Abstract
Statins lower plasma cholesterol by as much as 50%, thus reducing future cardiovascular events. However, the physiological effects of statins are diverse and not all are related to low density lipoprotein cholesterol (LDL-C) lowering. We performed a small clinical pilot study to assess the impact of statins on lipoprotein-associated proteins in healthy individuals (n = 10) with normal LDL-C (<130 mg/dL), who were treated with rosuvastatin (20 mg/day) for 28 days. Proteomic analysis of size-exclusion chromatography isolated LDL, large high density lipoprotein (HDL-L), and small HDL (HDL-S) fractions and spectral counting was used to compare relative protein detection before and after statin therapy. Significant protein changes were found in each lipoprotein pool and included both increases and decreases in several proteins involved in lipoprotein metabolism, complement regulation and acute phase response. The most dramatic effect of the rosuvastatin treatment was an increase in α-1-antirypsin (A1AT) spectral counts associated with HDL-L particles. Quantitative measurement by ELISA confirmed an average 5.7-fold increase in HDL-L associated A1AT. Molecular modeling predictions indicated that the hydrophobic reactive center loop of A1AT, the functional domain responsible for its protease inhibitor activity, is likely involved in lipid binding and association with HDL was found to protect A1AT against oxidative inactivation. Cell culture experiments, using J774 macrophages, demonstrated that the association of A1AT with HDL enhances its antiprotease activity, preventing elastase induced production of tumor necrosis factor α. In conclusion, we show that statins can significantly alter the protein composition of both LDL and HDL and our studies reveal a novel functional relationship between A1AT and HDL. The up-regulation of A1AT on HDL enhances its anti-inflammatory functionality, which may contribute to the non-lipid lowering beneficial effects of statins.
Epidemiological studies have clearly identified elevated plasma cholesterol as an independent risk factor for the development of cardiovascular disease (CVD)1 (1). Plasma cholesterol is carried in emulsions of lipid and protein called lipoproteins. Lipoproteins exist as a polydisperse distribution of distinct particle classes most commonly classified by density as very low, low, intermediate, and high-density lipoproteins. A perhaps overly simplistic but well accepted paradigm for the role of lipoproteins in the development CVD is that excess low density lipoproteins (LDL) promote CVD, by depositing cholesterol in atherosclerotic plaque, whereas high density lipoprotein (HDL) particles remove excess cholesterol and perhaps mediate other anti-atherogenic effects. This has been translated into the clinical setting in terms of diagnostic testing. The primary metric for assessment of CVD risk related to these lipoproteins is largely based on the cholesterol content of each of these lipoprotein particles (i.e. LDL-C and HDL-C).
Statins are a class of compounds that inhibit a key enzyme in cholesterol synthesis in the liver, namely HMG-CoA reductase (3-hydroxy-3-methyl-glutaryl-CoA reductase). These compounds also have indirect effects on cholesterol metabolism by upregulating hepatic expression of the LDL receptor and thus can lower circulating LDL-C by as much as 50% and provide significant protection against CVD (2). There is also growing evidence that the statins provide cardiovascular protection by mechanisms that are independent of their LDL-C lowering effect, including anti-inflammatory and anti-apoptotic activities and also by improving endothelial cell function (3). The mechanisms, however, by which statins mediate these so called pleiotropic effects on atherosclerosis are largely unknown.
The major lipoprotein classes contain distinct subclasses, with different physical and chemical properties and differ in their relationship with CVD. For example, total LDL is composed of at least two subclasses: large buoyant and small dense LDL, which is particularly proatherogenic (4). The subclass distribution of HDL is much more complex; it consists of numerous distinct subclasses with varying lipid and protein compositions. Modern mass spectrometry (MS) techniques have allowed for thorough characterizations of the lipoprotein proteomes of both LDL and HDL. While LDL typically contains only a few prototypical proteins, such as apoB, apoE, apoC's etc., HDL particles may contain as many as 90 different proteins among its particle subclasses (5, 6, 7, 8, 9). This proteomic diversity likely accounts for the dramatic functional diversity found in HDL, including numerous mechanisms for protection against inflammation and oxidation, as well as anticoagulant and pro-vasodilatory functions, to name only a few (10).
In the current study, we tested the hypothesis that statin therapy, in addition to its known LDL-C lowering effects, will also alter the proteome content of lipoproteins and that this may affect their functional properties. We performed shotgun proteomics on lipoprotein fractions isolated by size-exclusion liquid chromatography from patients before and during statin treatment. Our analysis revealed a dramatic elevation of HDL associated α-1-antitrypsin (A1AT) protein. In addition, we present functional studies of A1AT enriched HDL, which support a novel anti-inflammatory property of this HDL subfraction, and could also explain some of the pleiotropic effects of statins.
EXPERIMENTAL PROCEDURES
Subject Selection
Fasting blood samples were obtained from ten healthy volunteers participating in a study of the NIH Center for Human Immunology, intended to evaluate effects of a statin on the immune system. This protocol was approved by the institutional review board of the National Heart, Lung, and Blood Institute (NHLBI) and registered at clinicaltrials.gov (NCT01200836); all participants provided written informed consent. Three males and 7 females were enrolled, with an average age of 44.1 ± 11 years. Participants were selected to have normal LDL-C (<130 mg/dL) and were not on any lipid modification therapy prior to the study. Recruited participants were given rosuvastatin (20 mg/day) for 28 days. Blood was collected by venipuncture at the following time points: baseline, 14 and 28 days after rosuvastatin treatment, and 14 days after stopping treatment (washout). Samples were stored at −20° C. Because samples were taken from each participant at baseline and on-treatment, each subject acted as their own control.
Lipoprotein Analysis
Lipid and lipoprotein assays were performed on a Siemens Dimension Vista analyzer, using standard enzymatic assays. HDL-C was measured by a direct assay (Siemens; Munich, Germany) and LDL-C was determined by the Friedewald equation. Lipoprotein particle numbers and average particle sizes were determined from heparinized plasma on a Vantera Clinical Analyzer (LipoScience; Raleigh, NC).
Lipoprotein Isolation by Size-Exclusion Chromatography
Collected plasma from each subject at baseline (n = 10) and after 28 days of rosuvastatin treatment (n = 10) was applied to two Superose 6 columns (GE Healthcare) arranged in series on an Akta FPLC system. The flow rate was set to 0.5 ml/min and 0.5 ml fractions were collected. Fractions were assayed for phosphatidylcholine, total cholesterol, free cholesterol and triglyceride by enzymatic assays (Wako Diagnostics; Mountain View, CA) to determine position of elution for lipoproteins. For each subject, fractions were combined to make LDL (elution vol. 17.5 - 20 ml), HDL-large (HDL-L; elution vol. 21 - 23 ml) and HDL-small (HDL-S; elution vol. 23 - 25 ml) pools, generating a total of 60 samples for MS analysis.
Lipoprotein proteomics - experimental design and statistics
Pooled FPLC fractions were applied to a phospholipid binding resin and washed to isolate lipid bound protein components, as previously described (9). Resin bound proteins were then subjected to overnight trypsin digestion at 37 °C. Resulting peptides were collected and then reduced with dithiothreitol (200 mm; 30 min at 37 °C) and carbamidomethylated with iodoacetamide (800 mm; 30 min at 25 °C). Digest solutions were dried, reconstituted in 100 μl of water + 0.1% formic acid and desalted using ZipTips (Millipore; Billerica, MA), and stored at −20 °C until MS analysis.
