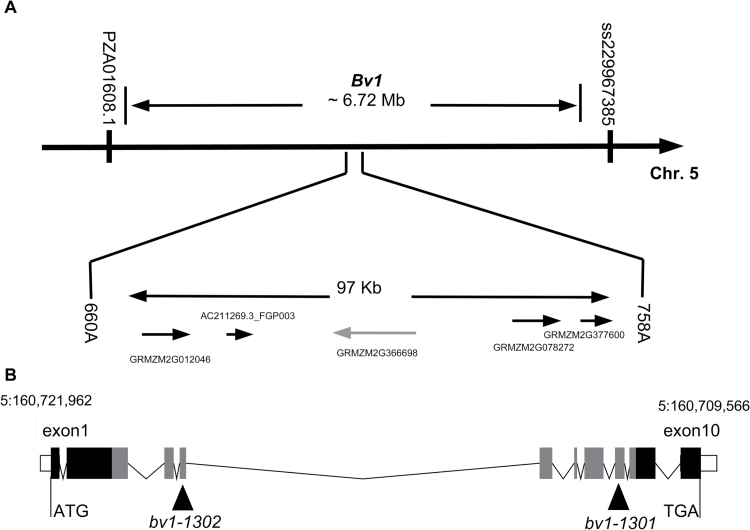
We assayed 491 F2 mutants and 3447 mutant and wild-type F2 plants of a bv1-1301×Mo17 cross to map bv1-1301 between nt 160 660 813 and 160 758 241 (~97kb) on the ZmB73 RefGen_v2 pseudochromosome 5 long arm (Fig. 5A and Supplementary Fig. S4 at JXB online). This map location coincides with the interval on chromosome 5 to which Beavis et al. (1991) genetically mapped bv1 and is less than 2Mb away from the region in the IBM2 2008 Neighbors map estimated for bv1 (5:158 262 770–159 134 168 between markers IDP4992 and IDP2401, available on MaizeGDB). Eight F2 plants had recombinant genotypes at the SNP markers flanking the ~97kb interval (Supplementary Fig. S4). There were five uncharacterized genes in this region (ZmB73_5b_FGS): GRMZM2G012046, AC211269.3_FGP003, GRMZM2G366698, GRMZM2G078272, and GRMZM2G377600. bv1-1301 mutant and Mo17 SNPs within GRMZM2G012046 and GRMZM2G377600 defined the outer boundaries of the ~97kb interval, and we observed recombinants between a SNP position within the gene and the mutant phenotype for both genes (Fig. 5). GRMZM2G366698 is a homologue of the rice dwarf50 gene (d50; Sato-Izawa et al., 2012). d50 (NCBI UniGene: Os.18140, Os02g0477700) is the closest rice homologue to GRMZM2G366698 (Protein BLAST against NCBI database, GenBank locus: BAJ61820.1, 99% query cover, e-value 0.0, 79% identity) and is located on rice chromosome 2: 16 360 196–16 364 326, a region syntenic to maize chromosome 5. Maize genes GRMZM2G012046 and GRMZM2G078272 flanking GRMZM2G366698 also had homologous genes in the same region of rice chromosome 2, Os02g0475300 (2:16, 259, 553–16, 264, 489) and Os02g0478550 (2:16, 426, 109–16, 430, 574), respectively. The putative gene spanned ~13kb and had three annotated splice variants, GRMZM2G366698_T01 (2648bp, 766 aa), GRMZM2G366698_T02 (2664bp, 213 aa), and GRMZM2G366698_T03 (3236bp, 308 aa). The putative GRMZM2G366698_P01 protein encoded a putative 5PTase. It contained an inositol polyphosphate phosphatase catalytic domain SMART: SM00128 (Letunic et al., 2012) and a Trp-Asp signature WD40 repeat-containing domain that may create a platform for stable protein–protein interactions (Ananieva et al., 2008; Stirnimann et al., 2010). bv1 has a similar phenotype to four d50 rice mutants (Sato-Izawa et al., 2012). We discovered two indel markers flanking GRMZM2G366698 (see Supplementary Table S1 at JXB online). Among the eight plants with recombinants within the ~97kb interval (Supplementary Fig. S4), seven wild-type individuals were heterozygous across the locus 1301/Mo17, and the one brachytic mutant was homozygous 1301/1301 for this region. Thus, the height phenotype and GRMZM2G366698 genotype were completely linked across more than 3900 F2 plants. We did not determine whether any of the other four genes within the interval were also completely linked with the bv1 phenotype.
Fig. 5.
(A) The bv1 locus genetically maps between SNP markers PZA01608.1 and ss229967385 with physical positions 158 599 491 and 165 319 744, respectively, on chromosome 5. Fine mapping of bv1 to an ~97kb region using SNP markers 660A and 758A with physical positions 160 660 813 and 160 758 241 (based on markers ss229964467 and ss229964577 from the HapMap project; Gore et al., 2009). The arrows represent the genes and their transcription orientation for this region, according to the B73 genome (AGPv2 release 5b.60). In grey is the candidate gene GRMZM2G366698. (B) Exons (solid boxes) and introns (lines) of transcript GRMZM2G366698_T01. The inositol phosphatase domain is shown in grey spanning from exon 2 to exon 9 (IPPc pfam PF03372, aa 249–595). White boxes are untranslated regions. Mutation points in bv1-1301 and bv1-1302 alleles are shown with arrowheads.
