Abstract
Objectives:
We investigated the correlation between polygenic risk of ischemic stroke (and its subtypes) and cognitive ability in 3 relatively healthy Scottish cohorts: the Lothian Birth Cohort 1936 (LBC1936), the Lothian Birth Cohort 1921 (LBC1921), and Generation Scotland: Scottish Family Health Study (GS).
Methods:
Polygenic risk scores for ischemic stroke were created in LBC1936 (n = 1005), LBC1921 (n = 517), and GS (n = 6,815) using genome-wide association study summary data from the METASTROKE collaboration. We investigated whether the polygenic risk scores correlate with cognitive ability in the 3 cohorts.
Results:
In the largest cohort, GS, polygenic risk of all ischemic stroke, small vessel disease stroke, and large vessel disease stroke, but not cardioembolic stroke, were correlated with both fluid and crystallized cognitive abilities. The highest correlation was between a polygenic risk score for all ischemic stroke and general cognitive ability (r = −0.070, p = 1.95 × 10−8). Few correlations were identified in LBC1936 and LBC1921, but a meta-analysis of all 3 cohorts supported the correlation between polygenic risk of ischemic stroke and cognitive ability.
Conclusions:
The findings from this study indicate that even in the absence of stroke, being at high polygenic risk of ischemic stroke is associated with lower cognitive ability.
The common cause theory of aging predicts that a proportion of the variance in both age-related cognitive and physical decline is attributable to common biological processes.1 Exploring the genetic links between cognitive function and diseases, such as ischemic stroke, may provide insight. Risk factors for cognitive decline and ischemic stroke overlap.1,2 Low cognitive ability in youth portends a greater risk of stroke in later life.3,4 Cognitive ability in later life is determined by cognitive ability in youth5 and rate of cognitive decline.
With the exception of an association between APOE and cognitive ability in later life and age-related cognitive decline,6,7 few individual genes have reliably been associated with variance in normal cognitive ability. However, Genome-wide Complex Trait Analyses indicates that 29% to 50% of the variance in cognition measured at a single time8,9 and about 24% of the variance in lifetime cognitive change10 can be attributed to common genetic variants. Heritability, calculated from genome-wide single-nucleotide polymorphism (SNP) data, is estimated at 38% for all ischemic stroke, 40% for large vessel disease (LVD), 33% for cardioembolic (CE) stroke, and 16% for small vessel disease (SVD) stroke.11
We used genome-wide association study (GWAS) summary data from the METASTROKE Collaboration12 to generate polygenic risk scores for ischemic stroke and its subtypes, SVD, LVD, and CE, in 3 Scottish cohorts consisting of relatively healthy participants—Generation Scotland: the Scottish Family Health Study (GS), Lothian Birth Cohort 1921 (LBC1921), and Lothian Birth Cohort 1936 (LBC1936). We investigated whether the polygenic risk scores predicted cognitive ability/cognitive decline in the 3 cohorts.
METHODS
Cohorts.
LBC1936.
LBC1936 comprises 1,091 (548 men) relatively healthy older participants, most of whom took part in the Scottish Mental Survey of 1947, when they were about 11 years old.13 At a mean age of 69.5 years (SD 0.8), they were enrolled in a study designed to determine factors that influence cognitive aging.14,15 They took a number of cognitive and physical tests including the Moray House IQ Test (MHT) No. 12 (a test of general cognitive function), which had been administered at age 11.
Medical history, including history of stroke, was recorded. For this study, a general fluid (gf) cognitive ability score was derived from principal components analysis of 6 Wechsler Adult Intelligence Scale–III UK16 nonverbal subtests (Matrix Reasoning, Letter Number Sequencing, Block Design, Symbol Search, Digit Symbol, and Digit Span Backward), as described previously.17 We also used MHT scores from age 11 and age 70 years, and the National Adult Reading Test (NART),18 taken at age 70 years, as a measure of crystallized cognitive ability. A general cognitive ability score was created as for the gf score plus the addition of NART. Cognitive measures were adjusted for age in days and sex before analysis. To obtain a measure of cognitive aging from age 11 to age 70, gf was corrected for age-adjusted age 11 MHT scores, age, and sex.
LBC1921.
LBC1921 comprises 550 (234 men) relatively healthy older participants, most of whom took part in the Scottish Mental Survey of 1932, when they were about 11 years old.19 At a mean age of 79.1 years (SD 0.6), they were enrolled in a study designed to determine factors that influence cognitive aging.5,15
They took a number of cognitive and physical tests including the MHT, which had been administered at age 11. For this study, a gf cognitive ability score was created from principal components analysis of MHT, Raven Matrices, Logical Memory, and Verbal Fluency, as described previously.8 We also used MHT scores from age 11 and age 79 years and the NART18 from age 79 years. A general cognitive ability score was created as for the gf score plus the addition of NART. Cognitive measures were adjusted for age in days and sex before analysis. To obtain a measure of cognitive aging from age 11 to age 79, gf was corrected for age-adjusted age 11 MHT scores, age, and sex.
Generation Scotland: The Scottish Family Health Study.
