To assess the importance of autophagy in prostate cancer, Santanam et al. generated a new autochthonous genetically engineered mouse model with inducible prostate-specific deficiency in the Pten tumor suppressor and autophagy-related-7 (Atg7) gene. Atg7 deficiency produced an autophagy-deficient phenotype and delayed Pten-deficient prostate tumor progression in both castrate-naïve and castrate-resistant cancers.
Keywords: prostate cancer, castrate resistant, autophagy, ER stress
Abstract
Understanding new therapeutic paradigms for both castrate-sensitive and more aggressive castrate-resistant prostate cancer is essential to improve clinical outcomes. As a critically important cellular process, autophagy promotes stress tolerance by recycling intracellular components to sustain metabolism important for tumor survival. To assess the importance of autophagy in prostate cancer, we generated a new autochthonous genetically engineered mouse model (GEMM) with inducible prostate-specific deficiency in the Pten tumor suppressor and autophagy-related-7 (Atg7) genes. Atg7 deficiency produced an autophagy-deficient phenotype and delayed Pten-deficient prostate tumor progression in both castrate-naïve and castrate-resistant cancers. Atg7-deficient tumors display evidence of endoplasmic reticulum (ER) stress, suggesting that autophagy may promote prostate tumorigenesis through management of protein homeostasis. Taken together, these data support the importance of autophagy for both castrate-naïve and castrate-resistant growth in a newly developed GEMM, suggesting a new paradigm and model to study approaches to inhibit autophagy in combination with known and new therapies for advanced prostate cancer.
With >220,000 estimated new cases and >27,500 estimated deaths in 2015, prostate cancer continues to be a major cause of cancer death in men in the United States (Siegel et al. 2015). Although many new agents are now available to treat advanced disease, with an initial high response rate with androgen receptor (AR) pathway inhibition, median survival is ∼4 yr overall (Alva and Hussain 2013). The emergence of a resistant and aggressive prostate cancer following multiple new therapies is a now particularly common and difficult clinical situation in need of new treatment paradigms. Currently approved therapies have focused on AR as a known pathway responsible for driving prostate cancer growth as well as more general cytotoxic therapies. Understanding the importance of other critical cellular processes that promote aggressive prostate cancer growth may lead to new therapeutic approaches.
In this regard, autophagy is a normal cellular process by which cells degrade and recycle damaged proteins and organelles to sustain metabolism and survival during periods of stress and starvation. Autophagy is thought to have a context-dependent role in cancer (Saleem et al. 2012). Autophagy eliminates damaged cellular components, thereby mitigating oxidative stress and tissue damage that could otherwise lead to chronic inflammation and tumor initiation (White 2012). Therefore, acting as a tumor promoter, autophagy facilitates the survival of tumor cells in hypoxic tumor regions (White 2012). Recent studies further demonstrate the dependence of multiple aggressive cancers on autophagy (Guo et al. 2013b). Tumors with activating mutations in K-ras and Braf up-regulate autophagy that is critical for their survival and growth, and genetically engineered mouse models (GEMMs) for lung and pancreatic cancers driven by these mutations show survival and growth defects with deletion of the autophagy-related 7 (Atg7) gene (White et al. 2010; Guo et al. 2013b). More recently, our group also demonstrated that Atg7 deficiency prevents melanoma development by BrafV600E and allelic Pten loss while also suppressing melanoma growth with activated BrafV600E and Pten deficiency (Xie et al. 2015). Finally, systemic, conditional genetic Atg7 deletion in a GEMM with pre-existing K-rasG12D-driven, Trp53-deficient lung cancer demonstrated marked anti-tumor activity, suggesting that tumors are more autophagy-dependent than most normal tissues (Karsli-Uzunbas et al. 2014). Thus, autophagy has an important role in tumor promotion and targeting autophagy may have therapeutic benefits. The role of autophagy in prostate cancer is less well understood.
Our prior investigations revealed elevated levels of autophagy markers in advanced cases of human prostate cancer (Gleason score 9) as compared with less advanced cases (Gleason score 7) as well as the therapeutic benefit of hydroxychloroquine, an autophagy inhibitor, in prostate cancer xenograft tumors (Saleem et al. 2012). Although suggestive of the importance of autophagy in prostate cancer, no published study has demonstrated the effects of a genetically defective autophagy pathway in mouse models of prostate cancer. Building on the success of prior studies of GEMMs, the development of an effective model in which autophagy is modulated would be important to investigate the role of autophagy and identify a novel pathway to target for therapeutic strategies in prostate cancer.
To test the hypothesis that autophagy is functionally important in the setting of prostate cancer, Atg7, an essential autophagy gene, was deleted in a GEMM to examine whether the resulting autophagy deficiency altered tumor growth and progression in naïve and hormone refractory mouse prostate cancers. We used a prostate-specific model with inducible deletion of the Pten tumor suppressor to initiate mouse prostate tumors and followed the effects of Atg7 deletion in the tumors of these mice that revealed the tumor-promoting effects of autophagy.