We performed RNA-Seq analyses of RNA harvested from the sixth internodes of wild-type and mutant plants shortly after the shoot had transitioned to generating reproductive tissues. Within the ~97kb interval, transcript abundances were similar between the three mutant and wild-type genes for which we detecteded transcripts (GRMZM2G366698, GRMZM2G012046, and GRMZM2G078272; P>0.05). The mean coverage of each gene’s transcript across genotypes was 15×, 0.51×, and 13×, respectively. Cufflinks predicted six different isoforms of GRMZM2G366698 within B73, Bv-1301, and the 1302 and 1301 bv1 mutants. These were the three splice variants reported in the reference genome annotation and three additional potentially novel splice variants (see Supplementary Tables S2 and S3 at JXB online). Although the isoform with highest abundance differed across genotypes (Supplementary Table S2), these isoform differences are likely stochastic. Across our sample of 22 446 expressed genes (FPKM >1), the most highly expressed isoform differed in 4223 (18.8%).
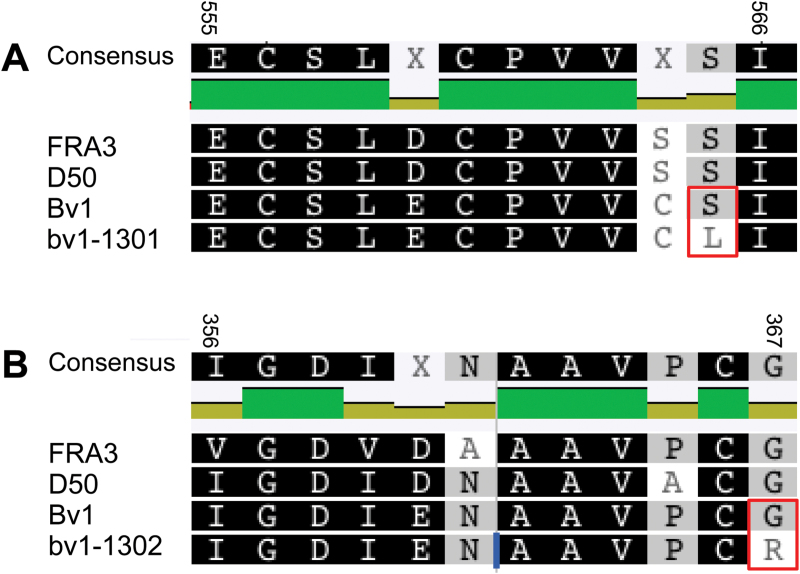
Reads derived from bv1-1301 mRNA reveal a G-to-A transition (on physical position 5:160 710 789) causing an S-to-L change at position 565 of 766 in GRMZM2G366698_P01 (Figs 5B and 6A). This nucleotide was found in 28 out of 28 RNA-Seq reads (10 unique reads) that overlapped this position. Transcript reads of bv1-1302 RNA identified a C-to-T (on physical position 5:160 716 570 ZmB73 RefGen_v2) transition in exon 4 that caused a putative G-to-R amino acid change at position 367 of GRMZM2G366698_P01 supported by 34 out of 34 RNA-Seq reads for this region (24 unique reads) (Figs 5B and 6B). Transcript reads of Bv1-1302 plants had only Cs at 5:160 716 570. Amino acid positions 367 and 565 were both within the inositol phosphatase domain of GRMZM2G366698. Amino acids at position 367 were invariant across diverse monocots and dicots. The amino acid at position 565 only differed within a single Ricinus communis homologue (see Supplementary Fig. S5 at JXB online). Using Sanger sequencing of amplified genomic DNA, we obtained 99% of all intronic sequences of GRMZM2G366698 from bv1-1301 and bv1-1302 plants and found no nucleotide changes relative to the B73 reference genome. We confirmed the bv1-1301 allele through Sanger sequencing, but were unable to amplify GC-rich exon 10, the location of the bv1-1302 mutation, from any genotype. Among the three expressed genes in the ~97kb interval, we identified no other nucleotide differences between bv1-1301, bv1-1302, B73, and Bv1-1302 reads and the B73 reference genome. We concluded that point mutations in GRMZM2G366698 caused the reduced-height phenotypes of bv1-1301 and bv1-1302 mutants.
Fig. 6.
A section of the protein alignments of Arabidopsis FRA3, rice D50, maize BV1 (as in the B73 reference genome), and maize mutants BV1-1301 (A) and BV1-1302 (B). The amino acid changes caused by EMS-induced base transitions are marked with a rectangle. Numbers indicate the amino acid positions in GRMZM2G366698_P1.
