Abstract
Identifying the genetic etiology of congenital heart disease (CHD) has been challenging despite being one of the most common congenital malformations in humans. We previously identified a microdeletion in a patient with a ventricular septal defect containing over 40 genes including MESP1 (mesoderm posterior bHLH transcription factor 1). Because of the importance of MESP1 as an early regulator of cardiac development in both in vivo and in vitro studies, we tested for MESP1 mutations in 647 patients with congenital conotruncal and related heart defects. We identified six rare, non-synonymous variants not seen in ethnically matched controls and one likely race-specific non-synonymous variant. Functional analyses revealed that three of these variants altered activation of transcription by MESP1. Two of the deleterious variants are located within the conserved HLH domain and thus impair the protein-protein interaction of MESP1 and E47. The third deleterious variant was a loss of function frameshift mutation. Our results suggest that pathologic variants in MESP1 may contribute to the development of CHD and that additional protein partners and downstream targets could likewise contribute to the wide range of causes for CHD.
Keywords: Mesoderm posterior bHLH transcription factor 1, Conotruncal heart defects, MESP1, Congenital heart disease
Introduction
Congenital heart defects (CHD) are the most common type of major birth defects with a complex and poorly understood etiology. Evidence strongly suggests a genetic component [Gelb and Chung, 2014] including chromosomal abnormalities, gene mutations and epigenetic insults [reviewed in Lalani and Belmont, 2014]. Chromosomal abnormalities including submicroscopic chromosomal aberrations (copy number variants, CNVs) contribute to disease risk for CHD in syndromic and seemingly non-syndromic cases [Lalani and Belmont, 2014]. Studies have found that sequence variants in genes disrupted by CNVs can likewise contribute to disease, including TBX1 (MIM# 602054)[Yagi et al., 2003], EMHT1 (MIM# 610253)[Kleefstra et al., 2006], JAG1 (MIM# 601920), NSD1 (MIM# 606681)[Cecconi et al., 2005], and ELN (MIM# 185500) [reviewed in Andersen et al., 2014]. To identify new disease-related genes, we have studied candidate genes disrupted by CNVs for potentially damaging variants in individuals with seemingly non-syndromic CHD. We previously reported a CNV spanning 4.3 Mbs on chromosome 15 in a case diagnosed with a posterior malalignment type ventricular septal defect VSD [Goldmuntz et al., 2011]. We hypothesized that mutations within genes contributing to cardiac development deleted by this CNV might contribute to CHD in humans.
Mesoderm posterior 1 (MESP1; MIM# 608689), a transcription factor of the basic helix-loop-helix (bHLH) family expressed in cardiac progenitors at different time points during their specification [Devine et al., 2014; Lescroart et al., 2014], was contained within this large CNV. MESP1 participates in the specification of the cardiac lineage and plays an essential role in early heart development in animal and in vitro studies [Bondue et al., 2008; David et al., 2008; Lindsley et al., 2008; Saga, 1998; Saga et al., 2000; Saga et al., 1999]. MESP1-null mice die in utero at 10.5 dpc due to anomalies in heart tube formation and heart looping, resulting in various degrees of cardiac bifida [Saga, 1998; Saga, et al., 1999]. Overexpression of MESP1 (mesoderm posterior protein 1) in two-cell Xenopus laevis embryos resulted in extra ectopic beating tissue in tadpoles [David, et al., 2008]. Similarly, overexpression of Mesp1 in mouse embryonic stem cells resulted in up- or down-regulation of specific gene sets and, subsequently, accelerated cardiovascular specification and premature appearance of beating cells. Among the up-regulated genes were core cardiovascular transcription factors such as HAND2 (MIM# 602407), GATA4 (MIM# 600576), NKX2-5 (MIM# 600584), TBX20 (MIM# 606061) and MYOCD (MIM# 606127)[Bondue et al., 2011; David, et al., 2008; He et al., 2011; Lindsley, et al., 2008]. Some of these genes, alone or in combination, are known to participate in human CHD [Garg et al., 2003; Granados-Riveron et al., 2012; Kirk et al., 2007; McElhinney et al., 2003; Posch et al., 2010; Reamon-Buettner and Borlak, 2010; Schott et al., 1998; Stallmeyer et al., 2010]. The potential upstream role of MESP1 in regulating these critical cardiac transcription factors suggests that mutations in this gene might also play a role in human CHD. Based on our CNV findings and the role of MESP1 in cardiogenesis, we evaluated a cohort with the same CHD as the original case harboring the CNV, followed by cases with etiologically related conotruncal defects for potentially deleterious variants.
Material and Methods
Sample collection and DNA isolation
We studied a cohort of 280 cases with a primary diagnosis of ventricular septal defect (VSD) including conoventricular, malalignment and conoseptal hypoplasia type VSDs, and a cohort of 367 cases with conotruncal defects (predominantly tetralogy of Fallot, Table 1). Subjects with a recognizable genetic syndrome at the time of enrollment were excluded [Peyvandi et al., 2013]. Informed consent was obtained from all cases and parents following protocols approved by the Institutional Review Board for Human Research at The Children’s Hospital of Philadelphia prior to collection of samples. DNA of ethnically matched control subjects was obtained from the Coriell Institute for Medical Research (Camden, NJ). DNA was extracted from whole blood or lymphoblastoid cell lines using standard methods (Gentra Puregene Blood kit by Qiagen, Valencia, CA).
Table 1.
