Abstract
Fungal infections are an increasing clinical problem. Decreased treatment effectiveness is associated with biofilm formation and drug recalcitrance is thought to be biofilm specific. However, no systematic investigations have tested whether resistance mechanisms are shared between biofilm and planktonic populations. We performed multiplexed barcode sequencing (Bar-seq) screening of a pooled collection of gene-deletion mutants cultivated as biofilm and planktonic cells. Screening for resistance to the ergosterol-targeting fungicide amphotericin B (AmB) revealed that the two growth modes had significant overlap in AmB-persistent mutants. Mutants defective in sterol metabolism, ribosome biosynthesis, and the TORC1 and Ras pathways showed increased persistence when treated with AmB. The ras1, ras2 and tor1 mutants had a high-persister phenotype similar to wild-type biofilm and planktonic cells exposed to the TORC1 pathway inhibitor rapamycin. Inhibition of TORC1 with rapamycin also increased the proportion of persisters in Candida albicans and Candida glabrata. We propose that decreased TORC1-mediated induction of ribosome biosynthesis via Ras can lead to formation of AmB-persister cells regardless of whether the cells are in planktonic or biofilm growth mode. Identification of common pathways leading to growth mode-independent persister formation is important for developing novel strategies for treating fungal infections.
Advances in medical procedures have increased the use of invasive devices and immunosuppressive treatments. This has led to increased numbers of patients susceptible to fungal infections1. The biomaterial of medical implants is suitable for fungal cell attachment and use of invasive devices is a risk factor for fungal biofilm infections2. Biofilm cells can survive high doses of antimicrobial agents and only echinocandins and polyenes have in vitro antibiofilm activity3,4. The polyene drug amphotericin B (AmB) targets ergosterol in the cell membrane and forms pores that rapidly lyse cells5. These fungicidal properties and broad spectrum of activity have made AmB the preferred agent for treatment of severe mycosis since its introduction in the late 1950s.
Clinical reports of fungal resistance to AmB are rare and known resistance mechanisms are limited to alterations in cell wall or sterol membrane patterns5,6. Nonetheless, biofilms are thought to become tolerant to AmB by sequestering the drug in the extracellular matrix7,8,9, decreasing membrane ergosterol levels10,11, or forming persister cells12. Persister cells remain viable after treatment with high doses of antimicrobial agents without heritable genetic changes. A persister subpopulation is typically about 1% of a population and consists of phenotypically tolerant variants of wild-type genotype. Once antimicrobial pressure is removed, this subpopulation can repopulate the infection site13. The clinical relevance of persister cells has been demonstrated in patients with oral candidiasis who receive antimicrobial therapy that selects for high-persister (hip) mutants. Persister mutants with hip phenotypes have minimal inhibitory concentrations (MICs) similar to wild-type, but generate a higher proportion of tolerant cells. Antifungal recalcitrance mediated by persister cells is a survival mechanism that might contribute significantly to treatment failure13, but cannot be detected by standard laboratory susceptibility tests. The mechanisms of persister formation in bacteria are well studied and involve toxin-antitoxin systems that inhibit protein synthesis and result in cellular dormancy13. Less is known about persistence in yeast, although histone deacetylases14 and superoxide dismutases15 are suggested to be involved.
Although biofilms and stationary phase planktonic populations share phenotypic properties including low metabolic activity, phenotypic heterogeneity, increased stress tolerance and persister formation16,17,18,19, biofilm research has mainly focused on differences between the two growth modes. Studies of tolerance mechanisms shared by planktonic and biofilm-forming cells could lead to discovery of novel treatment strategies that function independent of growth mode. One approach to characterizing general AmB-persister mechanisms is identifying mutants that have hip phenotypes under different growth modes. Saccharomyces cerevisiae is an experimental model for fungal biofilm studies20,21,22 and we have previously observed AmB-tolerant persisters in S. cerevisiae biofilm and planktonic populations17. Comprehensive barcoded gene-deletion strain collections are available for S. cerevisiae that enable the systematic study of protein function and genotype-to-phenotype correlations21,23,24. The unique barcode tags of each mutant and next generation sequencing facilitate multiplexed barcode-sequencing (Bar-seq) for high-throughput screens of pooled mutants25,26. To identify growth mode-independent persister mechanisms, we performed a genome-wide Bar-seq analysis of AmB-tolerance in S. cerevisiae biofilm and planktonic populations using a gene deletion collection in a biofilm competent strain21.
We found significant overlap in AmB-tolerance mutants between biofilm- and planktonic-growing cells, and many mutants uniquely identified in either growth mode had lost functions in metabolic and proliferative processes found to be important for tolerance in both growth modes. Several mutants were hip-phenotype mutants characterized by wild type minimal inhibitory concentration (MIC) values but with an increased subpopulation of AmB-persistent cells relative to wildtype. Many of these mutants were impaired in the regulation of growth and translation, suggesting a common mechanism underlying persister formation in planktonic growth and biofilms. In summary, we used genome-wide multiplexed Bar-seq analysis to identify hip-phenotype yeast mutants to understand how persistence arises in microbial populations. Our genetic analysis suggests that one mechanism leading to persistence is mediated by a general down-regulation of protein translation controlled by the TORC1 and Ras pathway activities.
