SUMMARY
Noncoding RNAs (ncRNAs) have long been known to play vital roles in eukaryotic gene regulation. Studies conducted over a decade ago revealed that maturation of spliced, polyadenylated coding mRNA occurs by reactions involving small nuclear RNAs and small nucleolar RNAs; mRNA translation depends on activities mediated by transfer RNAs and ribosomal RNAs, subject to negative regulation by micro RNAs; transcriptional competence of sex chromosomes and some imprinted genes is regulated in cis by ncRNAs that vary by species; and both small-interfering RNAs and piwi-interacting RNAs bound to Argonaute-family proteins regulate post-translational modifications on chromatin and local gene expression states. More recently, gene-regulating noncoding RNAs have been identified, such as long intergenic and long noncoding RNAs (collectively referred to as lncRNAs)—a class totaling more than 100,000 transcripts in humans, which include some of the previously mentioned RNAs that regulate dosage compensation and imprinted gene expression. Here, we provide an overview of lncRNA activities, and then review the role of lncRNAs in processes vital to reproduction, such as germ cell specification, sex determination and gonadogenesis, sex hormone responses, meiosis, gametogenesis, placenta-tion, non-genetic inheritance, and pathologies affecting reproductive tissues. Results from many species are presented to illustrate the evolutionary conserved processes lncRNAs are involved in.
While long noncoding RNAs are recognized as important mediators of cellular fate and function, their roles in the reproductive processes are only now being elucidated.
DISCOVERY OF lncRNAs
Initial efforts to comprehensively characterize the mammalian transcriptome revealed an abundance of RNAs that vastly exceeded what was expected from the coding genome. Many RNAs were identified as non-coding and distinct from previously known non-coding species, including small nuclear RNA, small nucleolar RNA, transfer RNA, ribosomal RNA, and Argonaute-family-associated small RNAs (Rinn et al., 2003; Shiraki et al., 2003; Bertone et al., 2004; Kampa et al., 2004; Carninci et al., 2005; Cheng et al., 2005; Kapranov et al., 2005). A very limited number of novel, noncoding RNAs had previously been characterized: Xist (X-chromosome inactive-specific transcript), transcribed from the inactive X-chromosome in female mammals (Borsani et al., 1991; Brockdorff et al., 1991; Brown et al., 1991) to regulate X-inactivation processes in cis (Penny et al., 1996; Marahrens et al., 1997); the imprinted transcripts H19 (Brannan et al., 1990; Sleutels et al., 2002), Airn (antisense of insulin-like growth factor two non-protein-coding RNA), which regulates IGF2R (insulin-like growth factor 2) expression in cis (Lyle et al., 2000; Sleutels et al., 2002), and Kcnq1ot1 (voltage-gated KQT-like potassium channel, subfamily Q member one opposite strand/antisense transcript 1), which controls the expression of genes in the KCNQ1 (voltage-gated potassium channel KQT-like subfamily Q) cluster (Lee et al., 1999; Fitzpatrick et al., 2002); and SRA (steroid receptor RNA activator), which enhances steroid hormone receptor responses (Lanz et al., 1999; Lanz et al., 2002). Other long non-coding RNAs (lncRNAs) remained uncharacterized, and it was not clear if they had a function or were simply byproducts of transcriptional noise. In 2007, 231 additional lncRNAs from the human HOX (homeobox) clusters were discovered. One of these, HOTAIR (HOX transcript antisense RNA), transcribed from the HOXC cluster, was shown to regulate coding transcripts from the HOXD cluster in trans (Rinn et al., 2007), thus revealing functions of lncRNAs beyond cis-regulation of the silent X-chromosome and a few imprinted genes.
The development of genome-wide chromatin-state maps using chromatin immunoprecipitation followed by sequencing (ChIP-seq) revealed that known genes actively transcribed by RNA polymerase II carried specific histone 3 (H3) lysine methylation marks: H3K4me3 (trimethylation on lysine 4) at the promoter and H3K36me3 (trimethylation on lysine 36) across the transcribed region. These so-called K4-K36 domains were also found at sites not previously annotated as genes, leading to the discovery of more than 1,600 intergenic, spliced non-coding transcripts; that these regions were evolutionary conserved, with many exhibiting coordinated regulation, argued against the notion that they represented transcriptional noise (Guttman et al., 2009). More than 100,000 lncRNAs have since been described for human alone (Volders et al., 2015a).
Accepted lncRNA properties, and practices for their identification and naming, are evolving, but lncRNAs generally exhibit the following features (Mattick and Rinn, 2015): (1) Lengths are >200nt with a median size of ~500nt—generally smaller than mRNAs, although some exceed 100 kb; 98% are spliced, with 80% having 2–4 exons, and the majority exist as a single isoform. (2) Most are polyadenylated (Poly[A]+), although proportionately more non-polyadenylated (Poly[A]−) forms exist than for mRNAs. (3) Many show nuclear enrichment and chromatin association, albeit cytoplasmic forms exist; coding potentials are low, as assayed by codon-substitution frequency scores (Guttman et al., 2009, 2010; Cabili et al., 2011), low ribosome association (Guttman et al., 2013), and an absence of open reading frames >100nt. (4) Cumulative abundance is lower than for mRNAs, and expression is more tissue-specific. (5) Purifying selection is common, but occurs with weaker constraints than coding transcripts, and in some cases, structure rather than sequence might be under selection (Derrien et al., 2012; Smith et al., 2013). Finally, (6) some InRNAs are circular in structure (Hansen et al., 2013; Memczak et al., 2013; Zhang et al., 2013; Petkovic and Muller, 2015).
Databases established to describe lncRNAs include lncRNA Disease (Chen et al., 2013), LncRBase (Chakraborty et al., 2014), NONCODE(Xie et al., 2014), LNCipedia (Volders et al., 2015b), lncRNAdb (Quek et al., 2015), lncRNAWiki (Ma et al., 2015), and RNAcentral (RNAcentral-Consortium, 2015). As with many similar databases, these examples are likely to include mis-annotated mRNAs. For example, some RNAs classified as lncRNAs associate with ribosomes (Ingolia et al., 2009; Chew et al., 2013; Ruiz-Orera et al., 2014). Additionally, mass spectrometry analyses of peptides in two cell lines revealed 69 of 9,640 so-called lncRNAs encode detectable peptides (Banfai et al., 2012; Derrien et al., 2012), and similar proteomic analysis of rat male germ cells identified peptide sequences derived from previously annotated lncRNAs (Chocu et al., 2014). It is not clear if these peptides are functional or represent translational noise. Nevertheless, the presence of a functional reading frame within an RNA does not exclude a non-coding function; indeed, SRY (sex-determining region Y), SRA, and oskar RNAs have both coding and non-coding functions.
lncRNA FUNCTIONS
It is likely that lncRNA classifications will be refined and that subtypes of lncRNAs will be identified (Tuck and Tollervey, 2013). Currently, distinctions may be made according to lncRNA interacting partners; their functioning in cis versus trans; whether they influence chromatin modification or organizational states; if activities are cytoplasmic or nuclear; lncRNA structural properties; or the kinds of sequences from which they originate. One example of the latter classification is represented by enhancer RNAs (eRNAs), which are enhancer-derived noncoding RNAs (ncRNAs) that are typically less than 2 kb in length and operate in cis (De Santa et al., 2010; Kim et al., 2010). Regulate transcript elongation by interacting with mediator complex (Lai et al., 2013) and recruiting NELF (negative-elongation factor) from RNA polymerase II pause sites (Schaukowitch et al., 2014). They also affect chromatin looping associated with enhancer function (Pefanis et al., 2015) and can regulate nucleosome remodeling (Mousavi et al., 2013). Impaired enhancer RNA accumulation, on the other hand, attenuates enhancer activity (Lam et al., 2013). lncRNAs ncRNA-a3, 4, 5, and 7 have enhancer-like functions (Oram et al., 2010), whereas other enhancer RNAs, such as LED (Leveille et al., 2015) and LUNAR1 (Trimarchi et al., 2014), augment enhancer activity in trans by mechanisms that include recruiting mediator and RNA Polymerase II to enhancers. Additional examples of this classification include extra-coding RNAs, which are Poly-(A)− RNAs that extend beyond the gene bodies of coding genes (Di Ruscio et al., 2013), and promoter-associated noncoding RNAs (Hamazaki et al., 2015). The following four sections provide details of various molecular processes controlled by lncRNAs (see also Table 1 and Fig. 1).
TABLE 1.
Processes Controlled by lncRNAs
| Examples | References | |
|---|---|---|
| Nuclear processes | ||
| DNA methylation inhibition in cis | Extra-coding RNA, CpG island R-loop RNAs | Ginno et al. (2012); Di Ruscio et al. (2013) |
| DNA methylation recruitment in cis | Ribosomal DNA, promoter-associated non-coding RNA, piRNA-targeted RNA | Schmitz et al. (2010); Watanabe et al. (2011); Hamazaki et al. (2015) |
| Histone modification recruitment in cis | Xist, Airn, HOTTIP | Sleutels et al. (2002); Nagano et al. (2008); Zhao et al. (2008); Wang et al. (2011) |
| Histone modification recruitment in trans | HOTAIR | Rinn et al. (2007) |
| Histone modification state maintenance | Firre | Yang et al. (2015) |
| Nucleosome positioning | SCHLAP1, MHRT, enhancer RNAs | Mousavi et al. (2013); Prensner et al. (2013); Han et al. (2014) |
| Transcription interference | Airn | Latos et al. (2012) |
| Transcription factor sequestration | Gas5, PANDA | Kino et al. (2010); Hung et al. (2011) |
| Transcription factor recruitment | BCAR4 | Xing et al. (2014) |
| Organize chromatin domains and nuclear bodies | HOTTIP, Firre, MALAT1 | Tripathi et al. (2010); Wang et al. (2011); Yang et al. (2015) |
| Enhancer control | ncRNA-a3,4,5,7, enhancer RNAs LED LUNAR | Oram et al. (2010); Lam et al. (2013); Trimarchi et al. (2014); Leveille et al. (2015) |
| Histone variant recruitment | centromeric lncRNA | Quenet and Dalai (2014) |
| Splicing control | MALAT1, FGF2R antisense lncRNA | Tripathi et al. (2010); Gonzalez et al. (2015) |
| Cytoplasmic processes | ||
| Source of mi RNAs | H19 | Gao et al. (2012); Keniry et al. (2012) |
| miRNA sequestration | competing endogenous RNAs: HULC, linc-MD1, HncRNA-RoR, H19 circular RNAs | Franco-Zorrilla et al. (2007); Wang et al. (2010, 2013); Cesana et al. (2011); Hansen et al. (2013); Kallen et al. (2013) |
| Translation control RNA stability control | Uchl1 antisense lncRNA, HncRNA-p21 TINCR | Carrieri et al. (2012); Yoon et al. (2012) Kretz et al. (2013) |
| Source of siRNAs and piRNAs | piRNA-targeted RNA, centromeric transcripts | Hall et al. (2002); Volpe et al. (2002); Watanabe et al. (2011) |
BCAR4, breast cancer anti-estrogen resistance 4; FGF2R, fibroblast growth factor receptor 2; Firre, Firre Intergenic Repeating RNA Element; HOTTIP, HOXA distal transcript antisense RNA; MHRT, Myheart; myosin heavy chain-associated RNA transcript; PANDA, promoter of CDKN1A antisense DNA damage activated; RoR, regulator of reprogramming; TINCR, tissue differentiation-inducing non-protein coding RNA; Uchl1, ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase).
