Abstract
Merkel cell carcinoma (MCC) is a virally associated cancer characterized by its aggressive behavior and strong immunogenicity. Both viral infection and malignant transformation induce expression of MHC class I chain-related protein (MIC) A and B, which signal stress to cells of the immune system via Natural Killer group 2D (NKG2D) resulting in elimination of target cells. However, despite transformation and the continued presence of virally-encoded proteins, MICs are only expressed in a minority of MCC tumors in situ and are completely absent on MCC cell lines in vitro. This lack of MIC expression was due to epigenetic silencing via MIC promoter hypo-acetylation; indeed, MIC expression was re-induced by pharmacological inhibition of histone deacetylases (HDACs) both in vitro and in vivo. This re-induction of MICs rendered MCC cells more sensitive to immune-mediated lysis. Thus, epigenetic silencing of MICs is an important immune escape mechanism of MCCs.
Merkel cell carcinoma (MCC) is an aggressive skin cancer with neuroendocrine features. The histogenesis of MCC remains controversial, with either epidermal stem cells or pre/pro-B cells as possible cells of origin1. Currently, there are no approved therapies for advanced MCC that is not surgically resectable, thus almost half of the patients diagnosed with MCC will die from the disease2. This situation is compounded because the reported incidence of MCC has more than tripled over the last few decades3. This increase in incidence is true across all age groups, indicating that the aging general population is not the sole reason for the increased incidence. Nevertheless, advanced age is still one of the relevant risk factors among UV exposure and immune suppression4.
The strong correlation of MCC and immune suppression prompted the discovery of a polyomavirus associated with MCC, termed Merkel cell polyomavirus (MCPyV)5. MCPyV is present and clonally integrated in at least 80% of MCCs6. Most MCPyV positive MCC cell lines critically depend on virally encoded transforming early genes, i.e. small and large T-antigen, in order to maintain the oncogenic phenotype7,8. The continuous expression of these viral proteins helps explain the exquisite immunogenicity of MCC. Notably, despite the highly aggressive phenotype of MCC, spontaneous regression or regression after cessation of immune suppressive measures are well established9. Moreover, we and others have recently demonstrated the presence of spontaneous adaptive cellular immune responses specific for epitopes derived from the MCPyV early genes in peripheral blood of MCC patients10,11. However, despite the continuous expression of the relevant antigens and the presence of respective specific cytotoxic T-cell responses, it is obvious that clinically manifest MCCs are able to escape destruction by the immune system. While this fact can be readily explained by a general immune suppression in approximately 10% of the patients12, the immune escape mechanisms of MCC are less clear for the remaining 90%13.
Natural Killer group 2D (NKG2D) is a lectin-like type 2 transmembrane receptor encoded by the gene Klrk1 (killer cell lectin-like receptor subfamily member 1) and is part of a critical pathway signaling cellular stresses to the innate and adaptive immune system. Charged residues in the transmembrane region enable NKG2D to pair with the signaling adaptor protein DAP10, which is essential for NKG2D surface expression and downstream signaling to PI3K and GRB214. These signaling molecules then stimulate proliferation, cytokine production, immune cell activation, and cytotoxic potential of NK and T cells14. A recent study suggested a link between the Natural Killer group 2D (NKG2D) receptor system and up-regulation of immune responses to MCC. Specifically, transcriptional analyses of MCC tumors revealed that NKG2D was among the highest expressed mRNAs in tumors obtained from patients with a good prognosis15. However, these tumors represented a minority of patients, suggesting most MCCs evade NKG2D signaling as a means of immune escape.
The NKG2D ligands include UL16-binding proteins (ULBPs) as well as the MHC class I chain-related protein (MIC) A and B family16. MICA and MICB are present at low to undetectable levels in normal cells, but are induced by cellular stresses including infectious agents and neoplastic transformation. Indeed, MICA and MICB are highly expressed in a number of solid tumors like carcinomas of the breast, colon, kidney, ovary, or prostate17, as well as in melanoma18. However, NKG2D expression renders tumor cells more susceptible to elimination by the immune system19. The importance of MICA and MICB induced NKG2D-signaling for immune surveillance of virally infected and transformed cells is highlighted by the fact that viruses and cancer cells have developed mechanisms to interfere with this interaction14. These mechanisms include shedding of surface expressed molecules, binding and retaining of MICA and MICB proteins in the cytoplasm, over-expression of MICA and MICB mRNA-targeting microRNAs, as well as other epigenetic mechanisms such as chromatin remodeling14,20.
Viral carcinogenesis should predispose MCC for induction of MICA and MICB expression; however, when screening for the respective mRNA expression using publicly available data from the Gene Expression Omnibus (GEO), we observed that both MICA and MICB mRNA were rarely present in MCC. Prompted by this observation, in the present study we confirmed these data in an independent set, and extended this notion to the protein level. Furthermore, we demonstrate that this lack of MICA and MICB expression in MCC is due to epigenetic silencing by promoter hypo-acetylation. Notably, MICA and MICB expression, particularly MICB expression, can be induced by histone deacetylase inhibitors, which in turn rendered the MCC cells more susceptible to lysis by cytotoxic lymphocytes. These findings open new avenues for therapy of advanced MCC particularly in combination with immune modulating molecules such as immune checkpoint blocking antibodies.
