Abstract
Recent studies demonstrate the functions of long non-coding RNAs (lncRNAs) in mediating gene expression at the transcriptional or translational level. Our previous study identified a Sirt1 antisense (AS) lncRNA transcribed from the Sirt1 AS strand. However, its role and regulatory mechanism is still unknown in myogenesis. Here, functional analyses showed that Sirt1 AS lncRNA overexpression promoted myoblast proliferation, but inhibited differentiation. Mechanistically, Sirt1 AS lncRNA was found to activate its sense gene, Sirt1. The luciferase assay provided evidences that Sirt1 AS lncRNA interacted with Sirt1 3′ UTR and rescued Sirt1 transcriptional suppression by competing with miR-34a. In addition, RNA stability assay showed that Sirt1 AS lncRNA prolonged Sirt1 mRNA half-life from 2 to 10 h. Ribonuclease protection assay further indicated that it fully bound to Sirt1 mRNA in the myoblast cytoplasm. Moreover, Sirt1 AS overexpression led to less mouse weight than the control because of less lean mass and greater levels of Sirt1, whereas the fat mass and levels of miR-34a were not altered. Based on the findings, a novel regulatory mechanism was found that Sirt1 AS lncRNA preferably interacted with Sirt1 mRNA forming RNA duplex to promote Sirt1 translation by competing with miR-34a, inhibiting muscle formation.
Recently, long noncoding RNA (lncRNA) has been shown to play important functional roles as regulators of gene expression through the recruitment of the complex epigenetic machinery that dictates distinctive chromatin signatures involved in active transcription and translation. One group of lncRNAs is the natural antisense transcripts (NATs), which are transcribed from the opposite DNA strand to their specific partner protein-coding genes. Antisense transcription is the pairing of a NAT with an overlapping protein-coding sense transcript, whereby NAT expression can lead to an increase or decrease in sense expression1.
Muscle development is orchestrated by a network of epigenetic regulators and transcription factors2,3,4,5. LncRNA emerges as an important novel player in muscle development regulation6. LncRNA regulates key function genes by numbers of mechanisms at different levels, including transcriptional, post-transcriptional and epigenetic7. At the transcriptional level, muscle-associated lincRNA (Yam-1) was decreased during muscle differentiation and served as a repressor of myogenesis8. In addition, endogenous RNA (eRNA) was another class of ncRNA which was transcribed from regulatory elements9. The eRNA transcribed from the regulatory elements of myogenic differentiation antigen (MyoD), which enhanced RNA polymerase II occupancy and transcription of MyoD and myogenin (MyoG), further activated the downstream myogenic effects10. At the post-transcriptional level, the muscle-specific lncRNA (linc-MD1) ‘‘sponges’’ miR-133 and miR-135 to regulate the expression of transcription factors mastermind-like protein 1 (MAML1) and myocyte-specific enhancer factor 2C (MEF2C) to activate muscle-specific gene expression11. At the epigenetic level, lncRNAs including metastasis-associated lung adenocarcinoma transcript 1 (Malat1), H19 and gene trap locus 2-maternally expressed gene 3 (Gtl2-Meg3) interacted with polycomb repressive complex 2 (PRC2) to modulate their target genes, resulting in myogenesis12,13,14. However, whether other novel lncRNAs are involved in muscle development needs to further explore.
According to sequence length, ncRNAs were classified as lncRNA (>200 bp) and microRNA (~22 bp)7. Increasing evidences verified that lncRNA or microRNA interacted with each other to fulfil the biological function. For example, linc-MD1 and H19 sponge miR-133, miR-135 and let7, respectively, inhibiting the function of miRNAs15. In addition, some lncRNAs directly regulated the processing of miRNAs. LncRNA (Uc.283 + A) bound to pri-miR-195 to prevent its cleavage and maturation16. Moreover, lncRNAs indirectly regulated the processing of miRNAs. MiR-361 targeted pri-miR-484 to suppress the processing of miR-484, but mitochondrial dynamic related lncRNA (MDRL) bound to miR-361 and indirectly promoted miR-484 maturation17. Therefore, it is necessary to further explore the novel mechanisms of interaction between lncRNA and miRNA.
More than 70% of mammalian transcripts show evidence of AS transcription not only indicates the biological importance of NATs but could also have various functional implications. Very recently, our group has successfully performed interference of PU.1 (also known as SPI1) AS lncRNA to substantially promote PU.1 mRNA translation, leading to inhibition of adipogenesis18. We have now identified a novel NAT that corresponds to Sirtuin type 1 (Sirt1) AS lncRNA.
Sirt1, a NAD-dependent class III protein deacetylases, is a member of sirtuins family19. Some studies suggested that Sirt1 regulated the balance between myoblast proliferation and differentiation20. Upregulation of Sirt1 promoted muscle precursor cell proliferation by inhibiting cell cycle inhibitor expression21,22. On the contrary, Sirt1 suppressed myoblasts differentiation through deacetylation of MEF223. Therefore, Sirt1 has opposite effects in myoblast proliferation and differentiation. Precise regulation of Sirt1 expression is crucial to balance myoblast proliferation and differentiation. Our previous study identified that Sirt1 AS lncRNA transcribed from Sirt1 AS strand and overlapped with Sirt1 mRNA 3′ untranslated region (3′ UTR)24, but its function and mechanism in myogenesis are still unclear.
In this study, the proliferation and differentiation were further detected in myoblasts of Sirt1 AS or/and miR-34a overexpression, and regulatory mechanism were explored using the dual luciferase reporter, RNA stability and ribonuclease protection assay, as well the mice of Sirt1 AS overexpression were studied by intraperitoneal injection of adenovirus on their weights, body composition and muscle fiber characteristics. Our findings provided a novel regulatory mechanism: it was that Sirt1 AS lncRNA preferably interacted with Sirt1 mRNA forming RNA duplex by competing with miR-34a to inhibit muscle formation.
Results
The expression patterns of Sirt1 mRNA, Sirt1 AS lncRNA and miR-34a during myoblast proliferation
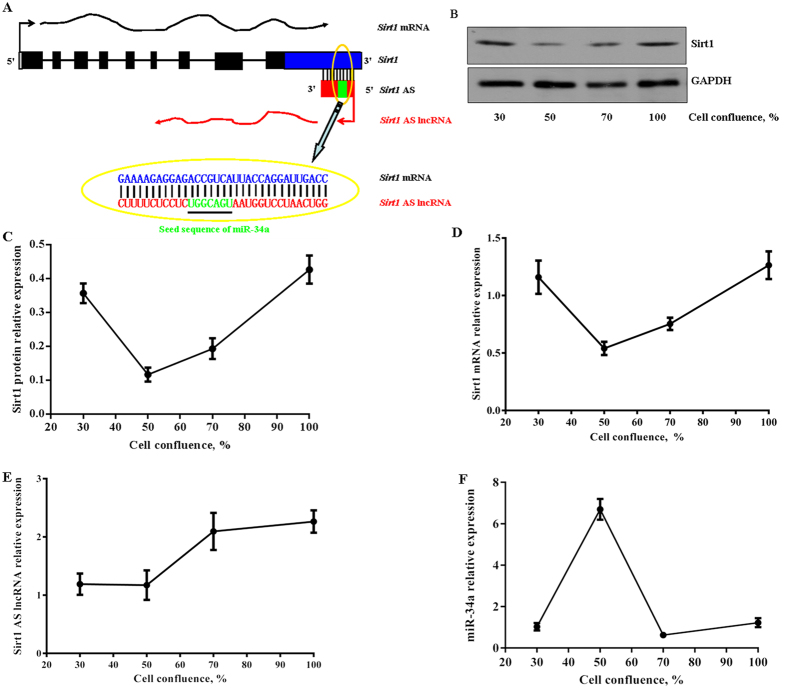
Our previous study showed the stability and expression of Sirt1 AS lncRNA overlapped with partial of Sirt1 3′ UTR24. Interestingly, in this study, we found that the target sequence of miR-34a to Sirt1 mRNA was in the overlapping region (Fig. 1A). To examine the expression patterns of Sirt1 AS lncRNA, Sirt1 mRNA and miR-34a, their levels were detected by qPCR or western blotting during myoblast proliferation, respectively. The results indicated that the levels of Sirt1 protein were concordant with Sirt1 mRNA, which decreased in 50% cell confluence and increased from 70% to 100% cell confluence (Fig. 1B–D). In addition, the levels of Sirt1 AS lncRNA gradually increased during myoblast proferation (Fig. 1E), whereas the levels of miR-34a increased in 50% cell confluence and decreased in 70% cell confluence (Fig. 1F).
