Abstract
Differentiated cells retain the genetic information of the donor but the extent to which phenotypic differences between donors or batches of differentiated cells are explained by variation introduced during the differentiation process is not fully understood. In this study, we evaluated four separate batches of commercially available neurons originating from the same iPSCs to investigate whether the differentiation process used in manufacturing iPSCs to neurons affected genome-wide gene expression, modified cytosines, or neuronal sensitivity to drugs. No significant changes in gene expression, as measured by RNA-Seq, or cytosine modification levels, as measured by the Illumina 450K arrays, were observed between batches relative to changes over time. As expected, neurotoxic chemotherapeutics affected neuronal outgrowth, but no inter-batch differences were observed in sensitivity to paclitaxel, vincristine and cisplatin. As a testament to the utility of the model for studies of neuropathy, we observed that genes involved in neuropathy had relatively higher expression levels in these samples across different time points. Our results suggest that the process used to differentiate iPSCs into neurons is consistent, resulting in minimal intra-individual variability across batches. Therefore, this model is reasonable for studies of human neuropathy, druggable targets to prevent neuropathy, and other neurological diseases.
Keywords: Induced Pluripotent Stem Cells, Neuropathy, neuron, chemotherapy
1. Introduction
Neurodegenerative diseases and neuropathy are difficult to study due to lack of relevant human models [1]. Cell culture systems and primary rodent cultures have proven to be indispensable to clarify disease mechanisms and provide insights into gene functions. However, the current models have not provided much in terms of therapy for inherited neuropathies (known collectively as Charcot-Marie-Tooth disease) [2], and the only effective treatments for diabetic neuropathy are glucose control and pain management [3]. Chemotherapy-induced peripheral neuropathy (CIPN) is a common neurotoxicity affecting 20–40% of patients receiving chemotherapy [4]. To truly understand and find relevant druggable targets that are causative, a cellular model that represents neuropathy is essential.
With recent advances in stem cell technology, the ability to differentiate human induced pluripotent stem cells (iPSCs) to neurons provides us with a new and potentially relevant human neuronal model. In addition, iPSC-differentiated neurons can be created from diseased individuals or individuals with severe sensitivity to neurotoxic chemotherapy to provide a model that will allow for the identification of in vitro phenotypic characteristics relevant to the disease or sensitivity to neurotoxic drug. These neurons may yield targets essential to overcoming and preventing heritable neuropathy or CIPN. Stem cell technology has revolutionized the field of “in vitro disease modeling” [5], as evidenced by the first set of drugs emerging into clinical trials from the use of iPSC derived neurons from patients with neurological diseases [6].
Human fibroblasts were reprogrammed from an individual into nociceptor neurons without creation of iPSCs and the neurons exhibited sensitization to the chemotherapeutic drug oxaliplatin, modeling the inherent mechanisms underlying painful CIPN [7]; however, the advantage to creating iPSCs as an intermediate is that they can grow indefinitely, thus providing a ready source to create additional neurons of the same genetic background. Recently, our laboratory developed a potential model to evaluate CIPN by employing commercially available human neurons differentiated from iPSCs [8]. We found reproducible differences in morphological characteristics including neurite outgrowth phenotypes, cellular viability and apoptosis following treatment with four distinct chemotherapeutic drugs: vincristine, paclitaxel, cisplatin and hydroxyurea. This model was also used to demonstrate functional consequences of gene knockdown on neuronal sensitivity to chemotherapeutics of genes identified through clinical genome-wide association studies (GWAS) of CIPN [8–11].
The potential of using the human iPSC-derived neuron model for larger genetic association studies requires an understanding of heterogeneity of cultures and to partition the variance associated with iPSC reprograming, culturing, and differentiation [12–14]. A major concern in the field of stem cell technology is that techniques to reprogram cells could introduce variation that masks important genetic differences between individuals. A recent study demonstrated that the genetic background of iPSCs generated from peripheral blood mononuclear cells or fibroblasts accounted for more of the variation in gene expression between iPSC lines than any other tested factors such as cell type of origin or reprogramming method [13]. These studies suggest that future studies should focus on collecting a large number of donors rather than generating large numbers of lines from the same donor. Since industrial grade cells can now be made, the evaluation of epigenetics, gene expression and phenotypic variation from batch to batch is an important consideration.
In this study, we obtained multiple batches of iCell® Neurons (iPSC-derived human cortical neurons) differentiated from a single iPSC originating from fibroblasts of an individual to evaluate inter-batch differences in gene expression, cytosine modification levels, and pharmacologic response to chemotherapeutics. To determine the utility of these cells for studies of neuropathy and other neurological diseases, we evaluated genes involved in hereditary neuropathy at different time points in culture as neurites were formed. We showed a consistent enrichment of genes with relatively higher expression levels among hereditary neuropathy associated genes over time.
2. Methods
2.1 iCell Neurons
Neurons (iCell Neurons®) were purchased from Cellular Dynamics International (CDI, Madison, WI, USA). iCell Neurons are a ~98% pure (Tuj1+/Nestin−) pan-neuronal population of GABAergic and to a lesser degree glutamatergic neurons produced from human induced pluripotent stem cells. All batches of iCell Neurons are tested for sterility, viability, purity and morphology, and released according to strict manufacturer’s standards. Four batches of iCell Neurons (Batch numbers: 1366431, 1366825, 1369525, 1362632) were thawed and maintained according to the manufacturer’s protocol. Each batch of iCell Neurons were mixed with 3.3µg/ml laminin (Sigma-Aldrich) prior to seeding on poly-D-lysine coated 96-well Greiner Bio-One plates at a density of 1.33 × 104 cells/well. Approximately 1.1 × 106 neuron cells were pelleted immediately for the 0 hour sample by spinning at 300 g for 10 minutes and either lysed using Quizol (Qiagen) for RNA extraction or media removed from pellet before storing both samples at −20°C. For the cell collections to evaluate modified cytosine and gene expression, cells were pelleted at 0, 4, 28, 76 hours and then extracted DNA and RNA.