Cardiac Diagnoses for Study Cohorts
| Diagnosis | Numbers |
|---|---|
| Tetralogy of Fallot | 318 |
| Ventricular septal defect | 292 |
| Conoventricular | 218 |
| Posterior malalignment | 51 |
| Conal septal hypoplasia | 19 |
| Others* | 4 |
| Truncus arteriosus | 21 |
| Interrupted aortic arch type-B | 13 |
| Interrupted aortic arch type-A | 3 |
|
| |
| Total | 647 |
Others included two unspecified VSD, one inlet VSD, and one VSD with virtually absent septum
Mutation analysis
The initial cohort of 280 cases was scanned for sequence variation in both coding exons of MESP1 (NM_018670.3) by high-resolution melting curve analysis using a 96-well LightScanner™ (Idaho Technology Inc, Salt Lake City, UTAH). Analyzed amplicons spanned exonic sequences as well as exon/intron boundaries with a size range of 222–404 bp (see Supp. Table S1). Samples were amplified in a volume of 10 μl containing 1X LightScanner® Master Mix (Idaho Technolgy Inc.), 20 ng of DNA and 0.25 μM of each primer following the suggested PCR protocol. For some amplicons, we added a touchdown cycling step at the beginning starting 3°C above the annealing temperature and then decreasing 1°C per cycle for 3 cycles. Primer sequences were designed using PrimerSelect from the Lasergene Core Suite (DNASTAR, Madison, WI) and are listed together with annealing temperatures, and product size in Supp. Table S1. PCR products were then transferred to the LightScanner for melting analysis and the melting data analyzed for sequence variations using the LightScanner Software. Samples that showed variation in the melting curves were chosen for Sanger sequencing. PCR products were re-amplified in a volume of 20μl using AmpliTaq Gold Polymerase (Applied Biosystems, ThermoFisher Scientific, Pittsburgh, PA), with 20ng of DNA and a final concentration of 0.2mM dNTP, 2 mM MgCl2, and 0.25 μM of each primer. PCR products were purified using the Agencourt AMPure XP system (Beckman Coulter Genomics, Danvers, MA) before sequencing using BigDye™ Terminator version 3.1 on an ABI 3100 Genetic Analyzer (Applied Biosystems, ThermoFisher Scientific). Sequences were analyzed using Sequencher™ (Gene Codes, Ann Arbor, MI). Case, parental and control DNA sequences were compared to the reference sequences for MESP1 (NM_018670.3) to identify sequence variations. Control samples were Sanger sequenced for exon 1 of MESP1 and exon 2 was analyzed using high-resolution melting curve analysis as described above.
A second cohort of 367 cases with conotruncal defects underwent targeted whole exome sequencing by the NHLBI Resequencing & Genotyping Service at the Northwest Genomics Center at the University of Washington. The VCF files were analyzed using SNP & Variation Suite v8.1 (Golden Helix, Inc. Bozeman, MT, http://www.goldenhelix.com) for variants in MESP1. We performed Sanger sequencing to validate non-synonymous variations. Previously unreported variants have been submitted to dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Site directed mutagenesis
A clone containing the mRNA sequence of MESP1 (NM_018670.3) was purchased from GenScript (Piscataway, NJ). The complete ORF sequence of MESP1 was cloned into an expression vector pcDNA3.1/V5-His© Topo® (Invitrogen, ThermoFisher). Mutations in MESP1 were introduced by site-directed mutagenesis using a QuikChange lightning site-directed mutagenesis Kit (Agilent Technologies, Inc., Santa Clara, CA) and mutagenic primers (Supp. Table S2) that were designed using the QuikChange Primer Design Program (Agilent Technologies, Inc). A V5 tag was incorporated at the N-terminus for the constructs pcDNA3.1-MESP1, pcDNA3.1-MESP1-2745, pcDNA3.1-MESP1-2365, and pcDNA3.1-MESP1-2034 (Supp. Table S2) for protein analysis. All expression vectors were transformed into One Shot TOP 10 competent cells, grown in appropriate antibiotic media and isolated using a QIAprep Spin Miniprep Kit (Qiagen), and sequenced for verification. Expression of MESP1 was verified by immunoblot analysis.
Dual luciferase reporter assay
Luciferase vector pGL4.23-DKK1-11: A triplicate of the E-box region acCATATGgt located approximately −11.6 kb upstream of DKK1 (MIM# 605189)[David, et al., 2008] was inserted into the luciferase vector pGL4.23 [luc2/minP] (Promega, Madison, WI). A pCMV vector containing the sequence of E47 (TCF3; MIM# 147141, X52078.1) was kindly provided by Dr. M. Atchinson, School of Veterinary Medicine, University of Pennsylvania. Human embryonic kidney (HEK) 293 cells were plated at a density of 60,000 cells per well on 24-well plates 24 hrs before transfection. Cells were co-transfected with 100 ng of empty pcDNA3.1 vector or pcDNA3.1 vector coding for either wildtype or mutant MESP1, 250 ng of pCMV-E47, 100 ng of either empty pGL4.23 or pGL4.23-DKK1-11 vector and 0.4 ng pGL4.75 (Renilla luciferase reporter vector) using FugeneHD (Promega) following the manufacturer’s protocol. Transfection efficiency was ascertained using a co-transfected GFP expressing vector. Transfected cells were incubated for 48 to 60 hrs at 37°C with 5% CO2 then washed and lysed using 1X passive lysis buffer for 15 min at RT provided by the Dual Luciferase Reporter Assay Kit (Promega). Dual luciferase assays (Promega) were performed according to the manufacture’s protocol using a GloMax® 96 Microplate Luminometer w/Dual Injectors (Promega). Firefly luciferase values were normalized relative to the Renilla luciferase values. At least three independent co-transfection experiments, all of which were done in triplicate, were performed to calculate average values and standard errors. A two-sample t-test was applied to assess statistical differences.
