Abstract
Azoles are widely used antifungal drugs. This family of compounds includes triazoles, mostly used in the treatment of systemic infections, and imidazoles, such as clotrimazole, often used in the case of superficial infections. Candida glabrata is the second most common cause of candidemia worldwide and presents higher levels of intrinsic azole resistance when compared with Candida albicans, thus being an interesting subject for the study of azole resistance mechanisms in fungal pathogens.
Since resistance often relies on the action of membrane transporters, including drug efflux pumps from the ATP-binding cassette family or from the Drug:H+ antiporter (DHA)1 family, an iTRAQ-based membrane proteomics analysis was performed to identify all the membrane-associated proteins whose abundance changes in C. glabrata cells exposed to the azole drug clotrimazole. Proteins found to have significant expression changes in this context were clustered into functional groups, namely: glucose metabolism, oxidative phosphorylation, mitochondrial import, ribosome components and translation machinery, lipid metabolism, multidrug resistance transporters, cell wall assembly, and stress response, comprising a total of 37 proteins. Among these, the DHA transporter CgTpo1_2 (ORF CAGL0E03674g) was identified as overexpressed in the C. glabrata membrane in response to clotrimazole. Functional characterization of this putative drug:H+ antiporter, and of its homolog CgTpo1_1 (ORF CAGL0G03927g), allowed the identification of these proteins as localized to the plasma membrane and conferring azole drug resistance in this fungal pathogen by actively extruding the drug to the external medium. The cell wall protein CgGas1 was also shown to confer azole drug resistance through cell wall remodeling. Finally, the transcription factor CgPdr1 in the clotrimazole response was observed to control the expression of 20 of the identified proteins, thus highlighting the existence of additional unforeseen targets of this transcription factor, recognized as a major regulator of azole drug resistance in clinical isolates.
Systemic fungal infections are a problem of increasing clinical significance since the extensive use of antifungal drugs, both as treatment and prophylaxis, has led to a huge increase in the number of intrinsically resistant infections with fungal pathogens (1, 2). This is particularly true for the non-albicans Candida species Candida glabrata.
Candidemia represents the fourth most common nosocomial infection in humans (3). Over the past two decades there has been an increase in the number of infections concerning non-albicans species, with C. glabrata arising as the second most frequent pathogenic yeast in mucosal and invasive fungal infections in humans, representing 15–20% of all infections caused by Candida species, depending on the geographical region (4–6). Together, C. albicans and C. glabrata represent 65–75% of all systemic candidiasis, followed by C. parapsilosis and C. tropicalis (5).
Azoles are one of the main families of drugs currently used to treat or prevent fungal infections. C. glabrata presents a higher level of intrinsic resistance to azoles than C. albicans and develops further resistance during prolonged azole therapy. The acquisition of azole drug resistance is commonly associated with the expression of multidrug efflux pumps such as those from the ATP-binding cassette family and from the major facilitator superfamily responsible for the efflux of structurally and functionally unrelated compounds (2, 7). Expression of several of these transporters was shown to be dependent on the transcription factor CgPdr1, recognized as a major pleiotropic drug resistance mediator in C. glabrata (8–10). Most studies conducted so far to study azole drug resistance focus mainly on fluconazole.
Despite the notion that azole resistance mechanisms are well established, this is not absolutely true for non-albicans Candida species, and certainly it is not true for azole drugs other than fluconazole. Since fluconazole is a triazole, it is possible to assume that information gathered for fluconazole may be extrapolated for other triazoles, like itraconazole and posaconazole. However, this would not be the case for imidazole drugs, which are widely used for topical applications in mucosal infections. Among these drugs, ketoconazole and miconazole are the most well known, but for clotrimazole very little information is available regarding its action and resistance mechanisms in C. glabrata. In a study by Calahorra et al. (11) the mode of action and resistance mechanisms of ketoconazole and miconazole (both imidazoles) were studied in the model S. cerevisiae. Although the primary mode of action involves the inhibition of sterol synthesis common to all azoles, these two drugs seem to also have an effect on cation transport systems. Ketoconazole and miconazole produced an efflux of K+ at low concentrations (∼200 μm) leading to an almost complete depletion of that ion in the cells. The drugs appear to bind to the surface of the cell due to their amphipathic and cationic nature, decreasing the surface charge of the membrane. As a consequence, both antifungals can stimulate efflux of K+ at low concentrations, once at higher concentrations, the uptake of H+ can be stimulated. The change of the surface charge was hypothesized to lead to a disruption of the membrane structure due to the interaction of the antifungals with lipid rafts. These results were not found with two triazole antifungals, fluconazole and itraconazole, indicating that the active group of the molecules is the imidazole moiety of the molecule, thus showing a clear difference between imidazole and triazole modes of action. Ketoconazole and miconazole were found to affect respiration, probably related to the cationic nature of the imidazolic portion of the molecule. More studies highlighted the importance of mitochondrial function for tolerance to antifungal drugs and virulence, with functions in lipid homeostasis likely to be the center of mitochondrial action in tolerance to antifungal drugs, with some crosstalk between mitochondria and cell wall integrity, but the exact molecular mechanisms are not fully understood (12, 13).
In this study, the C. glabrata response to clotrimazole, at the membrane proteome level was examined. Based on the identified proteins exhibiting altered concentrations in the membrane-enriched fraction, the effect of cell wall remodeling, in the dependence of CgGas1, in clotrimazole resistance was inspected. CgGas1 (ORF CAGL0G00286g) is a predicted glicosylphosphatidylinositol-anchored cell wall bound protein (14), homologue of the S. cerevisiae Gas1 β-1,3-glucanosyltransferase, required for cell wall assembly. The deletion of CgGAS1 was shown to lead to an aggregation phenotype and to lead to decreased growth rates (15).
Furthermore, the suggested role of the two uncharacterized homologs of S. cerevisiae Tpo1 drug:H+ antiporter in C. glabrata, encoded by ORFs CAGL0G03927g (CgTPO1_1) and CAGL0E03674g (CgTPO1_2) in clotrimazole drug resistance was also evaluated, given the observed up-regulation of CgTpo1_2 in C. glabrata membrane-enriched fractions exposed to clotrimazole. S. cerevisiae Tpo1 is one of the best characterized of the eukaryotic Drug:H+ Antiporters (16, 17). The primary physiological role attributed to Tpo1 has been the transport of polyamines (18), particularly spermidine (19). However, Tpo1 has been shown to confer resistance to numerous chemical stress agents, from herbicides (20) and agricultural fungicides (21) to the antifungal drug caspofungin (22), among many others (16, 17). Interestingly, the Tpo1 homolog in C. albicans, CaFlu1, was found to complement fluconazole hypersusceptibility in a S. cerevisiae Δpdr5 mutant but not to have a significant role in fluconazole resistance in C. albicans (23). More recently, Flu1 was shown to confer resistance to the salivary human antimicrobial peptide histatin 5, playing a direct role in its efflux from C. albicans cells, thus reducing histatin 5 toxicity (24). Based on the proteomics data, CgTPO1_1 and CgTPO1_2 were further analyzed, in this study. The subcellular localization of these transporters and their effect in antifungal drug resistance was assessed. Their action as clotrimazole resistance determinants was correlated with their action in the accumulation of radiolabeled clotrimazole in C. glabrata.
EXPERIMENTAL PROCEDURES
Strains and Growth Media
S. cerevisiae parental strain BY4741 (MATa, ura3Δ0, leu2Δ0, his3Δ1, met15Δ0) and the derived single deletion mutant BY4741_Δtpo1 were obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). Cells were batch-cultured at 30 °C, with orbital agitation (250 rpm) in basal medium (BM), with the following composition (per liter): 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco, Franklin Lakes, NJ), 20 g glucose (Merck, Kenilworth, NJ), and 2.65 g (NH4)2SO4 (Merck), supplemented with 20 mg/l methionine, 20 mg/L histidine, 60 mg/l leucine, 20 mg/l uracil (all from Sigma). C. glabrata parental strain KUE100 (25) and derived single deletion mutants KUE100_Δcggas1, KUE100_Δcgtpo1_1, or KUE100_Δcgtpo1_2, as well as the C. glabrata strains 66032u and 66032u_Δcgpdr1 (10), kindly provided by Thomas Edlind, from the Department of Microbiology and Immunology, Drexel University, College of Medicine, Philadelphia, PA, were batch-cultured at 30 °C, with orbital agitation (250 rpm) in BM medium, with the following composition (per liter): 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco), 20 g glucose (Merck), and 2.65 g (NH4)2SO4 (Merck). C. glabrata strain L5U1 (cgura3Δ0, cgleu2Δ0), kindly provided by John Bennett, (26) from the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, was grown in BM medium supplemented with 20 mg/l uracil and 60 mg/l leucine. Solid media contained, besides the above-indicated ingredients, 20 g/l agar (Iberagar, Coina, Portugal). The plasmid pGREG576 was obtained from the Drag & Drop collection.
Membrane Proteome-Wide Analysis of C. glabrata Response to Clotrimazole
Wild-type 66032 C. glabrata strain and the derived 66032_Δcgpdr1 deletion mutant were cultivated in liquid BM medium at 30 °C with orbital agitation (250 rpm) in the absence of stress until the standardized culture OD600 nm (optical density at 600 nm) of 0.8 ± 0.08 was reached. Cells were then transferred to fresh medium in the absence of stress (control conditions) or in the presence of 100 mg/l clotrimazole, with an initial OD600 nm of 0.4 ± 0.05. Upon 1 h of cultivation, cells were harvested by centrifugation and resuspended in A Buffer (50 mm Tris, pH 7.5, with 0.5 mm EDTA and 20% glycerol), with protease inhibitors (10 mg/l leupeptine, 1 mg/l pepstatine A, 20 mg/l aprotinine, 2 mg/l trypsin/quimotrypsin inhibitor, 1.5 mg/l benzamindine, and 1 mm phenylmethylsulfonyl fluoride (all obtained from Sigma). Cell lysis was accomplished by consecutive steps of vortexing and cooling in the presence of glass beads. The mixture was centrifuged for clarification (8000 rpm, 5 min, 4 °C) and the top phase collected. A Buffer was added to a final volume of 8 ml, the mix was ultracentrifuged on a Beckman XL-90 ultracentrifuge (24,000 rpm, 90 min, 4 °C), and the pellet washed with 8 ml of 0,1 M Na2CO3, and incubated on ice for 30 min with orbital agitation (60 rpm). The mixture was then ultracentrifuged (26,000 rpm, 60 min, 4 °C), and the pellet was washed with 8 ml of 50 mm tetraethylammonium bromide (Sigma) and ultracentrifuged again (26,000 rpm, 60 min, 4 °C). This procedure was repeated two more times and, finally, the pellet was resuspended in 325 μl of 50 mm tetraethylammonium bromide with 8 m urea (Sigma).
