Abstract
Protein–RNA cross-linking by UV irradiation at 254 nm wavelength has been established as an unbiased method to identify proteins in direct contact with RNA, and has been successfully applied to investigate the spatial arrangement of protein and RNA in large macromolecular assemblies, e.g. ribonucleoprotein-complex particles (RNPs). The mass spectrometric analysis of such peptide-RNA cross-links provides high resolution structural data to the point of mapping protein–RNA interactions to specific peptides or even amino acids. However, the approach suffers from the low yield of cross-linking products, which can be addressed by improving enrichment and analysis methods. In the present article, we introduce dithiothreitol (DTT) as a potent protein–RNA cross-linker. In order to evaluate the efficiency and specificity of DTT, we used two systems, a small synthetic peptide from smB protein incubated with U1 snRNA oligonucleotide and native ribonucleoprotein complexes from S. cerevisiae. Our results unambiguously show that DTT covalently participates in cysteine-uracil crosslinks, which is observable as a mass increment of 151.9966 Da (C4H8S2O2) upon mass spectrometric analysis. DTT presents advantages for cross-linking of cysteine containing regions of proteins. This is evidenced by comparison to experiments where (tris(2-carboxyethyl)phosphine) is used as reducing agent, and significantly less cross-links encompassing cysteine residues are found. We further propose insertion of DTT between the cysteine and uracil reactive sites as the most probable structure of the cross-linking products.
Cross-linking of biomolecules combined with mass spectrometry (MS) has emerged as a powerful tool to characterize not only the tertiary and quaternary arrangements of individual biomolecules, but especially their interaction sites in biologically active complexes. By MS-based identification of the cross-linked parts or even the exact cross-linking sites of the respective biomolecules, proximity information can be derived. This has proven highly useful for computational approaches to problems such as docking or the arrangement of subunits (1–3).
In principle, cross-linking can be achieved in two ways: (1) By using a chemical cross-linker that connects reactive groups of the respective biomolecules within a certain distance range, the range depending on the reagent used. (2) By generating a so-called zero-length cross-link that connects reactive groups of biomolecules that are already directly adjacent to one another. The latter is usually achieved by (UV) light-induced cross-linking, with or without the addition of compounds that induce the generation of radicals on reactive groups of the cross-linkable components or in close vicinity to them.
Cross-linking in combination with MS analysis is nowadays frequently used in protein-protein interaction studies (4–7) but can also be applied to protein-nucleic acid complexes. Indeed much attention is currently paid to their MS-based analysis owing to the crucial cellular function of many such complexes. A large variety of studies over decades have examined chemical cross-linking between proteins and DNA, using reagents such as the genotoxic diepoxybutane, endogenous aldehydes, transition metals, nitrogen mustard, and platinum compounds, alkynitrosoureas, and formaldehyde (8). In addition, UV irradiation has been used to establish cross-links between proteins and protein-bound single-stranded DNA (ssDNA), which can then be identified by mass spectrometry (9, 10). UV cross-linking makes use of the natural sensitivity of nucleobases to UV light (11, 12). The site of cross-linking can then be determined by mass spectrometric analysis (including gas phase fragmentation of the corresponding peptide-nucleic acid conjugates) and database searching. To enhance the low yield of protein-DNA cross-linking, derivatives with higher UV reactivity, such as halonucleotides, can be employed (13–15).
UV-induced cross-linking is more frequently used to monitor protein–RNA interactions. RNA is mainly present in its single-stranded form, and adopts a variety of tertiary structures in which the bases of the nucleotides are in close contact with amino acid residues of the proteins that are bound to the RNA. Several studies have used this technique to identify globally the proteins that directly interact with e.g. poly(A) mRNA in yeast and human systems, and have yielded insight into the large variety of RNA-binding proteins that exist within the cell (16–21). More detailed approaches apply UV-induced protein–RNA cross-linking in a similar manner, but extend the MS analysis toward the identification of the cross-linked amino acids together with the corresponding nucleotide moieties, allowing exact definition of the RNA-binding regions in the cross-linked proteins (22–24). To improve the yield of cross-linking, more reactive nucleoside derivatives (4-thiouridine and 6-thioguanosine) have been incorporated into RNA in growing cells. The cross-linking reaction of these derivatives with amino acids is not entirely additive, however, but is accompanied by the loss of H2S ((22), U.Z. and H.U, unpublished results). Other cross-linking reactions between proteins and RNA have made use of nucleotide derivatives that contain a cross-linkable function at the 2′ hydroxyl group or the phosphate moiety (25–27), but have not been characterized by MS yet.
Additional cross-linking agents for the analysis of protein–RNA interactions have been exploited, but have not yet found their way into modern MS-based proteome analyses. For instance, methylene blue has been described as a light-inducible cross-linker, in particular for mapping interactions of proteins with dsRNA (28). Similarly, protein–RNA interaction studies in ribosomal subunits have made use of diepoxybutane, or nitrogen mustard (29–31). The same holds true for the use of 2-iminothiolane (“Traut's reagent”), which is a protein–RNA cross-linking reagent that combines chemical and UV-inducible features and has been extensively applied to the analysis of protein–RNA contacts in ribosomal subunits (33, 32). Here, we introduce dithiothreitol (DTT)1 as a potent UV-inducible cross-linking reagent for the analysis of protein–RNA linkages following UV irradiation. By exhaustive mass-spectrometric analyses we found that upon UV light exposure DTT forms a covalent linkage between cysteine residues within proteins and uracil bases of RNA in close proximity. We applied this to protein–RNA complexes isolated from yeast cells and compared the protein–RNA cross-linking patterns that were obtained in the presence and absence of DTT. We found that the cross-linking reaction is surprisingly efficient and specific.
EXPERIMENTAL PROCEDURES
In Vitro Transcription of 13C-Labeled RNA Oligonucleotides
Human U4 snRNA 5′ stem loop oligonucleotide GCCAAUGAGGUUUAUCCGAGGC (position 26–47) was synthesized by in vitro transcription with T7 RNA polymerase (34). DNA template with T7 promoter sequence was obtained from Eurofins (Eurofins MWG Operon Ebersberg Germany). Annealing of DNA template (1 μm) with T7 promoter (1 μm) was performed by heating at 90 °C for 2 min. Four 13C labeled oligonucleotides were synthesized, each with one labeled ribonucleoside triphosphate (rNTP) and three unlabeled rNTPs, and final concentrations of 1 mm and 4 mm, respectively. Unlabeled rNTPs (Jena Bioscience, Jena, Germany) and 13C labeled rNTPs (Sigma-Aldrich, Taufkirchen, Germany) were obtained as sodium salts. Transcription was performed in 40 mm Tris-HCl pH 8.0, 10 mm DTT, 2 mm spermidine, 30 mm MgCl2, and 0.1 mg/ml T7 RNA polymerase. After incubation at 37 °C for 4 h the transcription product was purified using 20% polyacrylamide gels (0.7 mm, 20 × 30 cm) containing 7 m urea. RNA oligonucleotide bands on the gels were visualized by UV shadowing, excised and eluted using TEN buffer (10 mm Tris-HCl pH 8.0, 1 mm EDTA, 300 mm NaCl). Finally oligonucleotides were recovered by ethanol precipitation, dissolved in water and stored at −20 °C. Concentration was determined by UV absorption at 260 nm.
UV Cross-linking with Isotope-labeled RNA Oligonucleotides
Recombinant 15.5K protein was purified from Escherichia coli BL21 cells as described by Nottrott et al. (35). For UV cross-linking 1 nmol of 13C labeled U4 snRNA 5′ stem loop oligonucleotide was incubated with 1 nmol of 15.5K protein in buffer (20 mm HEPES, 50 mm NaCl, 1.5 mm MgCl2) on ice for 30 min. UV-irradiation was performed in microtiter plates at 254 nm for 10 min. After ethanol precipitation the sample was dissolved with 1 m urea in 50 mm Tris-HCl pH 7.9. The RNA moiety was hydrolyzed by addition of 1 μg of RNase A and 1 unit of RNase T1 for 2 h at 52 °C. Protein was digested with trypsin (protein/enzyme 50:1, w/w) overnight at 37 °C. Removal of noncross-linked RNA was achieved by solid phase extraction using reversed phase C18 microcolumns and cross-linked peptides enriched using TiO2 microcolumns, both prepared in-house (36). Noncross-linked controls were prepared and analyzed in parallel.
Samples were loaded onto a nanoflow liquid chromatography system (Agilent 1100 series, Agilent Technologies, Böblingen, Germany) coupled to an LTQ-Orbitrap Velos mass spectrometer equipped with a FlexIon nanoSpray source (Thermo Fisher Scientific, Bremen, Germany). The chromatography system was operated with a vented column setup (37) at an analytical flow rate of 300 nl/min achieved through passive splitting. Samples were desalted on a self-packed reversed phase-C18 pre-column (20 mm length, 0.15 mm inner diameter, ReproSil-Pur C18-AQ 5 μm resin, Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Separation was achieved on a self-packed reversed phase-C18 analytical column (150 mm length, 0.075 mm inner diameter, ReproSil-Pur C18-AQ 3 μm resin, Dr. Maisch GmbH) packed into a SilicaTip emitter (FS360–75-10-N, New Objective, Woburn, MA). Analytes were separated using a linear gradient of 3–36% acetonitrile over 37 min using 0.1% formic acid as modifier, followed by an increase to 95% acetonitrile versus 0.1% formic acid over 0.5 min and a 7.5 min hold at these conditions.
MS data was acquired in positive ion mode using a data-dependent top10 method dynamically choosing the most abundant precursor ions for higher energy collision-induced dissociation (HCD). MS survey spectra were acquired across the 350–1600 m/z range at a resolution setting of 30,000 full width half maximum (FWHM). MS/MS spectra were acquired at a resolution setting of 7500 FWHM with a normalized collision energy setting of 45%. A minimum of 5000 ion counts were required for triggering fragmentation events.