Although Sato-Izawa et al. (2012) noted that the putative protein encoded by the d50 gene has the highest homology with A. thaliana FRAGILE FIBER3 (FRA3; AT1G65580), a gene necessary for stem mechanical strength and proper cell wall development, we noted that bv1 and d50 were as similar to the putative A. thaliana inositol polyphosphate 5 phosphatase proteins At5PTase12 (At2g43900), At5PTase13 (At1g05630), and At5PTase14 (At2g31830) as they were to FRA3. In protein BLAST searches, over 97% of the bv1 and d50 protein sequences overlapped these putative Arabidopsis proteins, all with over 51% identity. For example, 99% of GRMZM2G366698_P01 aligned with FRA3 with 53% identity, and 97% of GRMZM2G366698_P01 aligned with At5PTase13 with 54% identity. At5PTase12 (At2g43900) and At5PTase13 (At1g05630) exhibited phosphatase activity towards Ins(1,4,5)P3, while At5PTase14 (At2g31830) and FRA3 exhibited phosphatase activity towards Ins(1,4,5)P3 and PtdIns(3,4,5) but had the highest affinity for PtdIns(4,5)P2 (Zhong et al., 2004; Zhong and Ye, 2004). The similarity of bv1 to At5PTase13 was especially interesting because At5PTase13 mutants have altered auxin accumulation and distribution, altered expression of several genes responsible for auxin biosynthesis and transport, and altered vesicle trafficking that may affect auxin transport (Lin and Wang, 2005; Wang et al., 2009).
Of 256 and 108 genes differentially expressed in bv1-1301 and bv1-1302 internode tissue relative to B73 internode tissue, respectively (FDR-adjusted P<0.05; see Supplementary Table S4 at JXB online), 42 genes were differentially expressed in the two mutants relative to B73 and were not differentially expressed between a Bv1/Bv1 bv1-1302 sister-line and B73 (see Supplementary Table S5 at JXB online). We assigned putative functions to 16 of these genes (Table 1). Genes with higher transcript levels in mutant plants compared with wild-type plants included: (i) GRMZM2G180659, which encodes a putative lysine histidine transporter; (ii) GRMZM2G099467 with similarity to a gibberellin 20 oxidase 2/flavonol synthase/flavanone 3-hydroxylase encoding gene; (iii) GRMZM2G063917 and (iv) GRMZM2G118345, both putative phenylalanine ammonia-lyase (PAL)-encoding genes; (v) GRMZM2G072529, encoding a putative aminocyclopropanecarboxylate oxidase (ACC oxidase), a key enzyme for the biosynthesis of ethylene (Kieber and Ecker, 1993); (vi) GRMZM2G127789, a glutathione S-transferase (GST 29); and (vii) GRMZM2G04902, an isochorismatase family protein rutB. Genes with lower transcript levels in mutant than in wild-type internode tissue included: (i) GRMZM2G118786, which encodes a putative actin-related protein 2/3 complex subunit 3; and (ii) GRMZM2G103273, which encodes a β-galactosidase. Functional relationships among genes can be inferred from co-expression. We tested for enrichment of the 27 differentially expressed genes (out of 68 genes only 27 were represented in the co-expression modules study; Supplementary Table S5) within groups of genes co-expressed across maize development (Downs et al., 2013). Nine (33.3%) differentially expressed genes were in one module, Zm_mod07. The nine genes included putative gibberellin 20 oxidase 2, LHT1, and PAL-encoding genes, as well as a putative endochitinase, GRMZM2G129189 (Table 1). Of the 15 527 expressed genes in Downs et al. (2013), 916 (5.9%) were in the Zm_mod7 module, a significantly smaller proportion (Table 1 and Supplementary Table S5, χ2=36.6, P<0.001). Because genes that are differentially expressed between wild-type and mutant plants are co-regulated across development, BV1 probably affects a discrete biological process that operates in diverse tissues. Genes within Zm_mod07 are more highly expressed in root tissues of different ages (P=0.0002) and mature leaf (P=0.003) relative to other tissues (Downs et al., 2013). Among the genes most overrepresented in Zm_mod07 are those involved in amine transmembrane transporter activity, glycolipid binding, glycolipid transporter activity, and glutathione transferase activity (Downs et al., 2013).
Table 1.