Mammalian Two-Hybrid assay
The ORFs of either the wildtype or mutant MESP1 were cloned into the pCMV-BD plasmid (GAL4 insert) from the mammalian two-hybrid assay kit (Stratagene, La Jolla, CA). The E47–ORF was cloned into pCMV-AD (NF-κB insert). The control vectors supplied by the assay kit (pFR-Luc, pBD-NF-kB, pBD-p53, pAD-sv40T, pAD-TRAF) as well as the cloned vectors were amplified, isolated and sequence-verified as described above. The mammalian two-hybrid assay was performed similarly to the dual luciferase reporter assay described above. The total amount of DNA transfected per well was 500 ng, 100 ng of which was either wildtype pCMV-BD-MESP1 or mutant pCMV-BD-MESP1, 200ng of pCMV-AD-E47, 200 ng pFR-Luc vectors and 0.5ng of pGL4.75 as the internal control. Cells were collected after 48h and luciferase activity measured using a GloMax® 96 Microplate Luminometer w/Dual Injectors (Promega). At least three independent co-transfection experiments, all of which were done in triplicate, were performed to calculate average values and standard errors. A two-sample t-test was applied to assess statistical differences.
Western Blot Assay
Human embryonic kidney (HEK) 293 cells were plated 24 hrs prior to transfection in 10 mm dishes or 6-well plates. Transfection of V5 or Gal4 tagged expression vectors was carried out using Fugene HD (Promega) according to the manufacturer’s protocol. After 48 hrs incubation at 37°C with 5% CO2, cells were washed with cold PBS and collected by scraping. Cells were then lysed in 100 μl 1 X cell lysis buffer (Cell Signaling Technology, Danvers, MA) collected, aliquotted and placed at −80°C. Single aliquots were then thawed on ice for the hours indicated. The supernatant was retained after 10 min centrifugation at 10,000 rpm at 4° C. 20μg of protein samples were separated by SDS-PAGE (NuPAGE 4–12% BT precast gels; Life Technologies, Carlsbad CA) and electrotransferred to Novex nitrocellulose (0.45μm; Life Technologies, ThermoFisher Scientific) or Immun-Blot PVDF membrane (BIO-RAD Laboratories, Inc., Hercules, CA). An Anti-V5 (Invitrogen, ThermoFisher Scientific) or Anti-GAL4 antibody (RK5C1, Santa Cruz Biotechnology, Inc, Dallas, Tx) was used for immunoblotting and visualized using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific, ThermoFisher Scientific).
Results
Identification of MESP1 variants in CHD
A previously reported subject harboring a large CNV encompassing MESP1 was diagnosed with a VSD [Goldmuntz, et al., 2011]. Therefore, we studied a cohort of 280 subjects with VSDs for potentially pathogenic sequence variants in MESP1. The majority of the VSDs were classified as conoventricular and a smaller number were malalignment VSDs (Table 1). The cases were of mixed ethnicity, were not recognized to have a genetic syndrome at the time of enrollment, did not carry a 22q11.2 deletion and were reported to have an affected first-degree relative in 6% of cases [Peyvandi, et al., 2013]. To identify sequence variants in MESP1 in the VSD cohort, we amplified short amplicons within exons and exon-intron boundaries and analyzed them for aberrant melting behavior using high resolution melting analysis. Due to the sequence variability in exon 1 (dbSNP, ExAC: exac.broadinstitute.org) this initial analysis resulted in a large number of variant curves. Therefore, a high number of cases were selected for subsequent Sanger sequencing of exon 1, which identified previously reported synonymous as well as non-synonymous variants also present in our control groups (dbSNP, ExAC, [Lahm et al., 2013]). In addition we identified three non-synonymous variants and one frameshift mutation resulting in a premature stop codon in four unrelated cases (p.P27_D29dup, p.G70D, p.L147Pfs*9, p.K268N) that were not present in ethnically matched controls. We also identified one non-synonymous variant (p.D168G) present in 6 out of 97 ethnically matched controls (Table 2).
Table 2.