Results
Prostate tumor-specific Atg7 deletion results in an autophagy-deficient phenotype
We used prostate-specific Pten deletion in GEMMs of prostate cancer to test the functional consequences of coordinate genetic ablation of Atg7 (Floc'h et al. 2012). Briefly, mice express a tamoxifen (TAM)-inducible Cre-ERT2 protein under the control of a prostate-specific promoter, Nkx3.1, resulting in prostate-specific Cre-recombinase activity (Wang et al. 2009). Systemic TAM administration to adult Nkx3.1CreERT2/+;Ptenflox/flox mice carrying floxed alleles of Pten results in biallelic deletion of Pten in the prostate gland. Following Pten deletion, mice develop high-grade prostatic intraepithelial neoplasia (PIN) and then prostate adenocarcinomas. The tumors progress relatively slowly, enabling the delineation of phases of progression in comparison with similar mice that additionally bear a prostate-specific autophagy deficiency. To introduce autophagy deficiency, the mice were crossed with Atg7 floxed (Atg7F/F) mice (Komatsu et al. 2005) so that TAM administration and Cre activation would simultaneously delete Pten and Atg7 in the prostate gland.
Confirmation of Atg7 deletion (PtenΔ/Δ;AtgΔ/Δ) was evaluated by immunohistochemistry (IHC) demonstrating significant elimination of ATG7 protein, indicative of efficient genetic deletion of the gene (Fig. 1A; Supplemental Fig. S1), also confirmed with PCR (data not shown). Defective cellular autophagy is characterized by accumulation of the autophagy substrate p62 and reduction of cleaved and lipidated LC3 (LC3-II) associated with autophagosome formation (Komatsu et al. 2005; Mathew et al. 2009; Moscat and Diaz-Meco 2009). The PtenΔ/Δ;AtgΔ/Δ prostate tumors had accumulation of p62 and reduction in LC3-II, as shown by IHC (Fig. 1A; Supplemental Fig. S1) and Western blot (Fig. 1B) as expected, indicative of defective autophagy initiation and substrate degradation.
Figure 1.

Atg7 deletion resulted in low ATG7 expression and an autophagy-deficient phenotype. (A) Representative Atg7 and p62 IHC of the Atg wild-type (Atg7+/+) and Atg-deleted (Atg7Δ/Δ) anterior mouse prostate at 3 mo after tumor induction. (B) Western blot analysis of p62 expression and LC3-I-to-LC3-II conversion in the Atg7+/+ and Atg7Δ/Δ prostate tumors at 3 mo after tumor initiation.
Atg7 deficiency delays prostate-specific tumor progression
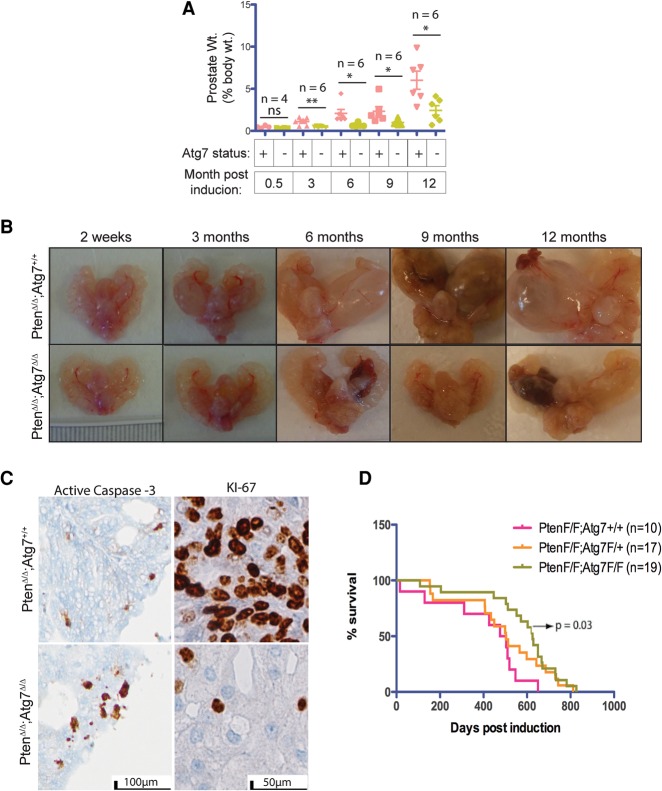
To investigate the functional consequences of autophagy deficiency, adult mice with the genotypes Nkx3.1CreERT2/+;PtenF/F;Atg7+/+ and Nkx3.1CreERT2/+;PtenF/F;AtgF/F were administered TAM, and tumorigenesis was monitored over time. Three months after TAM, there was a significant difference in the prostate tumor weights relative to total body weights between the Atg7+/+ and Atg7Δ/Δ groups. At 3, 6, 9, and 12 mo after tumor induction, the Atg7+/+ mouse prostate weights were 1.11%, 2.08%, 2.33%, and 6.02% of the total body weights, as compared with 0.48%, 0.69%, 0.99%, and 2.44% in Atg7Δ/Δ mouse prostates, respectively (P-values at 3, 6, 9, and 12 mo were 0.0087, 0.0171, 0.043, and 0.0147, respectively) (Fig. 2A). Thus, deletion of Atg7 reduced prostate tumor burden by approximately half.
