Abstract
Personalized medicine, in which individual tumor genetics drive the selection of targeted therapies and treatment plans for each patient, has recently emerged as the next generation of cancer therapy. Unfortunately, personalized medicine trials have had limited success in tumors that have complex combinations of disruptive genomic events, which drive differential responses to targeted therapies. Here, we will use head and neck squamous cell carcinoma as a model for genetically complex disease and discuss novel approaches to enhance personalized medicine trials for these complicated cases.
Keywords: HNSCC, Personalized Medicine, Sequencing
1. Introduction/Background
Head and neck squamous cell carcinoma (HNSCC) is the sixth-leading cancer worldwide, with an incidence of over 600,000 cases annually (Jemal et al. 2011). Despite advances in surgery, chemotherapy, and radiation, outcomes for advanced-stage cancers have remained dismal, with minimal improvement in survival (Pulte & Brenner 2010). In addition, treatment options for recurrent and metastatic HNSCC are often limited, with palliative treatment generally offered in these situations. In light of the current state of treatment for HNSCC, new paradigms for care are needed.
The concept of targeted treatment for cancer is well established. Targeted therapies have been approved and successfully employed in a wide array of other cancers. Imatinib (Gleevec; for chronic myeloid leukemia) and trastuzumab (Herceptin; for breast cancer) have been established as standard of care in their respective cancers, with impressive results (Druker et al. 2006, Slamon et al. 2001). Additional targeted therapies have been demonstrated to improve survival in many other cancers (Flaherty et al. 2010, Cappuzzo et al. 2010, Van Cutsem et al. 2011, Saltz et al. 2008).
In recent years, the rapid advancement of genomic sequencing techniques has allowed us to identify novel, targetable genes and genetic mutations in a variety of cancers (Brenner et al. 2011, Asangani et al. 2014, Robinson et al. 2013, Ryan et al. 2013). The rationale behind personalized, targeted therapy in cancer is an alluring one (Roychowdhury et al. 2011, Roychowdhury et al. 2014): specific genes or genetic pathways are frequently dysregulated in many cancers. In some instances, one specific genetic alteration is responsible for the cancer (e.g. the BCR-ABL fusion gene in chronic myeloid leukemia; Shtivelman et al. 1985). In such cases, developing personalized therapy against this specific mutation provides an ideal, highly targeted treatment regimen, as opposed to nonspecific and highly damaging chemotherapy or radiation options.
The implementation of personalized, targeted therapy in HNSCC has lagged behind other cancers. This is largely due to the high genomic complexity and high mutation rate in HNSCC compared to other cancers (Cancer Genome Atlas Network 2015). However, as we develop new and powerful genomic screening techniques (Li et al. 2014, Li et al. 2014, Shalem et al. 2014, Wang et al. 2014) to identify mutations in HNSCC, potential therapeutic implications are increasing. Unfortunately, single “magic bullet” agents like imatinib for chronic myeloid leukemia are highly unlikely in HNSCC given the variability, volume, and heterogeneity of mutations in these tumors. Rather, the future of HNSCC targeted therapy will more likely require incorporation of multiple agents and multimodality therapy.