Desalted samples were dried and reconstituted in 20 μl of water + 0.1% formic acid and 10 μl was analyzed on a Thermo Orbitrap Velos instrument. Blank runs were performed between each sample to prevent carry over. Peak lists were generated using Proteome Discoverer (version 1.3.0.339) and resulting spectra were searched against the SwissProt database (version 012214), using Mascot (version 2.4.0) to identify protein components of the lipoprotein fractions (fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 20 PPM). Search criteria included: human taxonomy, fixed modification: carbamidomethylation (C), variable modifications: oxidation (M), deamidation (N, Q) and up to 2 missed trypsin cleavage sites were allowed. Validation of peptide and protein identifications was performed using Scaffold software (version 4.1.1) and a 1.0% false discovery rate (FDR) for both peptide and protein thresholds and a minimum of 2 identified peptides were required per protein. Calculated decoy FDR for peptide and protein identifications were 0.03% and 0.7%, respectively. Spectral counting was used as a semi-quantitative comparison of protein abundance between baseline and on-treatment samples using normalized spectrum count calculated by the Scaffold software. Comparisons were only performed to estimate the relative abundance of the same protein in the same lipoprotein pool before and after rosuvastatin treatment; no comparisons were made between lipoprotein fractions. Data was analyzed by student's t test to identify proteins with statistically significant (p < 0.05) changes in normalized spectral counts. Because this MS analysis was intended primarily for screening purposes, to identify candidates for hypothesis driven functional experiments, the data analysis was not corrected for multiple comparisons. All MS data have been deposited to the ProteomeXchange Consortium (11) via the PRIDE partner repository with the dataset identifier PXD002633.
Preparation of α-1-antitrypsin Enriched HDL
Reconstituted HDL was prepared by cholate dialysis method, using human purified apolipoprotein A-I and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (Avanti; Alabaster, AL) as previously described (12). Native human HDL was isolated from plasma by sequential density gradient ultracentrifugation (13). Reconstituted or native HDL was co-incubated with α-1-antitrypsin protein (Human, Sigma Aldrich; St. Louis, MO) overnight at 37 °C. Unbound A1AT was removed by filtration, using Amicon Ultra 100 kDa centrifugal filter units (Millipore). Elastase activity assays were performed with the EnzChek® elastase assay kit (Life Technologies; Frederick, MD).
Cell Culture Experiments
J774 mouse macrophages were used to examine the potential functional role of α-1-antitrypsin enriched HDL in inflammation. For these experiments, cells were plated in 12-well culture plates at a density of 1 × 105 cells/well 2 days prior to the experiment. Cells were washed twice with PBS and placed in serum-free media for 1 h prior to addition of treatment. Treatments (PBS, nHDL, A1AT-HDL, or A1AT) were added to serum-free culture media: HDL protein was matched for nHDL and A1AT-nHDL at a final concentration of 200 μg/ml. A1AT concentration was matched between free A1AT and A1AT-nHDL at a final concentration of 2 μm. Cells were then incubated with elastase (porcine pancreas, Sigma Aldrich, 500 nm) for 4 h and media was collected and centrifuged at 3000 × g for 5 min to pellet any loose cells or debris and supernatant was transferred to fresh tubes and stored at −80 °C until further analysis. Elisa assays for mouse TNF-α (BioLegend; San Diego, CA) were performed on cell culture media.
RESULTS
Effect of Rosuvastatin on Lipoprotein Lipid Composition and Particle Numbers
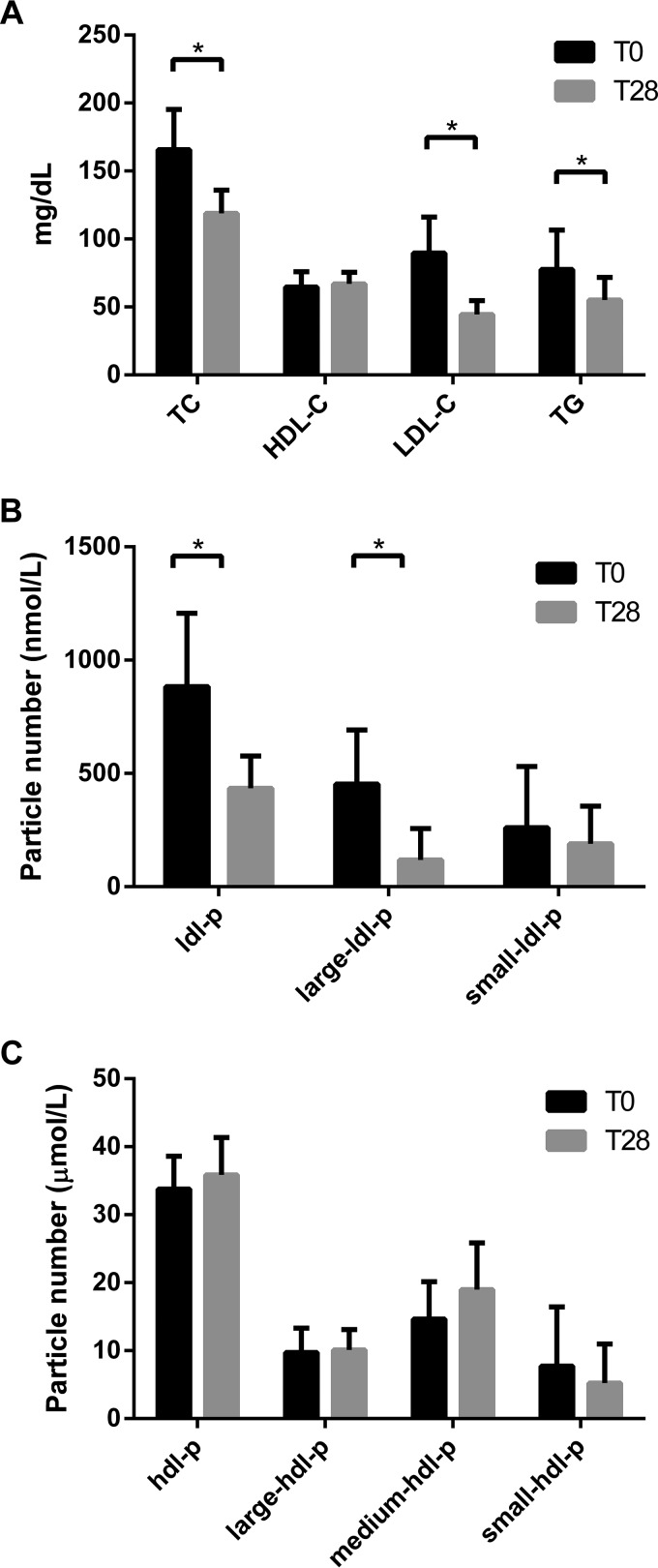
Serum samples from subjects at baseline and after 28 days of rosuvastatin therapy were analyzed for plasma lipids (Fig. 1A). As expected, treatment with rosuvastatin resulted in reductions in total cholesterol (-28%; p < 0.001), LDL-C (-50%; p < 0.001) and triglyceride (-29%; p < 0.01) (Fig. 1A). There was a modest 3.6% increase of HDL-C that was not statistically significant, although this effect of statins has been confirmed in larger studies (14). The effect of rosuvastatin on lipoprotein particle numbers mirrored the effects on the lipid levels. LDL particle number (LDL-p) was reduced by 51% (p < 0.001), which was predominantly due to reduction of large LDL (Fig. 1B). HDL-p was increased by 6.2% but was not statistically significant (Fig. 1C).
Fig. 1.
Effect of rosuvastatin on plasma lipids and lipoprotein particle numbers. A, Rosuvastatin effects on total plasma lipid levels (TC = total cholesterol; HDL-C = HDL cholesterol; LDL-C = LDL cholesterol; TG = triglyceride). The effect of rosuvastatin on LDL particle number (B) and HDL particle number (C) were measured by nuclear magnetic resonance. T0 and T28 are timepoints indicating baseline and after 28 days of rosuvastatin treatment, respectively. Data are mean ± standard deviation. * indicates p < 0.01.