UV-Cross-linking of Synthetic Peptide with RNA Oligonucleotides
UV-crosslinking of SmB/B′ specific peptides 9MLQHIDYRMRCILQDGR25 (kindly provided by Olaf Jahn, Mass Spectrometry Group at the Max Planck Institute for Experimental Medicine, Göttingen, Germany) with Sm site oligonucleotide 5′-AAUUUGUGG-3′ (Eurogentec Deutschland GmbH, Germany) was performed in four different conditions: (1) without DTT, (2) with 2 mm DTT, (3) with 2 mm perdeuterated DTT, (4) with 2 mm (tris(2-carboxyethyl).phosphine) (TCEP). For each experiment 1 nmol of synthetic peptide and 1 nmol of RNA oligonucleotide in binding buffer (20 mm HEPES, 50 mm NaCl, 1.5 mm MgCl2 and 0.01% formic acid) were incubated on ice for 30 min. UV irradiation was carried out for 5 min at 254 nm. RNA hydrolysis was performed by adding 1 μl of diluted RNase A (1:5, v/v) and 1 unit RNase T1 for 2 h at 52 °C. After peptide digestion by trypsin (protein/enzyme 50:1, w/w) overnight at 37 °C, samples were desalted using in house-packed reversed phase-C18 columns (Dr. Maisch, GmbH).
LC/MS/MS analysis was performed on a nanoflow liquid chromatography system (Easy nanoLC II System, Thermo Fisher Scientific) coupled to a Q Exactive mass spectrometer equipped with a FlexIon source (Thermo Scientific). Samples were desalted on a self-packed reversed phase-C18 pre-column (20 mm length, 0.10 mm inner diameter, ReproSil-Pur C18-AQ 5 μm resin, Dr. Maisch, GmbH). Separation was achieved on a self-packed reversed phase-C18 analytical column (100 mm length, 0.050 mm inner diameter, ReproSil-Pur C18-AQ 3 μm resin, Dr. Maisch, GmbH) packed into a SilicaTip emitter (FS360–50-8-N, New Objective).
Analytes were separated using a linear gradient of 4–36% acetonitrile over 22 min using 0.1% formic acid as modifier, followed by an increase to 95% acetonitrile versus 0.1% formic acid over 0.5 min and a 7.5 min hold at these conditions. A flow rate of 250 nl/min was used.
Data-dependent acquisition was performed in positive ion mode using a top15 method for analysis. MS survey spectra were acquired at a resolution setting of 70,000 FWHM across the 350–1600 m/z range. HCD with a normalized collision energy setting of 30% was applied for peptide fragmentation, and MS/MS spectra were acquired at a resolution setting of 17,500 FWHM. A minimum of 10,000 ion counts was required for triggering fragmentation events.
UV Cross-linking of Native Ribonucleoprotein Complex
Saccharomyces cerevisiae cells carrying a C-terminal TAP-tag on cbp20 were grown in YPD media (1% yeast extract, 2% bacto-peptone, 2% glucose) to an OD600 of 5.7 in a fermenter. Cells were harvested and lysate was prepared as described by Kramer et al. (22). TAP-Tag purification was done in two parallel experiments A and B with buffer containing either dithiothreitol (DTT) or TCEP. For both experiments the cell lysate (350 mg protein in 10 ml) was incubated with 600 μl of IgG SepharoseTM 6 Fast Flow beads (GE Healthcare, München, Germany) equilibrated with AGK buffer (10 mm HEPES, pH 7.5, 1.5 mm MgCl2, 200 mm KCl, 10% glycerol, EDTA-free protease inhibitor mixture tablet (Roche, Indianapolis, IN)). RNPs were bound to IgG beads by rotation for 2 h at 4 °C. After the flow through was collected, beads were washed with 5 ml of AGK buffer and equilibrated with 20 ml of CBB1 buffer (25 mm Tris-HCl pH 7.9, 150 mm sodium chloride, 1.5 mm magnesium acetate, 2 mm calcium chloride, 1 mm imidazole, 2 mm DTT) in experiment A and with CBB2 buffer (25 mm Tris-HCl pH 7.9, 150 mm sodium chloride, 1.5 mm magnesium acetate, 2 mm calcium chloride, 1 mm imidazole, 2 mm TCEP) in experiment B. RNPs were eluted from IgG by incubating with 12 μl PreScission protease and 1 μl RNasin overnight at 4 °C.
For UV cross-linking, purified RNPs from experiments A and B were divided into control and test samples containing 1 mg of protein each, respectively. Both test samples were UV-irradiated at 254 nm for 2 min on ice in Petri dishes (3.5 cm diameter). After cold ethanol precipitation, protein therein was digested with trypsin (Promega, protein/enzyme 50:1, w/w) overnight at 37 °C. Intact RNA and peptide-RNA cross-links were separated from the excess of noncross-linked peptides by size exclusion chromatography on a Superdex 200 column (3.2 mm × 300 mm) coupled to a SMART chromatography system (GE Healthcare; Uppsala, Sweden) with buffer containing 20 mm Tris pH 7.5, 150 mm NaCl, 1.5 mm MgCl2. Noncross-linked RNA was removed by reversed phase-C18 chromatography using in-house prepared columns (Dr. Maisch, GmbH) following hydrolysis with benzonase for 1 h at 37 °C and ribonucleases A/T1 (Ambion, Applied Biosystems, Darmstadt, Germany) as above for 2 h at 52 °C. Nonirradiated controls of experiments A and B were processed in the same manner.
LC/MS/MS analysis was performed in positive ion mode using nanoflow liquid chromatography coupled to an LTQ-Orbitrap Velos mass spectrometer with the setup described before, but with a gradient length of 118 min.
MS Data Analysis
Data analysis for identification of putative cross-links was performed by the novel RNPXL pipeline (22) integrated in the OpenMS software environment (38, 39). Briefly, the first step is the conversion of raw data (in *.raw format) to the mzML format using the ProteoWizard tool msconvert (40). Two different filters pipelines integrated in the RNPXL pipeline are applied: (1) the ID filter removes MS/MS spectra that match to noncross-linked peptides identified at high confidence by a database search with OMSSA (Open Mass Spectrometry Search Algorithm) (41). (2) the XIC filter reduces false positives by removing the MS/MS spectra of precursors from the UV irradiated sample if they are also present in the nonirradiated control at comparable intensity. The reduced data file is then submitted for generation of precursor mass variants that are obtained by subtracting all potential masses of cross-linked RNA moieties from the observed precursor masses in a combinatorial fashion. This list of precursor mass variants is then submitted for database search by OMSSA. The final output (in idXML format), which contains the true positive hits for cross-linked peptides, is used to annotate raw MS/MS data in TOPPView (42). For a detailed description of data analysis, we refer to the original publication by Kramer et al. and the corresponding tutorial (22).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (43) via the PRIDE partner repository with the data set identifier “PXD002656“, with the following file names:
XL_U4_Ex1_C13ATP
XL_U4_Ex1_C13UTP
XL_U4_Ex1_C13GTP
XL_U4_Ex1_C13CTP
XL_U4_Ex1_naturalrNTP
XL_peptide_Ex2_withoutDTT
XL_peptide_Ex2_withDTT
XL_peptide_Ex2_dDTT
XL_yeast_Ex3_TCEP
XL_yeast_Ex3_DTT
In addition theoretical masses of peptides, peptide fragments and oligonucleotides were calculated using Protein Prospector MS-Product (http://prospector.ucsf.edu), Peptide Mass Calculator (http://rna.rega.kuleuven.ac.be/masspec/pepcalc. htm), and Mongo Oligo Mass Calculator (http://mods.rna.albany.edu/masspec/Mongo-Oligo), respectively.
RESULTS
Working Hypothesis
In recent years we have established a purification strategy that allows for the isolation and subsequent mass spectrometric characterization of cross-linked peptide-RNA oligonucleotides derived from UV-irradiated protein–RNA particles of any kind (22). We were able to sequence a multitude of cross-linked peptides and identified their cross-linked amino acid and nucleotide parts, respectively. In most cases the determined cross-link was found to be of an additive nature, i.e. the mass of the entire cross-link is composed of the mass of its peptide moiety and its RNA moiety. Exceptions to this are observed when the RNA contains modified nucleosides, e.g. 4-thiouridine (44) or 6-thioguanosine (U.Z. and H.U., unpublished results). Another more frequently observed exception is the appearance of peptide-RNA cross-links with a nominal mass increase of 152 Da over the expected additive masses upon UV cross-linking with nonmodified RNAs. We first observed such cross-links in studies of the import factor Snurportin 1 bound to U1 snRNA (45). The peptide 346GSSHSPDHPGCLMEN360 derived from Snurportin 1 was found to be cross-linked to an oligonucleotide AAAU (position 102–105 in stem-loop III of U1 snRNA) plus the mass increment of 152 Da. MALDI-MS and ESI-MS/MS experiments revealed that the adduct causing the mass increment is directly attached to the peptide moiety of the cross-link (44).
We detected corresponding adducts upon cross-linking of other recombinant protein–RNA complexes, i.e. in cross-linking studies of the RNase H domain of the human spliceosomal hPrp8 with U4 snRNA (46), the spliceosome associated protein CWC2 with U6 and U4 snRNA (47), but also in cross-linked endogenous protein–RNA complexes where several ribosomal proteins from yeast were found to be cross-linked to RNA with the same mass increment of 152 Da (22). A common feature of all these modified cross-links is that cysteine can be identified as the cross-linked amino acid of the peptide sequence (22).
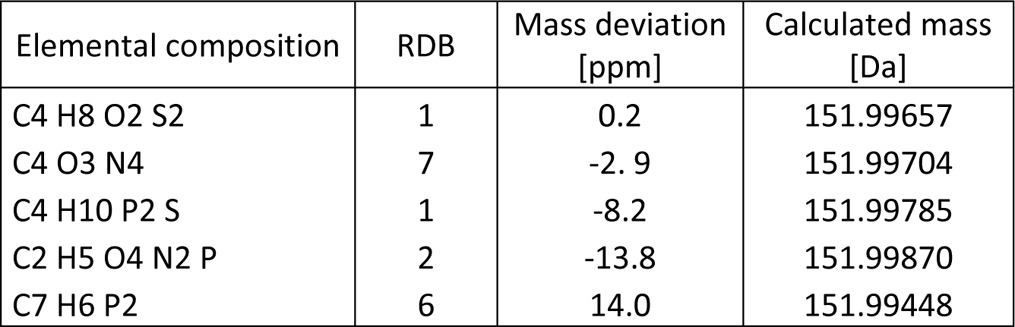
A comparison of the exact masses detected by high resolution mass spectrometry with the expected additive cross-link masses revealed an exact mass increment of 151.9966 Da (Table I) rather than the nominal value of 152 Da. The observed decimal deficiency of −0.0034 Da points to the presence of mass-deficient elements in these side products, i.e. 16O (15.9949 Da), 31P (30.9738 Da) or 32S (31.9721 Da) (main isotopic contributions, respectively). Individual mass errors observed in our data sets were ≤10 ppm. Following the rules of error propagation, the maximum expected error for the mass increment as the difference of two experimental values is ≤20 ppm. Allowing for sensible restrictions to the elemental composition and extending the nitrogen rule to nitrogen and phosphorous, a mass increment of 151.9966 ± 20 ppm allows only for a limited number of elemental compositions (Table I), the most plausible ones being C2H5O4N2P, C4O3N4, and C4H8O2S2. This led us to investigate two hypotheses for the origin of the mass increment: a derivative of a nucleobase formed e.g. by the irradiation used for cross-linking, or a small organic molecule from the pool of reagents used for isolation, purification and cross-linking of the complex. Because the 152 mass increment was observed neither in nonirradiated control samples nor in irradiated TCEP samples (see below), we did not pursue the third probability of a purely proteinaceous origin further.