Mutations Identified in MESP1
| Protein Q9BRJ9 |
Variant (based on cDNA) (NM_018670.3) |
hg19 | Region | Conserved Domain |
dbSNP MAF (ExAC#) |
Polyphen-2# | Sift# | CADD | Patient | Variant in Controls | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| Dx | Gender | Ethnicity | Father | Mother | CA | AA | Asian | |||||||||
| p.P27_D29dup | c.79_87dup9 | chr15:90294375_90294376insGTCGGAGGG | Exon1 | rs752416885 0.00007477 |
N/A | N/A | N/A | cVSD | M | CA (no HISP) | Carrier | WT | 0/95 | 0/87 | ||
| p.G70D | c.209G>A | chr15:90294254C>T | Exon1 | rs770840637* | benign (0.447) | Tolerated | 10.42 | cVSD | M | AAm (Hisp) | N/A | WT | 0/99 | 0/96 | ||
| p.E104K | c.310G>A | chr15:90294153C>T | Exon1 | bHLH | rs771329728 0.0002231 |
probably damaging, 0.981 | Tolerated | 16.35 | TOF | F | CA | WT | Carrier | 0/99 | 0/97 | |
| p.L120P | c.359T>C | chr15:90294104A>G | Exon1 | bHLH | rs565846523 0.000817 |
probably damaging, 1.0 | Damaging | 15.83 | TOF | F | CA | WT | WT | 0/99 | 0/97 | |
| p.L147Pfs*9 | c.436_437delAG | chr15:90294027_90294028delTC | Exon1 | rs777474430 2.106e-05 |
N/A | N/A | N/A | cVSD | F | AAm (no Hisp) | N/A | N/A | 0/99 | 0/97 | ||
| p.D168G | c.503A>G | chr15:90293960T>C | Exon1 | rs200810210 0.005209 |
possibly damaging (0.948) | Damaging | 13.68 | cVSD | M | AAm (no Hisp) | N/A | N/A | 0/99 | 6/97 | ||
| p.K268N | c.804G>C | chr15:90293378C>G | Exon2 | N/A | Benign (0.061) | Tolerated | 13.9 | mVSD/CoA/AA | M | AAm/CA/AS (Hisp) | WT | Carrier | 0/198 | 0/98 | 0/96 | |
N/A: Not available, Dx: diagnosis, CADD: Combined Annotation Dependent Depletion score (http://cadd.gs.washington.edu), cVSD: conoventricular VSD, mVSD: malalignment VSD, TOF: Tetralogy of Fallot, CoA: Coarctation of the Aorta, AA: Aortic atresia, AAm: African American, CA: Caucasian, WT: Wildtype; #ExAC:Exome Aggregation Consortium (exac.broadinstitue.org),
not listed in ExAC, MAF given in dbSNP: 0.000; hg19: GrCh37.p13 (GCF_000001405.25),
Values listed in Supp. Tables S3 and S4.
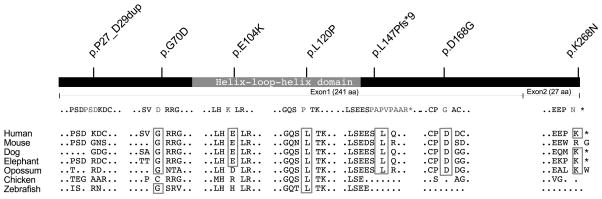
Thereafter, targeted exome sequence data for MESP1 from an additional 367 cases with conotruncal defects who did not have a recognized genetic syndrome at the time of enrollment, did not carry a 22q11.2 deletion and were reported to have an affected first-degree relative in 10% of cases [Peyvandi, et al., 2013] were studied for deleterious mutations (Table 2). We identified two additional non-synonymous variants (p.E104K, p.L120P) that were not present in ethnically matched controls (Table 2). Six of the seven non-synonymous variants have allele frequencies less than 0.0009 in dbSNP and ExAC (Table 2). Testing of available parents for four trios revealed that one variant was de novo while three were inherited; the inheritance of the three remaining variants could not be determined (Table 2). None of the parental mutation carriers were reported to have congenital heart disease, but screening for subclinical cardiac anomalies was not done. Of these seven non-synonymous variants, five resulted in amino acid substitutions (Table 2), one was a duplication of three amino acids (c.79_87dup9; p.P27_D29dup), and one was a deletion of two base pairs that resulted in a frameshift and premature stop codon at position 155 (c.436_437delAG, p.L147Pfs*9). Two of the missense variants were located in the conserved bHLH domain and both were predicted to be probably damaging by Polyphen-2 (v2.2.2r398, see Supp. Table S3). Variant p.D168G was also predicted to be possibly damaging (Table 2). Since the frameshift mutation results in a stop codon in the penultimate exon, we hypothesized that it could lead to either nonsense-mediated decay or a truncated protein retaining the complete bHLH domain (Figure 1).
Figure 1.
Schematic showing the MESP1 protein indicating the conserved domains and exon locations. Location and description of the mutations studied in our cases are noted above. Comparative alignment of mutated regions in homologous proteins in various species indicating the affected amino acids in boxes.
Assessment of variants on MESP1 transcriptional activity
Luciferase Reporter Assay
MESP1, a bHLH protein, is involved in the activation of several cellular processes via activation of transcription, including a cascade of cardiac-specific transcription factors [Bondue and Blanpain, 2010; Bondue, et al., 2008; Chan et al., 2013; David, et al., 2008; Lindsley, et al., 2008; Saga, et al., 2000; Wu, 2008]. Consequently, we studied whether the MESP1 variants affected activation of downstream target genes.