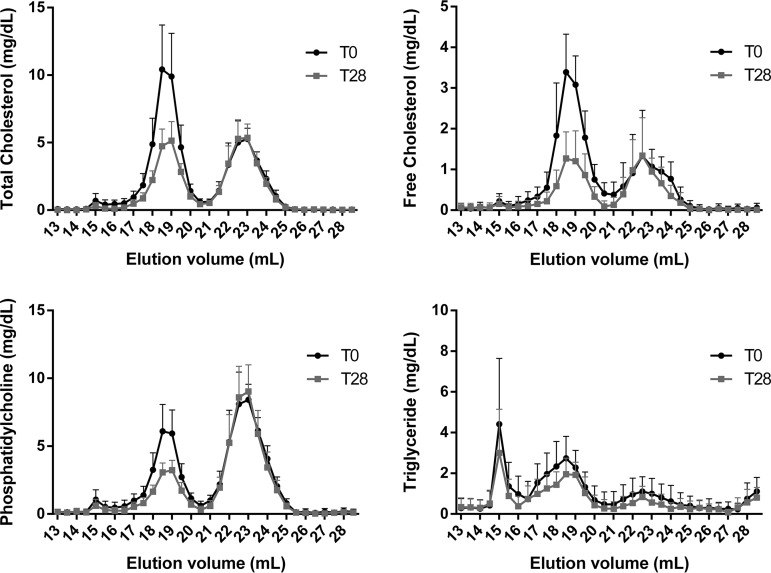
Serum was separated by size-exclusion chromatography to isolate lipoproteins by size. Collected fractions were analyzed for total cholesterol, free cholesterol, phosphatidylcholine, and triglycerides (Fig. 2). Results from this analysis, indicate a reduction in lipids associated with the LDL peak (Elution volume = 17–20 ml). This is consistent with the clinical lipid measures presented in Fig. 1. As before, no major effect on HDL lipids (Elution volume = 21–25 ml) was observed.
Fig. 2.
Effect of rosuvastatin on plasma lipid distributions by size-exclusion chromatography. Plasma from patients at baseline (T0) and after 28 days (T28) of rosuvastatin treatment was separated on two Superose 6 columns arranged in series. Collected fractions were analyzed for total cholesterol, free cholesterol, phosphatidylcholine and triglyceride. Data are mean ± standard deviation.
Mass Spectrometry Analysis of Lipoprotein Proteome
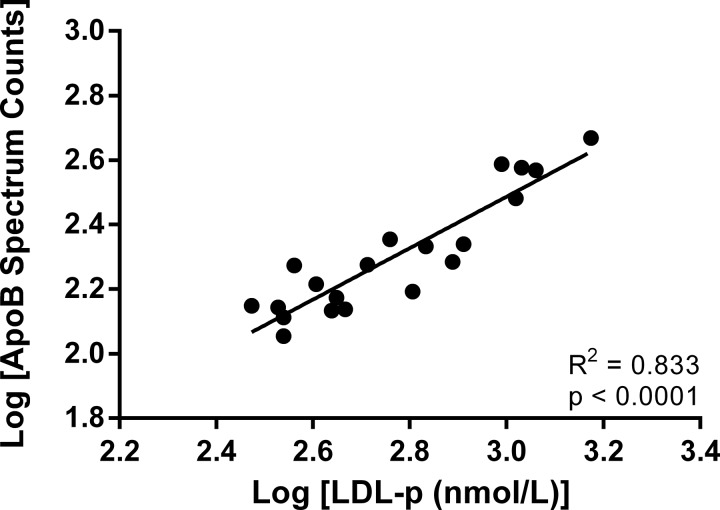
To test our hypothesis that rosuvastatin alters the lipoprotein proteome, we pooled fractions containing LDL or HDL and used mass spectrometry to analyze the lipid associated protein content of each of these lipoproteins in each subject at baseline and after 28 days on rosuvastatin. The HDL peak was divided into two halves, representing large and small HDL, to gain additional insight given the proteomic complexity of HDL. Spectral counting was used as a semi-quantitative screen to identify proteins whose abundance changed because of rosuvastatin treatment, an experimental approach that has been validated in previous studies (9, 15). As additional validation of the spectral counting approach, we also determined the correlation between spectral counts for apolipoprotein B and LDL-p. The stoichiometry on LDL is one molecule of apoB per particle so a linear relationship would be expected between a quantitative measure of apoB and LDL particle number, which was observed in Fig. 3 (R2 = 0.833, p < 0.0001) (Fig. 3).
Fig. 3.
ApoB spectral counts correlate with LDL particle number. As validation of the semi-quantitative potential of spectral counting under our experimental conditions we compared spectral counts for apolipoprotein B (apoB) versus LDL particle number. ApoB is a core protein of LDL and has a well-established 1:1 (mol apoB/mol LDL) stoichiometry.
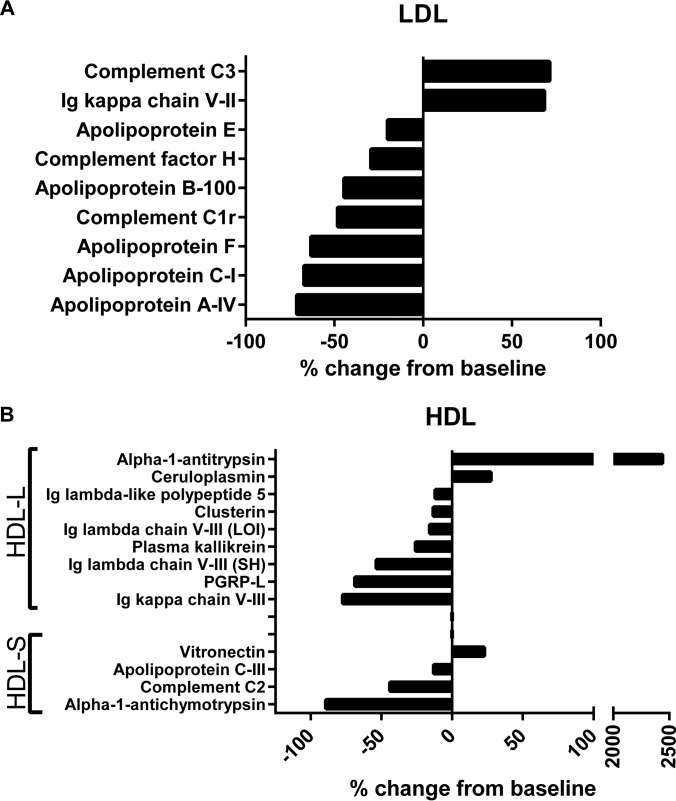
A total of 154 proteins were identified by MS analysis of all samples (supplemental Table S1). The large number of identified proteins and the relatively small sample size of this study resulted in few statistically significant results after correction for multiple comparisons; however, because the objective of this study was strictly focused on screening to identify candidates for hypothesis driven studies, statistical comparisons between spectral counts from baseline and rosuvastatin treated samples were performed without correction for multiple comparisons. In the LDL fraction, nine proteins were found to display changes in abundance by spectral counting as a result of the rosuvastatin treatment (Fig. 4A). The majority of these were decreases, indicating reductions in total protein mass of LDL from the treatment. Additionally, the magnitude of these reductions in spectral counts ranged from −20 to −71%, which is similar to the degree of LDL particle number lowering achieved in this study and suggests that these results are largely a result of the LDL lowering effect of the rosuvastatin. It is interesting, however, that complement C3 (+71%) and Ig κ chain V-II (+68%) demonstrated increased spectral counts in the LDL fraction, despite the significant reduction in LDL particle number and apoB. This relationship has not been previously described nor is the functional importance of these changes known.
Fig. 4.
