Abstract
Asthma is a complex airway allergic disease involving the interplay of various cell types, cytokines, and transcriptional factors. Though many factors contribute to disease etiology, the molecular control of disease phenotype and responsiveness is not well understood. Here we report an essential role of the matrix attachment region (MAR)-binding protein SMAR1 in regulating immune response during allergic airway disease. Conditional knockout of SMAR1 in T cells rendered the mice resistant to eosinophilic airway inflammation against ovalbumin (OVA) allergen with low immunoglobulin E (IgE) and interleukin-5 (IL-5) levels. Moreover, a lower IgE/IgG2a ratio and higher interferon-γ (IFN-γ) response suggested aberrant skewing of T-cell differentiation toward type 1 helper T cell (Th1) response. We show that SMAR1 functions as a negative regulator of Th1 and Th17 differentiation by interacting with two potential and similar MAR regions present on the promoters of T-bet and IL-17. Thus, we present SMAR1 as a regulator of T-cell differentiation that favors the establishment of Th2 cells by modulating Th1 and Th17 responses.
INTRODUCTION
Asthma is a chronic allergic disease of the airways. More than 235 million people currently suffer from asthma that is still a major socioeconomic burden.1 Although asthma correlates with allergic, eosinophilic, and type 2 helper T cell (Th2)-mediated disease with immunoglobulin E (IgE) response (corticosteroid responsive), steroid-resistant neutrophilic asthma with potential involvement of additional mediators such as interleukin-17 (IL-17) and interferon-γ (IFN-γ) as driving factors is being considered.2 Various allergens infiltrate the mucosal epithelium of the airways to stimulate the tissue-resident dendritic cells that in turn traffic to the lung-draining lymph nodes and activate the naive T cells, resulting in T-cell differentiation and cytokine production.3 Differentiation of T cells into Th2 lineage leads to production of inflammatory Th2 cytokines (IL-13, IL-5, and IL-4) and development of eosinophilic asthma accompanied by B-cell Ig class switching to IgE.4, 5, 6 Blockade in differentiation to Th2 lineage or function of Th2-specific cytokines has beneficial consequence to prevent the disease progression.7 Thus, T-cell differentiation programs directly influence the development of asthma, associated airway inflammation, and the phenotype of the disease.8, 9
Naive CD4+ T cells have the potential to differentiate into various effector subsets endowed with functional specificity in host defense.10 Depending on the type of antigen encountered and the cytokine milieu in the microenvironment, T cells differentiate to Th1, Th2, Th17, induced regulatory T cells, and so on.11, 12 Intracellular pathogens initiate Th1 cell differentiation program with the involvement of IFN-γ and IL12 signaling and concomitant activation of Th1-specific transcription factor, T-box protein expressed in T cells (T-bet).13 Extracellular pathogens or allergens promote Th2 cell lineage development that necessitates the induction of GATA-3, mediated by IL-4-dependent STAT6 (signal transducer and activator of transcription 6) activation.14 Similarly, combinatorial signals from transforming growth factor TGF-β and IL-6 induce expression of T helper-17 (Th17) specific transcription factor, retinoic acid receptor-related orphan receptor gamma-t (RORγt), which transactivate IL-17 gene expression.15, 16 Thus, each T-cell lineage is associated with distinct pathways, directed by lineage-specific transcription factors.17
Transcription factor-driven T-cell differentiation programs are associated with chromatin changes.18 Master regulators of transcription factors have to utilize various cis elements that interact with various chromatin-associated scaffold/matrix attachment region (MAR)-binding proteins to induce favorable chromatin changes.19, 20 MAR-binding proteins serve as the scaffold for the recruitment of transcriptional or chromatin remodeling factors that facilitate localized chromatin changes causing activation or repression of gene subsets.21, 22
In this report, we investigated the role of a MAR-binding protein, SMAR1, in progression of allergic airway disease through the regulation of T-cell differentiation programs. In previous studies, SMAR1 was identified as a MAR-binding protein attached to the MAR-β region of T cell receptor-β locus and overexpression of SMAR1 in transgenic mice resulted in perturbation of the peripheral T-cell repertoire.23, 24 Using T cell-specific conditional knockout mice (SMAR1cKO), we show that SMAR1 deficiency in T cells reduces airway inflammation. Compared with control littermate mice, SMAR1cKO mice exhibited significantly reduced eosinophilia and IgE response. The mice displayed increased IL-17 production with associated neutrophilia and also an increased IgG2a response. We show that GATA-3 directly promotes SMAR1 expression that in turn binds to the MAR elements present in the promoters of T-bet and IL-17, inhibiting Th1 and Th17 responses. SMAR1 deficiency in T cells caused severely compromised Th2 response and enhanced Th1 and Th17 differentiation in vitro. Taken together, we show an important role of SMAR1 in Th2 differentiation with concomitant repression of Th1 and Th17 differentiation.
RESULTS
GATA-3 promotes SMAR1 induction in Th2 cells
As SMAR1 transgenic mice exhibited perturbed immune responses,24 we investigated whether SMAR1 regulates CD4+ T-cell differentiation. Sera from SMAR1 transgenic mice were analyzed for immunoglobulin profile. We observed reduced IgG2a immunoglobulin in the sera of SMAR1 transgenic mice (Supplementary Figure S1a online). In addition, activation of CD4+ T cells from SMAR1 transgenic mice via T cell receptor stimulation resulted in reduced IFN-γ response (Supplementary Figure S1b) with a small increase in IL-4 production (Supplementary Figure S1c).
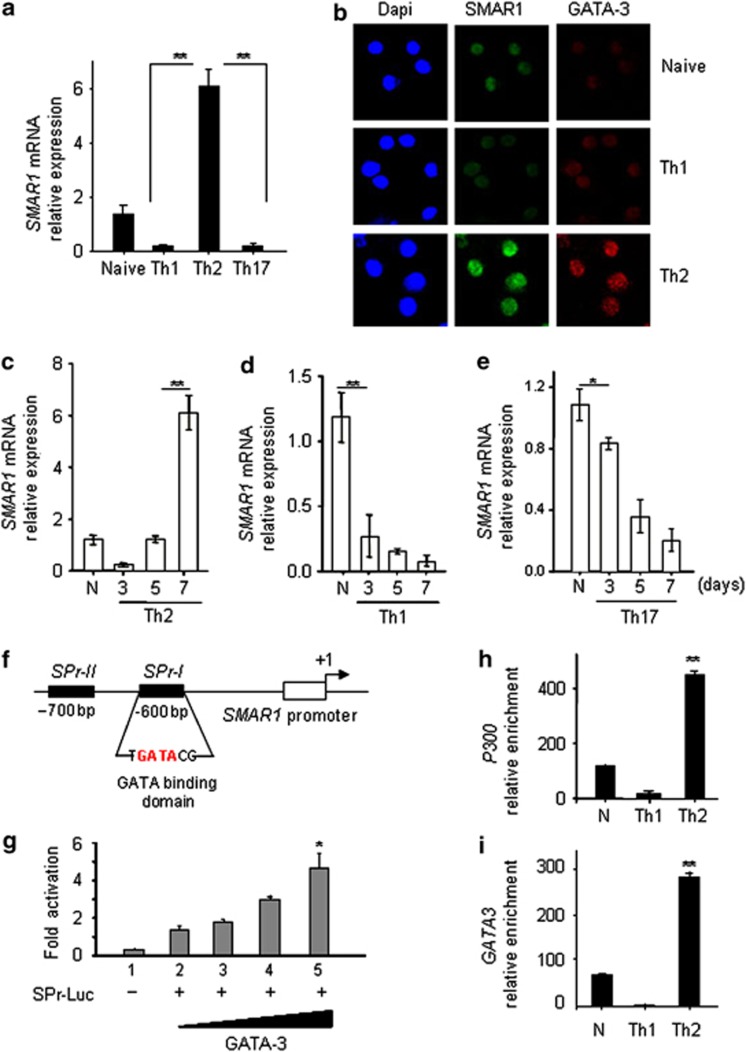
Next, CD4+ CD62Lhigh CD44low CD25- naive lymphocytes were purified from C57BL/6 mice and polarized in vitro into Th1, Th2, and Th17 cells and expression of SMAR1 was examined. Quantitative real-time PCR analysis revealed a sixfold induction of SMAR1 mRNA specifically in Th2 cells (Figure 1a). The selective expression of SMAR1 and GATA-3 in Th2 cells was also observed by imaging techniques in differentiated CD4+ T cells (Figure 1b). Kinetic studies revealed that SMAR1 expression is induced 5 days after initiation of T-cell differentiation (Figure 1c), suggesting its role in Th2 cell maintenance. However, in cells induced to differentiate along Th1 and Th17 lineages, SMAR1 expression was reduced during the course of differentiation (Figure 1d,e).