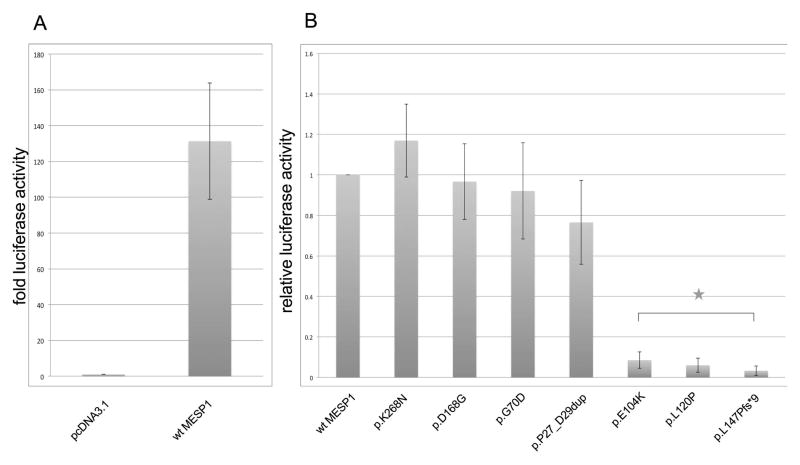
Bondue et al. [2008] has shown that MESP1 binds to promoter regions containing putative bHLH binding sites (Ebox) of several of the cardiac transcription factors. However, it has also been described that MESP1 does not bind to Eboxes alone [Chan, et al., 2013; Takahashi et al., 2007] but forms heterodimers with E12 and E47 in order to activate transcription [Chan, et al., 2013; Lindsley, et al., 2008; Takahashi, et al., 2007]. E12 and E47 are isoforms of TCF3 (MIM# 147141), a member of the ubiquitous E-protein family of bHLH transcription factors. We found that the MESP1/E47 heterodimer activated transcription significantly more using a previously identified MESP1 binding Ebox motif in the DKK1 enhancer [David, et al., 2008] as compared to either one alone (Supp. Figure S1, Figure 2, Table 3). Using this assay, three of the seven MESP1 variants, namely p.E104K, p.L120P and p.L147Pfs*9, abolished activation of transcription (p<0.005, Figure 2).
Figure 2.
Activation of transcription by the heterodimer MESP1/E47 using the DKK1-11 Ebox as enhancer. A: Activity of MESP1/E47 on DKK1-11 Ebox. Fold activation normalized to activation by empty expression vector with coexpression of E47. B: Relative luciferase activity of wildtype MESP1/E47 and mutant MESP1/E47 on DKK1-11 Ebox. * = p <0.005
Table 3.
Functional Consequences of Mutations Identified in MESP1
| Q9BRJ9 | Inheritance | Present in Controls | Relative Transcriptional Activation Compared to Wildtype MESP1 (Stdev)
|
|
|---|---|---|---|---|
| Luciferase Reporter Assay | Mammalian Two-Hybrid | |||
| p.P27_D29dup | Paternal | N | 0.77 (+/−0.21) | 1.02 (+/−0.13) |
| p.G70D | N/A | N | 0.92 (+/−0.24) | 1.19 (+/−0.21) |
| p.E104K | Maternal | N | 0.08 (+/−0.04)* | 0.56 (+/−0.08)* |
| p.L120P | De Novo | N | 0.06 (+/−0.03)* | 0.32 (+/−0.06)* |
| p.L147Pfs*9 | N/A | N | 0.03 (+/−0.02)* | 0.05 (+/−0.01)* |
| p.D168G | N/A | Y | 0.97 (+/−0.19) | 0.53 (+/−0.11)* |
| p.K268N | Maternal | N | 1.17 (+/−0.18) | 0.90 (+/−0.26) |
Significant (p<0.005)
Mammalian Two Hybrid Assay
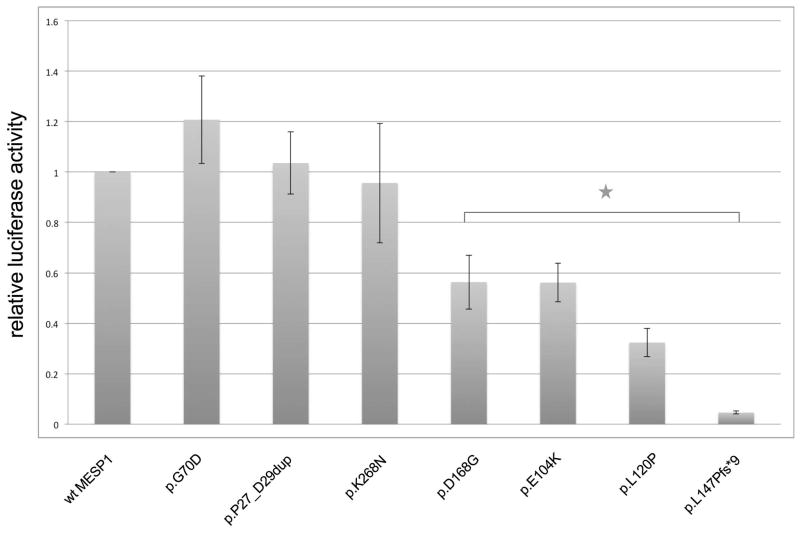
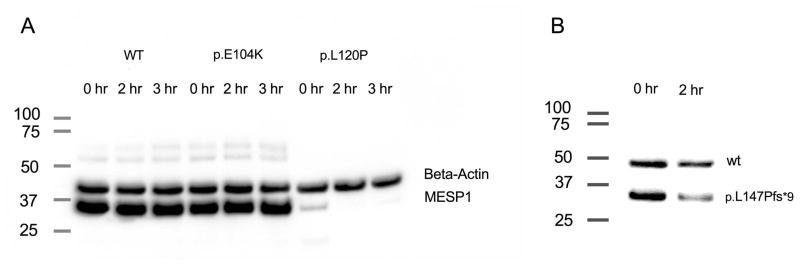
The observation that both MESP1 and E47 are needed to activate transcription suggests that MESP1 needs to form an active heterodimer with E47 in order to activate transcription using the DKK1-11 Ebox construct. MESP1 mutants might abolish transcriptional activation by disrupting binding to E47. Alternatively, the mutations could disrupt DNA binding. To test if the protein-protein interaction between E47 and MESP1 was impaired by the three variants, p.E104K, p.L120P and p.L147Pfs*9, we performed a mammalian two-hybrid assay for all variants. In this assay, co-expression of MESP1 and E47 resulted in strong expression of luciferase while each alone was insufficient to drive expression, confirming interaction of the two proteins (Supp. Figure S2). Significantly reduced interaction was noted with the three MESP1 variants, p.E104K, p.L120P and p.L147Pfs*9 (p<0.005, Figure 3, Table 3). In addition, the variant, p.D168G, showed significantly reduced interaction in the mammalian two-hybrid assay. The two variants, p.E104K, and p.L120P, might disrupt protein-protein interaction and hence transcriptional activation by virtue of their location within the HLH domain. Alternatively, the mutant proteins might be unstable and thus unavailable to activate transcriptional activity. To test this, we incubated lysates on ice for 2 to 3 hrs and then performed immunoblotting. Indeed, we found that two variants, p.L120P and p.L147Pfs*9, affected MESP1 protein stability (Figure 4 A, B).