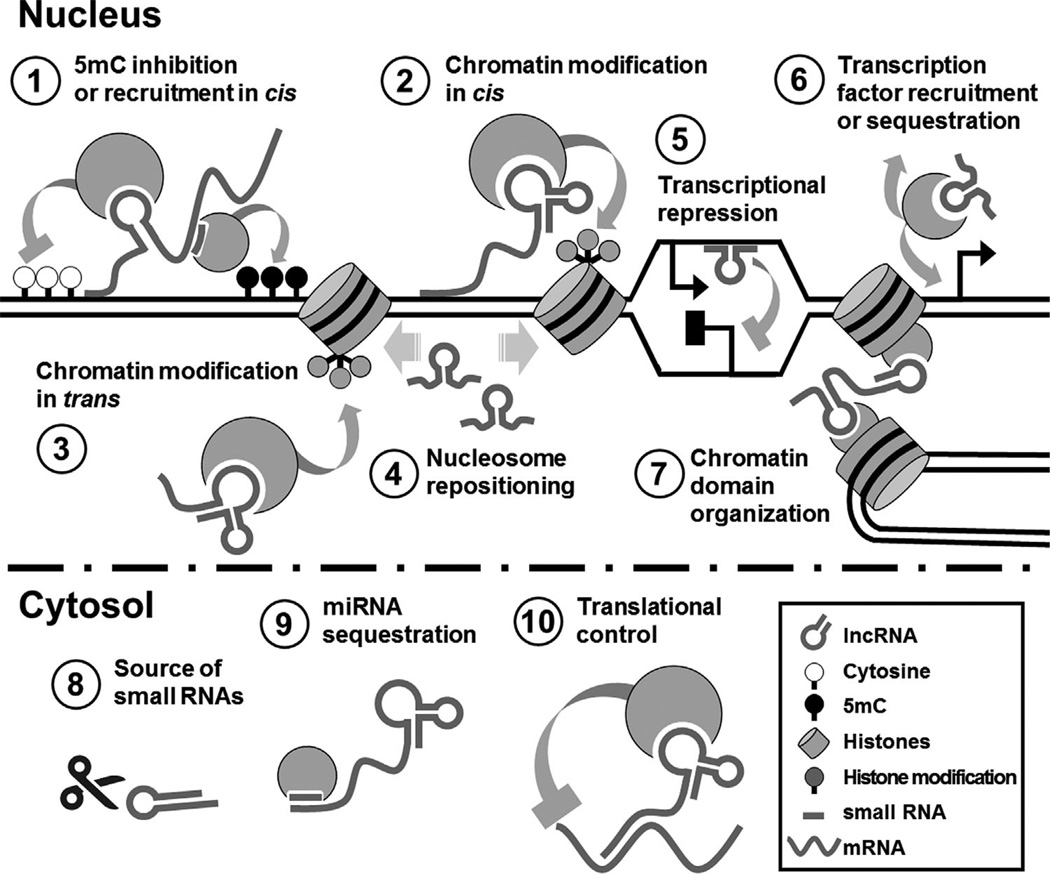
Figure 1.
Processes controlled by lncRNAs. In the nucleus (top), lncRNAs can either recruit or inhibit placement of a variety of chromatin modifications including DNA methylation (1) and histone modifications, either in cis (2) or in trans (3). They also control the positions of nucleosomes on the DNA by recruiting chromatin remodelers (4). While the simple act of antisense transcription (head-to-head orientation between a lncRNA and the gene it regulates) can directly inhibit gene expression (5), lncRNAs can also affect transcription by recruiting or sequestering transcription factors (6). The three-dimensional organization of the nucleus can be regulated by lncRNAs through chromatin looping (7), a phenomenon that brings distant regions of the DNA together. In the cytosol (bottom), lncRNAs can act as a source of miRNAs (8) or sponges to sequester them, inhibiting repression of translation by miRNAs (9). Additional small RNAs, including siRNAs and piRNAs, are also processed from lncRNAs outside the nucleus (8). Furthermore, lncRNAs can regulate protein accumulation either by inhibiting mRNA translation by the ribosome (10) or by changing the stability of target mRNAs. Specific examples of all these mechanisms can be found in Table 1.
lncRNA Control of Histone States
A common theme with lncRNAs is their regulation of chromatin states, including histone and DNA modifications, nucleosome positioning, and placement of histone variants. The HOXD-silencing lncRNA HOTAIR binds poly-comb repressive complex 2 (PRC2), the major H3K27 histone methyltransferase-containing complex, and is needed for deposition of H3K27me3 at HOXD (Rinn et al., 2007). The RepA (repeat A of Xist) lncRNA encoded within Xist, as well as Xist itself, also binds PRC2, and is necessary for initial deposition of H3K27me3 on the inactive X-chromosome (Zhao et al., 2008); conversely, maintenance of H3K27me3 on the inactive X-chromosome requires additional lncRNAs other than Xist. PRC2 seems to bind RNA promiscuously (Davidovich et al., 2013; Kaneko et al., 2013), yet some specificity exists according to immunoprecipitation experiments revealing that only 20% of ~3,300 lncRNAs queried were observed to bind PRC2 (Khalil et al., 2009); indeed, subsequent studies identified lncRNAs with a high affinity for PRC2 (Herzog et al., 2014; Davidovich et al., 2015). PRC2-interaction partners may further control specificity, as demonstrated by the partial regulation of Xist-PRC2 interaction by the nucleosome remodeler ATRX (alpha thalassemia/mental retardation syndrome, X-linked) (Sarma et al., 2014). PRC2 is a heterogeneous complex (Margueron and Reinberg, 2011), and various components were found to recruit it to lncRNAs: for example, lncRNA-PRC2 binding can occur through its component proteins JARID2 (Jumonji, AT Rich Interactive Domain 2) (Kaneko et al., 2014) or EZH2 (enhancer of Zeste 2 PRC2, subunit 2) (Zhao et al., 2008; Kaneko et al., 2014).
lncRNAs also bind a variety of writers, erasers, and readers of histone modifications, as well as other chromatin regulatory factors. In many cases, a given lncRNA can bind multiple chromatin regulatory factors (Guttman et al., 2011), although it is not yet known what the hierarchy of binding events is. Early findings reported that Airn (Nagano et al., 2008) and Kcnq1ot1 (Pandey et al., 2008) bind the H3K9 methyltransferase EHMT2/G9A (euchromatic histone-lysine N-methyltransferase 2), with Kcnq1ot1 also binding PRC2 (Pandey et al., 2008). By binding EHMT2/G9A, Airn directs this enzyme to the linked SLC22A3 (solute carrier family 22, member 3) promoter to silence it (Sleutels et al., 2002; Nagano et al., 2008). HOTTIP (HOXA distal transcript antisense RNA) is brought into proximity to other sites in the HOXA cluster by looping; it promotes H3K4me3 deposition and gene transcription within the HOXA cluster through its recruitment of WRD5-containing KMT2A/MLL (histone-lysine N-methyltransferase 2A/mixed-lineage leukemia) complexes (Wang et al., 2011). HOTAIR binds PRC2, and the KDM1A/LSD1 (lysine-specific demethylase 1A) through distinct domains (Tsai et al., 2010). It is possible that these two factors are functionally coordinated, with KDM1A/LSD1 removing activating marks on H3K4 and PRC2 placing silencing marks on H3K27.
In addition to promoting or removing chromatin modifications, lncRNAs can restrict them to specific domains. A lncRNA from a pericentromeric region in Schizosaccharo-myces pombe limits the spreading of H3K9me3 and binding of HP1 (heterochromatin protein 1), a reader of H3K9me3, beyond the centromeric region (Keller et al., 2013)—centromeric transcripts are the sources of small interfering RNAs (siRNAs) required for local placement of H3K9me3 (Hall et al., 2002; Volpe et al., 2002). Furthermore, Xist binds SMART/SPEN (SMART/HDAC1-associated transcriptional repressor protein), HNRNPU/SAF-A (heterogeneous nuclear ribonucleoprotein U/scaffold-attachment factor A), and LBR(lamin B receptor)—three factors necessary for X-inactivation. SHARP/SPEN interacts with NCOR2/SMRT (nuclear receptor corepressor 2) to activate HDAC3 (histone deacetylase 3), which likely participates in restricting silencing of the inactive X-chromosome (McHugh et al., 2015).
Beyond their influence on covalent modifications to histone proteins, lncRNAs can control nucleosome position and placement of histone variants. The lncRNA SCHLAP1 (Switch/Sucrose non-fermenter [SWI/SNF] complex antagonist-associated with prostate cancer 1) controls the localization and activity of SMARCB1/SNF5 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily B, member 1), a component of the SWI/SNF complex that repositions nucleosomes in an ATP-dependent manner (Prensner et al., 2013). The heart-specific lncRNA MHRT (Myheart; myosin heavy chain-associated RNA transcript) interacts with the nucleosome remodeler SMARCA4A/BRG1, restricting its activity at target sites (Han et al., 2014). Additionally, a centromeric lncRNA interacts with and is necessary for recruitment of the centromeric H3 variant, CENPA (centromere protein A) and its chaperone HJURP (Holliday junction recognition protein) to human centromeres (Quenet and Dalal, 2014).
lncRNA Control of DNA Methylation States
In addition to their effects on histone states, lncRNAs can provide signals for deposition of DNA methylation in cis. At the ribosomal DNA (rDNA) locus, a promoter-spanning antisense lncRNA forms an R-loop, a triplex structure between double-stranded DNA and a hybridized RNA, which recruits DNMT3B (DNA methyltransferase 3B) to the locus, leading to local methylation and rDNA silencing (Schmitz et al., 2010). At the imprinted RASGRF1 (Ras protein-specific guanine nucleotide-releasing factor 1) locus, P element-induced wimpy testes (piwi)-interacting RNA (piRNA)-targeted RNA, a lncRNA spanning the domain carrying the methylation imprint, is required for local DNA methylation (Watanabe et al., 2011). These piRNA-targeted RNA normally functions in cis (Park et al., 2012), but DNA methylation occurred in trans at the homologous locus when expression patterns were perturbed (Herman et al., 2003).
lncRNAs can also prevent the deposition of DNA methylation. In contrast to their role in placing DNA methylation at the rDNA locus, R-loops that form at CpG islands of other promoters have been implicated in preventing CpG island methylation (Ginno et al., 2012). The Poly(A)− extra-coding RNA that extends across the CEBPA (CCAAT/enhancer-binding protein [C/EBP], alpha) locus binds to DNMT1 (DNA methyltransferase 1). This binding sequesters DNMT1, limiting DNA methylation of the transcribed locus and enabling expression of the coding form of the Poly(A)+ mRNA (Di Ruscio et al., 2013). Many other Poly(A)− transcripts were identified by RNA immunoprecipitation studies using antibody against DNMT1, suggesting that extra-coding RNA control of DNMT1 might be commonplace. The domains from which extra-coding RNAs are transcribed tend to harbor less methylation and have more transcription relative to domains producing Poly(A)− transcripts that are unbound to DNMT1 (Di Ruscio et al., 2013); such observations are consistent with the notion that DNMT1 sequestration by extra-coding RNAs frequently limits DNA methylation.
lncRNA Control of Transcriptional States
lncRNAs can modify gene expression by influencing local chromatin states as well as through non-chromatin means. For example, lncRNAs such as GAS5 (growth arrest-specific 5) (Kino et al., 2010) and PANDA (promoter of CDKN1A antisense, DNA damage activated) (Hung et al., 2011) can limit access of transcription factors to their DNA targets by directly binding to these factors. Alternatively, the lncRNA BCAR4 (breast cancer anti-estrogen resistance 4) can enable transcription factor recruitment to DNA (Xing et al., 2014). In some cases, such transcription factor interactions are sensitive to extracellular signaling molecules (Trimarchi et al., 2014; Xing et al., 2014) or involve histone modifiers that affect local chromatin states (Wang et al., 2008; Xing et al., 2014). In addition to binding and recruiting EHMT2/ G9A to some target sites, Airn, the lncRNA that regulates imprinted IGF2R expression (Sleutels et al., 2002), exerts its effect by transcriptional interference at the silenced paternal IGF2R allele (Latos et al., 2012). At other loci, transcriptional interference by lncRNAs appears to be sufficient to control local gene expression (Martianov et al., 2007; Latos et al., 2012; Santoro et al., 2013).
lncRNAs may also respond to changes in chromatin and gene expression states. For example, the lncRNAs NEAT1 (nuclear paraspeckle assembly transcript 1) and MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) bind chromatin, but exhibited novel patterns of chromatin localization following treatments with the transcription elongation inhibitor flavopiridol. Thus, lncRNA localization can respond to and influence RNA polymerase II activity (West et al., 2014). Alternatively, lncRNAs might maintain chromatin or expression states, once they are established, as is the case for the lncRNA FIRRE (Firre intergenic repeating RNA element), which is required to maintain previously established H3K27me3 on the inactive X-chromosome (Yang et al., 2015).
lncRNA Control of Other Functions
lncRNAs influence a variety of other cellular functions beyond control of chromatin and transcriptional states, including nuclear architecture, splicing, and mRNA translation. In addition to its role in maintaining H3K27me3 on the inactive X-chromosome, FIRRE is required for nucleolar localization of the inactive X-chromosome in mammals. This occurs through a mechanism involving its own interaction with CTCF (CCCTC-binding factor zinc finger protein) (Yang et al., 2015). FIRRE also binds HNRNPU (heterogeneous nuclear ribonucleoprotein U), a nuclear matrix protein, through a sequence repeated within the lncRNA, and localizes to distinct regions in the genome in a manner dependent on HNRNPU expression (Hacisuleyman et al., 2014).
The lncRNA MALAT1 binds SRSFs (serine-arginine splicing factors) and influences their localization within nuclear speckles (Tripathi et al., 2010). MALAT1 also interacts with pre-messenger RNAs (Engreitz et al., 2014), and its depletion causes changes in alternative splicing (Tripathi et al., 2010). An additional example of splicing control by lncRNAs involves intron-encoded lncRNAs that are processed by small-nucleolar-RNA-dependent mechanisms. These so-called sno-lncRNAs influence splicing by their association with memberse of the FOX family of splicing factors (Yin et al., 2012). An anti-sense lncRNA from the human FGFR2 (fibroblast growth factor receptor 2) locus controls local alternative splicing choices by affecting local histone methylation state (Gonzalez et al., 2015).