Figure 1.
Expression of SMAR1 is regulated by GATA-3. Naive CD4+ T cells were isolated from C57BL/6 mice and activated under various polarizing conditions with plate-bound CD3 and CD28 antibodies. (a) Quantitative real-time mRNA expression of SMAR1 during type 1 helper T cell (Th1), Th2, and Th17 differentiation. (b) Confocal staining for SMAR1 (green) and GATA3 (red) in CD4+ T cells isolated from mice and cultured under Th1 and Th2 differentiation conditions. Relative mRNA expression of SMAR1 was quantified during the course of CD4+ T-cell differentiation to (c) Th2, (d) Th1, and (e) Th17 using real-time PCR. (f) Schematic diagram of the SMAR1 promoter and the regions (not to scale) amplified for the chromatin immunoprecipitation (ChIP) assay. SPr-I and -II correspond to the GATA-binding consensus motifs on SMAR1 promoter. (g) SMAR1 promoter luciferase assay performed by transient transfection of SMAR1 promoter luciferase construct (SPr-I Luc PGL-3 vector) along with GATA-3 expression plasmid in HEK 293 cells. Data represent relative fold change in luciferase activity. (h, i) ChIP assays for GATA-3 and CBP/p300 binding to the SMAR1 promoter SPr-I in CD4+ T cells in naive (N), Th1-, or Th2-polarizing conditions. *P<0.01, **P<0.005. Data are representative of three independent experiments.
Given that SMAR1 is induced selectively under Th2 conditions, we assumed GATA-3-mediated regulation of SMAR1 gene expression. In silico analysis of the SMAR1 promoter (−1 kb to +100 bp) revealed the presence of two putative GATA-3-binding sites 600 bp upstream of the transcription start site (Figure 1f). To determine whether GATA-3 directly mediates SMAR1 induction, a 400-bp SMAR1 promoter region containing the GATA-3-binding elements (SPr-1) was cloned in a luciferase reporter vector and reporter activity was followed after transient expression of GATA-3. We observed an approximately fourfold dose-dependent increase in SMAR1 promoter activity upon transfection of a GATA-3 expression plasmid (Figure 1g). Chromatin immunoprecipitation (ChIP) assays performed with differentiated T cells showed recruitment of GATA-3 and p300 on the SMAR1 promoter specifically under Th2 conditions (Figure 1h,i) showing lineage-specific induction of SMAR1 by GATA-3 under Th2 conditions. Collectively, these results showed that GATA-3 directly activates SMAR1 transcription in Th2 differentiated cells.
Conditional deletion of SMAR1 in T cells renders mice resistant to eosinophilic airway inflammation but promotes Th1 and Th17 responses
Different transcription factors regulate immune responses against pathogens and allergens that also involves cross-regulation of the different T helper cell differentiation programs.17 For example, T-bet deficiency in T cells causes spontaneous airway inflammation in mice,8 similar to that observed upon overexpression of GATA-3.14 As GATA-3 was found to directly induce SMAR1, we examined the consequence of SMAR1 deficiency in T cells on allergic immune response in the airways. Toward this end, SMAR1-floxed mice were crossed with Lck-Cre transgenic mice to generate T cell-specific conditional knockout mice (referred to as SMAR1cKO mice) (Supplementary Figure S2a). Ablation of SMAR1 in T cells was confirmed by analyzing its expression in purified CD4+ T cells (Figure 2a). Thymocytes (Supplementary Figure S2b), splenic T cells (Supplementary Figure S2c), and naive CD4+ T cells were comparable between SMAR1cKO mice and control littermate mice (Supplementary Figure S2d).
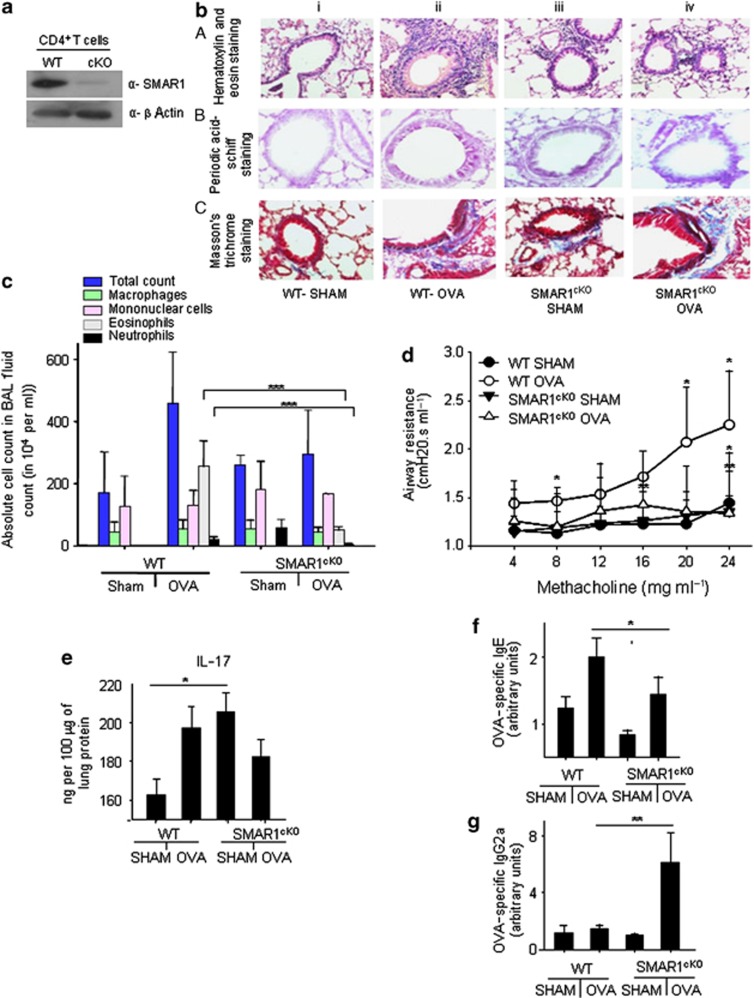
Figure 2.
SMAR1cKO mice exhibit attenuated allergic airway disease. (a) CD4+ T cells were isolated from control and SMAR1cKO mice and expression of SMAR1 was assessed by immunoblotting. β-Actin was used as the control for protein loading. SMAR1cKO, T cell-specific conditional knockout mice; WT, wild type. (b) SMAR1cKO and control mice were subjected to a protocol of allergic airway disease using ovalbumin (OVA). Lung sections of various groups of mice (n=3–6 mice per group) were examined histologically after staining with (b,A) hematoxylin and eosin (H&E), (b,B) periodic acid–Schiff, or (b,C) Masson's trichrome stains. (c) Differential cell counts in the bronchoalveolar lavage (BAL) fluid of mice. (d) Airway hyperresponsiveness in the form of airway resistance was assessed in response to increasing concentrations of methacholine (n=3–6 each group). Enzyme-linked immunosorbant assays (ELISAs) were performed in lung and sera to measure (e) interleukin-17A (IL-17A), (f) OVA-specific immunoglobulin E (IgE), and (g) OVA-specific IgG2a. Data are representative of two independent experiments, and results are shown as mean±s.e.m. Statistical significance was determined by unpaired Student's t-test or Mann–Whitney test. *P<0.05, **P<0.01, ***P<0.001.