2. Core Dysregulated Pathways in HNSCC
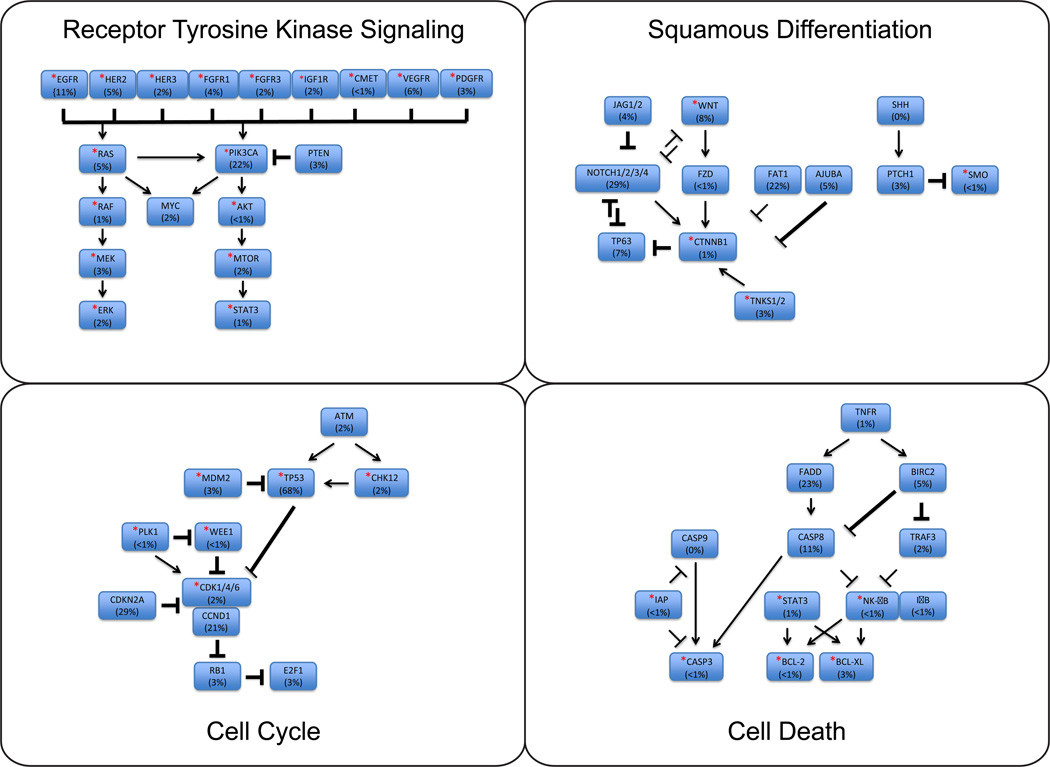
HNSCCs are broadly categorized into HPV positive and HPV negative disease due to the vast differences in outcomes associated with the two populations (Ang et al. 2010). Beyond HPV infection status, recent whole exome and whole genome sequencing data on HNSCC has identified multiple commonly mutated genes and genetic pathways. On average, data from The Cancer Genome Atlas (TCGA) identified 140 mutations per tumor (Cancer Genome Atlas Network 2015). Broadly, mutations in four key regulatory pathways are frequently identified, including: cell cycle control (96%), growth and proliferation (62%), death (44%), and squamous differentiation (64%) pathways (Cancer Genome Atlas Network 2015, Pickering et al. 2013; Figure 1). Importantly, each of these regulatory pathways currently has potentially targetable therapeutic options in development or early clinical testing (Table I; Bonner et al. 2010, Vermorken et al. 2008, Liu et al., 2013, Wang et al. 2014).
Figure 1. Key Dysregulated Pathways in HNSCC.
Core dysregulated pathways in HNSCC include receptor-tyrosine kinase mediated cell growth and proliferation, cell cycling, squamous differentiation, and cell death pathways. Mutation rates listed as obtained from the Oncomine program (Rhodes et al. 2004) using publically available sequencing results. Starred genes have targeted agents in development, as noted in Table I.
Table I. Targeted Agents in Clinical Trials for HNSCC.
Primary targeted pathways include growth/proliferation, cell cycling, squamous differentiation, cell death, and immune regulation pathways. Agents with FDA approval in other diseases are listed.
| Pathway/Gene | Agent | Phase in HNSCC | FDA Approval | |
|---|---|---|---|---|
| Growth/Proliferation | ||||
| AKT | MK2206 | II | -- | |
| BRAF/VEGFR/PDGFR | Sorafenib (Nexavar) | II | Renal, Liver, Thyroid | |
| CKIT | OSI930 | I | -- | |
| CMET | Tivantinib | II | -- | |
| INC280 | I/II | -- | ||
| LY2801653 | I | -- | ||
| CMET/VEGFR | Cabozantinib (Cometriq) | II | Medullary Thyroid | |
| Foretinib | II | -- | ||
| Golvatinib | I/II | -- | ||
| MGCD265 | I | -- | ||
| CMET/PDGFR/VEGFR | MGCD516 | I | -- | |
| EGFR | Cetuximab (Erbitux) | IV | HNSCC, Colorectal | |
| Gefitinib (Iressa) | III | NSCLC | ||
| Panitumumab (Vectibix) | III | Colorectal | ||
| Nimotuzumab (Theraloc) | II/III | Glioma | ||
| Erlotinib (Tarceva) | I/II | NSCLC, Pancreatic | ||
| Zalutumumab (HuMax) | I/II | -- | ||
| Dacomitinib | I/II | -- | ||
| MM151 | I | -- | ||
| EGFR/HER2 | Afatinib (Gilotrif) | III | NSCLC | |
| Lapatinib (Tykerb) | III | Breast | ||
| EGFR/HER2/HDAC | CUDC101 | I | -- | |
| EGFR/HER3 | MEHD7945A | I/II | -- | |
| EGFR/RET/VEGFR | Vandetanib (Caprelsa) | II | Medullary Thyroid | |
| FGFR | Dovitinib | II | -- | |
| AZD4547 | I | -- | ||
| BAY1163877 | I | -- | ||
| BGJ398 | -- | -- | ||
| FGFR/VEGFR | PD173074 | -- | -- | |
| FGFR/PDGFR/VEGFR | Pazopanib (Votrient) | I/II | Renal, Sarcoma | |
| FNTB/HRAS | Lonafarnib | I | -- | |
| L778123 | I | -- | ||
