Abstract
Regulatory T (Treg) cells are essential for self-tolerance and immune homeostasis. Transcription factor Foxp3, a positive regulator of Treg cell differentiation, has been studied to some extent. Signal transducer and activator of transcription factor 3 (STAT3) is known to negatively regulate Foxp3. It is not clear how STAT3 is regulated during Treg differentiation. We show that SMAR1, a known transcription factor and tumor suppressor, is directly involved in maintaining Treg cell fate decision. T-cell-specific conditional knockdown of SMAR1 exhibits increased susceptibility towards inflammatory disorders, such as colitis. The suppressive function of Treg cells is compromised in the absence of SMAR1 leading to increased T helper type 17 (Th17) differentiation and inflammation. Compared with wild-type, the SMAR1−/− Treg cells showed increased susceptibility of inflammatory bowel disease in Rag1−/− mice, indicating the role of SMAR1 in compromising Treg cell differentiation resulting in severe colitis. We show that SMAR1 negatively regulate STAT3 expression favoring Foxp3 expression and Treg cell differentiation. SMAR1 binds to the MAR element of STAT3 promoter, present adjacent to interleukin-6 response elements. Thus Foxp3, a major driver of Treg cell differentiation, is regulated by SMAR1 via STAT3 and a fine-tune balance between Treg and Th17 phenotype is maintained.
INTRODUCTION
Disruption of immune suppression contributes to progression of autoimmune diseases. Regulatory T (Treg) cells are essential for maintenance of immune homeostasis and organization of controlled immune responses.1 Dysregulated function of Treg cells could account for various immune disorders. In particular, it limits the magnitude of effector responses leading to failure to adequately control infection and inflammation.2 Treg cells also subside inflammation due to microbial immune responses, including commensals.3 Upon activation, naive CD4+ T cells differentiate into different lineages of helper T (Th) cells that are characterized by distinct developmental regulation and biological functions.4 Activation of naive T cells with immunoregulatory cytokine transforming growth factor (TGF)-β and pleotropic cytokine interleukin (IL)-2 in the absence of IL-6 induces a distinct transcriptional factor Foxp3, which dictates the cell toward induced Treg (iTreg) cells.5, 6 It suggests that signaling molecules and transcription factors downstream of TGF-β and IL-2 receptor must work together to induce Treg differentiation. TGF-β alone can generate Foxp3+ Treg cells both in vitro and in vivo, and in the presence of IL-2, a potent inhibition of TGF-β-driven Th17 cells is achieved.7 Moreover, STAT5 activation through IL-2 can restrict retinoic acid–related orphan receptor γt (RORγt), which is a transcriptional factor specific for Th17 lineage commitment.8 Th17 cells are one of the T-cell subtypes that secretes proinflammatory cytokine IL-17; it drives intraepithelial lymphocyte (IEL) accumulation and promotes Th1 response that enhances the inflammatory cascade.9 Th17 differentiation mostly necessitates the requirement of inflammatory cytokine IL-6, IL-23, and immunoregulatory cytokine TGF-β.10 STAT3 is a downstream signaling molecule that get activated by IL-6 and IL-23, which in turn transactivate RORγt. STAT3 and RORγt are also reported to bind to IL-17 promoter and activate its gene transcription.11, 12, 13 Moreover, it was reported that the STAT3 signaling pathway negatively regulates CD4+CD25+Foxp3+ Treg cells during the development of anti-inflammatory responses.14, 15, 16 STAT3 binds to CNS2 region of Foxp3 promoter and maintains its expression.17, 18 Thus Foxp3 gene transcription is under the tight control of specific cooperative external stimuli, contributed by positive regulators such as TGF-β and IL-2 and negative regulators such as IL-6 and IL-23.19
Thus T-cell polarization to different lineage commitment depends mostly on the induction of the genes for transcriptional factors and cytokines specific to particular cell type. This requires the coordination of nuclear matrix proteins at the chromatin level with the specific arrangement of chromatin domains distinct to the cell. Nuclear matrix proteins are integral part of the nucleus that has crucial roles in the maintenance and stability of chromatin conformation necessary for the functionality of a particular cell.20, 21 Such arrangement is facilitated by the anchorage of specific sequences of the DNA to the nuclear matrix. MARs (matrix-associated regions) are known to provide binding sites for specific proteins (MAR-binding proteins or MARBPs) that can influence the transcription of associated gene loci.22 Different modalities of this complex interaction can dictate the conformation of chromatin loop and thus the repression of a gene in the loci.23, 24
SMAR1 (scaffold/matrix attachment region binding protein 1), is one such nuclear matrix–associated protein that was initially isolated from the thymocyte library and was shown to be highly expressed in the double-positive T cells where it interacts with the MARβ sequence, located 400 bp upstream of the T-cell receptor β (TCRβ) enhancer and is highly expressed during the double-positive stage of thymocyte development.25 SMAR1 causes chromatin remodeling through the recruitment of histone deacetylases (HDACs) and de-acetylation of histones.24 These chromatin modifications favor the formation of repressor complex in the loci and thus suppress the gene transcription.26, 27 SMAR1 shares significant homology with other MARBPs such as Cux/CDP, Bright, and SATB1 in the MAR-binding domains. Among them, SATB1 is T lineage–enriched chromatin organizer and regulates T-cell differentiation.28 SMAR1 was identified as an immuno-modulator, and its role in immune responses was studied in detail. SMAR1 transgenic mice were developed, and it showed significant perturbation in the immune responses.29 Thus studying the chromatin changes during the differentiation of CD4+ T cells to their effector and regulatory phenotype by SMAR1 has relevance in understanding the molecular events that drive Foxp3 gene expression and stabilize Treg cell phenotype.
To elucidate the role of SMAR1 in regulation of Treg phenotype, we used Foxp3-expressing Treg cells from SMAR1F/F/Lck-Cre mice (T-cell-specific conditional knockout mice, represented as SMAR1−/−) and found that SMAR1 deletion in Treg cells lead to higher susceptibility toward inflammatory disorders. Adoptive transfer of SMAR1−/− Treg cells does not protect the colitis development in Rag1−/− mice. It exhibits compromised immune-suppressive function and homeostasis; establishing the role of SMAR1 in maintaining the Treg cell function. Perturbation of SMAR1 in T cells blocked the TGF-β1-induced generation of Foxp3+ Treg cells, and it promotes differentiation of Th17 cells in vitro and in vivo. SMAR1 has a pivotal role in maintaining STAT3 expression; it binds to the regulatory region of STAT3 promoter, which in turn favors Foxp3 expression leading to Treg cell differentiation. This study reveals a critical role of SMAR1 in maintaining the fine-tune balance between Treg and Th17 cell function, and it gives a novel insight into the immune-regulatory mechanisms.
RESULTS
SMAR1−/− mice are highly susceptible to acute dextran sodium sulfate (DSS)-induced colitis
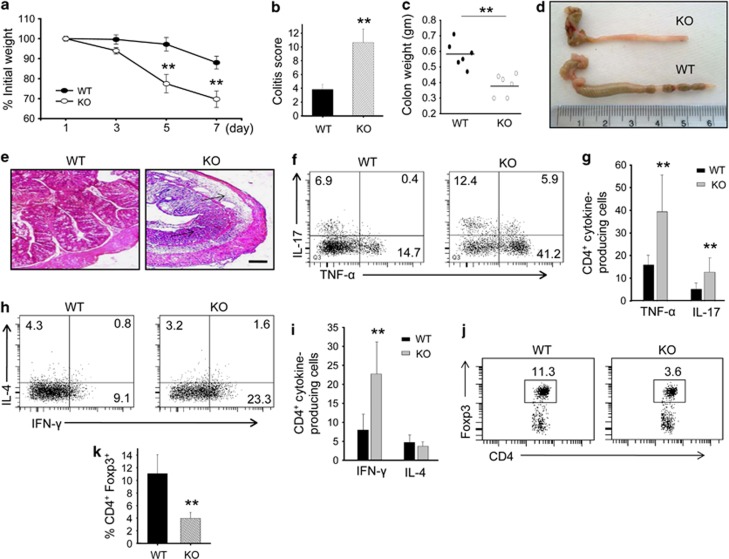
Using mouse model, we have shown that overexpression of SMAR1 perturbs both CD4+ and CD8+ T-cell differentiation. Additionally, these mice exhibited dysregulated V(D)J recombination associated with differential regulation of Vβ-specific T cells.29 It is known that CD4+ T effector lymphocytes have an important role during intestinal homeostasis.30, 31 With this in mind, we have generated Lck-Cre-driven T-cell-specific SMAR1 knockout mice. To confirm whether SMAR1 may have an influence on effector T-cell phenotype, we checked the SMAR1−/− CD4+ T lymphocytes and found that these lymphocytes are effector phenotype (CD62LlowCD44hi) in nature (Supplementary Figure S1a online). Further, we checked steady-state absolute number of T-cell (CD4+ and CD8+) count from the thymus, spleen and mesenteric lymph node from age- and gender-matched adult unmanipulated wild-type (WT) and SMAR1−/− mice and found that SMAR1−/− mice is not lymphopenic in nature (Supplementary Figure S1b). In order to evaluate the role of adaptive immunity on CD4+ T effector phenotype in intestinal homeostasis, we used SMAR1−/− and WT mice as experimental model of intestinal inflammation. DSS (3%) was administered through drinking water and monitored for disease progression. SMAR1−/− mice were more pronounced to DSS-induced colitis as compared with WT mice. The severity of progression of colitis was further judged by body weight loss, diarrhea, and rectal bleeding. Although there was progressive body weight loss in both WT and SMAR1−/− mice, it was even more severe in SMAR1−/− mice. After 7 day of DSS treatment, the weight loss of SMAR1−/− mice was 25±5% while the loss of body weight was about 5% for WT mice (Figure 1a). By day 7, SMAR1−/− mice showed severe diarrhea associated with rectal bleeding. Although SMAR1−/− mice showed colitis score of 10.5±1.5, for WT it was 3.5±0.5 (Figure 1b), indicating severity of disease progression upon reduced expression of SMAR1. Additionally, gross examination of colon showed features of severe colitis more pronounced in SMAR1−/− mice that includes atrophied, thin-walled, swollen, opaque proximal colon associated with severe bleeding and stool inconsistency. Both cecum and colon size were significantly shorter and quantified by measuring organ weights, which were notably less in SMAR1−/− mice (Figure 1c). The disease activity index based on rectal bleeding, stool consistency, and detection of blood in stool was significantly higher (P-value <0.001) in SMAR1−/− mice than in WT mice at day 7 of DSS treatment (Supplementary Figure S1c). Macroscopic evidence of colonic damage was not observed in the SMAR1−/− mice without DSS challenge. Colonic damage, especially effect in the transverse and descending segments of colon, increase in the ratio of colon length to weight, shortening of the colon, was examined only in mice with DSS treatment. In this report, compared with DSS-treated WT mice, SMAR1−/− mice had considerably shorter colon and elevated ratio of colon length to weight (Figure 1d). The level of colonic injury characterized by epithelial loss, crypt damage, ulceration, and inflammatory cell infiltration was considerably higher in SMAR1−/− mice (Figure 1e). DSS untreated SMAR1−/− mice did not show any inflammatory bowel disease symptoms that includes massive infiltrations of inflammatory leukocytes, loss of goblet cells as well as colonic inflammation. We then checked the levels of proinflammatory cytokine taking cells from colonic lamina propria (LP). We observed higher secretion of proinflammatory cytokine such as interferon (IFN)-γ, IL-17 and tumor necrosis factor (TNF)-α in SMAR1−/− mice. There was no apparent change in the level of IL-4 indicating that Th2 pathway remains unaltered (Figure 1f–i). In addition to increased colonic inflammation, SMAR1−/− mice showed more immunoglobulin A (IgA) in the circulation compared with WT mice (Supplementary Figure S1d). The colonic LP and IELs of SMAR1−/− mice had less CD4+Foxp3+ Treg cells than WT mice (Figure 1j,k). The CD4+CD8+ IELs of SMAR1−/− mice also had more proinflammatory Th1 and Th17 cells (Supplementary Figure S1e). Nevertheless, the overall ratio of Th17 cells to Foxp3+ Treg cells was significantly higher in SMAR1−/− mice than in WT mice. Thus the data demonstrate that SMAR1 regulates the reciprocal differentiation of Treg and Th17 cells in vivo in response to a chemical-induced experimental colitis.
