Abstract
The role of the unique proline-glutamic acid (PE)/proline-proline-glutamic acid (PPE) family of proteins in the pathophysiology and virulence of Mycobacterium tuberculosis is not clearly understood. One of the PE family proteins, PE11 (LipX or Rv1169c), specific to pathogenic mycobacteria is found to be over-expressed during infection of macrophages and in active TB patients. In this study, we report that M. smegmatis expressing PE11 (Msmeg-PE11) exhibited altered colony morphology and cell wall lipid composition leading to a marked increase in resistance against various environmental stressors and antibiotics. The cell envelope of Msmeg-PE11 also had greater amount of glycolipids and polar lipids. Msmeg-PE11 was found to have better survival rate in infected macrophages. Mice infected with Msmeg-PE11 had higher bacterial load, showed exacerbated organ pathology and mortality. The liver and lung of Msmeg-PE11-infected mice also had higher levels of IL-10, IL-4 and TNF-α cytokines, indicating a potential role of this protein in mycobacterial virulence.
Mycobacterium tuberculosis, the causative agent for tuberculosis (TB) employs several strategies to modulate the host immune responses to favor its intracellular survival1. It possesses a unique proline-glutamic acid (PE)/proline-proline-glutamic acid (PPE) family of proteins whose role in the pathophysiology and virulence is not clearly understood2. The M. tuberculosis PE/PPE family has a conserved PE and PPE motif at the N-terminal domain with variations in the C-terminal region2 and is found to be highly expanded in the pathogenic strains of mycobacteria3. The PE and PPE proteins have been thought to play important roles in generating antigenic variation and immune evasion, some of them are known to induce strong B cell response1. However, their exact pathophysiological roles are yet to be fully understood. A detail functional study of these genes is, therefore, crucial to understand the host-pathogen interactions and survival strategies adopted by mycobacteria.
The unique and highly complex cell wall of mycobacteria is implicated for its resistance to various anti-mycobacterial drugs and environmental stressors4. The cell wall contains distinctive lipids and glycolipids that contribute to extreme hydrophobicity to the outer surface. These lipids which include mycolic acids, phosphatidyl inositol mannosides, phthiocerol dimycocerosates and lipoglycans such as lipomannan and lipoarabinomannan play important roles in maintaining integrity of the cell envelope5. The mycobacterial lipids are also involved in modulating early immune responses of macrophages to infection6 like resistance to free radicals, prevention of phagosomal maturation and inhibition of synthesis of anti-mycobacterial cytokines by the host immune system7. Thus, the cell wall lipids are critical to the mycobacterial virulence process7,8. Due to its increasing resistance to current chemotherapeutic agents, it is important to identify newer generation anti-mycobacterial drugs and the processes/components involved in the cell wall lipid synthesis present an attractive target for this purpose9. Interestingly, some of the currently available drugs like isoniazid10 and ethambutol11 are known to inhibit mycobacterial cell wall synthesis.
The M. tuberculosis lipases/esterases play crucial roles in lipid metabolism and mycobacterial physiology12. Lipases/esterases and phospholipases are molecules that provide metabolic turnover of lipids and can be defined as essential biocatalysts for the hydrolysis of esters containing long-chain and short-chain fatty acids. These fatty acids, on one side provide energy for intracellular persistence of the dormant bacilli and its reactivation and on the other side they can act as precursors for the cell wall lipids and are thought to contribute to virulence and pathogenicity of the bacilli13. In silico analysis of M. tuberculosis genome predicted presence of about 24 putative lipase/esterase genes, belonging to the so called “lip” family (lipC to lipZ), based on the presence of a consensus ‘GXSXG’ motif, a characteristic feature of the members of the α/β hydrolase-fold family13. However, only few of these proteins like LipC (Rv0220)14, LipF (Rv3487c)15, LipH (Rv1399c)16 and LipY (Rv3097c)17,18 are functionally characterized to some extent.
One of the PE family proteins, PE11 (LipX or Rv1169c) is found to be specifically present in pathogenic strains like M. tuberculosis, M. bovis and clinical strain CDC1551 but is conspicuously absent in non-pathogenic M. smegmatis strain. PE11 is known to be upregulated in M. tuberculosis during acidic stress, starvation and adaptation to stationary phase as well as in hypoxic lipid-loaded macrophages12,19,20,21. Also its expression is found to be increased during palmitic acid induced stress conditions17. Presence of anti-PE11 antibody in TB patients22 and its up-regulation in human lung granulomas23 suggest that PE11 is possibly over-expressed during active tuberculosis infection. However, the exact functions of PE11 remains poorly understood. PE11 is predicted to be a putative lipase/esterase due to presence of ‘GXSXG’ motif, characteristic of the α/β hydrolase fold family13. However, whether PE11 is capable of modulating the cell wall architecture and lipid composition and whether these changes contribute to mycobacterial virulence are not well known. In this study, we demonstrate esterase activity of PE11 and its role in cell wall remodeling. Further, the data presented herein indicate that PE11 probably plays an important role in M. tuberculosis virulence and establishment of a successful infection.
Results
Expression of PE11 in M. smegmatis altered colony morphology and architecture of the bacilli
To understand how PE11 influence mycobacterial pathophysiology, we expressed pe11 gene in a non-pathogenic M. smegmatis strain mc2155 (Msmeg-PE11), which is widely used as a surrogate bacterium to characterize M. tuberculosis protein24. The immunoblot analysis indicates presence of PE11 in Msmeg-PE11 but not in M. smegmatis harboring the backbone vector (Msmeg-pVV) (Supplementary Fig. 1). When the transformants were grown on Middlebrook 7H10 agar plates containing 0.5% glycerol, 10% OADC (oleic acid-albumin-dextrose [glucose]-catalase), and 0.05% Tween 80 and incubated for 5–6 days at 37 °C, we observed a distinct colony morphology in PE11 positive transformants. While the colonies of Msmeg-pVV were found to be usual irregular wrinkled acne-like structures, those of Msmeg-PE11 were found to be rounded, shiny and smooth (Fig. 1a). Further, the control colonies were dry and fragile but the Msmeg-PE11 colonies were wetter and stickier. Since, PE11 was predicted to be a putative lipase/esterase like protein13, our observations are suggestive of a role of PE11 in changing the cell wall components of Msmeg-PE11.
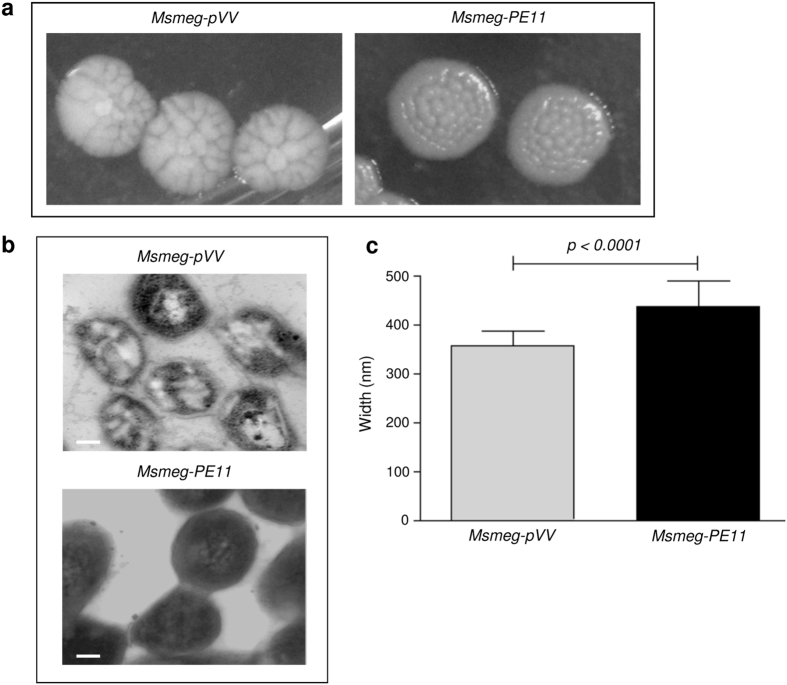
Figure 1. PE11 alters colony morphology, surface architecture and width of M. smegmatis.
(a) Msmeg-pVV or Msmeg-PE11 bacteria were plated on 7H10 agar plates supplemented with OADC and incubated for 5–6 days and the mycobacterial colonies were photographed. (b) For transmission electron microscopy (TEM) analysis, Msmeg-pVV or Msmeg-PE11 strains were cultured on 7H10 agar for 5–6 days and surface architecture of these bacteria were analyzed. Scale bar, 100 nm. (c) In another experiment, Msmeg-pVV or Msmeg-PE11 bacteria were harvested for scanning electron microscopy (SEM) analysis to measure the diameters of Msmeg-pVV or Msmeg-PE11 and mean width of 100 bacilli each of Msmeg-pVV and Msmeg-PE11 is graphically represented in nm.
We next compared the surface architecture of Msmeg-PE11 with Msmeg-pVV using transmission electron microscopy (TEM). In TEM analysis, we consistently observed poor contrast and hyper-staining of Msmeg-PE11 cells compared to Msmeg-pVV (Fig. 1b), which is indicative of alteration in the cell wall composition of M. smegmatis expressing PE11. Examination of the bacteria under a scanning electron microscope indicated that expression of PE11 in M. smegmatis could alter the cellular dimension of the bacilli (Fig. 1c and Supplementary Fig. 2). The Msmeg-PE11 cells were found to be significantly wider in diameter [437.2 ± 5.114 nm (mean ± SD)] as compared to Msmeg-pVV cells [356.1 ± 3.014 nm (mean ± SD)]. These results indicate that expression of PE11 alters the morphology and cellular dimensions of M. smegmatis bacilli.
PE11 increases hydrophobicity of M. smegmatis cell envelope
Several studies have proposed that changes in the cell wall lipid composition can significantly influence the surface hydrophobicity and various cell surface properties like biofilm/pellicle formation, congo red binding, cellular aggregation and sliding motility25,26. Since, earlier we observed conspicuously different colony morphology and increased electron-dense staining in TEM analysis, we next examined cell surface properties like cellular aggregation, pellicle/biofilm formation and congo red binding. When the pellicle formation at the air-liquid interface of the static 7H9 liquid cultures (without Tween 80) was examined as described elsewhere27, Msmeg-pVV was found to form thin visible surface pellicle, but cells from Msmeg-PE11 were found to be heavily aggregated and were able to form thick and robust pellicles at the air-liquid interface under similar experimental conditions (Fig. 2a). The pellicles are believed to be biofilm-like structures that assemble at the air-liquid interface. The biofilm formation was quantified using crystal violet stain as described by others27. The results shown in Fig. 2b indicate that compared to the control Msmeg-pVV, biofilm formation was significantly increased in Msmeg-PE11, indicating a role of PE11 in biofilm formation and possible changes in the cell surface properties.