Expression proteomics analysis of the obtained membrane-enriched fraction was carried out using and iTRAQ-MS procedure as a paid service at the Keck Foundation Biotechnology Resource Laboratory, Yale University (http://medicine.yale.edu/keck/proteomics/index.aspx). Briefly, samples were sonicated and proteins reduced by adding 50 mm tris(2-carboxyethyl)phosphine, followed by 200 mm methyl methane thiosulfonate. Protein digestion was achieved by adding 10 μl of a solution of 1 mg/ml Lys-C, followed by incubation at 37 °C for 3 h, and 10 μl of 1 mg/ml trypsin, followed by overnight incubation at 37 °C. Macrospin desalt of the digests with C18 spin columns for cleanup and quantitation was carried out, followed by dissolution in 65 μl of 500 mm tetraethylammonium bromide. iTRAQ labeling was carried out based on the AAA quant protocol. iTRAQ experiments were carried out through the SCX cartridge and experiments run on 5,600.
The search parameters and acceptance criteria used were the following: Peaklist generating software: ProteinPilot 4.5 and Mascot; search engine: Paragon Search Engine (ProteinPilot 4.2); sequence database/spectral library: Candida glabrata [5,478] from SwissProt (May 2013); The database used was downloaded from UniProt, with a total of 5,197 protein entries. Mass spectrometric analysis was done on an AB SCIEX TripleTOF® 5600 mass spectrometer with AB SCIEX ProteinPilot™ software used for protein identification and quantitation. ProteinPilot utilizes a Paragon™ algorithm with hybrid sequence tag and features probability database searches. Hence, specific details such as mass tolerances, specific modifications, etc. are not utilized. All iTRAQ results are uploaded into the Yale Protein Expression database for investigator viewing. Protein identification was considered reliable for a protein score > 2, corresponding to a confidence level of 99%. A reserve decoy database search, followed by filtering of all peptides above 1% false discovery rate was carried out before protein grouping.
Proteomics data analysis started from three iTRAQ sets (complete raw data are supplied as Tables S1-S3, corresponding to each iTRAQ set). The samples present in each of the sets were randomized to prevent bias, and in different sets, distinct labels were used to tag the samples, ensuring that protein identification in the MS step is not biased by the tags. For each sample in a given set, protein quantification was only considered for p value<.05. Protein expression changes above 1,4-fold or below 0,71-fold were considered relevant. Protein classification into functional groups was achieved based on their predicted function, according to the Candida Genome Database (www.candidagenome.org) or based on the function of their closest S. cerevisiae homolog, according to the Saccharomyces Genome Database (www.yeastgenome.org).
β-1,3-glucanase Susceptibility Assay
To monitor structural changes at the cell wall level, a lyticase (β-1,3-glucanase, Sigma) susceptibility assay was conducted as described before (27). C. glabrata KUE100 and KUE100_Δcggas1 cells were grown in BM medium, in the presence of 60 mg/l of clotrimazole, and harvested following 0 or 30 min of cell incubation, during the period of early adaptation to stress and at the exponential growth phase, when the standardized OD600 nm of 1.0 ± 0.1 was attained. The harvested cells were washed with distilled water and resuspended in 0.1 mm sodium phosphate buffer (pH 7). After the addition of 10 μg/ml lyticase, cell lysis was monitored by measuring the percentage decrease of the initial OD600 nm of the cell suspensions every 30 min for a total period of 180 min. Statistical analyses of the results were performed using analysis of variance, and differences were considered significant for p values < .05.
Disruption of the CgGAS1, CgTPO1_1, and CgTPO1_2 Genes
The deletion of the C. glabrata GAS1, TPO1_1, and TPO1_2 genes (ORFs CAGL0G00286g, CAGL0G03927g and CAGL0E03674g, respectively) was carried out in the parental strain KUE100, using the method described by Ueno et al. (28). The target genes CgGAS1, CgTPO1_1, and CgTPO1_2 were replaced by a DNA cassette including the CgHIS3 gene through homologous recombination. The replacement cassette was prepared by PCR using the primers 5′-AGCTGTATCAAACAACTCACTGTATCAATCACTATTTTACTATAACTAGATCAATAGGCCGCTGATCACG-3′ and 5′-CCCGTTGATCATATGAACATTAGGATTCATCAAATATTTAAATCATTCTGAAATTACATCGTGAGGCTGG-3 for the CgGAS1 gene, the primers 5′-GTTTTATCATTCGGTAGTCAACTGAATAAAAAAAATATATACATACATACAAAACGGGCCGCTGATCACG-3′ and 5′-TTGATTCTCTCTTTTAAAGGAAGAGTAGGAATCAGGATGTGGTCGCTTGAAGCTTACATCGTGAGGCTGG-3, for the CgTPO1_1 gene, and the primers 5′-CCAGTGCAGGAGAAGTAACAGCATATAATTCATCTCACGATAAGGAAGTTGGAGTGGGCCGCTGATCACG-3′ and 5′-AAAAGAGGGCACCAATTTCGTTAAGATATTATTAGTTTTTATTCTTTTTGGATTTACATCGTGAGGCTGG-3 for the CgTPO1_2 gene. The pHIS906 plasmid including CgHIS3 was used as a template and transformation was performed as described previously (25). Recombination locus and gene deletion were verified by PCR using the following pairs of primers: 5′-GTCTGGTTTCTTTCATAATAGC-3′ and 5′-TTGGAGTAGTTAGCGAAC-3′; 5′-CGCCAAAGTATACCAATG-3′ and 5′-CGGTGTTCCACAAATCTGT-3′; and 5′-GGCTCATCCATCTTCGCT-3′ and 5′-AGCGAAGATGGATGAGCC-3′, respectively.
For the disruption of CgTPO1_2 (ORF CAGL0E03674g) in the Δcggas1 derivative, the SAT1 cassette, encoding a nourseothricin selection marker was used for homologous recombination. The SAT1 cassette was obtained by PCR, using the pA83 plasmid as template and the following specific primers: 5′-GATCCCAGTGCAGGAGAAGTAACAGCATATAATTCATCTCACGATAAGGAAGTTGGAGTGATGGACGGTGGTATGTTTTA-3′ and 5′-ATATAACTCTAGTCCCATTTTTTAATTTGGAAGTTGGGATATCGAGATAAGGATAGGTGCTTAGGCGTCATCCTGTGCTC-3′. The designed primers contain, besides a region with homology to the first and last 20 nucleotides of the SAT1 coding region (italic), 60 nucleotide sequences with homology to the upstream and downstream regions of CgTPO1_2 (bold). The successful recombination of the SAT1 cassette was verified by PCR using the following specific primers: 5′-TTTGCTGCTTCGCCAGTTAT-3′ and 5′-TGTAAATGCACCTGCAATGG-3′. The designed primers are homologous to a sequence in the upstream region of CgTPO1_2 and to a sequence in the coding region of the SAT1 cassette, respectively.
Cloning of the C. glabrata CgTPO1_1 and CgTPO1_2 Genes (ORFs CAGL0G03927g and CAGL0E03674g, Respectively)
The pGREG576 plasmid from the Drag & Drop collection was used to clone and express the C. glabrata ORFs CAGL0G03927g and CAGL0E03674g in S. cerevisiae, as described before for other heterologous genes (29–31). pGREG576 was acquired from Euroscarf and contains a galactose-inducible promoter (GAL1), the yeast selectable marker URA3, and the GFP gene, encoding a green fluorescent protein (GFPS65T), which allows monitoring of the expression and subcellular localization of the cloned fusion protein. CAGL0G03927g or CAGL0E03674g DNA was generated by PCR, using genomic DNA extracted from the sequenced CBS138 C. glabrata strain, and the following specific primers: 5′-GAATTCGATATCAAGCTTATCGATACCGTCGACAATGGTGGAAGAGATATCGCC-3′ and 5′-GCGTGACATAACTAATTACATGACTCGAGGTCGACTTAAGCGTAGTAAGCATCC-3′; or 5′-GAATTCGATATCAAGCTTATCGATACCGTCGACAATGTCCTCCACTAGTAGCG-3′ and 5′-GCGTGACATAACTAATTACATGACTCGAGGTCGACTTATAACGAATATGCGTAC-3′, respectively. The designed primers contain, besides a region with homology to the first 20 and last 19 nucleotides of the CAGL0G03927g coding region (italic) and the first and last 19 nucleotides of the CAGL0E03674g coding region (italic), nucleotide sequences with homology to the cloning site flanking regions of the pGREG576 vector (underlined). The amplified fragments were cotransformed into the parental S. cerevisiae strain BY4741 with the pGREG576 vector, previously cut with the restriction enzyme SalI, to obtain the pGREG576_CgTPO1_1 or pGREG576_CgTPO1_2 plasmids. Since the GAL1 promoter only allows a slight expression of downstream genes in C. glabrata, to visualize by fluorescence microscopy the subcellular localization of the CgTpo1_1 or CgTpo1_2 proteins in C. glabrata, new constructs were obtained. The GAL1 promoter present in the pGREG576_CgTPO1_1 and pGREG576_CgTPO1_2 plasmids was replaced by the copper-induced MTI C. glabrata promoter, giving rise to the pGREG576_MTI_CgTPO1_1 and pGREG576_MTI_CgTPO1_2 plasmids. The MTI promoter DNA was generated by PCR, using genomic DNA extracted from the sequenced CBS138 C. glabrata strain, and the following specific primers: 5′-TTAACCCTCACTAAAGGGAACAAAAGCTGGAGCTCTGTACGACACGCATCATGTGGCAATC-3′ and 5′-GAAAAGTTCTTCTCCTTTACTCATACTAGTGCGGCTGTGTTTGTTTTTGTATGTGTTTGTTG-3′. The designed primers contain, besides a region with homology to the first 26 and last 27 nucleotides of the first 1,000 bp of the MTI promoter region (italic), nucleotide sequences with homology to the cloning site flanking regions of the pGREG576 vector (underlined). The amplified fragment was cotransformed into the parental strain BY4741 with the pGREG576_CgTPO1_1 or pGREG576_CgTPO1_2 plasmids, previously cut with SacI and NotI restriction enzymes to remove the GAL1 promoter, to generate the pGREG576_MTI_CgTPO1_1, and pGREG576_MTI_CgTPO1_2 plasmids. The recombinant plasmids pGREG576_CgTPO1_1, pGREG576_CgTPO1_2, pGREG576_MTI_CgTPO1_1, and pGREG576_MTI_CgTPO1_2 were obtained through homologous recombination in S. cerevisiae and verified by DNA sequencing.