GS is a family-structured, population-based cohort study. Between 2006 and 2011, 24,084 participants were recruited in Glasgow, Tayside, Ayrshire, Arran, and North East Scotland. Participants range between 18 and 100 years old and there are up to 4 generations per family.20,21
The cognitive tests applied were Logical Memory, Digit Symbol, Verbal Fluency, and Mill Hill Vocabulary. The Logical Memory test is a test of immediate verbal declarative memory from the Wechsler22 Memory Scale–III UK. It involves immediate and delayed recall of a story with 25 elements that is read aloud to the participant. The Digit Symbol test is from the Wechsler Adult Intelligence Scale–III UK and measures speed of information processing.16 The Verbal Fluency test23 measures executive function. The participant has to name as many words as possible beginning with the letters C, F, and L, and is given 1 minute for each letter. The sum of these 3 scores is taken as the overall measure. A gf cognitive ability score was created from principal components analysis of Logical Memory, Digit Symbol, and Verbal Fluency. The Mill Hill Vocabulary test24 (Junior and Senior Synonyms combined) was used as a measure of crystallized cognitive ability. A general cognitive ability score was derived as for the gf score plus the addition of the Mill Hill Vocabulary test. Cognitive measures were adjusted for age in years and sex.
Medical history, including history of stroke, was recorded. There were 10,000 white European participants, born in the UK and with near complete phenotype data, selected for genotyping. In the present study, 6,815 unrelated participants (2,813 men), with a mean age of 55.5 years (SD 11.4), were analyzed.
DNA extraction.
DNA was extracted from blood or buccal cells using standard procedures at MRC Technology Edinburgh (LBC1921) and the Wellcome Trust Clinical Research Facility Genetics Core Edinburgh (LBC1936 and Generation Scotland).
Standard protocol approvals, registrations, and patient consents.
Ethical approval was attained for LBC1921 and LBC1936 from the Lothian Research Ethics Committee and for LBC1936 from Scotland's Multicentre Research Ethics Committee and for Generation Scotland from the Tayside Research Ethics Committee.
Creating stroke polygenic risk scores.
DNA samples were genotyped at the Wellcome Trust Clinical Research Facility using the Illumina 610-Quadv1 array (San Diego) (LBC1936 and LBC1921)8 or the Illumina HumanOmniExpressExome (San Diego) (Generation Scotland). Individuals were excluded based on relatedness (n = 8 in LBC1936; n = 1 in LBC1921; n = 3,045 in GS), unresolved sex discrepancy (n = 12 in LBC1936; n = 1 in LBC1921; n = 14 in GS), low call rate (≤0.95 n = 16 in LBC1936; n = 5 in LBC1921; ≤0.97 n = 117 in GS), evidence of non-European descent (n = 1 in LBC1936; n = 2 in LBC1921), and pedigree mismatch (n = 8 in GS). SNPs were used in the analyses if they had a call rate ≥0.98, a minor allele frequency ≥0.01, and a Hardy-Weinberg equilibrium test with p ≥ 0.001. The first 4 components from a multidimensional scaling analysis of the SNP data were used as covariates in the analyses to control for population stratification.
GWAS summary data for ischemic stroke and its subtypes were obtained from the METASTROKE Consortium.12 Summary data included SNP name (rs number) and effect allele and size for imputed SNPs associated with all ischemic, SVD, LVD, or CE stroke subtypes. The information was obtained for 5 different p value thresholds: <0.8, <0.5, <0.1, <0.05, and <0.01. The ischemic stroke GWAS included 12,389 cases of all subtypes, SVD 1,894 cases, LVD 2,167 cases, and CE 2,365 cases. The same 62,004 controls were used in each GWAS. Twenty polygenic stroke risk scores (4 stroke phenotypes at 5 p value thresholds) were thus created for each participant of LBC1936, LBC1921, and GS as described in Ref. 25. A/T and G/C SNPs, SNPs with a minor allele frequency <0.02, and SNPs not present in METASTROKE were removed from LBC1936, LBC1921, and GS. Pruning was performed to remove those in linkage disequilibrium (r2 > 0.25 within a 200-SNP sliding window). Risk scores were then created in PLINK26 by summing the product of each of the betas obtained from METASTROKE and the number of effect alleles carried by the participant.
Statistical analyses.
For all cohorts, partial correlations were calculated between the 20 stroke polygenic risk scores and the cognitive phenotypes described above. Covariates included the number of nonmissing SNPs used to form the risk score, and 4 multidimensional scaling components. Where possible (in LBC1936 and GS), individuals with a self-reported history of stroke before the cognitive testing were removed from the cognitive analyses. Random-effects meta-analyses of the gf, general cognitive ability scores, and measures of crystallized cognitive ability (NART in LBC1936 and LBC1921, and Mill Hill Vocabulary in GS) were performed in R (MAc and Metafor packages). An omnibus effect size and standard error were derived and sample heterogeneity was investigated using Cochran Q statistic, which calculated the sum of squared deviations of each cohort's effect size from the overall meta-analytic estimate of significance.