Figure 1.
SMAR1−/− mice are highly susceptible to acute dextran sodium sulfate (DSS)-induced colitis. (a) Body weight changes shown as the percentage of initial weight of wild-type (WT), SMAR1−/− mice treated with DSS. Data represent mean±s.e.m. of n=6 mice/genotype. P-value were calculated with Student's t-test; **P<0.002 (mice are taken from same breed and co-caged). (b) Stool consistency and rectal bleeding were monitored at seventh day of DSS administration, and colitis was scored for each mouse. Colitis was graded on a scale of 0–12 as described in the Methods. Data represent the mean±s.e.m. P-value were calculated with Student's t-test; **P<0.001. (c) Colon weight of DSS-treated mice was measured on day 7. Data represent the mean±s.e.m. P-values were calculated with Student's t-test; **P<0.006. (d) Representative colon gross anatomy of SMAR1−/− and WT mice on seventh day after the start of DSS treatment. (e) Representative hematoxylin and eosin staining of colon section of a similar region (arrow) from SMAR1−/− and WT mice on seventh day after the start of DSS treatment. Bar=100 μm. (f–i) Flow cytometry of the intracellular expression of interleukin (IL)-17 and tumor necrosis factor (TNF)-α (f,g) or of interferon (IFN)-γ and IL-4 (h,i), in CD4+ T cells in colonic lamina propria (LP) of SMAR1−/− and WT mice on seventh day after the start of DSS treatment. (j,k) Flow cytometry of intracellular Foxp3 and surface CD4 in colonic LP of SMAR1−/− and WT mice on seventh day after the start of DSS treatment and frequency of CD+Foxp3+ cells (k) among CD4+ T cells. The numbers in the quadrants (f,h,j) indicate the percentage of cells in each. P-values were calculated with Student's t-test; **P<0.001. Data are a representative of three independent experiments with six mice per group. KO, knockout.
Compromised function of CD4+Foxp3+ Treg cells during development of acute intestinal inflammation in SMAR1−/− mice
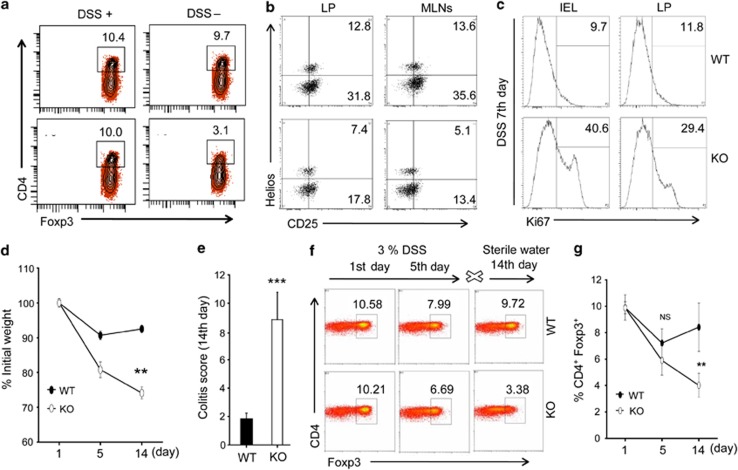
SMAR1−/− mice exhibited increased T-cell-dependent acute intestinal autoimmune inflammation. Treg cells are present in LP and involved in suppression of intestinal autoimmune inflammation.2 We therefore examined how the Treg cell population was affected in the absence of SMAR1 and whether these mice exhibit defective phenotype of CD4+Foxp3+ Treg cells. SMAR1−/− mice showed significantly 3–4-fold lower percentage of CD4+Foxp3+ Treg cells in the colonic LP than WT mice during development of acute colitis (Figure 2a). We observed that both CD25+Helios+ and CD25+Helios− Treg were lower in number both in colon LP and mesenteric lymph nodes of SMAR1−/− mice. Thus natural Treg (nTreg) and iTreg cells are affected in the absence of SMAR1 during colonic inflammation (Figure 2b). Interestingly, in the course of colonic inflammation, there was 3.5–4.5-fold higher numbers of Ki67+ cells among CD4+CD25− T-cell population in SMAR1−/− mice (Figure 2c). Thus SMAR1−/− mice showed increased effector T-cell activation along with lower number of Treg cell population. We presumed that in SMAR1−/− mice decreased Foxp3 expression might contribute to generation of increased proinflammatory responses. To prove this, we treated mice with 3% DSS for 5 days and left it to recover using normal water as detailed in Methods. SMAR1−/− mice were highly susceptible to this treatment by day 14 and showed disease symptoms as described before. In contrast, WT mice survived over this time course and maintained their body weight while the body weight of SMAR1−/− mice decreased gradually by 15–20% at day 5 and lost >25% of their initial body weight at day 14 (Figure 2d). SMAR1−/− mice developed rectal bleeding and diarrhea early on day 7 and showed severe symptoms of colitis (colitis score 10±2) and showed no improvement from day 6 to day 14. However, severe diarrhea and rectal bleeding was not observed by day 7 through day 14 in WT mice (Figure 2e). Next to determine whether SMAR1 deficiency affected generation and function of Foxp3+ Treg cells in vivo, we isolated colonic LP cells from SMAR1−/− and WT mice at initial stage, at fifth day, and at fourteenth day and checked the frequency of Foxp3+ Treg cells. Interestingly, the frequency and overall percentage of Foxp3+ Treg cells in SMAR1−/− mice progressively decreased and had lost >6% of their initial Treg cells at day 14. In contrast, the frequency and total number of Foxp3+ Treg cells in WT mice was gradually restored at day 14 (Figure 2f). The recovery of Treg cells in WT mice was mainly due to expansion of Treg population (Figure 2g). These results indicate that SMAR1 might be involved in stabilizing as well as maintaining Foxp3+ Treg response during acute intestinal inflammation.
Figure 2.
Compromised function of CD4+Foxp3+ Treg cells during development of acute intestinal inflammation. (a,b) Flow cytometry of CD4+ T cells in the colon lamina propria (LP) and mesenteric lymph nodes (MLNs) at seventh day of dextran sodium sulfate (DSS) administration of SMAR1−/− and wild-type (WT) mice. The numbers in the plot indicate the percentage of CD4+Foxp3+ (a) and CD25+Helios+ cells gated on CD4+ T cells (b). (c) Flow cytometry analysis of Ki67+ frequency within gated CD4+CD25− T cells in SMAR1−/− and WT mice after seventh day of DSS treatment. (d) Weights of SMAR1−/− and WT mice were recorded starting at first day, fifth day of DSS treatment and at fourteenth day of sterile water treatment. Asterisks indicate statistical significance determined by Student's t-test (**P<0.001). (e) Colitis scores at fourteenth day, using the criteria described in Methods. P-value were calculated using Student's t-test (***P<0.0005). (f,g) Frequency of CD4+Foxp3+ Treg cells in colon LP of SMAR1−/− and WT mice at first day, fifth day of DSS treatment and fourteenth day of sterile water treatment, the number in the plot indicate the percentage of CD4+Foxp3+ (f) and graphical representation of percentage of CD4+Foxp3+ cells (mean±s.e.m.) in the colon LP (g). **P<0.005 (Student's t-test). Data represent mean±s.e.m. of n=6 mice/genotype. Results are representative of three independent experiments. IEL, intraepithelial lymphocyte; KO, knockout.