2.2. Drug preparation and treatment
Paclitaxel (Sigma-Aldrich) was prepared in the semi-dark by dissolving powder in 100% DMSO and filtered to obtain a stock solution of 58.4 mM. Control wells were treated with 0.17% final concentration of DMSO to match drug treatments. Cisplatin (Sigma-Aldrich) was prepared in the semi-dark by dissolving powder in 100% DMSO and filtered to obtain a stock solution of 20 mM. Control wells were treated with 0.2% final concentration of DMSO to match drug treatment. Vincristine (Sigma-Aldrich) was prepared on ice in the dark by dissolving powder in cold PBS and filtered to obtain a stock solution of 100 mM. Vincristine stocks were each diluted independently then added into the media and onto the cells before proceeding to the next dilution. All stock drugs were serially diluted in media for a final dosing concentrations ranging from 0.01 µM to 100 µM, increasing by factors of ten. Cells were plated and 4 hours later treated with increasing concentrations of drug for 72 h.
2.3. High content imaging and neurite outgrowth analysis
After drug treatments of 72 h, neurons were stained for 15 min at 37°C with 1 µg/mL Hoechst 33342 (Sigma-Aldrich) and 2 µg/mL Calcein AM (Molecular Probes, Life Technologies Inc., Carlsbad, CA, USA) then washed twice using dPBS without calcium or magnesium (LifeTechnologies). Imaging was performed at 10× magnification using an ImageXpress Micro (Molecular Devices, LLC, Sunnyvale, CA, USA) at the University of Chicago Cellular Screening Core. Individual cell measurements of total neurite outgrowth (sum of the length of all processes), number of processes and number of branches were calculated using the MetaXpress software Neurite Outgrowth Application Module (Molecular Devices, LLC). At least 500 cells per dose were quantified in triplicate for three independent experiments.
2.4. Nucleic acid isolation
At each time point, DNA was extracted by adding 50 µl per well of tissue digestion buffer (100 mM NaCl, 50 mM Tris 8.0, 100 mM EDTA, 1% SDS) along with 2 mg/ml Proteinase K (Denville Scientific; South Plainfield, NJ, USA). The plate was sealed with parafilm and agitated overnight at 55°C using 200 RPM in an Innova orbital shaker (Eppendorf; Enfield, CT). The digested cells from each well were combined and DNA extracted with equal volume of phenol: chloroform (Invitrogen), vortexing for 10 minutes (Lab Vortex-setting of 4) and the samples were centrifuged for 5 minutes at 14,000 g. The aqueous phase was collected, extracted with equal volume of chloroform and repeated two times. The DNA was precipitated with equal volume 100% ethanol and centrifuged for 10 minutes at 14,000 g. Two washes with 70% ethanol produced a pellet that was air dried for 10 minutes at room temperature and finally dissolved in 50 µL of 10 mM Tris (pH 8.0) for 3 days at 4°C then stored at −80°C.
RNA isolation was performed after removal of media and addition of 50µL Qiazol per well plate for 5 minutes at room temperature to lyse cells. After vigorously pipetting each well several times, each batch was collected and stored for further processing at −80°C. When all the time points had been collected, the RNA was purified using the Ambion RNA protocol (15596026.PPS), substituting Qiazol as the lysing reagent after consulting with the company. The final pellet was resuspended in 50 µl of RNase-free water, dissolved at 55°C for 10 minutes, aliquoted, and stored at −80°C.
Nucleic acid quantification was performed using the Quibit dye for RNA or dsDNA kits (Molecular Probes, Life Technologies), as per manufacturer's specifications.
2.5. RNA-Seq and analysis
1 µg RNA from each time point was submitted to the University of Chicago Genomics Core. RNA quality was then checked on the Agilent Bio-analyzer 2100. RNA-Seq libraries were generated in the core using Illumina RS-122-2101 TruSeq® Stranded mRNA LT Libraries and the final libraries checked again on the Agilent bio-analyzer 2100, which was followed by sequencing on the Illumina HiSeq2500. High quality sequencing paired-end reads of 100bp in each (76.4–~83.7%) were mapped to the human genome reference (hg19) using TopHat2 [15]. Estimated genome coverage is 33.6%–~52.1%. Cufflinks [16] were then used to quantify gene expression levels of the assembled transcripts. Fragments per kilobase of transcript per Million mapped reads (FPKM) was calculated for each genes. Only genes with FPKM larger than one were remained for downstream analysis. The raw RNA-Seq reads have been deposited to the NIH Short Read Archive (Accession No. SRP056287).
2.6. 450K array profiling and analysis
1 µg purified DNA from each time point was submitted to the University of Chicago Genomics Core where it was converted with the EZ-96 DNA Methylation™ Kit, as per manufacturer's protocol, followed by array analysis using the Illumina Infinium HumanMethylation450 BeadChip (450K array) Kit, which interrogates >480,000 CpG sites in the human genome. The β-value (proportion of modified signals) values were quantile-normalized across all samples. CpGs probes that failed 450K profiling (detection p>0.01) in >5% samples were removed from consideration. The manufacturer’s annotations for CpGs sites were checked by aligning CpG probes to the human genome reference (hg19) following our previous publication [17]. The raw 450K array data have been deposited into the NIH Gene Expression Omnibus database (Accession No. GSE66330).
2.7. Batch effect evaluation
Since the conventional principal components analysis (PCA) is not effective for identifying batch effects if they are not the largest source of variation, we applied the gPCA method [18] to determine whether there was a batch effect observed in the RNA-Seq data and methylation data. Briefly gPCA guides the singular-value decomposition to look for batch effects in the data based on the batch indicator matrix. A statistic δ was derived from the ratio of the variance of the first PC from gPCA to the variance of the first PC from unguided PCA. A larger δ is expected if there exists a batch effect. To determine the significance, an empirical p-value was estimated by permuting the batch vector 1000 times. The percentage of total variation explained by batch was then calculated as a percentage of variance proportion increase of the first PC using guided PCA. In addition, for each gene, we performed a two-way ANOVA test to test the batch effect. False discovery rate (FDR) was estimated using the Benjamini-Hochberg procedure [19].