Table I.
Elemental compositions for 151.9966 Da ± 20 ppm
We set out to evaluate both these two working hypotheses in more detail.
UV-induced RNA-RNA cross-linking is a well-known phenomenon that can be exploited for elucidation of RNA-RNA interactions (48, 49). It is therefore conceivable that besides the covalent bond formed between cysteine and uracil upon UV cross-linking, another proximal nucleobase or its derivative might form an additional bond to the uracil moiety. To test this hypothesis we used a binary complex that consists of the human spliceosomal U4/U6 snRNP specific protein 15.5K bound to an RNA oligonucleotide harboring the canonical binding site of the 15.5K protein (35). Human 15.5K protein as part of U4/U6.U5 tri-snRNPs was found to be involved in the late stage of spliceosome assembly by binding to the 5′ stem loop of U4 snRNA (35). The used 22mer RNA oligonucleotide encompasses the U4 snRNA nucleotide positions 26–47 and was in vitro transcribed with 12C-rNTPs as well as with all four 13C-rNTP. If, upon UV cross-linking, the mass increment was induced by a derivative of a nucleobase, we expected to observe a shift of the 152 Da mass increment in the MS analysis corresponding to the incorporation of 13C-labeled nucleobases.
We also evaluated if the mass increment might be because of a small organic molecule either inserted in between, or added to the cysteine thiol and uracil moieties involved in the cross-links. The nominal mass increment of 152 Da e.g. corresponds to DTT, a reducing agent frequently used in cell lysis and subsequent protein purification protocols. To test this hypothesis UV cross-linking was performed with and without DTT, with TCEP and with perdeuterated DTT. For this purpose different experimental models were used. In initial studies a short synthetic peptide derived from U snRNP-specific SmB'/B protein and Sm site 9mer U1 snRNA oligonucleotide was used. Moreover cross-linking was performed in native RNP purified from S. cerevisiae in the presence of either DTT or TCEP.
Evaluation of Nucleoside Derivatives as the Origin of Unusual Mass Adducts
To assess our first hypothesis, namely if a nucleoside derivative could explain the observed mass increment, we first performed a UV cross-linking experiment with and without heavy isotope labeling of U4 snRNA oligonucleotide (nucleotide position 26–47 (35)) with human spliceosomal protein 15.5K.
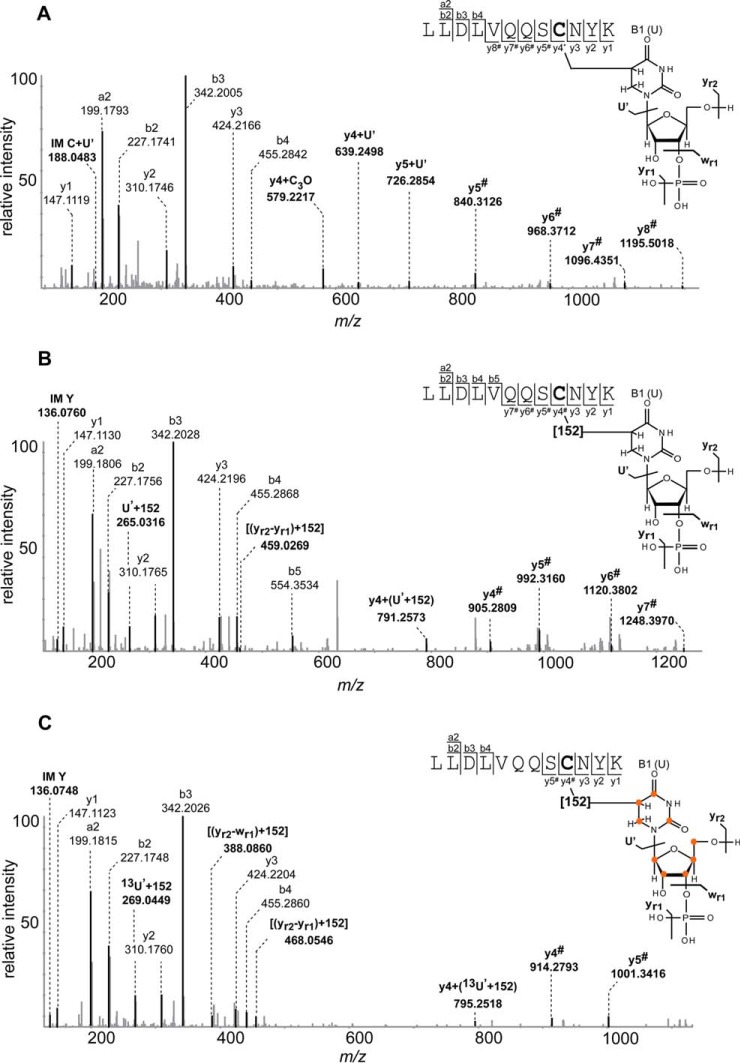
Using nonlabeled RNA for UV irradiation, we isolated and sequenced a peptide of the 15.5K protein, 22LLDLVQQSCNYK33, cross-linked to a U nucleotide without and with the mass increment of 152 Da (for data, see XL_U4_Ex1_naturalrNTP). Fig. 1A and 1B show the MS/MS spectra of the peptide-RNA oligonucleotide cross-link without and with mass increment, respectively. In Fig. 1A the y- and b-type fragment ions clearly reveal the sequence of the cross-linked peptide, and the mass difference between the peptide alone (1422.7177 Da) and the measured precursor mass (1746.7536 Da) corresponds exactly to a uridine nucleotide (324.0359 Da). The spectrum also shows a mass shift in the y-ion series starting from y4 corresponding to a fragment derived from gas phase dissociation of uridine (+C3O, +51.9949 Da, see below) identifying the cysteine residue as the cross-linked amino acid. The observed y5# to y8# peaks correspond to peptide fragment ions that still have the nucleoside attached. Fig. 1B shows the same peptide cross-linked to a U nucleotide, but additionally exhibiting the unexpected mass increment. The observed precursor at m/z 633.92343+ corresponds to a molecular weight of 1898.7502 Da, a difference of 476.0324 Da from the molecular weight of the peptide alone, which corresponds to a cross-link with [U+152 Da]. Manual interpretation of the spectrum reveals a +378.0556 shift of the y ion series starting with y4. The mass shift is consistent with a uridine fragment [+226.0590 Da] plus an additional 152 Da mass increment cross-linked to the cysteine residue (Fig. 1B).
Fig. 1.
Sequence analysis of peptide-RNA (oligo)nucleotide cross-links derived from human spliceosomal complex U4 specific 15.5K protein cross-linked to U4 snRNA. A, MS/MS spectrum of m/z 874.38562+ annotated as LLDLVQQSCNYK (positions 22–33) cross-linked to nucleotide U. The peptide fragment signals shifted by (U-H3PO4) or (226 Da) are indicated by #. B, MS/MS spectrum of m/z 663.92343+ annotated as LLDLVQQSCNYK (positions 22–33) cross-linked to nucleotide U with the additional mass of 152 Da. The peptide fragment signals shifted by [(U-H3PO4)+152 Da] or (378 Da) are indicated by #. C, MS/MS spectrum of m/z 954.89762+ annotated as LLDLVQQSCNYK (positions 22–33) cross-linked to nucleotide 13U with the additional mass of 152 Da. Fragment ions shifted by an additional mass of 387.0858 Da are indicated by #. Signals corresponding to uracil fragments with the 152 Da increment are annotated as U', (yr2-yr1) and (yr2-wr1). All signals corresponding to RNA and peptide-RNA adducts are highlighted by bold letter. RNA oligonucleotide fragmentation nomenclature is according to McLuckey et al. (61). A proposed schematic illustration of the peptide-RNA cross-link along with cross-link product of +152 Da is shown on the right. Cysteine as the cross-linked amino acid identified by shift in the y ion series (starting from the y4 ion), is highlighted by bold letter in the peptide sequence. y-type fragmentation of RNA is referred to as U', wr1, yr1, yr2. Positions of 13C on UTP are marked by orange circles in lower panel.
We next performed 15.5K-RNA cross-linking experiments with different 13C-labeled U4 snRNA oligonucleotides synthesized by in vitro transcription. In each experiment, a different 13C-rNTP was incorporated, i.e. 13C-A, 13C-G, 13C-U, 13C-C (for data, see XL_U4_Ex1_C13ATP, XL_U4_Ex1_C13GTP, XL_U4_Ex1_C13UTP, XL_U4_Ex1_C13CTP). If the mass increment was caused by a nucleobase derivative, then the cross-link would show an increased mass shift corresponding to its heavy isotope labeling, which is 4 Da for U and C; and 5 Da for A and G, respectively. None of the experiments employing heavy isotope labeling, however, produced any evidence of a heavy isotope-induced shift in the +152 Da mass increment. In all observed cross-links the observed mass shifts could be readily and conclusively rationalized by 13C labeling of the nucleobases directly involved in the actual cross-link, without any indication of a heavy isotope shift in the +152 Da mass increment. Fig. 1C shows an example MS/MS spectrum from the cross-linking experiment using 13C-UTP labeled oligonucleotide (for data, see XL_U4_Ex1_C13UTP), corresponding to a peptide-RNA (oligo)nucleotide cross-link 22LLDLVQQSCNYK33 + [13U+152]. The molecular weight of 13UTP is 333.0952 Da; consequently, the calculated molecular mass difference between the cross-link and the unmodified peptide is 485.0702 Da matching [13U + 152 Da]. Manual interpretation of the spectrum revealed a shift of the y-series starting with y4 by 387.0858, corresponding to [(13U-H3PO4) +152 Da]. The presence of several important signals in the lower m/z region such as m/z 269.0441, 388.0860, and 468.0548 can be attributed to uridine adducts [13U' +152 Da], [(13U-H3PO4) +152 Da], and [(13U-H2O) +152 Da], respectively, confirming that uracil is cross-linked to cysteine through the unusual mass adduct.