Figure 2.
Autophagy deficiency generates smaller prostate tumors. (A) Prostate weight as a percentage of body weight at various times after tumor induction. (B) Gross anatomy of urogenital systems of mice with Atg7+/+ and Atg7Δ/Δ prostates at various times after tumor induction showing changes in size and cyst formation over time. (C) Representative active caspase-3 and Ki-67 IHC of Atg7+/+ and Atg7Δ/Δ anterior mouse prostates at 3 mo after tumor induction. (D) Kaplan-Meier survival curve for mice with PtenΔ/Δ Atg7+/+, PtenΔ/Δ Atg7+/Δ, and PtenΔ/Δ Atg7Δ/Δ prostate tumors.
Examination of the gross anatomy of the mouse prostate lobes revealed the continuously striking increased size of the Atg7+/+ mouse prostates, especially the anterior lobe, whereas the Atg7Δ/Δ mouse prostates showed visibly smaller increments of growth at the times depicted (i.e., 2 wk and 3, 6, 9, and 12 mo) (Fig. 2B). Additionally, the Atg7+/+ anterior prostates developed cystic tumors roughly at 3 mo and grew rapidly thereafter. However, the Atg7Δ/Δ anterior prostate lobes developed cystic tumors with a much more delayed onset, which was usually visible only between 9 and 12 mo after TAM (Fig. 2B). This implicates autophagy in the promotion of prostate tumorigenesis.
To assess changes in cellular proliferation and apoptosis, we analyzed the harvested prostate tissue by IHC for Ki-67 and active caspase-3. Compared with the autophagy-competent tumors (Atg7+/+), the autophagy-deficient tumors (Atg7Δ/Δ) displayed lower Ki-67 staining, signifying decreased cellular proliferation, and increased active caspase-3 staining, indicative of increased apoptosis at 3 mo after induction (Fig. 2C) as well as through multiple additional time points (Supplemental Fig. S2). Assessment of Kaplan-Meier survival curves (Fig. 2D) demonstrated improved survival in mice with autophagy-deficient tumors (PtenΔ/Δ Atg7Δ/Δ) compared with autophagy-competent tumors (PtenΔ/Δ Atg7+/+) (P = 0.03).
Autophagy deficiency produces histologically distinct tumors
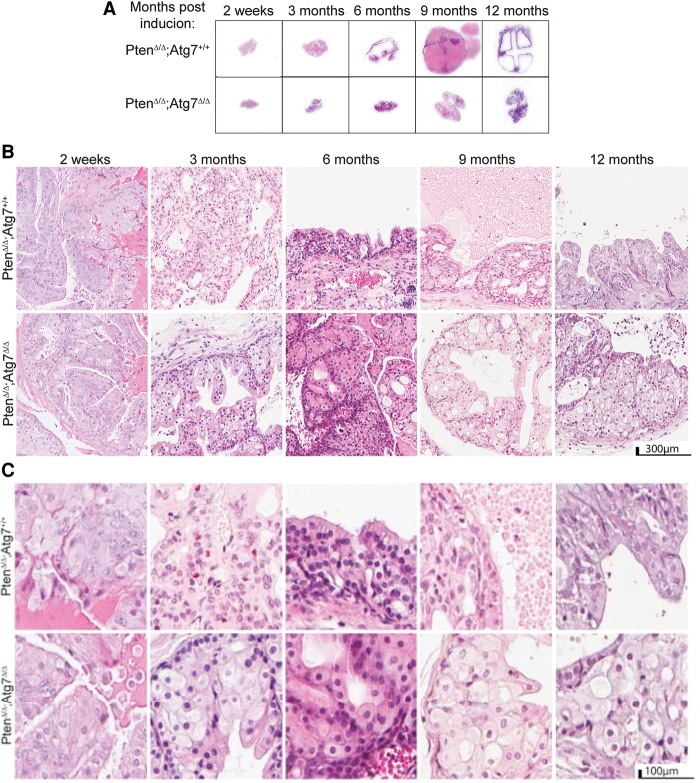
We assessed hematoxylin and eosin (H&E)-stained whole sections of the anterior prostate tissue in Nkx3.1CreERT2/+;PtenF/F;Atg7+/+ and Nkx3.1CreERT2/+;PtenF/F;Atg7Δ/Δ mouse prostates induced by TAM. As shown in Figure 3A, prostate tissue cross-sections demonstrated size increases from 2 wk to 12 mo after TAM that were greater in the Atg7+/+ tumors than the autophagy-deficient (Atg7Δ/Δ) tumors. In fact, the whole H&E section of the Atg7Δ/Δ anterior prostates at 9 mo after tumor induction resembled that of the Atg7+/+ anterior prostates at 3 mo after induction. Noticeably, there were dramatic increases in cyst sizes as the Atg7+/+ tumors progressed through time, and, in comparison, the Atg7Δ/Δ tumors remained smaller and less cystic.
Figure 3.