To test our hypothesis, we induced allergic airway disease in control and SMAR1cKO mice in an ovalbumin (OVA)-induced model as previously described.25, 26 OVA-sensitized and OVA-challenged wild-type (WT) mice showed increased airway inflammation (Figure 2b,c), increased mucus production (Figure 2b), and subepithelial fibrosis (Figure 2b). On the contrary, OVA-sensitized and -challenged SMAR1cKO mice were resistant to development of allergic airway inflammation (Figure 2b) as evidenced by minimal increase in a cellular infiltration in the airways and mucus production (Figure 2b and Supplementary Figure S3a). Subepithelial fibrosis was observed in OVA-challenged SMAR1cKO mice (Figure 2b,C,iv and Supplementary Figure S3b). Invasive Flexi-Vent measurements of airway hyperreactivity against increasing concentrations of methacholine showed significantly less airway resistance in OVA-challenged SMAR1cKO mice as compared with that in WT mice (Figure 2d and Supplementary Figure S3c, respectively). Furthermore, we noticed neutrophilic airway inflammation in SMAR1cKO SHAM mice (Figure 2b) with increased levels of IL-17 (Figure 2e), IFN-γ (Supplementary Figure S3d), IL-13, and IL-4 (Supplementary Figure 3ef) in the total lung protein, suggesting possible dysregulation of cytokine expressions from T cells because of SMAR1 deficiency. Levels of IgE were significantly lower in the sera of OVA-challenged SMAR1cKO mice as compared with that in WT mice (Figure 2f). Moreover, SMAR1cKO lung homogenates exhibited lower levels of IL-5 cytokine in the control or OVA-treated groups (Supplementary Figure S3g). This result correlates with the decreased eosinophilia and IgE response of SMAR1cKO mice. Notably, the IFN-γ levels (Supplementary Figure S3d) along with significant upregulation of OVA-specific IgG2a were observed in the SMAR1cKO mice (Figure 2g). These data showed reduced Th2-driven immune response in SMAR1cKO mice.
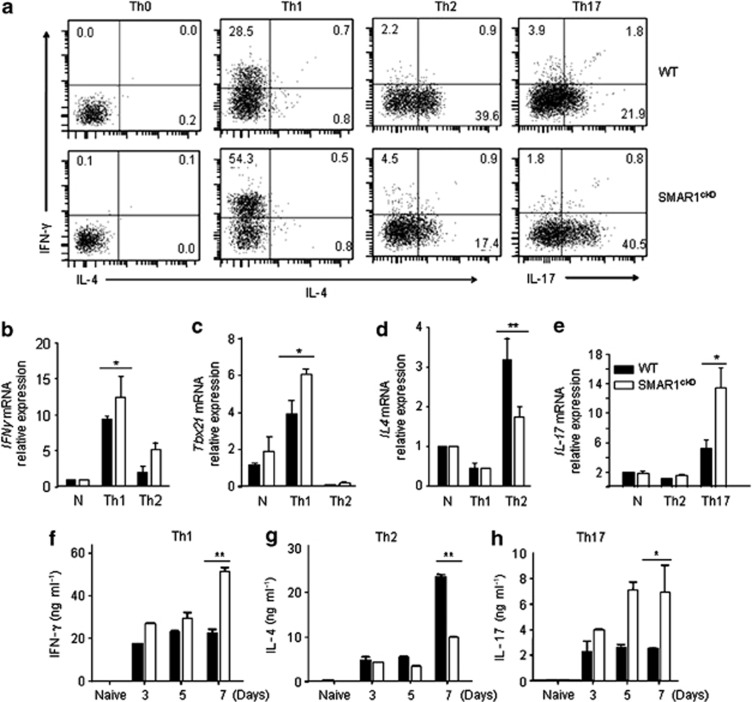
Given that the deficiency of SMAR1 in T cells inhibited Th2-driven T-cell responses, we next examined the effect of SMAR1 deficiency on Th1 and Th17 differentiation. Naive CD4+ T cells were isolated from control and SMAR1cKO mice and differentiated in vitro into Th1, Th2, and Th17 pathways. An increase in the frequency of IFN-γ (28.5% vs. 54.3%) and IL-17 (21.9% vs. 40.5%)-expressing CD4+ T cells was observed in SMAR1cKO mice as compared with that in WT mice (39.6% vs. 17.4% Figure 3a). Also, Th2 cell differentiation was inhibited in SMAR1-deficient T cells (Figure 3a). Accordingly, significant upregulation of IFN-γ and T-bet mRNA was observed in the SMAR1-deficient T cells as compared with that in cells from WT mice (Figure 3b,c). Notably, SMAR1cKO Th2 cells exhibited higher IFN-γ and T-bet expression compared with WT cells (Figure 3b,c). In contrast, the relative expression of IL-4 was twofold lower in SMAR1cKO T cells differentiated along the Th2 lineage (Figure 3d). An enhanced IL17 mRNA expression was noticed in SMAR1-deficient Th17 cells compared with SMAR1-sufficient WT cells (Figure 3e). Supernatants from the cultured T cells were collected on days 3, 5, and 7 after initiation of cell activation and cytokine levels were assayed. IFN-γ protein was twofold higher in day 7 culture supernatants of SMAR1cKO cells as compared with WT cells (Figure 3f), whereas IL-4 was lower in SMAR1cKO cells (Figure 3g). We also noticed enhanced IL-17 secretion (fourfold) from SMAR1cKO T cells as compared with that from WT cells (Figure 3h). Thus, our results showed that SMAR1 deficiency inhibits Th2 cell differentiation but increases Th1 and Th17 cell differentiation in vitro.
Figure 3.
T helper cell (Th) differentiation is altered in SMAR1-deficient T cells. Naive T cells were isolated from wild-type (WT) and SMAR1cKO mice and cultured in vitro under Th1, Th2, and Th17 differentiation conditions. (a) Intracellular cytokine staining of control and SMAR1-deficient T cells polarized for 4 days under Th1, Th2, and Th17 conditions, stained with anti-IFN-γ, anti-IL-4, and anti-IL-17 with naive CD4+ T cells (Th0) used as control. IL, interleukin; IFN-γ, interferon-γ SMAR1cKO, T cell-specific conditional knockout mice. (b–e) Quantitative analysis of relative mRNA expression for IFN-γ, T-bet, IL-4, and IL-17 respectively, by real-time PCR under naive (N), Th1-, Th2-, and Th17-polarized cells from SMAR1cKO and WT mice. Culture supernatants were collected at various time points as mentioned in the figures and cytokine secretion was analyzed by enzyme-linked immunosorbant assay (ELISA). T-bet, T-box protein expressed in T cells. (f–h) IFN-γ, IL-4, and IL-17 cytokine levels, respectively. All the results are representative of three independent experiments. *P<0.01, **P<0.005.
SMAR1 negatively regulates Th1 differentiation by inhibiting T-bet transactivation
SMAR1 is a MAR-binding protein that functions as a transcriptional repressor through its interaction with the MAR elements on target promoters and recruitment of histone-modifying complexes.27 As we observed that SMAR1 deficiency promotes Th1 and Th17 responses, to understand the mechanism of SMAR1 function, we carried out in silico analysis to identify the MAR elements on the promoters of T cell-expressed cytokine genes using the MAR-β sequence as the probe. We identified highly conserved AT-rich sequences on the promoters of T-bet and IL-17 genes (Supplementary Figure S4a), but not on IFN-γ and RORγt genes (data not shown).