RESULTS
Fifty LBC1936 members (5%) and 127 GS members (1.9%) reported having had a stroke before cognitive testing and were excluded from the analyses. The number of SNPs that made up the polygenic risk scores is shown in table e-1 on the Neurology® Web site at Neurology.org.
LBC1936.
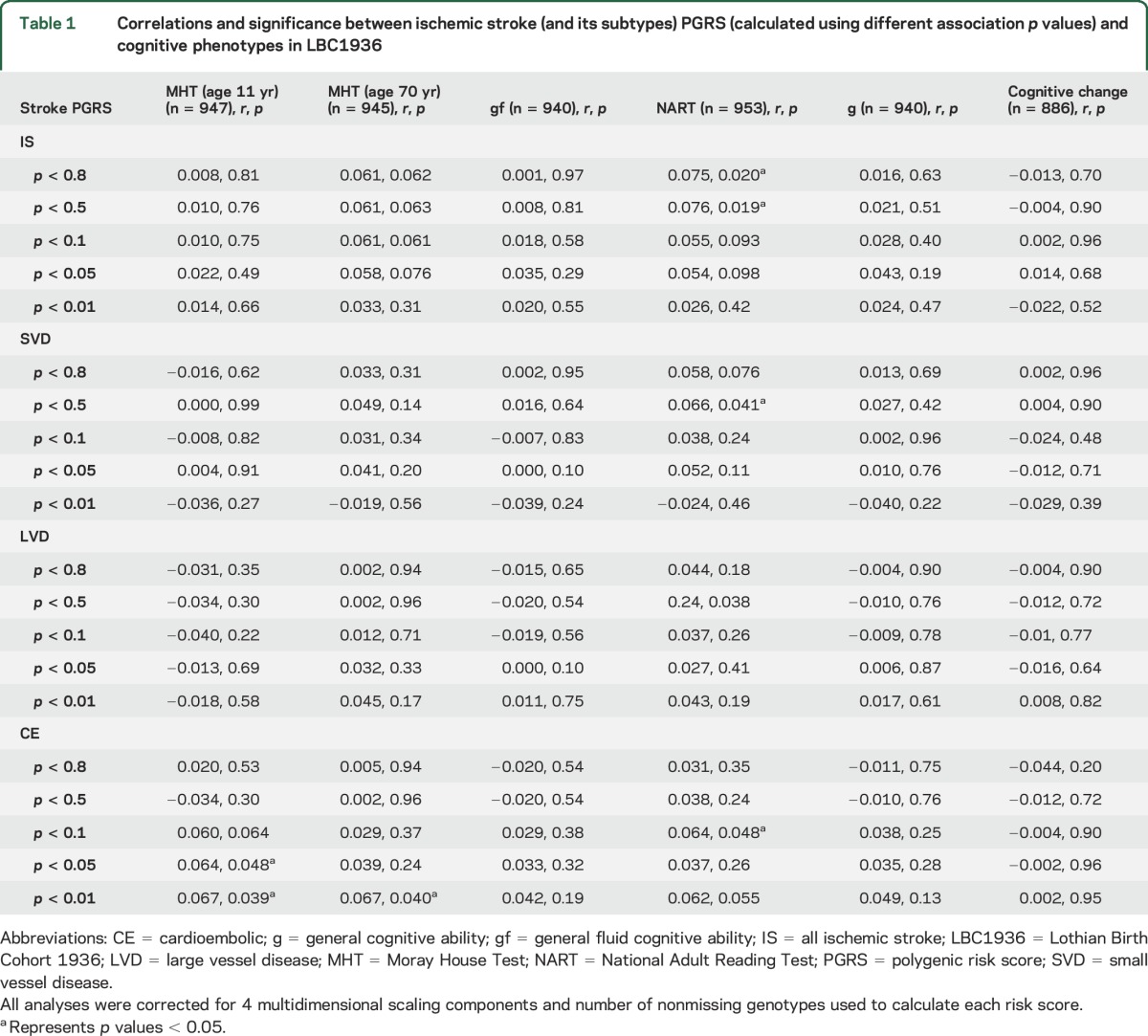
In LBC1936, 7 of 120 correlations between cognitive test scores and polygenic risk scores for ischemic stroke (and its subtypes) reached a significance level of p < 0.05 (table 1). All correlations indicated that higher polygenic risk of ischemic stroke (and its subtypes) was associated with higher cognitive ability.
Table 1.
Correlations and significance between ischemic stroke (and its subtypes) PGRS (calculated using different association p values) and cognitive phenotypes in LBC1936
LBC1921.
