Abstract
GM1-gangliosidosis is an inherited autosomal recessive disorder caused by mutations in the gene GLB1, which encodes acid β-galactosidase (β-gal). The lack of activity in this lysosomal enzyme leads to accumulation of GM1 gangliosides (GM1) in cells. We have developed a high-content-imaging method to assess GM1 levels in fibroblasts that can be used to evaluate substrate reduction in treated GLB1−/− cells [1]. This assay allows fluorescent quantification in a multi-well system which generates unbiased and statistically significant data. Fluorescently labeled Cholera Toxin B subunit (CTXB), which specifically binds to GM1 gangliosides, was used to detect in situ GM1 levels in a fixed monolayer of fibroblasts. This sensitive, rapid, and inexpensive method facilitates in vitro drug screening in a format that allows a high number of replicates using low working volumes.
Keywords: High-throughput imaging, GM1-gangliosidosis, Acid β-galactosidase, Cholera toxin B subunit
Specifications table
| Subject area | Biology. |
| More specific subject area | Inborn errors of metabolism. |
| Type of data | Fluorescence microscopy images, tables, figures. |
| How data was acquired | BD Pathway 855 High Content Bioimager (BD Biosciences). |
| Data format | Raw, segmented, analyzed. |
| Experimental factors | After attachment in a black wall, clear bottom 96 well plate, normal (GLB1+/+) and GM1-gangliosidosis (GLB1−/−) fibroblasts were incubated untreated or treated with recombinant β-gal for 24 h. |
| Experimental features | After treatment, cells were fixed, permeabilized and stained with fluorescently labeled CTXB. |
| Nuclei were counter stained with DAPI to allow cell count. | |
| Individual images were acquired in each well using both filters. | |
| Images were segmented using Attovision software. | |
| Segmentation data were analyzed using BD Data Explorer®. | |
| CTXB pixels per cell were calculated in each image-well. | |
| Treatment values were expressed as average pixel/cell. | |
| Data source location | Images collected in Jonesboro, Arkansas, USA. |
| Data accessibility | Data is with this article. |
Value of the data
-
•
We describe an imaging method that statistically differentiates levels of GM1 gangliosides in mammalian cells [1].
-
•
These data describe a sensitive, rapid, and unbiased high-throughput imaging method that allows quantification of GM1 gangliosides in situ.
-
•
This method can be easily used in primary compound screening or for the testing of post-primary treatment conditions, due to advantages of the low required level of sample processing and treatment volumes.
-
•
This assay can be adapted to multiple high-content-imaging instruments [2].
1. Data
High-content screening allows quantification of data obtained by fluorescence imaging in a multi-well format. Using this technology, we were able to statistically differentiate substrate levels (p<0.0001) between normal (GLB1+/+) and enzyme deficient (GLB1−/−) human fibroblasts. Reduction of substrate levels can be detected when GM1-gangliosidosis fibroblast (GLB1−/−) are treated with a corrective recombinant protein.
2. Experimental design, materials and methods
2.1. Conjugation of Dylight 594 to CTXB
For GM1 ganglioside detection, Dylight 594 (Thermo Cat #46412) was conjugated to the primary amines of Cholera Toxin B protein (CTXB) (List Biologicals Cat #104) following manufacture׳s protocol. Fluorophore conjugated CTXB has been previously used to detect GM1 gangliosides in cells (including macrophages and fibroblasts) using fluorescent microscopy [3], [4].
2.2. Cell treatment
Suspended Normal (GLB1+/+; Coriell GM-00010) and GM1-gangliosidosis (GLB1−/−; Coriell GM-10919) fibroblasts were diluted to 100,000 cells/ml and plated to a black-walled, clear bottom 96-well microtiter plates using 100 µl/well. Following a 24 h attachment period, media was replaced with 100 µl serum-free media (untreated) or serum-free media containing 6 nM of recombinant β-gal (R&D Systems). Cells were incubated for 24 h at 37 °C and 5% CO2. These parameters had been optimized to provide cell densities (impacted by cell type, cell line, growth rate, and treatment incubation times) with enough separation for accurate capture of cell count and resolution of the region of interest (ROI).