Figure 1. Expression of Sirt1 AS lncRNA, mRNA and miR-34a during C2C12 myoblast proliferation.
(A) Putative schematic of mouse miR-34a and bidirectional transcriptional Sirt1 transcripts including mRNA and AS lncRNA. (B,C) Expression of Sirt1 protein. (D) Expression of Sirt1 mRNA. (E) Expression of Sirt1AS lncRNA. (F) Expression of miR-34a.
Sirt1 AS lncRNA attenuated the inhibition of Sirt1 expression against miR-34a
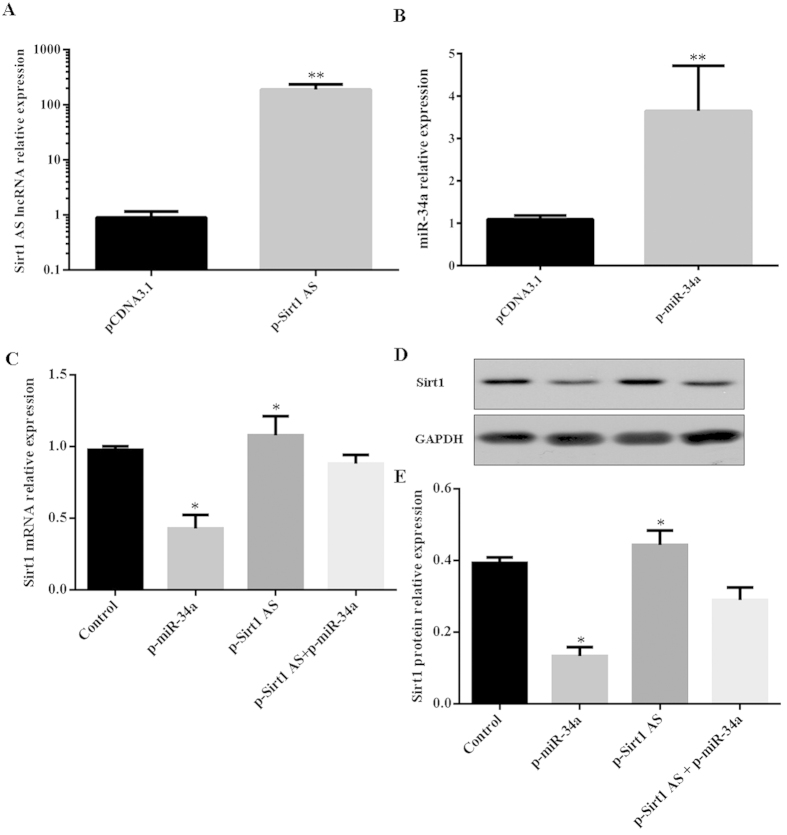
To explore the regulation of Sirt1 AS lncRNA and miR-34a to Sirt1 expression, Sirt1 AS and miR-34a expression plasmids were constructed. The results showed that expression efficiencies of Sirt1 AS and miR-34a were about 200-fold and 4-fold in myoblasts, respectively (Fig. 2A,B). In addition, Sirt1 AS lncRNA was elevated more than 40-fold by 100 μM resveratrol using strand-specific RT-PCR analysis (Fig. S1). Furthermore, miR-34a overexpression downregulated the levels of Sirt1 mRNA and protein, whereas Sirt1 AS overexpression upregulated the levels of Sirt1 mRNA and protein (Fig. 2C–E). Interestingly, the levels of Sirt1 mRNA and protein were rescued when Sirt1 AS and miR-34a were co-overexpressed (Fig. 2C–E).
Figure 2. Sirt1 AS overexpression recovered the levels of Sirt1 mRNA and protein against the function of miR-34a in C2C12 myoblasts.
(A) Sirt1 AS overexpression in myoblasts. GAPDH was employed as an internal reference. (B) miR-34a overexpression. (C) The levels of Sirt1 mRNA in myoblasts transfected with p-Sirt1 AS (pCDNA3.1-Sirt1 AS), p-miR-34a (pCMV-miR-34a) and p-Sirt1 AS plus p-miR-34a. (D,E) The levels of Sirt1 protein in myoblasts transfected with p-Sirt1 AS, p-miR-34a and p-Sirt1 AS plus p-miR-34a. The levels of Sirt1 protein were detected by western blotting. GAPDH protein was applied as internal reference. The data were presented as means ± SEM of 4 independent experiments. *P < 0.05 and **P < 0.01.
Sirt1 AS lncRNA promoted myoblast proliferation
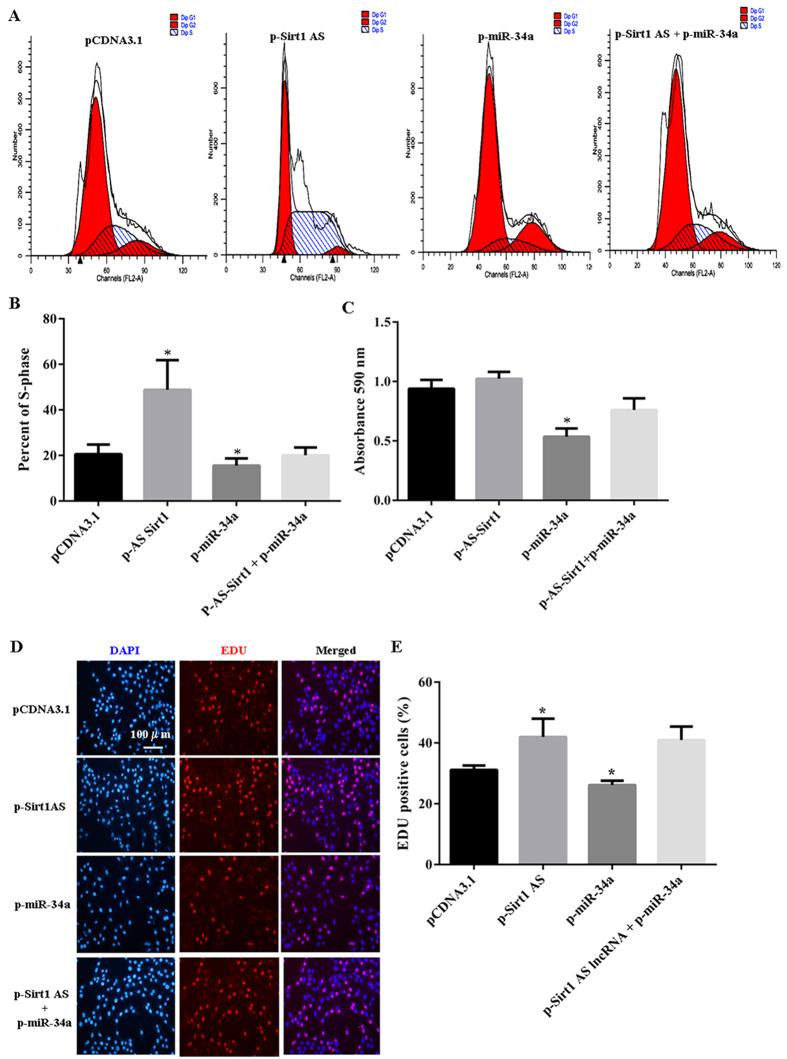
To discover the functional Sirt1 AS lncRNA associated with myoblast proliferation, we detected proliferation of the cells transfected with Sirt1 AS expression vector by flow cytometry, cell counting kit-8 (CCK-8) and 5-ethynyl-2′- deoxyuridine (EDU) analysis. As shown in Fig. 3A,B, Sirt1 AS overexpression increased the number of cells which were in DNA synthesis (S) phage of cell cycle, whereas miR-34a overexpression decreased this number. Furthermore, the ability of myoblasts to proliferation was rescued when Sirt1 AS and miR-34a were co-overexpressed (Fig. 3A,B). The CCK8 detection confirmed that Sirt1 AS lncRNA was against inhibiting myoblast proliferation of miR-34a (Fig. 3C). In addition, the above results were further verified by EDU assay as well. The ratio of EDU positive cells indicated the cell in DNA synthesis phage. Sirt1 AS overexpression increased the ratio of EDU positive cells and resisted the inhibitory myoblast proliferation of miR-34a (Fig. 3D,E).