Results
AmB-tolerant cells have a persister phenotype
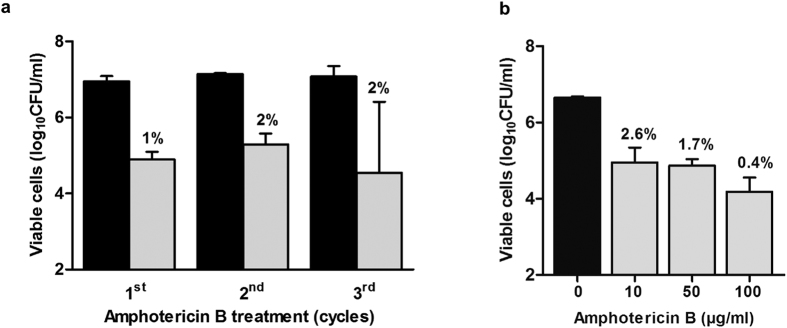
Biofilms formed by C. albicans, survive AmB treatment by the formation of persister cells that are characterized by noninherited resistance12. We investigated if AmB resistance was also noninherited in our S. cerevisiae biofilm model by treating 24-hour biofilms with AmB and re-treating new biofilms formed by persisters with AmB for a total of three treatment cycles (Fig. 1a). The first cycle killed most cells, with 1% surviving AmB treatment. The biofilm populations produced by the surviving cells showed similar AmB tolerance, with 2% viability. No significant differences in the sizes of the AmB-tolerant populations were observed during the cycles (P = 0.7314, one-way ANOVA), suggesting that the survival mechanism was not heritable. Since persister cells survive treatment with high concentrations of antifungal agents, we determined the correlation between drug concentration and cell viability of the AmB-tolerant subpopulation. Exposing biofilm cells to 10 μg/ml AmB for 24 hours (Fig. 1b) resulted in survival of 2.6% of the population. Exposure to 50 μg/ml led to survival of 1.7% and treatment with 100 μg/ml resulted in survival of 0.4%. Although there was a trend towards fewer persisters with increasing AmB treatment, it was not statistically significant (P = 0.0517, one-way ANOVA).
Figure 1. AmB persistence is not heritable.
(a) 48-hour wild-type biofilms treated with 10 μg/ml amphotericin B (AmB) for 24 hours (gray) or untreated (black). Viability was measured as colony-forming units (CFUs). Surviving populations were reinoculated to form new biofilms that were re-exposed to AmB for three cycles. (b) Survival of 48-hour wild type biofilms treated for 24 hours with 10, 50, or 100 μg/ml AmB. n = 3, error bars show standard deviation.
Screening for AmB-tolerant hip mutant strains using multiplexed barcode sequencing
We used Bar-seq to identify genes that contributed to the generation of AmB-tolerant persister cells in biofilm and planktonic populations. A barcoded mutant collection was pooled and four biological-replicate cultures were distributed to plastic surfaces for biofilm growth or glass flasks for planktonic growth. After 4 days, biofilm and planktonic cells were treated with AmB or left untreated. After another 12 hours, treated and untreated cell samples were outgrown for 24 hours to enrich for surviving mutants before barcode sequencing (Supplementary Fig. S1). Differential mutant abundance after AmB treatment was analyzed using the Bar-seq analysis pipeline developed by Robinson et al.27.
Dimensionality reduction of the mutant survival data showed that all biological replicates for the untreated control samples grouped together and untreated control samples for biofilm and planktonic growth were in the same cluster (Supplementary Fig. S2). This result indicated high similarity in the abundance of mutants in the biofilm and planktonically cultivated populations. For AmB-treated samples, three of four biological replicates were grouped for both the biofilm and planktonic grown cells. After treatment of the pooled biofilm population with AmB, the mean viable population decreased to 0.3% of initial population size (Fig. 2). AmB treatment of the planktonic population resulted in mean survival of 2% of initial population size. Hence, the tolerant population was approximately 10 times larger after planktonic growth compared to biofilm growth showing a higher proportion of tolerant cells among the planktonically grown cells (P = 0.0043, Student’s t-test).
Figure 2. Barcode sequencing screen.
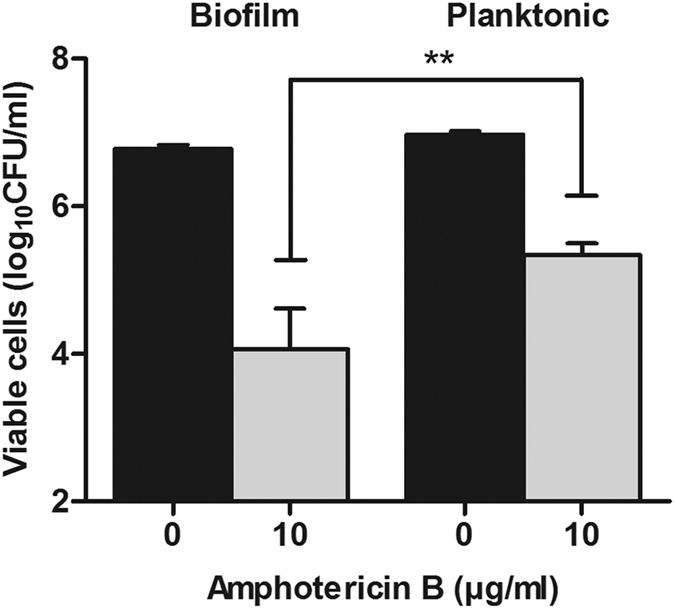
Number of viable cells in the pooled mutant collection measured as colony-forming units (CFUs) of untreated control (black bars) and amphotericin B-treated cells (grey bars). n = 4, error bars show standard deviation. Statistical significance between AmB treated samples of biofilm and planktonic cells was evaluated with Student’s t-test. **P < 0.005.
Mutants affected in lipid metabolism, protein synthesis and cell cycle genes increase AmB tolerance
Bar-seq analysis identified 69 mutants with significant (FDR < 0.005) differential abundance in biofilm cells after AmB treatment and 121 in planktonic cells (Supplementary Fig. S3 and Supplementary Table S1). Of these, 32 mutants were shared between the two growth modes representing a significant overlap (P < 0.0001, two-tailed Fisher’s exact test). No gene ontologies were significantly overrepresented among the mutants. However, following manual curation using GO Term Finder in SGD28 (Supplementary Table S2) mutants enriched in both biofilm and planktonic populations indicated similar persistence mechanisms in biofilm and planktonic cells (Table 1).