Figure 3.
Mammalian two-hybrid assays to evaluate effect of MESP1 mutations on MESP1-E47 interactions. Relative luciferase activity was normalized to wildtype MESP1-E47 and shown as an average over three independent experiments. * p < 0.005
Figure 4.
Western blot analysis of wildtype and mutant MESP1 proteins. A. The V5-tagged MESP1 proteins, wildtype and mutant (p.E104K, p.L120P) each of which was transfected separately into HEK293 cells. Mutant p.L120P-MESP1 protein is not stable on ice. Beta-actin is labeled as an internal control B. Similar experiments with GAL4-tagged wildtype and mutant MESP1 protein, p.L147Pfs*9, which were co-transfected. After 2 hrs on ice the GAL4-tagged truncated protein shows degradation.
Discussion
Many studies demonstrate that the bHLH transcription factor, MESP1, participates in heart development [Bondue, et al., 2008; David, et al., 2008; Lindsley, et al., 2008; Saga, 1998; Saga, et al., 2000; Saga, et al., 1999]. We previously identified a large deletion that included MESP1 in a patient with a VSD [Goldmuntz, et al., 2011]. We hypothesized that MESP1 sequence variants might contribute to disease-risk for VSD and related conotruncal defects. As have others, we identified marked sequence variability in exon 1 (dbSNP, ExAC, [Lahm, et al., 2013]). Lahm et al. [2013] also identified a novel non-synonymous variant in exon 1 of MESP1 in a patient population of 215 cases with a variety of congenital heart defects but it did not seem to have a deleterious effect. Our study of MESP1 sequence variations in 647 patients with conotruncal lesions resulted in the identification of six very rare, non-synonymous and nonsense variants (MAF < 0.0009 ExAC/dbSNP) not seen in ethnically matched controls, and one non-synomymous variant that may be race-specific. Of these seven variants, three conferred functional changes, of which one was de novo, one inherited and the parental status unknown for the last. None of the first-degree relatives were reported to have CHD. Inherited variants from a seemingly normal parent may yet contribute to disease risk given that CHD are likely complex traits where several genetic variants contribute incrementally to disease-risk in any one person. In addition, seemingly normal parents can harbor subtle, clinically insignificant cardiac anomalies that are only detected by echocardiograms. Overall, these results suggest that approximately 1 in 200 cases with a VSD or tetralogy of Fallot carry a potential disease-related variant.
It is important and challenging to test whether sequence variants in a candidate gene change protein function and thus likely contribute to disease risk. We developed several assays to test whether the sequence variants in our cases conferred any functional change. We found that MESP1 alone would not activate transcription using several Ebox or Ebox containing regions. With the dimerization partner E47 [Lindsley, et al., 2008; Nakajima et al., 2006; Takahashi, et al., 2007], only the heterodimer MESP1/E47 activated transcription using a specific Ebox of the DKK1 upstream region as a promoter [David, et al., 2008]. Using this assay three of the case-specific variants significantly affected MESP1-induced transcriptional activation. One of the variants (p.E104K) located in the bHLH region significantly affects the interaction of MESP1 and E47 as shown in the mammalian two-hybrid assay. It is difficult to say if the significantly lower activation of transcription with two other mutant MESP1 proteins (p.L120P, p.L147Pf*9) is due to protein instability or decreased dimerization, or a combination of both. Given that the premature stop codon of the variant p.L147Pfs*9 is in the penultimate exon, it is possible that the mRNA escapes nonsense-mediated decay and is translated into a truncated protein. Only one other variant (p.D168G) seemed to have a significant effect on dimerization of MESP1 and E47 but it did not affect the activation of transcription through the DKK1 Ebox. Possible explanations for this discrepancy might be found in the difference in sensitivity of the two assays. Whereas the mammalian two-hybrid assay is more sensitive to protein interaction, the luciferase reporter assay is more sensitive to promoter occupancy and transcriptional activation. Alternatively the MESP1/E47 interaction may be stabilized by binding to Ebox motif DNA, which would not be the case for the mammalian two-hybrid assay, which utilizes the GAL4 DNA binding site. Three of the variants did not alter protein stability or transcriptional activation in this particular assay but may confer changes in different circumstances.