A lncRNA corresponding to an antisense transcript from the coding gene UCHL1 (ubiquitin carboxyl-terminal esterase L1) regulates UCHL1 translation (Carrieri et al., 2012). Interestingly, this lncRNA exerts its translational control through sequences with similarity to the SINEB2 (short interspersed nuclear element B2) repetitive element. Long intergenic noncoding RNA p21 also regulates translation of specific transcripts, likely by a mechanism that involves physical interaction with its targets (Yoon et al., 2012). Although not controlling translation, the lncRNA TINCR (tissue differentiation-inducing non-protein coding RNA) can also affect protein levels after transcription and splicing by regulating mRNA stability (Kretz et al., 2013).
Effects of lncRNA may involve functional interactions with other regulatory ncRNAs as well. Micro RNAs (miRNAs) can be sequestered by lncRNAs, which are referred to as competing-endogenous RNAs, some of which are circular (Hansen et al., 2013; Memczak et al., 2013; Tay et al., 2014). Competing-endogenous RNAs limit the capacity of miRNAs to regulate translation of their mRNA targets. Originally reported in Arabidopsis (Franco-Zorrilla et al., 2007), this phenomenon was also found to occur in mammalian livercancercells (Wang et al., 2010), myoblasts (Cesana et al., 2011), and embryonic stem cells (Wang et al., 2013). Interestingly, H19 was shown to act as a miRNA sink as well (Kallen et al., 2013), yet also serves as a precursor for distinct miRNAs (Gao et al., 2012; Keniry et al., 2012). This is in addition to H19’s ability to bind the methylated DNA binding protein MBD1 (methyl-CpG binding domain protein 1) and to regulate other imprinted genes, both in cis and trans (Monnier et al., 2013). miR-9 can also target the lncRNA MALAT1 for degradation (Leucci et al., 2013). Beyond miRNAs, the piRNA pathway is necessary for lncRNA-mediated control of DNA methylation at the imprinted locus RASGRF1 (Watanabe et al., 2011).
The examples of genomic regulation provided here are illustrative of known lncRNA activities; additional studies are likely to reveal further activities and the necessity of individual lncRNAs for physiological processes in vivo. In one study of mice deficient for 18 lncRNAs, three were found to be essential for viability and two affected growth (Sauvageau et al., 2013). Important questions, whose answers are beginning to emerge, include the following: By what mechanisms are the lncRNAs themselves regulated (Amin et al., 2015)? What are the details of the mechanisms by which lncRNAs exert their effects? What health and disease-relevant phenotypes are controlled by lncRNAs? Understanding mechanisms of lncRNA action will require further knowledge of lncRNA structure and its impact on function (Brown et al., 2014a; Somarowthu et al., 2015); interacting factors, including proteins, other RNAs, and possibly metabolites; factors controlling lncRNA subcellular localization; and for chromatin-based phenomena, determining how lncRNAs localize to and/or restrict their activities at specific genomic domains. Many cis-acting lncRNAs are likely to function co-transcriptionally, while still tethered to their DNA template. On the other hand, how trans-acting lncRNAs become targeted to and act at specific loci is less clear. The transcription factor YY1 (yin and yang 1), for example, is important for recruiting Xist to the inactive X-chromosome(Jeon and Lee, 2011), buthow Xist is excluded from othergenomic locations is not clear. This might involve licensing enabled by X-chromosome pairing prior to X-inactivation (Xu et al., 2006), a mechanism that could be limited to X-inactivation. HOTAIR has many binding sites in the genome that are enriched for a GA-rich DNA motif, indicating DNA sequence-specific binding factors might recruit the RNA (Chu et al., 2011). In human cells, transcriptional targets of the lncRNA ANRIL/CDKN2B-AS1 (cyclin-dependent kinase inhibitor 2B antisense RNA 1) both contain and require Alu elements for their ANRIL/CDKN2B-AS1 response—yet not all Alu elements respond to ANRIL/CDKN2B-AS1, so it is not clear what provides specificity for Alu elements at ANRIL/CDKN2B-AS1 target genes (Holdt et al., 2013). Additional issues requiring more study include the functional importance, if any, of post-transcriptional modifications to lncRNAs (Kiani et al., 2013; Zheng et al., 2013b; Batista et al., 2014; Fu et al., 2014; Schwartz et al., 2014; Wang and He, 2014; Liu et al., 2015a), and the roles transposable elements have played in lncRNA diversity (Kelley and Rinn 2012; Liang et al., 2012; Kapusta et al., 2013). Approaches that systematically characterize proteins bound to specific lncRNAs will continue to be informative (McHugh et al.,2015).
lncRNAs IN REPRODUCTION AND DEVELOPMENT
Many excellent and recent reviews describe the discovery, cataloging, and activities controlled by lncRNAs, as well as approaches toward functional analysis (Wang and Chang 2011; Hu et al., 2012; Troy and Sharpless, 2012; Batista and Chang, 2013; Geisler and Coller, 2013; Ghosal et al., 2013; Sun and Kraus, 2013; Ulitsky and Bartel, 2013; Cech and Steitz, 2014; Fatica and Bozzoni, 2014; Flynn and Chang, 2014; Morris and Mattick, 2014; Quinodoz and Guttman, 2014; Chu et al., 2015; Engreitz et al., 2015; Holoch and Moazed, 2015; Iyer et al., 2015). Additional reviews focus on mechanisms of lncRNA control of sex-chromosome dosage compensation (Lee and Bartolomei, 2013; Autuoro et al., 2014; Briggs and Reijo Pera, 2014; Deng et al., 2014; Galupa and Heard, 2015), and control of development, including stem cell maintenance and differentiation (Batista and Chang, 2013; Ghosal et al., 2013; Fatica and Bozzoni, 2014; Flynn and Chang, 2014; Yao and Jin, 2014). Given the diverse roles lncRNAs play in essential biological processes common to many cell types, it should come as no surprise that lncRNAs play a vital role in reproduction, which is the focus of the remainder of this review. Though many ongoing studies are descriptive, functional and mechanistic studies exist and will be highlighted (see Table 2 and Fig. 2). Observations from a diversity of species will be presented, as they help define evolutionary conserved processes.
TABLE 2.
lncRNAs With Identified Functions in Reproduction
| Reproductive process |
lncRNAs involved | Mechanisms | References |
|---|---|---|---|
| Gonadogenesis | Sxl promoter proximal RNAs | Activates Sxl (sex-lethal) and recruits chromatin modifiers like Polycomb and Trithorax. | Mulvey et al. (2014) |
| Sex determination | SRY | Acts as a miRNA sponge that competitively binds miR-138 to potentially influence sex determination. | Hansen et al. (2013) |
| Dmr | Regulates splicing of DMRT1 (Doublesex and Mab-3 related transcription factor 1). | Zhang et al. (2010) | |
| Sex hormone responses | enhancer RNAs from ESR-bound enhancers | Controls estrogen receptor-regulated enhancer activity. | Li et al. (2013) |
| PCGEM1 | Binds androgen receptor and lncRNA PRNCR1 (prostate cancer- associated non-coding RNA 1); alters chromatin topology affecting androgen responses. | Yang et al. (2013) | |
| PRNCR1 | Binds androgen receptor and methyltransferase DOT1L (DOT1-like histone H3K79 methyltransferase); alters AR modification state and chromatin topology affecting androgen responses. | Yang et al. (2013) | |
| enhancer RNAs from the KLK3 enhancer cluster | Recruits mediator and androgen receptor to KLK3 and KLK2 (Kallikrein-related peptidases 3 and 2). | Hsieh et al. (2014) | |
| SRA | Binds to and modulates activity of androgen receptor, estrogen receptor, and progesterone receptor; binds additional transcriptional regulators. | Lanz et al. (1999); Shi et al. (2001); Watanabe et al. (2001); Lanz et al. (2002); Hatchell et al. (2006) | |
| Meiosis | UPGRADE2 | Expressed specifically in Boechera species capable of asexual reproduction. | Mau et al. (2013) |
| IRT1 | Increases nucleosome occupancy and repressive modifications to silence the meiotic regulator IME1. | van Werven et al. (2012) | |
| RME2 | Silences the oppositely transcribed meiotic regulator IME4, possibly by transcriptional interference. | Gelfand et al. (2011); Hongay et al. (2006); van Werven et al. (2012) | |
| meiRNA | Regulates nuclear import of the meiosis regulator Mei2 and formation of nuclear dots that are used to sequester Mmi1, which is capable of degrading meiosis regulating transcripts. | Yamashita et al. (1998); Harigaya et al. (2006); Shichino et al. (2014) | |
| Spermatogenesis | mhrl | Regulates Wnt signaling in spermatogonia! cells. | Arun et al. (2012) |
| Tsx | Suppresses apoptosis of pachytene spermatocytes. | Anguera et al. (2011) | |
| LDMAR | Required for anther development in rice. | Ding et al. (2012) | |
| Oogenesis | oskar | Non-coding functions of oskar mRNA required for early establishment of oocyte polarity. | Jenny et al. (2006) |
| Xlsirt and VegT | Required for cytoskeletal organization and oocyte polarity in Xenopus. | Kloc et al. (2005) | |
| XLOC_057324 | Regulates flowering time and seed setting in rice. | Zhang et al. (2014) | |
| Placentation | H19 | Controls placental growth regulated by NOM01 (NODAL modulator 1) by encoding miR-675 that controls NOM01 translation. | Gao et al. (2012); Keniry et al. (2012) |
| HELLP | Controls cell survival and migration of trophoblast cells. | van Dijk et al. (2012) | |
| SPRY4-IT1 | Controls cell survival and migration of trophoblast cells. | Zou et al. (2013) | |
| Airn | Regulates imprinted expression of Igf2r, Slc22a2, and Slc22a3, which influence placental growth. | Nagano et al. (2008); Zwart et al. (2001) | |
| Kcnq1ot1 | Regulates imprinted expression of eight linked genes that influence placental growth. | Pandey et al. (2008) | |
| Reproductive disease | NEAT1 | Required for corpus luteum formation and pregnancy maintenance | Nakagawa et al. (2014) |
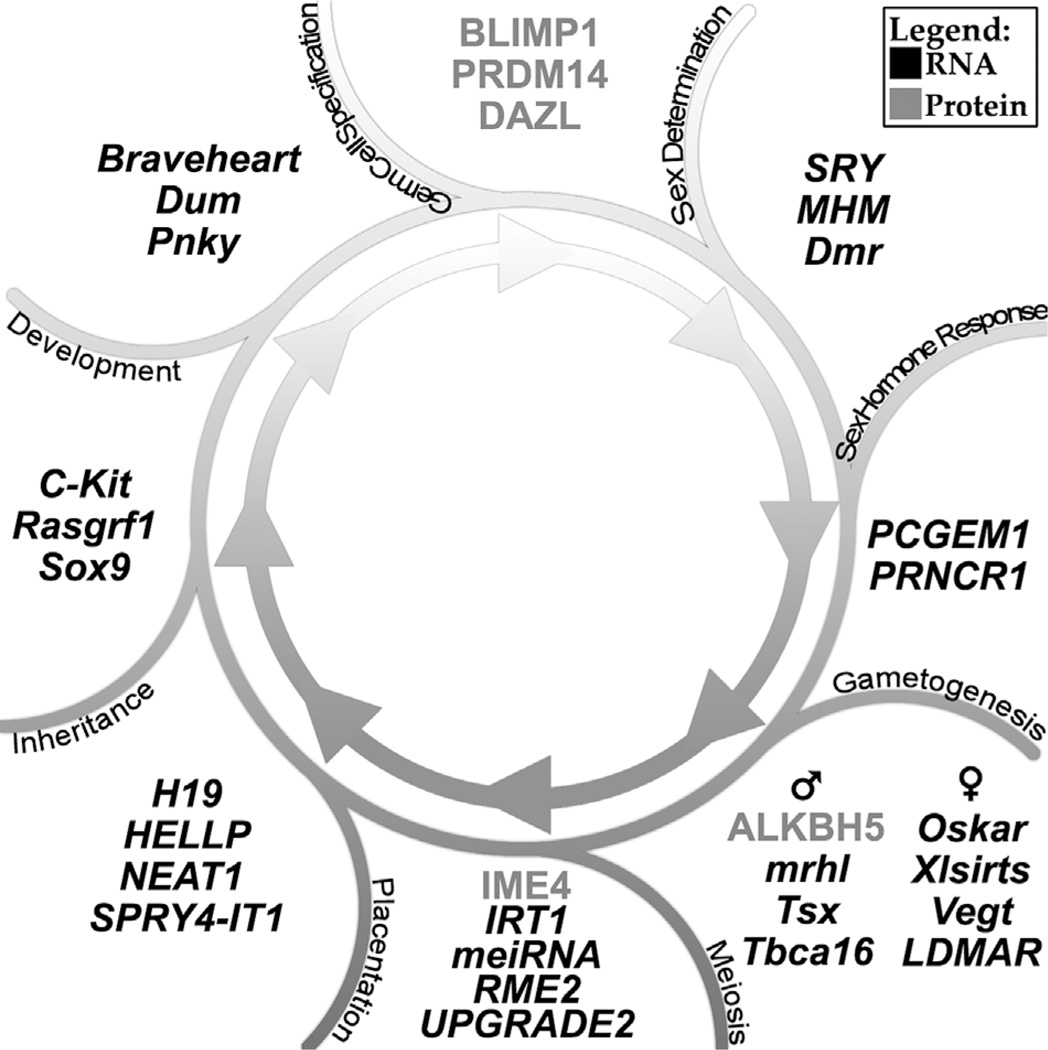
Figure 2.