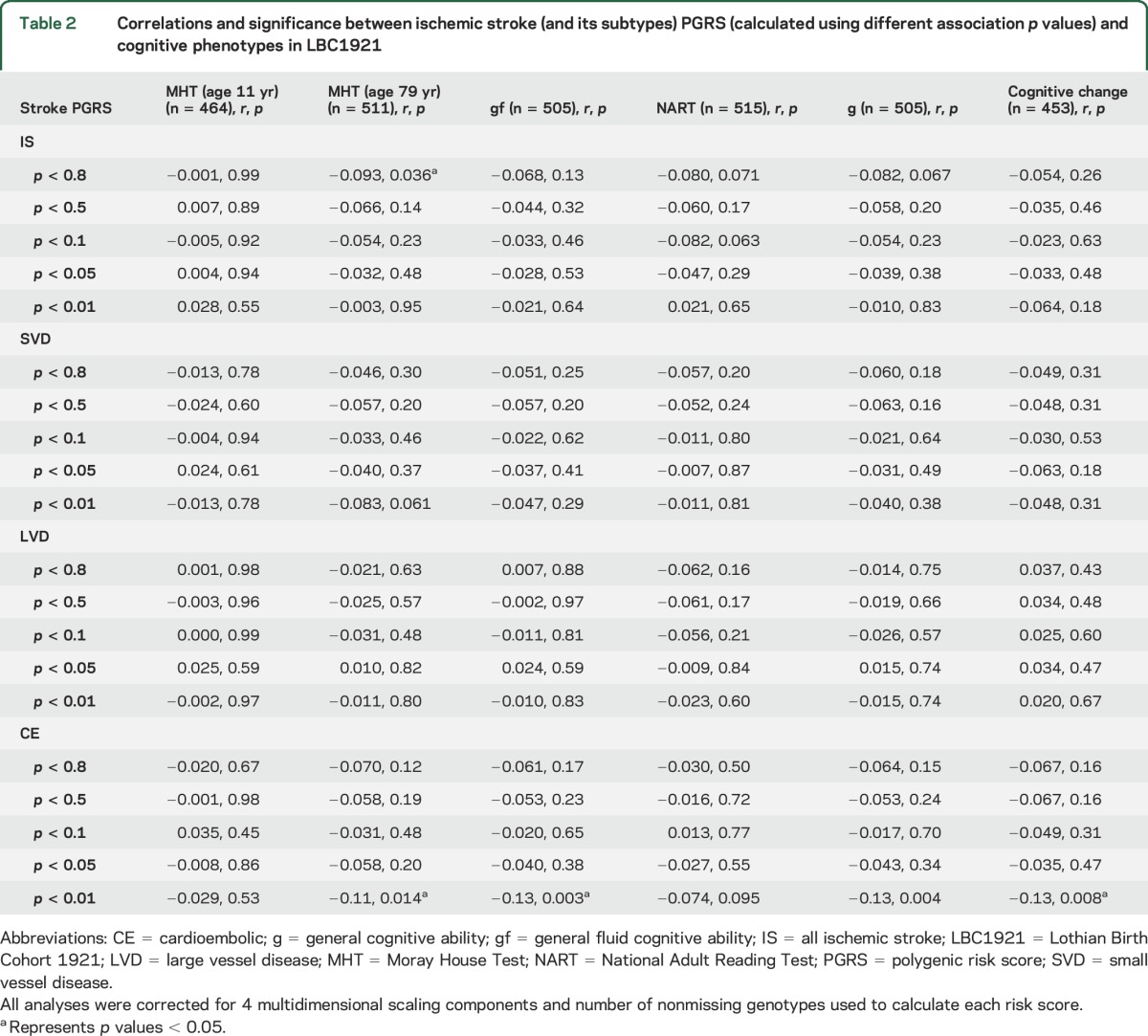
In the relatively small LBC1921, there is a general trend indicating that higher polygenic risk of ischemic stroke (and its subtypes) is associated with lower cognitive ability, with 4 of 120 reaching a significance level of p < 0.05 (table 2).
Table 2.
Correlations and significance between ischemic stroke (and its subtypes) PGRS (calculated using different association p values) and cognitive phenotypes in LBC1921
GS.