MESP1 is known to bind different Ebox-containing regulatory regions of known cardiac specific transcription factors including NKX2.5, HAND2, and MYOCD [Bondue, et al., 2008]. We evaluated several of these regions or single Eboxes contained within these regions for activation of transcription by MESP1 alone and the MESP1/E47 heterodimer but we did not see any activation (data not shown). Our findings suggest that the MESP1/E47 heterodimer likely prefers specific Ebox sequences, suggesting that MESP1 might use other dimerization partners to bind to other defined regulatory regions. Only a few dimerization partners of MESP1 are known so far, including Creb1 (MGI:88494) and E12 (MGI:98510)[Chan, et al., 2013; Shi et al., 2015]. However, recent studies suggest that additional cofactors influence MESP1 activity. For example embryonic stem cell-embryoid body differentiation differs depending upon the stage of differentiation of the cells and the signaling environment at the time point of induced MESP1 expression [Chan, et al., 2013]. Similarly, lineage-tracing experiments showed that the time point of MESP1 expression during development and the stage of specification of MESP1-expressing progenitor cells affects their cell fate. These findings suggest that different environmental cues during cardiac morphogenesis influence the ultimate fate of the progenitor cells [Devine, et al., 2014; Lescroart, et al., 2014]. Indeed, Lescroart et al. [2014] demonstrated differences in the molecular profile of the early and late expressing MESP1 progenitor cells in vivo. Further studies will show if co-expressed bHLH proteins, for example HAND1 protein, or other dimerization partners, interact or compete with MESP1 during early heart development [Lescroart, et al., 2014; Vincentz et al., 2011]. Of interest, studies suggest that the Ebox sequence we used for our assays, acCATATGgt, is also a target for TWIST1 (MIM# 601622), a bHLH protein interacting with HAND2 and involved in heart development [Conway et al., 2010; Vincentz et al., 2008; Vincentz et al., 2013].
We also found the Ebox flanking bases to be critical for MESP1/E47 binding [Fisher et al., 1993; Gould and Bresnick, 1998; Kophengnavong et al., 2000]. Assays using the same DKK1-11 Ebox sequence but with different flanking bases failed to activate transcription (data not shown). Since we only tested E47 as a dimerization partner with only one specific Ebox sequence, it is possible that the mutant MESP1 proteins that did not show any significant change in this assay might have an effect on other MESP1 interactions.
Our results suggest that potentially deleterious variants in MESP1 may contribute to the development of VSD and tetralogy of Fallot in humans. Additional studies testing a more extensive CHD population will help define the range of associated phenotypes. As with many genes implicated in CHD, the frequency of deleterious MESP1 sequence variants is likely low in the CHD population [Zaidi et al., 2013]. However, these findings further clarify the cardiac transcription factor network and demonstrate that additional protein partners and down stream targets could likewise be disease-related. In addition, alternative methods might identify additional variants, such as indels, that were not detected by our screening method, and thus our results represent the minimum prevalence in this population. Given the significant sequence variability detected in exon 1 and variable reporting by different groups in public datasets, gene burden studies may be challenging to perform but clearly MESP1 appears to be another important disease-related cardiac transcription factor.
Supplementary Material
Acknowledgments
Contract grant sponsors: US National Institutes of Health (P50-HL074731, P01-HD070454). The project described was also supported by the National Center for Research Resources, Grant UL1RR024134, and is now at the National Center for Advancing Translational Sciences, Grant UL1TR000003.
We thank Dr. K. Cole, Division of Oncology, Children’s Hospital of Philadelphia for the use of the Glomax Luminometer and the members of her and Dr. Maris’ Lab for assistance. We also thank Dr. M. Atchinson, School of Veterinary Medicine, University of Pennsylvania, for the E47 vector. Targeted exome sequencing services were provided through the RS&G Service by the Northwest Genomics Center at the University of Washington, Department of Genome Sciences, under U.S. Federal Government contract number HHSN268201100037C from the National Heart, Lung, and Blood Institute.
Footnotes
Conflict of interest
The authors do not have any conflicts of interest to disclose.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Contributor Information
Petra Werner, Division of Cardiology, Children’s Hospital of Philadelphia, 34th Street and Civic Center Boulevard, Philadelphia, PA 19104 USA.
Brande Latney, Division of Cardiology, Children’s Hospital of Philadelphia, 34th Street and Civic Center Boulevard, Philadelphia, PA 19104 USA.
Matthew A. Deardorff, Division of Genetics, Children’s Hospital of Philadelphia, Department of Pediatrics, University of Pennsylvania, Perelman School of Medicine, 34th Street and Civic Center Boulevard, Philadelphia, PA 19104 USA
Elizabeth Goldmuntz, Division of Cardiology, Children’s Hospital of Philadelphia, Department of Pediatrics, University of Pennsylvania, Perelman School of Medicine, 34th Street and Civic Center Boulevard, Philadelphia, PA 19104 USA.