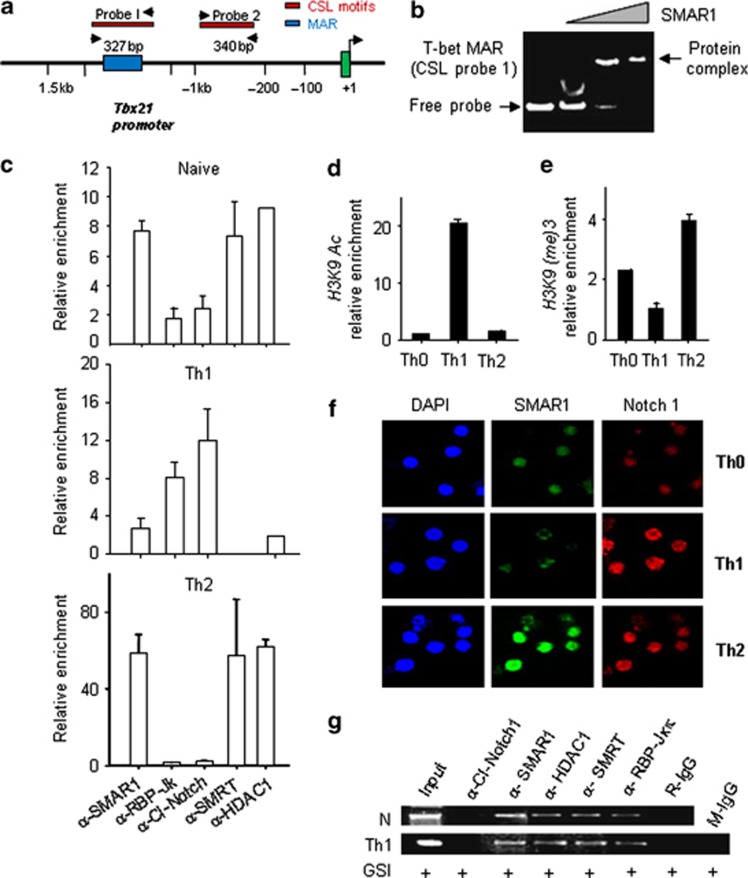
The cis regulatory MAR element on the T-bet promoter ∼800 bp upstream of the transcription start site coincided with a Notch-binding CSL element that has been shown to be important for T-bet expression28 (Figure 4a). To assess direct interaction of SMAR1 with the MAR region in the T-bet promoter, gel shift assays were performed using purified SMAR1 protein (harboring the DNA-binding domain; Supplementary Figure S4b) and a 327-bp T-bet MAR sequence as the probe. Generation of nucleoprotein complex was detected with increasing concentration of the purified protein showing direct binding of SMAR1 to the T-bet promoter (Figure 4b). Truncated SMAR1 protein lacking the DNA-binding domain did not bind to the probe (Supplementary Figure S4d). No binding of SMAR1 to the MAR sequence in IFN-γ promoter was detected (Supplementary Figure S4c). Next, a series of ChIP assays were carried out to determine the occupancy of various factors on the T-bet MAR region in Th1 and Th2 differentiated cells. As shown in Figure 4c, enrichment of SMAR1, SMRTe, and HDAC1 repressive complexes were observed on the T-bet promoter in naive and Th2 cells. In Th1 cells, T-bet MAR region was occupied by Notch 1 that should promote T-bet expression (Figure 4c, panel Th1).
Figure 4.
SMAR1 regulates type 1 helper T cell (Th1) differentiation through inhibition of T-bet (T-box protein expressed in T cells). (a) Schematic diagram of Tbx21 promoter and the regions (not to scale) amplified for chromatin immunoprecipitation (ChIP) assay by PCR. Probes I and II represent the two putative CSL consensus sites in the Tbx21 promoter. (b) Electrophoretic mobility shift assay (EMSA) experiment was carried out using T-bet matrix attachment region (MAR) probe (CSL probe I) and increasing concentrations (500 ng, 1 μg, and 2 μg) of purified SMAR1 protein. (c) ChIP of naive CD4+ T cells differentiated to Th1 or Th2 pathways. Figure represents relative enrichment of factors on the T-bet promoter under naive, Th1, and Th2 conditions. SMAR1, SMRTe, HDAC1, cleaved Notch-1, and RBPJκ antibodies were used to pull down the crosslinked DNA–protein complexes. DNA was precipitated and quantitative real-time PCR values were normalized with control IgGs to represent the data as relative enrichment (as described in Methods) of SMAR1, RBPJκ, SMRTe, Notch, and HDAC1 on the T- bet promoter. (d, e) Quantitative analysis by ChIP for enrichment of H3K9 acetylation and H3K9(me)3 respectively. (f) Analysis of Th1 and Th2 differentiated cells by confocal microscopy upon staining with anti-SMAR1 (green) and anti-Notch-1 (red). (g) ChIP assays performed on naive (N) CD4+ T cells or cultured under Th1 differentiation conditions±γ-secretase inhibitor (GSI; notch cleavage inhibitor).
To further study whether epigenetic modification of T-bet MAR region correlated with repressive complex, we characterized histone modifications at this locus during various T-cell differentiation programs. As expected, we observed enhanced occupancy of permissive H3K9 acetylation mark in Th1 cells (Figure 4d) and a repressive H3K9 (methylation)3 signature under naive and Th2 conditions (Figure 4e). These results collectively suggest that SMAR1 directly binds and represses T-bet gene expression by antagonizing Notch binding.
Our results suggest that SMAR1 is selectively induced in Th2 cells by GATA-3, but immunofluorescence analysis showed increased Notch expression in both Th1 and Th2 cells (Figure 4f). To understand the interplay of Notch 1 and SMAR1 on the T-bet promoter, the Notch pathway was inhibited by the γ-secretase inhibitor. The γ-secretase inhibitor may block the activation of cleaved Notch and its binding to the CSL region on T-bet promoter under Th1 condition. Under such condition, we assumed SMAR1 to occupy the T-bet promoter and repress the gene activation. As we hypothesized, lack of Notch activation mediated T-bet gene repression through SMAR1 and SMRT (Figure 4g). Taken together, these results suggest that SMAR1 regulates Th1 differentiation by fine-tuning the Notch-mediated transactivation of T-bet.