| HER2 | Trastuzumab (Herceptin) | I | Breast, Gastric | |
| HER3 | LJM716 | I | -- | |
| MM121 | I | -- | ||
| Patritumab | I | -- | ||
| HGF/CMET | Ficlatuzumab | I | -- | |
| IGF-1R | Cixutumumab | II | -- | |
| Figitumumab | II | -- | ||
| Linsitinib | II | -- | ||
| BMS754807 | I/II | -- | ||
| MEK1/2 | Trametinib (Mekinist) | I/II | Melanoma | |
| MTOR | Everolimus (Zortress) | II | Breast, Renal, SEGA, Pancreatic PNET | |
| Temsirolimus | II | Renal | ||
| Sirolimus | I/II | -- | ||
| Ridaforolimus | I | -- | ||
| PDGFR/VEGFR | Sunitinib (Sutent) | II | Renal, GIST | |
| Vatalanib | I | -- | ||
| PDGFR/RET/VEGFR | Axitinib (Inlyta) | II | Renal | |
| PI3K | Buparlisib | I/II | -- | |
| BYL719 | I/II | -- | ||
| PX866 | I/II | -- | ||
| Rigosertib | I | -- | ||
| VEGF-A | Bevacizumab (Avastin) | III | Colorectal, NSCLC, Renal, Glioblastoma | |
| VEGFR | Semaxanib | I | -- | |
| Lenvatinib (Lenvima) | -- | Thyroid | ||
| Cediranib | -- | -- | ||
| Cell Cycle | ||||
| CDK4/6 | LEE011 | I | -- | |
| Palbociclib (Ibrance) | I/II | Breast | ||
| Abemaciclib | -- | -- | ||
| CDK1/4/9 | P27600 | II | -- | |
| CDK9 | Alvocidib | II | -- | |
| CHK1/2 | LY2606368 | I | -- | |
| MDM2 | DS3032 | I | -- | |
| RO6839921 | I | -- | ||
| Nutlin3 | -- | -- | ||
| PLK1 | BI2536 | II | -- | |
| TP53 | Kevetrin | I | -- | |
| WEE1 | MK1775 | II | -- | |
| Squamous Differentiation | ||||
| CTNNB1 | PRI724 | I | -- | |
| ICG001 | -- | -- | ||
| FZD | OMP-54F28 | I | -- | |
| PORCN/WNT | LGK974/WNT974 | I | -- | |
| SMO | Vismodegib (Erivedge) | II | BCC | |
| Saridegib | I | -- | ||
| TNKS1/2 | WIKI4 | -- | -- | |
| XAV939 | -- | -- | ||
| Cell Death | ||||
| BCL-2/BCL-XL | AT101 | II | -- | |
| CASP3 | PAC1 | I | -- | |
| IAP1/IAP2/XIAP | D1143 | I/II | -- | |
| NF-κB | Bortezomib (Velcade) | II | Multiple Myeloma, Mantle Cell Lymphoma | |
| STAT3 | OPB31121 | I | -- | |
| OPB51602 | I | -- | ||
| Immune Regulation | ||||
| CCR4 | Mogamulizumab (Poteligeo) | I | -- | |
| CD27 | Varlilumab | I/II | -- | |
| CD134 | MEDI6469 | I | -- | |
| CD137 | Urelumab | I | -- | |
| CTLA-4 | Ipilimumab (Yervoy) | I | Melanoma | |
| Tremelimumab | I | -- | ||
| FLT3 | Quizaritinib | -- | -- | |
| IDO1 | INCB24360 | I/II | -- | |
| IL-6 | Siltuximab | I/II | Castleman’s Disease | |
| PD-1 | Nivolumab (Opdivo) | III | Melanoma | |
| Pembrolizumab (Keytruda) | II | Melanoma | ||
| MK3475 | II | -- | ||
| PD-L1 | MEDI4736 | II | -- | |
| MPDL3280A | I | -- | ||
| TLR8 | VTX2337 | II | -- | |
| TLR9 | EMD1201081 | II | -- | |
| Other | ||||
| ALK | Crizotinib (Xalkori) | II | NSCLC | |
| Ceritinib | I | NSCLC | ||
| AURKA | Alisertib | I | -- | |
| MLN8237 | I/II | -- | ||
| BCR-ABL | Ponatinib (Iclusig) | II/III | CML | |
| Imatinib (Gleevec) | II | CML | ||
| BCR-ABL/SRC | Dasatinib (Sprycel) | II | CML | |
| CD44 | Bivatuzumab | I | -- | |
| FAK | PF00562271 | I | -- | |
| HSP90 | Ganetespib | -- | -- | |
| KSP | Ispinesib | II | -- | |
| PARP1/2 | Veliparib | I/II | -- | |
| Olaparib | I | Ovarian | ||
| SRC | Saracatinib | II | -- | |
| SYK | Fostamatinib | II | -- | |
NSCLC = non-small cell lung carcinoma, SEGA = subependymal giant cell astrocytoma, PNET = primitive neuroectodermal tumor, GIST = gastrointestinal stromal tumor, BCC = basal cell carcinoma.