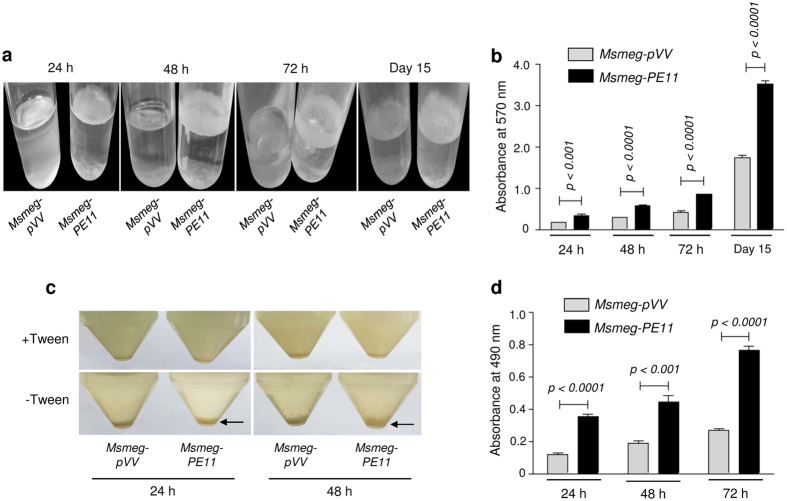
Figure 2. Cell surface of Msmeg-PE11 is highly hydrophobic.
(a) Pellicle formation of Msmeg-pVV or Msmeg-PE11 was monitored by growing standing cultures of the strains without shaking in Middlebrook 7H9 medium in absence of Tween 80 at 37 °C for various time points. (b) Biofilm formation was quantified by crystal violet staining for which Msmeg-pVV or Msmeg-PE11 cells were washed, stained with 1% crystal violet and ethanol extract was spectrophotometrically measured at 570 nm. (c) Msmeg-pVV or Msmeg-PE11 were cultured in 7H9 medium with (upper panel) or without (lower panel) 0.05% Tween-80 at 37 °C with shaking for 24 h and 48 h. Cultures were then allowed to settle at room temperature for 30 min. (d) In another experiment, Msmeg-pVV or Msmeg-PE11 were cultured in 7H9 broth with congo red (100 μg/ml) and 0.05% Tween 80 for 24 h, 48 h and 72 h at 37 °C. Cells were next washed and re-suspended in acetone. Congo red in the supernatant was spectrophotometrically measured at 490 nm. Data are representative of mean ± SD of three different experiments.
Detergent like Tween 80 (0.05% vol/vol) is added in liquid mycobacterial cultures to prevent clumping of bacterial cells. As expected, 24 h and 48 h old cultures of Msmeg-pVV and Msmeg-PE11 remained completely dispersed in medium in the presence of Tween 80 (Fig. 2c, upper panel, all tubes). However, growth in liquid medium without Tween 80 resulted in cell aggregation that led to reduced turbidity in 24 h and 48 h old cultures of Msmeg-pVV (Fig. 2c, lower panel, left tube) as well as Msmeg-PE11 (Fig. 2c, lower panel, right tubes) when allowed to settle at room temperature for 30 min. Interestingly, in Msmeg-PE11 cultures, the cells were found to be more aggregative and precipitated quicker in standing cultures than Msmeg-pVV at both 24 h and 48 h time points (Fig. 2c, lower panel). More aggregation of the Msmeg-PE11 cells in liquid cultures in the absence of Tween 80 is likely due to an increased surface hydrophobicity of these cells, indicating an alteration of the cell wall architecture in Msmeg-PE11.
To confirm the difference in hydrophobicity between Msmeg-pVV and Msmeg-PE11, congo red binding assay was carried out. Congo red, a planar and hydrophobic diazo-dye that binds to lipids, lipoproteins28, is widely used to characterize lipid content and modifications affecting the cell wall composition and hydrophobicity25,28. Cells from Msmeg-PE11 were found to retain three fold more congo red dye as compared to Msmeg-pVV (Fig. 2d) further confirming that Msmeg-PE11 cells had higher surface hydrophobicity.
Msmeg-PE11 cells are more resistant to cell wall stress
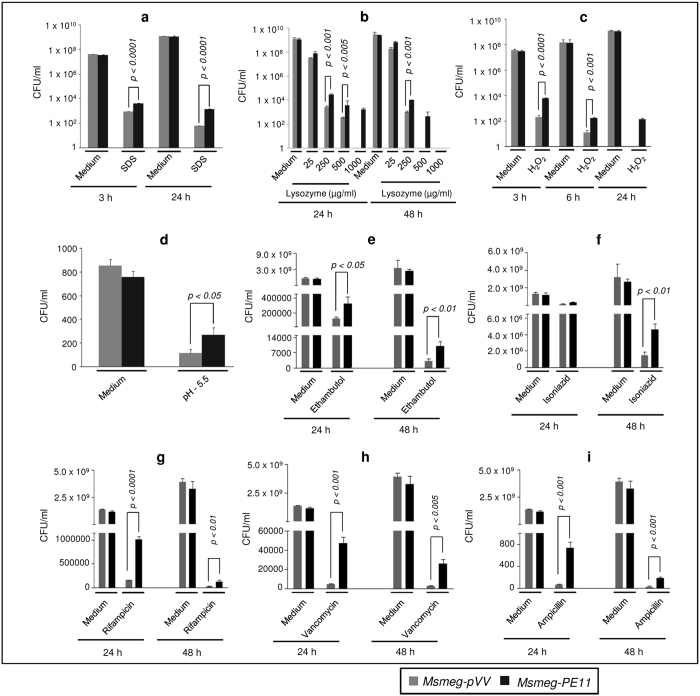
The mycobacterial cell wall is a rigid structure that enables the bacilli to survive under unfavorable intracellular environment. Conditions like low oxygen tension, chemical, oxidative, and acidic stress are encountered by M. tuberculosis during infection. Our earlier data indicate that PE11 is probably required to remodel cell wall architecture of mycobacteria. Since, PE11 is found to be present in the cell wall of virulent mycobacteria and is known to be upregulated during stress conditions19,20,21,29,30, it is possible that the protein plays an important role to protect the bacilli from various environmental stressors. Therefore, next we examined the ability of Msmeg-PE11 to resist antibiotics and other stress conditions that mimic the intracellular environment. The Msmeg-PE11 and the control Msmeg-pVV bacteria were either left untreated or treated for 3 h and 24 h with 0.1% SDS detergent that is known to disintegrate bacterial cell wall. Results shown in Fig. 3a suggest that Msmeg-PE11 is more resistant to 0.1% SDS detergent treatment than the control Msmeg-pVV at both 3 h and 24 h time points. These findings suggest that PE11 confers resistance to detergent-mediated damage to mycobacterial cell envelope.
Figure 3. Msmeg-PE11 are more resistant to SDS, lysozyme, hydrogen peroxide, low pH and antibiotics treatment when compared with Msmeg-pVV.
The Msmeg-pVV or Msmeg-PE11 were either treated with medium alone or subjected to (a) 0.1% SDS, (b) lysozyme, (c) 5 mM hydrogen peroxide, (d) low pH (5.5 for 24 h), (e) 20 μg/ml ethambutol, (f) 20 μg/ml isoniazid, (g) rifampicin (20 μg/ml), (h) vancomycin (5 μg/ml) and (i) ampicillin (750 μg/ml) treatment. Bacterial CFUs were counted on 7H10 agar plates. Data are representative of mean ± SD of three different experiments.
The antimicrobial agent lysozyme is secreted abundantly by macrophages, neutrophils, and lung epithelial cells. Lysozyme is known to damage mycobacterial cell wall by hydrolysing glycosidic bonds in peptidoglycan. When Msmeg-pVV and Msmeg-PE11 were treated with various concentrations of lysozyme for 24 h and 48 h, it was observed that Msmeg-PE11 was significantly more resistant to lysozyme treatment as compared to the control (Fig. 3b).
Next, we examined the ability of Msmeg-PE11 bacteria to resist oxidative (hydrogen peroxide) and acidic stress (pH 5.5), as macrophages produce reactive oxygen species (ROS) and low pH environment as defense against M. tuberculosis. When Msmeg-pVV and Msmeg-PE11 were treated with 5 mM hydrogen peroxide (H2O2) at various time points, it was found that Msmeg-PE11 was more resistant to H2O2 than Msmeg-pVV (Fig. 3c). Also, after exposure to pH 5.5 for 24 h, survival of Msmeg-PE11 was found to be 2.5 fold higher than the control Msmeg-pVV (Fig. 3d). We further evaluated the effect of PE11 on antibiotic sensitivity by comparing the survival of Msmeg-pVV and Msmeg-PE11 strains after treatment with various antibiotics like ethambutol, isoniazid, rifampicin, vancomycin and ampicillin for 24 h and 48 h. The results shown in Fig. 3e–i indicate that Msmeg-PE11 offered better resistance against all the drugs tested as compared to M. smegmatis harboring the vector control (Msmeg-pVV). We concluded that PE11-mediated changes in the cell wall architecture of Msmeg-PE11 resulted in lesser penetration of antibiotics. Expectedly, Msmeg-PE11 had better survival rates within primary macrophages (Supplementary Fig. 3). Taken together, these data indicate that PE11 may be important for maintaining a resilient cell wall structure which enables the bacteria to persist longer inside the macrophages.
PE11 shows esterase activity
We next characterized the enzyme activity of PE11 using esters of p-nitrophenyl (pNP), p-nitrophenylacetate (C2), p-nitrophenylbutyrate (C4), p-nitrophenyloctanoate (C8), p-nitrophenyldodecanoate (C12), p-nitrophenylmyristate (C14) p-nitrophenylpalmitate (C16) and p-nitrophenylstearate (C18) as substrates. We found that lysates prepared from Msmeg-PE11 had significantly higher hydrolyzing activities to a wide range of para-nitrophenyl (pNP) esters (C2–C14) with increased preferences to shorter carbon chain length derivatives at pH 7.0 and 37 °C (Fig. 4a). The pNP ester para-nitrophenylacetate containing the shortest carbon chain (C2) was found to be most efficiently hydrolyzed (Fig. 4a). Though significantly higher hydrolyzing activity was detected with intermediate carbon chain length pNP esters, no activity was found towards longer carbon chain pNP esters (Fig. 4a). These results suggest that PE11 protein is acting predominantly as an esterase rather than lipase. In contrast, the lysates prepared from Msmeg-pVV had very low hydrolyzing activity against C2–C14 pNP esters tested as compared to those prepared from Msmeg-PE11 (Fig. 4a), indicating that the esterase activity of Msmeg-PE11 lysate is attributable to PE11.