SMAR1−/− mice showed upregulation of the gut-homing markers in T cells
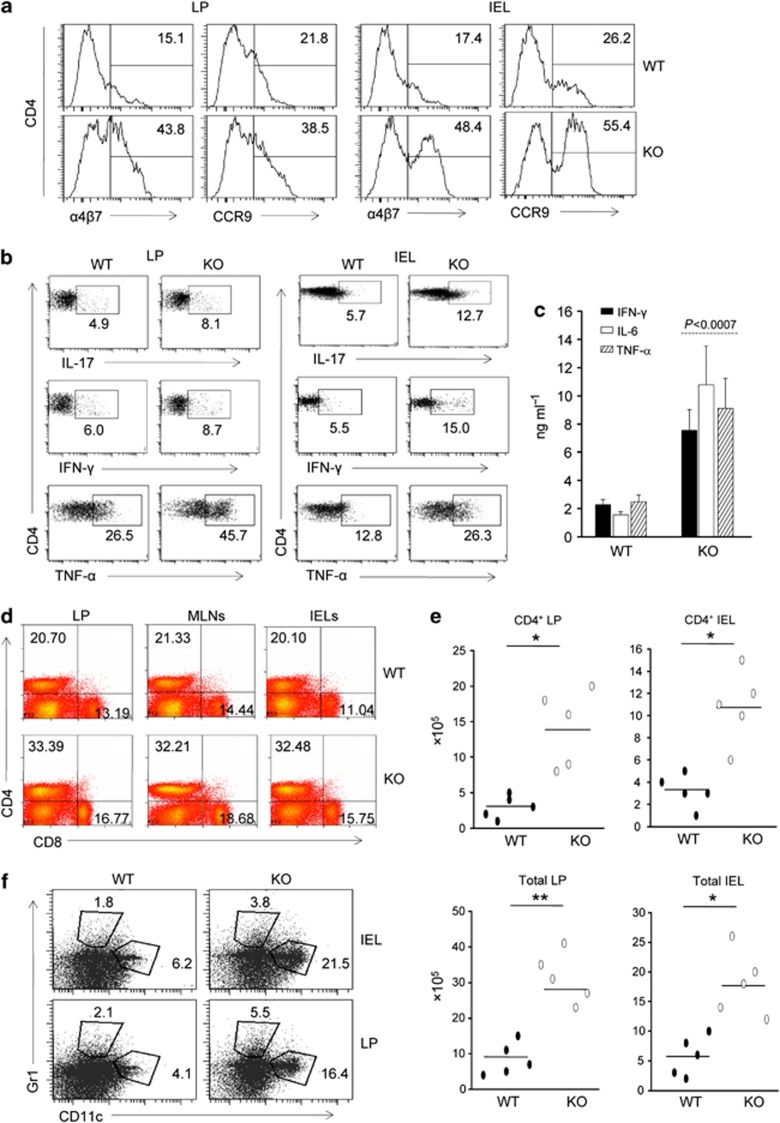
To find the cause of increase CD4+ T-cell accumulation in the colon of SMAR1−/− mice, we checked the level of gut-homing markers during development of acute colitis. In addition to CD44 (Supplementary Figure S2a), both integrin C-C motif chemokine receptor 9 (CCR9) and α4β7 in the colon LP and IELs were increased substantially to 2–3.5-fold in SMAR1−/− mice compared with WT mice (Figure 3a). Our data indicate that the increased accumulation of CD4+ T cells in SMAR1−/− mice is due to dysregulated expression of gut-homing markers. The proinflammatory Th1 and Th17 cytokines have a major impact on inflammatory bowel disease progression.31 We therefore checked the levels of proinflammatory cytokines in colonic T cells during disease progression. Noticeable increase in the Th1 and Th17 cytokine-producing cells in the colon of DSS-treated SMAR1−/− mice were observed (Figure 3b). At the same time, 5–6-fold increase in proinflammatory and inflammatory cytokines from total colonic LP cells from SMAR1−/− mice was observed (Figure 3c). Additionally, the SMAR1−/− mice shows increased IEL, LP, and mesenteric lymph node CD4+ CD8+ cell population and 3±1-fold increased number of CD4+ and total IEL as well as CD4+ and total LP cells (Figure 3d,e) in addition to CD19+ B-cell population (Supplementary Figure S2b) compared with WT mice. Hematoxylin and eosin staining of the diseased colon samples showed some of the polymorphonuclear leukocytes. The flow cytometry data showed that there was also major increase in the dendritic cell and neutrophil populations in the colon, both in colonic LP and IEL compartment (Figure 3f). All these observations clearly reveal that there was accumulation of CD4+ T lymphocytes and production of proinflammatory cytokines in the colonic LP in addition to neutrophils and dendritic cells, which collectively contribute to disease progression.
Figure 3.
Colons of SMAR1−/− mice showed upregulation of the gut-homing markers in T cells and presents large amount of infiltrating cells during acute dextran sodium sulfate (DSS) colitis. (a) Frequencies of CD4+ T cells expressing α4β7 integrin and C-C motif chemokine receptor 9 (CCR9) in the colonic lamin propria (LP) and intraepithelial lymphocytes (IELs) of SMAR1−/− and wild-type (WT) mice. (b) Flow cytometry of intracellular cytokine interleukin (IL)-17, interferon (IFN)-γ, and tumor necrosis factor (TNF)-α production by CD4+ T cells isolated from the colon LP and epithelium (IEL) of SMAR1−/− and WT mice after seventh day of DSS administration. (c) Enzyme-linked immunosorbent assay of IFN-γ, IL-6, and TNF-α in culture supernatant of total colonic LP cells of SMAR1−/− and WT mice. P-value were calculated with Student's t-test (P<0.0007). (d) Frequencies of CD4+ and CD8+ T cells among the colon LP, epithelium (IEL) and mesenteric lymph nodes (MLNs) evaluated by flow cytometry during acute DSS colitis. (e) Absolute number of CD4+ T cells and total cells in the colon LP and epithelium (IEL) compartment. Asterisks indicate statistical significance as determined by Student's t-test (*P<0.01, **P<0.007). (f) Frequencies of neutrophil, dendritic cells in the colon LP, and epithelial compartment (IEL) are shown. The numbers in the histogram indicate the percentage of CD11c+, Gr1+ population within the gated CD4−CD8− population. The data are representative of three independent experiments with six mice per group. KO, knockout.
SMAR1-deficient Treg cells showed altered suppressive activity in vitro
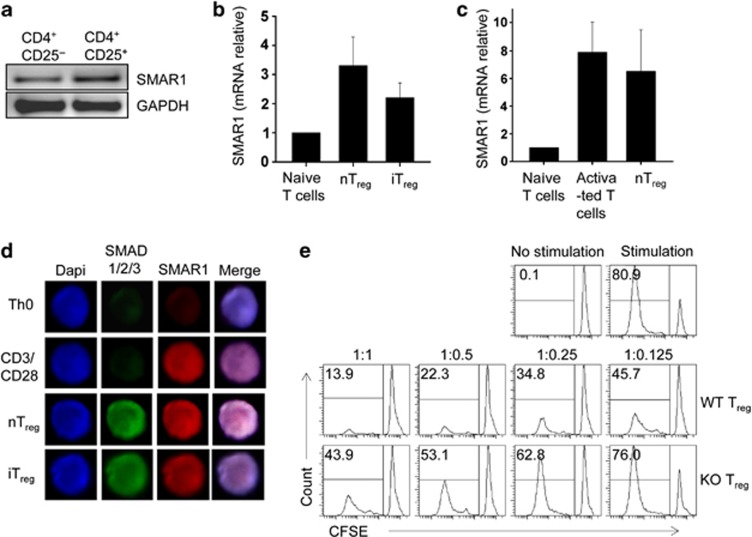
Transcription factors are important for directing the differentiation of proinflammatory Th1 and Th17 cells. Some of them were recently shown to have unique and essential roles in controlling Foxp3+ Treg cell function.32, 33 The polarization of naive CD4+ T cells toward Th1 and Th17 cells showed 4–5-fold downregulation of SMAR1 (Supplementary Figure S3a); therefore, to assess the involvement of SMAR1 in generation and function of Treg cells, SMAR1 expression was checked in purified CD4+CD25+ Treg cells and CD4+CD25− non-Treg cells isolated from WT mice. Treg cells expresses high level of SMAR1 compared with non-Treg cells (Figure 4a). iTreg cells and in vitro activated nTreg cells also showed high level of SMAR1 expression (Figure 4b). SMAR1 expression in nTreg was further confirmed by mRNA expression from both in vitro activated CD4+ T cells as well as nTreg. The results showed that in vitro activated nTreg expresses equivalent level of SMAR1 compared with in vitro activated CD4+ T cells (Figure 4c). Addition of IL-2 alone was sufficient to induce SMAR1 expression in Treg, upon TCR stimulation (Supplementary Figure S3b,c). Confocal imaging of in vitro generated iTreg and in vitro activated nTreg showed drastic increase in SMAD 1/2/3 expression and translocation of SMAD 1/2/3 to nucleus, demonstrating that SMAD 1/2/3 is only activated under Treg-polarizing condition. It is also found that SMAR1 is expressed more in Treg-inducing conditions compared with unstimulated control cells (Figure 4d), suggesting the strong possibility of epigenetic control of SMAR1 during Treg lineage commitment. Furthermore, we looked into what functions of Treg cells were altered by SMAR1 deletion. The number of conventional T cells expressing integrin α4β7 and CCR9 were found to be more in SMAR1−/− mice. This clearly suggests that the suppressive activity of Treg cell over conventional T cell is altered in the absence of SMAR1. Elevated levels of conventional CD4+ T cell produce high amount of proinflammatory cytokines in the colon leading to inflammation, even though Treg cells are present in the colon. To address this issue, we directly evaluated the immune-suppressive activity of SMAR1-deficient Treg cells; we used an in vitro suppression assay and observed that SMAR1-deficient Treg cells showed reduced suppressive activity and were unable to control proliferation of coexisting effector CD4+ T cells with the similar efficiency as WT Treg cells (Figure 4e). Moreover, this was not due to a major deficiency in their maintenance in vitro (Supplementary Figure S3d). Therefore, SMAR1 is important to maintain intact immune-suppressive function of Treg cells in vitro.
Figure 4.
SMAR1-deficient regulatory T (Treg) cells showed altered suppressive activity in vitro. (a) Quantitative real-time PCR (RT-PCR) analysis of SMAR1, control to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in freshly isolated CD4+CD25+ Treg cells and CD4+CD25− non-Treg cells in wild-type (WT) mice. (b) Quantitative RT-PCR analysis of SMAR1, presented as mRNA expression relative to β-actin, in in vitro activated natural Treg (nTreg) cells and naive CD4+ T cells isolated and differentiated under induced Treg (iTreg). (c) Naive CD4+ T cells and nTreg cells were cultured for 5 days in vitro with splenic dendritic cells in the presence of anti-CD3 antibody and 2 ng ml−1 of recombinant interleukin-2. The expression of SMAR1 was measured by quantitative RT-PCR. The relative expression was normalized by β-actin. Data are shown as mean±s.e.m. of normalized SMAR1 mRNA in three independent experiments. No statistical significance was observed. (d) SMAD1/2/3 and SMAR1 were indirectly fluorescein isothiocyanate or Cy3 labeled and observed by confocal microscopy in naive CD4+ T cells cultured in neutral, activated, transforming growth factor-β1 iTreg- and nTreg-inducing conditions. (e) Sorted CD4+CD45RBhiCD25− carboxyfluorescein succinimidyl ester (CFSE)-labeled WT T cells were activated in the presence of soluble anti-CD3 and anti-CD28 and co-cultured with sorted CD4+CD25+ Treg cells from WT and SMAR1−/− mice at the indicated ratio of conventional T cells to WT or knockout (KO) Treg cells (conventional T cell/Treg cells). The proliferation of CD4+ responder T cells was assessed by analyzing the CFSE dilution with flow cytometry. The results are representative of four independent experiments, each with three pair of mice.