2.1 Cell Growth and Proliferation
Overall, mutations and amplifications in cell growth and proliferation signaling pathways are identified in a significant portion of HNSCCs. Receptor tyrosine kinases (RTKs) play a key role in cell signaling for growth and proliferation. Numerous targeted agents against RTKs have been developed and successfully implemented in care in a number of cancers, including HNSCC (Table I). Currently, cetuximab (Erbitux) is the only approved targeted therapy for HNSCC. Importantly, cetuximab is most frequently used in combination with chemotherapy or radiation (Bonner et al. 2010, Vermorken et al. 2008). Monotherapy with cetuximab is indicated in select circumstances, but only shows a modest effect (Vermorken et al. 2007).
Despite its success, however, much remains to be studied in regards to anti-EGFR therapy, and therapy against other potential targets in HNSCC. For instance, cetuximab use is not dependent on any personalized analysis of EGFR expression levels or mutational status. Given the potential for resistance to cetuximab therapy, increased screening for genetic alterations that drive response to EGFR inhibition at the time of diagnosis may be of importance.
Cetuximab is far from the only RTK being explored for potential targeted therapy. Anti-HER2 therapy, which has been successful in breast and gastro-esophageal cancers, may have a role in HNSCC (Pollock et al. 2014). Interestingly, a subset of HNSCC patients demonstrates overexpression of HER2 (Birkeland et al. manuscript in review), suggesting that these patients could potentially benefit from trastuzumab or other anti-HER2 therapy. As the safety profile for trastuzumab has already been well established, it provides an intriguing option for future clinical trials in HNSCC patients with HER2-overexpressing tumors.
A potential role for FGFR inhibitors in HNSCC has just recently been identified, with findings of select tumors harboring amplifications of the FGFR1 gene and other members of the FGFR family (e.g. FGF3, FGF4 and FGF19 also reside on the 11q13 amplicon that is frequently amplified in HNSCC; Goke et al. 2013, Tillman et al. manuscript in review). Other inhibitors of RTKs are in various phases of clinical trials in HNSCC, including many combination inhibitors. Excitingly, some of these targeted therapies have already been FDA-approved in other cancers (Table I), suggesting a plausible role in HNSCC.
Downstream growth/proliferation targets of RTKs have been explored as drug targets (Figure 1). Specifically, inhibitors of the PI3K/Akt/mTOR pathway are being actively explored in HNSCC and other cancers (Simpson et al. 2014). Early phase clinical trials are in progress studying such inhibitors in HNSCC.
2.2 Cell Cycle Control
In regards to cell cycle control, inactivation of TP53, either through direct genetic mutation or increased expression of p53 regulatory proteins (e.g. MDM2 amplification or HPV E6), is identified almost universally in HNSCC cases (Cancer Genome Atlas Network 2015). Additionally commonly mutated genes regulating cell cycle include CCND1, CDKN2A, and CD4K4/6 (Figure 1). Currently, inhibitors against multiple agents in this pathway are in early clinical trials (Table I). In particular, the CDK inhibitor palbociclib (Ibrance) has been demonstrated to be effective in breast cancer (Finn et al. 2015), and recently achieved FDA approval. Overall, unlike the cell growth/proliferation pathway, where mutations often cause gene overactivity, mutations in cell cycle control include a significant amount loss of function mutations (especially in TP53). As we discuss below, these mutations can be more challenging to address with targeted therapies.
2.3 Cell Death
Anti-apoptotic and anti-cell death mechanisms are a key trait common to cancers (Hanahan & Weinberg 2000). Sequencing analysis of HNSCC has shown frequent mutations in cell death pathways, suggesting a role for targeted agents (Figure 1). Bortezomib (Velcade) is an inhibitor of NF-κB. It is a key regulator in cell death pathways, where it is a proteasome inhibitor, inhibiting anti-apoptotic genes (BCL-2, BCL-XL, and STAT3), and upregulating pro-apoptotic genes (NOXA; Qin et al. 2005). It has FDA approval for use in multiple myeloma and mantle cell lymphoma (Chen et al. 2011), and is being investigated in HNSCC and other cancers. Additionally, agents inhibiting other key anti-apoptotic genes (e.g. BCL-2, BCL-XL; Imai et al. 2012) are in early phase clinical trials in HNSCC and other cancers (Table I) including the anti-Bcl-xL compound AT-101 at our institution (NCT01633541).