Table 1. Mutants enriched in the barcode-sequencing screen after amphotericin B treatment.
| Biological Function | Growth Modes for Mutant Identification |
||
|---|---|---|---|
| Biofilm | Planktonic | Biofilm+Planktonic | |
| Lipid metabolism | tgl1 cki1 | ncp1 inp51 ura7 ecm22 | sac1 erg5 cyb5 tlg2 |
| Translation | rpp1b ssz1 | rps22b rpl40b rpl42a rps17b rpl6b rpl24a | rpl8b rps21a |
| Protein metabolism | hrt3 mnn4 | rpn10 ste14 glr1 ubc4 ptc7 | eug1 doa4 jem1 |
| RNA metabolism | brr1 pet127 isu1 ngl2 | set2 ssn2 sas2 gbp2 syf2 ski2 hbs1 ynr024w sfl1 tea1 vts1 nut1 edc3 pih1 pnc1 nsr1 trm11 yll029w nam7 | ssn3 |
| Cell cycle | ptc2 bud9 ndt80 mrc1 gic1 hsl7 | rme1 msc7 far8 pcl9 ama1 | elm1 gin4 |
| Intracellular transport | shr5 ypt7 pep3 age2 | srp101 aps3 pex21 tom5 jjj1 fmp37 vtc1 fmp43 btn2 vps13 arx1 yer128w xdj1 | osh2 vta1 apm3 |
| Small molecule metabolism | ypl033c | trp4 bna2 gcv1 | sah1 ykl151c |
| TORC1 signaling | gtr2 | gtr1 meh1 slm4 slm1 | |
| Other functions | gal83 mdh2 pck1 sak1 ynr064c srl2 pbp2 YBL055C | cis3 spo71 dit2 qcr8 rip1 yjl055w inp1 ylr073c pgm2 ymr099c ynl191w mms2 gyp1 arn1 sam3 | ras2 sma2 ybr238c rho3 izh3 vht1 zrt1 |
| Unknown | ylr283w om45 dlt1 ylr064w ylr278c ybr255w ypl068c | ecm27 pib2 ynl208w yal049c ylr125w rmd8 met12 mpa43 ytp1 ymr181c ylr407w ygr259c yor292c nrp1 ybr016w yor139c ybl081w tma16 | asg1 ygr182c ygr190c yhr159w |
Mutants significantly more abundant in mixed populations of knockout mutants after treatment with amphotericin B in biofilm and planktonic populations. Mutants are classified according to the gene ontology of the corresponding wild type alleles by biological processes. See Supplementary Table S2 for more details.
AmB binds to cell membrane ergosterol that result in pore formation and rapid cell lysis5. Thus it was expected to find mutants in the sterol biosynthesis genes in our screen. cyb5 and erg5 increased AmB survival in biofilm and planktonically cultivated cells, while mutation of two ergosterol biosynthesis genes, NCP1 and ECM22, conferred resistance in planktonic cells only (Table 1). Deletion of these genes likely resulted in structural modifications of sterols leading to reduced binding of AmB to the plasma membrane. Structural modification of ergosterol is a known resistance mechanism against AmB5,29 and the identification of ergosterol mutants positively validated the Bar-seq screen. In addition, deletion of the sterol transporter gene OSH2 and the lipid homeostasis regulator gene SAH1 increased AmB survival in both growth modes (Table 1). Sterol involvement in AmB survival was further supported by increased abundance of the sterol homeostasis mutant tgl1 in biofilm-cultivated cells. Identification of enriched cki1 in biofilm cells, inp51 in planktonic cells and sac1 in both cell types suggested involvement of phosphatidylcholine and sphingolipid membrane lipids in AmB sensitivity.
A large group of mutants had defects in genes involved in cell cycle progression, protein synthesis and protein turnover; we identified several ribosomal proteins and ubiquitination-associated genes in both biofilm and planktonic cells. Loss of the protein kinases Elm1 and Gin4, which are involved in novel daughter cell growth and septin ring formation30, led to enrichment in both planktonic and biofilm populations. Enrichment of bud9, gic1, hsl7 in biofilm and pcl9 in planktonic cultures after AmB treatment supported a role for bud growth and polarization in S. cerevisiae susceptibility to AmB.
Mutants produce a high proportion of AmB-tolerant persister cells
To investigate if AmB-tolerant mutants possessed a hip phenotype, we compared MIC values and persister cell levels in individually cultured mutants and wild-type. We included ssn3 (cyclin-dependent regulation of RNA polymerase II), erg5 (ergosterol biosynthesis) and sac1 (phosphatidylinositol dephosphorylation), which were the most significantly differentially abundant mutants with known biological processes and were AmB persistent in both biofilm and planktonic cultures. We included gin4 (bud growth and assembly of the septin ring) to represent the large group of mutants affected in cell cycle progression. One of the characteristics of hip mutants is that they have wild-type MIC values. Consistent with a hip phenotype, MIC determinations showed that only erg5 had reduced AmB susceptibility whereas all other tested mutants had MIC values similar to wild-type (Fig. 3). Because the MIC assay measures the ability to grow in the presence of an antifungal agent, these results suggested that the high level of AmB tolerance of the mutants could be attributed to a nongrowing state similar to persister cells.
Figure 3. Persister cell levels in enriched amphotericin B-tolerant mutants.