2.8. Evaluation of enrichment of neuropathy-associated genes
Hereditary neuropathy- (48 genes) and diabetes- (27 genes) associated gene lists were downloaded from Online Mendelian Inheritance in Man (OMIM) (accessed on Oct 30, 2014). In addition, 13 CIPN associated genes were extracted from previous publications [9, 20–28](Table 1). We further obtained a list of additional neuropathy-associated genes from Reyes-Gibby et al [29]. We calculated the median gene expression levels of neuropathy-associated genes (hereditary neuropathy or CIPN, separately) at each time point. To determine the null distribution of median gene expression, we randomly sampled the same number of genes across whole transcriptome according to the RefSeq annotations [30]. Empirical p-values of enrichment were calculated as the probability of getting at least the median expression value as neuropathy-associated genes under the null distribution. We performed the same analysis with diabetes-associated genes.
Table 1.
Published CIPN associated genes.
| HGNC symbol |
Cytogenetic band |
Description | References |
|---|---|---|---|
| CX3CL1 | 16q21 | chemokine (C-X3-C motif) ligand 1 | [24] |
| EPHA4 | 2q36.1 | Ephrin receptor A4 | [23] |
| EPHA5 | 4q13.2 | Ephrin receptor A5 | [22, 23] |
| EPHA6 | 3q11.2 | Ephrin receptor A6 | [23] |
| FGD4 | 12p11.21 | FYVE, RhoGEF and PH domain containing 4 | [22] |
| FZD3 | 8p21.1 | frizzled class receptor 3 | [29] |
| LIMK2 | 22q12.2 | LIM domain kinase 2 | [23] |
| PRX | 19q13.2 | periaxin | [26] |
| RFX2 | 19p13.3 | regulatory factor X, 2 (influences HLA class II expression) | [21] |
| TLR4 | 9q33.1 | toll-like receptor 4 | [25, 27] |
| TUBB2A | 6p25.2 | tubulin, beta 2A class IIa | [9] |
| XKR4 | 8q12.1 | XK, Kell blood group complex subunit-related family, member 4 | [22, 23] |
| ARHGEF10 | 8p23.3 | Rho guanine nucleotide exchange factor 10 | [26, 28] |
3. Results
3.1 Scheme of iPSC differentiation efficiency analysis in cortical neurons
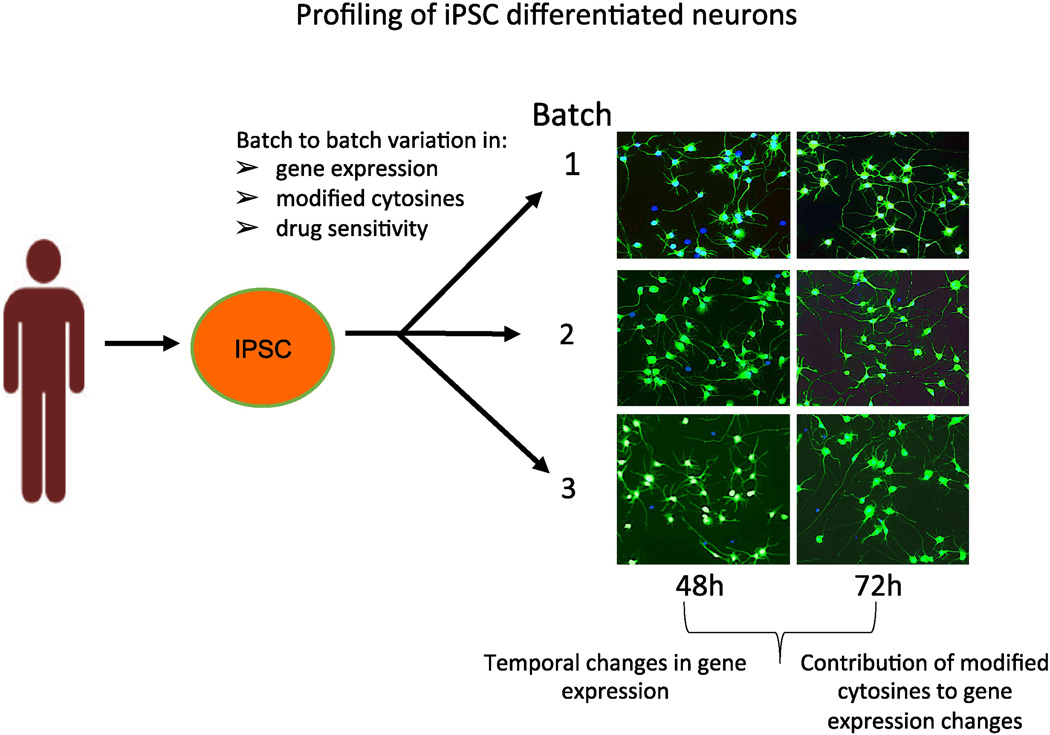
To investigate whether the differentiation process from iPSC to neurons introduces variation in epigenetics and gene expression, we evaluated these parameters in four distinct batches of neurons created from a single iPSC clone from a single individual (Figure 1, Table 2). iPSC-differentiated cortical neurons are a mixed population of neuronal cells consisting mainly of GABAergic and glutamatergic neurons [8]. These differentiated neurons, were shown to be Tuj1 positive with no obvious morphological differences between three neuronal batches imaged at 48 and 72 h after plating (Figure 1). We evaluated batch effect between lots by analyzing gene expression using RNA-Seq and modified cytosines using the Illumina 450K arrays, as well as temporal changes of these measurements following neuronal plating. These neurons quickly assumed a typical neuronal morphology with branching neurites that can be measured using an imaging system.
Fig 1. Overall schematic to investigate epigenetic and gene expression differences between batches and following time in culture.
Four independent batches of neurons reprogrammed from a single iPSC from a single human fibroblasts and were purchased from Cellular Dynamics, Int. These three neuron batches (shown) as well as a fourth batch (not shown) were pelleted to evaluate genome wide gene expression (RNA-Seq) and modified cytosines (Illumina 450K). In the phenotypic assay, three neuron batches out of the four were plated and neurite outgrowth was imaged at 48 and 76h, using MetaXPress.
Table 2.