Autophagy deficiency produces histologically distinct tumors with enlarged cells, delays tumor progression, and renders a survival advantage. (A) Representative H&E-stained whole-mount sections of anterior prostate lobes at various times after tumor induction. (B) Low-resolution anterior prostate histology at various times after tumor induction. (C) Representative high-resolution H&E sections of the anterior prostate at various times after tumor induction.
In H&E-stained sections at low magnification, we observed larger tumor areas in the Atg7+/+ prostates by 3 mo after TAM as well as increased necrosis, resulting in the development of well-formed cysts by 6 mo after induction (Fig. 3B). The Atg7Δ/Δ prostates progressed in a similar fashion but at a decreased rate. At higher magnification (Fig. 3C), the Atg7Δ/Δ tumor tissue had large areas of giant cells with an expansive cytoplasm most noticeable 3 mo after TAM and beyond in Nkx3.1CreERT2/+;PtenF/F;AtgF/F mouse prostates. As autophagy deficiency in tumors prevents the clearance and recycling of damaged proteins and organelles, these autophagy substrates accumulate, commonly producing a dramatic expansion in cytoplasmic volume (Guo et al. 2013a).
Autophagy deficiency results in endoplasmic reticulum (ER) stress and reduced AR signaling
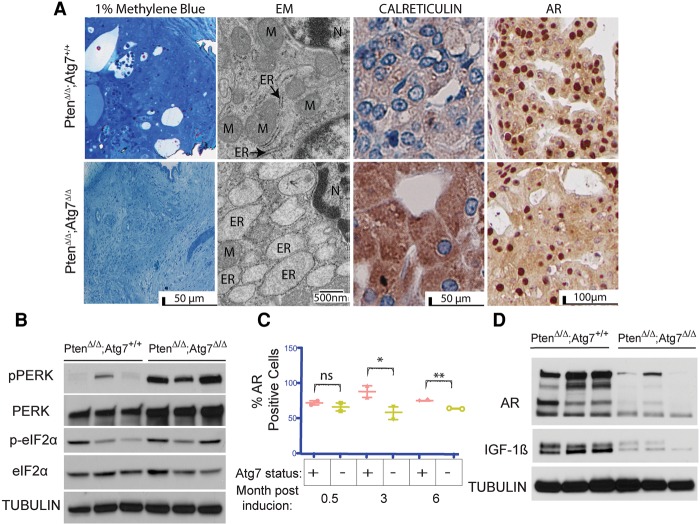
Our prior studies demonstrated that Atg7 deficiency in K-rasG12D- and BrafV600E-driven lung cancers caused the dramatic accumulation of defective mitochondria associated with metabolic impairment and defective tumor growth (Guo et al. 2013a). To gain insight into the nature of the autophagy substrates accumulating in the Atg7-deficient prostate tumors, we examined them by electron microscopy (EM). In striking contrast to Atg7-deficient lung cancers, the cytoplasmic contents of the giant autophagy-deficient Atg7Δ/Δ prostate tumor cells displayed a dramatic accumulation of swollen rough ER, indicative of ER stress (Fig. 4A). An increased abundance of ER was also indicated by an increase in the ER marker calreticulin by IHC (Fig. 4A). Thus, in contrast to lung tumors, prostate tumors appear to require autophagy for elimination of ER, perhaps to ameliorate accumulation of unfolded proteins and ER stress.
Figure 4.
Autophagy deficiency results in ER stress and decreased AR signaling. (A) One percent methylene blue-stained tissue showing representative areas used for EM analysis. Representative transmission EM (TEM) with ER and mitochondria (M) labeled as well as calreticulin and AR IHC of Atg7+/+ and Atg7Δ/Δ anterior mouse prostates at 3 mo after tumor induction. (B) Western blot analysis of pPERK, PERK, p-eIF2α, and eIF2α expression in tumor lysates from Atg7+/+ and Atg7Δ/Δ anterior mouse prostates at 3 mo after tumor induction. Tubulin served as a loading control. (C) Quantification of the percentage of cells with AR-positive nuclei in Atg7+/+ and Atg7Δ/Δ anterior mouse prostate tissue (n = 2) at various times after tumor induction. The Atg7+/+ tumors had 88.6% and 81.35% AR-positive nuclei at 3 and 6 mo, respectively, whereas the Atg7Δ/Δ tumors had only 58.2% and 65.45% positive nuclei at 3 and 6 mo after TAM, respectively. (D) Western blot analysis of AR and IGF-1β expression in Atg7+/+ and Atg7Δ/Δ mouse prostates at 3 mo after tumor induction.
ER stress is known to activate one of three arms of the unfolded protein response (UPR) pathway (Tsai and Weissman 2010), including dsRNA-activated protein kinase (PKR)-like ER kinase (PERK). As expected, Western blot analysis demonstrated significantly elevated P-PERK in the Atg7Δ/Δ as compared with Atg7+/+ tumor lysates from three individual mice at 3 mo after tumor induction (Fig. 4B). We also observed elevated expression of the downstream phospho-eIF2α (regulating initiation of the mRNA translation machinery) in the Atg7Δ/Δ as compared with the Atg7+/+ tumor lysates in two of the three mice assessed. However, one of the mice bearing Atg7+/+ prostate tumors had high p-eIF2α expression despite almost nondetectable levels of phospho-PERK (as seen in the first lane of Fig. 4B), which could be due to phosphorylation of eIF2α by kinases other than activated PERK.