Figure 3. Sirt1 AS overexpression promoted myoblast proliferation.
(A,B) Cell cycle analysis using flow cytometry for myoblasts of Sirt1 AS, miR-34a and Sirt1 AS plus miR-34a overexoression. (C) CCK-8 cell proliferation analysis. (D) EdU labeling and immunocytochemical staining of proliferating cells. (E) The quantification analysis of EdU-positive cells. Results were indicated as the mean ± SEM of 4 independent experiments. *P < 0.05.
Sirt1 AS lncRNA rescued the expression of proliferation genes by resisting miR-34a
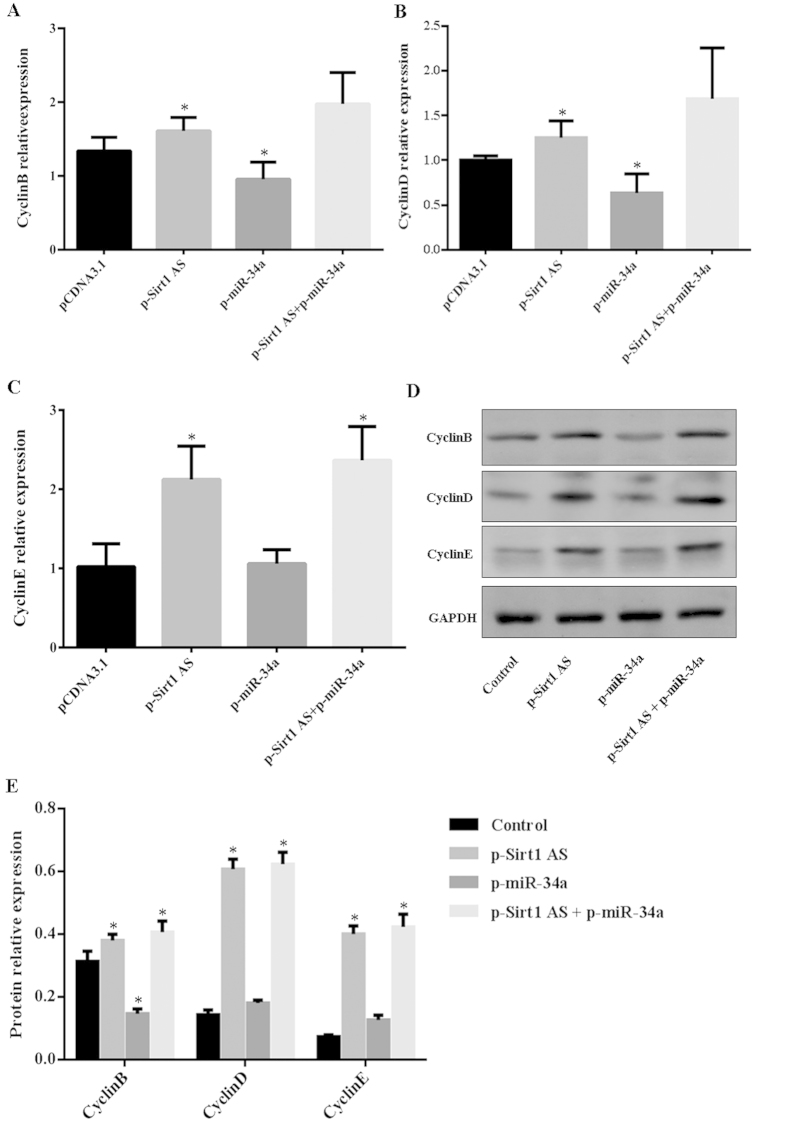
To further elucidate the regulation at gene level, the expression of proliferation genes were examined in C2C12 cells transfected with the expression vectors of Sirt1 AS, miR-34a and both of them, respectively. The results indicated that Sirt1 AS overexpression upregulated the mRNA levels of CyclinB, CyclinD and CyclinE, but miR-34a overexpression downregulated the mRNA levels of CyclinB and CyclinD (Fig. 4A–C). Moreover, Sirt1 AS overexpression rescued the levels of CyclinB, CyclinD and CyclinE when Sirt1 AS and miR-34a were co-overexpressed (Fig. 4A–C). In addition, western blotting analysis of CyclinB, CyclinD and CyclinE further confirmed that Sirt1 AS lncRNA promoted their expression and attenuated the inhibition of miR-34a on the expression of proliferation marker genes (Fig. 4D,E).
Figure 4. Sirt1 AS overexpression increased the levels of Cyclins.
(A–C) The mRNA levels of CyclinB, CyclinD and CyclinE were upregulated in C2C12 cells of Sirt1 AS overexpression, but downregulated in miR-34a overexpression at 24 hours after treatment with growth medium. (D,E) The levels of CyclinB, CyclinD and CyclinE proteins in C2C12 cells of Sirt1 AS, miR-34a and Sirt1 AS plus miR-34a overexoression at 24 hours after induced with growth medium. Results were indicated as the mean ± SEM of 4 independent experiments. *P < 0.05.
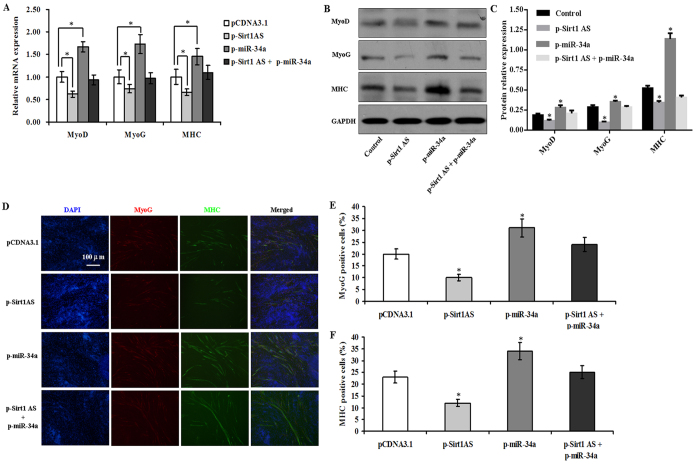
Sirt1 AS lncRNA inhibited myoblast differentiation
Upregulation of Sirt1 protein level by Sirt1 AS lncRNA suggested that it may be an inhibitory myogenic factor during myoblast differentiation. To investigate Sirt1 AS lncRNA relevancy in myogenesis, we overexpressed Sirt1 AS in C2C12 cells. The results showed that Sirt1 AS overexpression downregulated the levels of MyoD, MyoG and myosin heavy chain (MHC) mRNA and protein, whereas miR-34a overexpression upregulated the levels of above gene expression (Fig. 5A–C). Interestingly, Sirt1 AS overexpression decreased the levels of these genes when both Sirt1 AS and miR-34a were co-overexpressed (Fig. 5A–C). Moreover, the results were also strengthened by immunofluorescence staining for MHC and MyoG protein in differentiating myotubes on day 2 post-transfection (Fig. 5D). In addition, the number of positive myotubes was reduced by Sirt1 AS overexpression, but was increased by miR-34a overexpression (Fig. 5E,F). Interestingly, the number of positive myotubes was recovered by co-overexpression of both Sirt1 AS and miR-34a (Fig. 5E,F).