Four independently differentiated neurons and the experiments performed with each batch.
| Batch # Lot# |
Plating Time (h) |
Number of replicates | ||
|---|---|---|---|---|
| RNA-Seq | 450K Assay | Sensitivity Assay |
||
| 1 1369525 |
4 | 2 | 2 | 3 |
| 28 | 2 | 3 | - | |
| 76 | 2 | 3 | - | |
| 2 1366825 |
4 | 2 | 3 | 3 |
| 28 | 2 | 3 | - | |
| 76 | 2 | 3 | - | |
| 3 1366431 |
0 | 2 | 3 | - |
| 4 | 2 | 4 | 3 | |
| 28 | 2 | 4 | - | |
| 76 | - | 4 | - | |
| 4 1362632 |
0 | 2 | 3 | - |
3.2 Transcriptomic profiling of different batches of neurons
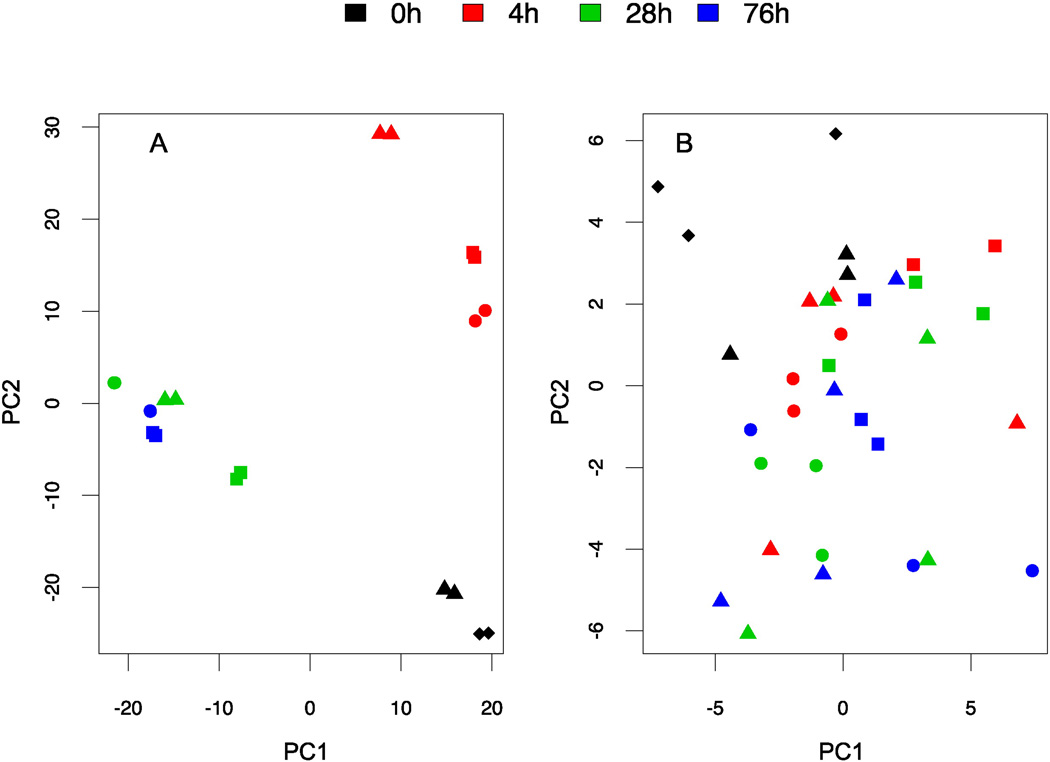
All four batches of cortical neurons differentiated from iPSCs were analyzed using RNA-Seq at 0, 4, 28 and 76h following plating. Traditional PCA of the log-transformed FPKM matrix was first plotted for visualization (Figure 2A). gPCA was then performed to derive statistical conclusions. Permutation test showed that there was no significant batch effect detected among RNA-Seq measurements. (δ= 0.49; P-value = 0.93). Of the total variances in gene expression among samples, which is represented by summations of squared deviation from mean value, 71.55% could be explained by temporal changes, while 27.05% of the total variance could be explained by different batches (see details in methods). Therefore, the temporal effect contributes predominantly to the observed variances. The sum of variances derived from these two major sources of variation may not be equal to 1 due to their possible interaction. Notably, two protein-coding genes (DNASE2B encoding deoxyribonuclease II beta and PALMD encoding palmdelphin) were found to be significant for the batch effect at a lenient cutoff of 20% FDR, with no detection of functional enrichment among batch effect genes. Furthermore, none of the neuropathy-associated genes [9, 21–29] was significantly (FDR < 0.2) affected by batch.
Fig 2. Analyzing batch effect using RNA-Seq and Illumina 450K array.
Four independent batches of neurons denoted by shapes (square lot# 1366825, triangle: lot # 1366431, circle: lot #1369525 and diamond: lot # 1362632) were collected at different time points (0, 4, 28, 76h) denoted by color. Lot# 1362632 had pellets collected only at time point 0. A) A PCA plot of the log-transformed RNA-Seq data in FPKM (fragments per kilobase of transcript per million mapped reads); B) A PCA plot of the β-values from the Illumina 450K array. PC1: the first principal component; PC2: the second principal component.
3.3 Modified cytosine profiling of different batches of neurons
We then analyzed cytosine modification levels using the Illumina 450K arrays for 4 neuron batches at the same 4 different plating time points. PCA plot of the β-value matrix was obtained from the 450K array data (Figure 2B). gPCA test indicated that no significant batch effect separating the batches (δ= 0.44; P-value = 0.50) was detected (Figure 2B). Similar to the RNA-Seq data, 89.28% of the variation was explained by temporal changes in the 450K array data, while 56.4% of the total variation could be explained by batch. These results suggested that both gene expression and cytosine modification measurements were consistent across different batches but changed as neurites were formed in culture, as expected.