Evaluation of DTT as the Origin of the Unusual Mass Adducts
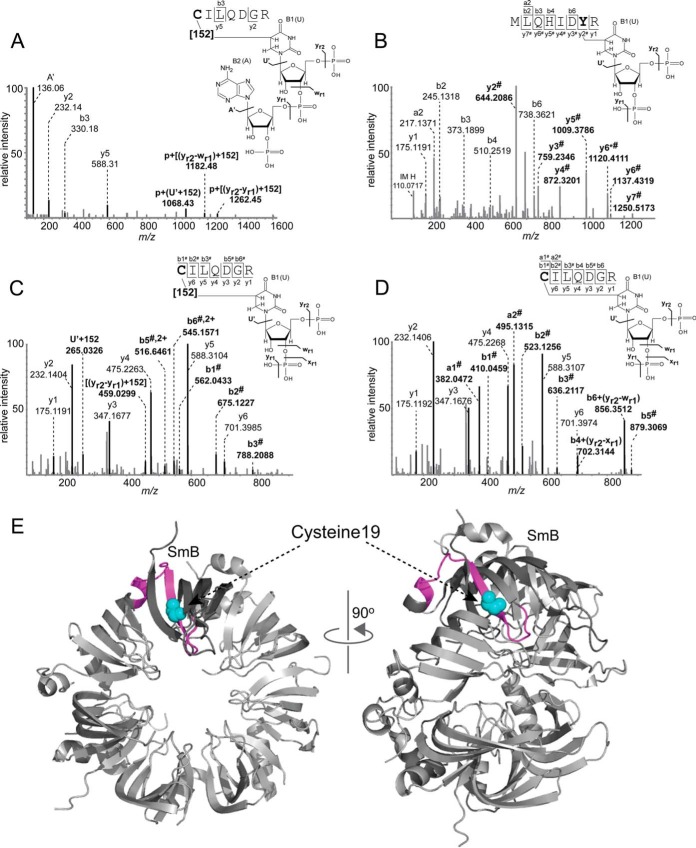
According to our second hypothesis, the unexpected mass increment might be caused by the addition or insertion of a small organic molecule, possibly dithiothreitol, which has an elemental composition matching the calculated exact mass difference of 151.9966 Da between regular and modified cross-links. To evaluate this hypothesis we again chose a simple system producing cross-links upon UV irradiation. We selected an SmB/B′ peptide 9MLQHIDYRMRCILQDGR25 from human spliceosomal U small nuclear ribonucleoprotein particles (snRNPs). Sm proteins consist of a highly conserved set of seven proteins (SmB/B′, D1, D2, D3, E, F, and G), which are common to all U snRNPs (U1, U2, U4, U5) and assemble as a seven-membered ring around a single stranded U-rich sequence called the Sm site. The selected peptide sequence was observed to cross-link with the Sm site within endogenous U snRNAs during the investigation of protein–RNA interactions in spliceosomal U1 small ribonucleoprotein particles (Fig. 2E, PDB ID: 4PJO, (50, 51)). Close inspection of peptide-RNA (oligo)nucleotide cross-links obtained in these studies of endogenous RNPs already showed that a cysteine within the C-terminal half of the peptide (19CILQDGR25) specifically cross-linked to uracil, again with a mass increment of 152 Da (Fig. 2A). We also identified peptide-RNA (oligo)nucleotide cross-links derived from the N-terminal half of the peptide 9MLQHIDYR17 but without the 152 Da adduct (Fig. 2B), even in the presence of DTT (for data, see XL_peptide_ Ex2_withDTT). Here, Tyr15 was found as the cross-linked amino acid (Fig. 2B). This result is consistent with previous observations that this mass increment is usually associated with cysteine containing peptide sequences, and strongly points toward a UV-induced cross-linking reaction that includes DTT.
Fig. 2.
Sequence analysis of peptide-RNA (oligo)nucleotide cross-links derived from human spliceosomal complex U1 SnRNP specific SmB/B′ peptide. A, MS/MS spectrum of m/z 805.24682+ showing the peptide CILQDGR (positions 19–25) cross-linked to AU with an additional mass of 152 Da in the presence of DTT. Spectrum obtained from LTQ-Orbitrap XL (HCD fragmentation). All signals corresponding to peptide-RNA adducts are highlighted by bold letter. B, MS/MS spectrum of m/z 691.28582+ shows the peptide MLQHIDYR (positions 9–16) cross-linked to U-H2O in the presence of DTT. A shifting of y ions by 306 Da corresponding to (yr2-yr1) is indicated by #. Tyrosine as the cross-linked amino acid, identified by a shift in y ion series from y2 to y7, is highlighted by bold letter. C, MS/MS spectrum of m/z 640.72022+ in the presence of DTT shows the peptide cross-linked to U with an additional mass of 152 Da. Shifting of a and b fragment ions of the peptide by different RNA fragments identified cysteine as cross-linked amino acid. D, MS/MS spectrum of m/z 564.72522+ shows the peptide cross-linked to U in the absence of DTT. In all spectra signal correspond to RNA and RNA fragments and shifting of the peptide signals by different RNA fragments masses are annotated and highlighted as bold letter. Proposed schematic illustration structures for peptide-RNA cross-links along with and without cross-link product of 152 Da mass are shown on the right side of each corresponding spectrum. Fragmentation of RNA refers as yr1, xr1, wr1 and yr2 by McLuckey et al. (61). E, Mapping of MLQHIDYRMRCILQDGR peptide (positions 9–25) on the crystal structure of Sm ring of U1 SnRNP (PDB ID: 4PJO, (62)). In SmB/B′ protein (both front and side view), the peptide is highlighted in pink color whereas the cross-linked cysteine is highlighted in cyan color as a sphere.
We performed in vitro UV cross-linking with this particular peptide with an Sm site RNA oligonucleotide AAUUUGUGG following our standard protocol, both without and with the addition of DTT (for data, see XL_peptide_Ex2_withoutDTT, XL_peptide_Ex2_withDTT). Peptide-RNA (oligo)nucleotide cross-links were observed corresponding to both peptide sequences MLQHIDYR and CILQDGR obtained by tryptic cleavage. Interestingly almost all cross-links derived from the tryptic peptide CILQDGR, but not those of from peptide MLQHIDYR, were observed both with and without the mass increment of 152 Da in the presence of DTT (supplemental Table S1). Without the addition of DTT, none of the side products were observed.
Fig. 2C and 2D shows the MS/MS spectra of two peptide-RNA oligonucleotide cross-links obtained from the peptide CILQDGR in the presence and absence of DTT (for data, see XL_peptide_ Ex2_withDTT, XL_peptide_ Ex2_withoutDTT), respectively. The calculated molecular weight difference between the cross-link (1279.4284 Da) and the corresponding peptide (803.3960 Da) is 476.0288 Da. This difference matches to a U nucleotide plus a mass increment of 151.9965 Da. An analysis of the product ion spectrum reveals a complete y ion series. In addition the spectrum contains a and b ion series shifted by a fragment of uracil plus the mass increment of 152 Da including an ion nominally corresponding to b1#, identifying the cysteine residue as the site of crosslinking (Fig. 2C). If a peptide's N-terminus is not modified by e.g. acetyl, then b1 ions are usually not observed under CID conditions as they cannot be stabilized via an oxazolone structure as is the case for higher order b ions (52). We postulate, however, that the large nucleotide moiety stabilizes the charge sufficiently. In the absence of DTT we found a cross-link of 1127.4318 Da corresponding to cross-linked U nucleotide (324.0359 Da) without the mass increment of 152 Da (Fig. 2D).
For further confirmation the UV cross-linking experiment was performed in the presence of perdeuterated DTT (d10-DTT). Of the ten deuterium atoms in d10-DTT, two belong to the thiol functions and two more to the weakly acidic hydroxyl functions, which are prone to hydrogen exchange in aqueous media. We would therefore expect a mass increment of 158.0342 Da if deuterium labeled DTT was incorporated in cross-links because of the remaining 6 Cα-deuteriums.
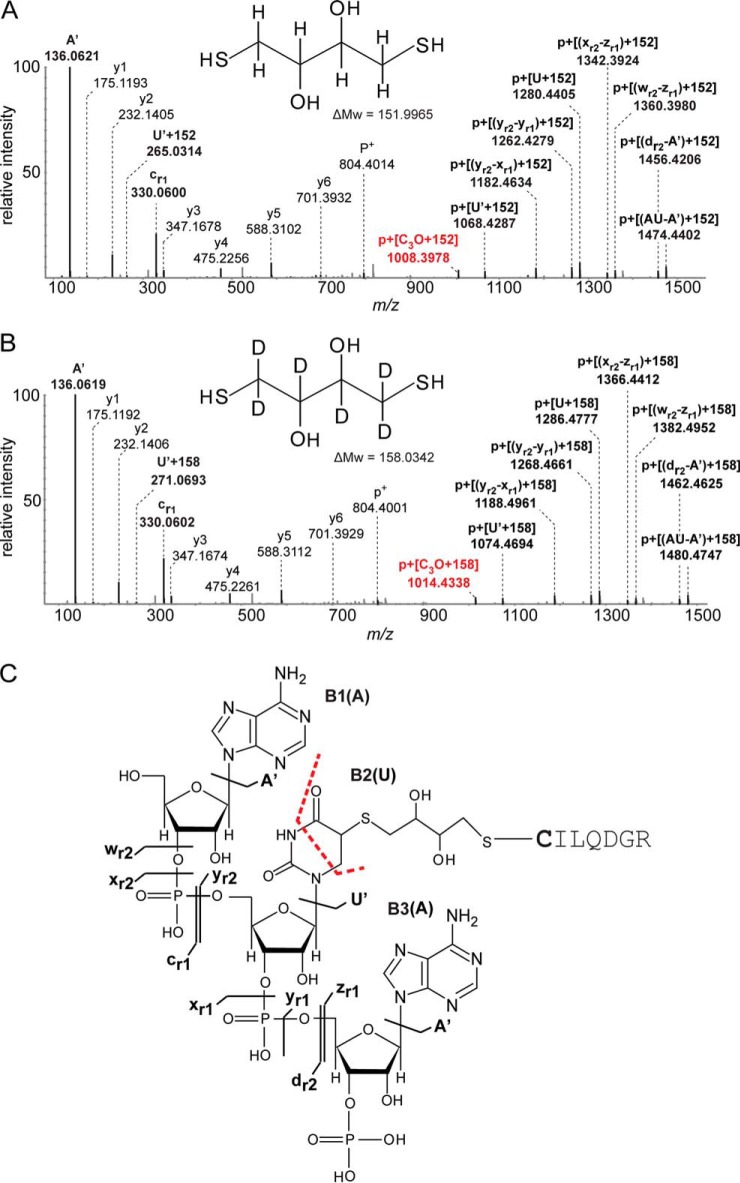
Fig. 3A shows the MS/MS spectrum of a cross-linked peptide-RNA (oligo)nucleotide obtained from the peptide CILQDGR in the presence of DTT (for data, see XL_peptide_ Ex2_withDTT). The calculated molecular weight difference between the cross-link (1937.5335 Da) and the corresponding peptide (803.3960 Da) is 1134.1375 Da. This difference matches the trinucleotide AAU (982.1410 Da) plus a mass increment of 151.9965 Da. An analysis of the fragment spectrum reveals a complete y ion series up to y6, again pointing to the cysteine residue as the site of cross-linking. In addition the spectrum contains a number of fragments in the upper m/z region that can be assigned to successive fragmentation of the RNA moiety of the cross-link.