Key steps in reproduction regulated by lncRNAs and lncRNA regulatory factors. lncRNAs denoted with italics; proteins denoted in upper case letters with no italics.
Germ Cell Specification
Formation of the animal germ line begins with specification of primordial germ cells (PGCs), pluripotent cells that, in mammals, are derived from a cluster of cells posterior to the definitive primitive streak in the extraembryonic mesoderm of mid-primitive-streak-stage embryos (7–7.5 days post-coitum in mouse). PGCs later migrate along the genital ridge, where they contribute to the developing gonad (Ginsburg et al., 1990). Three proteins required for PGC specification include BLIMP1/PRDM1 (PR domain-containing 1, with a zinc-finger domain), TFAP2C/AP2γ (transcription factor AP-2 gamma), and PRDM14 (PR domain-containing 14) (Magnusdottir et al., 2013). In mouse PGCs, BLIMP1/PRDM1 acts as a transcriptional repressor to block the expression of genes involved in somatic development (Keller and Maniatis, 1991; Gyory et al., 2003), but it also binds near other genes that are activated and important for PGC specification, such as Tcfap2c/Ap2γ and Cbx7 (chromobox homolog 7). Among the 5,046 BLIMP1/PRDM1 binding sites in mouse PGCs, 313 are associated with noncoding genes (Magnusdottir et al., 2013) whose functions in PGC specification are unknown. Given that lncRNAs are known to directly repress transcription, it is possible that BLIMP1/PRDM1 indirectly activates targets by negatively regulating repressive lncRNAs; further profiling of germ-line lncRNAs, and their interacting factors, could help resolve this.
The RNA-binding proteins DAZ1 (delete in azospermia 1), DAZL (deleted in azospermia-like), and BOLL/BOULE (Boule-like RNA-binding protein) are important for PGC specification and meiotic progression (Kee et al., 2009). These proteins bind RNA in the cytoplasm, regulating translation initiation (Collier et al., 2005), and have also been shown to translocate into and out of the nucleus during germ line development (Reijo et al., 2000). Within the nucleus, these proteins might functionally regulate coding and/or noncoding RNAs that are important for PGC formation or differentiation. These dual functions could be addressed by identifying and functionally characterizing RNAs associated with DAZ family members at different time points during germ line differentiation.
lncRNAs may also be involved in PGC specification via the mechanisms cited above. For example, Bvht (Brave-heart) is a lncRNA that controls the expression of MesP1 (mesoderm posterior basic helix-loop-helix transcription factor 1), a master regulator controlling differentiation of mesodermal precursors into cardiomyocytes (Klattenhoff et al., 2013). It is therefore possible that master regulators of PGC fate, like BLIMP1/PRDM1 or DAZL, may also be under the control of an unknown lncRNA. Thus, lncRNA regulation might be active at every level of PGC development, both as an initiator and as a downstream response element.
Sex Determination and Gonadogenesis
Several lines of evidence suggest the importance of lncRNAs for sex determination and sex-specific patterns of development. The number or identity of sex chromosomes is typically the genetic determinant of sex—although lncRNA involvement in this process is documented as well. For example, Drosophila become female upon the early expression of the X-encoded Sxl (Sex-lethal) gene. Sxl expression is dependent upon the ratio of X chromosomes to autosomes (A), involving a complex chromosome-counting mechanism that requires the competition of gene products from each chromosome. If X≥A, then the X transcription factors Sisterless-a and Sisterless-b activate Sxl expression; if X<A, then proteins including Deadpan and Extramacrochaetae directly or indirectly block binding to the Sxl promoter, resulting in male determination (Schutt and Nothiger, 2000). Interestingly, expression of a panoply of lncRNAs located ~1 kb upstream of the promoter add to the complexity of this counting mechanism, with RNAs from the R1 region repress whereas RNAs from the R2 region activate Sxl (Mulvey et al., 2014). These RNA species were also shown to recruit chromatin modifiers like Polycomb and Trithorax, indicating that Sxl expression is regulated by a complex interaction network involving many lncRNAs.
In mice, expression of the Y-chromosome-encoded Sry gene is sufficient to drive male sex determination (Koopman et al., 1991). SRY protein activates transcriptional cascades specific for male development (Kashimada and Koopman,2010), but its RNA independently functions as a circular-RNA miRNA sponge that competitively binds miR-138 in vitro, which can further positively regulate male specification (Hansen et al., 2013).
DMRT1 (Doublesex and Mab-3 related transcription factor 1) has been implicated in sex determination in a variety of vertebrate and invertebrate species, including humans, and acts both as a transcriptional repressor and activator (reviewed in Matson and Zarkower, 2012). In mice, the Dmrt1 transcript participates in trans splicing with the lncRNA Dmr, producing a transcript that encodes a protein with an altered carboxyl terminus. Overexpressing Dmr in primary Sertoli cell cultures increased the abundance of this altered form of DMRT1 protein, reduced the abundance of the cannonical DMRT1 isoform, and led to impaired expression of DMRT1 targetgenes, mimicking the Dmrt1 loss-of-function phenotype. It is clear that trans splicing negatively regulates DMRT1; what is unclear is whether or not the non-cannonical isoform has its own regulatory activity. Interestingly, reporters carrying a 3’ untranslated region (UTR) from Dmr exhibit enhanced expression (Zhang et al., 2010). It is not known if these contrasting results are due to idiosyncrasies of the specific assay system or if they reflect the range of regulation controlled by Dmr.
Sex Hormone Responses
The study of lncRNA activity in the context of sexhormone response has largely been restricted to the steroid sex hormones, which utilize nuclear receptors, including ESR1 (estrogen receptor 1), AR (androgen receptor), and PGR (progesterone receptor). On the other hand, roles for lncRNAs have not been demonstrated for signaling by follicle-stimulating hormone and luteinizing hormone, which utilize G-protein coupled receptors and cytosolic signal transduction cascades—although the cytosolic localization and activity of many lncRNAs still leave such roles plausible.
Functional studies of noncoding RNA involvement in AR and ESR1 responses include roles for enhancer RNAs and other lncRNAs. For example, enhancer RNAs transcribed from ESR1-bound enhancers recruit transcriptional activators to drive expression of nearby ESR1-responsive genes. Loss of the enhancer RNAs, by siRNA knockdown, reduced transcription of targetgenes without affecting ESR1 recruitment; moreover, tethering specific enhancer RNAs to a reporter gene enabled reporter activation(Li et al.,2013). In a similar manner, the lncRNAs PCGEM1 (prostate-specific transcript) and PRNCR1 (prostate cancer-associated non-coding RNA 1) associate with the AR. Knockdown of PCGEM1 or PRNCR1 reduced transcription of a number of canonical AR-targeted genes while enhancer-promoter interactions were reduced, based on chromatin-conformation capture assays (Yang et al., 2013). AR binds to an enhancer cluster 4 kb upstream of the AR-driven promoter of KLK3 (Kallikrein-related peptidase 3) in a hormone-dependent manner (Hsieh et al., 2014). This enhancer RNA acts as part of a scaffolding apparatus, which includes Mediator and AR, that enhances transcriptional activity at the endogenous KLK3 locus and at the downstream KLK2 locus. Additional AR and ESR1 target genes might also be influenced by lncRNAs.
The lncRNA SRA was originally identified in a yeast two-hybrid screen for human PGR-interacting factors, indicating it has a functional reading frame. Yet, its steroid-hormone-receptor-activator activity does not require SRA translation or an open reading frame (Lanz et al., 1999,2002). SRA has since been shown to modulate the activity of AR, ESR1, and PGR through direct association with hormone receptors (Lanz et al., 1999) and through recruitment of a variety of transcriptional activators and repressors (Shi et al., 2001; Watanabe et al., 2001; Hatchell et al., 2006). Furthermore, this lncRNA can be spliced and translated into SRAP/SRA1 (SRA protein 1), which also enhances steroid hormone-mediated gene expression (Kawashima et al., 2003). In breast cancer cells, SRA1 has also been shown to associate with a repressive histone-modifying complex containing unliganded PGR and with chromatin-binding and modifying factors, including CBX5/HP1 (chromobox homolog 5/het-erochromatin protein 1), KDM1A/LSD1, HDAC1 and 2 (histone deacetylase one and 2), and RCOR1/CoREST (RE1-silencing transcription factor corepressor 1). Unliganded PGR localizes this complex to approximately 20% of steroid-responsive genomic loci. Depletion of SRA led to destabilization of the complex and aberrant gene expression. Upon progesterone treatment, the repressive complex is evicted and replaced by ligand-bound PGR and basal transcription factors (Vicent et al., 2013). Yet, even though SRAP/SRA1 can enhance the activity of various steroid receptors—including, but not limited to, the steroid sex hormones—the regulation of SRAP/SRA1 transcription itself is unknown. Similarly, the factors that determine its differential splicing to become a transcript coding for SRAP/ SRA1 are also unknown.
Meiosis
lncRNAs have been implicated in the control of meiosis from studies in both plants and yeasts. In several plant species, seeds can form asexually through a variety of processes collectively referred to as apomixis (Koltunow and Grossniklaus, 2003). Shared features include female gamete formation in the absence of recombination or reductive division, which are normally seen in meiosis, followed by parthenogenic embryo development in the absence of fertilization. The resulting plants harbor their maternal genotype. In several apomictic species of the genus Boechera, microarray analyses identified a conserved lncRNA, UPGRADE2, that is present and highly upregulated in pollen mother cells. No homolog was found in sexually reproducing species of the same genus (Mau et al., 2013), so it remains to be determined if this lncRNA is simply associated with or is required for apomixis in Boechera.
In the budding yeast Saccharomyces cerevisiae, IME1 (inducer of meiosis 1) is kept transcriptionally silent by the repressor RME1 (regulator of meiosis 1) in vegetative cells growing in a nutrient-rich environment and in haploid cells encountering no partners of the opposite mating type. RME1 induces expression of a lncRNA, IRT1 (IME1 regulatory transcript), which spans the IME1 promoter and works in cis to increase local nucleosome occupancy and to recruit the SET3 complex that deposits repressive histone modifications at the promoter (van Werven et al., 2012). Interestingly, many SET3-repressed genes have overlapping lncRNA transcripts (Kim et al., 2012).
Another inducer of meiosis, IME4 (Shah and Clancy, 1992), is regulated in cis by the lncRNA RME2, which is transcribed antisense relative to IME4 and might block its expression by transcriptional interference rather than by recruiting chromatin-modifying factors (Hongay et al., 2006; Gelfand et al., 2011; van Werven et al., 2012). Antisense transcripts to these lncRNAs activate sporula-tion (van Werven et al., 2012). Interestingly, IME4 is a methyltransferase capable of placing N6-methyladenine (m6A) RNA modifications (Agarwala et al., 2012), a modification that makes the RNA that harbors it less stable than those lacking it (Batista et al., 2014). The Drosophila ortholog of IME4, METTL3 (methyltransferase-like 3), is essential for gametogenesis and embryo viability (Hongay and Orr-Weaver, 2011). These functions in Drosophila are mediated by IME4 through Notch signaling, although it is not clear how. Indeed, many RNAs with IME4-dependent m6A modifications have been described (Schwartz et al., 2013), and meiosis-specific lncRNAs beyond RMA2 have been described in S. cerevisiae, raising the likelihood that additional lncRNA-dependent mechanisms exist that regulate meiosis (Lardenois et al., 2011).
lncRNAs have also been implicated in meiotic control in the fission yeast S. pombe. Mei2 (meiosis RNA-binding protein 2) is the master regulator of S. pombe meiosis (Watanabe and Yamamoto, 1994; Watanabe et al., 1997). It is recruited to the nucleus by the lncRNA meiRNA (Yamashita et al., 1998; Shichino et al., 2014), forming a nuclear dot (Shimada et al., 2003) that includes the lncRNAs Mei2 and Mmi1 (meiotic mRNA interception 1). This occurs at the sme2 (suppressor of Mei2v) locus from which meiRNA is transcribed, which defines a cis-acting function for meiRNA. The nuclear dot promotes meiosis by sequestering Mmi1, an RNA-binding protein that degrades meiosis-promoting transcripts (Harigaya et al., 2006). Degradation requires polyadenylation and involves nuclear exosomes. Interestingly, Mmi1, whose function is antagonized by meiRNA, is required for meiRNA recruitment to sme2 (Shichino et al., 2014); therefore, although meiRNA functions in cis, its localization involves trans-acting factors.