3.3 Phenotypic measures of chemotherapeutic sensitivity of different batches of neurons
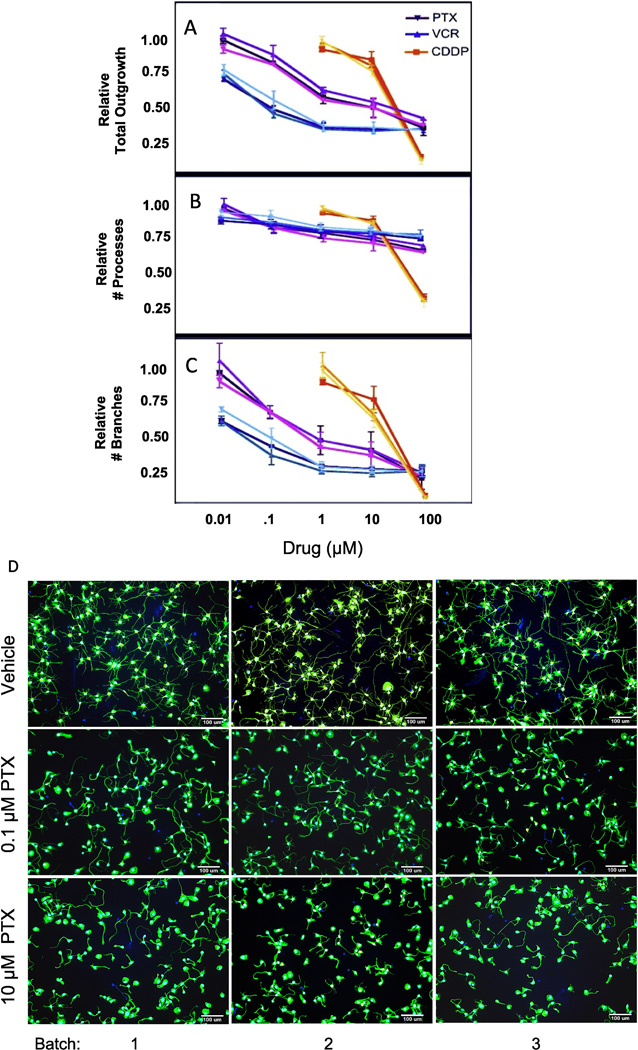
We also measured neuronal phenotype (i.e., pharmacologic response to chemotherapy), in three out of four batches of the neurons. Human neurons reprogrammed from fibroblasts have been used to study chemotherapeutics that cause CIPN [7, 8] and genes associated with clinical CIPN [8–11]. We identified no significant differences between batches in sensitivities to paclitaxel, vincristine and cisplatin as determined by total neurite outgrowth (Figure 3A), number of processes (Figure 3B) and branches (Figure 3C) and as illustrated morphologically for 0, 0.1 and 10 µM paclitaxel (Figure 3D), vincristine (Figure 3E) and cisplatin (Figure 3F).
Fig 3. Effect of chemotherapeutic agents on neuronal outgrowth.
Three batches of iCell Neurons out of the four batches were treated with increasing concentrations of paclitaxel (PTX, purple), vincristine (VCR, blue) or cisplatin (CDDP, orange) for 72 hours, and analyzed with the MetaXPress software for (A) relative total outgrowth; (B) relative number of processes; (C) relative number of branches. The three batches are represented as shaded differences in color for each drug response curve (lightest shade lot #1369525; medium shade lot# 1366825 and darkest shade lot # 1366431). Representative 10X images of 3 batches iCell Neurons treated with different concentrations vehicle (DMSO or PBS), 0.1 µM or 10 µM of (D) paclitaxel, (E) vincristine and (F) cisplatin at 72 hours using ImageXpress Micro.
3.4 Enrichment of highly expressed neuropathy related genes in the neuron model
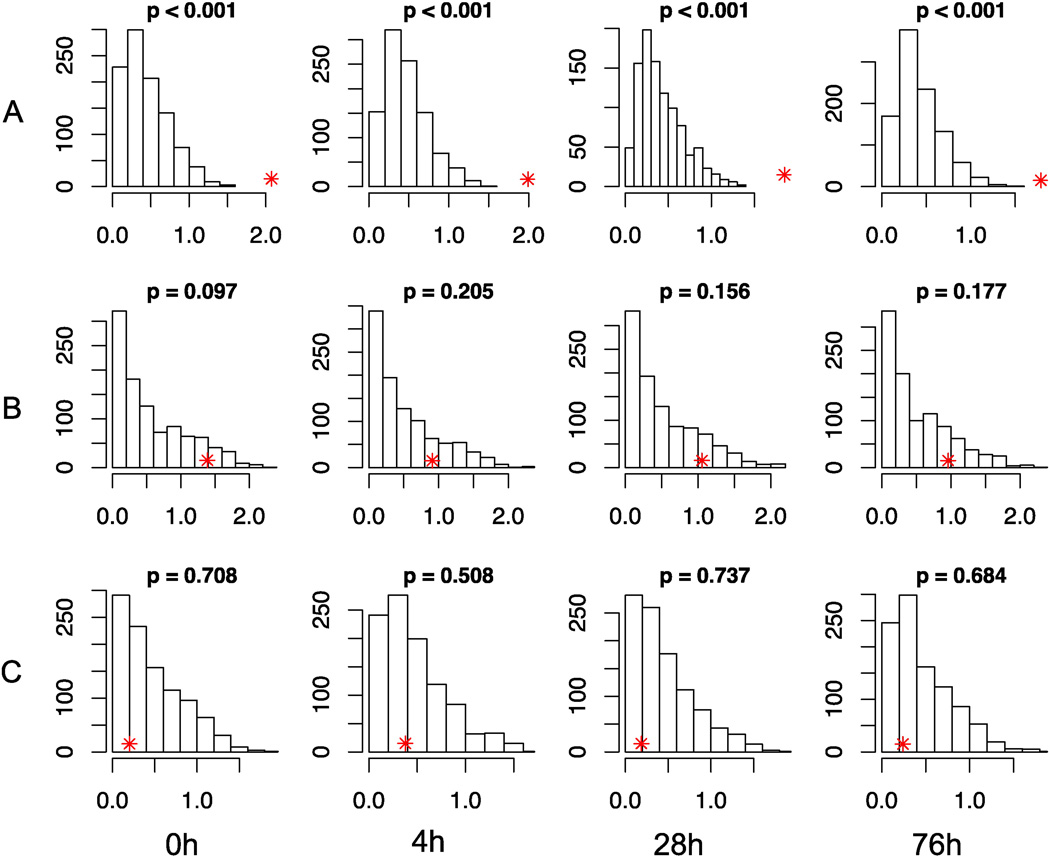
Tissue-specific models are essential in understanding mechanisms of disease onset and drug induced toxicity, however expression of genes critical to the disease is necessary for the model to be useful. We therefore sought to investigate the degree to which established neuropathy associated genes were enriched in this neuronal cell model. Figure 4 illustrates the median gene expression levels of neuropathy-associated genes at each time point. In the gene expression data, we found that at each time point (0, 4, 28 and 76h), hereditary neuropathy related genes from OMIM were highly expressed compared with background distribution across the human genome (empirical P-value < 0.001) (Figure 4A). In addition, a few known CIPN related genes from previous publications also showed a trend of enrichment of highly expressed genes relative to the background (Figure 4B, Table 1). To determine if this enrichment was specific to neuropathic disease but not other diseases, we evaluated diabetes-associated genes as a negative control and found no enrichment (Figure 4C). These results overall, suggest that this is a relevant neuronal model to study the pathophysiology of neuropathy.