Figure 5. Sirt1 AS overexpression inhibited myoblast differentiation.
(A–C) Sirt1 AS overexpression downregulated the levels of myogenic gene (MyoD, MyoG and MHC) mRNA and protein. (D) Immunocytochemical staining. (E,F) The quantification analysis of MyoG and MHC positive nuclei. The data were presented as means ± SEM of 4 independent experiments. *P < 0.05.
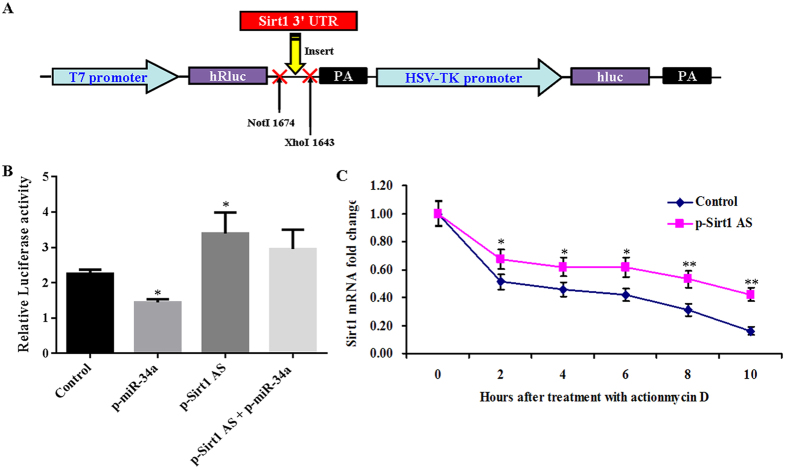
Sirt1 AS lncRNA increased the stability of Sirt1 mRNA by binding to its 3′ UTR via competing with miR-34a
To probe into the mechanism underlying the myogenic-mediated function of Sirt1 AS lncRNA, we constructed Sirt1 3′ UTR psiCHECK™-2 vector for luciferase reporter assay (Fig. 6A). The results indicated that Sirt1 AS lncRNA increased relative luciferase activity by directly interacting with Sirt1 3′ UTR in 293T cells transfected with Sirt1 AS expression vector, whereas miR-34a decreased relative luciferase activity by targeting Sirt1 3′ UTR in 293T cells transfected with miR-34a expression vector (Fig. 6B). Compare with miR-34a transfection, Sirt1 AS lncRNA partially rescued the relative luciferase activity in 293T cells co-transfected with Sirt1 AS lncRNA and miR-34a (Fig. 6B). Therefore, our results suggested that Sirt1 AS lncRNA bound to Sirt1 3′ UTR by competing with miR-34a to regulate Sirt1 gene transcription. Moreover, the RNA stability assay illustrated that Sirt1 AS overexpression prolonged Sirt1 mRNA half-life from 2 to 10 h after treatment with actionmy D (Fig. 6C), implying that Sirt1 AS lncRNA increased the stability of Sirt1 mRNA though interacting with Sirt1 3′ UTR by competing with miR-34a.
Figure 6. Sirt1 AS lncRNA promoted transcription of Sirt1 by attenuating the role of miR-34a.
(A) Schematic of Sirt1 3′ UTR inserted into psiCHECK™-2 vector. (B) Luciferase activity analysis for Sirt1 AS, Sirt1 and miR-34a. (C) Sirt1 AS lncRNA makes Sirt1 mRNA much more stable. The data were presented as means ± SEM of 4 independent experiments. *P < 0.05 and **P < 0.01.
To directly confirm the interaction between Sirt1 AS lncRNA and Sirt1 mRNA, ribonuclease protection assay (RPA) and RT-PCR were performed on the cytoplasmic RNA from C2C12 cells. We found that the ratio of cytoplasmic Sirt1 mRNA/nuclear Sirt1 mRNA did not changed after Sirt1 AS overexpression, but the ratio of cytoplasmic Sirt1 AS lncRNA/nuclear Sirt1 AS lncRNA increased (Fig. 7A). Furthermore, the RT-PCR primers were designed according to putative schematic of Sirt1 AS lncRNA and Sirt1 mRNA (Fig. 7B). RPA showed that cytoplasmic sense/antisense RNA duplex formation occured was fully overlapping between the two strands, resulting in protection (Fig. 7C).
Figure 7. Sirt1 AS lncRNA fully bound to Sirt1 mRNA on basement of basyl complementary pairing principle in myoblasts.
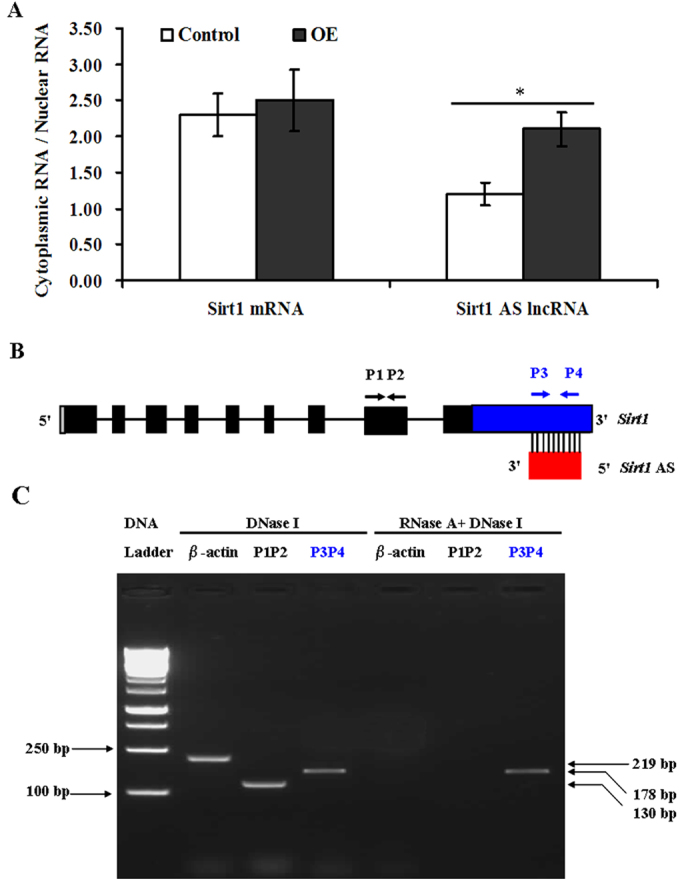
(A) Sirt1 AS lncRNA and Sirt1 mRNA existed in cytoplasm and nucleus. *P < 0.05. (B) Primer positions for ribonuclease protection assay (RPA). (C) Detection of the Sirt1 AS lncRNA/mRNA pairs. OE: Sirt1 AS overexpression.