While the inability of Atg7-deficient prostate tumor cells to mitigate ER stress could contribute to the anti-proliferative and cytotoxic effects observed, prostate cancers are known to be androgen-dependent for growth and survival (Heinlein and Chang 2004). In fact, it has been suggested that androgens assert their proliferative role by activating the autophagy pathway (Shi et al. 2013). Analysis of AR expression by IHC demonstrated a significant difference between the Atg7+/+ and Atg7Δ/Δ tumor tissues beyond 3 mo after TAM. The Atg7+/+ tumors had 88.6% and 81.35% AR-positive nuclei at 3 and 6 mo, respectively, whereas the Atg7Δ/Δ tumors had only 58.2% and 65.45% positive nuclei at 3 and 6 mo after TAM, respectively (P-values 0.013 and 0.001) (Fig. 4A, C; Supplemental Fig. S3A). Furthermore, Western blot analysis revealed decreased AR protein expression in the Atg7Δ/Δ tumor tissue lysates compared with that of the Atg7+/+ tumor tissue (Fig. 4D). In contrast, a decrease in AR protein expression was not seen in castrated animals (data described below; Supplemental Fig. S4C,D). IGF-1β is a known downstream effector of the AR in prostate cancer cells, and its expression signifies the presence of an active AR pathway (Pandini et al. 2005). Western blot analysis demonstrated high expression of IGF-1β in Atg7+/+ tumor lysates and low expression in Atg7Δ/Δ tumor lysates (Fig. 4D), also indicative of reduced AR signaling in the autophagy-deficient prostate tumors. Taken together, these data support a role for autophagy and modulation of ER stress and the AR receptor pathway signaling in castrate-sensitive tumors.
Atg7 deficiency delays castrate-resistant growth of prostate cancer
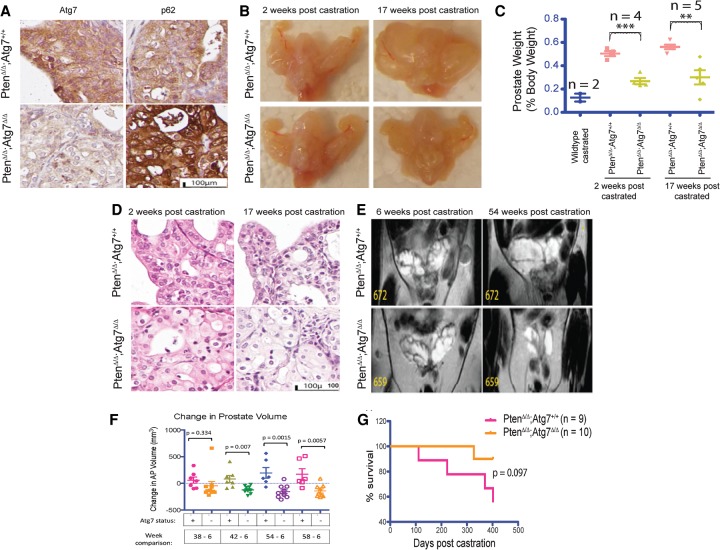
To study the effect of an autophagy deficiency on the progression of hormone refractory prostate cancer, Atg7+/+ and Atg7−/− prostate tumor-bearing mice were castrated 6 wk after TAM and monitored for tumor progression. Two weeks post-castration (PC), the tumors regressed, after which they relapsed independently of androgens, serving as a model for hormone refractory prostate cancer (Floc'h et al. 2012). As evidence of an autophagy-deficient phenotype, IHC staining demonstrated low ATG7 protein with concurrent p62 accumulation in the Atg7Δ/Δ tumors PC, characteristic of autophagy deficiency. In contrast, Atg7+/+ tumors demonstrated high levels of ATG7 and relatively low p62 staining (Fig. 5A; Supplemental Fig. S4A).
Figure 5.
Autophagy ablation delays progression of castrate-resistant prostate cancer. (A) Representative Atg7 and p62 IHC of anterior prostate tissue from castrated Atg7+/+ and Atg7Δ/Δ mice. (B) Representative gross anatomy of urogenital systems of mice with Atg7+/+ and Atg7Δ/Δ prostates at 2 and 17 wk PC. (C) Prostate weights as percent body weights from castrated mice. (D) Representative histology of H&E-stained sections of Atg7+/+ and Atg7Δ/Δ prostates at 2 and 17 wk PC. (E) Magnetic resonance imaging (MRI) analysis of Atg7+/+ and Atg7Δ/Δ mouse prostates PC. (F) MRI-guided volume quantification of individual mice with Atg7+/+ and Atg7Δ/Δ anterior prostates at various times from 6 through 58 wk PC. (G) Kaplan-Meier survival curve of castrated mice with Atg7+/+ and Atg7Δ/Δ prostates over time.