CgTpo1_1 and CgTpo1_2 Subcellular Localization Assessment
The subcellular localization of the CgTpo1_1 and CgTpo1_2 proteins was determined based on the observation of BY4741 S. cerevisiae or L5U1 C. glabrata cells transformed with the pGREG576-CgTPO1_1 and pGREG576-CgTPO1_2 or pGREG576-MTI-CgTPO1_1 and pGREG576-MTI-CgTPO1_2 plasmids, respectively. These cells express the CgTpo1_1_GFP or CgTpo1_2_GFP fusion proteins, whose localization may be determined using fluorescence microscopy. S. cerevisiae cell suspensions were prepared by cultivation in MM4-U medium, containing 0.5% glucose and 0.1% galactose, at 30 °C, with orbital shaking (250 rpm) until a standard culture OD600 nm = 0.4 ± 0.04 was reached. At this point, cells were transferred to the same medium containing 0.1% glucose and 1% galactose, to induce protein expression. C. glabrata cell suspensions were prepared in BM-U medium, until a standard culture OD600 nm = 0.4 ± 0.04 was reached, and transferred to the same medium supplemented with 50 μm CuSO4 (Sigma), to induce protein overexpression. After 5 h of incubation, the distribution of CgTpo1_1_GFP or CgTpo1_2_GFP fusion proteins in S. cerevisiae or in C. glabrata living cells was detected by fluorescence microscopy in a Zeiss Axioplan microscope (Carl Zeiss MicroImaging, Oberkochen, Germany), using excitation and emission wavelength of 395 and 509 nm, respectively. Fluorescence images were captured using a cooled CCD camera (Cool SNAPFX, Roper Scientific Photometrics, Sarasota, FL).
Antifungal Susceptibility Assays in C. glabrata
The susceptibility of the parental strain KUE100 toward toxic concentrations of the selected drugs was compared with that of the deletion mutants KUE100_Δcggas1, KUE100_Δcgtpo1_1, KUE100_Δcgtpo1_2, and KUE100_Δcgtpo1_2_Δcggas1 by spot assays or by comparing growth in liquid medium. The ability of CgTPO1_1 and CgTPO1_2 gene expression to increase wild-type resistance to the tested chemical stresses was also examined in the URA3- strain L5U1 C. glabrata strain, using the pGREG576_MTI_CgTPO1_1 and pGREG576_MTI_CgTPO1_2 centromeric plasmids.
KUE100 C. glabrata cell suspensions used to inoculate the agar plates or liquid medium were midexponential cells grown in basal BM medium, until culture OD600 nm = 0.4 ± 0.02 was reached and then diluted in sterile water to obtain suspensions with OD600 nm = 0.05 ± 0.005. These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 μl spots onto the surface of solid BM medium, supplemented with adequate chemical stress concentrations. L5U1 C. glabrata cell suspensions used to inoculate the agar plates were midexponential cells grown in BM medium, supplemented with 50 μm CuSO4 (Sigma), to induce protein overexpression, without uracil when using the L5U1 strain harboring the pGREG576-derived plasmids, until culture OD600 nm = 0.4 ± 0.02 was reached, and then diluted in sterile water to obtain suspensions with OD600 nm = 0.05 ± 0.005. These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 μl spots onto the surface of solid BM medium, without uracil for strains transformed with the pGREG576-derived plasmids, supplemented with 50 μm CuSO4 and with adequate chemical stress concentrations. The tested drugs included the following compounds, used in the specified concentration ranges: the azole antifungal drugs ketoconazole (10 to 60 mg/l), fluconazole (20 to 80 mg/l), miconazole (0.08 to 0.14 mg/l), tioconazole (0.2 to 0.9 mg/l), itraconazole (5 to 40 mg/l), and clotrimazole (2.5 to 10 mg/l), the polyene antifungal drug amphotericin B (0.12 to 0.19 mg/l), the fluoropyrimidine 5-flucytosine (0.010 to 0.017 mg/l), the pesticide mancozeb (0.5 to 2.5 mg/l), and the polyamines spermine (2 to 4.5 mm) and spermidine (3 mm to 5 mm) (all from Sigma).
Antifungal Susceptibility Assays in S. cerevisiae
The susceptibility of the parental strain BY4741 toward toxic concentrations of the selected drugs was compared with that of the deletion mutant BY4741_Δtpo1 by spot assays. The ability of CgTPO1_1 and CgTPO1_2 genes to increase wild-type resistance to the tested chemical stresses and to complement the susceptibility phenotype exhibited by the BY4741_Δtpo1 single deletion mutants was also examined, using the pGREG576_CgTPO1_1 and pGREG576_CgTPO1_2 centromeric plasmids through which the C. glabrata genes are expressed under the GAL1 promoter.
S. cerevisiae cell suspensions used to inoculate the agar plates were midexponential cells grown in MM4-U medium, containing 0.5% glucose and 0.1% galactose, until culture OD 600 nm = 0.4 ± 0.02 was reached, and then diluted in sterile water to obtain suspensions with OD600 nm = 0.05 ± 0.005. These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 μl spots onto the surface of solid MM4-U medium, containing 0.1% glucose and 1% galactose, supplemented with growth inhibitory chemical stress concentrations. The tested drugs and other xenobiotics included the following compounds, used in the specified concentration ranges: the polyamines spermidine (2 mm) and spermine (2.5 mm), and the fungicide mancozeb (1.25 mg/l).
[3H]-clotrimazole Accumulation Assays
3H-clotrimazole transport assays were carried out as described before (31). The internal accumulation of clotrimazole was determined by calculating the ratio between the radiolabeled clotrimazole measured within the yeast cells and in the external medium (Intracellular/Extracellular). The parental strain KUE100 and the mutant strains KUE100_Δcgtpo1_1 and KUE100_Δcgtpo1_2 were grown in BM medium until mid-exponential phase and harvested by filtration. Cells were washed and resuspended in BM medium, to obtain dense cell suspensions (OD600 nm = 0.5 ± 0.1, equivalent to ∼1.57 mg (dry weight) ml−1). Readily, 0.1 μm of 3H-clotrimazole (American Radiolabeled Chemicals, St. Louis, MO; 1mCi/ml) and 30 mg/l of unlabeled clotrimazole were added to the cell suspensions. Incubation proceeded for an additional period of 30 min. The intracellular accumulation of labeled clotrimazole was followed by filtering 200 μl of cell suspension, at adequate time intervals, through prewetted glass microfiber filters (Whatman GF/C, Pittsburgh, PA). The filters were washed with ice-cold TM buffer and the radioactivity measured in a Beckman LS 5000TD scintillation counter. Extracellular 3H-clotrimazole was estimated, by radioactivity assessment of 50 μl of the supernatant. Nonspecific 3H-clotrimazole adsorption to the filters and to the cells (less than 5% of the total radioactivity) was assessed and taken into consideration. To calculate the intracellular concentration of labeled clotrimazole, the internal cell volume (Vi) of the exponential cells, grown in the absence of drug and used for accumulation assays, was considered constant and equal to 2.5 μl (mg dry weight)-1 (32). Statistical analysis of the results were performed using analysis of variance, and differences were considered significant for p values < .05.
CgTPO1_1 and CgTPO1_2 Expression Measurements
The levels of CgTPO1_1 and CgTPO1_2 transcripts were assessed by real-time PCR. Synthesis of cDNA for real time RT-PCR experiments, from total RNA samples, was performed using the MultiscribeTM reverse transcriptase kit (Applied Biosystems, Carlsbad, CA) and the 7500 RT-PCR Thermal Cycler Block (Applied Biosystems), following the manufacturer's instructions. The quantity of cDNA for the following reactions was kept around 10 ng. The subsequent RT-PCR step was carried out using SYBR® Green reagents. Primers for the amplification of the CgTPO1_1, CgTPO1_2 and CgACT1 cDNA were designed using Primer Express Software (Applied Biosystems) and are 5′-CGCTGCTTCCCCAGTTATCT-3′ and 5′-CTAGCACACCACGTCTACCGTAA-3′; 5′-AGGACCCGCTCTATCGAAAAA-3′ and 5′-GCTGCGACTGCTGACTCAAC-3′; and 5′-AGAGCCGTCTTCCCTTCCAT-3′ and 5′-TTGACCCATACCGACCATGA-3′, respectively. The RT-PCR reaction was carried out using a thermal cycler block (7500 Real-Time PCR System, Applied Biosystems). Default parameters established by the manufacturer were used and fluorescence detected by the instrument and registered in an amplification plot (7500 System SDS Software, Applied Biosystems). The CgACT1 mRNA level was used as an internal control. The relative values obtained for the wild-type strain in control conditions were set as 1, and the remaining values are presented relative to that control. To avoid false positive signals, the absence of nonspecific amplification with the chosen primers was confirmed by the generation of a dissociation curve for each pair of primers. Statistical analysis of the results were performed using analysis of variance, and differences were considered significant for p values < .05.
RESULTS
Proteome-Wide Protein Identification in C. glabrata Membrane-Enriched Fraction
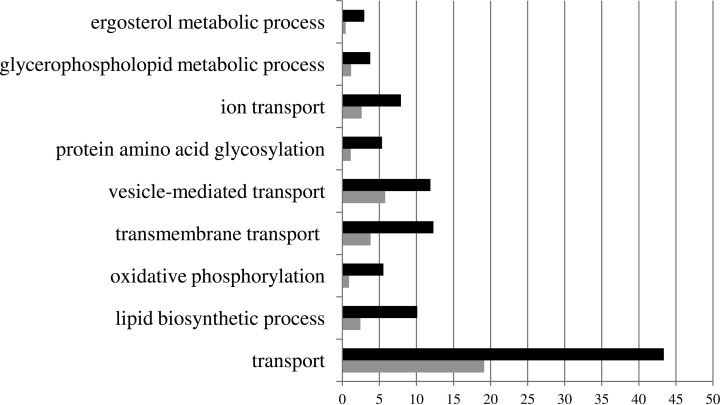
624 proteins were identified in the membrane-enriched fraction in C. glabrata, 131 encoded by characterized genes, while 493 are encoded by noncharacterized ORFs, comprising around 10% of the predicted C. glabrata proteome. To the best of our knowledge, this is the first membrane proteome-wide analysis carried out in C. glabrata, making this list of proteins an invaluable repository of information on the functional analysis of all of these 624 proteins. To obtain a global perspective of the functional distribution of these membrane-associated proteins, the online gene ontology term-based grouping tool GoToolBox (http://genome.crg.es/GOToolBox/) was used, considering their S. cerevisiae homologs. Using this tool, only 1.44% of the total proteins were found to have cytoplasm-localization associated gene ontology terms, attesting the high specificity of the applied approach for the yield of membrane proteins. Figure 1 highlights the most-enriched gene ontology terms associated to the C. glabrata membrane proteome. As expected, they are all related to transmembrane transport functions and membrane-associated metabolic processes and include proteins involved in the synthesis of ergosterol and phospholipids but also transmembrane transporters and proteins involved in cell trafficking. The whole list of positively identified membrane-associated proteins is available in Table S4. Details on the protein quantification can be assessed at the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository (http://massive.ucsd.edu/ProteoSAFe/datasets.jsp; MassIVE ID: MSV000079209). Annotated spectra for single peptide identification for each protein is provided in MS-viewer (http://prospector2.ucsf.edu/prospector/cgi-bin/msform.cgi?form = msviewer; search keys: qrvm65goix; xffxn22szm; tm0y9 × 0hqd).