SMAR1 is essential for TGF-β1-induced Foxp3 expression in Treg cells
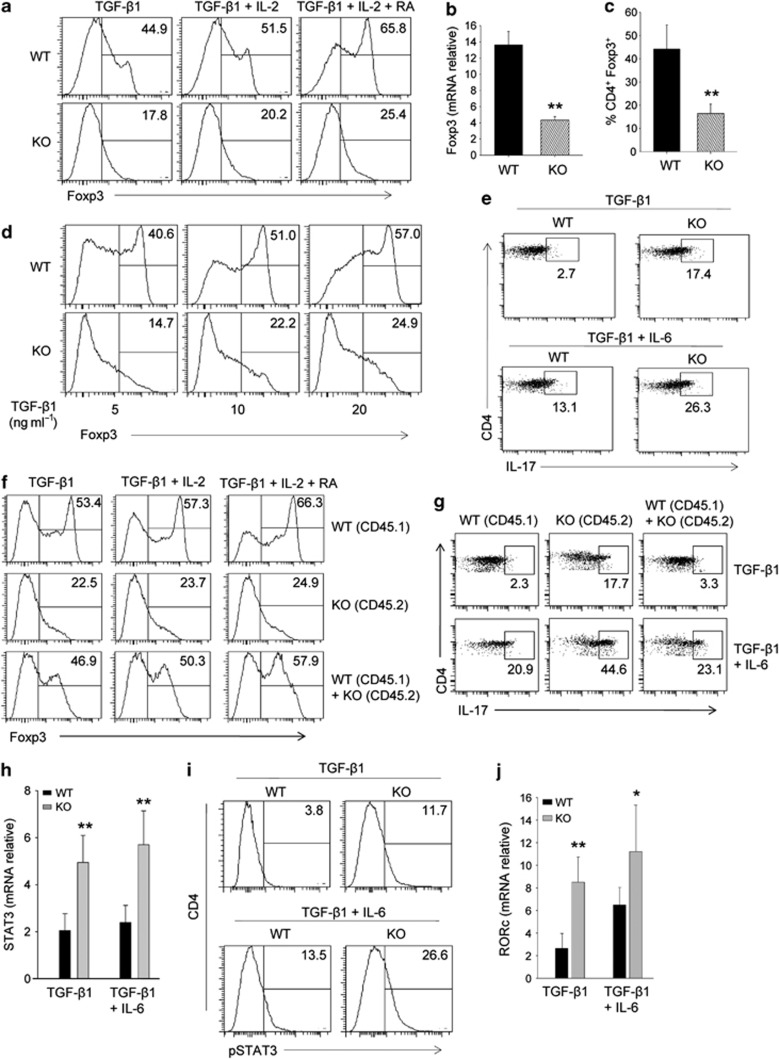
We assessed whether in the absence of SMAR1 the generation of Foxp3+ Treg cells is altered. Naive CD4+CD62LhiCD25− T lymphocytes were cultured in the presence of anti-CD3 and anti-CD28 along with TGF-β1 to polarize cells toward induced Foxp3+ Treg cells. Compared with WT, SMAR1−/− CD4+ T cells expressed about 3–4-fold reduced both Foxp3 mRNA and protein expression. (Figure 5a–c). The similar experiment was carried out by using different concentration of TGF-β1 (5, 10, 20 ng ml−1). Fluorescence-activated cell sorter (FACS) analysis showed 2.5–3.5-fold decreased Foxp3 expression in SMAR1−/− CD4+ T cells (Figure 5d). Further treatment with TGF-β1 along with either IL-2 or retinoic acid (RA) did not significantly alter the pattern, indicating that the major effect in the change of Foxp3 expression is primarily due to TGF-β1 (Figure 5a). We could not also detect any significant changes in the phosphorylation of SMAD2/3 that are the targets of TGF-β1 in SMAR1−/− CD4+ T cells (Supplementary Figure S4a). Thus, for the generation of Foxp3+ Treg cells, it is possible that SMAR1 has a role downstream of SMAD2/3. Several groups have reported the role of transcription factors in the regulation of balance between Treg and Th17 cells during inflammation. We therefore checked whether SMAR1 is involved in maintaining such balance. Although TGF-β promotes Foxp3 expression, IL-6 has been implicated in regulating IL-17 via STAT3. Naive CD4+CD62LhiCD25− T lymphocytes were cultured with TGF-β1 and/or IL-6. We speculate that TGF-β driven IL-17 production could be enriched in Treg cells in the absence of SMAR1. Treatment with TGF-β1 plus stimulation with anti-CD3 and anti-CD28 resulted in 5–6-fold increased number of IL-17-producing cells among SMAR1−/− naive CD4+ T cells compared with WT (Figure 5e). Treatment with IL-6 along with TGF-β1 increased the Th17 cells, with significantly high secretion of IL-17 in SMAR1−/− CD4+ T cells (Figure 5e). In order to further assess whether SMAR1 directly regulates T-cell fate, we studied the T cells from very young (3-week old) SMAR1−/− mice, which do not yet have effector/memory phenotype (Supplementary Figure S4b) and have not showed any surface upregulation of early activation marker CD69 (Supplementary Figure S4c). To this end, naive CD4+ T cells from congenically distinct 3-week-old WT (CD45.1) and SMAR1−/− (CD45.2) mice were differentiated in vitro towards Treg and Th17 lineage commitment and congenically distinct WT (CD45.1) and SMAR1−/− (CD45.2) T cells were co-cultured. At the conclusion of this experiment, TGF-β1 induced SMAR1−/− Treg cells in co-culture with WT Treg cells, expressed an equivalent level of Foxp3 and IL-17 compared with WT Treg cells alone (Figure 5f,g), and treatment with IL-2 and RA shows the similar trend. Similarly, treatment with IL-6 along with TGF-β1 co-cultured WT (CD45.1) and SMAR1−/− (CD45.2) T cells expressed a comparable level of IL-17 (Figure 5g). When we look in to upstream of IL-17 signaling pathway, treatment with TGF-β1 induces 3–4-fold higher expression of STAT3 and Rorc in SMAR1−/− CD4+ T cells (Figure 5h–j). Addition of IL-6 provides considerable enhancement (Figure 5h–j). However, TGF-β1 alone did not perturb the Th17 polarization of SMAR1−/− CD4+ T cells, because Foxp3 expression has not been restored. Therefore, SMAR1-deficient CD4+ T cells are more prone to express Th17 cell cytokines.
Figure 5.
SMAR1 is essential for transforming growth factor (TGF)-β1-induced Foxp3 expression in Treg cells. (a) Flow cytometry of intracellular expression of Foxp3 in naive CD4+ T cells cultured with TGF-β1 alone, TGF-β1 plus interleukin (IL)-2, TGF-β1 plus IL-2, and retinoic acid (RA) along with T-cell receptor (TCR) stimulation. The numbers in the plot indicate the percentage of Foxp3+ cells. Data are representative of one experiment representative of five. (b) Quantitative analysis of Foxp3 mRNA expression in naive CD4+ T cells in the presence of TGF-β1 plus IL-2 with TCR stimulation, presented relative to the expression of β-actin. P-values were calculated with Student's t-test (**P<0.001). (c) Frequencies of CD4+Foxp3+ T cells among TGF-β1-, IL-2-, and RA-treated cells in a. P-values were calculated with Student's t-test (**P<0.002). (d) Naive CD4+CD25− T cells were cultured with anti-CD3 and anti-CD28 in the presence of indicated doses of TGF-β1 for 5 days. Foxp3+ T cells were determined by flow cytometry. Data are shown as the percentage of CD4+Foxp3+ cells in one experiment representative of five. (e,i) Flow cytometry of intracellular IL-17 (e) and phosphorylated signal transducer and activator of transcription factor 3 (pSTAT3) (i) in naive CD4+ T cells from SMAR1−/− and wild-type (WT) mice 6 weeks of age and cultured for 5 days with TCR stimulation plus TGF-β1 with or without IL-6 (above plots). The numbers in plots indicate the percentage of IL-17+CD4+ and pSTAT3+CD4+ cells. (f) Flow cytometry of intracellular expression of Foxp3 from 3-week-old WT (CD45.1), SMAR1−/− (CD45.2), and co-cultured (WT+SMAR1−/−) naive CD4+ T cells treated with TGF-β1 alone, TGF-β1 plus IL-2, TGF-β1 plus IL-2, and RA along with TCR stimulation. The numbers in the plot indicate the percentage of Foxp3+ cells. Data are representative of one experiment representative of five. (g) Flow cytometry of intracellular IL-17 from 3-week-old WT (CD45.1), SMAR1−/− (CD45.2), and co-cultured (WT+SMAR1−/−) naive CD4+ T cells cultured for 5 days with TCR stimulation plus TGF-β1 with or without IL-6 (above plots). The numbers in the plots indicate the percentage of IL-17+CD4+ cells. Data are representative of one experiment representative of five. (h, j) Quantitative analysis of STAT3 (h) and Rorc (j) mRNA in naive CD4+ T cells 5 days after activation with TCR and TGF-β1 treatment with or without IL-6 (horizontal axis), presented relative to β-actin expression. **P<0.002, *P<0.01 (Student's t-test). KO, knock out.
SMAR1-deficient TGF-β1-induced Treg cells showed altered levels of cytokine genes expression
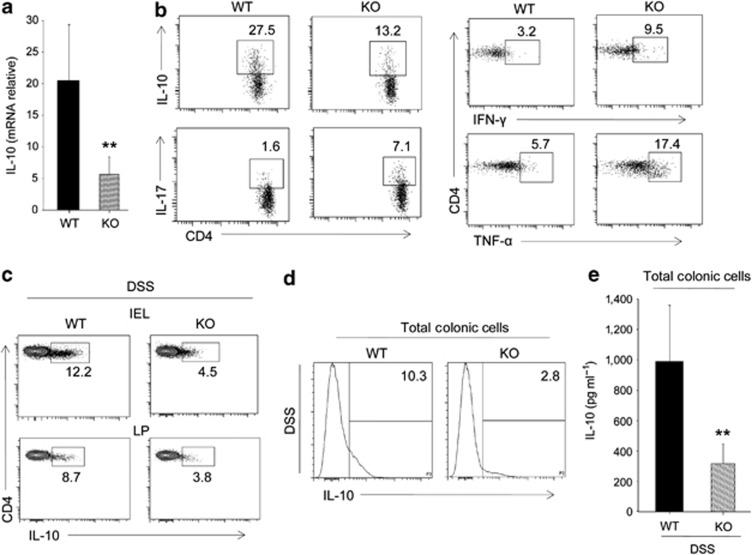
We checked the role of SMAR1 on TGF-β1-induced Treg cells. Notably, Foxp3 expression was downregulated along with genes that are important for Treg phenotype. The levels of surface marker CD25 remain unchanged, while CTLA-4, CD103, and Icos were decreased in SMAR1-deficient Treg cells (Supplementary Figure S5a). The result indicates SMAR1-deficient Treg cells have reduced suppressive function. Moreover, it has also been found that an IL-10 mRNA level was reduced by fourfold (Figure 6a). IL-10 is a well-known anti-inflammatory cytokine and has a significant role in Treg-suppressive function, especially in colitis. IL-10−/− mice develop spontaneous inflammation of the colon.34, 35 Not only mRNA level of IL-10 were downregulated in SMAR1-deficient Treg cells but notably the protein level were also reduced (Figure 6b). In addition, several effector Th1 and Th17 cell cytokines, including IL-17, IFN-γ, and TNF-α, were found to be upregulated (Figure 6b). Also Foxp3+ Treg cells from SMAR1−/− mice showed no difference in the level of NK1.1 compared with WT Treg cells (Supplementary Figure S5b). IL-10-producing CD4+ Treg cells are also essential to control immune responses during DSS-induced colitis.36 To investigate the role of IL-10 in colitis condition of SMAR1−/− mice, we checked IL-10 production. We found that the level of IL-10 in CD4+ T cells were less in SMAR1−/− mice compared with WT mice (Figure 6c). Moreover, there was 3.5-fold decrease in total colonic IL-10 level in SMAR1−/− mice as compared with WT mice (Figure 6d,e). These data collectively reveal that SMAR1-deficient Treg cells expressed altered levels of several effector cytokine genes and reduced level of IL-10, which are essential for its suppressive activity. It suggests that SMAR1 controls the expression of proinflammatory cytokine genes and maintain effector function of Treg cell lineage commitment.