A comparison of the gross anatomy of the urogenital systems of Atg7+/+ and Atg7−/− mice at 2 and 17 wk PC demonstrated larger cystic tumors by 17 wk PC in the autophagy-competent (Atg7+/+) tumors, while the Atg7Δ/Δ tumor-bearing anterior prostates were relatively smaller (Fig. 5B). Examination of prostate weights demonstrated a significant decrease in weight with autophagy deficiency by Atg deletion at 2 and 17 wk PC (Fig. 5C). Specifically, at 2 wk PC, Atg7+/+ tumor-bearing prostate weights (as percent body weights) were 0.50% of the body weight compared with 0.27% for the Atg7Δ/Δ tumor-bearing prostates (P-value = 0.0005). At 17 wk PC, the Atg7+/+ tumor-bearing prostates were 0.56% of the body weight, while the Atg7Δ/Δ tumor-bearing prostates were 0.3% of body weight (P-value = 0.0036) (Fig. 5C). Thus, an autophagy deficiency mediated by Atg7 deficiency resulted in significantly smaller tumors PC.
Upon histologic examination, H&E-stained whole-tissue sections showed a smaller anterior prostate in the Atg7Δ/Δ tumor-bearing mice as compared with their Atg7+/+ tumor-bearing counterparts at 2 and 17 wk PC. At 17 wk PC, the Atg7+/+ tumor-bearing anterior prostates also showed the development of clear cysts, and while the Atg7Δ/Δ tumor-bearing anterior prostates also show some cyst development, they were less pronounced. Low magnification of these tissue sections revealed the presence of giant cytoplasm-laden cells specifically in the Atg7Δ/Δ tumor-bearing anterior prostates (Fig. 5D).
To follow the effect of the autophagy deficiency over time, the Atg7+/+ and Atg7Δ/Δ mouse groups were monitored by periodic MRI analysis PC. A comparison of prostate MRI data from early and late periods PC revealed that while the Atg7+/+ tumor-bearing prostates had increased in size, as would be expected with castrate-resistant tumor growth, most Atg7Δ/Δ tumor-bearing prostates actually showed a decrease in size (Fig. 5E,F). A graph depicting the growth of each mouse prostate with time from 6 through 58 wk showed a general increase in the size of the Atg7+/+ tumor-bearing prostates that was less apparent in the Atg7Δ/Δ tumor-bearing prostates (Fig. 5F). Of note, one Atg7Δ/Δ tumor-bearing prostate (#656) not included in the analysis developed a large tumor ∼30 wk PC and demonstrated tissue ATG7 protein expression by IHC, indicating that Atg7 was not deleted, suggesting selection for retention of ATG7 expression among rare, fast-growing tumor clones (Supplemental Fig. S4E). Taken together, these data support a role for autophagy in castrate-resistant growth.
To examine whether autophagy ablation could improve prostate cancer prognosis and offer a survival benefit, the castrated mice were followed over time. A Kaplan-Meier curve depicting the percent survival of the Atg7+/+ and Atg7Δ/Δ tumor-bearing mice PC revealed a survival advantage trend in mice with Atg7-deleted prostate cancer (Fig. 5G). Although nonsignificant at 400 d PC (P-value = 0.097), it suggests a possible role for autophagy in the progression of hormone refractory prostate cancer, with a trend similar to that seen in noncastrate animals (Fig. 2D). Taken together, these data support a GEMM of growth inhibition through induced autophagy deficiency in both noncastrate and castrate-resistant disease states, potentially through reduced AR signaling and increased ER stress.
Discussion
Autophagy is a critical cellular function to promote stress tolerance by recycling intracellular components to sustain metabolism important for the growth and survival of aggressive malignancies (Bray et al. 2012; Karsli-Uzunbas et al. 2014). Additionally, in some aggressive cancers, it is now known that autophagy is used to promote tumor survival and can contribute to therapeutic drug resistance (Guo et al. 2013b). Given the often indolent nature of prostate cancer in many patients early in their course, in which the tumors may be present for many years before undergoing a more aggressive transformation, efforts to understand and modulate such a critical cellular survival mechanism may lead to larger long-term incremental improvements in disease control (Irshad et al. 2013). To assess the importance of autophagy in prostate cancer, we generated a new GEMM of prostate cancer driven by conditional prostate-specific deficiency in the Pten tumor suppressor without and with ablation of the essential autophagy gene Atg7 (Nkx3.1CreERT2/+;PtenF/F;Atg+/+ and AtgΔ/Δ).
Our new GEMM recapitulated the course of human prostate cancer with both castrate-sensitive and castrate-resistant growth and demonstrated that autophagy deficiency delayed prostate tumor progression. The effect of autophagy deficiency was more dramatic for castrate-naïve tumors with decreased growth and increased survival but also had a negative effect on the growth of castrate-resistant prostate cancer, a more aggressive form of the disease. Both castrate-naïve and castrate refractory Atg7Δ/Δ prostate tumors showed delayed progression marked by smaller, less cystic tumors and accumulation of characteristic cytoplasm-laden enlarged cells as compared with the Atg7+/+ prostate tumors. Atg7 deficiency delayed Pten-deficient prostate tumor progression and resulted in an autophagy-deficient phenotype typified by accumulation of p62, a large cytoplasm, swollen ER, and reduction in LC3-II. Evidence of ER stress in Atg7-deficient tumors was noted by increased calreticulin staining, P-PERK expression, and EM. As an assessment of the importance of autophagy in castration-resistant tumor growth, MRI demonstrated significantly decreased tumor volume in Atg7-deficient compared with wild-type tumors.