Fig. 1.
Categorization, based on the biological process taxonomy of gene ontology, of the proteins identified in membrane-enriched fractions of C. glabrata cells. These genes were clustered using the GoToolBox software (http://genome.crg.es/GOToolBox/), and the most highly ranked statistically significant (p value<10−10) gene ontology terms are displayed. The protein frequency within each class is indicated by the black bars, compared with the frequency registered for the C. glabrata whole genome, indicated by the gray bars, gene frequency being the percentage of the genes in a list associated to a specific GO term.
Membrane Proteome-Wide Changes Occurring in Response to Clotrimazole in C. glabrata
The analysis of the membrane-enriched fraction of the C. glabrata proteome obtained from cells exposed to clotrimazole when compared with control conditions allowed the identification of 12 proteins whose content increases and 25 proteins whose expression decreases upon exposure to the drug. Since only four of these 37 proteins had been previously characterized, their clustering was carried out based on the role of their predicted S. cerevisiae homologs (Table I and Fig. 2). Among the obtained clusters, the most populated one is ribosome components and translational machinery, with two ribosomal proteins being up-regulated, while the remaining 20 are down-regulated. This observation appears consistent with a decreased translation rate, which has been considered part of the so-called “environmental stress response” (33). This feature is consistent with growth arrest elicited by sudden exposure to clotrimazole-induced stress.
Table I. Set of 37 proteins found to have significant expression changes in C. glabrata wild-type cells in the presence of clotrimazole and correspondent fold changes in Δcgpdr1 mutant cells upon exposure to the drug. Protein clustering was performed based on the role of their predicted S. cerevisiae homologs.
| C. glabrata protein (ORF) name | S. cerevisiae homolog | Description of the function of the C. glabrata protein or of its S. cerevisiae homolog | Wild-type fold change (upon clotrimazole stress) | Δcgpdr1 fold change (upon clotrimazole stress) |
|---|---|---|---|---|
| Glucose metabolism | ||||
| CAGL0L01485g | GSF2 | Uncharacterized. S. cerevisiae homolog encodes a ER localized integral membrane protein that may promote secretion of certain hexose transporters, including Gal2 | 2.26 | 2.30a |
| PGK1 (CAGL0L07722g) | PGK1 | Uncharacterized. S. cerevisiae homolog encodes a 3-phosphoglycerate kinase; key enzyme in glycolysis and gluconeogenesis | 6.31 | 4.93a |
| Oxidative phosphorylation | ||||
| CAGL0H05489g | ATP4 | Uncharacterized. S. cerevisiae homolog encodes a subunit b of the stator stalk of mitochondrial F1F0 ATP synthase | 1.68 | 1.49 |
| CAGL0F04565g | COR1 | Uncharacterized. S. cerevisiae homolog encodes a core subunit of the ubiquinol-cytochrome c reductase complex, a component of the mitochondrial inner membrane electron transport chain | 0.32 | 4.79 |
| RIP1 (CAGL0I03190g) | RIP1 | Uncharacterized. S. cerevisiae homolog encodes a ubiquinol-cytochrome-c reductase; a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex | 0.10 | 1.96a |
| CAGL0G10131g | QCR2 | Uncharacterized. S. cerevisiae homolog encodes a subunit 2 of the ubiquinol cytochrome-c reductase complex, a component of the mitochondrial inner membrane electron transport chain | 0.40 | 0.68 |
| CAGL0G10153g | QCR7 | Uncharacterized. S. cerevisiae homolog encodes a subunit 7 of the ubiquinol cytochrome-c reductase complex, a component of the mitochondrial inner membrane electron transport chain | 0.21 | 1.24b |
| Mitochondrial import | ||||
| CAGL0I10472g | PHB1 | Uncharacterized. S. cerevisiae homolog encodes a subunit of the prohibitin complex (Phb1p-Phb2p), a 1,2 MDa ring-shaped inner mitochondrial membrane chaperone that stabilizes newly synthesized proteins | 3.09 | 2.18a |
| CAGL0L12936g | TOM70 | Uncharacterized. S. cerevisiae homolog encodes a component of the TOM (translocase of outer membrane) complex; involved in the recognition and initial import steps for all mitochondrially directed proteins | 0.57 | 0.37 |
| Ribosome components and translation machinery | ||||
| CAGL0A03388g | RPL15B | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L13B; not essential for viability | 2.22 | 1.78a |
| CAGL0E02013g | RPL18A | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 2.21 | 2.17 |
| CAGL0E03938g | RPL8B | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L8B | 0.16 | 0.13 |
| CAGL0F07073g | RPS2 | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) subunit | 0.45 | 0.64 |
| CAGL0F09031g | RPS4A | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.47 | 0.53 |
| CAGL0G00990g | RPP0 | Uncharacterized. S. cerevisiae homolog encodes a conserved ribosomal protein P0 of the ribosomal stalk | 0.59 | 1.03a |
| CAGL0G01078g | RPL26A | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L33B | 0.22 | 0.11 |
| CAGL0G06490g | RPS7A | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.22 | 0.36 |
| CAGL0H00462g | RPS5 | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.55 | 0.84b |
| CAGL0J03234g | RPS24B | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.33 | 0.31 |
| CAGL0K06567g | RPL27A | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L27A | 0.13 | 0.24 |
| CAGL0K07414g | RPL20B | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L20A | 0.36 | 0.33a |
| CAGL0K11748g | RPS11A | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.47 | 0.60 |
| CAGL0L08114g | RPS22A | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.52 | 0.40a |
| CAGL0L12870g | TMA19 | Uncharacterized. S. cerevisiae homolog encodes a protein that associates with ribosomes | 0.26 | 3.24a |
| CAGL0M02695g | RPL5 | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L5 | 0.45 | 0.58 |
| EFT2 (CAGL0A03234g) | EFT1 | Uncharacterized. S. cerevisiae homolog encodes an elongation factor 2 | 0.43 | 0.36 |
| CAGL0H08976g | RPL1A | Uncharacterized. S. cerevisiae homolog encodes a ribosomal 60S subunit protein L15A | 0.41 | 0.50a |
| CAGL0H03773g | RPL7 | Uncharacterized. S. cerevisiae homolog encodes a nucleolar protein with similarity to large ribosomal subunit L7 proteins; constituent of 66S pre-ribosomal particles | 0.36 | 0.47a |
| CAGL0I00792g | RPS16A | Uncharacterized. S. cerevisiae homolog encodes a protein component of the small (40S) ribosomal subunit | 0.20 | 0.31 |
| SSB1 (CAGL0C05379g) | SSB2 | Uncharacterized. S. cerevisiae homolog encodes a cytoplasmic ATPase that is a ribosome-associated molecular chaperone | 0.58 | 0.51a |
| TEF3 (CAGL0B03487g) | TEF3 | Uncharacterized. S. cerevisiae homolog encodes a gamma subunit of translational elongation factor eEF1B | 0.58 | 0.77 |
| Lipid and cell wall metabolism | ||||
| HFD1 (CAGL0K03509g) | HFD1 | Uncharacterized. Gene is upregulated in azole-resistant strain. S. cerevisiae homolog encodes a hexadecenal dehydrogenase | 6.74 | 1.19b |
| GAS1 (CAGL0G00286g) | GAS1 | Putative glycoside hydrolase of the Gas/Phr family; predicted GPI-anchor; S. cerevisiae homolog encodes a beta-1,3-glucanosyltransferase, required for cell wall assembly | 3.92 | 1.61a |
| Multidrug resistance transporters | ||||
| CAGL0E03674g | TPO1 | Uncharacterized. S. cerevisiae homolog encodes a polyamine transporter of drug:H(+) antiporter DHA1 family | 1.44 | 1.57 |
| CgQDR2 (CAGL0G08624g) | QDR2 | Drug:H+ antiporter of the Major Facilitator Superfamily, confers imidazole drug resistance; activated by Pdr1p and in azole-resistant strain | 4.29 | 3.11 |
| CgSNQ2 (CAGL0I04862g) | SNQ2 | Plasma membrane ATP-binding cassette (ABC) transporter; involved in Pdr1p-mediated azole resistance | 2.78 | 0.72b |
| CgCDR1 (CAGL0M01760g) | PDR5 | Multidrug transporter of ATP-binding cassette (ABC) superfamily; involved in Pdr1p-mediated azole resistance; increased abundance in azole resistant strains | 6.40 | 0.20 |
a Fold change quantification considered as not reliable (p value>.05).
b Fold change value outside of the chosen cut-offintervals (0.71 < fold change < 1.4).
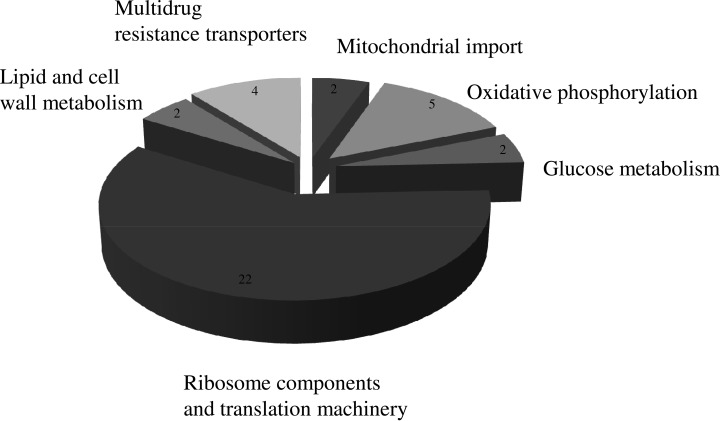
Fig. 2.
Major functional groups found to have significant expression changes in the membrane-enriched proteome upon exposure to clotrimazole in C. glabrata. Proteins with significant expression changes include ribosome components and translation machinery (22 proteins), lipid and cell wall metabolism (two proteins), multidrug resistance transporters (four proteins), mitochondrial import (two proteins), oxidative phosphorylation (five proteins), and glucose metabolism (two proteins).
Within the remaining classes, three are related to carbon and energy metabolism, comprising a total of nine proteins: glucose metabolism, oxidative phosphorylation, and mitochondrial import. These nine proteins include CgGsf2, an endoplasmic reticulum protein predicted to promote the traffic of hexose transporters.
All 4 proteins present in the multidrug resistance transporters are up-regulated upon exposure to clotrimazole. This is consistent with the characterization of several of these proteins as efflux pumps, despite a role in specific clotrimazole transport had only been described for CgQdr2 (31). The most up-regulated protein in this cluster is CgCdr1, widely characterized as an important ATP-binding cassette transporter. Accordingly, its S. cerevisiae homolog, ScPdr5, was previously found to be involved in clotrimazole extrusion (34). Additionally, CgSnq2 has been characterized as a multidrug transporter (35), as well as its S. cerevisiae homolog (36). The up-regulation of CgSnq2 observed in this study predicts a possible role for this transporter in clotrimazole extrusion. CgTpo1_2 is the only protein from this cluster uncharacterized in C. glabrata, presenting here a slight up-regulation upon clotrimazole exposure. Its S. cerevisiae homolog is known to confer resistance to spermine, putrescine, and spermidine; catalyzing the extrusion of polyamines in S. cerevisiae (18, 19). The up-regulation of CgTpo1_2 in clotrimazole-exposed C. glabrata cells raises the possibility that this predicted MDR transporter plays a role in imidazole transport in C. glabrata. This hypothesis will be addressed in this study.