2.4 Cell Differentiation
Mutations in the Notch pathway were identified in the initial HNSCC exome studies (Stransky et al. 2011, Agrawal et al. 2011); subsequently, it has been established that over 50% of HNSCC samples have mutations in the Notch pathway (Cancer Genome Atlas Network 2015; Figure 1). There are challenges to developing targeted agents in this pathway as a significant amount of these mutations are loss-of-function, for which inhibitors would not work for the mutated genes. However, inhibiting the consequently unchecked related cellular pathways is a viable therapeutic option. For example, we have recently shown that NOTCH1 deficient tumors are highly susceptible to inhibition of Wnt signaling by blocking PORCN, a protein critical for Wnt ligand secretion (Liu et al. 2013). Vismodegib, which targets the hedgehog pathway gene SMO, has FDA approval in basal cell carcinomas (Sekulic et al. 2012). Additional targets in these pathways, including β-catenin, are in early phase investigation in HNSCC and other cancers.
2.5 Other Pathways
Immune regulation in cancers is a complex and vast field. Robust research into immunotherapy in HNSCC is being performed, ranging from targeted small molecule inhibitors to tumor vaccines. Mutations in immune recognition pathways are identified in a subset of HNSCCs, making immune modulators an intriguing prospect. Currently, agents like ipilimumab (Yervoy) and Nivolumab (Opdivo) have shown survival benefit (Robert et al. 2011, Robert et al. 2015) and are FDA approved for use in melanoma.
Many other targeted agents for additional pathways or orphan genes are in development and early phase clinical trials (Table I). Some of these agents are already well established and FDA approved in other tumors (e.g. imatinib). Others suggest the potential for other genetic pathways or cellular phenotypes (e.g. CD44 inhibitors, which may suggest a role for targeting cancer stem cells, Tijink et al. 2006).
3. Combination Therapies
While targeting an individual gene may have limited success, combining targeted therapies against multiple genetic targets or pathways simultaneously could provide one strategy for improved outcomes in these genetically complex tumors. The concern for monotherapy is the development of resistance in cancer cells and the subsequent proliferation of these cells to repopulate the tumor. Indeed, as carcinogenesis is proposed to involved dysregulation of multiple key pathways (including growth, anti-apoptosis, invasion, and cell division), addressing as many of these dysregulated pathways as possible could lead to improved therapeutic outcomes, but may increase toxicity and morbidity.
3.1 Multiple Agents in a Single Pathway
Importantly, most tumors harbor mutations in multiple key regulatory pathways, as described above. As such, combining targeted therapies against multiple genetic targets or pathways simultaneously could provide improved outcomes. In cell growth/proliferation pathways, multiple RTKs can provide compensatory signaling, rendering monotherapy less efficacious (Janne et al. 2009). Additionally, genetic mutations in EGFR and other RTKs can confer resistance to either monoclonal antibodies or small molecule inhibitors (through resistance-driving EGFR mutations, or gene amplifications in compensatory RTKs; Kobayashi et al. 2005, Takezawa et al. 2012, Bean et al. 2007). Additionally, downstream activating mutations in the cell growth/proliferation pathway (e.g. in KRAS, BRAF, or PIK3CA) can render upstream RTK blockade ineffective (Lievre et al. 2006, Benvenuti et al. 2007). Newer antibodies and small molecule inhibitors in development are targeting multiple RTKs simultaneously (e.g. EGFR and HER2; Table I). Indeed, combining multiple agents against a signaling pathway may more effectively knock down signaling and provide improved response. Currently, there are limited trials studying combined therapy in HNSCC, although such regimens are being employed in other cancers (Geyer et al. 2006, Swain et al. 2015, Janjigian et al. 2014).
3.2 Combination Targeting of Multiple Pathways
An alternative combination treatment regimen is to target multiple key regulatory (e.g. cell growth/proliferation and cell apoptosis) pathways simultaneously. As it is well established that mutations in multiple pathways are required for carcinogenesis, using targeted agents against a combination of these dysregulated pathways could theoretically improve tumor response. Knocking down multiple pathways could also account for compensatory mechanisms of tumor cell resistance to monotherapy. An early trial targeting multiple pathways in melanoma (with ipilimumab and vemurafenib) had issues with toxicity (Ribas et al. 2013). Currently, inhibition of the growth/proliferation pathway is much more developed in regards to targeted therapy than other carcinogenic pathways. Nevertheless, consideration of combination therapies amongst different pathways will be crucial in establishing improved tumor response. Early combination therapy clinical trials are in progress on other solid cancers, including EGFR/BRAF/Wnt inhibition in colorectal cancer (NCT02278133), MEK/CD4/6 inhibition in solid tumors (NCT02065063), and NF-κB and VEGF inhibition in advanced malignancies (NCT00428545), among others.