SMAR1 regulates Th17 differentiation through cis regulatory MAR element
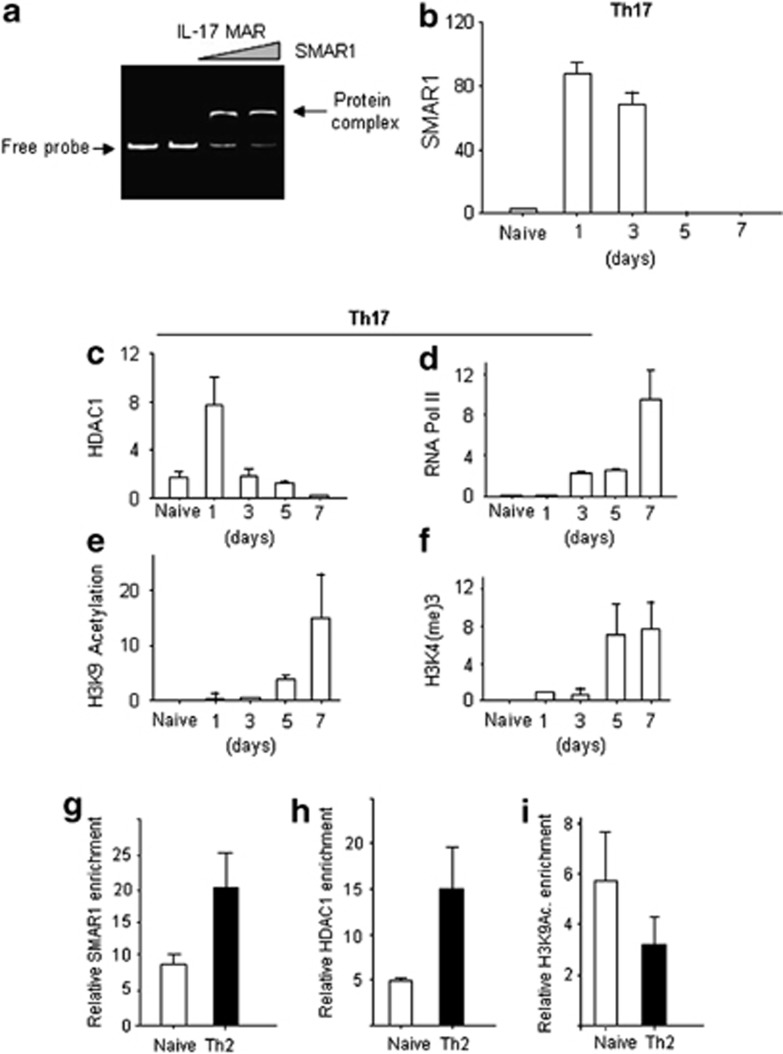
The MAR element on the IL-17 promoter is a highly conserved AT-rich region near the transcription start site (−1600 bp), similar to T-bet (Supplementary Figure S4a). To understand SMAR1-mediated regulation of IL-17 gene expression, the MAR region in the IL-17 gene was used as a probe for in vitro gel shift assays with purified SMAR1 protein. A specific shift was observed with IL-17 MAR DNA, suggesting that SMAR1 can bind to the IL-17 MAR region (Figure 5a). Next, ChIP assays were carried out using T cells at various time points after initiation of Th17 differentiation. SMAR1 recruitment on the IL-17 promoter was detected early after T-cell activation on day 1 (Figure 5b). We observed less enrichment of SMAR1 and HDAC1 (Figure 5b,c) on the MAR region during the course of Th17 differentiation. The release of SMAR1 after day 3 of polarization was inversely correlated with the transcriptional activation of the gene, as evidenced by enrichment of RNA Pol II (Figure 5d), positive epigenetic modifications, including H3K9 acetylation (Figure 5e) and H3K4(me)3 (Figure 5f). To validate the SMAR1 occupancy on the IL17 MAR region in Th2 cells, ChIP assays were carried out in Th0 (naive) and Th2 cells. Our data showed that SMAR1 (Figure 5g) and HDAC1 (Figure 5h) occupied the MAR region under Th2 condition. Moreover, SMAR1 enrichment was correlated with decreased epigenetic activation marker H3K9 acetylation (Figure 5i). These results suggest that SMAR1 directly binds to the IL-17 MAR region and removal of SMAR1 is necessary for the epigenetic activation of IL-17 promoter.
Figure 5.
SMAR1 controls type 17 helper T cell (Th17) differentiation. (a) Gel shift assay on 350 bp of interleukin-17 (IL-17) matrix attachment region (MAR) using increasing concentrations of purified SMAR1 protein. Naive T cells were isolated and differentiated under Th17 condition for various time points (1, 3, 5, and 7 days). Chromatin immunoprecipitation (ChIP) experiments were done on the MAR region of IL-17 gene with antibodies against (b) SMAR1, (c) RNA Pol II, (d) HDAC1, (e) H3K4(me)3, and (f) acetyl-H3K9. ChIP assay with the naive (Th0) and Th2 differentiated cells to show the relative enrichment of (g) SMAR1, (h) HDAC1, and (i) H3KAc. Relative enrichment was calculated by subtracting the Ct value with the control IgGs in the real-time PCR assays.
DISCUSSION
GATA-3 establishes and maintains Th2 pathway by activating Th2-related gene subsets and represses genes specific to other lineages.29 Although high-throughput genome studies have unraveled mechanisms of GATA-3-mediated global gene regulation by activation of the Th2 locus and repression of genes related to Th1 and Th17 pathways,30 what renders this specificity at target loci is not clearly known.11, 31 We report that the Th2-specific master regulator GATA-3 transcriptionally controls SMAR1 expression to facilitate the necessary chromatin orchestration specific to the Th2 subset. Thus, SMAR1 induction in Th2 cells might be a prerequisite for the maintenance of Th2 cells as SMAR1 deficiency compromised Th2 differentiation with increased Th1 and Th17 immune responses. Thus, SMAR1 might be functioning as a critical, cell-intrinsic molecular switch for the differentiation of T cells.
Since the discovery of SMAR1 from mouse double-positive thymocytes, SMAR1 has been shown to directly interact with various MAR elements and mediate transcriptional repression.32 It also facilitates epigenetic modulation of the chromatin by recruiting remodeling factors like Sin3A and HDAC1.27 Hence, we presumed SMAR1 to function as a transcriptional repressor by epigenetically silencing its target genes. We show that SMAR1 directly binds to MAR sequences present on T-bet and IL-17 promoters and represses the genes by recruiting SMRTe and HDAC1 complexes. On the T-bet promoter, the MAR sequence of SMAR1 binding coincides with a CSL element28 where Notch-1 signal induces T-bet expression. Thus, in Th2 differentiated cells, SMAR1 competes with Notch for its cognate site on T-bet promoter and forms transcriptional inactive complex.
A previous report showed regulation of the nuclear factor-κB pathway by SMAR1 by inhibition of IκB expression and accumulation of transactivation-deficient nuclear factor-κB in the nucleus.33 SMAR1 also inhibits tumor cell migration and metastasis by inhibiting tumor necrosis factor-α-mediated CD40 expression as well as its downstream target genes.34, 35 In line with this, we observed SMAR1 expression downregulated in Th1 and Th17 cells. It was also interesting to note that SMAR1 expression was induced with a delayed kinetics in developing Th2 cells that would allow nuclear factor-κB to promote GATA-3 expression early during Th2 differentiation.36 Thus, our results suggest that differentiation of naive T cells to Th1 and Th17 cells is correlated with reduced SMAR1 expression and its displacement from the promoter S/MAR sites of T-bet and IL-17 genes.
This report suggests SMAR1 as an important transcriptional repressor crucial for proper homeostatic regulation of T helper cell differentiation. In Th2 cells, GATA-3 induces SMAR1, and SMAR1 represses both T-bet and IL-17 gene expression through interactions with similar AT-rich MAR regions (Figure 6). It is important to note that global SMAR1 deficiency is embryonically lethal (data not shown), suggesting a fundamental role of SMAR1 in development. Our in vivo data show that SMAR1 deficiency in T cells attenuates eosinophilic airway inflammation in mice with significantly less cellular infiltration in the airways, mucus production, and airway hyperreactivity. The IgE/IgG2a profile showed a bias of T-cell response toward Th1 in these mice in response to antigen challenge. This study also put forward some unanswered questions that have to be addressed further in detail. We observed elevated cytokine response (increased IL-4, IL-13, and IFN-γ) and neutrophilic infiltration in the lungs of naive SMAR1cKO, suggesting the essential role of SMAR1 in the normal homeostasis of lung T-cell response. This could be possible because of the interaction between SMAR1-deficient T cells with other immune cells like basophils, mast cells, eosinophils, as well as other structural cells that secrete various cytokines and chemokines. However, the interplay of tissue-resident T cells with other immune/structural cells of the airway in the context of SMAR1 deficiency has to be explored further in detail. But why was this increased neutrophilia not observed in OVA-challenged mice also? This might be because of the antigen-specific skewing of T-cell response by OVA. OVA antigen is well known to stimulate type 2 allergic T-cell response in vivo with enhanced IL-4 and IL-13 production.4 The in vivo observations showed results in line with the previous reports that the balance between Th1 and Th2 cell types is critical for the establishment of OVA-specific antigen response and allergic airway disease3, 6, 13 over Th17 cell differentiation, necessitating distinct set of antigens under polarizing conditions.11, 16 Thus, in the context of a Th2-skewed condition of OVA-mediated antigenic allergic response, SMAR1 deficiency in T cells perturbs the Th1/Th2 balance. Notably, there was not much change in subepithelial fibrosis in spite of sufficient reduction of goblet cell metaplasia in SMAR1cKO OVA. This might be because of elevated levels of IL-4 and IL-5 and involvement of other recruited inflammatory cells such as monocytes, macrophages, and structural cells in the lungs in SMAR1cKO OVA that are also known to cause airway remodeling.37 Our preliminary data also suggest that SMAR1 plays important role in the regulation of Th17-mediated diseases (data not shown), and this needs further investigations. Taken together, we show a critical function of SMAR1 in allergic airway disease through the regulation of T-cell differentiation and its importance for further investigations to elucidate the stochastic interplay of SMAR1 and T cell-specific transcription factors. These findings have thrown some light on further understanding the molecular mechanism of asthma and other Th2-mediated diseases along with the possible SMAR1-targeted therapeutic implications.