The remaining cluster harbors two proteins related with lipid and cell wall metabolism. The putative cell wall remodeling protein CgGas1, up-regulated in this cluster, presumes some level of cell wall response against clotrimazole stress.
Effect of CgPdr1 Deletion in the Membrane Proteome-Wide Changes Occurring in Response to Clotrimazole in C. glabrata
The analysis of the membrane-enriched fraction of the C. glabrata proteome obtained from cells exposed to clotrimazole in the absence of the transcription factor CgPdr1 was assessed and compared with that of the C. glabrata wild-type cells exposed to clotrimazole. Among the 37 proteins whose expression was seen to change in the wild-type strain, six proteins were found to be repressed by CgPdr1, possibly in an indirect fashion, while six proteins were found to be activated by CgPdr1 (Table I). For the remaining 25 proteins, no statistically significant change could be detected in the current experiment.
Particularly interesting in this context are the six proteins that were found to be induced by clotrimazole in the dependence of CgPdr1: the multidrug transporters CgSnq2 and CgCdr1; the hexadecenal dehydrogenase Hfd1; the ribosomal protein Rpl26A; the component of the translocase of outer membrane complex Tom70; and the β-1,3-glucanosyltransferase Gas1. Interestingly, at least one CgPdr1-binding site is found in the promoter regions of the first four genes, suggesting that the action of CgPdr1 in their expression may be direct. These results are consistent with the characterization of several of these proteins as efflux pumps and of their expression to be dependent on CgPdr1 in response to other chemical stress inducers (30, 31, 35, 37). The expression changes here observed reinforce CgPdr1 as a major pleiotropic drug resistance mediator and highlight its role in mediating the expression of multidrug transporters in response to clotrimazole. The observation regarding the lipid metabolism related protein CgHfd1 is consistent with previous microarray studies, reporting the activation of CgHfd1 upon exposure to fluconazole induced stress in the dependence of CgPdr1 (9). It would be interesting to assess whether this result may relate to the imidazole mode of action on lipid raft binding, with Hfd1 possibly intervening in plasma membrane lipid destabilization as a resistance mechanism dependent on CgPdr1.
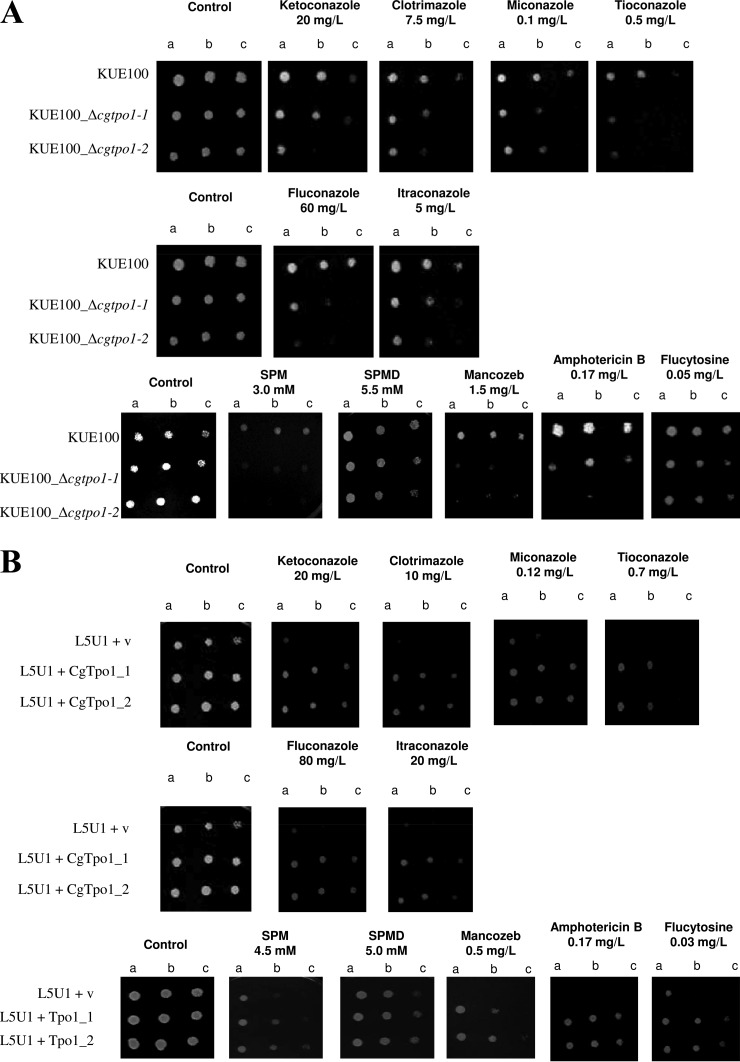
CgTpo1_1 and CgTpo1_2 Expression Confer Resistance to Azoles and Other Chemical Stress Inducers
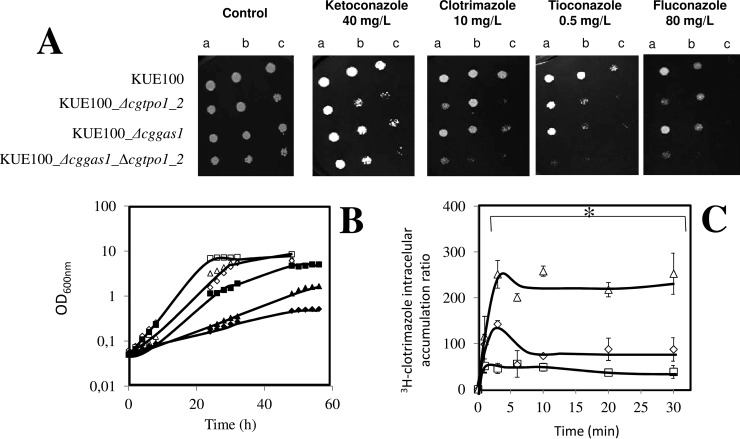
The deletion of CgTPO1_1 and especially CgTPO1_2 in C. glabrata was found to increase the susceptibility of this pathogen to clotrimazole but also to other imidazoles such as miconazole, ketoconazole, and tioconazole; as well as triazoles such as itraconazole and fluconazole (Fig. 3A). Additionally, an effect in susceptibility to other antifungal drug families was observed, namely to the polyene amphotericin B, the pyrimide analog flucytosine, the fungicide mancozeb, and to the polyamine spermine (Fig. 3A). As determined by spot assays, the wild-type strain KUE100 is capable of growing in the tested concentrations, while the Δcgtpo1_1 and especially Δcgtpo1_2 mutants display reduced growth when compared with wild type, suggesting a role of CgTPO1_1 and CgTPO1_2 as azole drug resistance determinants in C. glabrata. The overexpression of CgTPO1_1 or CgTPO1_2 in a wild-type strain was concordantly found to increase C. glabrata natural resistance toward the tested antifungal drugs (Fig. 3B). Consistent with the attributed role of S. cerevisiae Tpo1 in polyamine (18) and mancozeb (21) resistance, CgTPO1_1 or CgTPO1_2 expression was also found to increase C. glabrata resistance to the polyamine spermine and the fungicide mancozeb (Figs. 3A and 3B).
Fig. 3.
CgTpo1_1 and CgTpo1_2 confer resistance to azole antifungal drugs in C. glabrata cells. (A) Comparison of the susceptibility to inhibitory concentrations of several chemical stress inducers, at the indicated concentrations, of the C. glabrata KUE100, KUE100_Δcgtpo1_1 and KUE100_Δcgtpo1_2 strains, in YPD agar plates (SPM - spermine and SPMD - spermidine) and BM plates (remaining drugs) by spot assays. (B) Comparison of the susceptibility to several drug stress inducers, at the indicated concentrations, of the C. glabrata L5U1 strain, harboring the pGREG675 cloning vector (v) or the pGREG576_MTI_CgTPO1_1 or pGREG576_MTI_CgTPO1_2 plasmids in YPD agar plates (SPM and SPMD) and BM agar plates (remaining drugs), without uracil, by spot assays. (C) Comparison of the vector pGREG576 (v) or the derived CgTPO1_1 or CgTPO1_2 expression plasmids pGREG576_CgTPO1_1 or pGREG576_CgTPO1_2, on MM4 agar plates by spot assays. The inocula were prepared as described under “Experimental Procedures.” Cell suspensions used to prepare the spots were 1:5 (B) and 1:25 dilutions of the cell suspension used in (A). The displayed images are representative of at least three independent experiments.
Using S. cerevisiae has an heterologous expression system, the effect of CgTPO1_1 and CgTPO1_2 expression on yeast resistance to polyamines and mancozeb was further tested in order to assess if their genes are able to complement their S. cerevisiae homolog. The deletion of the S. cerevisiae TPO1 gene was found to increase the susceptibility to the polyamine spermine and the fungicide mancozeb. When expressed in the Δtpo1 S. cerevisiae background, the CgTPO1_1 and CgTPO1_2 genes were able to rescue the observed susceptibility phenotype to spermine and mancozeb (data not shown).
CgTpo1_1 and CgTpo1_2 Are Localized to the Plasma Membrane in C. glabrata and when Heterologously Expressed in S. cerevisiae
C. glabrata cells harboring the pGREG576_MTI_CgTPO1_1 and pGREG576_MTI_CgTPO1_2 plasmids were grown to midexponential phase in minimal medium and then transferred to the same medium containing 50 μm CuSO4 to induce fusion protein expression. At a standard OD600 nm of 0.5 ± 0.05, obtained after 5 h of incubation, cells were inspected by fluorescence microscopy. In C. glabrata cells, the CgTpo1_1_GFP and CgTpo1_2_GFP fusion proteins were found to be localized to the cell periphery (Fig. 4). In a similar approach, S. cerevisiae cells harboring the pGREG576_CgTPO1_1 and pGREG576_CgTPO1_2 plasmids were grown to mid-exponential phase in minimal medium containing 0.5% glucose and 0.1% galactose and then transferred to the same medium containing 0.1% glucose and 1% galactose, to promote protein overexpression. At a standard OD600 nm of 0.5 ± 0.05, cells were analyzed by fluorescence microscopy and the fusion proteins were found to be localized to the cell periphery (Fig. 4). These results strongly suggest plasma membrane localization for both CgTpo1_1 and CgTpo1_2, similar to that observed for their S. cerevisiae homolog Tpo1.
Fig. 4.