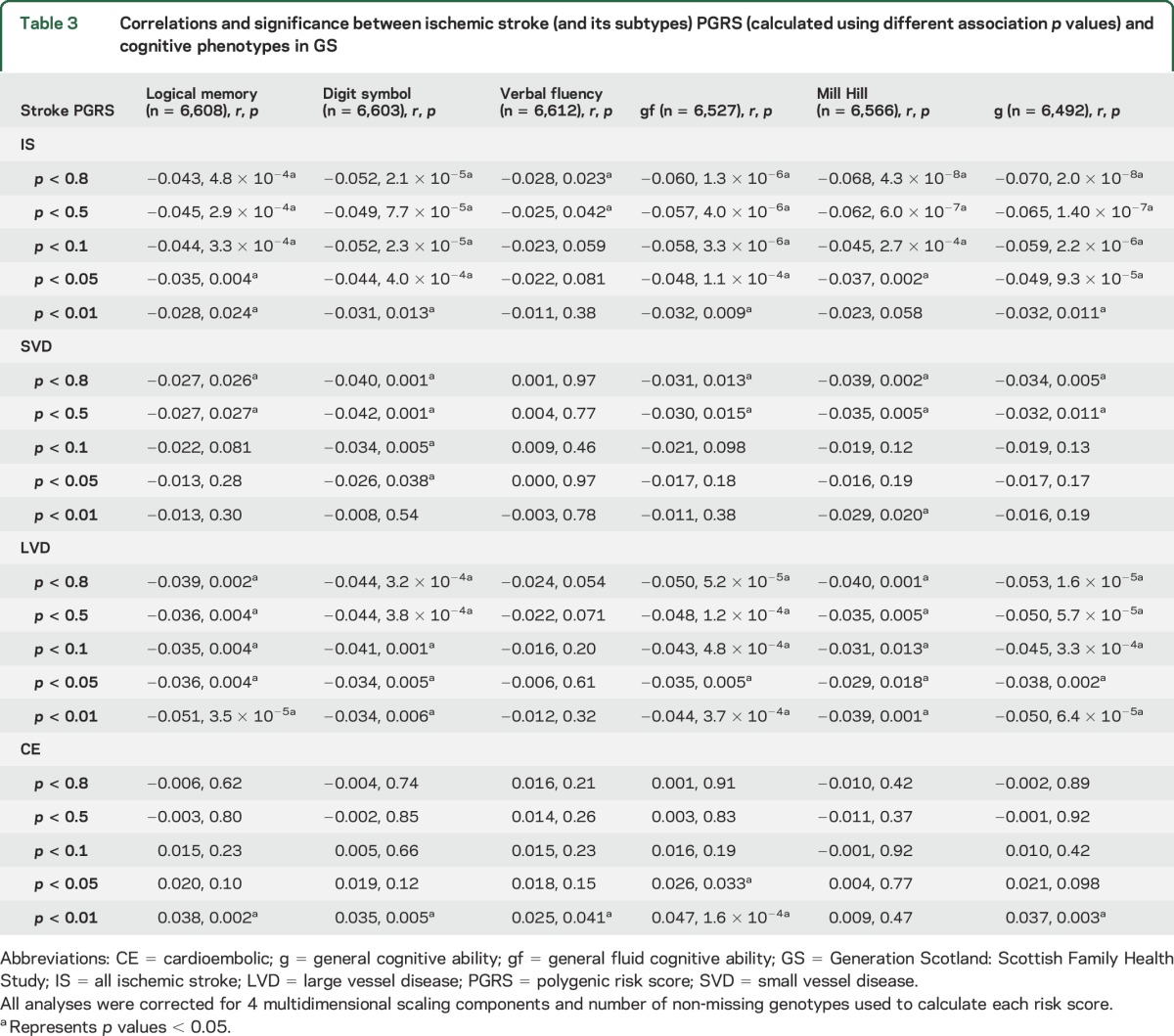
In GS, 71 of 120 correlations between cognitive test scores and polygenic risk scores for ischemic stroke (and its subtypes) reached a significance level of p < 0.05 (table 3). All correlations between polygenic risk of all ischemic stroke, SVD stroke, and LVD stroke and cognitive abilities indicated that higher polygenic risk is associated with lower cognitive abilities. Correlations between polygenic risk of CE stroke and cognitive abilities indicated that higher polygenic risk of CE stroke is associated with higher cognitive abilities.
Table 3.
Correlations and significance between ischemic stroke (and its subtypes) PGRS (calculated using different association p values) and cognitive phenotypes in GS
Meta-analysis: Cognition.
In the meta-analysis, 14 of 60 correlations between cognitive test scores and polygenic risk scores for ischemic stroke (and its subtypes) reached a significance level of p < 0.05 (table e-2). Sample heterogeneity was indicated for many of the crystallized and general cognitive ability analyses. All correlations between polygenic risk of all ischemic stroke, SVD stroke, and LVD stroke and cognitive abilities indicated that higher polygenic risk is associated with lower cognitive abilities. Correlations between polygenic risk of CE stroke and cognitive abilities indicated that higher polygenic risk of CE stroke is associated with higher cognitive abilities.
DISCUSSION
In the largest (by 5- and 10-fold over LBC1936 and LBC1921, respectively) of the cohorts (GS), we found an association between higher polygenic risk of all ischemic stroke and lower cognitive ability. Correlations were generally higher for general cognitive ability than the specific tests, suggesting that the greatest influence is on general cognitive ability. The lowest correlations were identified for Verbal Fluency. We found similar correlations between polygenic risk of SVD and LVD stroke and cognitive ability, albeit with smaller effects. For all ischemic and SVD stroke, the polygenic risk scores containing more SNPs generally correlated more strongly indicating that, as with polygenic risk of schizophrenia,27 many SNPs with very small effect sizes contribute to these risk scores. High polygenic risk of CE stroke was not associated with low cognition, perhaps because the mechanisms leading to CE stroke may involve blood clotting rather than neurovascular integrity, the former possibly having less of an effect on cognition.
In GS, polygenic risk of all ischemic stroke was a better predictor of cognitive ability than polygenic risk of specific subtypes, and had greater power. We note that the polygenic risk scores for all ischemic stroke were created from a GWAS of >12,000 stroke cases, whereas polygenic risk scores for each of the subtypes were created using data from only approximately 2,000 cases. Also, although METASTROKE used the Trial of Org 10172 in Acute Stroke Treatment (TOAST)28 to classify stroke subtypes, this is imprecise, depends on detailed investigations for accurate phenotyping, and therefore some cases may have been assigned incorrectly. Undetected vascular events within METASTROKE participants could also lead to incorrect classification of ischemic stroke subtype.