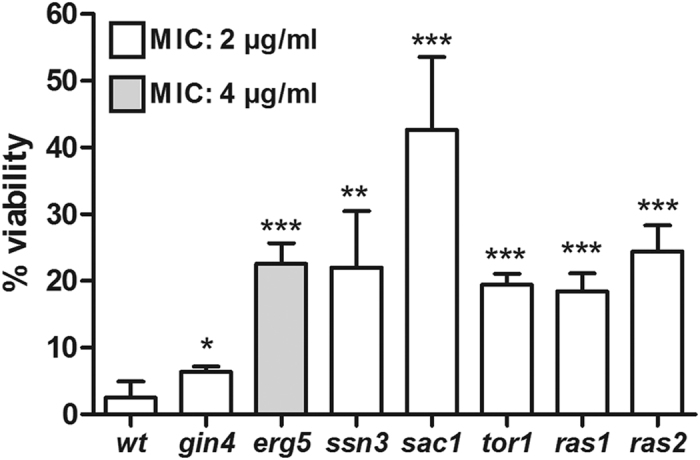
Viability was determined as colony-forming units (CFUs) after two hours exposure to amphotericin B. MIC, Minimal inhibitory concentration. wt, wild-type; n = 3–6; error bars show standard deviation. Statistical significance was evaluated with Student’s t-test. *P < 0.05, **P < 0.005, ***P < 0.0001.
To investigate AmB persister cell levels in the mutants, we treated S. cerevisiae with AmB and determined viability before and after treatment. Treatment of all tested mutants significantly increased the surviving subpopulations (Fig. 3) from 3% in the wildtype to 6% in gin4 (P = 0.0342, Student’s t-test), 23% in erg5 (P < 0.0001), 22% in ssn3 (P = 0.0009) and 43% in sac1 (P < 0.0001). These results showed that, with the exception of erg5, the mutants were not directly resistant to AmB, but were enriched in the screen because they produced more persister cells than wild-type. We therefore considered them hip mutants.
TORC1 and Ras pathways are involved in AmB tolerance
One group of mutants enriched in both planktonic and biofilm populations were deleted in the TORC1-activating EGO complex genes (gtr1, gtr2, meh1, and slm4)31,32 (Table 1). The highly conserved TORC1 pathway regulates cellular growth, is activated by nutrients, and inhibited by rapamycin33. The TORC1 pathway regulates cell cycle initiation and translation by transcriptional induction of ribosome protein genes34,35. The increased AmB tolerance in ribosomal protein and EGO mutants could indicate involvement of the TORC1 pathway in AmB tolerance for both biofilm and planktonic cells.
The tor1 and tor2 mutants were not represented in our Bar-seq screen because tor2 is essential and tor1 had low abundance and data from tor1 were removed during data analysis. To investigate if TORC1 itself is involved in AmB sensitivity, we exposed tor1 cells to AmB. The surviving mutant subpopulation was 19% and significantly (P < 0.0001, Student’s t-test) larger than the surviving wild-type population (Fig. 3). This increase was not caused by a change in MIC value (Fig. 3), suggesting a hip phenotype for tor1. TORC1 links nutrient sensing to induction of translation via the Ras pathway35. Among the 32 mutants with increased AmB tolerance in both biofilm and planktonic cultivated cells, the identification of ras2 supported a potential connection between TORC1 and protein synthesis in AmB tolerance. Like the tor1 mutant, the ras2 culture did not have an altered MIC (Fig. 3) but showed a significantly (P < 0.0001, Student’s t-test) increased number of cells that survived AmB treatment (24%, Fig. 3). We also analyzed mutations in RAS1, a RAS2 paralog, and found an AmB tolerance phenotype similar to tor1 and ras2 (Fig. 3).
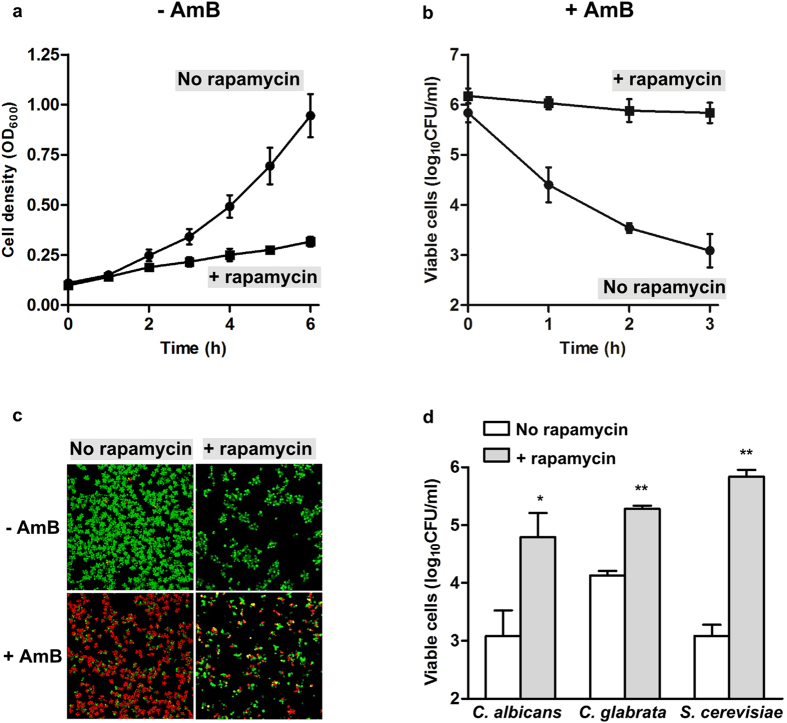
We investigated if growth arrest by chemical inhibition of TORC1 increased the proportion of AmB-tolerant persister cells. Rapid killing kinetics was observed after AmB treatment of exponentially growing cells resulting in killing of >99% of the cells after three hours (Fig. 4). Treatment of the wild-type with the TORC1-inhibiting agent rapamycin reduced growth (Fig. 4a) and resulted in slow AmB killing, with a ~500-fold increase in AmB tolerant cells compared to control (Fig. 4b). Rapamycin exposure also increased the proportion of biofilm cells that survived AmB treatment. Rapamycin-treated biofilm cells showed inhibited growth and a lower population density than untreated cells (Fig. 4c). Growth arrest of biofilm cells induced by rapamycin, followed by AmB treatment resulted in a higher proportion of surviving cells compared to cells not pre-exposed to rapamycin (Fig. 4c). This result supports our genetic results showing that inactivation of TORC1 in S. cerevisiae increased survival of both biofilm and planktonic cells after AmB treatment.