Fig. 3.
Sequence analysis of peptide-RNA (oligo)nucleotide cross-links derived from human spliceosomal complex U1 SnRNP specific SmB/B' peptide CILQDGR (positions 19–25). A, MS/MS spectrum of m/z 646.85103+ in the presence of DTT shows the peptide cross-linked to the trinucleotide AAU with an additional mass of 152 Da. The fragment nominally corresponding to the [M+H]+1 of the unmodified peptide is annotated as p+. All signals corresponding to RNA and RNA fragments and shifting of peptide signal fragment signals by different RNA fragments via 152 Da mass are annotated and highlighted by bold letters according to the nomenclature by McLuckey et al. (61). Marker ions of the adenine base and nucleotide with loss of water are annotated as A' and [yr1-(yr-1)] respectively. Similarly, uracil base and uracil nucleotide with the loss of water are annotated as U' and (yr2-yr1) respectively. Shifting in signal of uracil nucleobase (U') with 152 Da is also depicted. B, MS/MS spectrum of m/z 972.79572+ in the presence of d10-DTT shows the peptide cross-linked to the trinucleotide AAU with the additional mass of 158 Da. Signals in the high m/z range correspond to shifting of mass of peptide shifted peptide fragment masses by different RNA fragments along with 158 Da mass, and are annotated as in spectrum A. The difference of 6 Da is clearly observed between the two spectra in the high m/z region. C, Proposed structure for peptide-DTT-uracil cross-link formation. Upon UV irradiation at 254 nm, DTT can form covalent bonds with uracil and the thiol group of cysteine.
Interestingly the mass increment of 152 Da is present in all RNA-derived fragments down to the smallest units, suggesting that the structure causing the fragment is either close to, or actually part of the site of cross-linking. In the low m/z region the spectrum exhibits marker ions for the adenine nucleobase at m/z 136.0620 and the corresponding nucleotide at m/z 330.0600, respectively (Fig. 3A), corroborating that the cross-link is formed via the uracil nucleobase rather than one of the adenine bases. Uracil itself does not generate strong marker ions but ion series in the high m/z region confirmed that uracil is linked to cysteine either directly or via the 152 Da adduct. Moreover we observed a fragment ion at m/z 265.0314 because of the mass of the uracil base shifted by 151.9963 Da (Fig. 3A).
Fig. 3B shows the MS/MS spectrum of the peptide-RNA (oligo)nucleotide cross-link derived from the peptide sequence CILQDGR with the d10-DTT-AAU nucleotides (for data, see XL_peptide_Ex2_dDTT). Comparison of the spectrum with that of the nonheavy isotope-labeled species in panel A clearly shows that all fragments incorporating the oligonucleotide moiety are similarly shifted upward by 6 Da owing to the six deuterium atoms in DTT, whereas y-type fragments assigned to the peptide sequence do not exhibit this shift. The results obtained on this model system strongly support the hypothesis that DTT is indeed the origin of the 152 Da mass increment.
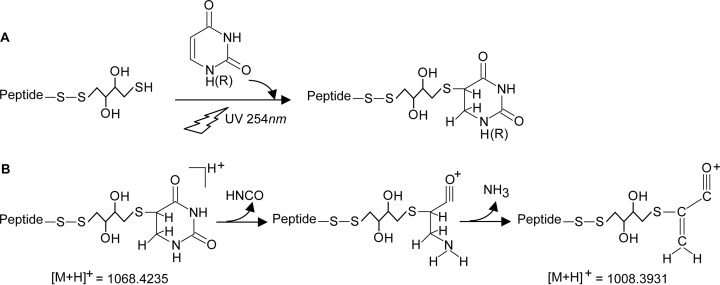
Importantly, we observed fragments at m/z 1008.3978 and 1014.4338 (Fig. 3A and 3B, respectively) corresponding to a 6 Da mass difference in these product ion spectra̧ the latter contains deuterated DTT (+6 Da). We assigned these values to the mass of the peptide (803.3960 Da) plus the DTT adduct (151.9965 Da and 158.0342 Da, respectively) plus C3O (51.9928 Da). We conclude that C3O must have derived from internal fragmentation of uracil as indicated in Fig. 4. According to Nelson and McCloskey (53) protonated uracil follows different fragmentation pathways under CID conditions. A neutral loss of HNCO leads to C3H4NO+ (70 Da) and further neutral loss of NH3 to C3HO+ with a nominal mass of 53 Da, which consists of the C4, C5, and C6 atoms from uracil. We note that the actual structure of this particular charged product ion is different as the cross-link to the DTT-peptide has to be considered. Therefore we postulate a structure of C3H2O+ covalently linked to the DTT-peptide as depicted in Fig. 4. If we assume a reduction of the double bond between positions 5 and 6 and the DTT-peptide moiety attached to the C5-atom, then a structure of C3H2O+ attached to the nonprotonated DTT-peptide appears highly plausible (Fig. 4). The exact masses of the product ions in Fig. 3A and 3B, with m/z 1008.3978 and 1014.4338, respectively, fit within less than 5 ppm to the predicted structure of the DTT-peptide to uracil and thus strongly support this structure. Note that similar product ions with the same mass increment of 52 Da were observed upon CID fragmentation of other UV-induced peptide-RNA (oligo)nucleotide cross-links containing the cross-linked amino acids glycine, lysine, proline, tyrosine, and valine (U.Z., H.U., unpublished observations) in the absence of DTT (i.e. mass increment of 151.9966 Da), which clearly shows that the product ion mass increment of 52 Da must have been derived from uracil.
Fig. 4.
Proposed scheme for the cross-linking reaction of uridine/uracil to cysteine/DTT. Panel (A) shows UV induced peptide-DTT-uridine cross-link formation. Panel (B) shows CID fragmentation of cross-linked uracil according to Nelson and McCloskey (53). Sequential loss of HNCO and NH3 from uracil result in formation of C3H2O+ covalently linked to DTT-peptide. Abbreviations used in the figure are “P” for peptide and “R” for ribose.
DTT-driven Protein–RNA Cross-linking in Intact RNP Complexes
To complement our in vitro experiments on low complexity model systems, we decided to investigate a more complex system to see if additional information could be gained from a more statistical approach evaluating a larger number of peptide-RNA (oligo)nucleotide cross-links. For this purpose a UV cross-linking experiment was performed on a large protein–RNA complex (RNP) from S. cerevisiae obtained by using fused TAP-Cbp20 tag. Because yeast Cbp20 together with Cbp80 binds to capped mRNA, the purified RNP represents a mixture of proteins that have direct or indirect interactions with mRNA. We have recently successfully combined this purification strategy with UV-induced cross-linking and several peptides cross-linked to RNA (22). Of note, in this preparation we used 2 mm DTT in the final buffer used for the purification of the yeast mRNPs, and most of the cysteine containing peptides that were found cross-linked showed the mass increment of 152 Da.
To study the role of DTT in cross-link formation in such a large-scale preparation, we performed TAP-tag purification of the yeast mRNPs according to Kramer et al. (22). The final step of mRNP purification was performed in buffer containing either DTT or TCEP in two parallel experiments (for data, see XL_yeast_Ex3_DTT, XL_yeast_Ex3_TCEP). After UV irradiation of the purified RNPs, proteins were hydrolyzed by trypsin under denaturing conditions and cross-linked peptide-RNA heteroconjugates separated from noncross-linked peptides by size exclusion chromatography. RNA was hydrolyzed to short fragments using RNases A/T1 and benzonase. After desalting by reverse phase chromatography, samples A and B were analyzed by nLC-ESI-MS/MS and data was analyzed using the RNPxl software pipeline (22). Product ion spectra assigned to cross-links were manually evaluated.
In total we identified 99 peptide-RNA (oligo)nucleotide cross-links containing 59 unique cross-linked peptide sequences (with various combination of cross-linked RNA nucleotides) derived from 43 different proteins (supplemental Table S2 and Table II). From these 59 unique cross-linked peptides, 26 were exclusively found in the preparation with DTT and 10 exclusively with TCEP, respectively. 17 cross-linked peptides were found in both preparations. The majority of peptide-RNA (oligo)nucleotide cross-links derived from the DTT experiment (24 out of 43) exhibit a mass increment of 152. Importantly, peptide-RNA (oligo)nucleotide cross-links containing the 152 Da mass increment were exclusively found in the presence of DTT in the buffer solutions and all these cross-linked peptides revealed a cysteine as cross-linked amino acid. In three cross-linked peptide RNA (oligo)nucleotides (60S ribosomal protein L35A/B, 40S ribosomal protein S6 A/B, Cruciform DNA recognizing-protein) the cysteine was found to be cross-linked to RNA with and without the 152 mass increment.
Table II.