Correlations between cognitive scores and stroke polygenic risk scores in the relatively small LBC1921 were generally very similar to those in GS but the majority had p > 0.05. In LBC1936, the majority of the correlations were in the opposite direction to our hypothesis. It is possible that, as the participants in LBC1936 are all aged about 70 years and still relatively healthy, they may, as a group, have a low polygenic risk of stroke. Only 5% of the cohort reported having experienced a stroke before cognitive testing and these were removed before the analyses. Including the 50 participants who had experienced a stroke made little difference to the results. LBC1921 is also a relatively healthy older cohort. However, incidence of stroke was not available and it is possible that the trend toward a correlation between cognitive ability at age 79 and stroke polygenic risk scores was driven by participants who had experienced cognitive decline following a stroke. Meta-analyses results also indicated that sample heterogeneity was present.
Both fluid and crystallized cognitive ability were associated with polygenic risk of all ischemic, SVD, and LVD stroke in GS. Although fluid cognitive ability tends to decline in later life, crystallized cognitive ability, as measured in this study by tests of vocabulary, remains relatively stable and is therefore a good proxy measure of cognitive ability earlier in life.29 A previous study with the LBC1936 cohort indicated that higher cognitive ability measured at age 11 years was associated with fewer white matter hyperintensities at age 73 years.30 These data suggest that genetic variants that predispose individuals to risk of ischemic stroke or to risk of brain damage should they have an ischemic trigger in later life, might act much earlier to influence cognitive ability, possibly through influencing brain integrity or brain circulation. It is also possible that certain genes influence cognitive ability through other pathways, for example, developmental pathways, oxidative stress pathways, neurotransmitter pathways,31 and, thereafter, individuals with lower cognitive ability might be more likely to lead lifestyles that predispose them to ischemic stroke. Finally, the same genes may be influencing both risk of ischemic stroke and cognitive ability through independent pathways.
One strength of this study is the large population-based cohort of GS. This allowed us to test the hypothesis that high polygenic risk of ischemic stroke is associated with lower cognitive ability even in the absence of stroke. A limitation was that LBC1921 and LBC1936 are relatively small older cohorts. Although the 3 cohorts consist of individuals born in different time periods, we have twin- and family-based and DNA-based evidence that the heritability of cognitive ability is similar over these eras, and the genetic variants influencing cognition will not have changed during this time period.8,9,32 However, cognitive ability in later life is an interaction between genetics and environmental factors and each cohort experienced very different situations, which may have had an important influence on cognition, and hence, may have influenced the results. All of the cohorts were composed of largely healthy individuals, so the effect sizes expected were small. A further limitation is that we lacked information on incidental vascular changes in the Scottish cohorts at the time of cognitive testing. The results may have been driven by individuals with undetected stroke. However, MRI data are available for about 700 members of LBC1936 at age 73 years (3 years after the cognitive testing data used in this report were collected). At a mean age of 73 years, as expected for an older population, some white matter hyperintensities were evident in 97% of LBC1936. However, only 11% of the cohort did not self-report having had a stroke, but did have imaging evidence of a stroke.30 Population-based studies indicate that white matter hyperintensities increase with age.33 Therefore, we expect that the percentage of individuals with undetected stroke and white matter hyperintensities will be lower in LBC1936 at mean age 70 years and lower still in GS, which has a mean age of 55 years, and slightly higher in LBC1921 at mean age 79 years.
We have presented uncorrected p values as all cognitive phenotypes are correlated (r range: 0.19–0.70, p < 0.001; tables e-3–e-5), as are many of the polygenic risk scores (tables e-6–e-8). However, it is possible that some of the findings may be attributable to type 1 error. Future studies on larger groups with more cognitive decline—especially where that is suspected to be of vascular origin—might show larger effect sizes. In the future, it would be helpful to be able to create subtype risk scores based on larger numbers of accurately phenotyped ischemic stroke patients.
The findings from this study indicate that even in the absence of stroke, being at high polygenic risk of ischemic stroke is associated with lower cognitive ability. This may be attributable to a genetic predisposition to defects in brain integrity or circulation, which increases the risk of stroke or reduces resilience to withstand the effect of ischemic triggers on brain damage. Alternatively, the genes may be acting independently to influence cognitive ability and stroke risk through different pathways.
Supplementary Material
ACKNOWLEDGMENT
METASTROKE: The authors thank all study participants, volunteers, and study personnel who made this consortium possible and the constituent studies for access to their data. LBC1936 and LBC1921: The authors thank the cohort participants who contributed to these studies. GS: The authors are grateful to all the families who took part, the general practitioners, and the Scottish School of Primary Care for their help in recruiting them, and the whole GS team.