Figure 4. TORC1 inhibition mediates amphotericin B tolerance in wild-type planktonic and biofilm cultures.
(a) Planktonic cells grown in YPD were treated with 1 μg/ml rapamycin (+rapamycin) or untreated (no rapamycin). Samples were extracted hourly for optical density measurements. n = 3, error bars show standard deviation. (b) Survival of exponential growing (No rapamycin) or rapamycin treated (+rapamycin, 1 μg/ml) planktonic cells exposed to 10 μg/ml amphotericin B (AmB). Viability was measured as colony-forming units (CFUs). n = 3, error bars show standard deviation. (c) Confocal laser scanning microscopy of AmB activity against four-hour biofilms after pre-exposure to 1 μg/ml rapamycin or no rapamycin. Survival is visualized with LIVE/DEAD staining. Green, live cells; red, dead cells. (d) Survival of exponential growing (No rapamycin) or rapamycin treated (+rapamycin, 1 μg/ml) planktonic cells exposed to five times MIC amphotericin B (AmB, 5 μg/ml against Candida spp. and 10 μg/ml against S. cerevisiae). Viability was measured as colony-forming units (CFUs) after 3 hours treatment. n = 3, error bars show standard deviation. Statistical significance between rapamycin treated and untreated samples was evaluated with Student’s t-test. *P < 0.05, **P < 0.001.
Inhibition of TORC1 significantly increases the proportion of AmB persisters in C. albicans and C. glabrata
To validate if the results obtained in the model organism S. cerevisiae can be translated into human fungal pathogens, we inhibited TORC1 with rapamycin in Candida albicans and Candida glabrata and investigated the level of AmB persister cells. Figure 4d shows the number of viable cells after AmB treatment, and compares persister levels in TORC1 inhibited cells (+rapamycin) with exponential growing controls (No rapamycin). The results show that inhibition of TORC1 generates a significantly larger proportion of AmB tolerant persister cells in C. albicans (P = 0.0485, Student’s t-test) and C. glabrata (P = 0.0003) compared to controls, similar to S. cerevisiae (P < 0.0003).
Discussion
Drug-tolerant persister cells survive high concentrations of antimicrobial agents while having MIC values similar to wild-type. Subpopulations of S. cerevisiae that tolerate AmB are phenotypically similar to AmB-tolerant persister cells in Candida biofilms12,17. Therefore, we performed a multiplexed Bar-seq screen using a S. cerevisiae gene deletion collection, to identify pathways and processes involved in AmB persistence in biofilm and planktonically cultivated cells. Our Bar-seq screen of S. cerevisiae led to identification of several mutants affected in (i) regulation of growth and in (ii) lipid metabolism, including several affected in ergosterol synthesis. Deletion of ergosterol synthesis genes likely resulted in decreased affinity of AmB to the plasma membrane, and AmB-tolerant persister cells might have lower levels of membrane ergosterol than susceptible cells. This conclusion is supported by the observation that Candida biofilms have decreased ergosterol levels12 and subpopulations of highly AmB-tolerant cells reduce transcription of ergosterol synthesis genes compared to the average biofilm11. The S. cerevisiae mutants that were enriched in the Bar-seq screen after AmB treatment had phenotypic properties similar to C. albicans hip mutants36: similar MIC value as the wild type, but a higher proportion of AmB-tolerant cells. hip mutants have been characterized in C. albicans, but the genetic basis for persistence is poorly understood.
We found that in S. cerevisiae, inhibition of the TORC1 complex, the TORC1-activating EGO complex and the TORC1-activated Ras pathway significantly increased the level of AmB tolerant cells, suggesting that reduced TORC1 and Ras activities were responsible for S. cerevisiae hip phenotypes. Ras signaling regulates programmed cell death in response to fungicidal exposure37,38, and persister cells might resist Ras-induced apoptosis that kills susceptible cells treated with antifungals14. In addition, ras1 and ras2 mutants have higher levels of AmB-tolerant persister cells than wild-type37, consistent with our results. Our finding of high AmB persistence in ras1, ras2, and tor1, and mutation of ribosomal protein genes suggests a general hip mechanism in S. cerevisiae caused by reduced ribosome synthesis through the TORC1-EGO-activated Ras pathway. TORC1 regulates ribosomal gene expression via Ras and the forkhead transcription factor Fhl135, therefore, this is likely to be one pathway for AmB persistence. Other Ras pathway members were not identified in our screen because they are essential (Ifl2, Esa1, Rap1), are not in the mutant collection (Fhl1), or have several isoforms that activate pathway steps (Tpk1, Tpk2, Tpk3).
How the TORC1-EGO-activated Ras response causes AmB persistence is not evident from our current data. However, a plausible hypothesis is that quiescent cells arrested in G0 contain persistent subpopulations that have reduced ability to produce ribosomes or another TORC1/Ras-regulated response. Quiescent cells survive extended periods of starvation and resume proliferation when growth conditions become favorable39,40 comparable to the phenotypic properties of persister cells. We suggest that TORC1-dependent or Ras-dependent quiescence leads to AmB persistence and that this is a mechanism for persister formation common to biofilm and planktonic growth modes. These results validate the use of S. cerevisiae as model organism for studying antifungal resistance in Candida spp.