Rosuvastatin alters the lipoprotein proteome. Statistically significant changes to the LDL (A) and HDL (B) proteomes resulting from rosuvastatin treatment are displayed as percent change compared with baseline. HDL-L = large HDL; HDL-S = small HDL; PGRP-L = N-acetylmuramoyl-l-alanine amidase. Statistical comparisons were made using student's t test. All displayed data are p < 0.05.
Although rosuvastatin had minimal effect on HDL lipids and caused only a small increase in HDL particle number, several changes were observed in the HDL proteome (Fig. 4B). Most notably, on the large HDL, there was a marked elevation in α-1-antitrypsin spectral counts (+2438%). In addition, ceruloplasmin also showed an increase in spectral counts (+27%). Reductions in spectral counts were also observed for the following proteins: several immunoglobulin chains (variable), N-acetylmuramoyl-l-alanine amidase (-69%), kallikrein (-26%), clusterin (-13%) and Ig lambda like polypeptide 5 (-12%). Overall, there were fewer changes in spectral counts in the small HDL fraction: (α-1-antichymotrypsin (-89%), complement C2 (-44%), apolipoprotein C-III (-13%), and vitronectin (+23%).
Quantitation of α-1-antitrypsin Raising Effect of Rosuvastatin
Of the numerous changes in the HDL proteome following rosuvastatin treatment, we decided to focus on A1AT because it showed the largest change and the function of A1AT is relatively well understood. In fact, the magnitude of the increase of HDL bound A1AT in this study was much greater than changes found in previous studies that examined the effect of various diseases or therapy on lipoprotein proteome (5, 15, 16). Additionally, A1AT has been shown to be involved in cardiovascular disease by modulating various inflammatory processes (17, 18).
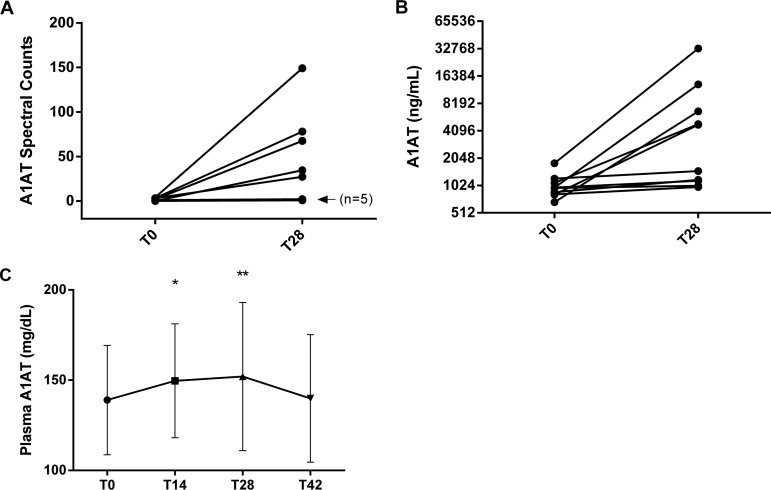
To more quantitatively evaluate the change in A1AT levels observed in the MS data, we performed ELISA assays to measure A1AT in the large HDL fractions from all subjects. We noted that there was a distinct segregation of the subjects into two groups: high responders that showed a marked elevation of A1AT (range: five- to 18-fold) in their large HDL fraction in response to rosuvastatin therapy and low-responders that only showed a modest increase in A1AT levels (range: 6 - 36%) after therapy. This segregation was consistent with our MS data (Fig. 5A and 5B) and did not appear to be associated with baseline plasma lipid measures or CRP (data not shown).
Fig. 5.
Quantitative measurement of α-1-antitrypsin on HDL and in plasma. A, Individual patient spectral counts for α-1-antitrypsin (A1AT) in large HDL at baseline (T0) and after 28 days (T28) of rosuvastatin treatment, n = 10 for each time point. The “n = 5” indicator points to data from 5 subjects with a high degree of overlap. B, Quantitative measurement of A1AT in large HDL by ELISA assay. C, Time course of plasma A1AT concentrations during rosuvastatin treatment and after two-week washout period (Day 42 time point). * indicates p < 0.05 and ** indicates p < 0.01 compared with T0.
In addition to measuring the A1AT content of large HDL, we also measured total A1AT in plasma. There was a clear trend of increasing plasma A1AT while on treatment and a return to baseline after a 2 week treatment washout, this effect was relatively small, ∼10% (Fig. 5C). The change in total A1AT did not appear to correlate with the change observed in A1AT on large HDL after rosuvastatin therapy. Based on the content of A1AT on HDL-L and in plasma, we estimate that on average ∼4.54 ± 1.73% of total A1AT was bound to HDL after rosuvastatin therapy, whereas only 0.84 ± 0.037% was bound at baseline before treatment. We calculated an A1AT/HDL-particle ratio (mol:mol) of about 1:100 at baseline and 1:25 after rosuvastatin treatment.
A1AT Binds to Lipids with its Reactive Center Loop
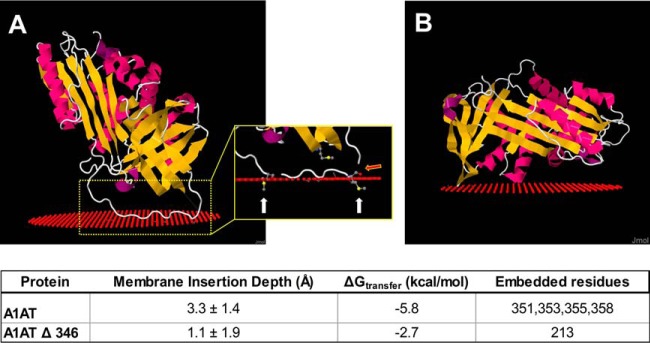
To examine the potential structural basis for the interaction of A1AT with lipoproteins, we used modeling software that calculated the optimum thermodynamic orientation for a protein to interact with a phospholipid bilayer, using their 3D structures. The crystal structure for A1AT was downloaded from Protein Data Bank (http://www.rcsb.org; structure ID: 3NE4) and used as input for the lipid binding prediction in the position of proteins in membranes server (http://opm.phar.umich.edu/) (19). From this analysis, an exposed random coil region of the A1AT protein was predicted to insert into the outer leaflet of a phospholipid bilayer (Fig. 6A). This region of A1AT corresponds to the reactive center loop (RCL) of A1AT, which contains the active site responsible for the ability of this protein to inhibit proteolytic activity and is relatively hydrophobic. We noted, in this binding model, that two methionine residues critical to the function of A1AT (Met351 and Met358) are predicted to be buried in the lipid surface. These methionine residues are highly susceptible to oxidation to methionine sulfoxide. Previous studies have shown that oxidative modification of either of these residues will result in the loss of anti-elastase activity (20). If the RCL region of the protein is removed from the protein structure (A1ATΔ346, Fig. 6B), the protein is predicted to have markedly reduced affinity for the lipid surface, as indicated by decreased calculated insertion depth and decreased ΔGtransfer energy.
Fig. 6.
Structural prediction of lipid binding by α-1-antitrypsin. A, Predicted binding of α-1-antitrypsin (A1AT) to a lipid surface (red spheres). Inset demonstrates that methionine residues (Met351 and Met358) are embedded in the lipid (white arrows) and indicates the cut site for neutrophil elastase (red arrow). B, Predicted lipid binding of A1AT structure with the reactive center loop removed (A1AT Δ 346).