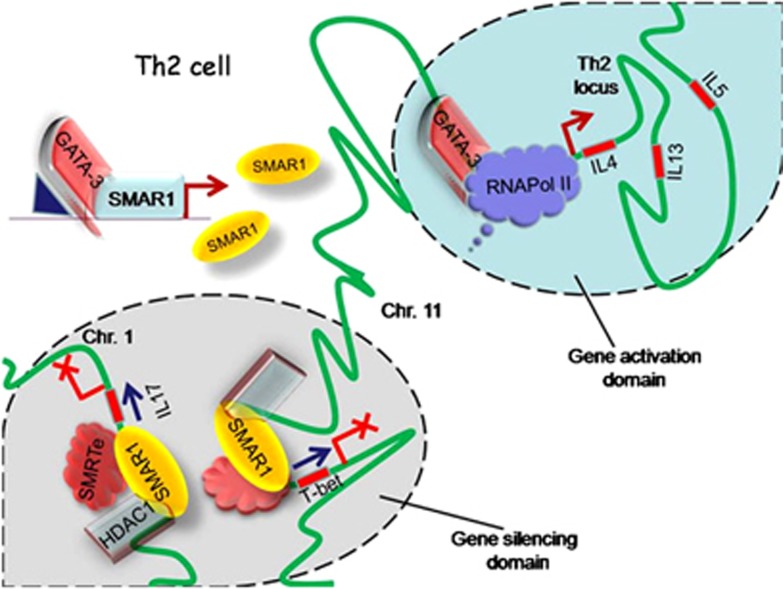
Figure 6.
Working model. In type 2 helper T (Th2) cells, GATA-3 induces SMAR1 expression. SMAR1 functions as a molecular switch in the maintenance of Th2 cells by inhibiting both Th1 and Th17 pathways by directly binding and suppressing the transcription of T-bet (T-box protein expressed in T cells) and IL-17 through specific matrix attachment region (MAR) sequences present on the promoters. Binding of SMAR1 recruits chromatin-modifying complexes that keep the gene silenced presumably by targeting these genes to specific silencing domains within the nucleus. This model also hypothesizes differential regulation of genes (T-bet and Th2 locus) present in the same chromosome (Chr). Chr. 11. In Th2 cells, T-bet is silenced and IL-4/IL-13/IL-5 genes are kept transcriptionally active possibly through spatial recruitment of genes to various subnuclear compartments. Thus, SMAR1 functions as a critical molecular switch in dictating the stability of Th2 cells by silencing Th1 and Th17 pathways.
Methods
Mice. T cell-specific (Lck (early)-Cre SMAR1f/f) conditional knockout mice (SMAR1cKO) were custom generated on C57/BL6 background at Ozgene (Betley DC, WA, Australia) using the strategy outlined in Supplementary Figure S2a. Exon 2 of SMAR1 was flanked by loxP sites. A phosphoglycerine kinase-neo cassette flanked by Flp recombinase target sites (data not shown) was used for selection. Following homologous recombination of the vector in C57BL/6 embryonic stem cells and establishment of germline transmission, the PGK-neo cassette was excised using the FLP recombinase, leaving exon 2 flanked by loxP sites. SMAR1f/f mice were crossed with early Lck-Cre transgenic mice to generate SMAR1cKO mice. The specificity of the deletion was further confirmed by western blotting techniques. SMAR1 transgenic mice (SMAR1 Tg) were generated38 at the National Institute of Immunology (New Delhi, India). WT C57BL/6J and all other mice were bred in the experimental animal facility of the National Centre for Cell Science (Pune, India). Animal experiments were done with 6–8-week-old mice using protocols approved by the Institutional Animal Ethical Committee of National Centre for Cell Science.
Flow cytometry. Anti-CD4, anti-CD8, anti-CD62L, anti-CD44, anti-CD25, anti-IFN-γ, anti-IL-17, and anti-IL-4 were purchased from BD Bioscience (San Jose, CA). For purification of T-cell subsets, CD4+ T cells were first enriched by magnetic-activated cell sorting beads (autoMACS; Miltenyi Biotec, San Jose, CA), and then further purified using FACS ARIA (BD Bioscience). Gating strategy is shown in Supplementary Figure S1d. Sorting of naive T cells was carried out with a flow rate of not more than 5,000 events per second, and purity of >97% was achieved.
In vitro T-cell polarization. Sorted naive CD4+CD25−CD62LhighCD44low T cells were activated with plate-bound anti-CD3e (1 μg ml−1; 2C11) and anti-CD28 (2 μg ml−1; 37.51) in the presence of polarizing cytokines and/or blocking antibodies. For Th1 polarization, IFN-γ (10 ng ml−1), IL-12 (5 ng ml−1), and anti-IL-4 (10 μg ml−1; 11B11) were used. For Th2 polarization, anti-IFN-γ (10 μg ml−1; XMG1.2) and IL-4 (10 ng ml−1 or doses indicated in figures) were used. For Th17 polarization, IL-6 (20 ng ml−1) and transforming growth factor-β (5 ng ml−1; R&D Systems, Minneapolis, MN) with anti-IFN-γ and anti- IL-4 were used. All the antibodies were obtained from BD Pharmingen (San Jose, CA) unless mentioned otherwise. Cells were cultured in RPMI-1640 medium (GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum and CD4+ T cells primed for 4 days in wells were expanded on day 4 with/without (for Th17 culture) 50 U ml−1 IL-2 until day 7.
Intracellular cytokine staining. For intracellular cytokine staining, in vitro cultured cells were restimulated with phorbol 12-myristate 13-acetate (Sigma, St. Louis, MO, 50 ng ml−1) and ionomycin (Sigma, 1 mM) for 6 h. Golgistop `or Monensin' (BD Bioscience) was added during the last 4 h of restimulation. After staining for extracellular markers, cells were fixed, permeabilized, and stained for specific cytokines. The cytokine fluorophores used were IFN-γ-FITC, IL-17-PE, CD4-PerCP Cy5.5, and IL-4-APC (BD Bioscience). Cells were acquired on a FACS CANTO flow cytometer (BD) and data analyzed with FACS DIVA software (San Jose, CA).