Figure 4. PE11 has esterase activity.
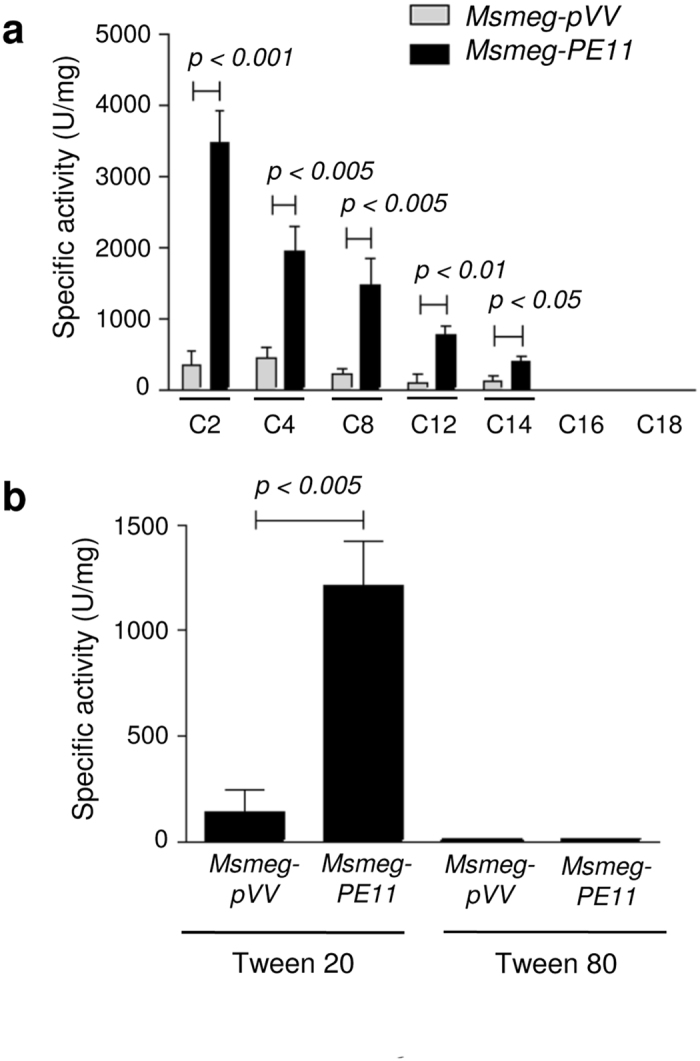
(a) Lipid hydrolyzing activity of PE11 was determined by measuring the release of para-nitrophenol (pNP) upon incubation of 20 μg of cell lysate prepared from either Msmeg-pVV or Msmeg-PE11 with p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), p-nitrophenyloctanoate (C8), p-nitrophenyldodecanoate (C12), p-nitrophenylmyristate (C14), p-nitrophenylpalmitate (C16) and p-nitrophenyl stearate (C18) at 37 °C and pH 7.0 for 10 min. The release of para-nitrophenol was measured spectrophotometrically at 405 nm. (b) Also, lipolytic activity of PE11 was monitored by cleavage of polyoxyethylene sorbitan monolaurate (Tween 20) and polyoxyethylene sorbitan monooleate (Tween 80) at pH 7.0. Data are representative of mean ± SD of three different experiments.
We further confirmed the esterase activity of PE11 by employing a turbidimetric esterase assay where the cleavage of polyoxyethylene sorbitans (Tweens) was measured31. This method involves hydrolysis of Tweens in the presence of CaCl2, and measuring the increased absorbance at 405 nm due to precipitation of the released fatty acids as calcium salts. Tween 80 is mostly hydrolyzed by lipase activity and rarely by esterase, as it contains esters of long chain oleic acid (C18). In contrast, Tween 20 is easily hydrolysable by an esterase as it contains esters of shorter chain lauric acid (C12)31. This assay is reported to be about 36-fold more sensitive than the titrimetric assay and at least 4-fold more sensitive than the colorimetric assay using pNP esters32. When Msmeg-PE11 lysates were tested for hydrolysis of polyoxyethylene sorbitan monolaurate (Tween 20), about 9 fold higher lipolytic activity was recorded compared to the control Msmeg-pVV lysates (Fig. 4b). The level of lipolytic activity of Msmeg-PE11 lysate on Tween 20 was 1215 ± 205.36 mU/mg at pH 7.0 whereas activity of the control lysate was limited to 138.33 ± 109.83 mU/mg. When polyoxyethylene sorbitan monooleate (Tween 80) was used as a substrate, negligible lipolytic activity was detected in both Msmeg-PE11 and Msmeg-pVV lysates. The observed data were in concordant with the previous results obtained using pNP esters as substrate indicating that PE11 is predominantly an esterase.
PE11 modulates the synthesis of glycolipids
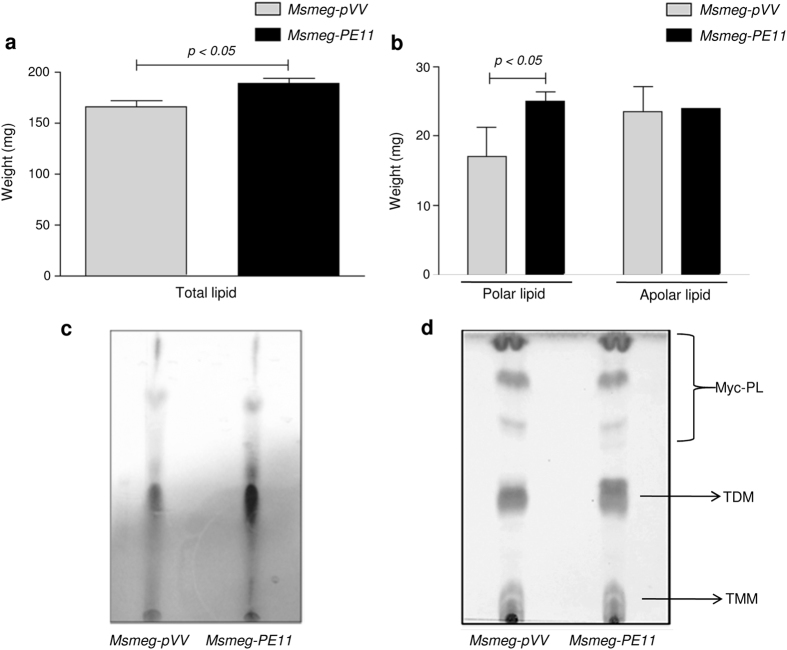
The changes in colony morphology and various cell surface properties like cellular aggregation, pellicle/biofilm formation, congo red binding and increased resistance to various stressors, suggested that the presence of PE11 resulted in modulation of components of the cell wall lipids. In order to confirm this hypothesis, we analyzed the lipid content of Msmeg-pVV and Msmeg-PE11 cells at the same stage of growth. Msmeg-PE11 was found to have increased quantities of both total and polar lipids in the cell envelope (Fig. 5a,b), however, the apolar lipid content was not found to be significantly changed (Fig. 5b). Mycobacteria defective in glycopeptidolipid (GPL) biosynthesis is often found to possess altered colony morphologies33 and biofilm formation26,34. Therefore, we examined whether the glycolipid composition and production was changed in M. smegmatis due to PE11 expression. Thus, total cell envelope lipids were extracted and separated by TLC (Thin layer chromatography), and glycolipids were visualized by α-naphthol/sulfuric acid staining. Interestingly, we found that the expression of PE11 led to an increase in glycolipid concentration in the cell wall (Fig. 5c). We also analyzed the cell wall to compare mycolate containing glycolipids [trehalose monomycolate (TMM), trehalose dimycolate (TDM) and mycolylmannosylphosphorylheptaprenol (Myc-PL)]. TLC results indicate an increase of TDM in Msmeg-PE11 cell wall (Fig. 5d). However, the relative amounts of TMM and Myc-PL were not significantly changed (Fig. 5d). These observations indicate that expression of PE11 led to an increased deposition of glycolipids in the cell wall of M. smegmatis.
Figure 5. Modulation of total, polar and glycolipids in Msmeg-PE11.
Quantification (% weight) of (a) total lipids, (b) polar and apolar lipids extracted from the Msmeg-pVV or Msmeg-PE11 wet pellets (equal weights), with chloroform-methanol (2:1, vol/vol). (c) Total glycolipid analysis of Msmeg-pVV or Msmeg-PE11 by analytical TLC. Methanol: Ammonium hydroxide (80:20, vol/vol) was used as mobile phase and the glycolipids were visualized by α-naphthol/sulfuric acid staining (d) Mycolic acid containing glycolipids were separated from the pool of total polar lipids using Chloroform:Methanol:Ammonium hydroxide (80:20:2, vol/vol/vol) as mobile phase. These glycolipids were identified by Iodine vaporization followed by charring with 5% phosphomolybdic acid.
Quantitative modulation of fatty acid composition of Msmeg-PE11
Next, we analyzed the fatty acid composition of Msmeg-pVV and Msmeg-PE11 strains using gas chromatography coupled with mass spectrometry (GC/MS). The total lipid fraction from each strain was transesterified and methylated, and the resulting fatty acid methyl esters (FAMEs) were analyzed by GC and GC/MS. GC chromatograms showed that the major FAMEs produced by Msmeg-pVV and the Msmeg-PE11 were C16:0, C16:1, C18:0, C18:1 and C18:0 (10-methyl) FAMEs based on their retention times and molecular masses (Supplementary Fig. 4). Interestingly, we observed an increase in C18:0 (10-methyl) FAME representing branched fatty acids, there was approximately 36% more abundance of C18:0 (10-methyl) FAME in Msmeg-PE11 from FAMEs isolated from total lipid and similar pattern was also observed in FAMEs isolated from polar lipid (Table 1). However, we noted 25% decrease in linear C18:0 FAME in Msmeg-PE11 (Table 1). A cluster of fatty acids longer than the C20 were more in the Msmeg-PE11 compared to Msmeg-pVV. These fatty acids were found to be between 18 and 24 min retention times (Supplementary Fig. 4 and Table 1). Further, these fatty acids mainly belong to a homologous series differing only in the acyl chain lengths. The identified FAMEs from total and polar lipid extracted from the Msmeg-PE11 strain were further validated by GC/MS, using the NIST02.L database. In case of apolar lipids, analyses of fatty acid composition did not reveal much difference between Msmeg-PE11 and Msmeg-pVV except there was approximately 27% more abundance of C24:0 FAME in Msmeg-PE11. Taken together, these data suggest that PE11 significantly modulated the fatty acid profiles extracted from the polar lipid of the cell wall.