The potential mechanisms of growth inhibition in our Atg7-deficient GEMM of prostate cancer are consistent with prior models of autophagy (Strohecker and White 2014a,b; White 2015). The ER-stress phenotype and decreased AR signaling was observed predominantly in the naïve autophagy-deficient tumors. Our data collectively suggest that autophagy deficiency in Pten-null naïve prostate tumors results in reduced AR signaling and the manifestation of an ER stress phenotype that activates the PERK arm of the UPR pathway in the ER. Upon activation, PERK autophosphorylates on its cytoplasmic domain and in turn phosphorylates eIF2α, which, under prolonged activation, is known to result in overall decreased protein translation, increased apoptosis, and decreased tumor growth. Our data revealed that autophagy ablation in prostate cancer results in the manifestation of ER stress, which correlated with activation of the PERK pathway. Future efforts to rigorously test other arms of the UPR pathway and understand and confirm whether these results are causal may provide an opportunity for targeting the PERK–eIF2α pathway in Pten-null prostate cancer.
Finally, as a new model for prostate cancer progression, our GEMM provides an opportunity to test therapeutics and modulate autophagy as a basic cancer survival mechanism. This is timely given the panoply of new therapies recently approved for prostate cancer (Hussain and DiPaola 2015). Additionally, given the bimodal nature of prostate cancer with more indolent castrate-naïve and more aggressive castrate-resistant stages, understanding the role of autophagy in treatment resistance will be important to the development of more impactful therapeutic paradigms. A recent large national phase III study that we conducted demonstrated a significant survival benefit of combining androgen ablation with chemotherapy in patients with castrate-naïve disease, although the mechanism for this combined benefit is not well understood (Sweeney et al. 2015). Our GEMM provides an opportunity to study the mechanistic effects of such combinations with and without castration as well as the role of autophagy. Given that prior studies in a xenograft model suggest that autophagy is also a mechanism of resistance to AR signaling inhibitors, future preclinical studies of autophagy modulation using our genetic model to further test the effect of Atg7 deficiency on castration or androgen axis targeting sensitivity and resistance with and without additional agents such as chemotherapy would be important. (Farrow et al. 2014; Nguyen et al. 2014).
Taken together, these data validate a new GEMM of prostate cancer driven by conditional prostate-specific deficiency in the Pten tumor suppressor without and with ablation of the essential autophagy gene Atg7 (Nkx3.1CreERT2/+;PtenF/F;Atg+/+ and AtgΔ/Δ) and support an important role of autophagy in aggressive prostate cancer and castrate-resistant growth. These data suggest a new paradigm and model for the study of therapeutic approaches in both less aggressive castrate-naïve as well as more lethal castrate refractory prostate cancer.
Materials and methods
Mouse lines and tumor inductions
Nkx3.1CreERT2/+;PtenF/F;Atg7+/+ mice, obtained from the Abate-Shen and Shen laboratories at Columbia University, were crossed with conditionally deleted Atg7F/F mice (Komatsu et al. 2005) to obtain Nkx3.1CreERT2/+;PtenF/F;Atg7+/+ and Nkx3.1CreERT2/+;PtenF/F;Atg7F/F mice. For tumor induction, both groups of mice were administered 200 μL of 20 mg/mL TAM on four consecutive days by oral gavage.
Histology and IHC
Harvested prostate tissue was fixed in 10% buffered formalin solution (Formalde-Fresh, Fisher Scientific) overnight and transferred to 70% ethanol before processing into paraffin blocks. Formalin-fixed paraffin-embedded tissue was deparaffinized, boiled in citrate buffer (pH 6) for 30 min at 95°, blocked in 10% goat serum for 30 min, and incubated in primary antibody and biotinylated anti-rabbit IgG (H+L) (Vector Laboratories, BA-1000) secondary antibody. Intermediate washes were performed with 0.1% Tween-20 in PBS. Further processing was conducted using a LSAB2 System-HRP kit (DAKO, K0673). The primary antibodies used were against Atg7 (Santa Cruz Biotechnology, sc-33211), p62 (Enzo Life Sciences, PW9860-0100), Ki-67 (Abcam, ab-15580), caspase-3 (Cell Signaling, 9661), calreticulin (Novus Biologicals, EPR3924), and AR (Sigma-Aldrich, A9853).