GLOSSARY
- CE
cardioembolic
- gf
general fluid
- GS
Generation Scotland: Scottish Family Health Study
- GWAS
genome-wide association study
- LBC1921
Lothian Birth Cohort 1921
- LBC1936
Lothian Birth Cohort 1936
- LVD
large vessel disease
- MHT
Moray House Test
- NART
National Adult Reading Test
- SNP
single-nucleotide polymorphism
- SVD
small vessel disease
Footnotes
Supplemental data at Neurology.org
Contributor Information
Collaborators: METASTROKE Consortium, Matthew Traylor, Martin Farrall, Elizabeth G Holliday, Jemma C Hopewell, Yu-Ching Cheng, Myriam Fornage, M Arfan Ikram, Steve Bevan, Alex P Reiner, Braxton D Mitchell, Robert Clarke, Giorgio B Boncoraglio, Pankaj Sharma, Joshua C Bis, Bruce M Psaty, Peter M Rothwell, Jonathan Rosand, James F Meschia, Kari Stefansson, Martin Dichgans, Unnur Thorsteinsdottir, Anita L DeStefano, Christopher Levi, Solveig Gretarsdottir, Peter Donnelly, Ines Barroso, Jenefer M Blackwell, Elvira Bramon, Matthew A Brown, Juan P Casas, Aiden Corvin, Panos Deloukas, Audrey Duncanson, Janusz Jankowski, Hugh S Markus, Christopher G Mathew, Colin NA Palmer, Robert Plomin, Anna Rautanen, Stephen J Sawcer, Richard C Trembath, Ananth C Viswanathan, Nicholas W Wood, Chris CA Spencer, Gavin Band, Celine Bellenguez, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, Matti Pirinen, Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, Cordelia Langford, Sarah E Hunt, Sarah Edkins, Rhian Gwilliam, Hannah Blackburn, Suzannah J Bumpstead, Serge Dronov, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T McCann, Jennifer Liddle, Simon C Potter, Radhi Ravindrarajah, Michelle Ricketts, Matthew Waller, Paul Weston, Sara Widaa, and Pamela Whittaker
AUTHOR CONTRIBUTIONS
Sarah E. Harris drafted and revised the manuscript, analyzed the data, contributed vital reagents, acquired data, and performed statistical analyses. Rainer Malik drafted and revised the manuscript, analyzed the data, and contributed vital reagents. Riccardo Marioni drafted and revised the manuscript, analyzed the data, and performed statistical analyses. Archie Campbell contributed vital reagents and acquired data. Sudha Seshadri drafted and revised the manuscript, analyzed the data, and contributed vital reagents. Bradford B. Worrall drafted and revised the manuscript, analyzed the data, and contributed vital reagents. Cathie L.M. Sudlow drafted and revised the manuscript, analyzed the data, and contributed vital reagents. Caroline Hayward drafted and revised the manuscript and contributed vital reagents. Mark E. Bastin drafted and revised the manuscript, acquired data, supervised the study, and obtained funding. John M. Starr drafted and revised the manuscript, analyzed the data, supervised the study, and obtained funding. David J. Porteous drafted and revised the manuscript, contributed vital reagents, supervised the study, and obtained funding. Joanna M. Wardlaw drafted and revised the manuscript, analyzed the data, supervised the study, and obtained funding. Ian J. Deary drafted and revised the manuscript, analyzed the data, contributed vital reagents, acquired data, supervised the study, and obtained funding.
STUDY FUNDING
The METASTROKE Consortium is supported by NINDS (NS017950). See Ref. 12 for detailed funding disclosure for the METASTROKE Consortium. LBC1936 and LBC1921: Genotyping was supported by the BBSRC. Phenotype collection in LBC1921 was supported by the BBSRC, The Royal Society, and The Chief Scientist Office of the Scottish Government. Phenotype collection in LBC1936 was supported by Research Into Ageing (continues as part of Age UK's The Disconnected Mind project). Generation Scotland has received core funding from the Chief Scientist Office of the Scottish Government Health Directorates CZD/16/6 and the Scottish Funding Council HR03006. The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross-council Lifelong Health and Wellbeing Initiative (MR/K026992/1).
DISCLOSURE
S. Harris is supported by the Medical Research Council and the UK's Biotechnology and Biological Sciences Research Council (BBSRC) (MR/K026992/1). R. Malik is supported by the Vascular Dementia Research Foundation. S. Seshadri is supported by the National Heart, Lung and Blood Institute N01HC2519. R. Marioni and A. Campbell report no disclosures relevant to the manuscript. B. Worrall is supported by the National Institute of Neurological Disorders and Stroke (NINDS) U01-NS069208. C. Sudlow is supported by UK Biobank (MRC and Wellcome Trust) and the Scottish Funding Council. C. Hayward and M. Bastin report no disclosures relevant to the manuscript. J. Starr is supported by the Medical Research Council and the UK's Biotechnology and Biological Sciences Research Council (BBSRC) (MR/K026992/1). D. Porteous and J. Wardlaw report no disclosures relevant to the manuscript. I. Deary is supported by the Medical Research Council and the UK's Biotechnology and Biological Sciences Research Council (BBSRC) (MR/K026992/1). Go to Neurology.org for full disclosures.