Genes differentially expressed in bv1-1301 and bv1-1302 relative to B73, with putative function annotation (values in FPKM)
| Function | Gene IDa | Putative function annotation | B73 | Bv1-1302 | bv1-1301 | bv1-1302 | Module membership; Score in moduleb |
|---|---|---|---|---|---|---|---|
| Actin polymerization | GRMZM2G118786 | Actin-related protein 2/3 complex subunit 3; uncharacterized | 0.90 | 1.31 | 0.00 | 0.00 | |
| Cellular component | GRMZM2G180659 | LHT1; lysine histidine transporter | 2.27 | 2.83 | 36.45 | 27.36 | Zm_mod07;0.82 |
| Hormones biosynthesis | GRMZM2G099467 | Gibberellin 20 oxidase 2; uncharacterized | 3.28 | 1.81 | 81.31 | 43.81 | Zm_mod07;0.70 |
| GRMZM2G072529 | ACC oxidase | 0.00 | 0.00 | 2.28 | 1.78 | ||
| Phenylpropanoid biosynthesis | GRMZM2G118345 | Phenylalanine ammonia-lyase | 2.62 | 3.34 | 21.66 | 16.10 | Zm_mod07;0.71 |
| GRMZM2G063917 | Phenylalanine ammonia-lyase | 0.00 | 0.00 | 8.59 | 2.83 | Zm_mod05;0.62 | |
| Metabolic process | GRMZM2G103273 | β-Galactosidase 5 | 3.39 | 5.94 | 0.00 | 0.00 | |
| GRMZM2G049021 | Isochorismatase family protein rutB | 12.61 | 0.00 | 42.65 | 39.69 | ||
| GRMZM2G129189 | Endochitinase PR4 putative uncharacterized protein | 0.00 | 0.00 | 2.82 | 1.93 | Zm_mod07;0.59 | |
| Signalling | GRMZM2G127789 | Glutathione S-transferase GST 29 | 0.00 | 0.00 | 4.16 | 2.81 | |
| Transcription factor | GRMZM2G169149 | WRKY62-superfamily of transcription factors having WRKY | 0.00 | 0.00 | 10.87 | 3.91 | |
| Transport | GRMZM2G153920 | Sorbitol transporter; uncharacterized | 0.00 | 0.00 | 1.15 | 0.98 | Zm_mod04;-0.49 |
| Transposon | GRMZM2G021020 | Transposable element | 22.24 | 22.90 | 0.00 | 0.00 | |
| GRMZM2G008283 | Transposable element | 15.17 | 33.16 | 0.0 | 0.0 | ||
| GRMZM2G020508 | Transposable element | 4.44 | 4.11 | 12.96 | 13.87 | ||
| GRMZM2G306371 | Transposable element | 2.53 | 5.26 | 0.0 | 0.0 |
a Although 42 genes were differentially expressed between bv1-1301 and bv1-1302 mutants relative to B73 and were not differentially expressed between Bv1-1302 and B73, 16 had functional annotations.
b Module membership refers to the co-expression module in which the gene is classified. The score refers to the correlation of the gene's expression level with the module’s eigengene. Gene module memberships and scores are from a transcriptome analysis of diverse maize tissues (Downs et al., 2013).
Discussion
We have presented four independent lines of evidence to support GRMZM2G366698 as Bv1/Br3. First, the bv1-1301 mutant mapped to a five-gene-containing, ~97kb chromosomal region on chromosome 5 (Fig. 5A). Fine mapping of the mutants was challenging. Gore et al. (2009) reported that this locus has low crossover frequency relative to other genomic regions, and we identified only eight crossovers between bv1 and flanking markers in a survey of over 3900 F2 individuals. Secondly, among all F2 plants, the GRMZM2G366698 genotype perfectly associated with plant height. Thirdly, GRMZM2G366698 encoded a putative 5PTase and was orthologous to the rice d50 gene, which is necessary for internode elongation. Fourthly, both mutants were homozygous for missense substitutions at highly conserved or invariant residues within the gene’s inositol phosphatase domain (Figs S5). One missense mutation converted serine to leucine in bv1-1301, and a second missense mutation converted glycine to arginine in bv1-1302. Among genes expressed within internodes and that were in the ~97kb interval, we detected no additional nucleotide differences between wild-type and mutant genes. Mutant and wild-type internodes have similar bv1 transcript abundances.
Auxin is a signal for cell elongation, and the developing grass inflorescence exports auxin to stem nodes and internodes to promote elongation (Wolbang et al., 2004). Protein(s) encoded by GRMZM2G366698 probably exhibit phosphatase activity towards the soluble inositol polyphosphate inositol 1,4,5 triphosphate [Ins(1,4,5)P3] and possibly phosphoinositides, including PtdIns(4,5)P2 (Zhong and Ye, 2004; Zhong et al., 2004). We propose that BV1 plays a central role in phosphoinositide/inositol phosphate auxin signal transduction in the maize stem. As noted above, the br2 gene encodes a P glycoprotein auxin transporter, and the mutant is defective in auxin transport (Multani et al., 2003; Knöller et al., 2010). The bv1 mutant is visually similar to the br2 mutant, and like the bv1 mutant, br2 mutants are insensitive to GA (Multani et al., 2003). In other contexts of plant growth, 5PTases affect auxin-associated cell expansion. Stems of maize and other grasses that are placed parallel to the ground will grow upward in a gravitropic response. This bending is caused by elongation of cells at the basal side of the pulvinus, a disc-shaped region of tissue immediately apical to the stem node. Auxin levels on the top of the pulvinus are lower than at the base (Wolbang et al., 2007), and auxin transport from the cells at the top of the pulvinus promote elongation of cells at the base of the pulvinus (Yun et al., 2006). PtdIns(4,5)P2 is generated by phosphatidylinositol 4-phosphate 5-kinase (PIP5K). In the initial responses of maize pulvini to gravity, PIP5K activity and Ins(1,4,5)P3 levels increase in the lower pulvinus (Perera et al., 1999). In A. thaliana, inositol signalling is also implicated in the auxin-mediated gravitropic response (Perera et al., 2006; Wang et al., 2009; Mei et al., 2012; Ischebeck et al., 2013).