Table 1. Fatty acid composition of Msmeg-pVV and Msmeg-PE11 estimated by GC, GC/MS analysis.
| Fatty Acid Compositions (Wt %) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14:0 | 15:0 | 16:0 | 16:1 | 17:0 | 17:1 | 18:0 | 18:1 | 18:0 (10-methyl) | 18:2 | 20:0 | 20:1 | 22:0 | 22:1 | 24:0 | 24:1 | |
| Msmeg-pVV Total | 2.6 | 0.3 | 18.5 | 13.8 | 0.4 | 1.1 | 7.4 | 44.2 | 6.3 | 0.8 | 1.0 | 0.4 | 0.9 | – | 2.3 | – |
| Msmeg-PE11 Total | 3.1 | 0.3 | 17.9 | 13.8 | 0.3 | 0.9 | 6.2 | 41.5 | 9.7 | 0.9 | 0.8 | 0.4 | 1.0 | – | 3.2 | – |
| Msmeg-pVV Polar | 2.2 | 0.5 | 26.3 | 10.7 | 0.5 | 0.6 | 10.1 | 35.4 | 11.6 | 1.5 | 0.2 | 0.2 | 0.1 | 0.1 | – | – |
| Msmeg- PE11 Polar | 2.3 | 0.5 | 25.6 | 9.8 | 0.5 | 0.7 | 7.6 | 35.9 | 15.2 | 0.4 | 0.2 | 0.2 | 0.2 | 0.9 | – | – |
| Msmeg-pVV Apolar | 2.8 | 0.3 | 15.4 | 14.9 | 0.4 | 1.0 | 6.5 | 46.0 | 3.6 | 2.6 | 1.2 | 0.5 | 1.2 | 0.1 | 3.2 | 0.3 |
| Msmeg-PE11 Apolar | 2.3 | 0.2 | 14.5 | 13.4 | 0.3 | 1.0 | 5.5 | 44.7 | 5.1 | 2.7 | 1.3 | 0.6 | 1.7 | 0.2 | 5.9 | 0.6 |
The number CX:Y, for example, indicates the carbon number X and Y double bond(s).
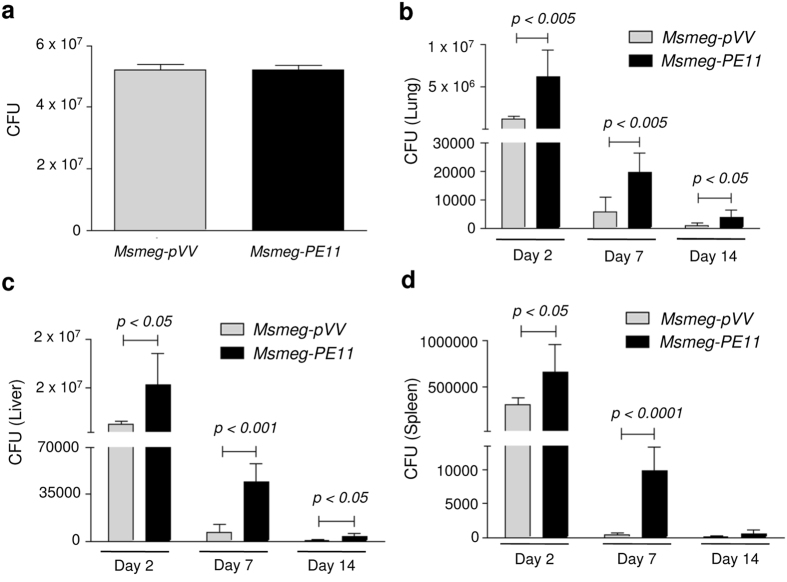
PE11 confers a growth advantage to M. smegmatis in a mouse model of infection
We next assessed the virulence property of PE11 in vivo using an animal infection model. The Balb/c mice were infected with 5 × 107 CFU of either Msmeg-pVV or Msmeg-PE11 via lateral tail vein route and the bacterial burden was estimated in lung, liver and spleen of the infected animals at day 2, 7 and 14 post-infection. Both Msmeg-pVV and Msmeg-PE11 were dispersed into single cell suspensions and plated to confirm administration of equal number of bacteria in mice (Fig. 6a). The CFU counts indicated that M. smegmatis expressing PE11 persisted significantly for longer time period than M. smegmatis harboring the pVV vector in all the organs at almost all the time points investigated. In lungs of Msmeg-pVV-infected mice, the mean CFU counts were significantly lower at day 2 post-infection as compared to those of infected with Msmeg-PE11 strain and this trend also continued to later time points (day 7 and 14) (Fig. 6b). Similar observations were also found in liver (Fig. 6c) as well as in spleen (Fig. 6d). These results indicate that expression of PE11 protein in non-pathogenic mycobacteria results in longer survival of the bacilli in host cells signifying that PE11 probably plays a role in mycobacterial virulence.
Figure 6. Better persistence of M. smegmatis expressing PE11 in mice.
Balb/c mice (n = 6) were infected intravenously with approximately (a) equal number (5 × 107) of Msmeg-pVV or Msmeg-PE11, and sacrificed after 2, 7 and 14 days post-infection for determination of CFU in (b) lung (c) liver and (d) spleen. Data represent mean ± SD of six mice per group for each time point.
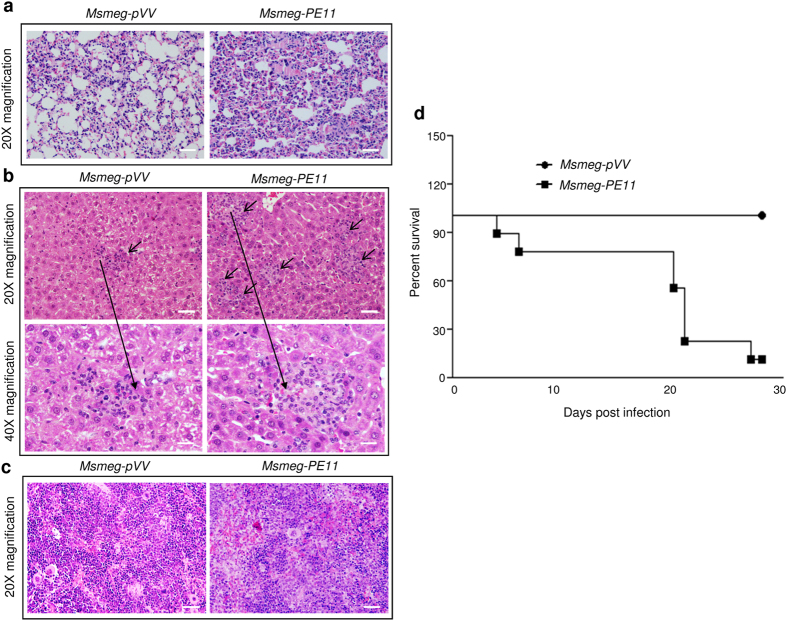
Mycobacterial burden in mice organs is generally well correlated with tissue pathology35,36. Therefore, we evaluated the extent of tissue damage in lung, liver and spleen in mice infected with Msmeg-PE11 or Msmeg-pVV strains by histopathological examination of hematoxylin and eosin (H&E) stained sections. It was found that Msmeg-PE11-infected mice had markedly pronounced tissue damage than that observed in Msmeg-pVV-infected mice (Fig. 7). Infiltration and lesions were more severe in the lungs of mice infected with Msmeg-PE11 by day 7 post-infection (Fig. 7a). Furthermore, extensive tissue necrosis, increased lymphocytic and phagocytic cellular infiltration and disappearance of alveolar spaces were recorded in the lungs of Msmeg-PE11-infected mice compared to lungs of Msmeg-pVV-infected mice. A similar pathology was also observed in the liver of Msmeg-PE11-infected mice. The lesions were found to be more severe with increased microgranuloma and more lymphohistiocytic in mice infected with Msmeg-PE11 than that of mice infected with Msmeg-pVV (Fig. 7b). Liver sections from Msmeg-pVV- infected mice showed mild lymphocytic infiltration and microgranuloma at day 7 post-infection and this was correlated well with lower CFU counts in the liver. Additionally, single cell necrosis began to appear in the liver of Msmeg-PE11-infected mice by day 7 post-infection (Fig. 7b). The histopathological analyses of spleen indicated presence of more white pulp in Msmeg-PE11-infected mice sacrificed at day 7 (Fig. 7c) and day 14 (Supplementary Fig. 5) post-infection. Also, there was an increase in the size and weight of spleen in Msmeg-PE11-infected mice as compared to Msmeg-pVV-infected mice sacrificed at day 14 (Supplementary Fig. 6a,b). To inquest the effect of PE11 on mortality, survival of Balb/c mice infected with Msmeg-pVV and Msmeg-PE11 was monitored. No deaths were registered in the group of mice infected with Msmeg-pVV strain during the entire study period of 30 days. However, in the group of mice infected with Msmeg-PE11, survival rate was found to be dropped to 20% by 28 days post-infection (Fig. 7d). Also, mice infected with Msmeg-PE11 had significant decrease in the total body weight at day 9 and day 18 post-infection as compared to mice infected with Msmeg-pVV (Supplementary Fig. 7). By day 27, mice infected with Msmeg-pVV returned to the baseline value whereas most of the Msmeg-PE11-infected mice were found to be dead.
Figure 7. Balb/c mice infected with Msmeg-PE11 showed more organ pathology and decreased survival rate as compared to mice infected with Msmeg-pVV.
The lung (a), liver (b) and spleen (c) sections of Balb/c mice infected with 5 × 107 of Msmeg-pVV or Msmeg-PE11 were stained with hematoxylin and eosin (H&E) stain at day 7 post-infection. The microgranuloma like structure in liver sections is marked with arrows and one of these is shown at 40X magnification. Photographs of representative sections from 2 mice were shown. Scale bar, 200 μm for lung, liver and spleen. (d) Survival of Balb/c mice (n = 10) following intravenous infection with 5 × 107 of Msmeg-pVV or Msmeg-PE11 was recorded for 30 days post-infection.