2.3. Fluorescence staining of cells
Cells were fixed with 4% paraformaldehyde for 8 min followed by 3X washes with PBS. Cells were permeabilized for 10 min using 0.25% Triton X-100 solution and blocked with 1% BSA+0.3 M glycine in PBS for 1 h.
Cells were incubated with conjugated CTXB-Dylight594 at 1:1600 dilution in PBS for 1 h at room temperature. Wells were washed 3X with PBS and cells were maintained in a PBS solution containing 600 nM DAPI and 0.03% sodium azide. Optimization of permeabilization time, Triton X-100 concentration, and CTXB-Dylight594 dilution was made in order to avoid signal saturation and improve assay sensitivity.
2.4. Image acquisition
Images were acquired with the BD Pathway 855 High Content Bioimager (BD Biosciences). A 20X NA 075 objective was used to acquire 2×2 montage images per well using DAPI (Ex=380 nm; Em=435 nm) and Texas Red (Ex=560 nm; Em=645 nm) filters. The well area acquired per each image yielded an average of 150 cells per image. All images were acquired using the same parameters summarized in Table 1 using the instrument׳s automated laser autofocus (Fig. 3).
Table 1.
Acquisition parameters.
| 1.1. DAPI | 1.2. Texas Red | ||
|---|---|---|---|
| Auto dynamic range | On | Auto dynamic range | On |
| Dynamic range min | 800 | Dynamic range min | 350 |
| Dynamic range max | 3200 | Dynamic range max | 1500 |
| Gain | 0 | Gain | 0 |
| Offset | 255 | Offset | 255 |
| Exposure | 0.3 | Exposure | 0.2 |
| Lamp intensity | 100 | Lamp intensity | 100 |
| Excitation position A | 380/10 | Excitation position B | 560/55 |
| Dichroic excitation position | 3 open | Dichroic excitation position | Mirror |
| Dichroic epifluorescence position | 400DCLP | Dichroic epifluorescence position | 595LP |
| Emission position A | 435LP | Emission position A | 645/75 |
| Confocal | No | Confocal | No |
| Background subtraction | Off | Background subtraction | Off |
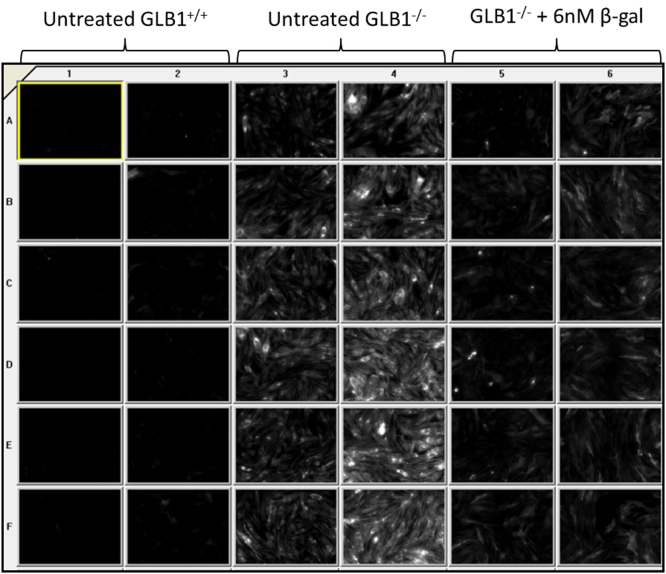
Fig. 3.
CTXB signal in different treatments. Image thumbnails of data acquired using Texas Red filter. Substrate accumulation differences are evident among treatments.