OVA-induced allergic asthma. Allergic airway inflammation was induced as described previously.25, 26 SMAR1cKO and control littermate mice (10–12 weeks old) were immunized intraperitoneally with 50 μg of OVA (Sigma) in 200 μl (4 mg) of alum (Sigma) on days 0, 7, and 14. At 1 week after immunization, mice were challenged with aerosolized 3% OVA dissolved in phosphate-buffered saline (PBS) or PBS only for 30 min per day for 7 days consecutively. Mice were assessed for airway hyperresponsiveness with invasive flexi-Vent method as previously described.26 At 24 h after the last challenge, mice were killed. Bronchoalveolar lavage fluid was collected by injecting 1 ml PBS into the lungs through a tracheal cannula and then gently aspirating the fluid. This was repeated three times. The bronchoalveolar lavage fluid was centrifuged and total cells in the pellet were counted by using a hemocytometer. Differential cell counts were performed on cytospins of cells stained with Leishman stain. For lung histology, the upper lobe of the left lung was fixed with 10% formaldehyde overnight, dehydrated, embedded in paraffin, cut into 4 μm sections, and stained with hematoxylin and eosin, periodic acid–Schiff, or Masson's trichrome stains. Morphometry was performed on periodic acid–Schiff- and Masson's trichrome-stained lung sections to estimate epithelial mucin content and subepithelial collagen content.25
OVA-specific IgE and IgG2a measurement. OVA-specific IgE and IgG2a were measured from the sera of the mice as described previously.25 In brief, each well of microtiter plate was coated with 2 μg of OVA (IgG2a) and 5 μg of OVA (IgE) (chicken egg OVA grade V; Sigma-Aldrich, St. Louis, MO) in 100 μl volume overnight at 4 °C. After three washes, nonspecific sites were blocked with 1% BSA in PBS. Mouse sera in duplicate were added to the Ag-coated wells, the plates were incubated, and bound IgE or IgGs were detected with biotinylated anti-mouse IgE or anti-mouse IgG2a (BD Pharmingen). Streptavidin-peroxidase conjugates were added; the bound enzymes were detected by addition of tetramethylbenzidine substrate system (BD Pharmingen); and absorbances were read at 450 nm. Absorbances were converted to arbitrary units.
Preparation of lung homogenates and cytokine analysis. Lungs were isolated and lung homogenates were prepared as previously described.26 In brief, lungs were homogenized in protein extraction buffer (Tris-HCl, NP-40, NaCl, EDTA, NaN3, and PMSF at pH 7.5) with freshly added protease inhibitors (dithiothreitol, leupeptin, and aprotinin) using mortar–pestle and lysate centrifuged at 27,000 g for 20 min. Supernatants were collected and protein concentration determined using protein assay BCA Kit (Bio-Rad Laboratories, Hercules, CA). Cytokines in the lung homogenates were measured by enzyme-linked immunosorbant assay (ELISA) and plotted against concentration of total protein. Similarly, cytokine levels in the supernatants were measured by ELISA. IL-4, IL-5, IL-13, and IFN-γ were measured with OptEIA kits (BD Pharmingen); IL-17 was measured with DuoSet ELISA kits (R&D Systems). ELISA plates were developed with TMB substrate (BD Pharmingen), and read with a microplate reader (Bio-Rad, Hercules, CA).
Quantitative real-time PCR and western blotting. Gene expression was examined using a thermocycler (Eppendorf, Hauppauge, NY) with an iQ SYBR Green Real-Time PCR kit (Bio-Rad Laboratories, Hercules, CA). Data were normalized to expression of gapdh (reference gene). Transcripts of Tbx21 (T-bet) Fwd: 5′-AGAGTCGCGCTCAGCAACCACCT-3′, Rev: 5′- TATCAACAGATGCGTACATGG-3′, IFN-γ Fwd: 5′AACGCTACACACTGCCATCTTGG-3′, Rev: 5′-CTCATGAATGCATCCTTTTTCG-3′, IL-4 Fwd: 5′-CTAGTTGTCATCCTGCTCTTCTTT-3′, Rev: 5′-CTTTAGGCTTTCCAGGAAGTCTTT-3′, IL-17 Fwd: 5′-CTCCAGAAGGCCCTCAGACTAC-3′, Rev: 5′-GGGTCTTCATTGCGGTGG-3′, GATA-3 Fwd: 5′-GAAGGCATCCAGACCCGAAAC-3′, Rev: 5′-ACCCATGGCGGTGACCATGC-3′, SMAR1 Fwd: 5′-GCATTGAGGCCAAGCTGCAAGCTC-3′, Rev: 5′-CGGAGTTCAGGGTGATGAGTGTGAC-3′, GAPDH Fwd: 5′-AATTCAACGGCACAGTCAAAGCCGAGAATG-3′, Rev: 5′-GCGGCACGTCAGATCCACGCAGGAC-3′ were amplified by PCR. For immunoblot analysis, 50 μg of protein amounts from whole T-cell lysates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and probed using T-bet (Santa Cruz, Dallas, TX), SMAD1/2/3 (Santa Cruz), SMAR1 (Bethyl, Montgomery, TX), and β-actin (Santa Cruz) antibodies using a standard protocol.27
ChIP assay. ChIP assays were done as previously described.38 In brief, naive CD4+ T cells polarized under various conditions were treated with 1% formaldehyde to crosslink protein to DNA. Cells were lysed and chromatins were sheared to ∼500 bp using a bioruptor (Diagenode, Rue Bois Saint-Jean, Seraing, Belgium). Cells were then subjected to immunoprecipitation with anti-HDAC1 (Santa Cruz), anti-SMAR1 (Bethyl), anti-Notch-1 (Santa Cruz), anti-cleaved Notch-1 (Santa Cruz), anti- GATA-3 (Santa Cruz), anti-SMRT (Santa Cruz), or anti-RBP-Jκ (Santa Cruz) that recognizes CSL. Isotype controls were as follows: for RBP-Jκ, anti-Notch-1, anti-SMAR1, anti-HDAC1, purified rabbit IgG; for cleaved Notch-1, CBP/p300, mouse IgG were used. After de- crosslinking, the immunoprecipitated chromatin DNA was PCR amplified for two regions of the Tbx21 promoter containing putative CSL-binding consensus motifs (Probe I, Fwd: 5′- CACAGCCCCTACCCCCAATACGAA-3′ and Rev: 5′-CCCCCAGCCCCGAGGATGATG-3′ Probe II, Fwd: 5′-CTGGGCATACAGGAGGCAGCAACAAAT-3′ and Rev: 5′- GTCCCCTCACCCCGCCACCTTG-3′). Also, two regions of Smar1 promoter (SPr I, Fwd: 5′- GCCACTGAACGAGCCCGGAA-3′ and Rev: 5′-AGTCTCTGCGGCCATGATTT-3′ SPrII, Fwd: 5′- GGCCTTCATCACCATCCTAG-3′ and Rev: 5′-ATTGTCTGGAAAGTTGCCTG-3′P) corresponding to GATA-binding consensus sequences was amplified. For IL-17 MAR region, Fwd: 5′- ATGGTGGCTCACAACCATTT-3′ and Rev: 5′-CAACATAGGTCTTCATGGATTACTTT-3′ were used. PCR conditions used were: 95 °C for 5 min, 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 45 s (40 cycles), and 72 °C for 10 min. The resulting DNA was also analyzed by real-time PCR to determine the relative enrichment (2^ (ΔCt of the control−ΔCt of the sample).39
Immunofluorescence staining. Purified naive CD4+ T cells were cultured under neutral (Th0), Th1, and Th2 differentiation conditions. Cells were harvested and fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and incubated with anti-SMAR1 (Bethyl), anti-GATA-3, or anti-Notch1 (Santa Cruz) antibodies. They were counterstained using the anti-mouse or anti-rabbit fluorescently labeled secondary antibodies (Millipore, Billerica, MA). For nuclear staining, cells were treated with 4,6-diamidino-2-phenylindole dihydrochloride (Fluka, St. Louis, MO). Cytospin was done to mount the cells onto the confocal slides. Coverslips were then mounted with fluorescent mounting media (Dako, Carpinteria, CA) and examined under a Zeiss LSM-510 Meta Confocal Laser Scanning Microscope (Carl Zeiss MicroImaging, Oberkochen, Germany).