REFERENCES
- 1.Deary IJ, Corley J, Gow AJ, et al. Age-associated cognitive decline. Br Med Bull 2009;92:135–152. [DOI] [PubMed] [Google Scholar]
- 2.Gorelick PB. Risk factors for vascular dementia and Alzheimer disease. Stroke 2004;35:2620–2622. [DOI] [PubMed] [Google Scholar]
- 3.Lawlor DA, Batty GD, Clark H, McIntyre S, Leon DA. Association of childhood intelligence with risk of coronary heart disease and stroke: findings from the Aberdeen Children of the 1950s Cohort Study. Eur J Epidemiol 2008;23:695–706. [DOI] [PubMed] [Google Scholar]
- 4.Modig WK, Silventoinen K, Tynelius P, Bergman L, Rasmussen F. Association between intelligence and type-specific stroke: a population-based cohort study of early fatal and non-fatal stroke in one million Swedish men. J Epidemiol Community Health 2010;64:908–912. [DOI] [PubMed] [Google Scholar]
- 5.Deary IJ, Whiteman MC, Starr JM, Whalley LJ, Fox HC. The impact of childhood intelligence on later life: following up the Scottish mental surveys of 1932 and 1947. J Pers Soc Psychol 2004;86:130–147. [DOI] [PubMed] [Google Scholar]
- 6.Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging 2011;32:63–74. [DOI] [PubMed] [Google Scholar]
- 7.Davies G, Harris SE, Reynolds CA, et al. A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Mol Psychiatry 2014;19:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies G, Tenesa A, Payton A, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 2011;16:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marioni RE, Davies G, Hayward C, et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence 2014;44:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deary IJ, Yang J, Davies G, et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature 2012;482:212–215. [DOI] [PubMed] [Google Scholar]
- 11.Bevan S, Traylor M, Adib-Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke 2012;43:3161–3167. [DOI] [PubMed] [Google Scholar]
- 12.Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 2012;11:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scottish Council for Research in Education. The Trend of Scottish Intelligence: A Comparison of the 1947 and 1932 Surveys of the Intelligence of Eleven-Year-Old Pupils. London: University of London Press; 1949. [Google Scholar]
- 14.Deary IJ, Gow AJ, Taylor MD, et al. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr 2007;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deary IJ, Gow AJ, Pattie A, Starr JM. Cohort profile: the Lothian Birth Cohorts of 1921 and 1936. Int J Epidemiol 2012;41:1576–1584. [DOI] [PubMed] [Google Scholar]
- 16.Wechsler D. WAIS-III UK Administration and Scoring Manual. London, UK: Psychological Corporation; 1998. [Google Scholar]
- 17.Luciano M, Gow AJ, Harris SE, et al. Cognitive ability at age 11 and 70 years, information processing speed, and APOE variation: the Lothian Birth Cohort 1936 Study. Psychol Aging 2009;24:129–138. [DOI] [PubMed] [Google Scholar]
- 18.Nelson HE, Willison JR. National Adult Reading Test (NART): Test Manual (Part 2). Windsor, UK: NFER-Nelson; 1991. [Google Scholar]
- 19.Scottish Council for Research in Education. The Intelligence of Scottish Children: A National Survey of an Age-Group. London: University of London Press; 1933. [Google Scholar]
- 20.Smith BH, Campbell H, Blackwood D, et al. Generation Scotland: the Scottish Family Health Study; a new resource for researching genes and heritability. BMC Med Genet 2006;7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith BH, Campbell A, Linksted P, et al. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS: SFHS). The study, its participants and their potential for genetic research on health and illness. Int J Epidemiol 2013;42:689–700. [DOI] [PubMed] [Google Scholar]
- 22.Wechsler D. WMS-III UK Administration and Scoring Manual. London, UK: Psychological Corporation; 1998. [Google Scholar]
- 23.Lezak M. Neuropsychological Testing. Oxford, England: Oxford University Press; 1995. [Google Scholar]
- 24.Raven JC. The Mill Hill Vocabulary Scale. London: H.K. Lewis; 1965. [Google Scholar]
- 25.Harris SE, Davies G, Luciano M, et al. Polygenic risk for Alzheimer's disease is not associated with cognitive ability or cognitive aging in non-demented older people. J Alzheimers Dis 2014;39:565–574. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIntosh AM, Gow A, Luciano M, et al. Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biol Psychiatry 2013;73:938–943. [DOI] [PubMed] [Google Scholar]
- 28.Adams HP, Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 29.Salthouse TA. Localizing age-related individual differences in a hierarchical structure. Intelligence 2004;32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdes Hernandez MC, Booth T, Murray C, et al. Brain white matter damage in aging and cognitive ability in youth and older age. Neurobiol Aging 2013;34:2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cogn Sci 2011;15:388–394. [DOI] [PubMed] [Google Scholar]
- 32.Benyamin B, Wilson V, Whalley LJ, Visscher PM, Deary IJ. Large, consistent estimates of the heritability of cognitive ability in two entire populations of 11-year-old twins from Scottish mental surveys of 1932 and 1947. Behav Genet 2005;35:525–534. [DOI] [PubMed] [Google Scholar]
- 33.Morris Z, Whiteley WN, Longstreth WT, Jr, et al. Incidental findings on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ 2009;339:b3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