PE11 induces T-helper 2 (Th2) cytokines in addition to tumor necrosis factor-α (TNF-α) in lung and liver of infected mice
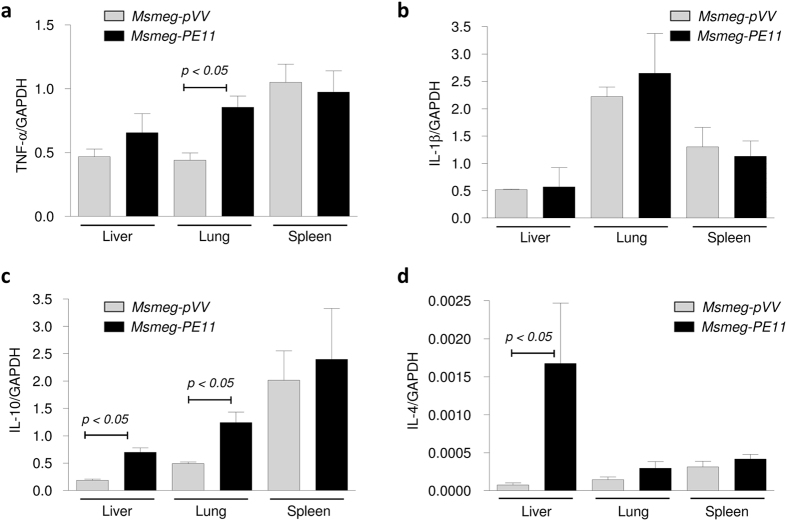
The levels of proinflammatory cytokines like TNF-α and interleukin (IL)-1β and Th2 cytokines like IL-10 and IL-4 were measured by reverse transcription qPCR (RT-qPCR) in the liver, lung and spleen of the infected mice at day 7 post-infection. It was found that the levels of TNF-α were considerably higher in liver and lungs (Fig. 8a) in the mice infected with Msmeg-PE11 as compared to those of mice infected with Msmeg-pVV, however, IL-1β levels did not differ significantly both at transcript and protein levels (Fig. 8B and Supplementary Fig. 8). On the other hand, the levels of IL-10 (Fig. 8c) was found to be significantly higher in both lungs and liver of the mice infected with Msmeg-PE11as compared to those infected with control Msmeg-pVV. Similarly, IL-4 cytokine levels were also found to be considerably higher in liver and lungs of mice infected with Msmeg-PE11 (Fig. 8d). Interestingly, none of the cytokines were found to differ significantly in the spleen of Msmeg-PE11-infected mice at day 7 post-infection.
Figure 8. Cytokine profile of Balb/c mice infected with Msmeg-PE11 and Msmeg-pVV at day 7 post-infection.
Balb/c mice were infected intravenously with 5 × 107 Msmeg-PE11 and Msmeg-pVV and at day 7 post-infection, the mice were sacrificed and total RNA was isolated from liver, lung and spleen. The levels of TNF-α (a), IL-1β (b), IL-10 (c) and IL-4 (d) in different tissues were measured by reverse-transcription qPCR and expression levels were normalized to corresponding GAPDH levels. The data are representative of 3 mice in each group.
Discussion
The unique cell envelope of M. tuberculosis has been implicated in the pathogenesis and resistance to anti-mycobacterial drugs. In the present study, we attempted to understand the possible role played by a Lip family protein of M. tuberculosis, PE11 which is also a member of the unique PE/PPE family proteins, in mycobacterial pathophysiology and virulence. PE11 is known to be induced under several stressful conditions those are frequently encountered by the invading M. tuberculosis inside the hostile cellular environment. Microarray analyses of the transcriptome responses under acidic conditions found in phagosomes revealed that a number of genes including those involved in fatty acid metabolism including pe11 were upregulated, possibly required for long term survival under such conditions19. pe11 is also found to be upregulated during nutrient starvation30 as well as in hypoxic conditions37 and in response to H2O2 treatment in M. tuberculosis38. In addition, pe11 is also found to be upregulated in response to stressors like SDS and diamide39,40. Further, data on the transcriptional responses of M. tuberculosis to bovine lung surfactant has shown that pe11 is significantly induced41. Also, pe11 is shown to be induced during non-replicating persistent conditions21. PE11 transcripts are known to be upregulated during macrophage infection and in granulomas of lungs of human pulmonary TB patients20,23,42. These data indicate that PE11 in conjunction with other lipases probably plays a role in release of fatty acids from triglycerides to be used as an energy source and may help the bacilli to adapt to the intracellular environment during infection and dormancy.
Bioinformatics analyses suggested that PE11 is a putative lipase/esterase13. From the published reports by at least two research groups, it appears that Rv1169c (pe11) is an essential gene for in vitro growth of M. tuberculosis43,44. Therefore, we used M. smegmatis, a widely used surrogate bacterium to investigate the pathophysiological role of PE11 in vitro and in vivo. We found that PE11 could significantly modify cell surface properties when expressed in M. smegmatis, and these modifications might be explained by its ability to alter the cell wall lipid composition. Sequence analysis of PE11 predicted presence of the ‘α/β hydrolase fold’ characteristic of lipase/esterase family of proteins. Biochemical assays of PE11 using a wide range of p-nitrophenyl esters with varying carbon chain lengths from C2 to C18 clearly showed that PE11 possesses esterase activity that could preferentially hydrolyzes shorter (C2–C4) to intermediate (C8–C14) carbon chain esters but failed to hydrolyze longer chain p-nitrophenyl esters (C16–C18) indicating that PE11 is predominantly an esterase rather than a lipase. The p-nitrophenyl acetate (C2) was found to be most efficiently hydrolyzed by PE11. The turbidimetric esterase assay using Tween 20 and Tween 80 as its substrates further confirmed the esterase activity of PE11. Interestingly, another protein of M. tuberculosis, Rv0045c was also reported to be an esterase45 that also hydrolyzes short chain p-nitrophenyl esters (C2–C8), but its preferred substrate was p-nitrophenyl caproate (C6). Similarly, another PE family protein, PE16 (Rv1430) could hydrolyze a wide range of p-nitrophenyl esters (C4–C12), of which pNPC6 was found to be most effectively hydrolyzed46. On the other hand, the lipase enzyme LipY is shown to hydrolyze mainly the triacylglycerol (TAG)17. It is believed that the host TAG serves as the power house for M. tuberculosis and gets catabolized by the M. tuberculosis lipolytic enzymes sequentially into DAG (diacylglycerol), then to MAG (monoacylglycerol) which is finally hydrolyzed to release free fatty acids to be used as the carbon source for energy as well as synthesis of cell wall components13. Thus, esterases play an important role in the M. tuberculosis pathophysiology as the free fatty acids act as building blocks for maintenance of M. tuberculosis cell wall in hostile environment and also act as energy source for persistence and reactivation. Since, PE11 specifically hydrolyzes shorter carbon chain pNP ester, we presumed that it probably acts on the final hydrolyzed product of TAG, i.e., MAG for release of free fatty acids. Our observations of increased total lipid content in Msmeg-PE11 cell wall as compared to the control Msmeg-pVV cell wall supports the hypothesis that PE11 is actually involved in mycobacterial lipid metabolism. Our data indicate that PE11 plays a structural role in modifying envelope composition through its esterase activity. Pertinent to this, PE11 appears to be capable of localizing on to the mycobacterial cell wall29. Therefore, it is tempting to speculate that the tubercle bacilli may employ PE11 for foraging and hydrolyzing shorter carbon chain lipids into fatty acids sourced from the local environment to be used for its energy requirement and structural function. Further, expression of PE11 during nutrient starvation30 suggests that PE11 might be required to maintain viability under nutritional stress.
Interestingly, Rv3036c of M. tuberculosis was also characterized as a novel cell wall-anchored esterase that hydrolyzes soluble p-nitrophenyl acetate47. However, its role in mycobacterial virulence and pathogenesis is not yet known. In this study, using a surrogate bacterium, we provide evidences that expression of PE11 can confer survival advantage to the bacilli inside the macrophages and further in vivo studies in a mice infection model supports a role for this protein in the mycobacterial pathogenesis and virulence. A putative carboxyl esterase, Rv2224c that preferentially hydrolyses ester bonds of substrates with intermediate carbon chain length is also found to be membrane associated and mutants had reduced pathology in mice lung indicating its role in the virulence of M. tuberculosis48. Also, LipY is demonstrated to play roles in M. tuberculosis virulence18, though the mechanisms are not clear. All these reports suggest an important role of the esterase/lipase in mycobacterial virulence.
Colony morphology of the mycobacterium species is known to be a complex phenotype associated with mycobacterial virulence. Colony morphology is also correlated to antibiotic susceptibility49, cytokine induction and MAP kinase signaling50,51. Earlier studies have indicated that changes in colony morphology are indicative of metabolic changes and virulence properties of M. avium52. Expression of pe11 gene in M. smegmatis led to a shift from a usual rough to unusual smooth morphology of the bacilli is suggestive of a structural role of this protein. Msmeg-PE11 cells were found to be wider in diameter as compared to Msmeg-pVV cells. We further observed an electron-dense staining of M. smegmatis expressing PE11 in TEM analysis. This implies that the changes in the composition and content of the surface exposed lipids of M. smegmatis due to expression of PE11 reacted more vigorously with uranyl acetate and lead citrate to give the more electron-dense appearance of the outermost cell envelope layer.
Changes in the cell wall lipid composition can also reflect marked variations in mycobacterial cell surface properties like pellicle/biofilm formation, congo red binding, cellular aggregation and sliding motility25,26,27. Mycobacteria grown in the absence of detergent tend to form surface pellicle, a biofilm like structure at the liquid-air interface when grown in synthetic media53 which can confer drug resistance and often harbor drug tolerant cells54. Pellicle formation may be regarded as an adaptive diversification of the organism in response to certain stimuli into a structured environment like biofilms which are considered to be surface-associated bacterial communities comprising micro colonies surrounded by extracellular matrix. We found that M. smegmatis expressing PE11 was able to form profuse pellicle as compared to the control cells (Msmeg-pVV). Similarly, PE11 was found to increase the cell surface hydrophobicity causing an increased tendency of Msmeg-PE11 to form cellular aggregates possibly due to an increase in the glycopeptidolipid content in the cell wall. We further found that Msmeg-PE11 is more resistant to antibiotics as well as various stressors like SDS, lysozyme, H2O2 and low pH (5.5) those mimicking the hostile macrophages environments encountered by the bacilli55. Thus, our data suggest that PE11 is actively involved in the cell wall remodeling that may confer increased drug resistance and survival advantages to the mycobacteria inside macrophages. Recently, Deng et al., (2015) also reported that PE11 (Rv1169c) can alter the composition of mycobacterial cell wall when expressed in M. smegmatis56. Interestingly, we found a different fatty acid profile from that reported by Deng et al., (2015). Also, we observed an increased resistance of the bacteria expressing PE11 to various environmental stressors and antibiotics in contrast to the report by Deng et al., (2015). This may be due to use of different promoters in different expression vectors as well as different positions of the Histidine tag used while expressing PE11 in M. smegmatis.