Additional lines of evidence indirectly suggest other mechanisms that potentially involve lncRNA in meiosis. Methyltransferases have been implicated in processes critical to RNA function in vertebrates, invertebrates, and plants (Zhong et al., 2008; Li and Mason 2014; Schwartz et al., 2013); the example of m6A controlled by IME4, cited above, is one. Indeed, more-thorough characterization of both m6A-modified RNAs and lncRNAs influencing meiosis may clarify the importance of m6A modifications to lncRNA function. Another potential mechanism influencing lncRNA action involves RNA-binding proteins known to be important in mammalian meiosis, including DAZL and DDX4/VASA (DEAD box polypeptide 4) (Medrano et al., 2012). The DAZ family of RNA-binding proteins, which are required for PGC specification, are found in the nucleus and cytoplasm of fetal germ cells, in the cytoplasm of developing oocytes, and in the nucleus of spermatogonia. Their translocation between the nucleus and cytoplasm during meiosis (reviewed in Brook et al., 2009; Smorag et al., 2014) implicates additional functions beyond translational control (Collier et al., 2005). DAZ and BOLL/BOULE are also required for later stages of meiosis (Kee et al., 2009). Immunoprecipitates of DAZL from rat testis homogenate contained many mRNAs, but as these data were detected by microarray, some recently characterized lncRNAs may have been overlooked. Indeed, associations between DAZ-family proteins and lncRNAs might reveal important participants in meiosis. DDX4/VASA is an RNA helicase that regulates mRNA translation and piRNA production (reviewed in Kotov et al., 2014). DDX4/VASA immunoprecipitates from mouse testicular cells contained 858 mRNAs (Nagamori et al., 2011), as identified by microarrays designed to detect mRNAs. RNA-sequencing (RNA-seq) analysis, however, would more reliably reveal if DDX4/VASA also binds and functionally regulates lncRNAs associated with meiosis. MOV10L1 (Mov10 RNA-induced silencing complex RNA helicase-like 1) is another RNA helicase expressed at increasing levels in germ cells between the gonocyte and pachytene spermatocyte stages. It binds the PIWI (P-element induced wimpy testes) proteins PIWIL1/MIWI and PIWIL2/MILI (Frost et al., 2010; Zheng et al., 2010) and piRNA precursor transcripts (Vourekas et al., 2015), which may formally be considered a class of lncRNAs. MOV10L1 is required for primary piRNA biogenesis (Zheng et al., 2010; Zheng and Wang 2012; Vourekas et al., 2015) and silencing retrotransposons in the male germ line (Frost et al., 2010). Male mice lacking MOV10L1, or carrying a point mutation in the ATP-binding domain of the helicase, exhibit meiotic arrest in prophase I (Frost et al., 2010; Zheng et al., 2010; Zheng and Wang 2012; Vourekas et al., 2015); on the other hand, females deficient for the protein are fertile. Special requirements for helicases during piRNA biogenesis may relate to G-quadruplex structures present in precursor transcripts (Vourekas et al., 2015).
Gametogenesis
Spermatogenesis
lncRNAs are dynamically expressed and appear to be highly regulated in spermatogenesis. Several studies have profiled the transcriptomes of the developing male germ line, revealing a clear pattern. First, transcript levels dramatically increase as spermatogonia enter meiosis. These increase further as spermatocytes give rise to spermatids, but is followed by a rapid depletion of RNA in spermatozoa (Bao et al., 2013; Laiho et al., 2013; Soumillon et al., 2013; Chalmel et al., 2014; Liang et al., 2014; Margolin et al., 2014). These total-RNA profiling studies revealed novel lncRNAs, most of which have not been functionally characterized. Recent RNA-seq profiling at different stages of spermatogenesis, however, highlight potential regulation of spermatogenesis by lncRNAs.
Stage-specific, differentially expressed lncRNAs have been found within 30 kb of coding-gene clusters by array-based profiling of lncRNAs and mRNAs during spermatogenesis. Positive and negative correlations between lncRNA expression and local mRNA expression were observed, depending on the gene cluster. The most pronounced changes in expression occurred after the onset of meiosis, with changes in lncRNA abundance correlating with expression of nearby mRNA clusters. A subset of these lncRNAs were characterized via cross-linking and immunoprecipitation (CLIP)-quantitative PCR (Bao et al., 2013); many were found to interact with EZH2 and KDM1A/ LSD1 in ways that potentially regulate nearby expression and methylation states. The coordinated change in expression of lncRNAs and corresponding gene clusters was also observed in an array-based profile (Liang et al., 2014). Such regulation is not surprising in the context of the promiscuous binding of PRC2 (Davidovich et al., 2015); the physiological relevance of these data, however, requires further characterization to clarify.
At birth, spermatogonia in mice possess a comparably low fraction of the total-testis lncRNA profile found in adults (Soumillon et al., 2013). mhrl is one interesting transcript detected at birth. This lncRNA resides in the nucleus, and has been shown suppress the WNT (wingless-type MMTV integration site family) signaling pathway in a spermatogonial cell line by regulating beta-catenin nuclear translocation (Arun et al.,2012). WNT signaling is a regulator of “stem cell-ness” and is implicated in maintaining a self-renewing population of spermatogonial stem cells (Golestaneh et al., 2009; Yeh et al., 2011). Although mhrl-mediated repression of WNT signaling suggests it influences spermatocyte differentiation, its specific function needs to be explored by in vivo manipulations of mhrl expression.
Upon induction of meiosis there is a considerable increase in lncRNA transcription in mouse spermatocytes (Soumillon et al., 2013). In pachytene spermatocytes, Tsx (testis-specific X-linked), a predominantly nuclear, testis-specific lncRNA, becomes highly expressed and escapes X-inactivation (Anguera et al., 2011). A Tsx-knockout produces viable and fertile offspring, although males have decreased testis size and exhibit increased apoptosis of pachytene spermatocytes. Interestingly, Tsx-knockout mice also showed deficiencies in learning and increased Xist expression. The nuclear localization and X-linked expression of Tsx therefore suggest a role in X-inactivation in pachytene spermatocytes that is far from understood.
The importance of RNA methyltransferases, such as IME4, was previously discussed in reference to gamete development. On the other hand, RNA demethylases, such as ALKBH5 (AlkB family member 5), are also vital during this time, specifically at the pachytene stage of spermatogenesis (Zheng et al., 2013a). AIkbh5-knockout mice exhibit decreased testis size, sterility, more m6A-modified mRNAs, altered RNA localization, and significant changes in gene expression. The increased half-life of demethylated RNAs at this stage (Batista et al., 2014) may contribute to the increased expression and overall abundance of lncRNAs in spermatocytes, which could potentially affect recruitment of other chromatin readers/writers such as PRC2 to specific loci. Alternatively, m6A may be regulating RNA-protein interactions or affinities via altered RNA base pairing (Liu et al., 2015a). m6A-seq has not yet been performed in developing testis.
TBCA13 (tubulin cofactor A chromosome isoform 13), a protein involved in tubulin assembly, increases in abundance from 14 to 25 days post partum in mouse testis. Transcription of Tbca13 in a spermatocyte cell line is regulated by a pseudogene, Tbca16, which originated from a duplication of Tbca13 with both sense and antisense transcription on chromosome 16. The antisense product of Tbca16 appears to negatively regulate Tbca13: When Tbca16 mRNA was depleted by short-hairpin RNA, Tbca13 escaped silencing (Nolasco et al., 2012). The mechanism behind Tbca16 silencing and the escape of Tbca13 during spermatogenesis has not been elucidated, although a corollary might be found at the 3’ actin pause site where antisense transcription and R-loop formation recruits AGO2 (Argonaute RNA-induced silencing complex catalytic component 2), EHMT2/G9A, and the repressive H3K9me2 mark to enhance mRNA termination (Skourti-Stathaki et al., 2014). The in vivo importance of such regulation and its involvement in spermatogenesis needs further exploration.
A majority of the transcriptome is depleted upon spermatozoa maturation. Most recently, the transcriptomes of the nucleus and periphery of mature spermatozoa were profiled, and revealed that the majority of spermatozoon RNA is localized to the cytoplasm while a minority (roughly 34%) localizes to the nucleus (Johnson et al., 2015). MALAT1 highlights the potential for lncRNA-mediated chromatin organization in the male germ line: Despite the expulsion of most RNAs, it is enriched in the sperm nucleus. Yet Malat1 knockouts do not exhibit defects in fertility, underlining the fact that its function at this stage is unclear (Zhang et al., 2012). Several RNAs are present in mature spermatozoa that are not present in unfertilized oocytes, but are delivered to the zygote upon fertilization (Johnson et al.,2015). It is currently not certain if spermatozoon- localized lncRNAs are vital for gamete formation or zygotic function after fertilization.
Oogenesis
The developing mammalian oocyte exists in a complex where in a network of cumulus cells surrounds the oocyte and remains in intimate communication with the oocyte through gap junctions. Early during oogenesis, cumulus cells form a compact layer around immature oocytes, which are arrested at prophasel. Surgesoffollicle-stimulating hormone and luteinizing hormone at ovulation cause the cumulus-oocyte complex to expand and detach from the follicle wall, coincident with the oocyte resuming meiosis (Yokoo and Sato, 2004). Although the transcriptional change in cumulus cells is considerable during expansion of the cumulus-oocyte complex, a small number of lncRNAs were detected as differentially expressed by RNA-seq (Yerushalmi et al., 2014). Ninety-six non-coding RNAs, 45 anti-sense, and 44 long-intergenic noncoding RNAs, were identified as differentially expressed between compact and expanded cumulus cells. While not evaluated functionally, the presence of antisense transcripts during this interval suggests a regulatory role for them. Another study investigated lncRNAs in “high-quality” versus “poor-quality” human cumulus cells by microarray (Xu et al., 2014). The samples were derived from in vitro fertilization, and quality was defined by their morphology. Of the 20,000 lncRNAs examined, 633 were identified as being differentially expressed between high-quality and poor-quality cumulus cells.
These cumulus-cell lncRNA profiles are especially important because of the evidence that the cytoplasm and its contents are shared in a limited way between cumulus cells and the oocyte. In mammals, cumulus cells that have been independently transfected with a GFP (green fluorescent protein) reporter allowed GFP mRNA to move into the oocytes, resulting in GFP-expressing oocytes that lack the reporter plasmid(Macaulay et al.,2014). In Drosophila, nurse cells transfer RNA and other cytoplasmic components to oocytes (Cha et al., 2001; Nicolas et al., 2009); similar phenomena are seen in hydra (Alexandrova et al., 2005) and in mouse (Cossetti et al., 2014), the latter of which might be an exosome-mediated process (Gezer et al., 2014; Pefanis et al., 2015). Such communication by cytoplasmic sharing is a perfect medium by which regulatory lncRNAs may be moved from a somatic cell type into the developing germ line.
In a single-cell RNA-seq profile of metaphase-II (MII) oocytes and preimplantation embryos, 8,700 maternal lncRNAs were identified in the preimplantation embryo (Yan et al., 2013). Six hundred sixty differentially expressed lncRNAs were identified between MII oocytes and zygotes, which are hypothesized to affect gene activation during the maternal-to-zygotic transition. Many lncRNAs with possible functional relevance in the transition from MII oocytes to 2-cell embryos have also been identified (Hamazaki et al., 2015). In an impressive screen using strand-specific RNA-seq, more than 1,000 potentially functional lncRNA/mRNA pairs have been identified, with a subset acting as promoter-associated noncoding RNAs in zygotes. A subsequent screen of these identified pairs may help elucidate their functions. Similarly, extensive antisense transcription was found near promoters in Drosophila oocytes (Brown et al., 2014b).