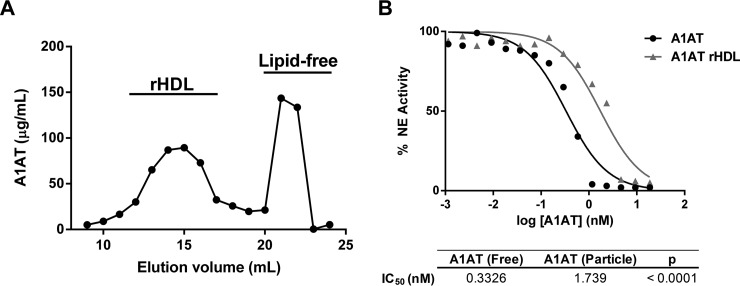
Based on this lipid binding model for A1AT, we hypothesized that the burying of the RCL domain into the lipids surface of HDL would affect its capacity to inhibit elastolytic activity. To test this, we made reconstituted HDL containing A1AT by cholate dialysis. As can be seen by the gel filtration profile, when A1AT was incubated with discoidal HDL, containing apoA-I and phospholipids, a significant fraction of A1AT associates with HDL (Fig. 7A). Similarly, and consistent with previous studies, when native HDL isolated from plasma was incubated with A1AT, it became enriched in A1AT (data not shown) (21).
Fig. 7.
α-1-antitrypsin has reduced anti-elastase activity when bound to reconstituted HDL. Reconstituted HDL (rHDL) were prepared from apoA-I and phospholipids by cholate dialysis and then co-incubated with α-1-antitrypsin (A1AT) to generate A1AT enriched rHDL. A, Size exclusion chromatography on tandem Superdex 200 columns was used to isolate HDL bound A1AT from lipid free protein. B, The ability of lipid free and rHDL bound A1AT to inhibit neutrophil elastase (NE) activity was measured by fluorometric assay.
Next, we compared the ability of A1AT bound to HDL versus free A1AT to inhibit elastase activity. The HDL particle bound A1AT showed a fivefold reduction in elastase inhibitor capacity (Fig. 7B), suggesting that the active site in the RCL is sterically blocked from interacting with elastase when bound to HDL. Overall, this data supports our model of lipid binding and suggests several possible functional implications of the interaction between A1AT and HDL, which we tested below.
HDL Binding Protects A1AT Anti-elastase Activity from Inactivation by H2O2
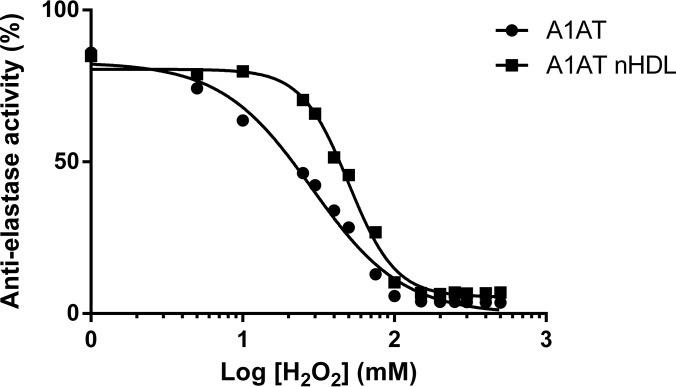
In atherosclerotic plaque, activated neutrophils and macrophages create an oxidizing environment by generating reactive oxygen species, such as hydrogen peroxide (H2O2) (22, 23). Based on the binding model of A1AT to HDL, we predicted that submersion of the critical Met351 and Met358 residues, which are susceptible to H2O2 oxidation, in the lipid surface may confer protection of these residues in the RCL against oxidation and therefore promote the preservation of anti-elastase activity in an oxidizing environment. To test this hypothesis, we generated A1AT enriched native HDL (nHDL) by co-incubating A1AT with isolated human HDL and then removed unbound A1AT. Either free A1AT or A1AT-nHDL was exposed to H2O2 at various concentrations and remaining anti-elastase activity was measured. Nonlinear regression analysis of the dose response curves of anti-elastase activity versus [H2O2] indicated a shift in IC50 (28.09 versus 46.42 mm H2O2 for A1AT and A1AT-HDL, respectively; p < 0.0001), indicating that HDL bound A1AT was more resistant to inactivation by H2O2 (Fig. 8). This finding suggested that although binding of A1AT to reconstituted HDL resulted in a reduction of A1AT activity, the interaction of A1AT with HDL may be beneficial in stabilizing its activity in a pro-oxidant environment, such as an atherosclerotic plaque.
Fig. 8.
Binding to HDL protects α-1-antitrypsin anti-elastase activity from oxidation by H2O2. HDL isolated from healthy human donors was co-incubated with α-1-antitrypsin (A1AT) to generate A1AT enriched nHDL. Lipid free A1AT and A1AT nHDL were exposed to varying concentrations of H2O2 for 30 min before measurement of anti-elastase activity by fluorometric assay. Nonlinear regression analysis was used for comparison of curve fits and found the two curves to be significantly different (p < 0.0001).
Effects of A1AT Enriched HDL on Elastase Induced Macrophage Activation
An early event in the initiation of atherosclerosis is the infiltration of circulating neutrophils and monocytes into the subendothelial space. Upon activation, neutrophils and macrophages in this environment produce oxygen radicals and also secrete neutrophil elastase, a proteolytic enzyme, which can cause degradation of the extracellular matrix and activation of proinflammatory signaling pathways in nearby cells via protease activated receptors. We used the J774 macrophage cell line to examine the proinflammatory response induced by elastase exposure and to determine the effect A1AT enriched HDL would have on this response.
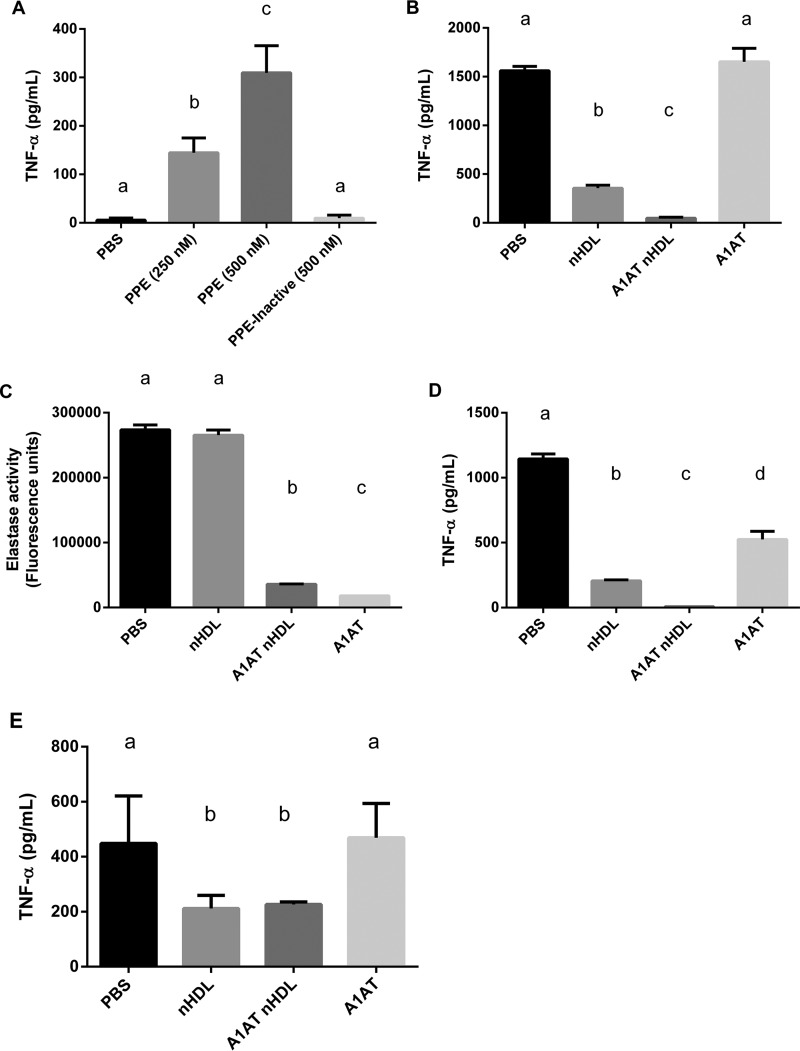
We first demonstrate that, in the J774 cell system, exposure to elastase induces a strong proinflammatory cytokine response, with increased TNF-α in the cell media in a dose-dependent manner and this effect is absent when treated with heat-inactivated elastase (Fig. 9A). Next, J774 cells were pre-incubated with PBS, native HDL (nHDL), A1AT enriched nHDL, or lipid-free A1AT for 1 h prior to the addition of elastase to the culture media to allow for inactivation of A1AT. Upon elastase addition, cells pretreated with PBS displayed a robust production of TNF-α. Native HDL had a strong anti-inflammatory effect, significantly reducing TNF-α production by −77.2% compared with PBS pretreatment (Fig. 9B). This already impressive effect was further amplified and TNF-α production was almost completely suppressed (-97%) when cells were pretreated with HDL further enriched in A1AT. In contrast, pretreatment with lipid-free A1AT had no significant protection against elastase induced TNF-α production. The almost complete lack of effect with lipid-free A1AT was unexpected but suggests that in the lipid-free form, A1AT may be quickly degraded or inactivated when cultured with macrophages. To determine if degradation was responsible for the loss of A1AT activity, we performed Western blot for A1AT on the cell culture media and found no evidence of proteolytic degradation of A1AT (supplemental Fig. S1). This suggested to us that the loss of antiprotease activity in the lipid-free A1AT treatment may be due to another inactivating protein modification. We then performed elastase inhibition assays on each of the treatments from Fig 9B, but in a cell-free system. The results of this experiment demonstrated that the PBS and nHDL treatments did not inhibit elastase activity, however, both A1AT HDL and lipid-free A1AT showed significant anti-elastase activity, indicating that the cell culture environment inactivated the lipid-free A1AT but did not affect the HDL bound A1AT (Fig. 9C). In further support of this model, when each of the treatments was pre-incubated with elastase prior to addition to the cells, free A1AT was then readily able to inhibit elastase induced TNF-α production (Fig. 9D).