Fig 4. Assessing enrichment of neuropathy associated genes in neuronal model.
Median gene expression levels measured as log FPKM (X-axis) of: A) hereditary neuropathy-; B) CIPN-; and C) diabetes-associated genes at each time point are indicated by red asterisks. The histograms show the null distributions (frequencies on the Y-axis) generated by random samplings of the same number of genes across the human genome. Empirical P-values are indicated for each situation. CIPN: chemotherapy-induced peripheral neuropathy; FPKM: fragments per kilobase of transcript per million mapped reads.
4. Discussion
We evaluated inter-batch variability of differentiated neurons in order to provide insight into variability associated with manufacturing neurons from iPSCs (i.e., the differentiation process). It is important to consider the magnitude of noise introduced by processing the cells relative to biological signal. High quality and stable iPSC-derived neurons depend on the robustness of the manufacturing protocol for both the iPSC-differentiated cells and the production of the iPSCs themselves [31]. Such standards of quality are most practically achieved by large core facilities and commercial manufacturers.
In our study, four independent productions of iCell Neurons were obtained from CDI; each production represents an independent differentiation from the same iPSC clone. We found through RNA-Seq analysis that gene expression variation due to different batches was not a major source of variance compared to changes over time. Modified cytosine changes also showed no significant variation across batches as compared to time dependent changes suggesting that the differentiation process results in minimal intra-individual variation. The lack of significant variation of neuronal sensitivity to neurotoxic chemotherapeutics for 3 separate batches provided further confidence that inter-batch variation is minimal. We next investigated whether the iPSC-derived neuronal model system is adequately enriched for the genes essential in identifying mechanisms of neuronal diseases and drug-induced neurotoxicities. Indeed, we found that the neurons were enriched for highly expressed genes implicated in hereditary neuropathy and showed an enrichment trend for a few genes known to be associated with chemotherapeutic-induced neuropathy. The lack of significance of CIPN related genes may be because of our limited knowledge of genes contributing to CIPN. The specificity of our model was further confirmed when we found that our model was not enriched for genes implicated in other disease types, such as diabetes.
An advantage of the creation of patient-derived iPSCs is that they retain the genetic "makeup" of the donor allowing for in vitro studies of neurons from patients with specific diseases that harbor the complex genetic background associated with the disorder [31]. The successful generation of iPSCs from patients' specific somatic cells, and differentiation to cortical neurons, have offered cell resources for disease modeling and potential cell transplantation therapy [32]. This revolutionary technology has already helped to advance our understanding of many diseases and inform mechanistically rationalized therapies, which are desperately needed in this arena [6, 33]
Charcot-Marie tooth (CMT) is one of the most common inherited neurological disorder, characterized by weakness of the foot and lower leg muscle [34] has no effective treatment. Therefore, human iPSCs can lead to the identification and optimization of potential drugs and thus move forward new pharmacological therapies for a wide range of neurodegenerative and neurodevelopmental conditions [32]. Critical to this field is the consistent manufacturing of differentiated neurons from iPSCs. Therefore, our evaluation of gene expression and modified cytosines at baseline, and changes over time following plating, will provide a framework for studies of these neurons. Furthermore, we show batch-to-batch consistency of iPSC-derived neurons in gene expression, modified cytosines and response to neurotoxic chemotherapeutics.
Creating a genetically diverse set of differentiated cells for the purposes of phenotype-genotype studies would have great value in pharmacogenomics, a field that has relied primarily on EBV-transformed lymphoblastoid cell lines [35–37]. Tissue specific cells will provide the appropriate model for functionally validating findings from large clinical genome wide association studies. Therefore, the development of human iPSC-derived neurons as a model system could have important implications for studies of neurotoxicity and developing new drugs to prevent or treat heritable neuropathy and/or CIPN, one of the most common and sometimes permanent side effects of chemotherapy [38, 39].
5. Conclusion
Our results indicate that batch-to-batch variation in gene expression, modified cytosines and drug sensitivity was not a major source of variances compared to the time effect. The enrichment of neuropathy associated genes in the neurons in contrast to other diseases such as diabetes, gives us a clinically relevant cell-based model that will allow us to understand the mechanisms of neurological disease and drug induced neurotoxicities. This model also provides a flexible platform to test potential neurotoxic and/or neuroprotectant drugs.
Highlights.
-
➢
Separate batches of commercially available neurons originating from the same iPSC were used to investigate whether the differentiation process used in manufacturing iPSCs to neurons affected genome-wide gene expression, modified cytosines, or neuronal sensitivity to drugs
-
➢
No significant changes between batches compared to changes over time were observed in gene expression (measured by RNA-Seq), cytosine modification levels (measured by Illumina 450K arrays) or neuronal sensitivity to several neurotoxic chemotherapeutic agents
-
➢
Genes involved in neuropathy had relatively higher expression levels in these samples across different time points than expected by chance
-
➢
Data supports using iPSC derived neurons for studies of human neuropathy, druggable targets to prevent neuropathy, and other neurological diseases
Acknowledgments
The authors are grateful for intellectual and technical advice from Ms. Susan DeLaura and Dr. Eugenia Jones at Cellular Dynamics International, Inc. (Madison, WI).