Figure 6.
SMAR1-deficient transforming growth factor (TGF)-β1-induced Treg cells showed reduced expression of interleukin (IL)-10 and elevated levels of proinflammatory cytokine genes. (a) Amount of IL-10 mRNA in TGF-β1 induced Treg cells from wild-type (WT) and SMAR1−/− mice evaluated by quantitative real-time PCR (**P<0.001, Student's t-test). (b) Flow cytometry of intracellular production of IL-10, IL-17, interferon (IFN)-γ, and tumor necrosis factor (TNF)-α in TGF-β1-induced Treg cells from WT and SMAR1−/− mice. The numbers in the plot indicate the percentage of CD4+ T cells. Data are representative of three independent experiments with similar results. (c,d) Flow cytometry analysis of intracellular IL-10 in CD4+ T cells (c) and total colonic cells (d) isolated from colon lamina propria (LP) and epithelium (intraepithelial lymphocyte (IEL)) at seventh day of dextran sodium sulfate (DSS)-treated WT and SMAR1−/− mice. The numbers in the plots indicate the percentage of cells in each; data are representative of three independent experiments with six mice per group. (e) Enzyme-linked immunosorbent assay analysis of IL-10 levels in total colonic LP cells at seventh day of DSS-treated WT and SMAR1−/− mice (mean±s.d. six mice per group). Data represent three independent experiments (**P<0.002, Student's t-test). KO, knock out.
SMAR1-deficient Treg cells do not protect colitis
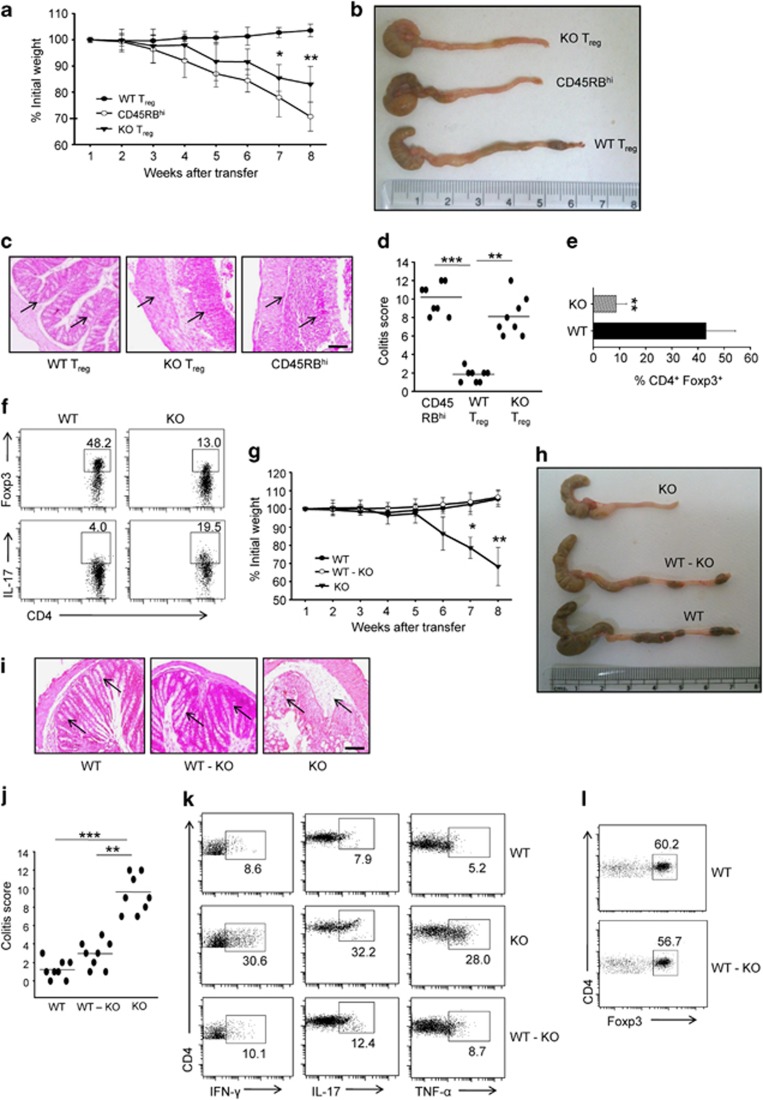
To evaluate whether deficiency of SMAR1 could have functional consequences on the capacity of Treg to control colitis, we used a well-known depicted model of T-cell transfer colitis. Colitogenic CD4+CD45RBhiCD25− T lymphocytes were adoptively transferred into Rag1−/− mice either alone or along with WT Treg and SMAR1−/− Treg. As anticipated, colitogenic T lymphocytes initiate severe colonic inflammation coupled with considerable weight loss, which was suppressed when WT Treg were co-transferred (Figure 7a). On the contrary, SMAR1−/− Treg when co-transferred with colitogenic T lymphocytes failed to suppress inflammation and weight loss (Figure 7a). Consistently, mice co-transferred with SMAR1−/− Treg showed augmented colonic inflammation associated with major infiltration of inflammatory cells as compared with mice co-transferred with WT Treg (Figure 7b–d). The Rag1−/− mice co-transferred with SMAR1−/− Treg cells fail to control inflammation due to downregulation of Foxp3 expression and elevated production of proinflammatory cytokine IL-17 compared with mice co-transferred with WT Treg cells (Figure 7e,f). SMAR1-deficient CD4+CD45RBhiCD25− T cells, when injected into Rag1−/− mice together with WT Treg cells, showed prevention of colonic inflammation (Figure 7g,h). Hematoxylin and eosin staining showed normal structure of the colon, with typical crypts containing profused goblet cells, and some negligible infiltrates (Figure 7i,j). Flow cytometric analysis confirmed noticeable decrease in the percentage of CD4+ T lymphocytes infiltrating the colon, in addition to CD4+ T lymphocytes producing IFN-γ, TNF-α, and IL-17 (Figure 7k). Collectively, these data suggest that SMAR1 control behavior of Treg cells in colonic inflammatory milieu. Control of disease consequently with WT Treg cell was caused by maintenance of suppressive function and stability of Foxp3 expression in the presence of SMAR1 (Figure 7l). These results reveal that provision of WT Treg cells along with SMAR1-deficient CD4+CD45RBhiCD25− T cells prohibited inflammatory bowel disease, and alteration in SMAR1-deficient Treg cells are especially the cause of inflammatory bowel disease.
Figure 7.
SMAR1-deficient regulatory T (Treg) cells do not protect colitis. (a) Rag1−/− mice received either CD4+CD45RBhiCD25− alone or together with CD4+CD25+ Treg from wild-type (WT) and SMAR1−/− mice, and weights of mice were assessed weekly. Asterisks specify statistical significance between WT and SMAR1−/− Treg cell transferred, *P<0.01, **P<0.003 (Student's t-test). (b) Colon gross anatomy representative for CD4+CD25+ Treg transferred from WT and SMAR1−/− mice to age- and sex-matched Rag1−/− mice. (c) Hematoxylin and eosin staining of colon sections of age- and sex-matched Rag1−/− mice. Section images are from similar areas along the length of colon. Arrow indicates infiltration of lymphocytes and monocytes. Bar=100 μm. (b,c) Data are representative of six pairs of mice of three independent experiments. (d) Colitis score of mice (see Methods for scoring). Each symbol represents one mouse, and crossbar depict the mean of eight mice analyzed, **P<0.003, WT Treg compared with SMAR1−/− Treg; ***P<0.0005 CD45RBhi compared with WT Treg. (e,f) WT and SMAR1−/− Treg were harvested from the colonic LP of Rag1−/− mice and stained for CD4, intracellular Foxp3, and interleukin (IL)-17; graph represents the percentage of CD4+Foxp3+ cells from colonic lamina propria (LP) of recipient Rag1−/− mice, **P<0.001 (Student's t-test). The numbers in the plots refer to the percentage of each subset. Data shown are representative of three independent experiments with similar results. Data are presented as mean±s.e.m. (g) Sorted CD4+CD45RBhiCD25− T cells from WT and SMAR1−/− mice were transferred into 6-week-old Rag1−/− mice in combination with CD4+CD25+ WT Treg and SMAR1−/− Treg, as described in Methods, and the weights of the mice were assessed weekly. Asterisks show statistical significance between the Treg cells transferred from WT and SMAR1−/− mice. (*P<0.005, **P<0.001; determined by Student's t-test). (h) Colon gross anatomy of SMAR1−/− CD4+CD45RBhiCD25− T cell transferred along with SMAR1−/− Treg and WT Treg (WT-KO (knockout)) transferred into Rag1−/− mice. (i) Representative hematoxylin and eosin–stained photomicrographs of colon sections from mice in h, arrow indicates same region and infiltration of lymphocytes and monocytes. Bar=100 μm. (j) Graphical representation of colitis score, conducted as described in Methods. (**P<0.003, ***P<0.0005; determined by Student's t-test) Each symbol represents one mouse, and crossbar depicts the mean of eight mice analyzed. (k) Flow cytometry of intracellular cytokine, namely, interferon (IFN)-γ, IL-17, and tumor necrosis factor (TNF)-α production by CD4+ T cells isolated from the colon LP of the indicated group of Rag1−/− mice. The numbers in the plots refer to the percentage of each subset. (l) Sorted WT Treg cells were transferred along with CD4+CD45RBhiCD25− T cells from WT and SMAR1−/− mice into Rag1−/− mice, and WT CD4+Foxp3+ Treg cells were evaluated 7–8 weeks after the transfer. WT Treg cells were harvested from the colon LP and stained for surface CD4 and intracellular Foxp3 and analyzed by flow cytometry. The numbers in the plot refer to the percentage of each subset. Data are representative of three independent experiments each with eight mice per group. Data are presented as mean±s.e.m.