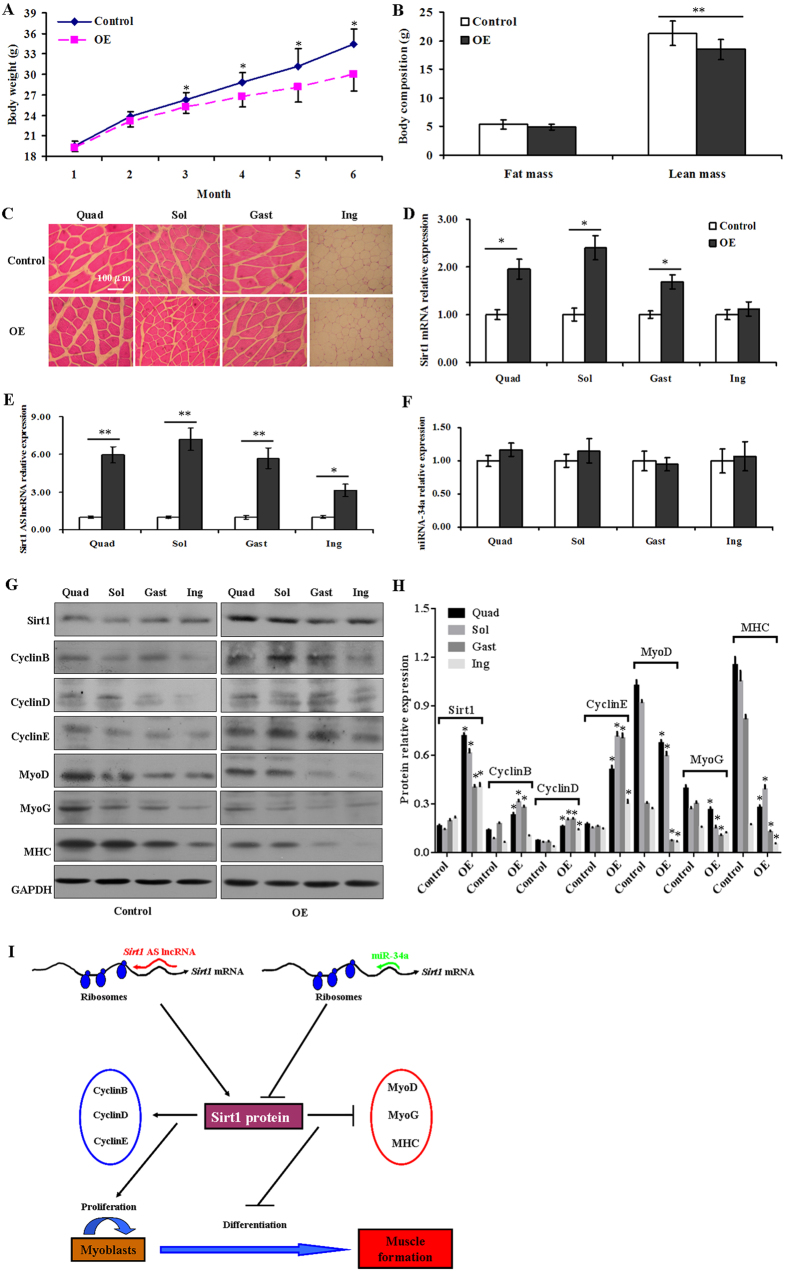
Sirt1 AS lncRNA inhibited muscle formation
To further confirm whether Sirt1 AS lncRNA was implicated in muscle formation in vivo, mouse intraperitoneal injection of adenovirus-mediated Sirt1 AS overexpression was performed. The results showed that mouse body weights decreased in Sirt1 AS overexpression treatment than in the control at month 3, 4, 5 and 6 (Fig. 8A). Moreover, compared with the control, the lean mass was less in Sirt1 AS overexpression treatment, whereas the fat mass did not change at month 6 (Fig. 8B). Muscle fiber size of quadriceps (Quad), soleus (Sol) and gastrocnemius (Gast) were apparently greater in Sirt1 AS overexpression treatment than in the control, but fat cell size of inguinal fat (Ing) did not alter at month 6 (Fig. 8C). The levels of Sirt1 mRNA and Sirt1 AS lncRNA were upregulated in Quad, Sol and Gast in Sirt1 AS overexpression treatment at month 6 (Fig. 8D,E). Interestingly, the levels of miR-34a did not change in Quad, Sol, Gast and Ing between the treatment and control (Fig. 8F). In addition, the protein levels of Sirt1 and the proliferation genes including CyclinB, CyclinD and CyclinE increased, but protein levels of the differentiation gene including MyoD, MyoG and MHC decreased (Fig. 8G,H). Collectively, the above results suggested that Sirt1 AS lncRNA was associated with myogenesis in vivo. Therefore, Sirt1 AS lncRNA fully bound to Sirt1 mRNA forming RNA duplex to regulate muscle formation by competing with miR34a (Fig. 8I).
Figure 8. Sirt1 AS overexpression inhibited muscle development.
(A) Body weight. (B) Body composition analysis. (C) HE staining. (D) Levels of Sirt1 mRNA. (E) Levels of Sirt1 AS lncRNA. (F) Levels of miR-34a. (G/H) Levels of Sirt1, CyclinB, CyclinD, CyclinE, MyoD, MyoG and MHC proteins. The results were presented as means ± SEM of at least 4 independent experiments. *P < 0.05, **P < 0.01. (I) Model for Sirt1 AS lncRNA fully bound to Sirt1 mRNA to regulate muscle formation by competing with miR34a. OE: Sirt1 AS overexpression, Quad: quadriceps, Sol: soleus, Gast: gastrocnemius, Ing: inguinal fat.
Discussion
The intrinsic nature and complex secondary structures of lncRNAs enable them to specifically interact with DNA, RNA and proteins. On the basement of relationship of Sirt1 AS lncRNA, Sirt1 mRNA and miR-34a sequences, we speculated that Sirt1 AS lncRNA bound to Sirt1 mRNA by competing with miR-34a to regulate the efficiency of Sirt1 protein expression in myoblast proliferation and differentiation. Our results supported the above hypothesis. In this study, Sirt1 AS lncRNA promoted myoblast proliferation by upregulating the expression of cell cyle genes including CyclinB, CyclinD and CyclinE, whereas it inhibited myoblast differentiation though repressing the expression of the myogenic factors MyoD, MyoG and MHC. The results were further confirmed by overexpression of Sirt1 AS in vivo. Therefore, we uncovered the novel functional roles of Sirt1 AS lncRNA in mediating muscle formation. As shown in Fig. 8I, functionally and mechanistically, Sirt1 AS lncRNA fully bound to Sirt1 mRNA to regulate muscle formation by competing with miR34a.
Although a large number of mammalian lncRNAs were pervasively identified by lncRNA-seq, only a minority were functionally explored. Our previous studies found that PU.1 AS lncRNA was involved in adipogenesis25 and immunifaction26. In this study, our results clearly demonstrated that the expression of Sirt1 AS lncRNA in myoblast proliferation was increased, which was similar with Sirt1 mRNA, but opposite to miR-34a. It has been reported that miR-34a targeted Sirt1 to inhibit Sirt1 expression in embryonic stem cells27, neural stem cell28 and colon cancer cells29. Therefore, the expression patterns of Sirt1 and miR-34a implied that miR-34a targets Sirt1 in C2C12 cells. By luciferase reporter assay, we proved that both miR-34a and Sirt1 AS lncRNA directly targeted Sirt1 3′ UTR. Interestingly, the levels of Sirt1 and miR-34a were not strictly opposite, because more than 16 miRNAs regulate Sirt1 expression and activity30, we thought that the other miRNAs as well were implicated in regulation of Sirt1 expression in C2C12 proliferation. Moreover, miR-34a was not only targets Sirt1 but also other genes31 to exert its biological functions. Surprisingly, Sirt1 AS lncRNA resisted the roles of miR-34a in myoblast proliferation and differentiation, implying that interaction existed among Sirt1 AS lncRNA, Sirt1 mRNA and miR-34a.
In this study, we focused on Sirt1 AS lncRNA and investigated relationship among Sirt1 AS lncRNA, Sirt1 mRNA and miRNA-34a in myogenesis. Based on our results, we found that Sirt1 AS lncRNA preferably interacted with Sirt1 mRNA forming RNA duplex to promote Sirt1 translation by competing with miR-34a, inhibiting muscle formation. Up to now, the effects of Sirt1 protein on myoblast proliferation and differentiation had been investigated very well21,32,33,34,35,36,37. In addition, our previous studies indicated the expression of Sirt1 AS lncRNA in mouse various tissues including heart, kidney, liver, spleen, brain, muscle and white adipose tissue24. Therefore, we thought that the expression of Sirt1 AS lncRNA was not localized to specific cell type.