List of peptide-RNA cross-links purified from S. cerevisiae. Protein IDs are shown by UniProt accession. Proteins identified in the previous study of Kramer et al. (22) are marked by * sign. Abbreviations used in the table are, TCEP: Tris(carboxyethyl) phosphine-hydrochloride, DTT: dithiothreitol. “-” is denoted for cross-links not identified in the corresponding experiment. “0” is denoted for the nucleotide minus water and “-p” for the loss of metaphosphoric acid (HPO3). “152” is denoted for mass of DTT adduct. Where possible, identified cross-linked amino acids are highlighted as bold letter within the cross-linked peptide sequences. Green color indicate cross-links identified only in the presence of DTT, red color indicates cross-links identified only in the presence of TCEP and yellow color shows cross-links common in both experiments. Further details of cross-links are given in supplementary Table S2
| Protein ID | Protein | Cross-linked Peptide Sequence | Cross-linked RNA |
|
|---|---|---|---|---|
| TCEP | DTT | |||
| P0CX43/P0CX44* | 60S ribosomal protein L1-A/B | 79SCGVDAMSVDDLKK92 | U0+152 | |
| P0CX45/P0CX46* | 60S ribosomal protein L2-A/B | 201GVAMNPVDHPHGGGNHQHIGK221 | AAGU0 | AAGU0 |
| P14126 | 60S ribosomal protein L3 | 326GCIPGNR332 | – | U0+152 |
| P10664* | 60S ribosomal protein L4-A | 347TGTKPAAVFTETLK360 | AU0, AAAU-P | AAAU-P |
| P49626* | 60S ribosomal protein L4-B | 347TGTKPAAVFAETLK360 | AU0, AAAU-P | – |
| P10664/P49626* | 60S ribosomal protein L4-A/-B | 85SGQGAFGNMCR95 | – | U0+152 |
| P26321* | 60S ribosomal protein L5 | 9SSAYSSRFQTPFR21 | AGU-P | – |
| 16FQTPFR21 | – | AGU-P | ||
| 197SYIFGGHVSQYMEELADDDEER218 | U0, U | U0 | ||
| Q02326/P05739* | 60S ribosomal protein L6-A/B | 32ASLVPGTVLILLAGRFR48 | GU-P, GU0, U0 | GU-P, GU0, U0 |
| P05739* | 60S ribosomal protein L6-B | 9WYPSEDVAAPK19 | AU0 | – |
| P05739* | 60S ribosomal protein L6-B | 57HLEDNTLLVTGPFK70 | U0 | U0 |
| P29453* | 60S ribosomal protein L8-B | 134YGLNHVVSLIENK146 | UU0, GU0 | GU0 |
| P05748* | 60S ribosomal protein L15-A | 148YNWICDPVHK157 | – | U0+152 |
| P26785* | 60S ribosomal protein L16-B | 37AEALNISGEFFR48 | ACU | – |
| 37AEALNISGEFFRNK50 | ACU, ACU-P, AU0 | ACU, ACU-P | ||
| P0CX49/ P0CX50* | 60S ribosomal protein L18-A/-B | 51ALFLSK56 | AU0 | AU0 |
| P0CX82/ P0CX83* | 60S ribosomal protein L19-A/-B | 115IDK(Carbamyl)HLYHVLYK125 | ACUU | ACUU |
| P0C2H8/P0C2H9* | 60S ribosomal protein L31-A/-B | 20LHGVSFK26 | UU, UU0, UUU | UU, UU0, UUU |
| P87262/P40525 | 60S ribosomal protein L34-A/-B | 92AFLIEEQK99 | U0 | -– |
| P0CX84/P0CX85* | 60S ribosomal protein L35-A/-B | 50SIACVLTVINEQQR63 | U0 | U0, U0+152 |
| P49166/P51402 | 60S ribosomal protein L37-A/-B | 15SHTLCNR21 | – | U0+152 |
| P0CH08 | 60S ribosomal protein L40A | 115CGHTNQLRPK124, 98KCYAR102 | – | U0+152, U+152 |
| P0CX27/P0CX28* | 60S ribosomal protein L42-A/-B | 88CKHFELGGEK97 | – | U0 |
| P33442/P23248* | 40S ribosomal protein S1-A/-B | 116KWQTLIEANVTVK128 | AU-P, U0 | U0 |
| P25443 | 40S ribosomal protein S2 | 162CGSVTVR168 | – | U0+152 |
| P26783* | 40S ribosomal protein S5 | 189TIAETLAEELINAAK203 | GU-P, GU0, U0, U | GU-P, GU0, U0 |
| 85AQCPIIER92 | – | U0+152 | ||
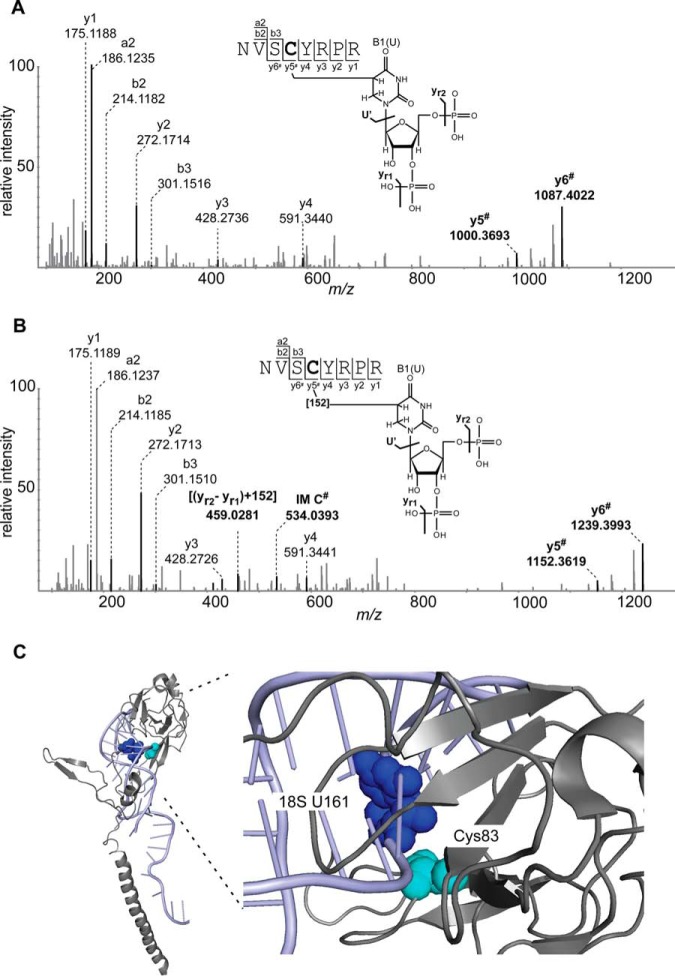
| P0CX37/P0CX38 | 40S ribosomal protein S6-A/B | 80NVSCYRPR87 | U0 | U0+152 |
| 168TYTKAPK174 | – | AU0 | ||
| O13516/P05755 | 40S ribosomal protein S9-A/B | 92KLDYVLALK100 | U0, U | – |
| P0CX47/P0CX48 | 40S ribosomal protein S11-A/B | 16QPHIFNNPK24 | U0 | U0 |
| P0CX51/P0CX52* | 40S ribosomal protein S16-A/-B | 69VTGGGHVSQVYAIR82 | U0 | – |
| P02407/P14127* | 40S ribosomal protein S17-A/-B | 34LCDEIATIQSK44 | – | U0+152 |
| 50IAGYTTHLMK59 | U0 | – | ||
| P0CX31/P0CX32* | 40S ribosomal protein S24-A/-B | 44LAEVYKAEK52 | U0 | – |
| 115DKKIFGTGK123 | – | CU | ||
| P41057* | 40S ribosomal protein S29-A | 23VCSSHTGLIR32 | – | U0+152 |
| P41058* | 40S ribosomal protein S29-B | 23VCSSHTGLVR32 | – | U+152, U0+152 |
| P38845* | Cruciform DNA-recognizing protein | 105IPEAGGLLCGKPPR118 | AU0 | U0+152 |
| P04147* | Polyadenylate-binding protein | 319YQGVNLFVK327 | – | U0 |
| P02994* | Elongation factor 1-alpha | 320RGNVCGDAK328, 402FVPSKPMCVEAFSEYPPLGR421 | – | U0+152 |
| P41920 | Ran-specific protein 1 | 134ICANHIIAPEYTLKPNVGSDR154 | – | U0+152 |
| P10080* | SS-Nucleic acid binding protein | 184SKDTLYINNVPFK196, 186DTLYINNVPFK196 | U0 | U0 |
| Q01560* | Nucleolar protein 3 | 156ILNGFAFVEFEEAESAAK173, 222ENSLETTFSSVNTR235 | U, U0 | – |
| P00549* | Pyruvate kinase 1 | 370NCTPKPTSTTETVAASAVAAVFEQK394, 414YRPNCPIILVTR425 | – | U+152, U0+152 |
| P38013* | Peroxiredoxin type-2 | 114FASDPGCAFTK124 | – | U+152, U0+152 |
| P41277 | Glycerol phosphohydrolase | 210GCDIIVK216, 99LCNALNALPK108 | – | U0+152 |
| P38720 | Phosphogluconate dehydrogenase | 363GGCIIR368 | – | U0+152 |
| P00924* | Enolase 1 | 244IGLDCASSEFFK255 | – | U+152 |
| P38999 | Saccharopine dehydrogenase | 333GNALDTLCAR342 | – | U0+152 |
| P00817* | Inorganic pyrophosphatase | 83NCFPHHGYIHNYGAFPQTWEDPNVSHPETK112 | – | U0+152 |
| Q07163 | Transposon TyH3 polyprotein | 308VACQLIMR315 | – | U0+152 |
Fig. 5 shows two MS/MS examples for peptide-RNA (oligo)nucleotide cross-links with and without the DTT increment, both derived from 80NVSCYRPR87 of S6–40S ribosomal protein. The (U-H2O) cross-linked RNA moiety was identified both in the DTT and TCEP preparations (for data, see XL_yeast_Ex3_DTT, XL_yeast_Ex3_TCEP). Subtraction of the peptide molecular weight (993.4814 Da) from the calculated molecular weight of the cross-link (1299.5067 Da) corresponds to an RNA moiety of 306.0253 Da (U-H2O). In the corresponding DTT preparation we identify the cross-link with the 152 Da mass increment, with a calculated molecular weight of 1451.5033 Da and an RNA moiety increment of 458.0219 Da [(U-H2O)+152 Da], respectively (Fig. 5B). In both cases the fragment spectra are highly similar with almost complete y-ion series. Shifting of the y5 and y6 ions by 306 Da and 458 Da, respectively, identified Cys83 as the cross-linked amino acid, which lies within the flexible region of protein. Mapping of the cross-linked peptide to the crystal structure of S6–40S ribosomal protein (PDB ID: 3J78, (54)) also reveals the presence of U161 of 18S rRNA in close spatial proximity to Cys83 (Fig. 5C).