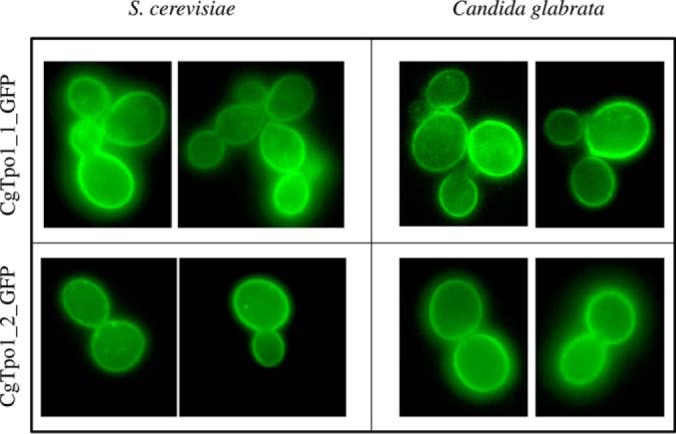
Fluorescence of exponential phase BY4741 S. cerevisiae and L5U1 C. glabrata cells, harboring the expression plasmids pGREG576_CgTPO1_1 and pGREG576_CgTPO1_2 or pGREG576_MTI_CgTPO1_1 and pGREG576_MTI_CgTPO1_2, after galactose or copper-induced recombinant protein production, respectively.
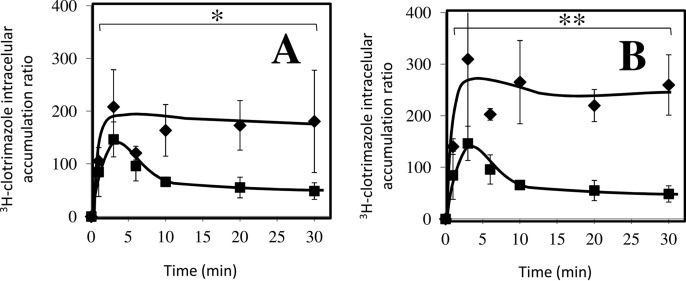
CgTpo1_1 and CgTpo1_2 Mediate 3H-clotrimazole Efflux in C. glabrata
Based on the identification of CgTpo1_1 and CgTpo1_2 as plasma membrane MDR transporters conferring resistance to azole drugs, their possible involvement in reducing clotrimazole accumulation in stressed yeast cells was examined. Under these conditions, the deletion of the CgTPO1_1 gene, and especially the deletion of the CgTPO1_2 leads to a very significant decrease in the exponential growth rate when compared with the parental strain (Fig. 5). 3H-clotrimazole accumulation assays were carried out in the absence or presence of the encoding genes. Consistent with the observed susceptibility phenotypes, Δcgtpo1_1 and Δcgtpo1_2 deletion mutants were found to accumulate four- and fivefold more radiolabeled clotrimazole than the corresponding parental KUE100 strain, respectively (Figs. 6A and 6B). These results strongly suggest that CgTpo1_1 and CgTpo1_2 activities increase C. glabrata resistance toward clotrimazole by reducing its accumulation within yeast cells.
Fig. 5.
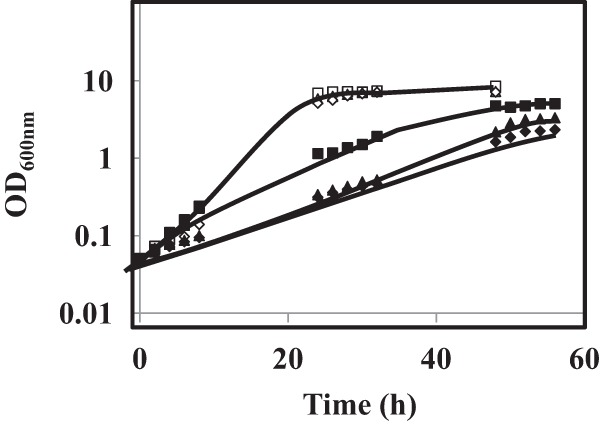
Comparison of growth curves of C. glabrata KUE100 (■,□), KUE100_Δcgtpo1_1 (▴,▵) and KUE100_Δcgtpo1_2 (♦,♢) cell populations, in liquid BM medium, in the absence (open symbols) or presence of 90 mg/l clotrimazole (filled symbols), measured in terms of variation in OD600. The displayed growth curves are representative of at least three independent experiments.
Fig. 6.
Time-course accumulation of radiolabeled 3H-clotrimazole in strains KUE100 (■) wild-type and KUE100_Δcgtpo1_1 (♦) (A) and KUE100 (■) and KUE100_Δcgtpo1_2 (♦) (B), during cultivation in BM liquid medium in the presence of 30 mg/L unlabeled clotrimazole. Accumulation values are the average of at least three independent experiments. Error bars represent the corresponding standard deviations. *p < .05. **p < .01.
CgTPO1_1 and CgTPO1_2 Transcript Levels Are Up-Regulated under Clotrimazole Stress
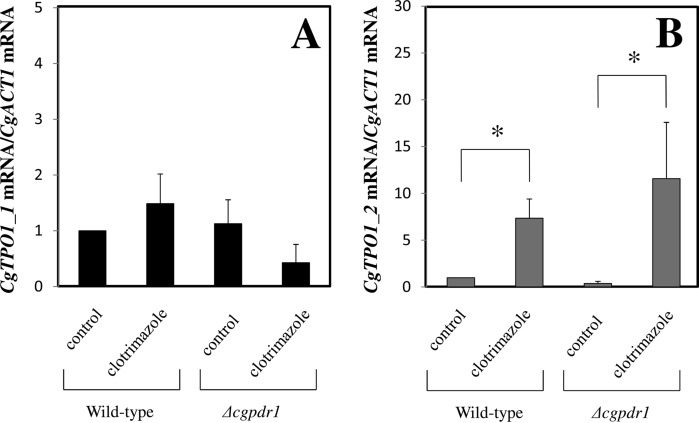
The effect of C. glabrata cell challenge with inhibitory concentrations of clotrimazole in CgTPO1_1 and CgTPO1_2 transcript levels was evaluated. The transcript levels of CgTPO1_1 were seen to have no significant change upon 1h of exposure to inhibitory concentrations of clotrimazole (Fig. 7A), whereas the transcript levels of CgTPO1_2 were found to have a sevenfold increase upon clotrimazole exposure (Fig. 7B). These results show CgTPO1_2 transcript levels to be responsive to clotrimazole exposure, consistent with the observed increase of CgTpo1_2 concentration in the membrane of clotrimazole-exposed C. glabrata cells (Table I). Expression values attained in Δcgpdr1 mutant cells show a slight decrease in the case of CgTPO1_1; however, this decrease was not found to be statistically relevant (Fig. 7A). On the other hand, the attained expression values confirm that CgPdr1 is not controlling the observed CgTPO1_2 up-regulation (Fig. 7B).
Fig. 7.
Comparison of the variation of the CgTPO1_1 (A) and CgTPO1_2 (B) transcript levels in the 66032u C. glabrata wild-type strain and in the derived 66032u_ Δcgpdr1 deletion mutant, before (control) and after 1 h of exposure 60 mg/l clotrimazole. The presented transcript levels were obtained by quantitative RT-PCR and are relative CgTPO1_1/CgACT1 or CgTPO1_2/CgACT1 mRNA,relative to the values registered in the 66032 parental strain in control conditions. The indicated values are averages of at least three independent experiments. Error bars represent the corresponding standard deviations. *p < .05.
CgGas1 Expression Confers resistance to Azoles
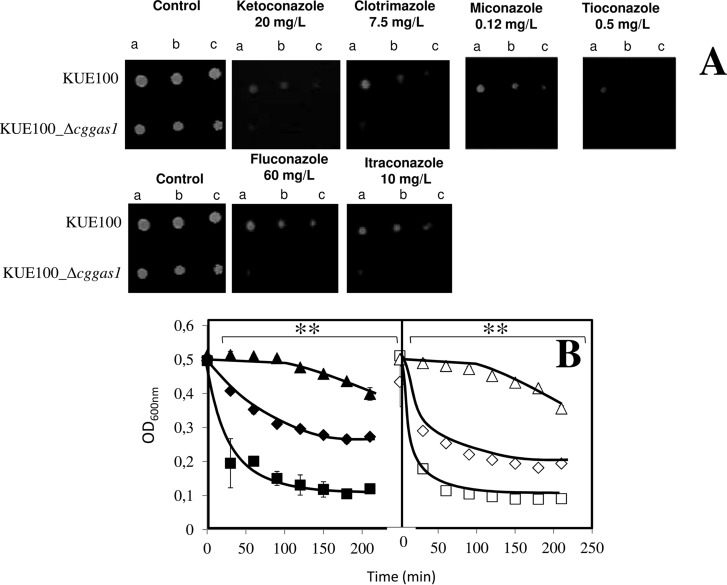
The deletion of CgGAS1 in C. glabrata was found, based on spot assays, to increase the susceptibility of this pathogen to several azole antifungal drugs, such as clotrimazole, miconazole, ketoconazole, tioconazole (imidazoles), and fluconazole and itraconazole (triazoles). The wild-type strain (KUE100) is capable of growing in the tested concentrations, while the Δcggas1 mutant displays very limited growth when compared with the wild type, therefore showing a higher degree of susceptibility toward the tested azole drugs (Fig. 8A).
Fig. 8.
CgGas1 confers resistance to azole antifungal drugs in C. glabrata cells. (A) Comparison of the susceptibility to azole antifungal drugs, at the indicated concentrations, of the C. glabrata KUE100 wild-type and KUE100_Δcggas1 strains, in BM agar plates by spot assays. The inocula were prepared as described under “Experimental Procedures.” Cell suspensions used to prepare the spots were 1:5 (B) and 1:25 (C) dilutions of the cell suspension used in (A). The displayed images are representative of at least three independent experiments. (B) Lyticase susceptibility in Candida glabrata KUE100 (♦,■,▴) and KUE100_Δcggas1 (♢,□,▵) cells harvested in the exponential phase of growth in the absence of stress (♦, ♢) or upon 30 min of exposure to 30 mg/l clotrimazole (■,□), or in the exponential phase of growth reached upon adaptation to 30 mg/l clotrimazole (▴,▵). After addition of 10 mg/l lyticase, the decrease in the OD600 nm of the cell suspension was measured periodically and indicated as a percentage of the initial OD600 nm. The indicated values are averages of at least three independent experiments. Error bars represent the corresponding standard deviations. **p < .01.