Funding Source: This work is supported, in part, by the NIH/NIGMS Pharmacogenomics of Anticancer Agents Research Grant U01 GM61393 (M.E.D.), R21 HG006367 (W.Z. and M.E.D.), and The Robert H. Lurie Comprehensive Cancer Center-Developmental Funds P30 CA060553 (to W.Z.). Gladys Morrison is supported by the NIH/NIGMS Clinical Therapeutics training grant T32 GM007019.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Gladys Morrison: Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing
Cong Liu: Data analysis and interpretation, Manuscript writing
Claudia Wing: Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing (Methods)
Shannon M Delaney: Collection and/or assembly of data
Wei Zhang: Conception and design, Financial support, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
M. Eileen Dolan: Conception and design, Financial support, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Disclosure of Potential Conflict of Interest: The authors do not have any potential conflict of interest.
References
- 1.Phillips W, Michell A, Pruess H, Barker RA. Animal models of neurodegenerative diseases. Methods Mol Biol. 2009;549:137–155. doi: 10.1007/978-1-60327-931-4_10. [DOI] [PubMed] [Google Scholar]
- 2.Ekins S, Litterman NK, Arnold RJ, Burgess RW, Freundlich JS, Gray SJ, Higgins JJ, Langley B, Willis DE, Notterpek L, Pleasure D, Sereda MW, Moore A. A brief review of recent Charcot-Marie-Tooth research and priorities. F1000Res. 2015;4:53. doi: 10.12688/f1000research.6160.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callaghan BC, Cheng HT, Stables CL, Smith AL, Feldman EL. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol. 2012;11:521–534. doi: 10.1016/S1474-4422(12)70065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith EM, Pang H, Cirrincione C, Fleishman S, Paskett ED, Ahles T, Bressler LR, Fadul CE, Knox C, Le-Lindqwister N, Gilman PB, Shapiro CL O. Alliance for Clinical Trials. Effect of duloxetine on pain, function, and quality of life among patients with chemotherapy-induced painful peripheral neuropathy: a randomized clinical trial. Jama. 2013;309:1359–1367. doi: 10.1001/jama.2013.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandoe J, Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nature neuroscience. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 6.Mullard A. Stem-cell discovery platforms yield first clinical candidates. Nat Rev Drug Discov. 2015;14:589–591. doi: 10.1038/nrd4708. [DOI] [PubMed] [Google Scholar]
- 7.Wainger BJ, Buttermore ED, Oliveira JT, Mellin C, Lee S, Saber WA, Wang AJ, Ichida JK, Chiu IM, Barrett L, Huebner EA, Bilgin C, Tsujimoto N, Brenneis C, Kapur K, Rubin LL, Eggan K, Woolf CJ. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nature neuroscience. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler HE, Wing C, Delaney SM, Komatsu M, Dolan ME. Modeling chemotherapeutic neurotoxicity with human induced pluripotent stem cell-derived neuronal cells. PloS one. 2015;10:e0118020. doi: 10.1371/journal.pone.0118020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leandro-Garcia LJ, Leskela S, Jara C, Green H, Avall-Lundqvist E, Wheeler HE, Dolan ME, Inglada-Perez L, Maliszewska A, de Cubas AA, Comino-Mendez I, Mancikova V, Cascon A, Robledo M, Rodriguez-Antona C. Regulatory polymorphisms in beta-tubulin IIa are associated with paclitaxel-induced peripheral neuropathy. Clin Cancer Res. 2012;18:4441–4448. doi: 10.1158/1078-0432.CCR-12-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diouf B, Crews KR, Lew G, Pei D, Cheng C, Bao J, Zheng JJ, Yang W, Fan Y, Wheeler HE, Wing C, Delaney SM, Komatsu M, Paugh SW, McCorkle JR, Lu X, Winick NJ, Carroll WL, Loh ML, Hunger SP, Devidas M, Pui CH, Dolan ME, Relling MV, Evans WE. Association of an inherited genetic variant with vincristine-related peripheral neuropathy in children with acute lymphoblastic leukemia. Jama. 2015;313:815–823. doi: 10.1001/jama.2015.0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komatsu M, Wheeler HE, Chung S, Low SK, Wing C, Delaney SM, Gorsic LK, Takahashi A, Kubo M, Kroetz D, Zhang W, Nakamura Y, Dolan ME. Pharmacoethnicity in Paclitaxel-Induced Sensory Peripheral Neuropathy. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, Rodolfa CT, Dimos JT, Mikkilineni S, MacDermott AB, Woolf CJ, Henderson CE, Wichterle H, Eggan K. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas SM, Kagan C, Pavlovic BJ, Burnett J, Patterson K, Pritchard JK, Gilad Y. Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature. PLoS Genet. 2015;11:e1005216. doi: 10.1371/journal.pgen.1005216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, Myers J, Godley LA, Dolan ME, Zhang W. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194:987–996. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reese SE, Archer KJ, Therneau TM, Atkinson EJ, Vachon CM, de Andrade M, Kocher JP, Eckel-Passow JE. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics. 2013;29:2877–2883. doi: 10.1093/bioinformatics/btt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hotchberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;51:289–300. [Google Scholar]