Previous studies have shown that changes in cell hydrophobicity and cell wall integrity are related to alteration in the lipid composition of the mycobacterium cell envelope27,52. In this context, the glycopeptidolipids (GPLs), which are species-specific glycolipids, are characterized by a high variability in glycosylation patterns. GPLs are present on the cell wall surface of pathogenic and non-pathogenic mycobacteria and are implicated to play critical roles in the colony morphology. Glycosylation of the lipopeptide core of GPLs is found to be a prerequisite for colony smoothness of the members of the M. avium complex33. In addition, the mps mutants of M. smegmatis which are devoid of GPLs also exhibited a rough colony morphotype33. Interestingly, we observed enhanced synthesis of GPLs in Msmeg-PE11, which at least partly explains the observed smooth colony morphology and altered surface properties in these cells. Interestingly, when we quantified the cell wall fatty acids as methyl esters (FAMEs) using a high throughput gas chromatography coupled with mass spectrometry (GC/MS), we found almost similar fatty acid composition in both the strains, however, expression of PE11 caused a noticeable decrease in the amount of linear C18:0 polar fatty acids (Table 1), along with an increase in the branched chain polar fatty acid content (C18:10-methyl) which may increase the membrane fluidity and the ability of Msmeg-PE11 to tolerate environmental stress. Interestingly, branched chain fatty acids have been implicated in the development of pH-stress tolerance in Listeria monocytogenes57. From our lipid analysis data (Fig. 5), it is apparent that PE11 also significantly increased total polar lipid content in M. smegmatis without significantly affecting the total apolar lipid content. This may well be correlated with the observed persistence and virulence of Msmeg-PE11 in mice infection models. The polar lipid fractions of pathogenic M. bovis were found to induce higher IL-10 production and antigen presenting cells had significant decrease in the levels of costimulatory molecules associated with antigen presentation including MHC-II and CD8658. In fact, we were able to detect higher levels of IL-10 and IL-4 cytokines in the liver and lung of Msmeg-PE11 infected mice as compared to those infected with control Msmeg-pVV (Fig. 8c,d). On the other hand, Msmeg-PE11 infected mice had higher induction of pro-inflammatory cytokines like TNF-α in the lung and liver (Fig. 8a) though the levels of IL-1β did not significantly differ (Fig. 8b and Supplementary Fig. 8). The high expression of IL-4 has been implicated as a virulence factor, both for its anti-inflammatory ability and also for its apparent capacity to promote tissue damage in association with TNF-α59. These observations are well correlated with the exacerbated pathology found in liver and lung and consequently mortality in Msmeg-PE11-infected mice similar to those observed in a Balb/c mouse model of pulmonary tuberculosis infection where appearance of IL-4 in the lung lesions coincides temporally and spatially with the appearance of areas of pneumonia and necrosis, leading to rapid clinical deterioration and death60.
In summary, we found that expression of PE11 in a non-pathogenic surrogate M. smegmatis could confer its properties akin to typical virulent mycobacteria including increased cell wall integrity, resistance to environmental stress, improved survival inside macrophages and mice. PE11 actively participates in remodeling the cell wall and may also provide energy by hydrolyzing acyl esters through its esterase activity for intracellular persistence of the pathogen. The possible localization of the M. tuberculosis PE11 at the cell surface29 and its involvement in the mycobacterial cell wall alteration prompted us to speculate that this protein may also participate in the active modulation of the immune responses. Accordingly, we found that PE11 can induce a pronounced Th2 environment by inducing cytokines like IL-10 and IL-4 in the lung and liver of mice infected with M. smegmatis expressing PE11, in addition to TNF-α which correlated well with the observed pathology in these tissues and also explains their higher CFU as compared to mice infected with control strain harboring the vector alone. Our findings suggest PE11 is a putative virulence factor for M. tuberculosis and may represent a promising target for future development of anti-mycobacterial therapeutics.
Methods
M. smegmatis culture and transformation
M. smegmatis mc2155 bacteria were grown in Middlebrook 7H9 medium (BD Difco, USA) supplemented with 10% OADC (HiMedia, India), 0.5% glycerol, and 0.05% Tween 80 (Fisher Scientific, USA). For preparing competent cells, the culture was allowed to grow until mid-log phase. The culture was centrifuged at 3000 rpm for 10 min, washed four times with 10% glycerol, and re-suspended in 1/100th of the culture volume in sterile deionized water with 10% glycerol and 100 μl aliquots of the cells were snap-frozen and stored at −80 °C until further use. Prior to transformation, the cells were thawed on ice, and 1 μg of DNA was added. The cells were incubated on ice for 10 min and transferred to a pre-chilled 1 mm gap width cuvette. One ml of the culture medium was added immediately, and the cells were incubated at 37 °C for 4 h. The transformants were selected on 7H10 (BD Difco, USA) agar plates containing 50 μg/ml kanamycin (Amresco, USA) and 50 μg/ml hygromycin B (Invitrogen, USA).
Generation of recombinant M. smegmatis expressing PE11
For generation of recombinant M. smegmatis expressing PE11, the pe11 gene was PCR amplified from M. tuberculosis H37Rv genomic DNA using specific primers (forward primer, F-5′-AATCGTGCATATGTCTTTTGTCACCACACGGCCCGATTC-3′ and reverse primer, R-5′CGGGATCCGGTGGAGGTGCCCGCGCGG-3′) encompassing the pe11 ORF. The PCR amplicon was cloned into the NdeI and BamHI sites of pVV16 expression vector harbouring hsp60 promoter system (pVV-pe11) with a C-terminal His-tag. All the clones were confirmed by restriction digestion and the sequence of the inserts was validated by automated DNA sequencing. M. smegmatis underwent electroporation with pVV-pe11 to generate recombinant M. smegmatis (Msmeg-PE11) and transformants were selected on 7H10 agar plate containing 10% OADC, 25 μg/ml kanamycin and 50 μg/ml hygromycin B. Expression of PE11 was confirmed by immunoblotting using anti-Histidine-tag antibody (Sigma-Aldrich, USA). M. smegmatis transformed with pVV16 vector (Msmeg-pVV) was used as control group.
Preparation of M. smegmatis cell lysate
The Msmeg-pVV and Msmeg-PE11 cultures were grown in Middlebrook 7H9 medium supplemented with 10% OADC, 0.5% of glycerol and 0.05% Tween 80 till the absorbance at 600 nm reached 0.6. Bacterial cell pellets were harvested and washed thrice with phosphate buffer saline with Tween 80 (PBS-T). The resulting pellets were then re-suspended in lysis buffer (50 mM Tris-Cl of pH 8.0, 300 mM NaCl and protease inhibitor) and vortexed. Bacterial cells were lysed using an ultrasonic sonicator and the lysed cells were centrifuged at 14000 rpm to collect the supernatant. Protein concentration was estimated using bicinchoninic acid method (Thermo scientific, USA).
Reverse Transcription qPCR for cytokine
Total RNA was isolated from the tissues using TRI Reagent (Sigma-Aldrich, USA) as per manufacturer’s instructions. Briefly, the tissues were homogenized for 30 second in 0.5 ml TRI Reagent using a Mini BeadBeater (BioSpec Products, Inc., USA) and the aqueous phase was separated by addition of 0.1 ml chloroform. The total RNA in the aqueous phase was precipitated by addition of equal volume of isopropanol and the RNA pellet was washed with 75% chilled ethanol. After air drying the pellet was dissolved in RNAse-free water and treated with amplification grade DNAse I (Sigma-Aldrich, USA) to remove any genomic DNA contamination.
RNA concentration and purity was assessed using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies,USA). RNA integrity was assessed using the RNA 6000 Nano LabChip Series II Assay with the 2100 Bioanalyzer System (Agilent Technologies, USA). About 2 μg total RNA with RIN (RNA integrity number) value of >7.5 were used for generation of first strand cDNA using First Strand cDNA synthesis kit (Thermo Scientific, USA) and using OligodT18 primers as per manufacturer’s instructions. The cDNA reactions were diluted to 50 μl in nuclease-free water, aliquoted and stored at −20 °C till further use.
Realtime qPCR reactions were carried out in duplicate for each sample using SYBR Premix Ex Taq (Tli RNaseH Plus, DSS Takara Bio India Pvt. Ltd, India) in a 20 μl reaction volume containing each primers at 500 nM final concentration. Reactions were temperature cycled, and SYBR green levels were measured using a CFX-96 system (Bio-Rad Laboratories, USA). The initial target concentration for each gene except IL-4 was calculated by relative standard curve method using a pool of experimental sample cDNA as calibrator. For IL-4, the calibrator for the relative standard curve was prepared by using a mixture of purified IL-4 and GAPDH PCR products in an equimolar ratio due to its low copy number and low levels of expression in most of the samples. The level of expression for each gene in each sample was normalized to corresponding GAPDH levels. All the primers were synthesized from Eurofins Genomics India Pvt Ltd (Bangalore, India). The sequences of the primers used for RT-qPCR were given in Supplementary Fig. 9.
Colony morphology
For colony morphology, Msmeg-pVV and Msmeg-PE11 strains were plated onto Middlebrook 7H10 (BD, Difco, USA) plates with 0.5% glycerol, and 10% OADC and incubated for 5–6 days at 37 °C. A Canon PowerShot G11 Digital camera (Japan) was used to take photomicrographs of colonies.
Enzyme activity
The enzyme activity of PE11 was measured, as described elsewhere45, using different esters of p-nitrophenol (pNP) (Sigma-Aldrich, USA) such as acetate (pNPC2), butyrate (pNPC4), octanoate (pNPC8), laurate (pNPC12), myristate (pNPC14), palimitate (pNPC16) and stearate (pNPC18). Enzymatic reactions were performed at 37 °C for 10 min in a 100 mM Tris-HCl buffer (pH 7.0) containing mycobacterial cell lysate (20 μg) and 2 mM of pNP substrate, solubilized in acetonitrile (ACN). Final volume was fixed to 200 μl in each microtiter well. The enzymatic hydrolysis was quantified by measuring the absorbance of the hydrolyzed products of all the substrates (p-nitrophenol) at 405 nm using a 96-well plate spectrophotometer (Ultra Microplate Reader EL808, Bio-Tek, USA) using a standard pNP calibration curve. Activity was expressed in International units (U, corresponding to 1 μmol of pNP released per minute). Specific activities were expressed as U/mg of cell lysate. Lipolytic activity as determined by Tween cleavage was performed as previously reported31. Briefly, a reaction mixture was prepared in Tris (50 mM) buffer containing 33 mM CaCl2 and 0.33% Tween 20 (Ameresco, USA) or 0.33% Tween 80 (pH 7.0) and 40 μg cell lysate and incubated at 37 °C. Reaction mixture (200 μl) was transferred to a 96-well microwell plate, and turbidity was assessed by measuring the absorbance at 405 nm. Activity in this assay is reported as U/mg of lysate, where 1U is defined as the amount of enzyme required to induce a change in absorbance (405 nm) of 0.01 under assay conditions.