The bv1 mutant internodes may have different phosphoinositide abundances relative to wild-type plants. Within the mutant, active auxin may be misdistributed but retain or exceed wild-type levels. In pip5k1 pip5k2 double-mutant A. thaliana root tips, PtdIns(1,4)P2 levels are reduced. Although the abundance of auxin is unchanged, the distribution and polar transport of auxin are affected (Ischebeck et al., 2013). Over-expression of PIP5K1 and PIP5K2 increase PtdIns(4,5)P2 levels. Roots have non-directional growth and have lost gravitropism, again suggesting perturbed auxin distribution (Ischebeck et al., 2013). We suggest that active auxin levels are high within mutant internodes because during gravistimulation, auxin increases precede ethylene production within grass nodes (Wright et al., 1978). The key ethylene biosynthetic enzyme, ACC oxidase (Lin et al., 2009), is putatively encoded by GRMZM2G072529 and is up-regulated in the bv1 mutants relative to wild-type plants. Possibly, apolar auxin distribution could contribute to the rectangular shapes of bv1 internode parenchyma cells.
Genes misregulated within mutant internodes are notable for encoding cytoskeleton, cell wall, and membrane-associated proteins. GRMZM2G1187786 probably functions to maintain cytoskeleton function. It is expressed within wild-type internodes and silenced within the bv1 mutants. GRMZM2G1187786 encodes a putative protein with 75% identity to the actin-related protein C3 (ARPC3; AT1G60430), which is one of seven subunits of the actin-related protein complex (Hussey et al., 2006). In A. thaliana, the absence of functional ARP proteins causes shortened cells. Expanding cells contain dense actin bundles and exhibit pronounced morphological aberrations. (Li et al., 2003; Mathur et al., 2003a,b; El-Assal et al., 2004; Saedler et al., 2004). Shortened d50 mutant cells have actin thickened in longitudinally aligned bundles while wild-type rice internode parenchyma cells have a fine network of actin filaments (Zhong and Ye, 2004; Sato-Izawa et al., 2012). Cell wall-related genes include GRMZM2G11834, GRMZM2G063917, and GRMZM2G103273. Polysaccharide-linked hydroxycinnamoyl esters ferulic acid and p-coumaric acid are important cell wall components and are produced by the phenylpropanoid pathway, of which PAL is the first committed step. Transcripts of two putative PAL-encoding genes, GRMZM2G118345 and GRMZM2G063917, are highly abundant in the bv1 mutant relative to wild-type stems. The activity of PAL also greatly increases in d50 (Nishikubo et al., 2000; Sato-Izawa et al., 2012), and the middle lamellae of d50 cells have high levels of polysaccharide-linked ferulic acid and p-coumaric acids (Sato-Izawa et al., 2012). GRMZM2G103273, a putative β-galactosidase, has low transcript abundance in mutant internodes relative to wild-type internodes. β-Galactosidases can target xyloglucan to regulate cellulose fibre separation during wall extension, and a blockage of xyloglucan digestion results in shortened plant organs within Arabidopsis (Sampedro et al., 2012). GRMZM2G103273 is highly similar to the A. thaliana protein ATBGAL10 (BLASTP e-value 2e–25), which is responsible for the majority of β-galactose activity against xyloglucan. Misexpressed genes encoding membrane-associated transporters include GRMZM2G153920 and GRMZM2G180659. The sorbitol transporter-like protein GRMZM2G153920 is up-regulated in bv1 relative to wild-type internodes. Sorbitol transporter-like proteins can transport pentoses as well as polyols and different hexoses (Klepek et al., 2005). Cell wall-derived xylose and arabinose are high, and glucose is low in the Fukei 71/d50 rice mutant relative to wild type (Nishikubo et al., 2000). GRMZM2G180659 encodes a putative lysine histidine transporter, LHT1, and is similar to AUX transporters.
Although the substrates of BV1 are unknown, the putative functions of misexpressed genes suggest that BV1 contributes to plasma membrane-associated molecular transport. BV1 could affect transport by altering the patterns of vesicular cycling (Ischebeck et al., 2013) and/or by altering the interaction of a phosphoinositide with an actin-binding protein, thereby affecting actin polymerization (Stevenson et al., 2000). A high proportion of transcripts misexpressed within the bv1 mutant internodes are co-expressed across maize development with genes whose functions are enriched for membrane-associated transport processes including glycolipid transport and amine transmembrane transport. Both bv1 transcripts and many genes misregulated in bv1 mutants accumulate in diverse maize tissues (Sekhon et al., 2011; Downs et al., 2013), indicating a broad function for BV1. We expect that paralogous 5PTases such as GRMZM2G065225 can compensate for the loss of BV1 function in tissues other than the expanding internode.