We found 158 gene deletions affected survival of yeast cells grown as biofilms or planktonically. The diversity of processes involved in AmB tolerance was similar to the range of mechanisms that generate antimicrobial persisters in bacteria, which is hypothesized to result from fluctuating variability in cellular processes41,42. Many of the genes we identified in the Bar-seq screen might not be directly involved in ergosterol syntesis, TORC1-mediated or Ras-mediated AmB resistance but affect other processes that impact cell cycle progression. For example, we also identified high levels of AmB-tolerant persister cells compared to wild-type in mutants with defects in small molecule metabolism and intracellular transport in addition to a large group of genes with unknown protein function. It is therefore important to note that yet undiscovered mechanisms might be equally important as TORC1 and Ras for generation of AmB persister cells.
We found that biofilm and planktonic populations share AmB-tolerance mechanisms and that a common S. cerevisiae hip response involves the TORC1 and Ras pathways. Although we identified 37 mutants uniquely involved in AmB tolerance in biofilm and 89 uniquely involved in planktonic cells, several mutants are affected in overlapping biological processes. A significant number of genes involved in AmB tolerance overlapped between the two growth modes and 17 of the 20 most significant contributors to tolerance in biofilm cells were also significant contributors to planktonic cell survival. This finding does not exclude specific biofilm AmB-tolerance mechanisms in S. cerevisiae, but shows the existence of common tolerance mechanisms between biofilm and planktonic populations, which has also been observed in bacteria18,19,43.
Diagnosing patients with biofilm infections can be difficult and laborious44. Therapies for microbial infections that are independent of growth mode and target both biofilm and planktonic cells could lead to faster patient treatment. However, growth-independent treatment initiatives require knowledge of common drug-tolerance mechanisms and shared targets between biofilm and planktonic cells. Our findings identified shared drug-tolerance mechanisms that could serve as a foundation for developing novel treatment strategies against fungal infections that are independent of growth mode. Our data suggest that inducing exit from TORC1-mediated quiescence could be a strategy to reverse the tolerance phenotype of persister cells, but further studies are needed to investigate this. Organ transplant recipients treated with rapamycin as an immunosuppressant agent have increased risk of being infected with fungi, which often requires antifungal drug therapy to eradicate the infection45,46. Furthermore, observations of in vitro synergistic activity of rapamycin in combination with AmB suggests a potential as novel drug combination against systemic fungal infections47,48. However, our results raise the possibility that the use of rapamycin in combination with conventional antifungal agents may increase the probability of generating persister cells.
Materials and Methods
Strains
Saccharomyces cerevisiae Σ1278b YS-11 (MATa can1Δ::STE2p-spHIS5 lyp1Δ::STE3p-LEU2 his3::HisG leu2Δ ura3Δ) was used as reference wild-type strain21. The S. cerevisiae Σ1278b haploid gene-deletion collection (MATa can1Δ::STE2p-spHIS5 lyp1Δ::STE3p-LEU2 his3::HisG leu2Δ ura3Δ)21 was transferred from 96-well glycerol stock plates to yeast extract peptone dextrose (YPD) agar and incubated at 30 °C for three days. Colonies were subsequently pooled in YPD medium and incubated overnight in glass flasks with agitation at 30 °C to obtain a mixed population of haploid mutants. Candida albicans ATCC 90028 and Candida glabrata ATCC 90030 were obtained from American Type Culture Collection.
Media and growth conditions
All Bar-seq and all biofilm experiments were performed in minimal medium with 6.7 g/l yeast nitrogen base without amino acids (BD Difco) and 2.0 g/l glucose supplemented with amino acids (Supplementary Table S3). In all other experiments YPD was used to cultivate cells to verify the general applicability of our findings. All S. cerevisiae experiments were performed at 30 °C and all Candida spp. experiments were performed at 37 °C. AmB (A2411, Sigma) was dissolved in DMSO in 5 mg/ml stock solution and kept at −20 °C. Final DMSO concentrations in all assays were <1%49.
Minimal inhibitory concentration (MIC)
MIC was determined as previously described17. In short, a two-fold dilution series of AmB was prepared in fresh medium and distributed into 96-well microtiter plates. Visibly turbid overnight cultures were diluted in fresh medium and transferred to the microtiter plates in OD600 0.05 final cell densities. The microtiter plates were incubated with agitation (60 rpm) at 30 °C for 24 h. Biomass was measured with a microplate spectrophotometer (BioTek PowerWave 340). Growth inhibition of ≥90% was determined as MIC as recommended by EUCAST50. MIC values were determined in YPD medium and minimal medium with 2% glucose.
Phenotypic characterization of drug-tolerant biofilm subpopulation
Yeast cultures at OD600 0.1 were distributed into microtiter plates with a polystyrene surface. After 48 hours static incubation, medium with planktonic and loosely adherent cells was removed and centrifuged (2 min at 10,000 × g). Biofilm cells attached to the microtiter plate surfaces were challenged for 24 hours with the indicated concentrations of AmB in the spent medium. Biofilm cells were washed twice in saline and serially diluted in saline before plating on YPD agar. Inheritance of the drug-tolerant phenotype was investigated as previously described12 with modifications. In short, biofilm cells exposed to AmB at 10 times the MIC were suspended in fresh medium and transferred to a new microtiter plate. After two days, the biofilm cells were exposed to AmB at 10 times MIC. This process was repeated in a total of three re-inoculation and treatment cycles.