3.3 Combination Therapy with Chemoradiation
To overcome issues with tumor heterogeneity and unknown compensatory responses, one alternative approach is to merge conventional and/or non-specific therapies with individually tailored targeted inhibitors. Tailoring conventional therapy, including established cetuximab, radiation, and chemotherapy options to fit genetic profiles may be important for identifying rational improvements in survival in genetically complex and highly heterogeneous tumors. Conversely, understanding the biomarkers that predict tumor components that will be unresponsive to chemotherapy, radiation, or cetuximab will be equally important for identifying patients that require more aggressive therapy.
Current standard of care for HNSCC incorporates a combination of surgery, radiation and chemotherapy. Particularly for advanced and recurrent disease, multimodality therapy is important. Traditional chemotherapy and radiation therapies have a significant amount of well-described off-target effects and toxicities (Givens et al. 2009). It is unlikely targeted therapies will completely replace these established regimens. Rather, they can act as adjuvants in combination with these established treatments to enhance care rates and reduce toxicity. Studies with cetuximab in HNSCC have demonstrated a beneficial effect when added to current standard-of-care chemotherapy and radiation (Bonner et al. 2010, Vermorken et al. 2008). Addition of targeted therapy could be investigated in combination with reduced doses of radiation and traditional chemotherapy. The balance of maintaining treatment efficacy while reducing radiation and chemotherapy toxicity will be important to investigate.
4. Caveats to the Personalized Medicine Paradigm and Future Directions
4.1 Predicting Driver Mutations and Compensatory Pathways
Personalized medicine is not without it’s limitations, however (Stahel et al. 2014). In fact, a major question for personalized medicine tumor boards is how to predict which patients will respond to targeted therapies (i.e. Is the presence of a mutated driver in the majority of tumor cells sufficient to warrant intervention with a companion therapeutic?). Cancers, particularly HNSCC, are often heterogeneous, with multiple, variable genetic drivers (Fisher et al. 2013). Thus, successfully targeting of only one of the deregulated pathways in these genetically complex diseases may not be sufficient for treatment.
At this stage, concern for tumors not responding to targeted therapies are largely attributed to either additional innate genetic lesions (e.g. multiple pathway disruption), or the emergence of compensatory pathways. Initially, clarification of the specific genetic mutations or alterations that drive HNSCC pathogenesis will be important in developing combination strategies against innate genetic lesions. Streamlining the identification of actionable targets, possibly through select genetic panels of commonly altered genes, will be useful in driving down cost and providing valuable information without inundation of data. However, this will not overcome issues with compensatory response mechanisms of tumors to targeted therapies, which can differ from tumor to tumor.
4.2 Unpredictability of Response
Indeed, biologically plausible targets may not demonstrate successful clinical response. This is evidenced in the case of trastuzumab against HER2 positive ovarian cancers, in which clinical trials did not identify a survival benefit (Bookman et al. 2003). Similarly, in HNSCC, application of the EGFR inhibitor gefitinib (Iressa) did not show improved outcomes, despite acting on the same genetic pathway as cetuximab (Argiris et al. 2013). This highlights that each agent needs to be assessed individually, regardless of previous outcomes with similar compounds, and brings in to question the value of negative responses in precision medicine trials. In the same fashion that gefitinib does not appear to show benefit despite cetuximab’s success, other compounds may ultimately demonstrate benefit despite lack of success for similar compounds. Thus, a major concern for personalized medicine trials is that a few failed responses may drive negative bias towards a particular class of compounds/lesions. Moreover, initially sensitive tumors can subsequently develop resistance to targeted therapy, as detailed above. Thus, continual reassessment of tumor response will be important.
4.3 HNSCC Tumor and Population Heterogeneity
Further characterization of the interplay between common drivers and lost suppressors in HNSCC will be important for the molecular stratification of disease. Unfortunately these efforts are currently limited by the small number of HNSCC cases that have been sequenced. For example, although sequencing data from over 400 HNSCC cases are now publicly available, these cases are from a variety of head and neck subsites, often lack associated clinical outcome data, and have a drastically skewed percentage of cases that favor Caucasian-Americans with a history of smoking and alcohol use. With the recent increase in HPV-related HNSCCs (Chaturvedi et al. 2008), much increased focus should be paid to understanding the disruptive genetic events that are co-altered in HPV positive tumors to understand the molecular stratification of these tumors. In the future, sequencing studies of tumors with associated therapeutic and clinical response will further define the subsets of patients that require more aggressive approaches to increase survival.