Transmission electron microscopy (TEM)
For, TEM, cells were washed, fixed with 3% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 24 h at 4 °C and then fixed with 1% osmium tetroxide, dehydrated in a graded series of alcohol, and embedded in araldite 6005 resin. Ultra-thin sections (50–70 nm) were made with a glass knife on ultra-microtome (Leica Ultra cut UCT-GA-D/E-1/00), mounted on a copper grid, and stained with saturated uranyl acetate and counter stained with Reynold’s lead citrate. Specimens were examined using a Hitachi, H-7500 transmission electron microscope.
Scanning electron microscopy (SEM)
For scanning electron microscopy, the Msmeg-PE11 or Msmeg-pVV bacilli were harvested by low-speed centrifugation when the absorbance was about 0.6 at 600 nm. The bacterial pellets were washed in sterile PBS, and fixed for 24 h at 4 °C in a modified Karnovsky’s fixative in which cacodylate buffer was replaced with 0.2 M phoshphate buffer pH 7.4. Cells were then dehydrated through series of graded ethanol (30%, 50%, 70%, 80%, 90% and 100%) and dried by vacuum desiccation. The samples were then attached to stubs, sputter coated with gold (Model E-1010, Hitachi, Japan) and were examined using a Hitachi S3400N Scanning Electron Microscope at 15.0 kV. The diameter of the bacteria was measured by using ImageJ, an image analysis software available in the public domain.
Mycobacterial cell surface properties
Mycobacterial cell surface properties were determined as described elsewhere25. For studying cellular aggregation, the bacilli were grown to an absorbance of 0.6 at 600 nm in 7H9 medium with or without 0.05% Tween 80 in a 37 °C shaker incubator. The cultures were kept at room temperature for 30 min to allow the cell aggregates to settle down, which were photographed with Canon PowerShot G11 Digital camera (Japan).
An assay for pellicle and biofilm formation was carried out as described earlier26,27. Briefly, pellicle formation was monitored by growing the cultures of M. smegmatis without shaking at 37 °C for 24 h, 48 h, 72 h and day 15 in Middlebrook 7H9 medium devoid of Tween 80 and the cultures were then photographed. To measure M. smegmatis biofilm formation, 200 μl of medium was added to 96-well polystyrene Costar plate (Corning, NY, USA) and inoculated with mycobacterial cells to an absorbance of 0.02 at 600 nm. The plates were then incubated at 37 °C for 24 h, 48 h, 72 h and day 15 for formation of biofilm. Next, the medium was discarded and the biofilm was washed with deionized water and stained with 1% crystal violet (Fisher Scientific, USA) for 30 min. Quantitation of biofilm formation was performed by extracting the biofilm associated crystal violet with 100% ethanol for 1 h and measuring the optical density at 570 nm. Congo red accumulation was determined by cultivating Msmeg-pVV or Msmeg-PE11 for 24 h, 48 h and 72 h at 37 °C with shaking (200 rpm) in 7H9 broth in presence of 100 μg/ml congo red (Sigma-Aldrich, USA) and 0.05% Tween 80. Cells were collected by centrifugation (3000 g, 30 min) and washed extensively with distilled water until the supernatant was colorless. Cells were re-suspended in 1 ml acetone, vortexed and gently shaken for 2 h at room temperature, cells were then removed by high speed microcentrifugation and congo red in the supernatants was quantified spectrophotometrically by measuring absorbance at 488 nm (Ultra Microplate Reader EL808, Bio-Tek, USA).
Measurement of sensitivity to SDS, lysozyme, hydrogen peroxide, low pH and antibiotics
Bacterial cultures were centrifuged and washed with 7H9 medium containing 0.05% Tween 80. Mycobacterial cell concentration was adjusted to an absorbance of 0.02 at 600 nm. To measure sensitivity towards low pH (pH 5.5), the bacteria were inoculated in 7H9 medium, pH 5.5 for 24 h at 37 °C. Sensitivity of bacterial culture towards 0.1% SDS (Sigma-Aldrich, USA) for 3 h and 24 h, and lysozyme (Sigma-Aldrich, USA) (25 μg/ml, 250 μg/ml, 500 μg/ml and 1000 μg/ml) for 24 h and 48 h, was measured at 37 °C. Further, to measure sensitivity to hydrogen peroxide, the bacteria were incubated at 37 °C with 5 mM hydrogen peroxide (Merck, USA) for 3 h, 6 h and 24 h. Susceptibility of M. smegmatis to antibiotics like ethambutol, isoniazid and rifampicin (20 μg/ml) (all from Sigma-Aldrich, USA) and vancomycin (5 μg/ml) and ampicillin (750 μg/ml) (both from Amresco, USA) was determined in 7H9 broth cultured for 24 h and 48 h. Numbers of CFU for each stress and antibiotics were determined by plating serial dilutions onto 7H10 agar plates.
Mycobacterial lipid extraction and TLC analysis
M. smegmatis strains (Msmeg-pVV and Msmeg-PE11) were grown at 37 °C in 500 ml Middlebrook 7H9 broth supplemented with 0.5% glycerol, 0.05% Tween 80 and 10% OADC until an absorbance of 0.8 at 600 nm was reached. Cells recovered by centrifugation at 5,000 g for 10 min and total lipids from the mycobacterial wet pellets (equal weights of both strains) were extracted at room temperature, with 50 ml chloroform-methanol at a ratio of 2:1 (vol/vol). The organic phase containing the whole-cell lipid extracts were evaporated to dryness using rotary evaporator (Heidolph Instruments, India). Lipids were re-suspended in a minimal volume of chloroform and identified by thin layer chromatography (TLC) on Silica Gel 60 plates (Macherey-Nagel, Germany). Polar and apolar lipids were fractionated on a preparative TLC using specific solvent systems. Polar lipids were extracted using chloroform-methanol-water (65:25:4, vol/vol/vol), whereas apolar lipids were extracted using Hexane-ethylacetate (90:10, vol/vol). Glycolipids were extracted from Msmeg-PE11 or Msmeg-pVV using Methanol: Ammonium hydroxide (80:20, vol/vol) as mobile phase and detected by α-naphthol/sulfuric acid staining. The Mycolic acid containing glycolipids were separated from the pool of total polar lipids using Chloroform: Methanol: Ammonium hydroxide (80:20:2 vol/vol/vol) mobile phase and visualized by Iodine vaporization, followed by charring with 5% phosphomolybdic acid.
Fatty acid composition by gas liquid chromatography
The fatty acid compositions of the lipid fractions were determined by gas liquid chromatography (GLC). The lipid fractions were converted to their corresponding methyl esters and GLC analysis of the fatty acid methyl esters (FAMEs) was performed using a Agilent 6890 gas chromatograph coupled to a flame ionization detector (FID) equipped with a DB 225 capillary column (30 m × 0.25 mm × 0.25 μm; 50% cyanopropylphenyl dimethyl polysiloxane stationary phase material, J & W Scientific, USA), a split injector (split ratio, 10:1), and a FID. The column temperature program was set for 2 min at 160 °C, 5 °C/min to 230 °C and 20 min at 230 °C, respectively. The injector was adjusted to 230 °C with a split ratio of 10:1 using nitrogen carrier gas at a flow rate of 1 ml/min. The detector temperature was 270 °C with air and hydrogen flow rates of 300 ml/min and 30 ml/min, respectively. The fatty acid peaks were identified by comparing the retention times with those of a mixture of standard FAMEs, C4–C24 (Supelco, USA).
GC/MS studies of fatty acid methyl esters
GC/MS studies were carried out for the identification of FAMEs formed during the transesterification reaction. The GC/MS detection was performed with Agilent 6890 N gas chromatograph connected to Agilent 5973 mass selective detector at 70 eV (m/z 50–800; source at 230 °C and quadruple at 150 °C) in the EI mode with a HP-1 ms capillary column (30 m × 0.25 mm I.D. × 0.25 μM film thickness). The oven temperature was kept at 100 °C for 2 min and finally raised to 300 °C at 10 °C/min and maintained for 20 min at 300 °C. The carrier gas, helium was used at a flow rate of 1.0 ml/min. The inlet temperature was maintained at 300 °C and split ratio was 50:1. Structural assignments were based on interpretation of mass spectrometric fragmentation and confirmed by comparison of retention times as well as fragmentation pattern of authentic compounds.
Infection of mice
Six weeks old male Balb/c mice (six per group, housed in the Centre for DNA Fingerprinting and Diagnostics animal facility located within the premises of Vimta Labs, Hyderabad) were infected with 5 × 107 CFUs of either Msmeg-pVV or Msmeg-PE11 by intravenous injection. All the strains were dispersed in PBS-T and no clumping was observed. The inoculums were cultured on 7H10 plates containing kanamycin and hygromycin B to confirm that equal numbers of bacteria were administered intravenously for each strain. At each time point (day 2, 7 and 14), survival of the recombinant M. smegmatis strains in mouse organs was determined using methods as described previously36. Briefly, the mice were sacrificed and lungs, liver and spleens were removed aseptically and homogenized in sterile PBS-T. The mouse tissue homogenates were diluted in PBS-T, and 100 μl aliquots from each dilution were plated on Middlebrook 7H10 agar plate containing 10% OADC, 25 μg/ml kanamycin and 50 μg/ml of hygromycin B. The plates were incubated at 37 °C, and the CFUs were determined after 3–4 days. Animal experiments were carried out in accordance with the approved guidelines of the Institutional Animal Ethics Committee (IAEC) of the Vimta Labs Ltd. (IAEC protocol approval number: PCD/CDFD/15).
Histopathology of infected organs
The histopathology was carried out as described earlier36. Briefly, lung, liver and spleen of mice was aseptically removed from euthanized animals and were fixed in 10% formalin and then embedded in paraffin wax. Sections were then stained with hematoxylin and eosin stain for visualizing under an Olympus CX21 microscope (Olympus, Japan). Also, microphotographs were taken using an Olympus DP72 CCD camera, (Japan) attached to the microscope. DP2-BSW software was used for image analysis. A group of 10 mice were used for determining the survival index infected with either Msmeg-pVV or Msmeg-PE11 strain for 30 days post-infection.
Statistical analysis
Data were expressed as mean ± SD and Student’s t test was used to determine statistical differences between the groups and p < 0.05 was considered to be significant. GraphPad Prism 5.02 software (GraphPad software) was used for statistical analysis.
Additional Information
How to cite this article: Singh, P. et al. PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci. Rep. 6, 21624; doi: 10.1038/srep21624 (2016).