Bar-seq screen
The pooled mutant collection was inoculated in four separate cultures of 700 ml minimal medium at a cell density of 105 colony-forming units per ml (CFU/ml). After 5 hours with agitation at 30 °C, 50-ml samples were distributed in CELLSTAR Cell Culture Dishes (Greiner Bio) and sealed with parafilm to prevent evaporation for biofilm growth, or in glass flasks for planktonic growth. After 96 hours, the mean biofilm cell density was 6.1 × 106 CFU/ml and the mean planktonic cell density was 9.4 × 106 CFU/ml. AmB was added to the biofilm and planktonic populations at 10 μg/ml final concentration, corresponding to 10 times the MIC in minimal medium. AmB was added directly into planktonic cultures. For biofilm-growing cells, nonattached cells were removed from the medium by centrifugation (10 minutes at 4500 × g) and AmB was added to the spent medium to 10 μg/ml final concentration before reintroduction to biofilm cells. Drug-treated samples were protected from light with foil and incubated for 12 hours.
DNA extraction from viable mutants
Biofilm-growing cells were harvested by removing media with nonadhering cells, and washing the remaining biofilm population in saline. Biofilm cells were scraped off the surface with a Drigalski spatula and resuspended in saline. Planktonic-growing cells were centrifuged for 10 minutes at 4500 × g and pellets were washed once in saline. One ml harvested cell culture was transferred to 25 ml YPD and outgrown at 30 °C with agitation to enrich for viable mutants. Genomic DNA was extracted with Wizard Genomic DNA Purification kits (A1125, Promega).
Library preparation
Amplification of molecular barcodes from genomic DNA and incorporation of the Illumina adaptor sequences was performed in a two-step PCR protocol as previously described26 with a few modifications. In short, UPTAGs and DNTAGs were amplified in the first PCR reaction with unique index primers (Supplementary Table S4) and purified PCR concentrations were estimated from gel band intensities. UPTAGs and DNTAGs were combined in separate pools at 10 ng for each library. In a second PCR step, the Illumina adaptor sequence was incorporated into UPTAGs and DNTAGs in two separate PCR reactions. The two PCR products were combined in equimolar amounts and diluted to 2 nM final concentration. The library was sequenced in a single lane (HiSeq 2000, Illumina).
Data analysis
Sequence reads were matched to a library of known barcode sequences using BarNone with default settings25. The HO knockout strain was present multiple times in the 96-well stock plates that the deletion collection was pooled from. This strain accounted for 30% of total sequence reads and was removed from the dataset. UPTAG and DNTAG reads (Supplementary Fig. S4) within each biological replicate were summed and normalized to total reads per sample. Low-abundance mutants present at fewer than 100 counts per million in fewer than four samples were filtered, leaving 2051 mutants for analysis. The dataset followed an overdispersed Poisson distribution (Supplementary Fig. S5). EdgeR (version 3.2.4)27 was used for differential abundance analysis as previously described26,51. False discovery rate (FDR) < 0.005 was considered significant. Dimensionality reduction analysis was generated by EdgeR and compares the relationship between all pairs of samples, using a count-specific pairwise distance measure. Distances correspond to leading log-fold change between each pair of sample27,51.
Gene ontology
Genes significantly overrepresented after AmB treatment were manually assigned a biological process and grouped based on gene ontology (Supplementary Table S2) as annotated by the Saccharomyces Genome Database (SGD)28.
Persister level determination
Exponentially growing cells were dissolved in fresh, preheated YPD medium to OD600 0.1 in glass flasks. Cultures were incubated with agitation (200 rpm) and growth curves were performed by measuring optical density at absorbance 600 nm hourly. Exponentially growing cells were treated with five times the AmB MIC and viability was determined as CFUs. Rapamycin-induced target of rapamycin (TOR) inhibition was performed by exposing exponential growing populations to 1 μg/ml rapamycin (R0395, Sigma) prior to AmB treatment. AmB was added after four hours of rapamycin pre-exposure.
Confocal microscopy of biofilm
Visibly turbid cultures were diluted to OD600 0.1 and transferred to biofilm chambers (Technical University of Denmark17) with a polyvinyl chloride coverslip surface (Rinzl, Electron Microscopy Sciences) and cultivated for 4 hours before adding AmB for 3 hours. TORC1 inhibition of biofilm cells was by incubation of fresh cultures with 1 μg/ml rapamycin for 4 hours followed by 3 hours AmB treatment at 10 times the MIC. Staining for 15 minutes with 5 μM Syto9 (Invitrogen) was used to visualize live cells and 20 μM propidium iodide (Sigma-Aldrich) was used to stain dead cells. Confocal laser scanning microscopy (CLSM) was performed with a Zeiss LSM710 microscope equipped with excitation lasers at 488 nm and 514 nm. Imaging used an EC Plan-Neofluar 40x/1.30 oil lens.
Additional Information
How to cite this article: Bojsen, R. et al. A common mechanism involving the TORC1 pathway can lead to amphotericin B-persistence in biofilm and planktonic Saccharomyces cerevisiae populations. Sci. Rep. 6, 21874; doi: 10.1038/srep21874 (2016).
Supplementary Material
Acknowledgments
We thank Charlie Boone and Owen Ryan (University of Toronto) for the Σ1278b strains. This work was supported by the Danish Agency for Science Technology and Innovation is (FTP 10-084027) and the National Institutes of Health (NIH 1R01GM107466-01).
Footnotes
Author Contributions R.B. performed all experiments; R.B., B.R., D.G. and A.F. designed experiments; R.B., B.R., D.G. and A.F. analyzed data; R.B., B.R., D.G. and A.F. wrote the manuscript.