SMAR1 control STAT3 expression in Foxp3+ Treg cells
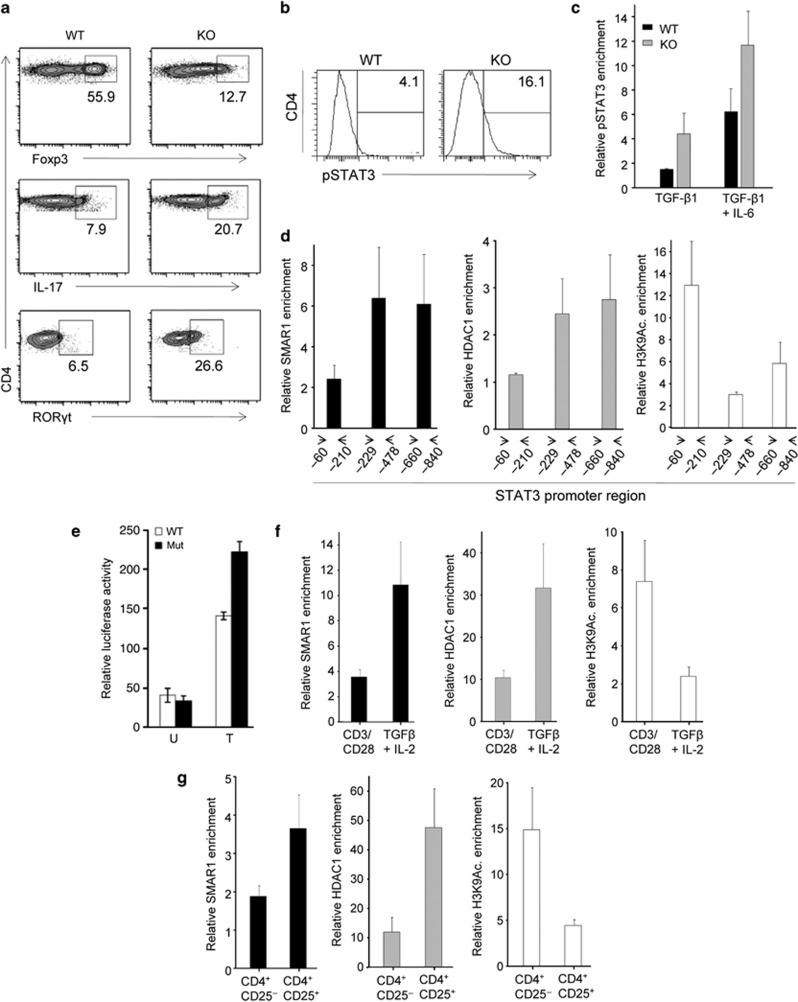
We further explored the mechanism responsible for altered suppressor activity of SMAR1-deficient Treg cells. In our initial study, we observed a lesser amount of Foxp3 mRNA as well as protein expression in SMAR1-deficient Treg cells compared with WT Treg cells. Furthermore, we observed that, during the course of DSS-induced colitis in WT scenario, the expression of SMAR1 in colonic CD4+ T cells was drastically downregulated and inversely the expression of STAT3 is upregulated, which leads to generation of more proinflammatory Th17 cells (Supplementary Figure S6a,b). As we have shown previously, after deletion of SMAR1 in T cells, the number of Th17 cells goes high in colonic LP and 2.5-fold higher STAT3 level in CD4+ T cells at the seventh day of DSS treatment (Supplementary Figure S6c), hence an altered Foxp3 expression was observed. In order to rule out the possibility that TCR signaling is not a cause of the observed altered Foxp3 expression, we checked the level of TCRβ and CD69 on the surface of CD4+ T lymphocytes. CD69 is the early activation marker and usually upregulated upon TCR stimulation. The data showed no major difference in TCRβ (Supplementary Figure S6d) and CD69 (Supplementary Figure S6e) level. It clearly showed that response to TCR stimulation of SMAR1-deficient Treg cells was similar to WT Treg cells. Thus reduction in Foxp3 level is not due to altered TCR signaling in SMAR1-deficient Treg cells. Therefore, SMAR1 might regulate the expression of STAT3 in Foxp3+ Treg cells. STAT3, a key transcription factor for Th17 differentiation, binds to the CNS2 region (silencer site) of Foxp3 promoter and represses its transcription.17, 18 As Foxp3 inhibits IL-17 and RORγt expression in CD4+ T cells,5 we first compared IL-17 and RORγt expression in SMAR1−/− and WT naive CD4+ T cells. Even if naive CD4+ T cells isolated from WT and SMAR1−/− mice showed unnoticeable IL-17 and RORγt (data not shown), SMAR1−/− CD4+ T cells express 4–5-fold more amount of RORγt mRNA as well as protein after TGF-β1 treatment (Figures 5j and 8a). TGF-β1 stimulation particularly eliminates IL-17 production in WT CD4+ T cells. As SMAR1−/− CD4+ T cells fail to suppress IL-17 production upon TGF-β1 treatment, it indicates that Foxp3 was unable to inhibit IL-17 expression. Higher IL-17 production in SMAR1−/− CD4+ T cells was possibly due to high level of pSTAT3 in these T cells after TGF-β1 treatment (Supplementary Figure S6f; Figure 8a). Thus we speculate SMAR1 controls STAT3 expression, which in turn regulates Foxp3 production in CD4+ T cells. Next we examined the effect of SMAR1 deficiency on the binding of STAT3 in Foxp3 promoter. SMAR1−/− CD4+ T cells showed higher IL-17 expression and that was probably due to upregulation of STAT3 upon TGF-β1 treatment (Figures 5h and 8b). As mentioned above, STAT3 binds directly to Foxp3 promoter and act as a repressor for its transcription. CD4+ T cells treated with TGF-β1 inhibit the STAT3 expression and resulted in less binding of STAT3 to the Foxp3 promoter in WT CD4+ T cells (Figure 8c). Because of elevated STAT3 level in SMAR1−/− CD4+ T cells, (Figure 8b) more binding of STAT3 to Foxp3 promoter was observed after TGF-β1 plus IL-6 treatment compared with WT CD4+ T cells (Figure 8c). On the other hand, treatment with only TGF-β1 was not able to reduce the binding of STAT3 to Foxp3 promoter in SMAR1−/− CD4+ T cells (Figure 8c). These results showed that TGF-β1 is not able to elevate Foxp3 expression owing to upregulation of STAT3, which represses Foxp3 transcription in SMAR1−/− CD4+ T cells.
Figure 8.
SMAR1 control signal transducer and activator of transcription factor 3 (STAT3) expression in Foxp3+ regulatory t (Treg) cells. (a,b) Flow cytometry analysis of naive CD4+ T cells at 5 days after treatment with transforming growth factor (TGF)-β1 and interleukin (IL)-2 together with T-cell receptor (TCR) stimulation. Cells were harvested and restimulated for 4 h with phorbol myristate acetate and ionomycin and stained for markers CD4, Foxp3, IL-17, retinoic acid–related orphan receptor γt (RORγt), and pSTAT3 (b). The number in the plots indicate the percentage of CD4+Foxp3+, CD4+IL-17+, CD4+RORγt+ (a), and CD4+pSTAT3+ (b) cells. Data shown are representative of three independent experiments with similar results. (c) Binding of pSTAT3 to Foxp3 promoter in wild-type (WT) and SMAR1−/− naive CD4+ T cells cultured for 5 days (treatment, horizontal axis), evaluated with anti-pSTAT3, and showed relative to binding with control immunoglobulin G (IgG). (d) Quantitative chromatin immunoprecipitation (ChIP) analyses of the enrichment of SMAR1 at STAT3 gene promoter at different positions (horizontal axis) in naive CD4+ T cells after treatment with TGF-β1 and IL-2 together with TCR stimulation and evaluated with anti-SMAR1 and gene activation and repressor markers H3K9 Ac. and HDAC1, respectively; data are presented relative to binding with control IgG. (e) Mutation of predicted SMAR1-binding site at the STAT3 promoter increases the luciferase reporter activity. HEK 293T cells were transfected with STAT3 promoter reporter firefly Luciferase construct (WT) and the same reporter containing the deletion mutation of a SMAR1-binding site at −321 to −337 from the transcription start site in the STAT3 promoter (Mut). Cells were treated (T) or untreated (U) with IL-6 for 24 h, and the luciferase activity was estimated. The data represent triplicates within same experiment and are representative of three independent experiments. Error bar indicates s.d. *P<0.02 (Student's t-test). (f) Quantitative ChIP analyses of the enrichment of SMAR1 and gene activation and repression marker H3K9 Ac. and HDAC1, respectively, in the region between positions −229 to −478 of the STAT3 gene promoter in WT naive CD4+ T cells 5 days after treatment with TGF-β1 and IL-2 together with TCR stimulation, assessed in d and presented relative to results obtained without TGF-β1 and IL-2 (with anti-CD3 and anti-CD28). (g) ChIP coupled quantitative PCR analyses of SMAR1, HDAC1, and H3K9 Ac. enrichment as in d in purified CD4+CD25+ Treg cells, presented relative to results obtained with purified CD4+CD25− T cells. Data are representative of four independent experiments (mean and s.d. of duplicate wells). KO, knock out.
As SMAR1 is an MARBP and as these proteins have the propensity to bind at AT-rich sequences, which frequently flank different promoters, we analyzed the promoter sequence of STAT3-inducible gene. Computational analysis using MARFINDER program predicted potential MARs in the STAT3 promoter within ∼400 bp upstream of IL-6 response element and ∼300 bp upstream of transcription start site (TSS; Supplementary Figure S6g). We assessed whether SMAR1 binds to these MAR regions in STAT3 gene promoter. We treated naive CD4+ T cells from the spleen of WT mice using TGF-β1 plus IL-2 along with TCR-specific stimulation and analyzed whether it could influence the binding of SMAR1 to STAT3 promoter during generation of iTreg. We processed iTreg cells for chromatin immunoprecipitation coupled quantitative PCR to assess the binding of SMAR1 to STAT3 promoter. We precisely investigated the promoter and IL-6 response elements, which are known to have an important role in activation of STAT3 gene expression.37, 38, 39, 40 We found that SMAR1 strongly binds to MAR region within the IL-6 response elements of STAT3 gene. The potential binding sites of SMAR1 at positions −660 to −840 and −229 to −478, belonging to IL-6 response elements were found in the proximal region of the promoter (Figure 8d; Supplementary Figure S6g). To further verify this, SMAR1-binding site were deleted between −321 to −337 from the TSS in a construct containing STAT3 promoter upstream of luciferase. The deleted region contains a strong MAR site, and it belongs to IL-6 response element where SMAR1 binds. The deletion considerably increases the reporter activity in response to IL-6 activation (Figure 8e) and it suggests that this site is important for STAT3 promoter activity. Next, we checked the binding of SMAR1 to STAT3 promoter in CD4+ T cells, treated with anti-CD3 and anti-CD28 alone or with anti-CD3 anti-CD28 along with TGF-β1 plus IL-2 and found that TGF-β1 plus IL-2 treatment considerably enhanced the binding of SMAR1 to the promoter compared with the control condition (Figure 8f). In addition, the enrichment of SMAR1 to STAT3 promoter in CD4+CD25+ Treg cells from the spleen of WT mice is relatively high compared with the CD4+CD25− T cells (Figure 8g). The binding of SMAR1 to STAT3 promoter in Foxp3+ Treg cells is directly correlated with the transcriptional repression of gene evidenced by the enrichment of HDAC1 and positive epigenetic modifications, including H3K9 acetylation (Figure 8d,f,g). However, TGF-β1 and IL-2 treatment enhanced the binding of SMAD2/3 to the Foxp3 promoter in WT as well as SMAR1−/− CD4+ T cells (Supplementary Figure S6h). Our data showed that SMAR1 binds to the transcriptional regulatory regions of STAT3 and modulates its expression, which in turn governs the Foxp3 expression upon TGF-β1 and IL-2 treatment and maintains the Treg phenotype, whereas these events were defective in SMAR1−/− T cells, leading to the generation of proinflammatory T cells. These results demonstrated a pivotal role of SMAR1 in the generation of Foxp3+ Treg cells.