Fig. 9.
α-1-antitrypsin enriched HDL prevents elastase induced TNF-α production by macrophages. A, J774 mouse macrophages treated with increasing amounts of porcine pancreatic elastase (PPE) or heat inactivated elastase for 4 h, TNF-α in the culture media was measured by ELISA. B, J774 cells pretreated with PBS, isolated human HDL (nHDL), the same HDL enriched with α-1-antitrypsin (A1AT nHDL), or lipid free A1AT for 1 h prior to PPE addition. C, The ability of each of the cell treatments to inhibit elastase activity was measured in a cell-free assay. D, Treatments were pre-incubated with PPE prior to addition to cells and TNF- α was measured in the culture media after 4 h. E, J774 cells were pre-incubated with each treatment for 1 h; cells were then washed twice with PBS and placed in fresh media containing PPE and TNF- α was measured in the culture media after 4 h. All experiments were repeated at least 3 times and were done in triplicates. Treatments were compared using one-way ANOVA and Tukey's multiple comparisons test, p < 0.05 was considered significant. The letters above each treatment indicate statistical significance; within each graph, bars bearing different letters were statistically different from each other.
In another experiment, J774 cells were pretreated in the same manner as for Fig. 9B, except that the cell media was removed and the cells were washed prior to addition of fresh serum free media containing elastase. Under these conditions, the nHDL pretreatment still provided significant protection against elastase induced TNF-α production (Fig. 9E), indicating that the effect of native HDL is occurring via an intracellular or signaling pathway. However, the additional protection conferred by A1AT enrichment was lost, indicating that the mechanism of action for A1AT HDL is by direct inhibition of elastase in the culture media. Treatment with lipid-free A1AT still had no effect.
Overall, this data supports a mechanism whereby lipid-free A1AT is rapidly inactivated in macrophage culture media and is unable to inhibit elastase induced cytokine production by macrophage cells. However, when associated with HDL, A1AT retains its anti-elastase activity and efficiently inhibits elastase induced TNF-α production. Based on our lipid binding model, we predict that the preservative effect of HDL binding on A1AT activity may be a result of the interaction of the RCL with the lipid surface conferring protection in an oxidizing environment.
DISCUSSION
In this study, we report the impact of rosuvastatin treatment on the lipoprotein proteome in humans and found that several protein components of both LDL and HDL particles were affected by treatment. The majority of proteome effects on LDL were reductions in protein content, which likely reflect the overall reduction in LDL particle number associated with statin therapy. However, despite this reduction in particle number, there were two proteins that displayed increased abundance on LDL with statin treatment, namely complement C3 and Ig kappa chain V-II. We found no existing reports of direct functional relationships between these proteins and LDL, but their elevation on the background of dramatic LDL lowering may indicate the presence of a subpopulation of LDL that is either resistant to lowering by rosuvastatin or perhaps is generated as a consequence of the treatment. Whether or not these particles have any influence on lipoprotein metabolism or cardiovascular disease is an area of interest for future studies.
Although rosuvastatin treatment had little or no impact on HDL particle number, total HDL-C, or HDL lipid profile by FPLC, there were several major proteome changes observed. Among these was a dramatic 24-fold increase in A1AT spectral counts associated with large HDL, a protein change much greater than typically observed in other similar lipoprotein proteome studies. Because of this and the known function of A1AT, we chose to focus our attention on this protein change and to investigate its functional consequences. A1AT is an acute phase reactive plasma protein with a typical plasma concentration of about 1.5 mg/ml. It belongs to the serine protease inhibitor (SERPIN) family of proteins of which there are 36 members in humans, 29 of which have antiprotease activity. A1AT is one of the most abundant SERPINs and is the primary physiological inhibitor of neutrophil elastase (NE) (24). NE is produced by activated neutrophils and macrophages in atherosclerotic lesions where it degrades components of the extracellular matrix (i.e. elastin, collagen and fibronectin) (25, 26). Additionally, cholesterol loaded monocyte-derived macrophages, such as those found in atherosclerotic plaque, express elevated elastolytic activity (27). Besides degrading extracellular matrix, elastase can also stimulate production of pro-inflammatory cytokines and trigger pro-apoptotic signaling via protease activated receptors expressed on most cells (28, 29). These combined activities of elastase within the vessel wall likely contribute to atherosclerotic progression by promoting smooth muscle cell migration, endothelial cell apoptosis, and plaque instability (30).
It has been demonstrated that HDL is actively transported across the vascular endothelium at the site of atherosclerotic plaque (31), thus delivering HDL to this inflammatory, proteolytic environment. The fate of the protein cargo (i.e. non-apoA-I proteins) of HDL during this process has not yet been examined. However, it seems possible that the process of endothelial trancytosis of HDL may act as a mechanism for delivery of HDL's largely anti-inflammatory and antiproteolytic cargo to a site of inflammatory distress. A1AT is fairly abundant in plasma, but it may need to reach the subendothelial space, where elastolytic proteases are produced by activated neutrophils and macrophages, to effectively inhibit protease mediated damage to the vessel wall. Although some small plasma proteins are capable of relatively rapid movement across the vascular endothelium, A1AT is similar to albumin in size and negative charge (pI ≈ 4), and thus it is likely not capable of passive diffusion across the endothelium and may require active transport or a chaperone such as HDL.
Our findings expand significantly upon existing studies by first showing that A1AT binding to HDL is induced by rosuvastatin and by identifying a novel functional benefit of this association. Our data support a protective role for HDL, whereby the anti-elastase activity of A1AT is preserved by interaction with HDL. Although our experiments with reconstituted HDL demonstrate that A1AT activity is reduced upon binding to HDL, a significant amount of antiprotease activity remains. Furthermore, it has recently been shown, in atherosclerotic plaques, that HDL particles eventually disassemble when they enter tissue and therefore may release free A1AT (32). Based on our HDL binding model and our experiments with hydrogen peroxide, one possible mechanism for the preservation of A1AT activity when bound to HDL may be through shielding of oxidation susceptible methionine residues in the RCL, which when oxidized result in loss of A1AT activity. Other possible mechanisms may involve the protection of A1AT from degradation by other macrophage secreted proteases or HDL-directed cellular uptake and intracellular antiproteolytic activity. Detailed mechanistic and structural investigations of these hypotheses are underway.