Several lncRNAs have been characterized in oogenesis in non-mammalian systems. Much like the previously mentioned SRA and SRY gene products, Drosophila oskar is a good example of an RNA with coding and noncoding functions. Loss of the oskar protein causes defects in oocyte polarity, embryonic germ line specification, and abdominal development; the loss of the oskar RNA, however, caused early arrest in oogenesis (Jenny et al., 2006). This lncRNA is only translated when localized to the posterior pole, where its 3’UTR is necessary to recruitment other factors involved in the establishment of cell polarity (Kugler and Lasko, 2009). The independent activity of oskar 3’UTR was further supported when it was identified years later in a genome-wide profile of 3’UTR-associated RNAs (Mercer et al., 2011). A similar RNA scaffolding function is found in Xenopus, in which two RNAs are required for proper cytoskeletal organization and oocyte polarity (Kloc et al., 2005). XIsirt is a lncRNA composed of short tandem repeats that are suspected to form stem-loop structures for its correct localization (Allen et al., 2003). VegT (vegetal T-box protein) mRNA, another dual-purpose transcript, is necessary for cytokeratin network assembly whereas the VegT protein is a transcription factor required for mesoderm and endoderm differentiation (Kofron et al., 1999; Xanthos et al., 2001). Mammals do not have the same asymmetric distribution of molecules associated with oocyte development as either Drosophila or Xenopus, so the structural functions described above are not likely applicable in these animals—but this does not preclude similar lncRNA function in mammalian oocytes.
Studies in plants are revealing additional lncRNA-based mechanisms essential for gamete formation. An RNA-seq screen for lncRNAs expressed in reproductive tissues of rice identified a number of transcripts. One,XLOC_057324, was expressed exclusively in young panicles and pistils. Strains with a transfer-DNA insertion in XLOC_057324 flowered prematurely and set fewer seeds (Zhang et al., 2014b). In another study, rice hybrids exhibiting long-day-specific male sterility were shown to carry a mutation in the lncRNA LDMAR (long-day-specific male-fertility-associated RNA). A point mutation in LDMAR increased DNA methylation of the locus, reduced LDMAR expression under long daylight conditions, and caused premature apoptosis of developing anthers (Ding et al., 2012). The mechanisms underlying these effects are not known.
Placentation
Initial data, though limited, are consistent with a role for lncRNAs in placenta formation and function, with some of the strongest results coming from studies of H19. H19 is a source for miR-675, a miRNA that directly down-regulates NOMO1 (Nodal modulator 1) and inhibit its ability to stimulate proliferation of a human trophoblast cell line (Gao et al., 2012; Keniry et al., 2012). In normal placentae, H19 and its miR-675 repress NOMO1-mediated proliferation, but in preeclamptic placentas, H19 and the miRNA are repressed, allowing NOMO1 mis-regulation to cause placental overgrowth.
The lncRNA SPRY4-IT1 (Sprouty homolog 4, intronic transcript 1), which is expressed in placenta, was overexpressed in preeclamptic placentae. siRNA knockdown of SPRY4-IT1 in a transformed human trophoblast line increased cell migration and reduced apoptosis, whereas overexpressing SPRY4-IT1 had the opposite effects (Zou et al., 2013). Additional lncRNAs were reported to exhibit differential expression in preeclamptic versus control placentae, although the functional relevance has not be tested (He et al., 2013). In vivo manipulations are necessary to assess directly the importance of these lncRNAs in placenta function.
A disease-associated locus was mapped to an intergenic region harboring a lncRNA that is expressed in several trophoblast subtypes in human placentae in studies of HELLP syndrome—a maternal condition of hemolysis, elevated liver enzymes, and low platelets (hence its abbreviation) that has its origins in placental insufficiency. When knocked down in extravillous trophoblast cells, gene expression changes were associated with increased G1/S and cell death functions, as well as decreased G2/M, cell survival, and migration. Accumulation of the HELLP lncRNA had the opposite effects, specifically decreasing cell invasion (van Dijk et al., 2012). The mechanisms by which the HELLP lncRNA exerts these effects are unknown.
Intrauterine growth restriction (IUGR) is associated with a fourfold enrichment in NEAT1 (nuclear paraspeckle assembly transcript 1) lncRNA compared to control placentae at term pregnancies (Gremlich et al.,2014). This lncRNA is present in nuclear paraspeckles, and is essential for their assembly (Clemson et al., 2009). Unfortunately, it is not clear if increased NEAT1 contributes to, or is a consequence of, IUGR.
Unique lncRNA-mediated control of some previously mentioned imprinted loci occurs in the placenta as well. For example, Airn controls imprinted expression of IGF2R globally, and controls the placenta-specific imprinted expression of two additional adjacent genes, SLC22A2 and SLC22A3 (Zwart et al., 2001; Nagano et al., 2008). The lncRNA Kcnq1ot1 similarly regulates imprinting of four nearby genes in all tissues, but controls four additional, more distantly located genes in placental tissue (Pandey et al., 2008). Both Airn and Kcnq1ot1 directly interact with chromatin-modifying machinery in a lineage-specific way, suggesting that other lncRNAs might work similarly. These imprinting mechanisms are probably more tightly regulated in the placenta due to the tissue’s direct role in embryonic growth.
Inheritance
While DNA is responsible for genetic inheritance, non-genetic transmission of traits through meiosis—a phenomenon referred to as trans-generational epigenetic inheritance (TEI)—has been observed (Rakyan and Whitelaw, 2003). Mechanisms underlying TEI are mediated by his-tone modifications, DNA methylation, prions, and RNA species. The first evidence for the involvement of RNA in TEI came from studies of paramutation, a form of TEI involving the b1 locus in maize. Two alleles of b1 exist, B-I and B’, which are genetically identical. B’, however, is silent and harbors DNA methylation in a repeat region necessary for paramutation, whereas B-I is active and lacks methylation in the region (Haring et al., 2010). When present in the same plant, B’ converts B-I to its own state; this conversion is stable through meiosis for several generations. The role of RNA in paramutation was demonstrated when a 6-kb tandem repeat 100 kb upstream of the locus that has enhancer activity was shown to be transcribed and processed into small RNAs—a process that requires the RNA-dependent RNA polymerase MOP1 (modifier of paramutation 1) (Dorweiler et al., 2000; Stam et al., 2002; Alleman et al., 2006; Arteaga-Vazquez et al., 2010).
Animal systems display similar RNA-mediated paramutation inheritance. Studies of murine Kit+ (Rassoulzadegan et al., 2006), Sox9 (SRY box 9) (Grandjean et al., 2009), Cdk9 (cyclin-dependent kinase 9) (Wagner et al., 2008), Rasgrf1 (Herman et al., 2003), and work in stressed mice (Gapp et al., 2014) revealed paramutation-like effects that are consistent with RNA-mediated mechanisms. Some of these early studies sought to prove the sufficiency of small RNAs to recapitulate a phenotype-of-interest by injecting miRNA species into wild-type zygotes. While making a strong argument for sufficiency, these studies do not answer all of the questions. For instance, elimination of the miRNA pathway by Drosha knockout or the piRNA pathway by Mov10I1 knockout increased the penetrance of the Kit phenotype, suggesting that miRNAs and piRNAs act as suppressors rather than activators of paramutation (Yuan et al., 2015). The mechanisms controlling TEI in mammals are unknown, but one would be in error to rule out either the importance of indirect lncRNA control via small RNA regulation or of direct lncRNA transmission upon fertilization. Indeed, RNA modifications appear to play a role, as the Kit and Sox9 phenotypes are dependent on the RNA methyltransferase DNMT2 (transfer RNA aspartic acid methyltransferase 1) (Kiani et al., 2013), although the universality of the effect must be studied further.
Development
lncRNAs associated with preimplantation development have been characterized by RNA-seq of zygotes and other preimplantation stages of development (Paranjpe et al., 2013; Yan et al., 2013; Caballero et al., 2014; Zhang et al., 2014a; Hamazaki et al., 2015). By comparing lncRNA profiles of the zygotes with those of its parental gametes, it is possible to identify lncRNAs arising immediately after zygote activation. Evaluating the importance of these lncRNAs for early embryonic events will require their experimental manipulation (Sauvageau et al., 2013).
Some of the most-extensive findings related to preim-plantation embryos come from studies of embryonic stem cells. Cultured embryonic stem cells express at least 226 lncRNAs, 137 of which have been shown to affect gene expression and 26 of which are necessary to repress differentiation and to maintain pluripotency (Guttman et al., 2011). These lncRNAs contribute to many activities. For instance, Meg3 (maternally expressed gene 3) interacts with JARID2 (Jumonji, AT-rich interactive domain 2) to specifically recruit PRC2, and its repressive activity, to embryonic development genes in trans (Kaneko et al., 2014). On the other hand, six lncRNAs were shown to interact with WDR5 (WD repeat domain 5), a protein that actively recruits KMT2A/MLL and its H3K4me3 activity (Wang et al., 2011; Yang et al., 2014). By a distinct approach, the lncRNA RoR (regulator of reprogramming) does not control chromatin remodelers, instead maintaining expression of the core pluripotency factors by acting as a sponge to titrate out repressive miRNAs that would down-regulate their translation (Wang et al., 2013). Through a variety of mechanisms, the central role for lncRNAs at this stage of development is to maintain self-renewal characteristics (reviewed in Flynn and Chang, 2014).
Many critical steps in post-implantation somatic development are regulated by lncRNAs (see Table 3). The reader is referred to recent reviews addressing this issue, including stem cell maintenance and differentiation (Batista and Chang, 2013; Ghosal et al., 2013; Fatica and Bozzoni, 2014; Flynn and Chang, 2014; Yao and Jin, 2014).
TABLE 3.
lncRNAs With Identified Functions in Development
| Tissue | lncRNA | Function | References |
|---|---|---|---|
| Cardiovascular | Fendrr | Regulates cardiac development | Grote and Herrmann (2013); Grote et al. (2013) |
| Braveheart | Regulates cardiovascular development | Klattenhoff et al. (2013) | |
| tie 1AS | Regulates TIE1 (tyrosine kinase with immunoglobulin-like and EGF-like domains 1) and vascular development | Li et al. (2010) | |
| Hematopoietic | H19 | Maintains hematopoietic stem cell quiescence | Venkatraman et al. (2013) |
| lncRNA-αGT | Necessary for embryonic to adult alpha-globin switching | Arriaga-Canon et al. (2014) | |
| 7 species | Controls terminal erythroid differentiation | Paralkar et al. (2014) | |
| Musculoskeletal | SRA | Enhances myogenic differentiation and myogenic conversion of non-muscle cells | Hube et al. (2011) |
| lincMD1 | Enhances myoblast differentiation | Cesana et al. (2011) | |
| Dum | Regulates myoblast differentiation | Wang et al. (2015) | |
| MUNC | Induces myoblast differentiation | Mueller et al. (2015) | |
| Neural | Six3OS | Regulates SIX3 (SIX homeobox 3) and retinal development | Rapicavoli et al. (2011) |
| TUNAR | Regulates pluripotency and neural differentiation | Lin et al. (2014) | |
| Evf2 | Represses DLX5 (distal-less homeobox 5) and controlling GABA circuitry | Berghoffet al. (2013) | |
| Pnky | Regulates neurogenesis from neural stem cells | Ramos et al. (2015) | |
| Mammary | Pinc1 | Regulates alveolar development | Shore et al. (2012) |
| Zfas1 | Regulates alveolar development | Askarian-Amiri et al. (2011) | |
| Endoderm | DEANR1/LINC00261 | Regulates endoderm differentiation | Jiang et al. (2015) |
| Adipose | HOTAIR | Regulates preadipocyte differentiation | Divoux et al. (2014) |
| 10 species | Regulates preadipocyte differentiation | Sun et al. (2013) |
REPRODUCTIVE DISEASE
Beyond placental insufficiencies that are associated with perturbations in lncRNA regulatory mechanisms, several lines of evidence document additional roles for lncRNAs in various reproductive pathologies. A study of nineteen men with idiopathic infertility and histologically confirmed meiotic arrest revealed copy-number variants of three genes, including the lncRNA LOC100507205, that are unique to the meiotic-arrest patients as compared to 95 fertile controls (Eggers et al., 2015). Similarly, a screen in women for lncRNAs associated with premature rupture of the placental membranes (PPROM) identified thirteen lncRNAs that were differentially expressed in PPROM versus full-term placentae. These lncRNAs appear to play roles in the inflammatory response, smooth muscle contraction, and ligand-receptor interactions (Luo et al., 2013, 2015). In a third study, women suffering from polycystic ovary syndrome—characterized by high serum androgens, absence or irregular menstruation, and infertility—the lncRNAs SRAP/SRA1 and CTBP1-AS1 (carboxy-terminal binding protein 1, antisense transcript 1) were overexpressed compared to healthy controls (Liu et al., 2014,2015b).