Electrophoretic mobility shift assay. Probes were prepared from mouse genomic DNA by PCR-amplifying the T-bet and IL-17 MAR regions. Each binding reaction contained 50 ng probe, 1 μg poly(deoxyinosinic-deoxycytidylic) acid as nonspecific competitor, 1 × binding buffer (20 mM Hepes, pH 7.9, 60 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, and 4% glycerol) in a final volume of 10 μl with increasing concentration (500 ng, 1 μg, and 1.5 μg) of purified His-tag SMAR1 protein. His-tag protein binding domain of SMAR1 was purified using the His-tag system. Binding was carried at room temperature for 30 min and then reactions were loaded onto a 7.5% polyacrylamide gel and exposed to ethidium bromide. Excess ethidium bromide was then washed off vigorously and picture was taken using Versa Doc gel imager (Bio-Rad).
Statistical analysis. For all experiments unless stated otherwise, the unpaired Student's t-test was applied with Graph Pad Prism software (La Jolla, CA) to all data points. Correlation among the data in the animal studies was examined using Pearson's correlation test. P-values of <0.05 were considered significant.
Acknowledgments
We thank Dr Sekhar Mande, Director, National Centre for Cell Science. We acknowledge Rakhshinda Rehman, IGIB, New Delhi for technical support. This work is funded by NCCS, Department of Biotechnology (DBT), University Grants Commission (UGC), Indian Council of Medical Research (ICMR), and Council for Scientific and Industrial Research (CSIR), Government of India. We thank Professor Anuradha Ray, Professor of Medicine and Immunology, UPMC Montefiore Hospital, Pittsburgh, for the critical review of the manuscript.
The authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
Supplementary Material
References
- 1Description of the global burden of NCDs, their risk factors and determinants. Global status report on noncommunicable diseases, WHO, 2011, ISBN: 978 92 4 156422 9.
- 2Alcorn, J.F. et al. TH17 cells in asthma and COPD. Ann. Rev. Physiol. 72, 495–516 (2010). [DOI] [PubMed] [Google Scholar]
- 3Cohn, L. et al. Asthma; mechanisms of disease persistence and progression. Ann. Rev. Immunol. 22, 789–815 (2004). [DOI] [PubMed] [Google Scholar]
- 4Galli, S.J. & Tsai, M. IgE and mast cells in allergic disease. Nat. Med. 18, 693–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Licona-Limón, P. et al. TH2, allergy and group 2 innate lymphoid cells. Nat. Immunol. 6, 536–542 (2013). [DOI] [PubMed] [Google Scholar]
- 6Rosenberg, H.F. et al. Eosinophils: changing perspectives in health and disease. Nat. Rev. Immunol. 13, 9–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Holgate, S.T. et al. Treatment strategies of allergy and asthma. Nat. Rev. Immunol. 8, 218–230 (2008). [DOI] [PubMed] [Google Scholar]
- 8Zhu, J., Yamane, H. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Nakayamada, S., Takahashi, H., Kanno, Y. & O'Shea, J.J. Helper T cell diversity and plasticity. Curr. Opin. Immunol. 3, 297–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Murphy, K.M. & Reiner, S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2, 933–944 (2002). [DOI] [PubMed] [Google Scholar]
- 11Kanno, Y. et al. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 30, 707–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12O'Shea, J.J. & Paul, W.E. Mechanism underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Szabo, S.J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000). [DOI] [PubMed] [Google Scholar]
- 14Zheng, W. & Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997). [DOI] [PubMed] [Google Scholar]
- 15Ivanov, I.I. et al. The orphan nuclear receptor ROR-γ directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- 16Manel, N. et al. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 9, 641–649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Zhou, L., Chong, M.M. & Littman, D.R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009). [DOI] [PubMed] [Google Scholar]
- 18Spilianakis, C.G., Lalioti, M.D., Town, T., Lee, G.R. & Flavell, R.A. Interchromosomal associations between alternatively expressed loci. Nature 435, 637–645 (2005). [DOI] [PubMed] [Google Scholar]
- 19Mirkovitch, J., Mirault, M.E. & Laemmli, U.K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39, 223–232 (1984). [DOI] [PubMed] [Google Scholar]
- 20Gasser, S.M. & Laemmli, U.K. The organization of chromatin loops: characterization of a scaffold attachment site. EMBO J. 5, 511–518 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Heng, H.H. et al. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 117, 999–1008 (2004). [DOI] [PubMed] [Google Scholar]
- 22Chattopadhyay, S. & Pavithra, L. MARs and MARBPs: key modulators of gene regulation and disease manifestation. Subcell. Biochem. 41, 213–230 (2007). [PubMed] [Google Scholar]
- 23Chattopadhyay, S., Kaul, R., Charest, A., Housman, D. & Chen, J. SMAR1, a novel, alternatively spliced gene product, binds the Scaffold/Matrix-associated region at the T cell receptor beta locus. Genomics 1, 93–96 (2000). [DOI] [PubMed] [Google Scholar]
- 24Kaul-Ghanekar, R. et al. Abnormal V(D)J recombination of T cell receptor beta locus in SMAR1 transgenic mice. J. Biol. Chem. 280, 9450–9459 (2005). [DOI] [PubMed] [Google Scholar]
- 25Mabalirajan, U. et al. Mitochondrial structural changes and dysfunction are associated with experimental allergic asthma. J. Immunol. 181, 3540–3548 (2008). [DOI] [PubMed] [Google Scholar]
- 26Aich, J. et al. Loss-of-function of inositol polyphosphate-4-phosphatase reversibly increases the severity of allergic airway inflammation. Nat. Commun. 3, 877 (2012). [DOI] [PubMed] [Google Scholar]
- 27Rampalli, S. et al. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell Biol. 25, 8415–8429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Minter, L.M. et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 7, 680–688 (2005). [PubMed] [Google Scholar]
- 29Zhu, J. et al. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 1, 3–10 (2006). [DOI] [PubMed] [Google Scholar]
- 30Wei, G. et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Smale, S.T. & Fisher, A.G. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 20, 427–462 (2002). [DOI] [PubMed] [Google Scholar]
- 32Malonia, S.K. et al. Gene regulation by SMAR1: role in cellular homeostasis and cancer. Biochim. Biophys. Acta. 1815, 1–12 (2011). [DOI] [PubMed] [Google Scholar]
- 33Singh, K. et al. Tumor suppressor SMAR1 represses IκB expression and inhibits p65 transactivation through matrix attachment regions. J. Biol. Chem. 284, 1267–1278 (2009). [DOI] [PubMed] [Google Scholar]
- 34Singh, K. et al. p53 target gene SMAR1 is dysregulated in breast cancer: its role in cancer cell migration and invasion. PLoS One 2, 8.e660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Singh, K., Sinha, S., Malonia, S.K. & Chattopadhyay, S. Tumor necrosis factor alpha (TNFalpha) regulates CD40 expression through SMAR1phosphorylation. Biochem. Biophys. Res. Commun. 391, 1255–1261 (2010). [DOI] [PubMed] [Google Scholar]
- 36Das, J. et al. A critical role for NF-kappa B in GATA-3 expression and TH2 differentiation in allergic airway inflammation. Nat. Immunol. 1, 45–50 (2001). [DOI] [PubMed] [Google Scholar]
- 37Cho, J.Y. et al. Inhibition of airway remodeling in IL-5 deficient mice. J. Clin. Invest. 113, 551–560 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Suveera, D. & Subeer, S.M. Transgenesis via permanent integration of genes in repopulating spermatogonial cells in vivo. Nat. Methods 5, 601–603 (2008). [DOI] [PubMed] [Google Scholar]
- 39Nelson, J.D. et al. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protocols 1, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.