Because lncRNAs are enriched in the nucleus or the cytosol, they can act at virtually every level of gene expression38,39. In this study, we found that Sirt1 AS lncRNA was in both nucleus and cytoplasm, but its levels increased in myoblasts of Sirt1 AS overexpression. Moreover, the RNA stability assay indicated Sirt1 AS overexpression prolonged Sirt1 mRNA half-life. The RPA assay further confirmed that Sirt1 AS lncRNA bound to its mRNA forming mRNA/AS lncRNA compound in the myoblast cytoplasm. Taken together, we suggested that Sirt1 AS lncRNA elevated translational efficiency of Sirt1 mRNA by RNA duplex. However, whether nuclear Sirt1 mRNA/AS lncRNA duplex affects sense RNA processing including capping, polyadenylation, nuclear localization and transport, will need further exploration.
Here, we need point out that interference of Sirt1 AS lncRNA using RNAi will influence sense mRNA as well40, because Sirt1 AS lncRNA is fully overlapping with Sirt1 3′ UTR by base complementary pairing principle. Additionally, AS transcripts including Sirt1 AS lncRNA are generally low in abundance and are, on average, more than 10-fold lower in abundance than sense expression40,41,42. It was why we overexpressed Sirt1 AS lncRNA to explore its myogenic function, but not interfered in this study.
Because the target site of miR-34a located in Sirt1 mRNA and AS lncRNA overlapping region, the function of miR-34a was studied as well. As expected, miR-34a overexpression decreased the levels of Sirt1 expression and suppressed C2C12 cell proliferation but promoted the cell differentiation. Whether Sirt1 AS lncRNA interacted with miR-34a regulates Sirt1 expression was still unknown. Recent studies showed that lncRNAs involved in miRNAs processing regulation16,17. In addition, lncRNA also influenced miRNAs function by directly “sponging”11,15. Interestingly, we here found that Sirt1 AS lncRNA bound to Sirt1 3′ UTR by competing with miR-34a to attenuate the functional role of miR-34a. Therefore, we thought that AS lncRNA regulated cognate gene mRNA through competing with miRNA may be an extensive novel mechanism.
AS lncRNA is an important class of lncRNAs, which transcripts from antisense strand40. AS lncRNAs which were partially or fully complementary to sense mRNA in the nucleus or/and cytoplasm, decided their specific functional mechanisms1. For example, apolipoprotein A1 (APOA1) AS lncRNA can regulate the histone methylation patterns of sense APO gene cluster through guiding histone-modifying enzymes lysine (K)-specific demethylase 1 (LSD1) or suppressor of zeste 12 homolog (SUZ12) to APO gene cluster42. Ubiquitin carboxy-terminal hydrolase L1 (Uchl1) AS lncRNA regulated Uchl1 translation through an embedded inverted short interspersed nuclear element B2 (SINEB2) element which activated polysomes43. AS lncRNA and mRNA formed double strand duplex to recruit Staufen 1 (STAU1) protein to mRNAs and mediate their degradation44. In our study, Sirt1 AS overexpression increased Sirt1 mRNA stability, implying that Sirt1 AS lncRNA regulated Sirt1 expression at translational level. Moreover, we found that Sirt1 AS lncRNA bound to Sirt1 3′ UTR though competing with miR-34a to resist downregulation of Sirt1 level resulting from miR-34a.
In the cytoplasm, lncRNAs affect translational output in different ways. They can control gene expression by reducing or stimulating mRNA decay44,45. A particular class of cytoplasmic lncRNAs, the competing endogenous RNAs (ceRNA), regulates both the translation and the degradation rates of mRNAs by acting as molecular “sponges” for miRNAs, thus modulating the repressive activity of miRNA on their mRNA targets46,47,48,49,50. On the basement of our above findings, we here defined a novel regulatory mechanism of AS lncRNA, which is that AS lncRNA can also make mRNA stability and compete with miRNA to influence translational output to regulate muscle formation.
Intraperitoneal injection of adenoviral vectors was validated as an efficient gene manipulation tool for overexpressing recombinant proteins in vivo51,52. Of course, gene editing (CRISP/Cas9) and transgenic technologies may be also the good methods. Moreover, our lab first identified Sirt1 AS lncRNA and investigated its stability and expression. Fortunately, we also found that Sirt1 AS lncRNA was elevated more than 40-fold by resveratrol (an agonist of Sirt153,54). Based on our findings, we think that Sirt1 AS lncRNA may be a biomarker which indicates muscle formation.
Collectively, our findings unravel a novel molecular mechanism on muscle formation by Sirt1 AS lncRNA. Mechanically, full complementary binding of Sirt1 AS lncRNA to Sirt1 mRNA forming RNA duplex in the cytoplasm facilitated Sirt1 translation output by competing with miR-34a, inhibiting myogenesis via promoting myoblast proliferation and inhibiting differentiation. The elucidation of mechanism on Sirt1 protein output by Sirt1 AS lncRNA will provide insight into novel pathways of regulatory myogenesis.
Methods
Cell culture and transfection
C2C12 cell line was purchased from ATCC (Rockville, MD, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM: Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS: Hyclone Laboratories, Logan, UT) at 37 °C and 5% CO2. After cells reached confluence, they were induced to myogenic differentiation by replacing 10% FBS with 2% horse serum (Gibco, Carlsbad, CA). For cell proliferation detection, cells were transfected with Sirt1 AS lncRNA, miR-34a, Sirt1 AS lncRNA plus miR-34a and pCDNA3.1 empty plasmids using Lipofectamine 2000 (Life Technologies, Shanghai, China) according to the manufacturer’s instructions when the cells reached 50–60% confluence after seeding for 12 hours. Twenty-four hours post-transfection, the cells were harvested and preserved at −80 °C for further proliferation assay. For cell differentiation analysis, cells were seeded in six-well plates with culture medium and transfected with above plasmids on the first day of myogenic differentiation. Forty-eight hours post-transfection, they were harvested and preserved at −80 °C for further myogenic differentiation detection.
Real-time qPCR
Cell total RNA was extracted according to the standard method with a TRIzol reagent (Takara, Kyoto, Japan). Reverse transcription was performed to synthesize cDNA by using the RT Kit (Takara, Kyoto, Japan). The primers used for real-time qPCR were designed and synthesized (Table S1). Real time qRT-PCR was carried out in IQ5 real-time PCR detection system (Bio-Rad, Hercules, CA) using SYBR-Green (Vazyme). The mRNA data was quantified using the comparative threshold cycle (ΔΔCT) methods.
Strand-specific RT-PCR
Strand-specific RT-PCR was performed as our previous publications18,26. Briefly, before reverse transcription reactions, 1μg cells total RNA was performed to wipe out DNA contamination by gDNA wiper Mix (Vazyme) at 42 °C for 2 min. Reverse transcription reactions were carried out using RT kit (Vazyme). The strand-specific primers, reverse primer of Sirt1 AS lncRNA or miR-34a RT primer as the reverse transcription primers. The RT was at 50 °C for 30 min, 85 °C for 5 s in 10 μl volumes.
Cytoplasmic and nuclear lncRNA
RNAs were extracted from C2C12 cells according to merchant guide using Nuclear or Cytoplasmic RNA Purification Kit (Fisher BioReagents) as previously described55. Briefly, cell pellet was resuspended in buffers of RNA Purification Kit and followed by centrifugation at 4 °C twice. The supernatant was saved as cytoplasmic fraction and the pellet was used as nuclear fraction. RNAs were extracted from both fractions using Trizol. The relative levels of Sirt1 AS lncRNA and mRNA were analyzed by real-time qPCR.