Response to Clotrimazole Includes Cell Wall Remodeling, Mostly Independently of CgGas1
A possible role of CgGas1 in cell wall resistance was assessed through the evaluation of lyticase susceptibility in yeast cells before and after adaptation to clotrimazole. The susceptibility to lyticase of exponential wild-type (KUE100) cells was seen to be lower than that exhibited by Δcggas1 deletion mutant cells (Fig. 8B) grown in the absence of clotrimazole. This result indicates that, in the absence of stress, the cell wall of Δcggas1 cells is more susceptible than that of wild-type cells. In wild-type or Δcggas1 cells, sudden exposure to clotrimazole during 30 min leads to similarly increased susceptibility to lyticase, showing that clotrimazole appears to have a drastic effect at the level of the cell wall structure. However, once adapted to exponential growth in the presence of clotrimazole, either wild-type or Δcggas1 cells exhibited levels of lyticase resistance that are even higher than those exhibited by nonstressed cells. This result suggests that adaptation to clotrimazole includes cell wall remodeling. Altogether, the lack of the CgGas1 putative cell wall assembly protein increases lyticase susceptibility at the level of cell wall in control conditions, eventually helping the C. glabrata cells to cope with sudden stress exposure. However, even in the absence of this protein cell wall remodeling taking place during adaptation to clotrimazole still takes place but at a slower rate than observed in the wild-type cell population.
CgGas1 Reduces the Intracellular Accumulation of 3H-clotrimazole in C. glabrata
Once the C. glabrata CgGas1 was identified as conferring resistance to azole drugs, its possible involvement in reducing clotrimazole accumulation in yeast cells was examined. The accumulation of radiolabeled clotrimazole in nonadapted C. glabrata cells suddenly exposed to 30 mg/l clotrimazole was seen to be ∼2 times higher in cells devoid of CgGas1 when compared with the KUE100 wild-type cells (Fig. 9C). These findings strongly suggest that CgGas1 contributes to C. glabrata tolerance toward clotrimazole also by reducing its accumulation within yeast cells. These results show CgGas1 to be an important factor for cell wall composition, apparently necessary for clotrimazole resistance by catalyzing glucan linkage and chain elongation, thus reducing drug diffusion through the cell wall and into the cytosol.
Fig. 9.
The combined action of CgTpo1_2 and CgGas1 confers resistance to azole antifungal drugs in C. glabrata cells. (A) Comparison of the susceptibility to azole antifungal drugs, at the indicated concentrations, of the C. glabrata KUE100 wild-type and KUE100_Δcggas1 strains, in BM agar plates by spot assays. The inocula were prepared as described under “Experimental Procedures.” Cell suspensions used to prepare the spots were 1:5 (B) and 1:25 (C) dilutions of the cell suspension used in (A). The displayed images are representative of at least three independent experiments. (B) Comparison of growth curves of C. glabrata KUE100 (■,□), KUE100_Δcggas1 (▴,Δ) and KUE100_Δcggas1_Δcgtpo1_2 (♦, ♢), in liquid BM, in the absence (open symbols) or presence of 90 mg/l clotrimazole (filled symbols), measured in terms of variation in OD600. The displayed growth curves are representative of at least three independent experiments. (C) Time-course accumulation of clotrimazole in KUE100 (), KUE100_Δcggas1 (224) and KUE100_Δcggas1_Δcgtpo1_2 (Δ) strains, during cultivation in BM liquid medium in the presence of radiolabeled 3H-clotrimazole. Accumulation values are the average of at least three independent experiments. Error bars represent the corresponding standard deviations. *p < .05.
Given that the deletion of the CgTPO1_2 gene was previously found to significantly increase the susceptibility of this pathogen to azole drugs as well, a double deletion mutant was constructed in order to assess if both genes contribute in a cumulative manner to azole drug resistance. The deletion of both genes in C. glabrata was found, based on spot assays and growth curves, to further increase the susceptibility to the antifungal drugs clotrimazole, miconazole, tioconazole (imidazoles), and fluconazole (triazole) when compared with the susceptibility attained for the correspondent single mutants, leading to a drastic decrease in the growth rate of C. glabrata cells in the presence of clotrimazole (Figs. 9A and 9B). The accumulation of radiolabeled clotrimazole was also tested in a KUE100_Δcggas1_Δcgtpo1_2 double mutant. The accumulation of radiolabeled clotrimazole in nonadapted C. glabrata cells suddenly exposed to 30 mg/l clotrimazole was seen to be ∼5 times higher in double mutant cells when compared with wild-type cells (Fig. 9C). However, the deletion of CgTPO1_2 in the Δcggas1 background increases the amount of clotrimazole accumulation in C. glabrata cells but only to levels close to the ones registered in the KUE100_Δcgtpo1_2 single mutant.
DISCUSSION
In this work, the first iTRAQ-based membrane proteomics study focused on the fungal pathogen C. glabrata was undertaken, leading to functional characterization of the C. glabrata CgTpo1_1 and CgTpo1_2 drug:H+ antiporters, and of the cell wall assembly protein CgGas1 in the context of clotrimazole drug resistance.
Using a membrane proteomics analysis, several proteins from distinct functional groups were found to be differentially expressed in C. glabrata clotrimazole response. Ribosomal proteins were among the down-regulated ones, in accordance with the environmental stress response described by Gasch et al. (33), in which ribosomal proteins have a stress-dependent repression as a mechanism to conserve mass and energy while redirecting transcription to genes whose expression is induced by stress. The up-regulated proteins encompass glucose metabolism, also in accordance with the predicted environmental stress response (33), and therefore were considered to be part of a general response. The more specific roles of CgCdr1 and CgSnq2 in clotrimazole response, registered in this study, could also eventually be predicted based on their known role in fluconazole adaptive response (38). Interestingly, in a distinct proteomics study using fluconazole-resistant C. glabrata strains and consistent with our work, several proteins involved in energy transfer and various metabolic pathways were identified (39). In the referred study, resistant strains have been described to exhibit several up-regulated membrane proteins, in contrast with the down-regulation verified for several intracellular proteins in response to the drug. These data reinforce the relevance for directed membrane fraction proteomics studies, such as the work presented herein, as these proteins can reveal important factors and mechanisms of azole drug resistance in this pathogenic yeast.
More interesting was the observation that the multidrug transporter CgTpo1_2 and the cell wall related protein CgGas1 appear to be implicated in clotrimazole response. So far, CgTpo3 was the only transporter from the Tpo1–4 group to be associated with azole drug resistance in C. glabrata (30). CgTpo1_1 and CgTpo1_2 (ORFs CAGL0G03927g and CAGL0E03674g, respectively) are described herein as the fourth and fifth members of the DHA1 family to be associated with azole drug resistance, after CgQdr2, CgAqr1, and CgTpo3 (29–31). Azole antifungal drugs, to which CgTpo1_1 and cgTpo1_2 confer resistance, were found to include the imidazoles clotrimazole, miconazole, tioconazole, and ketoconazole, used in the treatment of superficial skin and mucosal infections but also the triazoles itraconazole and fluconazole, used against systemic infections. Given the observation that clotrimazole accumulation is three to five times higher in the absence of CgTPO1_1 or CgTPO1_2 than observed in wild-type C. glabrata cells, the mechanism underlying the role of CgTpo1_1 and CgTpo1_2 in clotrimazole resistance appears to be direct transport of the drug. Nonetheless and similarly to what was observed for their S. cerevisiae homolog, Tpo1, CgTPO1_1, and CgTPO1_2 deletion was also found to increase susceptibility to spermine, suggesting a physiological role for these transporters in polyamine homeostasis. Interestingly, the homolog of these transporters in C. albicans, FLU1, was previously found to confer fluconazole resistance when expressed in S. cerevisiae (23). These transporters were further found to confer resistance to the polyene antifungal drug amphotericin B and to the fungicide mancozeb. Interestingly, the related DHA transporter CgTpo3 does not appear to confer resistance to flucytosine or amphotericin B (30), while CgAqr1 is involved in flucytosine resistance (29). These observations seem to imply that CgTpo1_1 and CgTpo1_2 are gifted with extraordinary substrate variety, even within their own DHA1 family (7). Consistent with these results, their S. cerevisiae homolog was demonstrated to confer resistance to at least, five different drugs besides polyamines, including the fungicide cycloheximide, the antiarrythmic drug quinidine, the polyene nystatin, and the herbicides 2-methyl-4-chlorophenoxyacetic acid and 2,4-dichlorophenoxyacetic acid (16, 17).
The possibility that these transporters could be regulated by the major controller of the MDR phenomenon in C. glabrata, CgPdr1, was further investigated. As far as we could determine, the expression of these transporters appears to be independent on CgPdr1. Consistently, an analysis of the CgTPO1_1 and CgTPO1_2 promoter regions, performed in the Regulatory Sequence Analysis Tools web site (http://rsat.ulb.ac.be/), was unable to identify any of the CgPdr1-binding elements (BCCRYYRGD and TCCRYGGA) (9), supporting the possibility that these transporters may not be under direct control of CgPdr1.
This study further highlights the importance the cell wall protein CgGas1 in yeast resistance to clotrimazole. So far, C. glabrata Gas1 is described to be constitutively expressed, probably due to an important role in cell wall homeostasis since its deletion was found to result in the formation of cell aggregates and growth defects, much in tune with the observed S. cerevisiae mutant phenotype (15). The formation of aggregates has been also reported for the C. albicans homolog gene deletion mutant (40). Based on the obtained data, the changes undergone by the cell wall upon sudden clotrimazole challenge were studied using a lyticase susceptibility screening assay. It is remarkable to realize that just upon 30 min of clotrimazole exposure C. glabrata cell walls become more susceptible to lyticase, suggesting that this drug has a deleterious effect at the cell wall level. Also consistent with the harmful effect of clotrimazole in the cell wall is the observation that C. glabrata cells adapted to exponential growth in the presence of clotrimazole exhibit cell walls that are clearly more resistant to lyticase. The cell wall remodeling that underlies this observation is expected to depend on the cell-wall-related genes found to confer resistance to clotrimazole. Interestingly, the observed strengthening of the cell wall makes the clotrimazole-adapted cells even more lyticase tolerant than nonstressed exponentially growing cells. The fact that clotrimazole has such an effect over the cell wall, a structure targeted directly by the new class of antifungal drugs, the echinocandins, suggests that a combined therapy using echinocandins and azoles may be a promising approach that, to the best of our knowledge, has not been attempted so far. The cell-wall-related protein CgGas1 was found to be required for clotrimazole resistance, with the correspondent deletion mutant displaying higher susceptibility to azole drugs and showing increased clotrimazole intracellular accumulation, suggesting a role in making the cell wall less permeable to this compound. The obtained results suggest that CgGas1 may have a protective effect in sudden exposure to clotrimazole, but it appears to have a limited role in the observed clotrimazole-induced cell wall remodeling.