Fig. 5.
Comparison of MS/MS fragment spectra of a peptide derived from S. cerevisiae mRNPs in two cross-linking experiments. A, MS/MS spectrum of m/z 650.76092+ shows the peptide NVSCYRPR (positions 80–87) cross-linked to U-H2O without the additional mass of 152 Da observed in the presence of TCEP. Shifts of the y5 and y6 ions by 306 Da corresponding to (yr2-yr1) are indicated by #. B, MS/MS spectrum of m/z 726.76052+ shows peptide cross-linked to U-H2O with the increment of 152 Da, obtained in the presence of DTT. Signals corresponding to shifts of the y5 and y6 ions by 458 Da [(yr2-yr1) + 152 Da] are highlighted by bold letters and indicated by # according to the nomenclature by McLuckey et al. (61). Proposed schematic structures for peptide-RNA (oligo)nucleotide cross-links with and without the 152 Da increment are also shown. Cysteine is identified as the cross-linked amino acid by the shifting of the y4 ion, highlighted as bold letter in the sequence. C, Crystal structure of S6–40S ribosomal protein in complex with 18S rRNA (PDB ID: 3J78, (54)). The cross-linked peptide NVSCYRPR (positions 80–87) is mapped on the crystal structure of S6–40S ribosomal protein. Cross-linked cysteine (position 83) and uracil161 (18S-rRNA), which are in close spatial proximity, are shown as cyan and blue spheres within the structure, respectively.
For the other peptide-RNA (oligo)nucleotide cross-links it was not always possible to determine the position of the cross-linked amino acid on the peptide sequence accurately owing to the labile nature of the cross-links under CID-MS/MS conditions (22).
From 43 identified cross-linked proteins, 32 have been identified in the study of Kramer et al. (22) as well. The majority of the cross-linked proteins derived from ribosomal subunits, 14 are nonribosomal proteins. Among these are cross-linked peptides derived from proteins with a described RNA-binding motif: nucleolar protein 3, polyadenylate-binding protein, single-stranded nucleic acid-binding protein, and Transposon TyH3 polyprotein with cross-linked peptides located in the RNA-binding motif. Further cross-links derived from a nonribosomal protein that does not contain a “classical” RNA binding domain (RBD), i.e. elongation factor 1-alpha and metabolic enzymes including glycerol phosphate phosphohydrolase, phosphogluconate dehydrogenase, pyruvate kinase 1, peroxiredoxin type-2, enolase 1, saccharopine dehydrogenase, and inorganic prophosphatase. Indeed, cross-linking of metabolic enzymes to RNA has been previously described (16, 17, 22) and the sites we have identified here in enolase 1, pyruvate kinase 1, and inorganic pyrophosphatase were also previously identified by Kramer et al. (22).
DISCUSSION
We have shown that DTT specifically cross-links cysteine residues within RNA-binding proteins to the uridine nucleobase, and that this reaction requires activation by UV irradiation. DTT acts in a surprisingly efficient manner when compared with “pure” UV-light based photo-cross-linking and thus can be actively used as a protein–RNA cross-linking enhancer in UV light induced cross-linking. This is an entirely novel finding and expands the use of DTT, which is used mainly in the isolation and purification of biochemical preparations or assays. As a strong reducing agent, DTT usually reacts to form a stable six-membered ring with an internal disulfide bond (55). In its reduced state it promotes protein denaturation by disruption of S-S bonds and prevents protein aggregation by preventing or reversing oxidation of cysteine side chains.
We observed that UV-induced cross-links are purely additive in nature, including cysteine cross-links to uracil both without and with DTT. This suggests that the double bond of uracil between positions 5 and 6 is reduced when the free sulfhydryl group (of DTT or cysteine) reacts under UV light with uracil. This finding is in line with earlier and recent detailed investigations of the UV-induced reaction of amino acids within proteins to RNA (22). In previous studies it has been suggested that sulfhydryl groups are photoreactive toward uracil and add preferentially at the 5 position of uracil in its triplet excited state (56, 57). This is further corroborated by the observation that UV irradiation of 5-bromouracil in the presence of cysteine or glutathione results in the formation of 5-S-cysteinyl uracil or 5-S-glutathione uracil, respectively (58, 59). Addition of DTT at position 6 of uracil is not favored (60).
We therefore have postulated a plausible reaction scheme outlined in Fig. 4 to explain the cross-linking reaction of uridine/uracil to cysteine/DTT. However, our mass spectrometric results do not allow for any conclusions as to how the reaction exactly occurs mechanistically.
In earlier and recent studies we found that the UV-induced protein–RNA cross-linking reaction is not specific to a certain amino acid, as all amino acids except for aspartic acid and glutamic acid have been found to be cross-linkable by UV light to bases of nucleotides ((22), U.Z. and H.U., unpublished results). However, no precise conclusion can be drawn about the cross-linking yield in these studies, owing to a lack of appropriate standards for quantification. Interestingly, when compared with pure UV-induced zero-length cross-links, the UV-induced DTT cross-linking reaction seems to be surprisingly efficient for identification of cross-linked peptides containing cysteine residues. When we compared the identified cross-linked peptide-RNA oligonucleotides derived from the purified yeast pre-mRNA complexes in the presence of DTT or TCEP, we found that 21 peptide-RNA (oligo)nucleotide cross-links contained cysteine residues with DTT, demonstrating that DTT enhances efficiently the cross-linking of peptides containing cysteine residues to RNA. Even in previous studies where DTT was only present in cell lysis and/or RNP purification buffers, we found a relatively large proportion (19 out of 24) of cross-linked peptides containing cysteine residues that also contained DTT (22).
The DTT-driven cysteine photo-cross-linking of proteins to nucleotides is specific, although the presence of the DTT increment suggests a “spacer-distance” of ca. 10 Å between the C-α atom of cysteine and C-5 atom of uracil. Because we show here that cross-linking to nucleobases occurs at the same cysteine with and without DTT adduct in ribosomal proteins (see Table II and Fig. 5) it identifies those cysteine residues that are in close proximity to the RNA. Moreover, in a recent study of the crystal structure of the RNase H domain (without its cognate RNA) of the spliceosomal protein Prp8, two peptides derived from Prp8 were found to be cross-linked to U4 snRNA (46). The apparent cross-linked amino acids Tyr1858 (peptide 1) and Cys1878 (peptide 2) are both found at the base of the same β-hairpin loop. The peptide-RNA (oligo)nucleotide cross-links including Cys1878 showed the DTT mass increment.
In summary, we introduce here DTT as a potent protein–RNA cross-linker that makes it possible to link cysteine residues in RNA-binding regions to RNA, and is superior over pure UV light cross-linking of cysteine residues. The DTT-mediated protein–RNA cross-linking reveals RNA-binding proteins with their regions that have not been brought to attention in the context of RNA-binding before. Its suitability for similar approaches, e.g. in protein-DNA cross-linking, however, still needs to be evaluated. In addition, a detailed understanding of the reaction mechanism might help overcome the limited reaction yield achieved by allowing for further optimization of the cross-linking conditions.
Supplementary Material
Acknowledgments
We thank Monika Raabe and Uwe Pleβmann for technical support, and Prof. Dr. Olaf Jahn (Mass Spectrometry Group at the Max Planck Institute for Experimental Medicine) for providing synthetic peptides. We are also thankful to Klaus Hartmuth and Claudia Höbartner for helpful discussions.
Footnotes
Author contributions: U.Z., and H.U., designed research; U.Z., and F.R., performed research; K.K., T.S., and O.K., contributed new analytic tools; U.Z., analyzed data; U.Z., C.L., and H.U., wrote the paper.
* The work was supported by DFG grant (SFB860) to H.U.
 This article contains supplemental Table S1 and S2.
This article contains supplemental Table S1 and S2.
1 The abbreviations used are:
- DTT
- dithiothreitol
- RNPs
- ribonucleoprotein complexes
- rNTP
- ribonucleoside triphosphate
- 13C-rNTP
- 13C labelled ribonucleoside triphosphate
- d10-DTT
- perdeuterated dithiothreitol
- TCEP
- (tris(2-carboxyethyl)phosphine)
- HCD
- higher energy collision-induced dissociation
- MS/MS
- product ion scan mass spectrum
- FWHM
- full width half maximum
- 13UTP
- 13C labelled uridine triphosphate
- 13C-A
- 13C-G, 13C-U 13C-C, 13C labelled adenine, guanine, uracil and cytosine.