DISCUSSION
Effector CD4+ T cells are responsible for the production of the proinflammatory cytokines that causes tissue damage. Treg cells are responsible for maintaining peripheral tolerance of effector T cells and keeping these cells in check.3 In this study, we showed for the first time the role of a MARBP SMAR1 in maintaining the balance between Th17 and Treg cells and its role in inflammatory diseases. Our results demonstrate that, in the absence of SMAR1, Treg cells lose their suppressive activity that leads to increased production of proinflammatory cytokine-producing T cells in the colon, which showed upregulation of gut-homing markers integrin α4β7 and CCR9 that helps to accumulate in the gut during colonic inflammation. This study revealed an indispensible role of SMAR1 in regulating Treg cell function and immune tolerance and maintaining the balance between Treg cells and Th17 cells.
Deletion of SMAR1 in T cells enhances Th17 cells in DSS-induced colitis. This preferential increase of Th17 cells were not limited to DSS-induced colitis; it has also been observed in T-cell transfer model of colitis and in vitro from TGF-β1-treated naive CD4+ T cells. Collectively, these observations point toward the fact that the differentiation of Th17 cells is favored during the absence of SMAR1 and increased level of Th17 cells is the reason for disease progression. It can also be inferred here that Treg cells are not able to gain control on Th17 cell proliferation. Transfer of Treg cells along with CD4+CD45RBhi T cells from SMAR1−/− mice into syngeneic Rag1−/− mice was not able to prevent the disease, although transfer of WT Treg cells successfully prevented the disease, indicating that Treg cells are responsible for the disease progression.
We found that CD25, CTLA-4, Icos, and CD103 surface levels were either equivalent or elevated in SMAR1-deficient Treg cells, suggesting that suppressive function of Treg cell is severely compromised. SMAR1-deficient Treg have no defect in proliferation as shown in in vitro proliferation assay (Supplementary Figure S3d). This eliminates the possibility that SMAR1 has no role in Treg cell proliferation, rather it has a crucial role in suppressive function of Treg cells. However, SMAR1-deficient Treg cells showed reduced levels of IL-10 and showed upregulation of proinflammatory gene expression, including TNF-α, IL-17, and IFN-γ. Studies have shown that IL-10-deficient mice lack Treg cells and are capable of controlling inflammatory responses in the intestine.39, 41 That led us to argue reduced suppressive function of SMAR1-deficient Treg cells. We have demonstrated that SMAR1-deficient Treg cells showed enhanced ability to produce inflammatory cytokines, indicating that SMAR1 downregulation under inflammatory condition could allow Treg cell to acquire effector function that contribute to inflammation. Our study therefore suggests that SMAR1 expression in Treg cells is important for the modulation of Treg cell function and immune response. Considering above observations, it is clear that SMAR1 is having a crucial role in suppressive function of Treg cells. Because of the absence of SMAR1, Treg cells fail to suppress the reactive CD4+ T cells, resulting in severe damage of the whole balance among CD4+ T cells and consequent development of the disease.
CD4+ regulatory T cells have an essential role in intestinal homeostasis. Treg cells are characterized by constitutively higher level of transcription factor Foxp3,6, 42 which is considered to confer their regulatory activity. Treg cells have the ability to block colitis upon transfer in vivo. We found constitutive expression of SMAR1 in nTreg cells as well as in iTreg cells. Our data also support the idea that IL-2 contributes to the expression of SMAR1 by Treg. Recent reports suggest that the Treg cell required the acquisition of specific transcription factors to exhibit control in defined polarized situation.43, 44 Previous reports demonstrated that increased expression RORγt in Treg can produce IL-17A45 that leads to compromised Treg function. We found that, in the colon, the expression of RORγt was influenced by SMAR1 in Treg and increased expression of IL-17A compared with WT. The expression of Foxp3 is reduced by genetic alteration causing upregulation of RORγt, followed by increased level of IL-17A production and generation of effector Th17 cells.8, 46, 47 We observed high IL-17 expression in SMAR1-deficient Treg cells. We also found that Foxp3 level was also reduced. Therefore, we speculate that the loss of SMAR1 has severe effects on Foxp3 expression, which leads to the loss of Foxp3 and induction of IL-17A conferring a Treg phenotype to Th17 phenotype.
SMAR1 is a member of MARBP family that interacts with regulatory regions (promoters/enhancers) of the gene and potentially controls the transcriptional activity.25 MAR, also known as Scaffold/matrix attachment regions (S/MARs), are sequences in the DNA of eukaryotic chromosomes where the nuclear matrix attaches.48, 49 Studies on specific genes lead to the outcome that the dynamic and complex organization of the chromatin mediated by S/MAR elements has an important role in the regulation of gene expression.50 Therefore, SMAR1 exerts diverse function to control gene expression in a cell type–specific manner.51 Nevertheless, the understanding of how SMAR1 regulates immune function is far from complete.
Indeed, our studies open a new role of SMAR1 in controlling Treg cell function. The function of Treg cell is dependent on Foxp3 expression. Foxp3 is an important transcription factor that regulates many essential genes that governs the Treg cell development.42 The predominant T-cell type that expressed Foxp3 was CD4+CD25+ T cells, the same population that has been reported to suppress proliferation and cytokine production in conventional CD4+ T cells. Foxp3 appears to exert its function through the transcriptional repression of many genes, including that of the effector cytokines.33 The factors mediating the transactivation or transrepression are critical to delineate the molecular mechanisms involved in controlling regulation of transcription factor Foxp3. Previous reports suggest that TGF-β mediates enrichment of SMAD2/3 at the Foxp3 promoter and the activation of Foxp3 transcription.52, 53 On the other hand, STAT3 is reported to bind to regulatory (silencer) regions of Foxp3 promoter15, 53 and suppress its expression.
Deficiency of SMAR1 in Treg cells leads to uncontrolled STAT3 production and, in turn, production of IL-17. Additionally, IL-6-mediated suppression of SMAR1 has a direct effect on the enrichment of STAT3 to the Foxp3 promoter, although inhibition of SMAR1 could restore the STAT3 enrichment in Foxp3 promoter in response to TGF-β1 in SMAR1−/− CD4+ T cells. Finally, IL-6 could influence Foxp3 epigenetically by losing the chromatin, allowing more access of STAT3 to the Foxp3 promoter. This data support that overexpression of STAT3 is a key factor in defective Foxp3 induction in SMAR1−/− CD4+ T cells. Thus we speculate that SMAR1 controls transcriptional activity of STAT3 and in turn regulates the Foxp3 expression and maintain the Treg cell phenotype. It is now necessary to answer how SMAR1 is regulating the STAT3 expression. It is known from earlier studies that SMAR1 exerts its transcriptional activity mainly through DNA binding.24, 54 Presence of several MAR-binding region at the promoter of STAT3 makes us believe that SMAR1 can bind to these site and influence the STAT3 expression. SMAR1 bound to regulatory regions of STAT3 locus and inhibits the activity of STAT3, a negative regulator for Foxp3.55, 56 In support, we observed that in Th17 cells SMAR1 expression was very low and it is not able to bind to STAT3 locus. However, in Treg cells, SMAR1 binds at a position −660 to −840 associated with strong MAR and −229 to −478 associated with IL-6 response elements from the TSS of STAT3 locus. We found that deletion of SMAR1-binding MAR site located at −321 to −337 from the TSS in STAT3 promoter attenuated the promoter activity, suggesting that this region is essential. These observations conforms the fact that SMAR1 controlled Foxp3 expression via STAT3 dependent manner and governs the overall balance between Th17 and Treg cells.
Our results showed that in WT scenario treatment with TGF-β1, SMAD2/3 bind to the Foxp3 promoter at the same time SMAR1 bind to STAT3 promoter, which further suggested a positive role for SMAR1 in the transcriptional regulation of STAT3, this activity was regulated through TGF-β signaling. It suggests that SMAR1 is involved in regulating Foxp3 expression in TGF-β1-induced Treg cells, and ultimately it decides the plasticity of Treg cells. Here we found that SMAR1 has an important role in switching on STAT3 and IL-17 in WT CD4+ T cells, which indicates a function for SMAR1 in inhibiting the binding of STAT3 to the Foxp3 promoter. The inability of CD4+ T cells lacking SMAR1 to induce Foxp3 resulted in an intrinsic preference for Th17 differentiation. On the basis of data presented here, we propose that SMAR1 induces Foxp3 expression by promoting the binding of SMAD2/3 to the Foxp3 promoter (enhancer) and by removing the negative factor STAT3 to the Foxp3 promoter (silencer).
Further studies are required in order to understand the complex interplay of these transcription factors in the control of Treg adaptation vs. pathogenicity during inflammation. Accumulation of Treg is critical for their ability to maintain tissue homeostasis. A role of SMAR1 in promoting Treg fitness/survival was revealed under the inflammation or homeostasis proliferative situation. Finally, our results support the idea that the role of SMAR1 in Treg is complex and affects many physiological functions. In the present study, we uncover potential roles; in particular, SMAR1 controls excessive effector polarization by Treg cells to Th17 cells and their suppressive activity during inflammation. Although identification of the factors involved in the stability of Treg is likely to offer important therapeutic interventions, the role of SMAR1 in controlling of Treg fate during inflammation will be equally critical to our understanding of peripheral tolerance and development of safe therapeutic approaches.
METHODS
Mice. T-cell-specific (LCK-Cre/SMAR1f/f) conditional knockout mice (SMAR1−/−) were generated at Ozgene, Bentley, Australia. Rag1−/− mice were a gift from S Rath (National Institute of Immunology, New Delhi, India). Mice were maintained under specific pathogen-free conditions in the animal resource facility of National Centre for Cell Science. C57BL/6J and all other mice were inbred in the experimental animal facility of the NCCS, Pune, India. SMAR1−/− mice were maintained as colony by homozygous × homozygous (brother × sister) and the WT mice were C57BL/6J. The weanling of both WT (C57BL/6J) and SMAR1−/− (LCK-Cre/SMAR1f/f) mice were housed together during the course of experiment. Mice were killed between 6 and 8 weeks of age, except for the Rag1−/− mice, which were killed at 13–15 weeks of age. The experiments were conducted according to protocols approved by the Institutional Animal Ethical Committee of NCCS.