References
- Andersen TA, de Troelsen KL, Larsen LA. Of mice and men: molecular genetics of congenital heart disease. Cell Mol Life Sci. 2014;71(8):1327–1352. doi: 10.1007/s00018-013-1430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res. 2010;107(12):1414–1427. doi: 10.1161/CIRCRESAHA.110.227058. [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3(1):69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. The Journal of cell biology. 2011;192(5):751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi M, Forzano F, Milani D, Cavani S, Baldo C, Selicorni A, Pantaleoni C, Silengo M, Ferrero GB, Scarano G, Della Monica M, Fischetto R, et al. Mutation analysis of the NSD1 gene in a group of 59 patients with congenital overgrowth. Am J Med Genet A. 2005;134(3):247–253. doi: 10.1002/ajmg.a.30492. [DOI] [PubMed] [Google Scholar]
- Chan SS, Shi X, Toyama A, Arpke RW, Dandapat A, Iacovino M, Kang J, Le G, Hagen HR, Garry DJ, Kyba M. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12(5):587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Firulli B, Firulli AB. A bHLH code for cardiac morphogenesis. Pediatr Cardiol. 2010;31(3):318–324. doi: 10.1007/s00246-009-9608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Muller-Hocker J, Kitajima S, Lickert H, Rupp R, Franz WM. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10(3):338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife. 2014:3. doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher F, Crouch DH, Jayaraman PS, Clark W, Gillespie DA, Goding CR. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993;12(13):5075–5082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Chung WK. Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb Perspect Med. 2014;4(7):a013953. doi: 10.1101/cshperspect.a013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS, Gai X, Shaikh TH. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6(6):592–602. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KA, Bresnick EH. Sequence determinants of DNA binding by the hematopoietic helix-loop-helix transcription factor TAL1: importance of sequences flanking the E-box core. Gene Expr. 1998;7(2):87–101. [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron JT, Pope M, Bu’lock FA, Thornborough C, Eason J, Setchfield K, Ketley A, Kirk EP, Fatkin D, Feneley MP, Harvey RP, Brook JD. Combined mutation screening of NKX2-5, GATA4, and TBX5 in congenital heart disease: multiple heterozygosity and novel mutations. Congenital heart disease. 2012;7(2):151–159. doi: 10.1111/j.1747-0803.2011.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP, Waddell LB, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81(2):280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Genevieve D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79(2):370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kophengnavong T, Michnowicz JE, Blackwell TK. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Mol Cell Biol. 2000;20(1):261–272. doi: 10.1128/mcb.20.1.261-272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm H, Deutsch MA, Dressen M, Doppler S, Werner A, Horer J, Cleuziou J, Schreiber C, Bohm J, Laugwitz KL, Lange R, Krane M. Mutational analysis of the human MESP1 gene in patients with congenital heart disease reveals a highly variable sequence in exon 1. Eur J Med Genet. 2013;56(11):591–598. doi: 10.1016/j.ejmg.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Lalani SR, Belmont JW. Genetic basis of congenital cardiovascular malformations. Eur J Med Genet. 2014;57(8):402–413. doi: 10.1016/j.ejmg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A, Auer H, Achouri Y, Dubois C, Bondue A, Simons BD, Blanpain C. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014;16(9):829–840. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, Murphy KM. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3(1):55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42(9):1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Morimoto M, Takahashi Y, Koseki H, Saga Y. Identification of Epha4 enhancer required for segmental expression and the regulation by Mesp2. Development. 2006;133(13):2517–2525. doi: 10.1242/dev.02422. [DOI] [PubMed] [Google Scholar]
- Peyvandi S, Lupo PJ, Garbarini J, Woyciechowski S, Edman S, Emanuel BS, Mitchell LE, Goldmuntz E. 22q11.2 deletions in patients with conotruncal defects: data from 1,610 consecutive cases. Pediatr Cardiol. 2013;34(7):1687–1694. doi: 10.1007/s00246-013-0694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch MG, Gramlich M, Sunde M, Schmitt KR, Lee SH, Richter S, Kersten A, Perrot A, Panek AN, Al Khatib IH, Nemer G, Megarbane A, et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet. 2010;47(4):230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Borlak J. NKX2-5: an update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD) Hum Mutat. 2010;31(11):1185–1194. doi: 10.1002/humu.21345. [DOI] [PubMed] [Google Scholar]
- Saga Y. Genetic rescue of segmentation defect in MesP2-deficient mice by MesP1 gene replacement. Mech Dev. 1998;75(1–2):53–66. doi: 10.1016/s0925-4773(98)00077-x. [DOI] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10(8):345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126(15):3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281(5373):108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Shi X, Zirbes KM, Rasmussen TL, Ferdous A, Garry MG, Koyano-Nakagawa N, Garry DJ. The Transcription Factor Mesp1 Interacts with cAMP-responsive Element Binding Protein 1 (Creb1) and Coactivates Ets Variant 2 (Etv2) Gene Expression. J Biol Chem. 2015;290(15):9614–9625. doi: 10.1074/jbc.M114.614628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallmeyer B, Fenge H, Nowak-Gottl U, Schulze-Bahr E. Mutational spectrum in the cardiac transcription factor gene NKX2.5 (CSX) associated with congenital heart disease. Clin Genet. 2010;78(6):533–540. doi: 10.1111/j.1399-0004.2010.01422.x. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Takagi A, Hiraoka S, Koseki H, Kanno J, Rawls A, Saga Y. Transcription factors Mesp2 and Paraxis have critical roles in axial musculoskeletal formation. Dev Dyn. 2007;236(6):1484–1494. doi: 10.1002/dvdy.21178. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Firulli AB. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birt Defects Res A Clin Mol Teratol. 2011;91(6):485–494. doi: 10.1002/bdra.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Rodgers R, Firulli BA, Conway SJ, Firulli AB. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol. 2008;320(1):131–139. doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Firulli BA, Lin A, Spicer DB, Howard MJ, Firulli AB. Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest. PLoS Genet. 2013;9(3):e1003405. doi: 10.1371/journal.pgen.1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM. Mesp1 at the heart of mesoderm lineage specification. Cell Stem Cell. 2008;3(1):1–2. doi: 10.1016/j.stem.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362(9393):1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.