Variation among genes identified through the study of induced, large-effect mutations can contribute to quantitative trait variation within a population, and bv1 may be allelic to plant architecture quantitative trait loci (QTLs) identified in mapping studies. In a segregating population derived from crosses between an Illinois High Oil population and an Illinois Low Oil population, Berke and Rocheford (1995) identified a QTL for plant height between markers npi449a and npi295a. This locus corresponds to a region between 151 and 169Mb on the maize physical map. Beavis et al. (1991) also identified a QTL for plant height in an F2 of inbred lines K05×W65 that maps close to bv1. Nonetheless, in an association mapping panel, Pfeiffer et al. (2014) found little correlation between bv1 alleles and plant height. Similarly, SNP variation flanking the sorghum gene model homologous to bv1, Sobic.004G145500, was uncorrelated with plant height variation in a set of 377 diverse sorghum accessions (Casa et al., 2008; X. Li and J. Yu, personal communication, 2015). We note that although BV1 affects the transcript abundance of GRMZM2G099467, a putative gibberellin 20 oxidase, this gene is not the orthologue of Os3g0122300, the gene responsible for the ‘green revolution’ dwarf rice. When using the rice green revolution protein in a BLASTP query, GRMZM2G180659 is the fifth best hit in the maize genome with 37.5% indentity over 315 aa, compared with 84% over 340 aa for the top hit.
Maize bv1 alleles may have come under artificial selection, as has a sorghum allele of a br2 homologue (Multani et al., 2003), because stem lodging can significantly reduce maize yields (Elmore and Ferguson, 1999). Although our bv1 alleles were very short and had smaller ears than wild-type plants, weak bv1 alleles would be appealing targets for selection for a number of reasons. bv1 mutants are indistinguishable from wild-type plants until after the transition of the apical meristem from the vegetative to the reproductive state and respond similarly to neighbouring plants and low R:FR light (Fig. 2 and Supplementary Figs S1 and S2). Although maize height and flowering time are consistently genetically correlated (Salas Fernandez et al., 2009; Khanal et al., 2011), bv1 mutations do not change flowering times (Fig. 3C), a key trait for crop adaptation. bv1 has normal flower development (data not shown) in contrast to other reduced-height maize mutants such as anther ear1 (an1), dwarf plant1 (d1), dwarf plant3 (d3), dwarf plant5 (d5), dwarf plant8 (d8) and dwarf plant11 (d11) (Fujioka et al., 1988a, b; Bensen et al., 1995; Lawit et al., 2010; Wang et al., 2013). The bv1 genomic region is not within the 256 regions identified by Heerwaarden et al. (2012) with evidence for directional selection as a product of modern, North America maize breeding. Nonetheless, we note that plant height in maize is a highly polygenic trait (Peiffer et al., 2014), and selection could rapidly change height (or reduce lodging) with only moderate allele frequency shifts at many existing polymorphic loci, including bv1. In this case, association and selective sweep analyses would not identify these loci (Pritchard et al., 2010). Suitable bv1 alleles are candidates for future crop improvement.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Plant height of bv1-1302 (bv1) and B73 (wt) plants grown with low R:FR reflected light (surrounding turfgrass) and high R:FR reflected light (no turfgrass) at different times after planting.
Fig. S2. Number of visible leaf tips of bv1-1302 (bv1) and B73 (wt) plants grown with low R:FR reflected light (turfgrass) and high R:FR reflected light (no turfgrass).
Fig. S3. Internode lengths of field grown bv1-1301, bv1-1302 and B73 plants measured 66 d after planting.
Fig. S4. Genotypes of plants with crossovers that mapped the bv1 mutation to a ~97kb region on chromosome 5 (5:160 660 813–160 758 241).
Fig. S5. Multiple sequence alignment of maize GRMZM 2G366698_P01 subsequence and 20 homologous proteins across diverse plant species showing amino acid changes (in red rectangle) caused by the mutations in bv1-1301 (A) and bv1-1302 (B).
Table S1. Markers designed.
Table S2. Transcript abundance of GRMZM2G366698 across sample pools.
Table S3. Exon coding regions for the three potentially novel splice variants identified in the RNA-Seq reads.
Table S4. Number of differentially expressed genes between mutants bv1-1301 and bv1-1302 and wild-type genotypes Bv1-1302 and B73.
Table S5. All 68 genes differentially expressed in bv1-1301 and bv1-1302 relative to B73 (values in FPKM).
Acknowledgements
This work was funded by the Ontario Research Fund and the Natural Sciences and Engineering Research Council of Canada. We thank Dr Kosala Ranathunge for providing training for the light microscopy methodology used in this article. We thank Dr Joseph Colasanti for his productive discussions, and Hollie Rowlands and Sophie Tayler for field and laboratory assistance.