Western blotting
The cellular protein was extracted in the RIPA lysis buffer (Appligen, Beijing, China) with 1 mM PMSF. Peterson’s method was used to quantify the protein contents. Western blotting were performed as our previously described23. Following primary antibodies were used: Sirt1 (1:500; Santa Cruz Biotechnology, Dallas, TX), CyclinD (1:500; Boster, Wuhan, China), CyclinB (1:500; Santa Cruz Biotechnology, Dallas, TX), CyclinE (1:1000; Santa Cruz Biotechnology, Dallas, TX), MyoD (1:1000; Santa Cruz Biotechnology, Dallas, TX), MyoG (1:1000; Santa Cruz Biotechnology, Dallas, TX), MHC (1:2000; Sigma, St. Louis, MO) and GAPDH (1:1000; Santa Cruz Biotechnology, Dallas, TX). The secondary antibodies were anti-rabbit, anti-goat and anti-mouse antibodies (Santa Cruz Biotechnology, Dallas, TX). The targeted proteins were detected using the Gel Doc XR System and analysis software Quantity One (Bio-Rad, Hercules, CA) as per the instructions of the manufacturer.
Flow cytometric analysis
Flow cytometric analysis was carried out as our previous publication56. Twenty-four hours post-transfection, the cells were washed 3 times with cold PBS and fixed in 70% (v/v) cold ethanol overnight. Then centrifugation and the supernatants were discarded, the cells were washed with PBS again and resuspended in 500μl propidium iodide (50 μg/ml; DOJINDO, Shanghai, China) solution with 100 μg/ml RNase A and 0.2% (v/v) TritonX-100 at 4 °C for 30 min. The cells were detected with a flow cytometry instrument (Becton Dickinson, FACSCa-libur, Franklin, GA). Cell cycle phases were assigned to G0/G1, S, and G2/M according to the amount of DNA. The proportion of cells in each cell cycle phase was statistically analyzed.
CCK-8 proliferation detection
C2C12 cells were seeded in 96-well plates at 4 × 103 cells per well with 100 μl DMEM. The Cells were transfected with Sirt1 AS lncRNA, miR-34a, Sirt1 AS lncRNA plus miR-34a and pCDNA3.1 empty plasmids. Twenty-four hours post-transfection, cell proliferation index was detected using CCK-8 kit (Beyotime, Shanghai, China) according to manufacturer’s instructions.
EDU proliferation analysis
Twenty-four hours post-transfection, C2C12 cells were treated with 10 μM EdU (Ribobio) and incubated for 3 h. EDU staining were processed according to the manufacturer’s protocol. Cell nucleus was stained with 5 μg/ml DAPI (Roche, Penzberg, Germany) for 10 min. The cells were visualized by a fluorescence microscope (Nikon, Tokyo, Japan). The ratio of positive cells (EdU-staining cells/the total of cells) was calculated.
Immunocytochemistry
Immunocytochemistry were performed according to the recently published method55. Briefly, cultured cells were fixed with 4% paraformaldehyde for 10 min at 4 °C, washed 3 times with PBS, and permeabilized using 0.3% TritonX-100 in TBS. Following blocking with 2% goat serum in TBS for 1 h at room temperature, the cells were incubated with primary antibodies MHC (1:200) or MyoG (1:200) at 4 °C overnight with gentle shaking, washed, and then incubated with secondary antibodies (AlexaFluor-488 conjugated mouse IgG1 and AlexaFluor-568 conjugated mouse IgG2b (1:1000, Invitrogen)) for 1 h at room temperature. Cells were then washed 3 times with TBST, stained with DAPI, and photographed using a NIKON TE2000-U fluorescent microscope (Nikon, Tokyo, Japan).
Luciferase reporter assay
The partial 3′ UTR of mouse Sirt1 mRNA was amplified by PCR and inserted into the psiCHECK™-2 Vector (Promega, Madison, USA). The primers used to construct plasmids for luciferase reporter assay were shown in Table S2. Sirt1 AS lncRNA and miR-34a expression plasmids were co-transfected with Sirt1 3′ UTR psiCHECK™-2 vector into 293T cells by Transfection Reagent (Roche, Penzberg, Germany), respectively. Forty-eight hours post-transfection, the cells were harvested, incubated with cell lysis buffer, and the luciferase activity was then measured using ELIASA (PerkinElmer, Waltham, MA) according to the manufacturer’s instructions.
RNA stability assay
To detect the stability of Sirt1 mRNA, C2C12 cells were treated with 2μg/ml Actinomycin D (Sigma-Aldrich, Louis, MO) which could suppress transcription. The cells were harvested at 0, 1, 2, 6 and 10 h post treatment, and total cellular RNA was extracted to detect the residual mRNAs by real time qPCR. GAPDH mRNA was acted as internal control, because was stable within 32 h.
Ribonuclease protection assay
To detect the sense-antisense RNA duplex, ribonuclease protection assay (RPA) and RT-PCR were performed on the total RNA from C2C12 cells. This step was conducted according to the previous studies57,58 with a few changes. Sequence of oligonucleotides used for RPA was listed in Table S3. Cytoplasmic RNA was orderly digested by DNaseI (Fermentas) and RPA-grade RNaseA (Applied Biosystems) to remove all the genomic DNA contamination and single-strand RNAs. Then the residues, endogenous double-strand RNAs (dsRNA) were applied in the RT reaction catalyzed by the reverse transcriptase Superscript III (Invitrogen). The reaction system was incubated at 55 °C for 60 min and terminated at 75 °C for 10 min. Finally, the double-strand cDNA was amplified in 25 μl PCR reaction system. After 35-cycle amplification, the products were checked by electrophoresis on 1.5 % agarose gel with ethidium bromide staining.
Animal studies
C57B/L6 mice were housed in the animal facilities of Northwest A&F University under conventional conditions with constant temperature and humidity and fed a standard diet. All mice experiments were carried out in accordance with the protocol approved by the Animal Ethics Committee of Northwest A&F University and the experimental protocol was performed in accordance with applicable guidelines and regulations. Intraperitoneal injection of adenoviral vectors was validated as an efficient gene manipulation tool for overexpressing recombinant proteins in vivo51. For treatment with adenovirus-mediated Sirt1 AS in vivo according to our previous publication52, 2-month-old mice were injected with 500 μl adenovirus (Ad-Sirt1 AS or Ad-EGFP) into the cavitas abdominalis every 10 days until 180 days of age. Whole body composition analysis was carried out per month using EchoMRI-100 (Houston, TX, USA). Mice were sacrificed and muscle and fat tissues were harvested on day 180, and total RNAs and proteins were extracted for real-time RT-PCR and western blotting analyses. Mice (4~6) were used in each group.
Frozen section and HE staining
For Frozen section and HE staining of mouse Quad, Sol, Gast, Ing sections were collected on day 180 for Sirt1 AS lncRNA overexpression. Frozen section and HE staining were performed according to the recently published method59. The sections were observed and taken through pictures with microscope (Olympus, New York, NY).
Statistical analysis
All experimental data were obtained through at least four independent experiments. Values were showed as means ± SEM. Statistical analysis was performed in GraphPad Prism version 6 (GraphPad software, La Jolla). Student’s t test is used for individual comparisons. Multiple comparisons are assessed by one-way ANOVA followed by Dunnett’s tests. Difference between groups was considered statistically significant if P < 0.05.
Additional Information
How to cite this article: Wang, G.-q. et al. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 6, 21865; doi: 10.1038/srep21865 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation (31572366 and U1201213), from the National Basic Research Programs of China (2015CB943102 and 2012CB124705), from the Natural Science Foundation of Shaanxi Province (2015JM3096), and from NWAFU Basic Science Research Programme (2452015149).