Flow cytometry. Surface and intracellular staining was performed on FcR-blocked lymphocytes using the antibodies listed in Supplementary Material and Methods. For intracellular cytokine determination, up to 1 × 106 lymphocytes per well were stimulated with 50 ng ml−1 phorbol myristate acetate, 200 ng ml−1 ionomycin, 10 μg ml−1 brefeldin, and 2 μM monesin in complete media for 6 h at 37 °C. Cells were first stained with surface markers, followed by fixation in 2% formaldehyde and permeabilization with 0.05% saponin, and later followed by intracellular staining for cytokines with the fluorescence-labeled antibodies listed in Supplementary Material and Methods. Flow cytometry analysis was conducted on an upgraded five-color FACS caliber, FACS canto, or LSR II flow cytometer (BD, San Jose, CA), and data were analyzed using the FACS DIVA software (San Jose, CA). Sorting of CD4+CD62LhiCD25−, CD4+CD45RBhiCD25−, and CD4+CD25+ cells was performed using a FACS area instrument (BD).
Induction of DSS colitis. Experimental colitis was induced by adding DSS (36,000–55,000 kDa, MP Biomedicals (Solon, OH); Cat.: 160110) to the drinking water at concentration of 3% (wt/vol). Animals were treated either for 5 days and then allowed to recover by normal drinking water for additional 9 days or were treated continuously with DSS for 7–11 days. Colon was assessed for weight, length, and histology. The animal were weighed daily and monitored for signs of distress, rectal bleeding, stool consistency, detection of blood in stool, and body weight loss measurement. An individual score was given for each one of this parameter, and disease activity index ranging from 0 to 4 was calculated by combining all the three scores.57 In order to get the uniform results, mice were taken from the same breed and were co-caged from the time of weaning all throughout the experiment.
Data analysis. Groups were compared with the Prism software (GraphPad, La Jolla, CA) using two-tailed unpaired Student's t-test. Data are presented as mean±s.e.m. P<0.05 was considered significant.
Acknowledgments
We thank our Director Dr Shekhar Mande for providing us opportunity to work on this project. We thank Dr L.S. Limaye, Mrs Trupti Joshi for FACS facility, and Dr Rahul Bankar and Mr M.L. Shaikh for assistance with animal husbandry. We thank Dr S. Rath, NII, New Delhi, India for providing Rag1−/− mice. This work is funded by NCCS, Department of Biotechnology (DBT), Indian Council of Medical Research (ICMR), New Delhi, India. B.M. is a recipient of fellowship from ICMR.
The authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
Supplementary Material
References
- 1Belkaid, Y. & Tarbell, K. Regulatory T cells in the control of host-microorganism interactions. Annu. Rev. Immunol. 27, 551–589 (2009). [DOI] [PubMed] [Google Scholar]
- 2Izcue, A., Coombes, J.L. & Powrie, F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol. Rev. 212, 256–271 (2006). [DOI] [PubMed] [Google Scholar]
- 3Vignali, D.A., Collison, L.W. & Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Zhu, J., Hidehiro, Y. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 7Laurence, A. et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007). [DOI] [PubMed] [Google Scholar]
- 8Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector Th17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 9Feng, T. et al. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J. Immunol. 186, 6313–6318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Lee, Y. et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Mathur, A.N. et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 178, 4901–4907 (2007). [DOI] [PubMed] [Google Scholar]
- 12Ruan, Q. et al. The Th17 immune response is controlled by the Rel-RORγ-RORγ T transcriptional axis. J. Exp. Med. 208, 2321–2333 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Zhou, L. et al. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 (2007). [DOI] [PubMed] [Google Scholar]
- 14Fujino, M. & Xiao-Kang, L. Role of STAT3 in regulatory T lymphocyte plasticity during acute graft-vs.-host disease. JAK-STAT 2, e24529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Laurence, A. et al. STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity 37, 209–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Lee, Y.K., Ryuta, M., Robin, D.H. & Weaver, C.T. Developmental plasticity of Th17 and Treg cells. Curr. Opin. Immunol. 21, 274–280 (2009). [DOI] [PubMed] [Google Scholar]
- 17Masahide, T. & Greene, M.I. Cooperative regulatory events and Foxp3 expression. Nat. Immunol. 12, 15–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Maruyama, T., Konkel, J.E., Zamarron, B.F. & Chen, W. The molecular mechanisms of Foxp3 gene regulation. Semin. Immunol. 23, 418–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Weaver, C.T. & Hatton, R.D. Interplay between the TH17 and Treg cell lineages: a (co-)evolutionary perspective. Nat. rev. immunol 9, 883–889 (2009). [DOI] [PubMed] [Google Scholar]
- 20Kosak, T.S. & Groudine, M. The undiscovered country: chromosome territories and the organization of transcription. Dev. Cell. 2, 690–692 (2002). [DOI] [PubMed] [Google Scholar]
- 21Schneider, R.R. & Grosschedl, R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 21, 3027–3043 (2007). [DOI] [PubMed] [Google Scholar]
- 22Chattopadhyay, S. & Pavithra, L. MARs and MARBPs: key modulators of gene regulation and disease manifestation. Subcell. Biochem. 41, 213–230 (2007). [PubMed] [Google Scholar]
- 23Harris, S.L. & Levine, A.J. The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 (2005). [DOI] [PubMed] [Google Scholar]
- 24Rampalli, S., Pavithra, L., Bhatt, A., Kundu, T.K. & Chattopadhyay, S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25, 8415–8429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Chattopadhyay, S., Kaul, R., Charest, A., Housman, D. & Chen, J. SMAR1, a novel, alternatively spliced gene product, binds the scaffold/matrix-associated region at the T cell receptor beta locus. Genomics 68, 93–96 (2000). [DOI] [PubMed] [Google Scholar]
- 26Sinha, S. et al. Coordinated regulation of p53 apoptotic targets BAX and PUMA by SMAR1 through an identical MAR element. EMBO J. 29, 830–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Sreenath, K. et al. Nuclear matrix protein SMAR1 represses HIV-1 LTR mediated transcription through chromatin remodeling. Virology 25, 76–85 (2010). [DOI] [PubMed] [Google Scholar]
- 28Burute, M., Gottimukkala, K. & Galande, S. Chromatin organizer SATB1 is an important determinant of T-cell differentiation. Immunol. Cell Biol. 90, 852–859 (2012). [DOI] [PubMed] [Google Scholar]
- 29Kaul-Ghanekar, R. et al. Abnormal V(D)J recombination of T cell receptor β locus in SMAR1 transgenic mice. J. Biol. Chem. 280, 9450–9459 (2005). [DOI] [PubMed] [Google Scholar]
- 30Eastaff-Leung, N., Mabarrack, N., Barbour, A., Cummins, A. & Barry, S. Foxp3+ regulatory T cells, Th17 effector cells, and cytokine environment in inflammatory bowel disease. J. Clin. Immunol. 30, 80–89 (2010). [DOI] [PubMed] [Google Scholar]
- 31Zenewicz, L.A., Antov, A. & Flavell, R.A. CD4 T-cell differentiation and inflammatory bowel disease. Trends Mol. Med. 15, 199–207 (2009). [DOI] [PubMed] [Google Scholar]
- 32Lee, G.R., Kim, S.T., Spilianakis, C.G., Fields, P.E. & Flavell, R.A. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24, 369–379 (2006). [DOI] [PubMed] [Google Scholar]
- 33Ouyang, W., Beckett, O., Ma, Q., Paik, J., DePinho, R.A. & Li, M.O. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 (2010). [DOI] [PubMed] [Google Scholar]
- 34Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K. & Muller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). [DOI] [PubMed] [Google Scholar]
- 35Rubtsov, Y.P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008). [DOI] [PubMed] [Google Scholar]
- 36Unutmaz, D. & Pulendran, B. The gut feeling of Treg cells: IL-10 is the silver lining during colitis. Nat. Immunol. 10, 1141–1143 (2009). [DOI] [PubMed] [Google Scholar]
- 37Makoto, I., Nakajima, K., Yamanaka, Y., Kiuchi, N. & Hirano, T. Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein. J. Bio. Chem. 273, 6132–6138 (1998). [DOI] [PubMed] [Google Scholar]
- 38Wei, S. et al. The genomic structure and chromosomal localization of the mouse STAT3 gene. Int. Immunol. 8, 1205–1211 (1996). [DOI] [PubMed] [Google Scholar]
- 39Ken, K. et al. Structure and functional analysis of the human STAT3 gene promoter: alteration of chromatin structure as a possible mechanism for the up-regulation in cisplatin-resistant cells. Biochim. Biophys. Acta 1493, 91–100 (2000). [DOI] [PubMed] [Google Scholar]
- 40Kiyoshi, T. et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. 94, 3801–3804 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Roers, A. et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J. Exp. Med. 200, 1289–1297 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Fontenot, J.D., Gavin, M.A. & Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 43Chaudhry, A. et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 458, 351–356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Osorio, F. et al. DC activated via dectin-1 convert Treg into IL-17 producers. Eur. J. Immunol. 38, 3274–3281 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Wan, Y.Y. & Flavell, R.A. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007). [DOI] [PubMed] [Google Scholar]
- 47Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Berezney, R., Mortillaro, M.J., Ma, H., Wei, X. & Samarabandu, J. The nuclear matrix: a structural milieu for genomic function. Int. Rev. Cytol. 162A, 1–65 (1995). [DOI] [PubMed] [Google Scholar]
- 49Zink, D., Fischer, A.H. & Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687 (2004). [DOI] [PubMed] [Google Scholar]
- 50Zaidi, S.K. et al. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 6, 128–133 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Malonia, S.K. et al. Gene regulation by SMAR1: role in cellular homeostasis and cancer. Biochim. Biophys. Acta 1815, 1–12 (2011). [DOI] [PubMed] [Google Scholar]
- 52Tone, Y., Furuuchi, K., Kojima, Y., Tykocinski, M.L., Greene, M.I. & Tone, M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9, 194–202 (2008). [DOI] [PubMed] [Google Scholar]
- 53Xu, L., Kitani, A., Stuelten, C., McGrady, G., Fuss, I. & Strober, W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity 33, 313–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Kaul-Ghanekar, R., Jalota, A., Pavithra, L., Tucker, P. & Chattopadhyay, S. SMAR1 and Cux/CDP modulate chromatin and act as negative regulators of the TCRbeta enhancer (Ebeta). Nucleic Acids Res. 32, 4862–4875 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Zorn, E. et al. IL-2 regulates Foxp3 expression in human CD4+CD25+ regulatory T cells through a STAT dependent mechanism and induces the expansion of these cells in vivo. Blood 108, 1571–1579 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Yang, X.P. et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12, 247–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57Laukoetter, M.G. et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J. Exp. Med. 204, 3067–3076 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.