2.5. Image segmentation
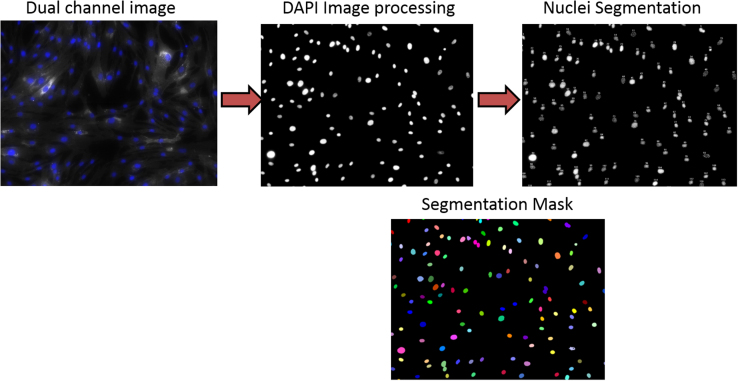
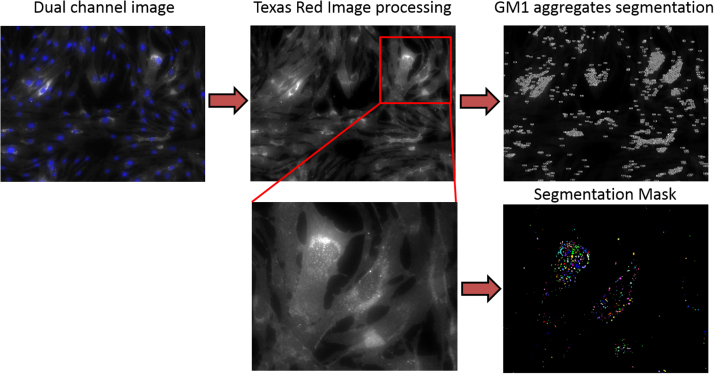
Images were segmented using Attovision software. Nuclei were counted in DAPI images by using polygon segmentation parameters described in Table 2.1. GM1 aggregates within cells were defined in Texas Red images using polygon segmentation parameters described in Table 2.2. All images were segmented using the same segmentation parameters defined for each filter channel. The segmentation process is depicted in Fig. 1, Fig. 2.
Table 2.
Segmentation parameters for nuclei and GM1 aggregates definition.
| 2.1. Nuclei segmentation | 2.2. GM1 aggregates segmentation | ||
|---|---|---|---|
| Threshold mode: | Automatic | Threshold mode: | Manual |
| Number of threshold steps | 1 | Min threshold: | 294 |
| Level 1 offset mode | Percent | Max threshold: | 4095 |
| Level 1 offset percent | 0.000000 | Scrap min pixels: | 10 |
| Scrap min pixels: | 500 | Scrap max pixels: | 50,000 |
| Scrap max pixels: | No maximum | Scrap mode: | Normal |
| Scrap mode: | Normal | Shape: | Polygon |
| Shape: | Polygon | Dilation: | 0 |
| Dilation: | 0 | ROI output: | Whole cell |
| ROI output: | Nucleus | Watershed: | Off |
| Watershed: | Off | Preprocessing filters: | On |
| Preprocessing filters: | On | ||
| Filter2 (A) | Erode 3×3 | Filter4 (A): | Top hat (7×7) |
| Filter3 (A) | Sharpen hat | ||
| Filter4 (A) | RB 75×75 | ||
Fig. 1.
Nuclei segmentation process. After defining dye signal threshold, every image was segmented using the same parameters described in Table 2.1. Nuclei segmentation was used for cell counting.
Fig. 2.
GM1 aggregates segmentation process. After defining dye signal threshold and applying filters, every image was segmented using the same parameters described in Table 2.2. Pixel area of each aggregate was used to calculate total pixels per image.
2.6. Data analysis
Data analysis was performed using BD Data Explorer® software (BD Biosciences). Total pixels, corresponding to GM1 gangliosides, were calculated by adding the area of one of each GM1 aggregate polygons in each image/repetition. Total pixels were divided by number of cells (nucleus count) in the corresponding image. Data were expressed as the average of CTXB-Dylight594 pixels per cell ratios in each treatment. Twelve images (n=12) per treatment were used to determine the average value (Table 3). Considering that each image included an average 150 cells at this magnification, the total amount of cells analyzed for each treatment was approximately 1800.
Table 3.