References
- Ananieva EA, Gillaspy GE, Ely A, Burnette RN, Erickson FL. 2008. Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiology 148, 1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero Farfan ID, Bergsma BR, Johal G, Tuinstra MR. 2012. A stable dw3 allele in sorghum and a molecular marker to facilitate selection. Crop Science 52, 2063–2069. [Google Scholar]
- Beavis WD, Grant D, Albertsen M, Fincher R. 1991. Quantitative trait loci for plant height in four maize populations and their associations with qualitative genetic loci. Theoretical and Applied Genetics 83, 141–145. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57, 289–300. [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. 1995. Cloning and characterization of the maize An1 gene. The Plant Cell 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke TG, Rocheford TR. 1995. Quantitative trait loci for flowering, plant and ear height, and kernel traits in maize. Crop Science 35, 1542–1549. [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S. 2008. Community resources and strategies for association mapping in Sorghum. Crop Science 48, 30–40. [Google Scholar]
- Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, Kobayashi M, McCarty DR, Tan B-C. 2014. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual-localized to the nucleus and cytosol. Plant Physiology 166, 2028–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Muszynski M. 2009. The maize floral transition. In: Handbook of maize: its biology. New York: Springer, 41– 55. [Google Scholar]
- Downs GS, Bi Y-M, Colasanti J, Wu W, Chen X, Zhu T, Rothstein SJ, Lukens LN. 2013. A developmental transcriptional network for maize defines coexpression modules. Plant Physiology 161, 1830–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal SED, Le J, Basu D, Mallery EL, Szymanski DB. 2004. Distorted2 encodes an ARPC2 subunit of the putative arabidopsis ARP2/3 complex. The Plant Journal 38, 526–538. [DOI] [PubMed] [Google Scholar]
- Elmore RW, Ferguson RB. 1999. Mid-Season stalk breakage in corn: hybrid and environmental factors. Journal of Production Agriculture 12, 293–299. [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Gaskin P, Macmillan J, Phinney B, Takahashi N. 1988a Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiology 88, 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Katsumi M, Phinney BO, Gaskin P, MacMillan J, Takahashi N. 1988b The dominant non-gibberellin-responding dwarf mutant (D8) of maize accumulates native gibberellins. Proceedings of the National Academy of Sciences, USA 85, 9031–9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, et al. 2005. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. The Plant Journal 44, 179–194. [DOI] [PubMed] [Google Scholar]
- Gore MA, Chia J-M, Elshire RJ, et al. 2009. A first-generation haplotype map of maize. Science 326, 1115–1117. [DOI] [PubMed] [Google Scholar]
- Heerwaarden J Van, Hufford MB, Ross-Ibarra J. 2012. Historical genomics of North American maize. Proceedings of the National Academy of Sciences, USA 109, 12420–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ. 2006. Control of the actin cytoskeleton in plant cell growth. Annual Review of Plant Biology 57, 109–125. [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Werner S, Krishnamoorthy P, et al. 2013. Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. The Plant Cell 25, 4894–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. 2009. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempton JH. 1941. Heritable characters in maize. III. Brachytic culms. Journal of Heredity 3, 111–115. [Google Scholar]
- Khanal R, Earl H, Lee EA, Lukens L. 2011. The genetic architecture of flowering time and related traits in two early flowering maize lines. Crop Science 51, 146–156. [Google Scholar]
- Khush GS. 2001. Green revolution: the way forward. Nature Reviews Genetics 2, 815–822. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Ecker JR. 1993. Ethylene gas: it’s not just for ripening any more! Trends in Genetics 9, 356–362. [DOI] [PubMed] [Google Scholar]
- Klepek Y-S, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. 2005. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribose. The Plant Cell 17, 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöller AS, Blakeslee JJ, Richards EL, Peer WA, Murphy AS. 2010. Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. Journal of Experimental Botany 61, 3689–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawit SJ, Wych HM, Xu D, Kundu S, Tomes DT. 2010. Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant and Cell Physiology 51, 1854–1868. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Research 40, D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HW. 1931. Heritable characters in maize. XXXVII. Brevis. Journal of Heredity 22, 14–16. [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM. 2003. The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiology 132, 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Wang Y. 2005. At5PTase13 modulates cotyledon vein development through regulating auxin homeostasis. Plant Physiology, 1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. 2009. Recent advances in ethylene research. Journal of Experimental Botany 60, 3311–3336. [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M. 2003a Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. The Plant Cell 15, 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hülskamp M. 2003b Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130, 3137–3146. [DOI] [PubMed] [Google Scholar]
- Mei Y, Jia W-J, Chu Y-J, Xue H-W. 2012. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Research 22, 581–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meintjes P, Duran C, Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. 2002. Positional cloning of rice semidwarfing gene, sd-1: rice ‘green revolution gene’ encodes a mutant enzyme involved in gibberellin synthesis. DNA Research 9, 11–7. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Moser LE, Vogel KP, Waller SS, Johnson BE, Pedersen JF. 1991. Describing and quantifying growth stages of perennial forage grasses. Agronomy Journal 83, 1073–1077. [Google Scholar]