Many studies have focused on the role of lncRNAs in various reproductive cancers. For example, SRA, already discussed as a regulator of nuclear hormone responses, is elevated in estrogen-responsive ovarian and breast cancer (Leygue et al., 1999; Murphy et al., 2000; Hussein-Fikret and Fuller, 2005). PCGEM1 and PCNR1 lncRNAs were first identified in aggressive prostate adenocarcinomas due to their overexpression (Yang et al., 2013), while NEAT1 was implicated in the progression of androgen-insensitive prostate tumors (Chakravarty et al., 2014).
Extensive phenotypes are also linked to H19 expression. Two related genital malformation syndromes are associated with epigenetic alterations at H19, which is methylated on the paternal allele and thereby silenced. In addition to silencing the paternal copy of H19, methylation is required for expression of the paternal copy of IGF2, to which H19 is linked. Silver-Russell syndrome is clinically and genetically heterogeneous, with some patients exhibiting hypomethylation of H19. The most-severely hypomethylated females show congenital aplasia of the uterus and upper vagina, and severely hypomethylated males exhibit cryptorchidism and testicular agenesis (Bliek et al., 2006; Bruce et al., 2009). H19 hypomethylation is also associated with some Müllerian aplasia patients, whose congenital abnormalities of the female genital tract produce vaginal and uterine malformations that limit reproduction to methods involving surrogacy (Sandbacka et al., 2011). Because paternal silencing of H19 and paternal expression of IGF2R are coupled, it is not clear whether aberrant expression of either or both loci is responsible for the reproductive phenotypes of these patients.
Evidence beyond associations are required to demonstrate the importance of candidate lncRNAs, identified from human clinical studies, in reproductive processes, and further investigation is necessary to reveal their mechanisms of action. Animal studies will be important in this regard, such as those demonstrating that mice deficient for the lncRNA NEAT1 have impaired corpus luteum formation and failure to maintain pregnancy (Nakagawa et al., 2014).
CONCLUSION
While lncRNAs are recognized as important mediators of cellular fate and function, their roles in the reproductive processes are only now being elucidated. Descriptive, hypothesis-generating studies that characterize lncRNAs associated with reproduction represent the low-hanging fruit in the field. These studies are heavily concentrated in specific reproductive events, but remain sparse in others, and have been applied to a limited number of organisms. The more challenging studies entail identifying which of the discovered lncRNAs influence reproductive processes and how they do so. Detailed mechanistic studies will require manipulating lncRNA expression and evaluating reproductive phenotypes; characterizing lncRNA structures, ideally in vivo; identifying proteins and other factors interacting with lncRNAs; cataloging the chemical modifications present on lncRNAs and their interacting partners; and assessing the importance of those modifications for structure and function. Expanding such analyses across many species and a diversity of individuals within human populations will help reveal the evolutionary conservation of those lncRNA-mediated mechanisms that affect reproduction and the genetic variants that are important for reproductive health. Given the vast array of lncRNAs transcribed from complex genomes and their range of activities, such studies will rival the complexity and importance of functional genomic analyses of the coding genome.
Acknowledgments
Grant sponsor: United States National Institutes of Health; Grant number: R01GM105243
Abbreviations
- Airn
antisense of IGF2R non protein-coding RNA
- BOLL/BOULE
Boule-like RNA-binding protein
- DAZ[L]
deleted In azoospermia [-like]
- EHMT2/G9A
euchromatic histone-lysine N-methyltransferase 2
- EZH2
enhancer of Zeste 2 polycomb repressive comlpex 2 subunit
- H3H#me1/2/3
histone H3 lysine & methylation mono-/ di-/ or tri-methylation
- HOX
homeobox
- HOTAIR
HOX transcript antisense RNA
- IGF2R
insulin-like growth factor 2 receptor
- IME
inducer of meiosis
- Kcnq1ot1
voltage-gated KQT-like potassium channel, subfamily Q member 1 KCNQ1 opposite strand/antisense transcript 1
- KDM1A/LSD1
lysine-specific demethylase 1A
- KMT2A/MLL
lysine-specific methyltransferase 2A/mixed-lineage leukemia
- lncRNA
long noncoding RNA
- m6A
N6-methyladenine
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- miRNA
micro RNA
- MOV1OL
Mov10 RNA-induced silencing complex RNA helicase-like 1
- NEAT1
nuclear paraspeckle assembly transcript 1
- PCGEM1
prostate-specific transcript
- PGC
primordial germ cell
- piRNA
piwi-interacting RNA
- Poly(A)
polyadenylation
- PRC2
polycomb repressive complex 2
- Rasgrfl
Ras protein-specific guanine nucleotide-releasing factor 1
- RNA-seq
RNA sequencing
- siRNA
small-interfering RNA
- SLC22A
solute-carrierfamily 22 member
- SRA [SRAP/SRA1]
steroid receptor RNA activator [protein]
- SRY
sex-determining region Y
- UTR
untranslated region
- Xist
X-chromosome inactive-specific transcript
REFERENCES
- Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova O, Schade M, Bottger A, David CN. Oogenesis in Hydra: Nurse cells transfer cytoplasm directly to the growing oocyte. Dev Biol. 2005;281:91–101. doi: 10.1016/j.ydbio.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- Allen L, Kloc M, Etkin LD. Identification and characterization of the Xlsirt cis-acting RNA localization element. Differentiation. 2003;71:311–321. doi: 10.1046/j.1432-0436.2003.7106003.x. [DOI] [PubMed] [Google Scholar]
- Amin V, Harris RA, Onuchic V, Jackson AR, Charnecki T, Pai-thankar S, Lakshmi Subramanian S, Riehle K, Coarfa C, Milo-savljevic A. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nature Commun. 2015;6:6370. doi: 10.1038/ncomms7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, 3rd, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7:e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Vazquez M, Sidorenko L, Rabanal FA, Shrivistava R, Nobuta K, Green PJ, Meyers BC, Chandler VL. RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc Natl Acad Sci USA. 2010;107:12986–12991. doi: 10.1073/pnas.1007972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G, Akhade VS, Donakonda S, Rao MR. Mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell Biol. 2012;32:3140–3152. doi: 10.1128/MCB.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autuoro JM, Pirnie SP, Carmichael GG. Long noncoding RNAs in imprinting and X chromosome inactivation. Biomole-cules. 2014;4:76–100. doi: 10.3390/biom4010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Kundaje A, Gunawardena HP, Yu YB, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Wu J, Schuster AS, Hennig GW, Yan W. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol Reprod. 2013;89:107. doi: 10.1095/biolreprod.113.113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, Carter AC, Flynn RA, Zhou C, Lim KS, Dedon P, Wernig M, Mullen AC, Xing Y, Giallourakis CC, Chang HY. M(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bliek J, Terhal P, van den Bogaard MJ, Maas S, Hamel B, Salieb-Beugelaar G, Simon M, Letteboer T, van der Smagt J, Kroes H, Mannens M. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet. 2006;78:604–614. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard HF, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SF, Reijo Pera RA. X chromosome inactivation: Recent advances and a look forward. Curr Opin Genet Dev. 2014;28:78–82. doi: 10.1016/j.gde.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brook M, Smith JW, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014a;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, Wan KH, Yu C, Zhang D, Carlson JW, Cherbas L, Eads BD, Miller D, Mockaitis K, Roberts J, Davis CA, Frise E, Hammonds AS, Olson S, Shenker S, Sturgill D, Samsonova AA, Weiszmann R, Robinson G, Hernandez J, Andrews J, Bickel PJ, Carninci P, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Lai EC, Oliver B, Perrimon N, Graveley BR, Celniker SE. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014b;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce S, Hannula-Jouppi K, Peltonen J, Kere J, Lipsanen-Nyman M. Clinically distinct epigenetic subgroups in Silver-Russell syndrome: The degree of H19 hypomethylation associates with phenotype severity and genital and skeletal anomalies. J Clin Endocrinol Metab. 2009;94:579–587. doi: 10.1210/jc.2008-1805. [DOI] [PubMed] [Google Scholar]
- Caballero J, Gilbert I, Fournier E, Gagne D, Scantland S, Macaulay A, Robert C. Exploring the function of long non-coding RNA in the development of bovine early embryos. Reprod Fertil Dev. 2014;27:40–52. doi: 10.1071/RD14338. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-lmpiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojo-bori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, lacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kum-merfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, lida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawa-shima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding anti-sense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106:35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Deb A, Maji RK, Saha S, Ghosh Z. LncRBase: An enriched resource for lncRNA information. PLoS ONE. 2014;9:e108010. doi: 10.1371/journal.pone.0108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, Vergara IA, Ghadessi M, Davicioni E, Jenkins RB, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander NH, Beltran H, Fox AH, Elemento O, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nature Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F, Lardenois A, Evrard B, Rolland AD, Sallou O, Dumargne MC, Coiffec I, Collin O, Primig M, Jegou B. High-resolution profiling of novel transcribed regions during rat spermatogenesis. Biol Reprod. 2014;91:1–13. doi: 10.1095/biolreprod.114.118166. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5’ leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chocu S, Evrard B, Lavigne R, Rolland AD, Aubry F, Jegou B, Chalmel F, Pineau C. Forty-four novel protein-coding loci discovered using a proteomics informed by transcriptomics (PIT) approach in rat male germ cells. Biol Reprod. 2014;91:1–14. doi: 10.1095/biolreprod.114.122416. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. Embo J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossetti C, Lugini L, Astrologo L, Saggio I, Fais S, Spadafora C. Soma-to-germline transmission of RNA in mice xeno-grafted with human tumour cells: Possible transport by exo-somes. PLoS ONE. 2014;9:e101629. doi: 10.1371/journal.pone.0101629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D/’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci USA. 2012;109:2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL. Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S, DeBoer KD, van den Bergen J, Gordon L, White SJ, Jamsai D, McLachlan Rl, Sinclair AH, O’Bryan MK. Copy number variation associated with meiotic arrest in idiopathic male infertility. Fertility Steril. 2015;103:214–219. doi: 10.1016/j.fertnstert.2014.09.030. [DOI] [PubMed] [Google Scholar]
- Engreitz J, Lander ES, Guttman M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol Biol. 2015;1262:183–197. doi: 10.1007/978-1-4939-2253-6_11. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of non-coding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvD MR1. Nat Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga Ml, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)ARNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Galupa R, Heard E. X-chromosome inactivation: New insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Gao WL, Liu M, Yang Y, Yang H, Liao Q, Bai Y, Li YX, Li D, Peng C, Wang YL. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1) RNA Biol. 2012;9:1002–1010. doi: 10.4161/rna.20807. [DOI] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand B, Mead J, Bruning A, Apostolopoulos N, Tadigotla V, Nagaraj V, Sengupta AM, Vershon AK. Regulated anti-sense transcription controls expression of cell-type-specific genes in yeast. Mol Cell Biol. 2011;31:1701–1709. doi: 10.1128/MCB.01071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: New players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction. 2009;138:151–162. doi: 10.1530/REP-08-0510. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22:370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- Gremlich S, Damnon F, Reymondin D, Braissant O, Schittny JC, Baud D, Gerber S, Roth-Kleiner M. The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development. Placenta. 2014;35:44–49. doi: 10.1016/j.placenta.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I, Fejer G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J Immunol. 2003;170:3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hamazaki N, Uesaka M, Nakashima K, Agata K, Imamura T. Gene activation-associated long noncoding RNAs function in mouse preimplantation development. Development. 2015;142:910–920. doi: 10.1242/dev.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HS, Quertermous T, Chang CP. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen Tl, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Haring M, Bader R, Louwers M, Schwabe A, van Driel R, Stam M. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04245.x. [DOI] [PubMed] [Google Scholar]
- Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, Redfern AD, Miles LE, Barker A, MacDonald LM, Arthur PG, Lui JC, Golding JL, McCulloch RK, Metcalf CB, Wilce JA, Wilce MC, Lanz RB, O’Malley BW, Leedman PJ. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- He X, He Y, Xi B, Zheng J, Zeng X, Cai Q, Ouyang Y, Wang C, Zhou X, Huang H, Deng W, Xin S, Huang Q, Liu H. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS ONE. 2013;8:e81437. doi: 10.1371/journal.pone.0081437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman H, Lu M, Anggraini M, Sikora A, Chang Y, Yoon BJ, Soloway PD. Trans allele methylation and paramutation-like effects in mice. Nat Genet. 2003;34:199–202. doi: 10.1038/ng1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Lempradl A, Trupke J, Okulski H, Altmutter C, Ruge F, Boidol B, Kubicek S, Schmauss G, Aumayr K, Ruf M, Pospisilik A, Dimond A, Senergin HB, Vargas ML, Simon JA, Ringrose L. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet. 2014;46:973–981. doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gabel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through transregulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci USA. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, Balk SP, Liu XS, Brown M, Kantoff PW. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein-Fikret S, Fuller PJ. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol Cell Endocrinol. 2005;229:149–160. doi: 10.1016/j.mce.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Hachet O, Zavorszky P, Cyrklaff A, Weston MD, Johnston DS, Erdelyi M, Ephrussi A. Atranslation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Mackie P, Jodar M, Moskovtsev S, Krawetz SA. Chromatin and extracellular vesicle associated sperm RNAs. Nucleic Acids Res. 2015;43:6847–6859. doi: 10.1093/nar/gkv591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory Rl, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, Tammana H, Gingeras TR. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashimada K, Koopman P. Sry: The master switch in mammalian sex determination. Development. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: Expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. 2003;369:163–171. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Buhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013;20:994–1000. doi: 10.1038/nsmb.2619. [DOI] [PubMed] [Google Scholar]
- Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales Thomas, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. RNA-Mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Kofron M, Demel T, Xanthos J, Lohr J, Sun B, Sive H, Osada S, Wright C, Wylie C, Heasman J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development. 1999;126:5759–5770. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U. Apomixis: A developmental perspective. Ann Rev Plant Biol. 2003;54:547–574. doi: 10.1146/annurev.arplant.54.110901.160842. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Kotov AA, Akulenko NV, Kibanov MV, Olenina LV. DEAD-Box RNA helicases in animal gametogenesis. Mol Biol. 2014;48:16–28. [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiho A, Kotaja N, Gyenesei A, Sironen A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS ONE. 2013;8:e61558. doi: 10.1371/journal.pone.0061558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lanz RB, Razani B, Goldberg AD, O’Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc Natl Acad Sci USA. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenois A, Liu Y, Walther T, Chalmel F, Evrard B, Granovskaia M, Chu A, Davis RW, Steinmetz LM, Primig M. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc Natl Acad Sci USA. 2011;108:1058–1063. doi: 10.1073/pnas.1016459108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, Porse B, Kauppinen S, Lund AH. MicroRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Akbari Moqadam F, Maia AR, Wijchers PJ, Geeven G, den Boer ML, Kalluri R, de Laat W, Esteller M, Agami R. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59:4190–4193. [PubMed] [Google Scholar]
- Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim H-S, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Li W, Tian H, Hu T, Wang L, Lin Y, Li Y, Huang H, Sun F. Sequential expression of long noncoding RNA as mRNA gene expression in specific stages of mouse spermatogenesis. Sci Rep. 2014;4:5966. doi: 10.1038/srep05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS ONE. 2012;7:e29950. doi: 10.1371/journal.pone.0029950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hao C, Song D, Zhang N, Bao H, Qu Q. Androgen receptor coregulator CTBP1-AS is associated with polycystic ovary syndrome in chinese women: A preliminary study. Reprod Sci. 2014;22:829–837. doi: 10.1177/1933719114565037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015a;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hao C, Huang X, Zhang N, Bao H, Qu Q. Peripheral blood leukocyte expression level of lncRNA steroid receptor RNA activator (SRA) and its association with polycystic ovary syndrome: A case control study. Gynecol Endocrinol. 2015b;31:363–368. doi: 10.3109/09513590.2014.999763. [DOI] [PubMed] [Google Scholar]
- Luo X, Pan J, Wang L, Wang P, Zhang M, Liu M, Dong Z, Meng Q, Tao X, Zhao X, Zhong J, Ju W, Gu Y, Jenkins EC, Brown WT, Shi Q, Zhong N. Epigenetic regulation of lncRNA connects ubiquitin-proteasome system with infection-inflammation in preterm births and preterm premature rupture of membranes. BMC Pregnancy Childbirth. 2015;15:35. doi: 10.1186/s12884-015-0460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Shi Q, Gu Y, Pan J, Hua M, Liu M, Dong Z, Zhang M, Wang L, Gu Y, Zhong J, Zhao X, Jenkins EC, Brown WT, Zhong N. LncRNA pathway involved in premature preterm rupture of membrane (PPROM): An epigenomic approach to study the pathogenesis of reproductive disorders. PLoS ONE. 2013;8:e79897. doi: 10.1371/journal.pone.0079897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, teVruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- Ma L, Li A, Zou D, Xu X, Xia L, Yu J, Bajic VB, Zhang Z. LncRNAWiki: Harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015;43:D187–D192. doi: 10.1093/nar/gku1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay AD, Gilbert I, Caballero J, Barreto R, Fournier E, Tossou P, Sirard MA, Clarke HJ, Khandjian EW, Richard FJ, Hyttel P, Robert C. The gametic synapse: RNA transfer to the bovine oocyte. Biol Reprod. 2014;91:90. doi: 10.1095/biolreprod.114.119867. [DOI] [PubMed] [Google Scholar]
- Magnusdottir E, Dietmann S, Murakami K, Gunesdogan U, Tang F, Bao S, Diamanti E, Lao K, Gottgens B, Azim Surani M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Margolin G, Khil PP, Kim J, Bellani MA, Camerini-Otero RD. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics. 2014;15:39. doi: 10.1186/1471-2164-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Matson CK, Zarkower D. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- Mau M, Corral JM, Vogel H, Melzer M, Fuchs J, Kuhlmann M, de Storme N, Geelen D, Sharbel TF. The conserved chimeric transcript UPGRADE2 is associated with unreduced pollen formation and is exclusively found in apomictic Boechera species. Plant Physiol. 2013;163:1640–1659. doi: 10.1104/pp.113.222448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Swere-doski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. TheXist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano JV, Ramathal C, Nguyen HN, Simon C, Reijo Pera RA. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells. 2012;30:441–451. doi: 10.1002/stem.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, Saint R, Koopman P, Mattick JS. Expression of distinct RNAs from 3’ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci USA. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. ERNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey BB, Olcese U, Cabrera JR, Horabin JI. An interactive network of long non-coding RNAs facilitates the Drosophila sex determination decision. Biochim Biophys Acta. 2014;1839:773–784. doi: 10.1016/j.bbagrm.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. 2000;60:6266–6271. [PubMed] [Google Scholar]
- Nagamori I, Cruickshank VA, Sassone-Corsi P. Regulation of an RNA granule during spermatogenesis: Acetylation of MVH in the chromatoid body of germ cells. J Cell Sci. 2011;124:4346–4355. doi: 10.1242/jcs.096461. [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, Hirose T. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Chenouard N, Olivo-Marin JC, Guichet A. A dual role for actin and microtubule cytoskeleton in the transport of Golgi units from the nurse cells to the oocyte across ring canals. Mol Biol Cell. 2009;20:556–568. doi: 10.1091/mbc.E08-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolasco S, Bellido J, Goncalves J, Tavares A, Zabala JC, Soares H. The expression of tubulin cofactor A (TBCA) is regulated by a noncoding antisense Tbca RNA during testis maturation. PLoS ONE. 2012;7:e42536. doi: 10.1371/journal.pone.0042536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shie-khattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJ. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics. 2013;14:762. doi: 10.1186/1471-2164-14-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Herman H, Gao Y, Lindroth AM, Hu BY, Murphy PJ, Putnam JR, Soloway PD. Sequences sufficient for programming imprinted germline DNA methylation defined. PLoS ONE. 2012;7:e33024. doi: 10.1371/journal.pone.0033024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. RNAexosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME. LncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife. 2014;3:e03254. doi: 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz S, Guttman M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V, Whitelaw E. Transgenerational epigenetic inheritance. Curr Biol. 2003;13:R6. doi: 10.1016/s0960-9822(02)01377-5. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Reijo RA, Dorfman DM, Slee R, Renshaw AA, Loughlin KR, Cooke H, Page DC. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, Harrison PM, Nelson FK, Miller P, Gerstein M, Weissman S, Snyder M. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RNAcentral-Consortium RNAcentral: An international database of ncRNA sequences. Nucleic Acids Res. 2015;43:D123–D129. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. eLife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbacka M, Bruce S, Halttunen M, Puhakka M, Lahermo P, Hannula-Jouppi K, Lipsanen-Nyman M, Kere J, Aittomaki K, Laivuori H. Methylation of H19 and its imprinted control region (H19 ICR1) in Mullerian aplasia. Fertil Steril. 2011;95:2703–2706. doi: 10.1016/j.fertnstert.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, Pauler FM. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development. 2013;140:1184–1195. doi: 10.1242/dev.088849. [DOI] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhar-dinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt C, Nothiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 2000;127:667–677. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, Carr SA, Lander ES, Fink GR, Regev A. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, Regev A. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudour-idylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichino Y, Yamashita A, Yamamoto M. Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Biol. 2014;4:140022. doi: 10.1098/rsob.140022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamashita A, Yamamoto M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell. 2003;14:2461–2469. doi: 10.1091/mbc.E02-11-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smith MA, Gesell T, Stadler PF, Mattick JS. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 2013;41:8220–8236. doi: 10.1093/nar/gkt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smorag L, Xu X, Engel W, Pantakani DV. The roles of DAZL in RNA biology and development. Wiley Interdiscip Rev RNA. 2014;5:527–535. doi: 10.1002/wrna.1228. [DOI] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, Dym M, de Massy B, Mikkelsen TS, Kaessmann H. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Kraus WL. Minireview: Long noncoding RNAs: New “links” between gene expression and cellular outcomes in endocrinology. Mol Endocrinol. 2013;27:1390–1402. doi: 10.1210/me.2013-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy A, Sharpless NE. Genetic “lnc”-age of noncoding RNAs to human disease. J Clin Invest. 2012;122:3837–3840. doi: 10.1172/JCI66645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. LincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk M, Thulluru HK, Mulders J, Michel OJ, Poutsma A, Windhorst S, Kleiverda G, Sie D, Lachmeijer AM, Oudejans CB. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J Clin Invest. 2012;122:4003–4011. doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, Reyes D, Beato M. Unliganded progesterone receptor-mediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev. 2013;27:1179–1197. doi: 10.1101/gad.215293.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCi-pedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015a;43:4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCi-pedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015b;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, Wang PJ. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29:617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell. 2014;56:5–12. doi: 10.1016/j.molcel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, Masuhiro Y, Kato S. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. Embo J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway PD, Sasaki H. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Wylie C, Heasman J. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development. 2001;128:167–180. doi: 10.1242/dev.128.2.167. [DOI] [PubMed] [Google Scholar]
- Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, Hung MC, Lin C, Yang L. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Xu XF, Li J, Cao YX, Chen DW, Zhang ZG, He XJ, Ji DM, Chen BL. Differential expression of long noncoding RNAsin human cumulus cells related to embryo developmental potential: A microarray analysis. Reprod Sci. 2014;22:672–678. doi: 10.1177/1933719114561562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Watanabe Y, Nukina N, Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, Tang F. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, Noble WS, Shendure J, Disteche CM. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG. LncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Jin P. Unlocking epigenetic codes in neurogenesis. Genes Dev. 2014;28:1253–1271. doi: 10.1101/gad.241547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JR, Zhang X, Nagano MC. Wnt5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. J Cell Sci. 2011;124:2357–2366. doi: 10.1242/jcs.080903. [DOI] [PubMed] [Google Scholar]
- Yerushalmi GM, Salmon-Divon M, Yung Y, Maman E, Kedem A, Ophir L, Elemento O, Coticchio G, Dal Canto M, Mignini Renzinu M, Fadini R, Hourvitz A. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol Hum Reprod. 2014;20:719–735. doi: 10.1093/molehr/gau031. [DOI] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Yokoo M, Sato E. Cumulus-oocyte complex interactions during oocyte maturation. Int Rev Cytol. 2004;235:251–291. doi: 10.1016/S0074-7696(04)35006-0. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Oliver D, Schuster A, Zheng H, Yan W. Breeding scheme and maternal small RNAs affect the efficiency of trans-generational inheritance of a paramutation in mice. Sci Rep. 2015;5:9266. doi: 10.1038/srep09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu H, Xin D, Cheng H, Zhou R. A novel ncRNA gene from mouse chromosome 5 trans-splices with Dmrt1 on chromosome 19. Biochem Biophys Res Commun. 2010;400:696–700. doi: 10.1016/j.bbrc.2010.08.130. [DOI] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Zhang K, Huang K, Luo Y, Li S. Identification and functional analysis of long non-coding RNAs in mouse cleavage stage embryonic development based on single cell transcriptome data. BMC Genomics. 2014a;15:845. doi: 10.1186/1471-2164-15-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, Li QF, Qu LH, Shu WS, Chen YQ. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014b;15:512. doi: 10.1186/s13059-014-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013a;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fu Y, Klungland A, Yang YG, He C. Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biol. 2013b;10:915–918. doi: 10.4161/rna.24711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Wang PJ. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012;8:e1003038. doi: 10.1371/journal.pgen.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Jiang Z, Yu X, Sun M, Zhang Y, Zuo Q, Zhou J, Yang N, Han P, Ge Z, De W, Sun L. Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo. PLoS ONE. 2013;8:e79598. doi: 10.1371/journal.pone.0079598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001;15:2361–2366. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]