Segmentation quantitative data in untreated normal fibroblast, GM1-gangliosidosis fibroblast and GM1-gangliosidosis fibroblast treated with 6 nM of recombinant human β-galactosidase for 24 h.
|
Untreated GLB1+/+fibroblasts |
Untreated GLB1−/−fibroblasts |
GLB1−/−fibroblasts+6 nM β-gal |
||||||||||||
| Well ID | GM1 aggregates count | Total GM1 pixels | Cell count | Pixels/cell | Well ID | GM1 aggregates count | Total GM1 pixels | Cell count | Pixels/cell | Well ID | GM1 aggregates count | Total GM1 pixels | Cell count | Pixels/cell |
| A001 | 5 | 115 | 50 | 2 | A003 | 570 | 10,652 | 94 | 113 | A005 | 131 | 2997 | 99 | 30 |
| A002 | 18 | 383 | 201 | 2 | A004 | 1195 | 24,797 | 110 | 225 | A006 | 649 | 12,237 | 120 | 102 |
| B001 | 4 | 114 | 54 | 2 | B003 | 2544 | 45,589 | 168 | 271 | B005 | 277 | 5456 | 147 | 37 |
| B002 | 62 | 975 | 161 | 6 | B004 | 2053 | 43,573 | 124 | 351 | B006 | 613 | 11,481 | 178 | 65 |
| C001 | 6 | 129 | 84 | 2 | C003 | 782 | 14,401 | 119 | 121 | C005 | 117 | 2856 | 158 | 18 |
| C002 | 48 | 840 | 122 | 7 | C004 | 1616 | 30,804 | 150 | 205 | C006 | 315 | 5520 | 150 | 37 |
| D001 | 1 | 19 | 21 | 1 | D003 | 1031 | 19,449 | 151 | 129 | D005 | 291 | 6819 | 142 | 48 |
| D002 | 9 | 197 | 140 | 1 | D004 | 3687 | 67,779 | 186 | 364 | D006 | 378 | 6944 | 193 | 36 |
| E001 | 6 | 95 | 131 | 1 | E003 | 1138 | 20,604 | 129 | 160 | E005 | 386 | 7300 | 158 | 46 |
| E002 | 26 | 457 | 115 | 4 | E004 | 3746 | 75,338 | 155 | 486 | E006 | 578 | 10,198 | 159 | 64 |
| F001 | 68 | 1125 | 143 | 8 | F003 | 1299 | 25,129 | 133 | 189 | F005 | 421 | 7850 | 177 | 44 |
| F002 | 16 | 217 | 119 | 2 | F004 | 3356 | 63,919 | 217 | 295 | F006 | 415 | 6854 | 170 | 40 |
| Ave | 22 | 389 | 112 | 3 | Ave | 1918 | 36,836 | 145 | 243 | Ave | 381 | 7209 | 154 | 47 |
| StDev | 24 | 382 | 51 | 2 | StDev | 1146 | 22,092 | 34 | 115 | StDev | 171 | 2952 | 26 | 22 |
| StErr | 6 | 102 | 14 | 1 | StErr | 306 | 5904 | 9 | 31 | StErr | 46 | 789 | 7 | 6 |
Acknowledgments
This research was supported by funding to BioStrategies LC from an NIH/NINDS Phase I SBIR grant (1R43NS084565-01). The BD pathway was purchased through a NSF MRI grant (DBI-0960089) to C.L.C, Arkansas State University.
References
- 1.Condori J., Acosta W., Ayala J., Katta V., Flory A., Martin R. Enzyme replacement for GM1-gangliosidosis: uptake, lysosomal activation, and cellular disease correction using a novel β-galactosidase: RTB lectin fusion. Mol. Genet. Metab. 2016 doi: 10.1016/j.ymgme.2015.12.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanella F., Lorens J.B., Link W. High content screening: seeing is believing. Trends Biotechnol. 2010;28(5):237–245. doi: 10.1016/j.tibtech.2010.02.005. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 3.Beer C., Pedersen L., Wirth M. Amphotropic murine leukaemia virus envelope protein is associated with cholesterol-rich microdomains. Virol J. 2005;2:36. doi: 10.1186/1743-422X-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers K.A., Szaszi K., Khadaroo R.G., Tawadros P.S., Marshall J.C., Kapus A. Oxidative stress generated by hemorrhagic shock recruits Toll-like receptor 4 to the plasma membrane in macrophages. J. Exp. Med. 2006;203(8):1951–1961. doi: 10.1084/jem.20060943. [DOI] [PMC free article] [PubMed] [Google Scholar]