Supplementary Material
Acknowledgments
The authors thank Mr. A. Harikrishna, CCMB, Hyderabad for TEM experiments; Mr. P. Madhusudhana Chary, NIN, Hyderabad for technical assistance in performing scanning electron microscopy; Mr. V. Srikanth and Mr. V. Vijayender, IICT, Hyderabad for helping in lipid experiments; Dr. Jayant Hole, Mr. K. Suman, Mr. K. Sridhar and Ms. Sravani CDFD, Hyderabad for kind help in mice experiments and Mr. Niteen Pathak, CDFD, Hyderabad for technical help. The laboratory of SM is supported by the grant from Department of Biotechnology (DBT), Govt of India (BT/PR5496/MED/29/512/2012) and also core grant from CDFD by DBT. PS was supported by a fellowship from University Grant Commission (UGC). SKK was supported by a fellowship from University Grant Commission (UGC).
Footnotes
Author Contributions P.S., R.N.R. and S.M. designed and performed experiments. S.M. analyzed the data. J.R.C.R. and R.B.N.P. designed and analyzed lipid experiments. S.K.K. and S.G. performed RT-qPCR. S.M. and S.G. wrote and edited the manuscript with input from P.S. and R.N.R.
References
- Mukhopadhyay S., Nair S. & Ghosh S. Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol. Rev. 36, 463–485 (2011). [DOI] [PubMed] [Google Scholar]
- Cole S. T. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998). [DOI] [PubMed] [Google Scholar]
- Gey van Pittius N. C. et al. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 6, 95 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarlier V. & Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123, 11–18 (1994). [DOI] [PubMed] [Google Scholar]
- Neyrolles O. & Guilhot C. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis (Edinb) 91, 187–195 (2011). [DOI] [PubMed] [Google Scholar]
- Rousseau C. et al. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell. Microbiol. 6, 277–287 (2004). [DOI] [PubMed] [Google Scholar]
- Passemar C. et al. Multiple deletions in the polyketide synthase gene repertoire of Mycobacterium tuberculosis reveal functional overlap of cell envelope lipids in host-pathogen interactions. Cell. Microbiol. 16, 195–213 (2014). [DOI] [PubMed] [Google Scholar]
- Reed M. B. et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431, 84–87 (2004). [DOI] [PubMed] [Google Scholar]
- McNeil M. Target preclinical drug development for Mycobacterium avium complex: a biochemical approach. In Mycobacterium avium-complex infection (eds Korvick J. A. & Benson C. A.) (New York, NY: Marcel Dekker., 1996). [Google Scholar]
- Alland D., Steyn A. J., Weisbrod T., Aldrich K. & Jacobs W. R. Jr. Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J. Bacteriol. 182, 1802–1811 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L. et al. Recognition of multiple effects of ethambutol on metabolism of mycobacterial cell envelope. Antimicrob. Agents Chemother. 39, 694–701 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J., Maamar H., Deb C., Sirakova T. D. & Kolattukudy P. E. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 7, e1002093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Singh G., Jadeja D. & Kaur J. Lipid hydrolizing enzymes in virulence: Mycobacterium tuberculosis as a model system. Crit. Rev. Microbiol. 36, 259–269 (2010). [DOI] [PubMed] [Google Scholar]
- Shen G. et al. LipC (Rv0220) is an immunogenic cell surface esterase of Mycobacterium tuberculosis. Infect. Immun. 80, 243–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. Expression and characterization of the carboxyl esterase Rv3487c from Mycobacterium tuberculosis. Protein Expr. Purif. 42, 59–66 (2005). [DOI] [PubMed] [Google Scholar]
- Canaan S. et al. Expression and characterization of the protein Rv1399c from Mycobacterium tuberculosis. A novel carboxylesterase structurally related to the HSL family. Eur. J. Biochem. 271, 3953–3961 (2004). [DOI] [PubMed] [Google Scholar]
- Deb C. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 281, 3866–3875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V. K. et al. Increased virulence of Mycobacterium tuberculosis H37Rv overexpressing LipY in a murine model. Tuberculosis (Edinb) 94, 252–261 (2014). [DOI] [PubMed] [Google Scholar]
- Fisher M. A., Plikaytis B. B. & Shinnick T. M. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184, 4025–4032 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D. et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J. Exp. Med. 198, 693–704 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil M. I., Visconti K. C. & Schoolnik G. K. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 84, 218–227 (2004). [DOI] [PubMed] [Google Scholar]
- Narayana Y., Joshi B., Katoch V. M., Mishra K. C. & Balaji K. N. Differential B cell responses are induced by Mycobacterium tuberculosis PE antigens Rv1169c, Rv0978c, and Rv1818c. Clin. Vaccine Immunol. 14, 1334–1341 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson S. L. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin. Dev. Immunol. 2011, 497203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A. et al. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62, 132–147 (2006). [DOI] [PubMed] [Google Scholar]
- Etienne G. et al. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148, 3089–3100 (2002). [DOI] [PubMed] [Google Scholar]
- Recht J., Martinez A., Torello S. & Kolter R. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182, 4348–4351 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. M. et al. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 188, 633–641 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi G. A., Palermo C. O., Laurent J. P., Hamlin A. M. & Brabant W. H. Colony morphotypes on congo red agar segregate along species and drug susceptibility lines in the Mycobacterium avium-intracellulare complex. Microbiology 145, 1317–1324 (1999). [DOI] [PubMed] [Google Scholar]
- Cascioferro A. et al. PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol. Microbiol. 66, 1536–1547 (2007). [DOI] [PubMed] [Google Scholar]
- Betts J. C., Lukey P. T., Robb L. C., McAdam R. A. & Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731 (2002). [DOI] [PubMed] [Google Scholar]
- West N. P. et al. Cutinase-like proteins of Mycobacterium tuberculosis: characterization of their variable enzymatic functions and active site identification. FASEB J. 23, 1694–1704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Rathi P., Gupta N. & Bradoo S. Lipase assays for conventional and molecular screening: an overview. Biotechnol. Appl. Biochem. 37, 63–71(2003). [DOI] [PubMed] [Google Scholar]
- Belisle J. T., McNeil M. R., Chatterjee D., Inamine J. M. & Brennan P. J. Expression of the core lipopeptide of the glycopeptidolipid surface antigens in rough mutants of Mycobacterium avium. J. Biol. Chem. 268, 10510–10516 (1993). [PubMed] [Google Scholar]
- Howard S. T. et al. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152, 1581–1590 (2006). [DOI] [PubMed] [Google Scholar]
- Lazar-Molnar E. et al. Programmed death-1 (PD-1)-deficient mice are extraordinarily sensitive to tuberculosis. Proc. Natl. Acad. Sci. USA 107, 13402–13407 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. H., Ahmed A., Kumar S., Sharma P. & Mukhopadhyay S. Role of PPE18 protein in intracellular survival and pathogenicity of Mycobacterium tuberculosis in mice. PLoS One 7, e52601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad T. R., Harrell M. I., Liao R. & Sherman D. R. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3, e1502 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil M. I., Bartek I. L., Visconti K. & Schoolnik G. K. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2, 105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontán P. A. et al. The Mycobacterium tuberculosis sigma factor σB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J. Bacteriol. 191, 5628–5633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli R., Voskuil M. I., Schoolnik G. K. & Smith I. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41, 423–437 (2001). [DOI] [PubMed] [Google Scholar]
- Schwab U. et al. Transcriptional responses of Mycobacterium tuberculosis to lung surfactant. Microb. Pathog. 46, 185–193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachman H. et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 74, 1233–1242 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H. & Rubin E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 (2003). [DOI] [PubMed] [Google Scholar]
- Griffin J. E. et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Characterization of a novel esterase Rv0045c from Mycobacterium tuberculosis. PLoS One 5, e13143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R., Vemula M. H., Banerjee S. & Guruprasad L. The PE16 (Rv1430) of Mycobacterium tuberculosis is an esterase belonging to serine hydrolase superfamily of proteins. PLoS One 8, e55320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. et al. Characterization of a novel exported esterase Rv3036c from Mycobacterium tuberculosis. Protein. Expr. Purif. 104, 50–56 (2014). [DOI] [PubMed] [Google Scholar]
- Lun S. & Bishai W. R. Characterization of a novel cell wall-anchored protein with carboxylesterase activity required for virulence in Mycobacterium tuberculosis. J. Biol. Chem. 282, 18348–18356 (2007). [DOI] [PubMed] [Google Scholar]
- Kuze F. & Uchihira F. Various colony-formers of Mycobacterium avium-intracellulare. Eur. J. Respir. Dis. 65, 402–410 (1984). [PubMed] [Google Scholar]
- Shiratsuchi H., Toosi Z., Mettler M. A. & Ellner J. J. Colonial morphotype as a determinant of cytokine expression by human monocytes infected with Mycobacterium avium. J. Immunol. 150, 2945–2954 (1993). [PubMed] [Google Scholar]
- Tse H. M., Josephy S. I., Chan E. D., Fouts D. & Cooper A. M. Activation of the mitogen-activated protein kinase signaling pathway is instrumental in determining the ability of Mycobacterium avium to grow in murine macrophages. J. Immunol. 168, 825–833 (2002). [DOI] [PubMed] [Google Scholar]
- Kansal R. G., Gomez-Flores R. & Mehta R. T. Change in colony morphology influences the virulence as well as the biochemical properties of the Mycobacterium avium complex. Microb. Pathog. 25, 203–214 (1998). [DOI] [PubMed] [Google Scholar]
- Zambrano M. M. & Kolter R. Mycobacterial biofilms: a greasy way to hold it together. Cell 123, 762–764 (2005). [DOI] [PubMed] [Google Scholar]
- Sambandan D. et al. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. MBio. 4, e00222–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal O. H. et al. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J. Bacteriol. 191, 625–631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Zeng J., Xiang X., Li P. & Xie J. PE11 (Rv1169c) selectively alters fatty acid components of Mycobacterium smegmatis and host cell interleukin-6 level accompanied with cell death. Front. Microbiol. 6, 613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giotis E. S., McDowell D. A., Blair I. S. & Wilkinson B. J. Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 73, 997–1001 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirson C., Jones G. J., Steinbach S., Besra G. S. & Vordermeier H. M. Differential effects of Mycobacterium bovis derived polar and apolar lipid fractions on bovine innate immune cells. Vet. Res. 43, 54 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah G. T. & Rook G. A. W. IL-4 influences apoptosis of mycobacterium-reactive lymphocytes in the presence of TNF-α. J. Immunol. 167, 1230–1237 (2001). [DOI] [PubMed] [Google Scholar]
- Hernandez-Pando R. et al. Correlation between the kinetics of Th1/Th2 cells and pathology in a murine model of experimental pulmonary tuberculosis. Immunology 89, 26–33 (1996). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.