Since two distinct mechanisms for antifungal resistance in C. glabrata were addressed in this study, namely drug efflux mediated by the transporter proteins CgTpo1_1 and CgTpo1_2, and cell wall integrity mediated by cell wall assembly protein CgGas1, the eventual cross-talk between these two mechanisms was assessed. In fact, C. glabrata cells devoid of both CgTPO1_2 and CgGAS1 were found to accumulate similar levels of clotrimazole to the ones attained for the single mutant Δcgtpo1_2. These results indicate that despite the fact that C. glabrata cells accumulate two times more drug in the absence of CgGAS1, the absence of CgTPO1_2 appears to have a more deleterious effect in terms of drug accumulation. Despite this observation, the fact that the double deletion of CgTPO1_2 and CgGAS1 genes leads to an increased susceptibility to clotrimazole, when compared with each of the individual single mutants, highlights the cooperative action of these proteins in providing protection against this drug. Indeed, the obtained results suggest that the effect of CgGas1 in clotrimazole resistance goes beyond cell wall remodeling and decreased drug accumulation. Interestingly, a very recent study by Eustice and Pillus (41) reported new functions for this protein in DNA damage response upon exposure to genotoxins in the model S. cerevisiae, apparently through an unforeseen role in regulating transcriptional silencing. This line of evidence, still to be fully explored, expands immensely the possible effects of CgGas1 deletion that may contribute to the increased susceptibility exhibited by Δcggas1 cells.
Altogether, the results described in this study highlight the importance of multidrug transporters from the major facilitator superfamily in antifungal resistance phenotypes. The characterization of C. glabrata CgTpo1_1 and CgTpo1_2 multidrug transporters involved in azole drug resistance reinforces the need to study remaining members of this family in this increasingly relevant pathogenic yeast, given that these transporters are likely to have clinical impact. This work also highlights the importance of genome/proteome-wide approaches in the identification of new antifungal resistance mechanisms.
Supplementary Material
Acknowledgments
We acknowledge Thomas Edlind, from the Department of Microbiology & Immunology, Drexel University, College of Medicine, Philadelphia, and John Bennett, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, for kindly providing the 66032u and L5U1 derived strains, respectively. We further acknowledge the contribution of Mary LoPresti, Kathrin Wilczak, Thomas Abbott, Christopher Colangelo, and TuKiet Lam, from the MS & Proteomics Resource at Yale, for their role in the iTRAQ-based proteomics analysis described in the paper.
Footnotes
Author contributions: C.C., H.C., and M.C.T. designed the research; and P.P., C.C., C.P., and K.S. performed research.
* This work was supported by FEDER and “Fundação para a Ciência e a Tecnologia” (FCT) (Contracts PTDC/EBB-BIO/119356/2010, UID/BIO/04565/2013 and PhD grants to PP and CC).
 This article contains supplemental material Tables S1-S4.
This article contains supplemental material Tables S1-S4.
1 The abbreviations used are:
- DHA
- drug:H+ antiporter
- iTRAQ
- isobaric tag for relative and absolute quantitation
- ORF
- open reading frame OD600nm, optical density at 600nm
- PCR
- polymerase chain reaction
- GFP
- green fluorescent protein
- cDNA
- complementary DNA
- RT-PCR
- real-time PCR.
REFERENCES
- 1.Fidel P. L. Jr., Vazquez J. A., and Sobel J. D. (1999) Candida glabrata: Review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12, 80–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra N. N., Prasad T., Sharma N., Payasi A., Prasad R., Gupta D. K., and Singh R. (2007) Pathogenicity and drug resistance in Candida albicans and other yeast species. A review. Acta. Microbiol. Immunol. Hung. 54, p. 201–235 [DOI] [PubMed] [Google Scholar]
- 3.Pfaller M. A., and Diekema D. J. (2007) Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jandric Z., and Schüller C. (2011) Stress response in Candida glabrata: Pieces of a fragmented picture. Future Microbiol. 6, 1475–1484 [DOI] [PubMed] [Google Scholar]
- 5.Perlroth J., Choi B., and Spellberg B. (2007) Nosocomial fungal infections: Epidemiology, diagnosis, and treatment. Med. Mycol. 45, 321–446 [DOI] [PubMed] [Google Scholar]
- 6.Roetzer A., Gabaldón T. and Schüller C. (2011) From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa C., Dias P. J., Sá-Correia I., and Teixeira M. C. (2014) MFS multidrug transporters in pathogenic fungi: Do they have real clinical impact? Front Physiol. 5, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caudle K. E., Barker K. S., Wiederhold N. P., Xu L., Homayouni R., and Rogers P. D. (2011) Genome-wide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot. Cell 10, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai H. F., Krol A. A., Sarti K. E., and Bennett J. E. (2006) Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50, 1384–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermitsky J.P., and Edlind T. D. (2004) Azole resistance in Candida glabrata: Coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48, 3773–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calahorra M., Lozano C., Sánchez N. S., and Peña A. (2011) Ketoconazole and miconazole alter potassium homeostasis in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1808, 433-r45 [DOI] [PubMed] [Google Scholar]
- 12.Shingu-Vazquez M., and Traven A. (2011) Mitochondria and fungal pathogenesis: Drug tolerance, virulence, and potential for antifungal therapy. Eukaryot. Cell 10, 1376–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh A., Yadav V., and Prasad R. (2012) Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PLoS ONE 7, e39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weig M., Jänsch L., Gross U., De Koster C. G., Klis F. M., and De Groot P. W. (2004) Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology 150, 3129–3144 [DOI] [PubMed] [Google Scholar]
- 15.Weig M., Haynes K., Rogers T. R., Kurzai O., Frosch M., and Mühlschlegel F. A. (2001) A GAS-like gene family in the pathogenic fungus Candida glabrata. Microbiology 147, 2007–2019 [DOI] [PubMed] [Google Scholar]
- 16.Dos Santos S. C., Teixeira M. C., Dias P. J., and Sá-Correia I. (2014) MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: Understanding their physiological function through post-genomic approaches. Front Physiol. 5, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sá-Correia I, dos Santos S. C., Teixeira M. C., Cabrito T. R., and Mira N. P. (2009) Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 17, 22–31 [DOI] [PubMed] [Google Scholar]
- 18.Tomitori H., Kashiwagi K., Sakata K., Kakinuma Y., and Igarashi K. (1999) Identification of a gene for a polyamine transport protein in yeast. J. Biol. Chem. 274, 3265–3267 [DOI] [PubMed] [Google Scholar]
- 19.Albertsen M., Bellahn I., Krämer R., and Waffenschmidt S. (2003) Localization and function of the yeast multidrug transporter Tpo1p. J. Biol. Chem. 278, 12820–12825 [DOI] [PubMed] [Google Scholar]
- 20.Teixeira M.C., and Sá-Correia I. (2002) Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 292, 530–537 [DOI] [PubMed] [Google Scholar]
- 21.Cabrito T. R., Teixeira M. C., Duarte A. A., Duque P., and Sá-Correia I. (2009) Heterologous expression of a Tpo1 homolog from Arabidopsis thaliana confers resistance to the herbicide 2,4-D and other chemical stresses in yeast. Appl. Microbiol. Biotechnol. 84, 927–936 [DOI] [PubMed] [Google Scholar]
- 22.Markovich S., Yekutiel A., Shalit I., Shadkchan Y., and Osherov N. (2004) Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 48, 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calabrese D., Bille J., and Sanglard D. (2000) A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146, 2743–2754 [DOI] [PubMed] [Google Scholar]
- 24.Li R., Kumar R., Tati S., Puri S., and Edgerton M. (2013) Candida albicans flu1-mediated efflux of salivary histatin 5 reduces its cytosolic concentration and fungicidal activity. Antimicrob. Agents Chemother. 57, 1832–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueno K., Uno J., Nakayama H., Sasamoto K., Mikami Y., and Chibana H. (2007) Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryot. Cell 6, 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K. H., Miyazaki T., Tsai H. F., and Bennett J. E. (2007) The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 386, 63–72 [DOI] [PubMed] [Google Scholar]
- 27.Simões T., Teixeira M. C., Fernandes A. R., and Sá-Correia I. (2003) Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes: Important role of SPI1. Appl. Environ. Microbiol. 69, 4019–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueno K., Matsumoto Y., Uno J., Sasamoto K., Sekimizu K., Kinjo Y., and Chibana H. (2011) Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS ONE 6, p. e24759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa C., Henriques A., Pires C., Nunes J., Ohno M., Chibana H., Sá-Correia I., and Teixeira M. C. (2013) The dual role of Candida glabrata drug:H+ antiporter CgAqr1 (ORF CAGL0J09944g) in antifungal drug and acetic acid resistance. Front Microbiol. 4, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa C., Nunes J., Henriques A., Mira N. P., Nakayama H., Chibana H., and Teixeira M. C. (2014) Candida glabrata drug:H+ antiporter CgTpo3 (ORF CAGL0I10384g): Role in azole drug resistance and polyamine homeostasis. J. Antimicrob. Chemother. 69, 1767–1776 [DOI] [PubMed] [Google Scholar]
- 31.Costa C., Pires C., Cabrito T. R., Renaudin A., Ohno M., Chibana H., Sá-Correia I., and Teixeira M. C. (2013) Candida glabrata drug:H+ antiporter CgQdr2 confers imidazole drug resistance, being activated by transcription factor CgPdr1. Antimicrob. Agents Chemother. 57, p. 3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa M. F., and Sá-Correia I. (1996) Intracellular acidification does not account for inhibition of Saccharomyces cerevisiae growth in the presence of ethanol. FEMS Microbiol. Lett. 135, p. 271–274 [DOI] [PubMed] [Google Scholar]
- 33.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., and Brown P. O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golin J., Kon Z. N., Wu C. P., Martello J., Hanson L., Supernavage S., Ambudkar S. V., and Sauna Z. E. (2007) Complete inhibition of the Pdr5p multidrug efflux pump ATPase activity by its transport substrate clotrimazole suggests that GTP as well as ATP may be used as an energy source. Biochemistry 46, p. 13109–13119 [DOI] [PubMed] [Google Scholar]
- 35.Torelli R., Posteraro B., Ferrari S., La Sorda M., Fadda G., Sanglard D., and Sanguinetti M. (2008) The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol. Microbiol. 68, 186–201 [DOI] [PubMed] [Google Scholar]
- 36.Decottignies A., Lambert L., Catty P., Degand H., Epping E. A., Moye-Rowley W. S., Balzi E., and Goffeau A. (1995) Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J. Biol. Chem. 270, 18150–18157 [DOI] [PubMed] [Google Scholar]
- 37.Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., and Bille J. (1999) The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43, 2753–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cannon R. D., Lamping E., Holmes A. R., Niimi K., Baret P. V., Keniya M. V., Tanabe K., Niimi M., Goffeau A., and Monk B. C. (2009) Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22, 291–321, Table of Contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo J. I., Choi C. W., Kim H. S., Yoo J. S., Jeong Y. H., Lee Y. S. (2012) Proteomic analysis of cellular and membrane proteins in fluconazole-resistant Candida glabrata. Osong Public Health Res. Perspect. 3, p. 74–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhlschlegel F.A., and Fonzi W. A., (1997) PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17, 5960–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eustice M., and Pillus L. (2014) Unexpected function of the glucanosyltransferase Gas1 in the DNA damage response linked to histone H3 acetyltransferases in Saccharomyces cerevisiae. Genetics 196, 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.