Footnotes
Author Contributions Conceived and designed the experiments: W.J.P. Performed the experiments: G.Q.W., Y.W., Y.X., X.C.C., M.L.M., Y.G., Y.M.S. and R.C. Analyzed the data: G.Q.W., Y.W., Y.X., X.C.C. and W.J.P. Contributed reagents/materials/analysis tools: W.J.P. and G.S.Y. Wrote the paper: G.Q.W., Y.W., Y.X., X.C.C. and W.J.P. All authors have read and approved the final manuscript.
References
- Faghihi M. A. & Wahlestedt C. Regulatory roles of natural antisense transcripts. J. Nat Rev Mol Cell Biol. 10, 637–643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. J. Development. 139, 3543–3552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y. et al. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. J. Cell Stem Cell. 11, 231–241 (2012). [DOI] [PubMed] [Google Scholar]
- Potthoff M. J. & Olson E. N. MEF2: a central regulator of diverse developmental programs. J. Development. 134, 4131–4140 (2007). [DOI] [PubMed] [Google Scholar]
- Luo W., Nie Q. & Zhang X. MicroRNAs involved in skeletal muscle differentiation. J. J Genet Genomics. 40, 107–116 (2013). [DOI] [PubMed] [Google Scholar]
- Neguembor M. V., Jothi M. & Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. J. Skelet Muscle. 4, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. & Steitz J. A. The noncoding RNA revolution—trashing old rules to forge new ones. J. Cell. 157, 77–94 (2014). [DOI] [PubMed] [Google Scholar]
- Lu L. et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. J. Embo J. 32, 2575–2588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U. A. & Shiekhattar R. Noncoding RNAs and enhancers: complications of a long-distance relationship. J. Trends Genet. 27, 433–439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K. et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. J. Mol Cell. 51, 606–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. J. Cell. 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V. G. et al. H19 antisense RNA can up-regulate Igf2 transcription by activation of a novel promoter in mouse myoblasts. J. PLoS One. 7, e37923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts R. et al. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. J. Am J Physiol Cell Physiol. 304, C995–1001 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou Y. et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. J. Development. 137, 2643–2652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen A. N. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. J. Mol Cell. 52, 101–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liz J. et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. J. Mol Cell. 55, 138–147 (2014). [DOI] [PubMed] [Google Scholar]
- Wang, K.et al. MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. J. PLoS Genet, 10, e1004467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N. et al. PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. J. Animal genetics. 46, 133–140 (2015). [DOI] [PubMed] [Google Scholar]
- North B. J. & Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. J. Genome Biol. 5, 224 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo P. S. & Boriek A. M. The physiological roles of Sirt1 in skeletal muscle. J. Aging (Albany NY). 3, 430–437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbone C. R., Booth F. W. & Lees S. J. Sirt1 increases skeletal muscle precursor cell proliferation. J. Eur J Cell Biol. 88, 35–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar K. S. & Hubbard B. P. Lifespan and healthspan extension by resveratrol. J. Biochim Biophys Acta. 1852, 1209–1218 (2015). [DOI] [PubMed] [Google Scholar]
- Fulco M. et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. J. Mol Cell. 12, 51–62 (2003). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Identification, stability and expression of Sirt1 antisense long non-coding RNA. J. Gene. 539, 117–124 (2014). [DOI] [PubMed] [Google Scholar]
- Pang W. J. et al. Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation. J. J Cell Biochem. 114, 2500–2512 (2013). [DOI] [PubMed] [Google Scholar]
- Wei N. et al. Knockdown of PU.1 mRNA and AS lncRNA regulates expression of immune-related genes in zebrafish Danio rerio. J. Dev Comp Immunol. 44, 315–319 (2014). [DOI] [PubMed] [Google Scholar]
- Tarantino C. et al. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. J. Faseb J. 24, 3255– 3263 (2010). [DOI] [PubMed] [Google Scholar]
- Aranha M. M. et al. miR-34a regulates mouse neural stem cell differentiation. J. PLoS One. 6, e21396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M., Ferlito M. & Lowenstein C. J. miR-34a repression of SIRT1 regulates apoptosis. J. Proc Natl Acad Sci USA 105, 13421–13426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M. MicroRNA Regulation of SIRT1. J. Front Physiol. 3, 68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. C. et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. J. Mol Cell. 26, 745–752 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M. et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. J. Dev Cell. 14, 661–673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. GATA-binding protein 4 (GATA-4) and T-cell acute leukemia 1 (TAL1) regulate myogenic differentiation and erythropoietin response via cross-talk with Sirtuin1 (Sirt1). J. J Biol Chem. 287, 30157–30169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. & Goldberg A. L. SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscleatrophy and promotes muscle growth. J. J Biol Chem. 288, 30515–30526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre A. et al. Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts during muscle cell differentiation. J. Cell Cycle. 13, 78–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubicic V. et al. Resveratrol induces expression of the slow, oxidative phenotype in mdx mouse muscle together with enhanced activity of the SIRT1-PGC-1α axis. J. Am J Physiol Cell Physiol. 307, C66–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. et al. Mitochondrial complex I deficiency enhances skeletal myogenesis but impairs insulin signaling through SIRT1 inactivation. J Biol Chem. 289, 20012–20025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P. J. & Chang H. Y. Long noncoding RNAs: cellular address codes in development and disease. J. Cell. 152, 1298–1307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S. et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. J. Genome Biol. 15, R6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V. & Steinmetz L. M. Gene regulation by antisense transcription. J. Nat Rev Genet. 14, 880–893 (2013). [DOI] [PubMed] [Google Scholar]
- Ozsolak F. et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. J. Cell. 143, 1018–1029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley P. et al. Regulation of the Apolipoprotein Gene Cluster by a Long Noncoding RNA. J. Cell Reports. 6, 222–230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. J. Nature. 491, 454–457 (2012). [DOI] [PubMed] [Google Scholar]
- Gong C. & Maquat L. E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. J. Nature. 470, 284–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M. et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. J. Nature. 493, 231–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. J. Nature. 465, 1033–1038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. J. Cell. 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. J. Dev Cell. 25, 69–80 (2013). [DOI] [PubMed] [Google Scholar]
- Hansen T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. J. Embo J. 30, 4414–4422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. J. Nature. 495, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- Saimura M. et al. Intraperitoneal injection of adenovirus-mediated NK4 gene suppresses peritoneal dissemination of pancreatic cancer cell line AsPC-1 in nude mice. J. Cancer Gene Ther. 9, 799–806 (2002). [DOI] [PubMed] [Google Scholar]
- Cheng J. et al. Ectopic expression of RBP4 impairs the insulin pathway and inguinal fat deposition in mice. J. J Physiol Biochem. 70, 479–486 (2014). [DOI] [PubMed] [Google Scholar]
- Bai L. et al. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. J. Mol Cell Biochem. 307, 129–140 (2008). [DOI] [PubMed] [Google Scholar]
- Pang W. J. et al. Lentivirus-mediated Sirt1 shRNA and resveratrol independently induce porcine preadipocyte apoptosis by canonical apoptotic pathway. J. Mol Biol Rep. 40, 129–139 (2013). [DOI] [PubMed] [Google Scholar]
- Lu L. et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. J. EMBO J. 32, 2575–2588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H. et al. microRNA-151-3p regulates slow muscle gene expression by targeting ATP2a2 in skeletal muscle cells. J. J Cell Physiol. 230, 1003–1012 (2015). [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Sun B. K. & Lee J. T. Intersection of the RNA interference and X-inactivation pathways. J. Science. 320, 1336–1341 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. G. et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. J. Blood. 115, 133–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Q. et al. Mulberry 1-Deoxynojirimycin Inhibits Adipogenesis by Repression of the ERK/PPARγ Signaling Pathway in Porcine Intramuscular Adipocytes. J. J Agric Food Chem. 63, 6212–6220 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.