The association of A1AT with both LDL and HDL particles has been previously demonstrated by biochemical and proteomics based techniques. Binding to LDL occurs when A1AT has been oxidized and is inactivated (33), but a change in the oxidation status of A1AT has not been previously implicated in altering its binding to HDL. There have been no reports on the mechanism of A1AT binding to HDL. It is currently unclear whether A1AT becomes associated with HDL during particle generation or is in a state of equilibrium, constantly exchanging to and from preformed HDL particles. Our data suggest only a minimal increase in total plasma A1AT while on rosuvastatin, an amount that is likely insufficient to account for a simple equilibrium driven shift onto HDL of the magnitude seen in this study. This suggests the possibility of increased de novo generation of A1AT containing HDL or that an additional plasma factor or a change in some physical property of HDL in response to statin treatment that may be responsible for the increased binding of A1AT. There have been only a limited number of functional investigations of A1AT on HDL. Studies by Meilhac et al. first suggested a possible functional role for HDL bound A1AT in preventing apoptosis in vascular smooth muscle cells (34). A second study from the same group demonstrated a role for HDL in the selective transport of functional A1AT from the circulation to the lung, where it can prevent elastase induced morphological and functional damage in a rat model (21). These findings suggest a possible important role of A1AT bound HDL for the treatment of chronic obstructive pulmonary disease (COPD) or emphysema, where excessive elastolytic activity causes destruction of lung tissue and is often not adequately treated by replacement therapy with just A1AT (35).
Although the focus of this study is on A1AT, 9 other members of the SERPIN family are consistently identified in proteomics studies of HDL (8). The complete structural information is not available for all of these, but all of the well characterized SERPINs have structural organizations similar to that of A1AT with an exposed hydrophobic meta-stable RCL domain. It may be that many of these other SERPINs also bind HDL by a similar mechanism involving the RCL and similar functional relationships exist for this entire class of proteins.
Additional questions raised by the present report include whether the observed effects of rosuvastatin on the lipoprotein proteome are similar among all statins and whether this accounts for some of the known anti-inflammatory effect of statins (36). One study of the HDL proteome, in patients receiving combination therapy of atorvastatin and niacin, found reduced apoE content in HDL3, however no effect on A1AT was detected (16). The absence of effect on A1AT in this study may be explained by several factors including the use of a different statin, addition of niacin treatment, or differences in HDL isolation (ultracentrifugation versus gel filtration). Some of the anti-inflammatory effects of statins have been attributed to the ability to block the Rho signaling pathway (37) but based on the results of this study their ability to modulate A1AT levels on HDL may also be relevant. It is interesting to note that there have been several recent reports of statins (i.e. simvastatin and atorvastatin) having protective effects on lung injury induced by elastase (38, 39) or by cigarette smoke (40, 41), which could potentially also be mediated by A1AT enriched HDL.
In summary, studies of the lipoprotein proteome have gained increasing interest due to their clear potential for identifying functionally relevant lipoprotein subspecies. However, the list of lipoprotein associated proteins is growing faster than our understanding of the functional relevance of these complexes. New insight into the diagnostic or therapeutic implications of HDL subfractions with specific protein content will likely require direct experimental investigation as was done in this study. Although much still needs to be done in regard to the role of A1AT interaction with HDL, to our knowledge this is the first report of a newly identified protein on HDL by LC-MS that has been shown to have a possible functional role in the pathogenesis of atherosclerosis.
Supplementary Material
Acknowledgments
We thank the National Heart Lung and Blood Institute Proteomics Core for advice and use of facilities.
Footnotes
Author contributions: S.M.G., N.S.Y., M.B.F., and A.T.R. designed research; S.M.G., B.M., G.K., M.S., and S.P. performed research; S.M.G. analyzed data; S.M.G. wrote the paper; A.T.R. edited manuscript.
* This work was supported by the National Institutes of Health, National Heart Lung and Blood Institute. Intramural Research Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental Fig. S1, Table S1, Peptide report, Protein report.
This article contains supplemental Fig. S1, Table S1, Peptide report, Protein report.
1 The abbreviations used are:
- CVD
- Cardiovascular disease
- A1AT
- α-1-antitrypsin
- FDR
- False discovery rate
- FPLC
- Fast protein liquid chromatography
- HDL
- High density lipoprotein
- LDL
- Low density lipoprotein
- NE
- Neutrophil elastase
- nHDL
- Native HDL
- PPE
- Porcine pancreatic elastase
- RCL
- Reactive center loop
- rHDL
- Reconstituted HDL
- SERPIN
- Serine protease inhibitor
- TNF-α
- Tumor necrosis factor α.
REFERENCES
- 1.Kannel W. B., Castelli W. P., and Gordon T. (1979) Cholesterol in the prediction of atherosclerotic disease. New perspectives based on the Framingham study. Ann. Intern. Med. 90, 85–91 [DOI] [PubMed] [Google Scholar]
- 2.Baigent C., Keech A., Kearney P. M., Blackwell L., Buck G., Pollicino C., Kirby A., Sourjina T., Peto R., Collins R., and Simes R. (2005) Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 366, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 3.Jain M. K., and Ridker P. M. (2005) Anti-inflammatory effects of statins: clinical evidence and basic mechanisms. Nat. Rev. Drug. Discov. 4, 977–987 [DOI] [PubMed] [Google Scholar]
- 4.Chapman M. J., Guerin M., and Bruckert E. (1998) Atherogenic, dense low-density lipoproteins. Pathophysiology and new therapeutic approaches. Eur. Heart J. 19, A24–30 [PubMed] [Google Scholar]
- 5.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., Chea H., Knopp R. H., Brunzell J., Geary R., Chait A., Zhao X. Q., Elkon K., Marcovina S., Ridker P., Oram J. F., and Heinecke J. W. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlsson H., Leanderson P., Tagesson C., and Lindahl M. (2005) Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 5, 551–565 [DOI] [PubMed] [Google Scholar]
- 7.Karlsson H., Leanderson P., Tagesson C., and Lindahl M. (2005) Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 5, 1431–1445 [DOI] [PubMed] [Google Scholar]
- 8.Davidson W. S. (2015) The HDL proteome watch. www.davidsonlab.com [DOI] [PMC free article] [PubMed]
- 9.Gordon S. M., Deng J., Lu L. J., and Davidson W. S. (2010) Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J. Proteome Res. 9, 5239–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon S. M., Hofmann S., Askew D. S., and Davidson W. S. (2011) High density lipoprotein: it's not just about lipid transport anymore. Trends Endocrinol. Metab. 22, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matz C. E., and Jonas A. (1982) Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. J. Biol. Chem. 257, 4535–4540 [PubMed] [Google Scholar]