REFERENCES
- 1.Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 2.Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., and Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappsilber J. (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrotchenko E. V., and Borchers C. H. (2010) Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 29, 862–876 [DOI] [PubMed] [Google Scholar]
- 5.Paramelle D., Miralles G., Subra G., and Martinez J. (2013) Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics 13, 438–456 [DOI] [PubMed] [Google Scholar]
- 6.Holding A. N. (2015) XL-MS. Protein cross-linking coupled with mass spectrometry. Methods pii: S1046–2023(15)00252–2 [DOI] [PubMed] [Google Scholar]
- 7.Srinivasa S., Ding X., and Kast J. (2015) Formaldehyde cross-linking and structural proteomics: Bridging the gap. Methods ii: S1046–2023(15)00197–8 [DOI] [PubMed] [Google Scholar]
- 8.Trytayakova N. Y., Groehler A. 4th, Ji S. (2015) DNA-Protein Cross-Links: Formation, Structural Identities, and Biological Outcomes. Acc. Chem. Res. 48, 1631–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen O.N., Barofsky D. F., Young M. C., von Hippel P. H., Swenson S., and Seifried S. E. (1993) Direct observation of UV-crosslinked protein-nucleic acid complexes by matrix-assisted laser desorption ionization mass spectrometry. Rapid Commun. Mass Spectrom. 7, 496–501 [DOI] [PubMed] [Google Scholar]
- 10.Steen H., Petersen J., Mann M., and Jensen O. N. (2001) Mass spectrometric analysis of a UV-cross-linked protein-DNA complex: tryptophans 54 and 88 of E. coli SSB cross-link to DNA. Protein Sci. 10, 1989–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görner H. (1994) Photochemistry of DNA and related biomolecules: quantum yields and consequences of photoionization. J. Photochem. Photobiol. B 26, 117–139 [DOI] [PubMed] [Google Scholar]
- 12.Gurzadyan G. G., and Görner H. (1994) Damage to uracil- and adenine-containing bases, nucleosides, nucleotides and polynucleotides: quantum yields on irradiation at 193 and 254 nm. Photochem. Photobiol. 60, 323–332 [DOI] [PubMed] [Google Scholar]
- 13.Blatter E. E., Ebright Y. W., and Ebright R. H. (1992) Identification of an amino acid-base contact in the GCN4-DNA complex by bromouracil-mediated photocrosslinking. Nature 359, 650–652 [DOI] [PubMed] [Google Scholar]
- 14.Hicke B. J., Willis M. C., Koch T. H., and Cech T. R. (1994) Telomeric protein-DNA point contacts identified by photo-cross-linking using 5-bromodeoxyuridine. Biochemistry 33, 3364–3373 [DOI] [PubMed] [Google Scholar]
- 15.Golden M. C., Resing K. A., Collins B. D., Willis M. C., and Koch T. H. (1999) Mass spectral characterization of a protein-nucleic acid photocrosslink. Protein Sci. 8, 2806–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M. Krijgsveld J., Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 17.Baltz A. G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. Wyler E., Bonneau R., Selbach M., Dieterich C., and Landthaler M. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 18.Kwon S. C., Yi H., Eichelbaum K., Fohr S., Fischer B., You K. T., Castello A., Krijgsveld J., Hentze M. W., and Kim V. N. (2013) The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122–1130 [DOI] [PubMed] [Google Scholar]
- 19.Mitchell S. F., Jain S., She M., and Parker R. (2013) Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ascano M., Gerstberger S., and Tuschl T. (2013) Multi-disciplinary methods to define RNA-protein interactions and regulatory networks. Curr. Opin. Gen. Dev. 23, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerstberger S., Hafner M., and Tuschl T. (2014) A census of human RNA-binding proteins. Nat. Rev. Gen. 15, 829–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.L., Hentze M.W., Kohlbacher O., and Urlaub H. (2014) Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Meth. 11, 1064–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qamar S., and Urlaub H. (2015) Studying RNA–Protein Interactions of Pre-mRNA Complexes by Mass Spectrometry. Methods Enzymol. 558, 417–463 [DOI] [PubMed] [Google Scholar]
- 24.Sharma K., Kramer K., Sachsenberg T., Staals R.H.J., Randau L., Marchfelder A., van der Oost J., Kohlbacher O., Conti E., and Urlaub H. (2015) Analysis of protein-RNA interactions by UV-induced cross-linking and mass spectrometry. Methods pii: S1046–2023(15)00246–7 [DOI] [PubMed] [Google Scholar]
- 25.Konarska M. M. (1999) Site-specific derivatization of RNA with photocrosslinkable groups. Methods 18, 22–28 [DOI] [PubMed] [Google Scholar]
- 26.Reed R., and Chiara M. D. (1999) Identification of RNA-protein contacts within functional ribonucleoprotein complexes by RNA site-specific labeling and UV crosslinking. Methods 18, 3–12 [DOI] [PubMed] [Google Scholar]
- 27.Rhode B. M., Hartmuth K., Urlaub H., and Luhrmann R. (2003) Analysis of site-specific protein-RNA cross-links in isolated RNP complexes, combining affinity selection and mass spectrometry. RNA 9, 1542–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z. R., Wilkie A. M., Clemens M. J., and Smith C. W. (1996) Detection of double-stranded RNA-protein interactions by methylene blue-mediated photo-crosslinking. RNA 2, 611–621 [PMC free article] [PubMed] [Google Scholar]
- 29.Baumert H. G., Skold S. E., and Kurland C. G. (1978) RNA-protein neighbourhoods of the ribosome obtained by crosslinking. Eur. J. Biochem. 89, 353–359 [DOI] [PubMed] [Google Scholar]
- 30.Nygård O., and Nika H. (1982) Identification by RNA-protein cross-linking of ribosomal proteins located at the interface between the small and the large subunits of mammalian ribosomes. EMBO J. 1, 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brimacombe R., Stiege W., Kyriatsoulis A., and Maly P. (1988) Intra-RNA and RNA-protein cross-linking techniques in Escherichia coli ribosomes. Methods Enzymol. 164, 287–309 [DOI] [PubMed] [Google Scholar]
- 32.Wower I., Wower J., Meinke M., and Brimacombe R. (1981) The use of 2-iminothiolane as an RNA-protein cross-linking agent in Escherichia coli ribosomes, and the localisation on 23S RNA of sites cross-linked to proteins L4, L6, L21, L23, L27 and L29. Nucleic Acids Res. 9, 4285–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urlaub H., Kruft V., Bischof O., Müller E. C., and Wittmann-Liebold B. (1995) Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 14, 4578–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milligan J. F., Groebe D. R., Witherell G. W., and Uhlenbeck O. C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nottrott S., Hartmuth K., Fabrizio P., Urlaub H., Vidovic I., Ficner R., and Luhrmann R. (1999) Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18, 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X., Hsiao H. H., Bubunenko M., Weber G., Court D. L., Gottesman M. E., Urlaub H., and Wahl M.C. (2008) Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell 32, 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licklider L. J., Thoreen C. C., Peng J., and Gygi S. P. (2002) Automation of nanoscale microcapillary liquid chromatography-tandem mass spectromentry with a vented column. Anal. Chem. 74, 3076–3083 [DOI] [PubMed] [Google Scholar]
- 38.Kohlbacher O., Reinert K., Gröpl C., Lange E., Pfeifer N., Schulz-Trieglaff O., and Sturm M. (2007) TOPP - the OpenMS proteomics pipeline. Bioinformatics 23, E191-E197 [DOI] [PubMed] [Google Scholar]
- 39.Sturm M., Bertsch A., Gröpl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., and Kohlbacher O. (2008) OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P.(2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., and Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 42.Sturm M., and Kohlbacher O. (2009) TOPPView: an open-source viewer for mass spectrometry data. J. Proteome Res. 8, 3760–3763 [DOI] [PubMed] [Google Scholar]
- 43.Mayer G., Jones A. R., Binz P. A., Deutsch E. W., Orchard S., Montecchi-Palazzi L., Vizcaíno J. A., Hermjakob H., Oveillero D., Julian R. Stephan C., Meyer H. E., Eisenacher M. (2014) Controlled vocabularies and ontologies in proteomics: overview, principles and practice. Biochim. Biophys. Acta 1844, 98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer K., Hummel P., Hsiao H. H., Luo X., Wahl M., and Urlaub H. (2011) Mass-spectrometric analysis of proteins cross-linked to 4-thio-uracil- and 5-bromo-uracil-substituted RNA. Int. J. Mass Spectrom. 304, 184–194 [Google Scholar]
- 45.Kühn-Hölsken E., Lenz C., Dickmanns A., Hsiao H. H., Richter F. M., Kastner B., Ficner R., and Urlaub H. (2010) Mapping the binding site of snurportin 1 on native U1 snRNP by cross-linking and mass spectrometry. Nucleic Acids Res. 38, 5581–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mozaffari-Jovin S., Santos K. F., Hsiao H. H., Will C. L., Urlaub H., Wahl M. C., and Lührmann R. (2012) The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Devel. 26, 2422–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitzová J., Rasche N., Dybkov O., Kramer K., Fabrizio P., Urlaub H., Lührmann R., and Pena V. (2012) Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein. EMBO J. 31, 2222–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ofengand J., Liou R., Kohut J. 3rd, Schwartz I., and Zimmermann R. A. (1979) Covalent cross-linking of transfer ribonucleic acid to the ribosomal P site. Mechanism and site of reaction in transfer ribonucleic acid. Biochemistry 18, 4322–4332 [DOI] [PubMed] [Google Scholar]
- 49.Downs W. D., and Cech T. R. (1990) An ultraviolet-inducible adenosine-adenosine cross-link reflects the catalytic structure of the Tetrahymena ribozyme. Biochemistry 29, 5605–5613 [DOI] [PubMed] [Google Scholar]
- 50.Pomeranz Krummel D. A., Oubridge C., Leung A. K., Li J., and Nagai K. (2009) Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458, 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber G., Trowitzsch S., Kastner B., Lührmann R., and Wahl M. C. (2010) Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 29, 4172–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paizs B., Suhai S., and Harrison A. G. (2003) Experimental and theoretical investigation of the main fragmentation pathways of protonated H-Gly-Gly-Sar-OH and H-Gly-Sar-Sar-OH. J. Am. Soc. Mass Spectrom. 14, 1454–1469 [DOI] [PubMed] [Google Scholar]
- 53.Nelson C. C., and Mccloskey J. A. (1994) Collision-Induced Dissociation of Uracil and Its Derivatives. J. Am. Soc. Mass Spectrom. 5, 339–349 [DOI] [PubMed] [Google Scholar]
- 54.Svidritskiy E., Brilot A. F., Koh C. S., Grigorieff N., and Korostelev A. A. (2014) Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure 22, 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cleland W. W. (1964) Dithiothreitol, a New Protective Reagent for SH Groups. Biochemistry 3, 480–482 [DOI] [PubMed] [Google Scholar]
- 56.Jellinek T., and Johns R. B. (1970) The mechanism of photochemical addition of cysteine to uracil and formation of dihydrouracil. Photochem. Photobiol. 11, 349–359 [DOI] [PubMed] [Google Scholar]
- 57.Varghese A. J. (1974c) Photoaddition products of uracil and cysteine. Biochim. Biophys. Acta 374, 109–114 [DOI] [PubMed] [Google Scholar]
- 58.Varghese A. J. (1974b) Photoreactions of 5-bromouracil in the presence of cysteine and glutathione. Photochem. Photobiol. 20, 461–464 [Google Scholar]
- 59.Varghese A. J. (1974a) Photochemical addition of glutathione to uracil and thymine. Photochem. Photobiol. 20, 339–343 [DOI] [PubMed] [Google Scholar]
- 60.Pullman B. (1965) Some recent developments in the quantum-mechanical studies on the electronic structure of the nucleic acids. J. Chem. Phys. 43, Suppl:S233+. [DOI] [PubMed] [Google Scholar]
- 61.McLuckey S. A., Van Berkel G. J., and Glish G. L. (1992) Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 3, 60–70 [DOI] [PubMed] [Google Scholar]
- 62.Kondo Y., Oubridge C., van Roon A. M., and Nagai K. (2015) Crystal structure of human U1 snRNP, as mall nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.