- 20.Seretny M, Currie GL, Sena ES, Ramnarine S, Grant R, MacLeod MR, Colvin LA, Fallon M. Incidence, prevalence, and predictors of chemotherapy-induced peripheral neuropathy: A systematic review and meta-analysis. Pain. 2014;155:2461–2470. doi: 10.1016/j.pain.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler HE, Gamazon ER, Wing C, Njiaju UO, Njoku C, Baldwin RM, Owzar K, Jiang C, Watson D, Shterev I, Kubo M, Zembutsu H, Winer EP, Hudis CA, Shulman LN, Nakamura Y, Ratain MJ, Kroetz DL, Cancer, B. Leukemia Group. Cox NJ, Dolan ME. Integration of cell line and clinical trial genome-wide analyses supports a polygenic architecture of Paclitaxel-induced sensory peripheral neuropathy. Clin Cancer Res. 2013;19:491–499. doi: 10.1158/1078-0432.CCR-12-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, Watson D, Eclov RJ, Mefford J, McLeod HL, Friedman PN, Hudis CA, Winer EP, Jorgenson EM, Witte JS, Shulman LN, Nakamura Y, Ratain MJ, Kroetz DL. A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res. 2012;18:5099–5109. doi: 10.1158/1078-0432.CCR-12-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leandro-Garcia LJ, Inglada-Perez L, Pita G, Hjerpe E, Leskela S, Jara C, Mielgo X, Gonzalez-Neira A, Robledo M, Avall-Lundqvist E, Green H, Rodriguez-Antona C. Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. J Med Genet. 2013;50:599–605. doi: 10.1136/jmedgenet-2012-101466. [DOI] [PubMed] [Google Scholar]
- 24.Li D, Huang ZZ, Ling YZ, Wei JY, Cui Y, Zhang XZ, Zhu HQ, Xin WJ. Up-regulation of CX3CL1 via Nuclear Factor-kappaB-dependent Histone Acetylation Is Involved in Paclitaxel-induced Peripheral Neuropathy. Anesthesiology. 2015;122:1142–1151. doi: 10.1097/ALN.0000000000000560. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zhang H, Zhang H, Kosturakis AK, Jawad AB, Dougherty PM. Toll-like receptor 4 signaling contributes to Paclitaxel-induced peripheral neuropathy. J Pain. 2014;15:712–725. doi: 10.1016/j.jpain.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beutler AS, Kulkarni AA, Kanwar R, Klein CJ, Therneau TM, Qin R, Banck MS, Boora GK, Ruddy KJ, Wu Y, Smalley RL, Cunningham JM, Le-Lindqwister NA, Beyerlein P, Schroth GP, Windebank AJ, Zuchner S, Loprinzi CL. Sequencing of Charcot-Marie-Tooth disease genes in a toxic polyneuropathy. Ann Neurol. 2014;76:727–737. doi: 10.1002/ana.24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Zhang H, Kosturakis AK, Cassidy RM, Zhang H, Kennamer-Chapman RM, Jawad AB, Colomand CM, Harrison DS, Dougherty PM. MAPK signaling downstream to TLR4 contributes to paclitaxel-induced peripheral neuropathy. Brain Behav Immun. 2015 doi: 10.1016/j.bbi.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boora GK, Kulkarni AA, Kanwar R, Beyerlein P, Qin R, Banck MS, Ruddy KJ, Pleticha J, Lynch CA, Behrens RJ, Zuchner S, Loprinzi CL, Beutler AS. Association of the Charcot-Marie-Tooth disease gene ARHGEF10 with paclitaxel induced peripheral neuropathy in NCCTG N08CA (Alliance) J Neurol Sci. 2015 doi: 10.1016/j.jns.2015.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes-Gibby CC, Wang J, Yeung SC, Shete S. Informative gene network for chemotherapy-induced peripheral neuropathy. BioData Min. 2015;8:24. doi: 10.1186/s13040-015-0058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A, Yamanaka S, Takahashi K. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci U S A. 2014;111:12426–12431. doi: 10.1073/pnas.1413299111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corti S, Faravelli I, Cardano M, Conti L. Human pluripotent stem cells as tools for neurodegenerative and neurodevelopmental disease modeling and drug discovery. Expert Opin Drug Discov. 2015;10:615–629. doi: 10.1517/17460441.2015.1037737. [DOI] [PubMed] [Google Scholar]
- 33.Wiethoff S, Arber C, Li A, Wray S, Houlden H, Patani R. Using human induced pluripotent stem cells (hiPSC) to model cerebellar disease: Hope and hype. J Neurogenet. 2015:1–25. doi: 10.3109/01677063.2015.1053478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman CJ, Walsh M, O'Sullivan R, Jenkinson A, Bennett D, Lynch B, O'Brien T. The characteristics of gait in Charcot-Marie-Tooth disease types I and II. Gait Posture. 2007;26:120–127. doi: 10.1016/j.gaitpost.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Moen EL, Godley LA, Zhang W, Dolan ME. Pharmacogenomics of chemotherapeutic susceptibility and toxicity. Genome Med. 2012;4:90. doi: 10.1186/gm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler HE, Dolan ME. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics. 2012;13:55–70. doi: 10.2217/pgs.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welsh M, Mangravite L, Medina MW, Tantisira K, Zhang W, Huang RS, McLeod H, Dolan ME. Pharmacogenomic discovery using cell-based models. Pharmacol Rev. 2009;61:413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alberti P, Rossi E, Cornblath DR, Merkies IS, Postma TJ, Frigeni B, Bruna J, Velasco R, Argyriou AA, Kalofonos HP, Psimaras D, Ricard D, Pace A, Galie E, Briani C, Dalla Torre C, Faber CG, Lalisang RI, Boogerd W, Brandsma D, Koeppen S, Hense J, Storey D, Kerrigan S, Schenone A, Fabbri S, Valsecchi MG, Cavaletti G C.I.-P. Group. Physician-assessed and patient-reported outcome measures in chemotherapy-induced sensory peripheral neurotoxicity: two sides of the same coin. Ann Oncol. 2014;25:257–264. doi: 10.1093/annonc/mdt409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavaletti G, Cornblath DR, Merkies IS, Postma TJ, Rossi E, Frigeni B, Alberti P, Bruna J, Velasco R, Argyriou AA, Kalofonos HP, Psimaras D, Ricard D, Pace A, Galie E, Briani C, Dalla Torre C, Faber CG, Lalisang RI, Boogerd W, Brandsma D, Koeppen S, Hense J, Storey D, Kerrigan S, Schenone A, Fabbri S, Valsecchi MG C.I.-P. Group. The chemotherapy-induced peripheral neuropathy outcome measures standardization study: from consensus to the first validity and reliability findings. Ann Oncol. 2013;24:454–462. doi: 10.1093/annonc/mds329. [DOI] [PMC free article] [PubMed] [Google Scholar]