- 13.Chapman M. J., Goldstein S., Lagrange D., and Laplaud P. M. (1981) A density gradient ultracentrifugal procedure for the isolation of the major lipoprotein classes from human serum. J. Lipid Res. 22, 339–358 [PubMed] [Google Scholar]
- 14.McTaggart F., and Jones P. (2008) Effects of statins on high-density lipoproteins: a potential contribution to cardiovascular benefit. Cardiovasc. Drugs Ther. 22, 321–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon S. M., Davidson W. S., Urbina E. M., Dolan L. M., Heink A., Zang H., Lu L. J., and Shah A. S. (2013) The effects of type 2 diabetes on lipoprotein composition and arterial stiffness in male youth. Diabetes 62, 2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green P. S., Vaisar T., Pennathur S., Kulstad J. J., Moore A. B., Marcovina S., Brunzell J., Knopp R. H., Zhao X. Q., and Heinecke J. W. (2008) Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation 118, 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilutz H., Siegel Y., Paran E., Cristal N., and Quastel M. R. (1983) Alpha 1-antitrypsin in acute myocardial infarction. Br. Heart J. 49, 26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duckers J. M., Shale D. J., Stockley R. A., Gale N. S., Evans B. A., Cockcroft J. R., and Bolton C. E. (2010) Cardiovascular and musculskeletal co-morbidities in patients with alpha 1 antitrypsin deficiency. Respir. Res. 11, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., and Lomize A. L. (2012) OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taggart C., Cervantes-Laurean D., Kim G., McElvaney N. G., Wehr N., Moss J., and Levine R. L. (2000) Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J. Biol. Chem. 275, 27258–27265 [DOI] [PubMed] [Google Scholar]
- 21.Moreno J. A., Ortega-Gomez A., Rubio-Navarro A., Louedec L., Ho-Tin-Noe B., Caligiuri G., Nicoletti A., Levoye A., Plantier L., and Meilhac O. (2014) High-density lipoproteins potentiate alpha1-antitrypsin therapy in elastase-induced pulmonary emphysema. Am. J. Respir. Cell. Mol. Biol. 51, 536–549 [DOI] [PubMed] [Google Scholar]
- 22.Nathan C. F., and Root R. K. (1977) Hydrogen peroxide release from mouse peritoneal macrophages: dependence on sequential activation and triggering. J. Exp. Med. 146, 1648–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan S., Meng X. P., Ramasamy S., Harrison D. G., and Galis Z. S. (1996) Reactive oxygen species produced by macrophage-derived foam cells regulate the activity of vascular matrix metalloproteinases in vitro. Implications for atherosclerotic plaque stability. J. Clin. Invest. 98, 2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gettins P. G. (2002) Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4804 [DOI] [PubMed] [Google Scholar]
- 25.Weiss S. J. (1989) Tissue destruction by neutrophils. N. Engl. J. Med. 320, 365–376 [DOI] [PubMed] [Google Scholar]
- 26.Dollery C.M., Owen C.A., Sukhova G.K., Krettek A., Shapiro S.D., and Libby P. (2003) Neutrophil elastase in human atherosclerotic plaques: production by macrophages. Circulation 107, 2829–2836 [DOI] [PubMed] [Google Scholar]
- 27.Rouis M., Nigon F., Lafuma C., Hornebeck W., and Chapman M.J. (1990) Expression of elastase activity by human monocyte-macrophages is modulated by cellular cholesterol content, inflammatory mediators, and phorbol myristate acetate. Arteriosclerosis 10, 246–255 [DOI] [PubMed] [Google Scholar]
- 28.Shpacovitch V., Feld M., Hollenberg M. D., Luger T. A., and Steinhoff M. (2008) Role of protease-activated receptors in inflammatory responses, innate and adaptive immunity. J. Leukoc. Biol. 83, 1309–1322 [DOI] [PubMed] [Google Scholar]
- 29.Ramachandran R., Mihara K., Chung H., Renaux B., Lau C. S., Muruve D. A., DeFea K. A., Bouvier M., and Hollenberg M. D. (2011) Neutrophil elastase acts as a biased agonist for proteinase-activated receptor-2 (PAR2). J. Biol. Chem. 286, 24638–24648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Touchard A., Henry T. D., Sangiorgi G., Spagnoli L. G., Mauriello A., Conover C., and Schwartz R. S. (2005) Extracellular proteases in atherosclerosis and restenosis. Arterioscler. Thromb. Vasc. Biol. 25, 1119–1127 [DOI] [PubMed] [Google Scholar]
- 31.Rohrer L., Ohnsorg P. M., Lehner M., Landolt F., Rinninger F., and von Eckardstein A. (2009) High-density lipoprotein transport through aortic endothelial cells involves scavenger receptor BI and ATP-binding cassette transporter G1. Circ. Res. 104: 1142–1150 [DOI] [PubMed] [Google Scholar]
- 32.Huang Y., DiDonato J. A., Levison B. S., Schmitt D., Li L., Wu Y., Buffa J., Kim T., Gerstenecker G. S., Gu X., Kadiyala C. S., Wang Z., Culley M. K., Hazen J. E., Didonato A. J., Fu X., Berisha S. Z., Peng D., Nguyen T. T, Liang S., Chuang C. C., Cho L., Plow E. F., Fox P. L., Gogonea V., Tang W. H., Parks J. S., Fisher E. A., Smith J. D., and Hazen S. L. (2014) An abundant dysfunctional apolipoprotein A1 in human atheroma. Nat. Med. 20, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashiba S., Wada Y., Takeya M., Sugiyama A., Hamakubo T., Nakamura A., Noguchi N., Niki E., Izumi A., Kobayashi M., Uchida K., and Kodama T. (2001) In vivo complex formation of oxidized alpha(1)-antitrypsin and LDL. Arterioscler. Thromb. Vasc. Biol. 21, 1801–1808 [DOI] [PubMed] [Google Scholar]
- 34.Ortiz-Munoz G., Houard X., Martin-Ventura J.L., Ishida B.Y., Loyau S., Rossignol P., Moreno J.A., Kane J.P., Chalkley R.J., Burlingame A.L., Michel J.B., and Meilhac O. (2009) HDL antielastase activity prevents smooth muscle cell anoikis, a potential new antiatherogenic property. FASEB J. 23, 3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abusriwil H., and Stockley R. A. (2006) Alpha-1-antitrypsin replacement therapy: current status. Curr. Opin. Pulm. Med. 12, 125–131 [DOI] [PubMed] [Google Scholar]
- 36.Antonopoulos A. S., Margaritis M., Lee R., Channon K., and Antoniades C. (2012) Statins as anti-inflammatory agents in atherogenesis: molecular mechanisms and lessons from the recent clinical trials. Curr. Pharm. Des. 18, 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin G., Duez H., Blanquart C., Berezowski V., Poulain P., Fruchart J. C., Najib-Fruchart J., Glineur C., and Staels B. (2001) Statin-induced inhibition of the Rho-signaling pathway activates PPARalpha and induces HDL apoA-I. J. Clin. Invest. 107, 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi S., Nakamura H., Seki M., Shiraishi Y., Yamamoto M., Furuuchi M., Nakajima T., Tsujimura S., Shirahata T., Nakamura M., Minematsu N., Yamasaki M., Tateno H., and Ishizaka A. (2008) Reversal of elastase-induced pulmonary emphysema and promotion of alveolar epithelial cell proliferation by simvastatin in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L882–890 [DOI] [PubMed] [Google Scholar]
- 39.Boiati R. F., Manchini M. T., Jacobsen O., Dos Santos Batista J. G., Silva J. A. Jr., and Nascimento J. W. (2014) Evaluation of the anti-inflammatory activity of atorvastatin and its effect on alveolar diameter in a model of elastase-induced emphysema in rats. Drug Res. [DOI] [PubMed] [Google Scholar]
- 40.Lee S. D., Lee J. H., Kim E. K., Choi K. H., Oh Y. M., and Shim T. S. (2005) Effects of simvastatin on cigarette smoking-induced structural and functional changes in rat lungs. Chest 128, 574S. [DOI] [PubMed] [Google Scholar]
- 41.Wright J. L., Zhou S., Preobrazhenska O., Marshall C., Sin D. D., Laher I., Golbidi S., and Churg A. M. (2011) Statin reverses smoke-induced pulmonary hypertension and prevents emphysema but not airway remodeling. Am. J. Respir. Crit. Care Med. 183, 50–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.