- Multani DS, Briggs SP, Chamberlin M a, Blakeslee JJ, Murphy AS, Johal GS. 2003. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302, 81–84. [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Araki T, Kajita S, Kuroda K, Kitano H, Katayama Y. 2000. Specific accumulation of polysaccharide-linked hydroxycinnamoyl esters in the cell walls of irregularly shaped and collapsed internode parenchyma cells of the dwarf rice mutant Fukei 71. Plant & Cell Physiology 41, 776–784. [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy A, Spalding E. 2001. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. The Plant Cell 13, 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page ER, Tollenaar M, Lee E a, Lukens L, Swanton CJ. 2010. Shade avoidance: an integral component of crop-weed competition. Weed Research, 281–288. [Google Scholar]
- Peiffer JA, Romay MC, Gore MA, et al. 2014. The genetic architecture of maize height. Genetics 196, 1337–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Boss WF. 1999. Transient and sustained increases in inositol 1,4,5-triphosphate precede the differential growth response in gravistimulated maize pulvini. Proceedings of the National Academy of Sciences, USA 96, 5838–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung C, Brady S, Muday GK, Boss WF. 2006. A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiology 140, 746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney BO. 1956. Growth response of single-gene dwarf mutants in maize to gibberellic acid. Proceedings of the National Academy of Sciences, USA 42, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilu R, Cassani E, Villa D, Curiale S, Panzeri D, Badone FC, Landoni M. 2007. Isolation and characterization of a new mutant allele of brachytic 2 maize gene. Molecular Breeding 20, 83–91. [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Current Biology 20, R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rajcan I, Chandler K, Swanton C. 2004. Red–far-red ratio of reflected light: a hypothesis of why early-season weed control is important in corn. Weed Science 52, 774–778. [Google Scholar]
- Robertson MJ. 1994. Relationships between internode elongation, plant height and leaf appearance in maize. Field Crops Research 38, 135–145. [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nature Biotechnology 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Mathur N, Srinivas BP, Kernebeck B, Hülskamp M, Mathur J. 2004. Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant and Cell Physiology 45, 813–822. [DOI] [PubMed] [Google Scholar]
- Salas Fernandez MG, Becraft PW, Yin Y, Lübberstedt T. 2009. From dwarves to giants? Plant height manipulation for biomass yield. Trends in Plant Science 14, 454–461. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Gianzo C, Iglesias N, Guitian E, Revilla G, Zarra I. 2012. AtBGAL10 is the main xyloglucan-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiology 158, 1146–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Izawa K, Nakaba S, Tamura K, et al. 2012. DWARF50 (D50), a rice (Oryza sativa L.) gene encoding inositol polyphosphate 5-phosphatase, is required for proper development of intercalary meristem. Plant, Cell and Environment 35, 2031–2044. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. 2011. Genome-wide atlas of transcription during maize development. The Plant Journal 66, 553–563. [DOI] [PubMed] [Google Scholar]
- Siemer E, Leng E, Bonnett O. 1969. Timing and correlation of major developmental events in maize, Zea mays L. Agronomy Journal 61, 14–17. [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. 2002. Semidwarf (sd-1), ‘green revolution’ rice, contains a defective gibberellin 20-oxidase gene. Proceedings of the National Academy of Sciences, USA 99, 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray CR, Kobayashi M, Suzuki Y, Phinney BO, Gaskin P, MacMillan J. 1996. The dwarf-1 (d1) mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway. Proceedings of the National Academy of Sciences, USA 93, 10515–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J, Perera I, Heilmann I, Persson S, Boss W. 2000. Inositol signaling and plant growth. Trends in Plant Science 5, 357. [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Müller CW. 2010. WD40 proteins propel cellular networks. Trends in Biochemical Sciences 35, 565–574. [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Weil C, et al. 2004. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biology 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Deng D, Ding H, Xu X, Zhang R, Wang S, Bian Y, Yin Z, Chen Y. 2013. Gibberellin biosynthetic deficiency is responsible for maize dominant Dwarf11 (D11) mutant phenotype: physiological and transcriptomic evidence. PloS One 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lin W-H, Chen X, Xue H-W. 2009. The role of Arabidopsis 5PTase13 in root gravitropism through modulation of vesicle trafficking. Cell Research 19, 1191–1204. [DOI] [PubMed] [Google Scholar]
- Winkler R, Helentjaris T. 1995. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. The Plant Cell 7, 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ. 2004. Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiology 134, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Davies NW, Taylor S a., Ross JJ. 2007. Gravistimulation leads to asymmetry of both auxin and gibberellin levels in barley pulvini. Physiologia Plantarum 131, 140–148. [DOI] [PubMed] [Google Scholar]
- Wright M, Mousdale DMA, Osborne DJ. 1978. Evidence for a gravity-regulated level of endogenous auxin controlling cell elongation and ethylene production during geotropic bending in grass nodes. Biochemie und Physiologie der Pflanzen 172, 581–596. [Google Scholar]
- Yun HS, Joo SH, Kaufman PB, Kim TW, Kirakosyan A, Philosoph-Hadas S, Kim SK, Chang SC. 2006. Changes in starch and inositol 1,4,5-trisphosphate levels and auxin transport are interrelated in graviresponding oat (Avena sativa) shoots. Plant, Cell and Environment 29, 2100–2111. [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH, Ye ZH. 2004. FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. The Plant Cell 16, 3242–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z. 2004. Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana . Plant & Cell Physiology 45, 1720–1728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.