4.4 Addressing Tumor Suppressor Genes
Not all genetic targets are created equally, however. This majority of the agents developed have been designed to inhibit function in oncogenes (e.g. kinase inhibition). Restoring lost function of tumor suppressor genes remains a challenging process. For instance, although TP53 expression is lost in almost all HNSCC, we have not developed successful targeted therapies to restore TP53 function. Interestingly, virus-restored p53 clinical trials are currently being performed (Liu et al. 2013). Titrating to restore physiologic function of lost tumor suppressor genes, however, will remain a challenging prospect.
4.5 Future Goals of Care
Consequently, to move forward in the current design of personalized medicine trials, evaluation of successful targeted therapies against mutated or deregulated targets from other cancers will be an important initial study in HNSCC. These agents have already been studied for safety and dosing, they have the potential for earlier application against HNSCC, if warranted. However, these classes of therapies need to be carefully evaluated in multiple “N of 1” trials to fully understand the response of tumors to each therapy.
Long-term, we believe that the development of tumor-site specific Personalized Medicine Tumor Boards will be crucial to adequately investigate patient options with ongoing clinical trials and experimental therapies, especially for tumors with complex mutational loads. In these early stages, many personalized medicine tumor boards being run at many large academic institutions focus on advanced or non-resectable disease from multiple tumor sites (Everett et al. 2014). Integrating the experiences learned from these multi-tumor trials with tissue-specific experiences will be critical in the long run.
In these precision medicine tumor boards, open discussion amongst surgeons, medical oncologists, radiation oncologists, geneticists, and ethicists can lead to identifying the ideal treatment regimen or study for patients, and foster discussion on proper patient counseling. In this nascent stage of personalized medicine, ethical issues regarding incidental findings, counseling patients, and disclosure are in flux (Birkeland et al. manuscript in review); we will need to continually assess and modify our discussions with patients as our technology and ability to understand genomic alterations advances.
New clinical trials and treatment paradigms are considering cancers based on their mutational profile rather than anatomic location (Conley & Doroshow 2014). While this may be too simplistic a view on cancers (e.g. anti-HER2 treatment has variable effects on breast vs. ovarian cancer), it does point to the importance of identifying and classifying tumors based on their mutational profiles.
5. Conclusion
We have entered an exciting era in personalized medicine for cancer therapy and the development of targeted therapies and novel approaches for HNSCC will assuredly play an important role in the ongoing battle against this deadly disease. Already, we are introducing targeted therapy concepts into HNSCC through ongoing precision medicine trials, where we expect the greatest impact to come for patients with relatively few genetic drivers. In the future, as our understanding of the interplay of disruptive genomic events driving HNSCC pathogenesis and response to therapy increases, so too will our ability to design optimal therapeutic approaches for patients with genetically complex disease.
References
- Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiris A, Ghebremichael M, Gilbert J, et al. Phase III randomized, placebo-controlled trial of docetaxel with or without gefitinib in recurrent or metastatic head and neck cancer: an eastern cooperative oncology group trial. J Clin Oncol. 2013;31(11):1405–1414. doi: 10.1200/JCO.2012.45.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104(52):20392–20397. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67(6):2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- Birkeland AC, Uhlmann WR, Brenner JC, Shuman AG. Getting personal: Head and neck cancer management in the era of personalized medicine. doi: 10.1002/hed.24132. Manuscript in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland AC, Yanik M, Tillman BN, et al. Identification of HER2 amplifications in head and neck squamous cell carcinoma. Manuscript in review. [Google Scholar]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11(1):21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- Bookman MA, Darcy KM, Clarke-Pearson D, et al. Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2; a phase II trial of the Gynecologic Oncology Group. J Clin Oncol. 2003;21(2):283–290. doi: 10.1200/JCO.2003.10.104. [DOI] [PubMed] [Google Scholar]
- Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19(5):664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Ciuleanu T, Stehlmakh L, et al. Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol. 2010;11(6):521–529. doi: 10.1016/S1470-2045(10)70112-1. [DOI] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and –unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26(4):612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11(3):239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41(3):297–299. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- D’Amato V, Rosa R, D’Amato C, et al. The dual PI3K/mTOR inhibitor PKI-587 enhances sensitivity to cetuximab in EGFR-resistant human head and neck cancer models. Br J Cancer. 2014;110(12):2887–2895. doi: 10.1038/bjc.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Everett JN, Gustafson SL, Raymond VM. Traditional roles in non-traditional setting: genetic counseling in precision oncology. J Genet Couns. 2014;23(4):655–660. doi: 10.1007/s10897-014-9698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Crown JP, Lang I, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as a first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355(26):2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- Givens DJ, Karnell LH, Gupta AK, et al. Adverse events associated with concurrent chemoradiation therapy in patients with head and neck cancer. Arch Otolaryngol Head Neck Surg. 2009;135(12):1209–1217. doi: 10.1001/archoto.2009.174. [DOI] [PubMed] [Google Scholar]