References
- Richardson M. D. Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother 56 Suppl 1, i5–i11 (2005). [DOI] [PubMed] [Google Scholar]
- Ramage G., Martinez J. P. & Lopez-Ribot J. L. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res 6, 979–86 (2006). [DOI] [PubMed] [Google Scholar]
- Bachmann S. P. et al. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob Agents Chemother 46, 3591–6 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn D. M., George T., Chandra J., Mukherjee P. K. & Ghannoum M. A. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Chemother 46, 1773–80 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Arango A. C., Scorzoni L. & Zaragoza O. It only takes one to do many jobs: Amphotericin B as antifungal and immunomodulatory drug. Front Microbiol 3, 286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock C. A., Barrett-Bee K. J. & Russell N. J. The lipid composition and permeability to azole of an azole- and polyene-resistant mutant of Candida albicans. J Med Vet Mycol 25, 29–37 (1987). [DOI] [PubMed] [Google Scholar]
- Nett J. et al. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother 51, 510–20 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J. E., Crawford K., Marchillo K. & Andes D. R. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother 54, 3505–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M., Henriques M., Lopez-Ribot J. L. & Oliveira R. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 55, 80–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Chandra J., Kuhn D. M. & Ghannoum M. A. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 71, 4333–40 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khot P. D., Suci P. A., Miller R. L., Nelson R. D. & Tyler B. J. A small subpopulation of blastospores in candida albicans biofilms exhibit resistance to amphotericin B associated with differential regulation of ergosterol and beta-1,6-glucan pathway genes. Antimicrob Agents Chemother 50, 3708–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur M. D., Kumamoto C. A. & Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother 50, 3839–46 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol 64, 357–72 (2010). [DOI] [PubMed] [Google Scholar]
- Al-Dhaheri R. S. & Douglas L. J. Apoptosis in Candida biofilms exposed to amphotericin B. J Med Microbiol 59, 149–57 (2010). [DOI] [PubMed] [Google Scholar]
- Bink A. et al. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob Agents Chemother 55, 4033–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates A. R. M. Dormancy and Low-Growth States in Microbial Disease, 292 (Cambridge University Press 2003). [Google Scholar]
- Bojsen R., Regenberg B. & Folkesson A. Saccharomyces cerevisiae biofilm tolerance towards systemic antifungals depends on growth phase. BMC Microbiol 14, 305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering A. L. & Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183, 6746–51 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl J. N., Zahller J., Roe F. & Stewart P. S. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 47, 1251–6 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T. B. & Fink G. R. Bakers’ yeast, a model for fungal biofilm formation. Science 291, 878–81 (2001). [DOI] [PubMed] [Google Scholar]
- Ryan O. et al. Global gene deletion analysis exploring yeast filamentous growth. Science 337, 1353–6 (2012). [DOI] [PubMed] [Google Scholar]
- Vandenbosch D. et al. Genomewide screening for genes involved in biofilm formation and miconazole susceptibility in Saccharomyces cerevisiae. FEMS Yeast Res 13, 720–30 (2013). [DOI] [PubMed] [Google Scholar]
- Giaever G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–91 (2002). [DOI] [PubMed] [Google Scholar]
- Andersen K. S. et al. Genetic Basis for Saccharomyces cerevisiae Biofilm in Liquid Medium. G3 (Bethesda) 4, 1671–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M. et al. Quantitative phenotyping via deep barcode sequencing. Genome Res 19, 1836–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. G., Chen W., Storey J. D. & Gresham D. Design and analysis of Bar-seq experiments. G3 (Bethesda) 4, 11–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40, D700–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C. M. et al. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 54, 3578–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan A. & Kellogg D. The elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol 19, 7983–94 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R. & Hall M. N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E. & De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 19, 15–26 (2005). [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R. & Hall M. N. TOR signaling in growth and metabolism. Cell 124, 471–84 (2006). [DOI] [PubMed] [Google Scholar]
- Barbet N. C. et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7, 25–42 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Soulard A. & Hall M. N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–79 (2004). [DOI] [PubMed] [Google Scholar]
- Lafleur M. D., Qi Q. & Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother 54, 39–44 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P., Camacho D. & Collins J. J. Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep 3, 350–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A. J., Crowe J. D. & Ramsdale M. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA 103, 726–31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C. et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol 174, 89–100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon A. D. et al. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol Biol Cell 19, 1271–80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison K. R., Brynildsen M. P. & Collins J. J. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol 14, 593–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. J. & Levin B. R. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet 9, e1003123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G., Robertson S. N. & Williams C. Strength in numbers: antifungal strategies against fungal biofilms. Int J Antimicrob Agents 43, 114–20 (2014). [DOI] [PubMed] [Google Scholar]
- Dahlan R., Patel A. & Haider S. Successful use of posaconazole to treat invasive cutaneous fungal infection in a liver transplant patient on sirolimus. Can J Infect Dis Med Microbiol 23, e44–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi S. B. et al. Endogenous Aspergillus endophthalmitis occurring after liver transplantation: a case report. Transplant Proc 41, 2933–5 (2009). [DOI] [PubMed] [Google Scholar]
- Vakil R., Knilans K., Andes D. & Kwon G. S. Combination antifungal therapy involving amphotericin B, rapamycin and 5-fluorocytosine using PEG-phospholipid micelles. Pharm Res 25, 2056–64 (2008). [DOI] [PubMed] [Google Scholar]
- Dannaoui E., Schwarz P. & Lortholary O. In vitro interactions between antifungals and immunosuppressive drugs against zygomycetes. Antimicrob Agents Chemother 53, 3549–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst E. J., Yodoi K., Roling E. E. & Klepser M. E. Rates and extents of antifungal activities of amphotericin B, flucytosine, fluconazole, and voriconazole against Candida lusitaniae determined by microdilution, Etest, and time-kill methods. Antimicrob Agents Chemother 46, 578–81 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin Microbiol Infect 14, 398–405 (2008). [DOI] [PubMed] [Google Scholar]
- Anders S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8, 1765–86 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.