Western blotting
Freshly harvested or −80°C frozen tissue was ground in liquid nitrogen and lysed using Tris lysis buffer (1 M Tris HCl, 1 M NaCl, 0.1 M EDTA, 10% NP-40). Lysate was run through PAGE (30 μg of protein per well), and blots were probed with antibodies against p62 (Progen, GP62-C), LC3 (Novus Biologicals, NB600-1384), p-PERK (Bioss, BS-3330R), PERK (Cell Signaling, 3192), p-eIF2α (Cell Signaling, 9721), eIF2α (Cell Signaling, 9722), AR, IGF-1β (Cell Signaling, 3027), p-Akt (Cell Signaling, 9271), Akt (Cell Signaling, 9272), p-S6 (Cell Signaling, 4858), S6 (Cell Signaling, 2217), and α-tubulin (Cell Signaling, 2125).
EM
Lung tumors were fixed in Karnovsky fixative (Electron Microcsopy Sciences, 15732-10) and processed as previously described. Images were taken using an AMT XR41 digital camera at 80 Kv on a JEOL 1200EX transmission electron microscope.
Castration
Twelve-week-old to 14-wk-old mice were induced by TAM and castrated 6 wk after induction. The castrated mice were then monitored regularly by MRI for tumor development and growth.
MRI
MRIs were acquired using a 1 tesla M2-High-Performance MRI system (Aspect Magnet Technologies Ltd.) and performed under inhalation anesthesia using 4% isoflurane initially and 1%–2% for maintenance during the procedure. Animals were placed prone within the animal bed in the RF coil. Images were analyzed using vivoQuant MRI analysis software.
Competing interest statement
E.W. is an advisor to Forma Therapeutics.
Supplementary Material
Acknowledgments
This work was supported in part by P30 CA072720, UM1 CA186716, R01 CA163591, R01 CA130893, R01 CA188096, the Rutgers Rutgers Cancer Institute of New Jersey Histopathology Service Core, the Biostatistics Core, and the Rutgers Molecular Imaging Core.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.274134.115.
References
- Alva A, Hussain M. 2013. The changing natural history of metastatic prostate cancer. Cancer J 19: 19–24. [DOI] [PubMed] [Google Scholar]
- Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, Chen HY, Ghavami A, Stein M, DiPaola RS, et al. 2012. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One 7: e41831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow JM, Yang JC, Evans CP. 2014. Autophagy as a modulator and target in prostate cancer. Nat Rev Urol 11: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floc'h N, Kinkade CW, Kobayashi T, Aytes A, Lefebvre C, Mitrofanova A, Cardiff RD, Califano A, Shen MM, Abate-Shen C. 2012. Dual targeting of the Akt/mTOR signaling pathway inhibits castration-resistant prostate cancer in a genetically engineered mouse model. Cancer Res 72: 4483–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. 2013a. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27: 1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Xia B, White E. 2013b. Autophagy-mediated tumor promotion. Cell 155: 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. 2004. Androgen receptor in prostate cancer. Endocr Rev 25: 276–308. [DOI] [PubMed] [Google Scholar]
- Hussain M, DiPaola RS. 2015. Clinical research in metastatic prostate cancer: a focus on impact and value. Am Soc Clin Oncol Educ Book 35: 17–21. [DOI] [PubMed] [Google Scholar]
- Irshad S, Bansal M, Castillo-Martin M, Zheng T, Aytes A, Wenske S, Le Magnen C, Guarnieri P, Sumazin P, Benson MC, et al. 2013. A molecular signature predictive of indolent prostate cancer. Sci Transl Med 5: 202ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. 2014. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4: 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137: 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HG, Yang JC, Kung HJ, Shi XB, Tilki D, Lara PN Jr, DeVere White RW, Gao AC, Evans CP. 2014. Targeting autophagy overcomes Enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene 33: 4521–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandini G, Mineo R, Frasca F, Roberts CT Jr, Marcelli M, Vigneri R, Belfiore A. 2005. Androgens up-regulate the insulin-like growth factor-I receptor in prostate cancer cells. Cancer Res 65: 1849–1857. [DOI] [PubMed] [Google Scholar]
- Saleem A, Dvorzhinski D, Santanam U, Mathew R, Bray K, Stein M, White E, DiPaola RS. 2012. Effect of dual inhibition of apoptosis and autophagy in prostate cancer. Prostate 72: 1374–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Han JJ, Tennakoon JB, Mehta FF, Merchant FA, Burns AR, Howe MK, McDonnell DP, Frigo DE. 2013. Androgens promote prostate cancer cell growth through induction of autophagy. Mol Endocrinol 27: 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2015. Cancer statistics, 2015. CA Cancer J Clin 65: 5–29. [DOI] [PubMed] [Google Scholar]
- Strohecker AM, White E. 2014a. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy 10: 384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, White E. 2014b. Targeting mitochondrial metabolism by inhibiting autophagy in Braf-driven cancers. Cancer Discov 4: 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney CJ, Chen YH, Carducci M, Liu G, Jarrard DF, Eisenberger M, Wong YN, Hahn N, Kohli M, Cooney MM, et al. 2015. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N Engl J Med 373: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Weissman AM. 2010. The unfolded protein response, degradation from endoplasmic reticulum and cancer. Genes Cancer 1: 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. 2009. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2015. The role for autophagy in cancer. J Clin Invest 125: 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. 2010. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 22: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Koh JY, Price S, White E, Mehnert JM. 2015. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov 5: 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.