- Goke F, Bode M, Franzen A, et al. Fibroblast growth factor receptor 1 amplification is a common event in squamous cell carcinoma of the head and neck. Mod Pathol. 2013;26(10):1298–1306. doi: 10.1038/modpathol.2013.58. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Imai A, Zeitlin BD, Visoli F, et al. Metronomic dosing of BH3 mimetic small molecule yields robust antiangiogenic and antitumor effects. Cancer Res. 2012;72(3):716–725. doi: 10.1158/0008-5472.CAN-10-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjigian YY, Smit EF, Groen HJ, et al. Dual inhibition with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov. 2014;4(9):1036–1045. doi: 10.1158/2159-8290.CD-14-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8(9):709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Li H, Wawrose JS, Gooding WE, et al. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol Cancer Res. 2014;12(4):571–582. doi: 10.1158/1541-7786.MCR-13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Conley A, Zhang H, Kim HL. Whole-transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics. 2014;15(1):1087. doi: 10.1186/1471-2164-15-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Liu J, Pan S, Hsieh MH, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 2013;110(50):20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen P, Hu M, et al. Randomized, controlled phase II study of post-surgery radiotherapy combined with recombinant human p53 gene therapy in treatment of oral cancer. Cancer Gene Ther. 2013;20(6):375–378. doi: 10.1038/cgt.2013.30. [DOI] [PubMed] [Google Scholar]
- Pickering CR, Zhang J, Yoo SY, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3(7):770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock NI, Grandis JR. HER2 as a therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-14-1432. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncologist. 2010;15(9):994–1001. doi: 10.1634/theoncologist.2009-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JZ, Ziffra J, Stennett L, et al. Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res. 2005;65(14):6282–6293. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, Hodi FS, Callahan M, Konto C, Wolchok J. Hepatotoxicity with combination of vemurafenib and ipilimumab. N Engl J Med. 2013;368(14):1365–1366. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- Robert C, Thomas L, Bondarenko I, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364(26):2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Y M, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, Chinnaiyan AM. Translating genomics for precision cancer medicine. Annu Rev Genomics Hum Genet. 2014;15:395–415. doi: 10.1146/annurev-genom-090413-025552. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3(111):111–121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368(2):138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltz LB, Clarke S, Diaz-Rubio E, et al. Bevacizumab in combination with oxaplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomised phase III study. J Clin Oncol. 2008;26(12):2013–2019. doi: 10.1200/JCO.2007.14.9930. [DOI] [PubMed] [Google Scholar]
- Sekulic A, Migden MR, Oro AE, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366(23):2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukemia. Nature. 1985;315(6020):550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- Simpson DR, Mell LK, Cohen EE. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2014 doi: 10.1016/j.oraloncology.2014.11.012. epub. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Stahel R, Bogaerts J, Ciardiello F, et al. Optimising translational oncology in clinical practice: Strategies to accelerate progress in drug development. Cancer Treat Rev. 2014 doi: 10.1016/j.ctrv.2014.12.004. epub. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Baselga J, Kim SB, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2(10):922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijink BM, Buter J, de Bree R, et al. A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin Cancer Res. 2006;12(20 Pt 1):6064–6072. doi: 10.1158/1078-0432.CCR-06-0910. [DOI] [PubMed] [Google Scholar]
- Tillman BN, et al. Targeted sequencing of an epidemiologically low risk patient defines fibroblast growth factor family aberrations as a driver of head and neck squamous cell carcinoma. doi: 10.1002/hed.24292. Manuscript in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- Vermorken JB, Trigo J, Hitt R, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. L Clin Oncol. 2007;25(16):2171–2177. doi: 10.1200/JCO.2006.06.7447. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Martin D, Molinolo AA, et al. mTor co-targeting in cetuximab resistance in head and neck cancers harboring PIK3CA and RAS mutations. J Natl Cancer Inst. 2014;106(9) doi: 10.1093/jnci/dju215. [DOI] [PMC free article] [PubMed] [Google Scholar]