Results
MCC tumors express low levels of MICA and MICB mRNAs
Since both viral infection and malignant transformation induce expression of the immune activating NKG2D ligands MICA and MICB, we screened for the respective mRNA expression among 75 MCC tumors from 61 patients and a number of MCC cell lines. For this, we employed 2 publicly available gene expression arrays obtained online from GEO (accession numbers GSE2239615 and GSE 3961221; Supplementary Fig. 1). Somewhat unexpectedly, MICA mRNA was expressed only at very low levels in the MCC tumors and cell lines when compared to genes commonly expressed in MCCs such as RB1, E2F2, ENO2 or RPLP0. The MICB mRNA expression level was also low compared to those genes, but generally higher than for MICA. On the GSE22396 Array a subset of tumors (24%) were characterized by moderate to high levels of MICB mRNA. Notably, patients with higher levels of MICB mRNA in their tumors where characterized by better outcomes. In line with this, in MCPyV positive tumors, MICB mRNA expression correlated with the gene expression signature for infiltrating immune competent cells, a feature that had been associated with a good prognosis previously15 (Supplementary Fig. 2).
MCC tumors largely lack MICA and MICB expression in situ
Next, we analyzed tissue microarrays (TMA) comprising 134 FFPE fixed paraffin embedded MCC tumors of 99 patients by immunohistochemistry (IHC) using an antibody reacting with both MICA and MICB to determine MICA/B protein expression18 (Fig. 1a). MICA/B protein was present in only 18% of MCC tumors, while 82% were negative (Supplementary Fig. 3).
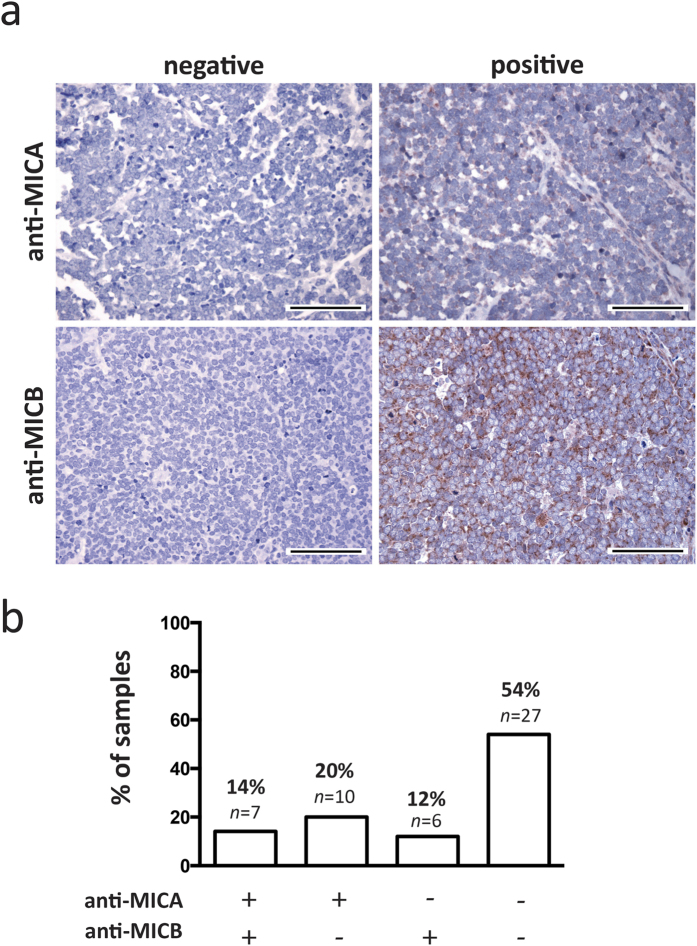
Figure 1. MICA and MICB expression of MCC tumors in situ.
MCC tumor samples from 34 patients were analyzed by immunohistochemistry for expression of MICA and MICB. (a) Samples were classified as positive or negative. Representative negative and positive samples are depicted at 40× magnification, scale bars are 100 μm. The positive examples represent the strongest obtained signal with the respective antibody. (b) Tumor samples were stratified into 4 groups: Double positive for MICA and MICB (+/+, 14%, n = 7), only positive for MICA (+/−, 20%, n = 10), only positive for MICB (−/+, 12%, n = 6), or double negative (−/−, 54%, n = 27).
The samples used in the cDNA microarray and the TMA were partly overlapping. Thus, to confirm the lack of MICA and MICB expression in a completely independent set of samples, we analyzed 50 additional MCC tumor samples of 34 patients for their MICA and MICB expression by IHC in conventional tumor sections. The use of sections of the whole tumor also allowed us to determine the expression pattern of MICA and MICB. A heterogeneous expression pattern of other immune modulatory molecules such as CD274 (aka PD-L1) had recently been described for MCC22.
Tumors classified as negative or positive for MICA and MICB expression are exemplarily depicted in Fig. 1a; notably, the strongest staining observed for the respective antibody is depicted. In line with higher MICB mRNA expression, MICB staining intensity was stronger than that of MICA. Overall we observed that more than half (54%) of the lesions expressed neither MICA nor MICB protein, 20% were weakly positive for only MICA, and 12% were classified positive for only MICB; and only 14% of the tumors stained positive for both MICA and MICB (Fig. 1b). The effective frequency of MICA and MICB expression remained essentially the same when calculated for each patient instead of the individual lesions (Supplementary Fig. 4). Indeed, when examining multiple lesions, i.e. primary tumors and metastatic lesions obtained from the same patients, the expression pattern of the MICA and MICB was concordant (data not shown). Similarly, with respect to a possible heterogeneity of MICA and MICB expression within the tumor, our analyses revealed that both the expression as well as the lack of it was homogenous throughout the tumor. This homogenous intralesional and intraindividual expression pattern suggests a rather robust mechanism for suppressed protein expression, such as epigenetic silencing.
MICA and MICB expression in MCC cell lines
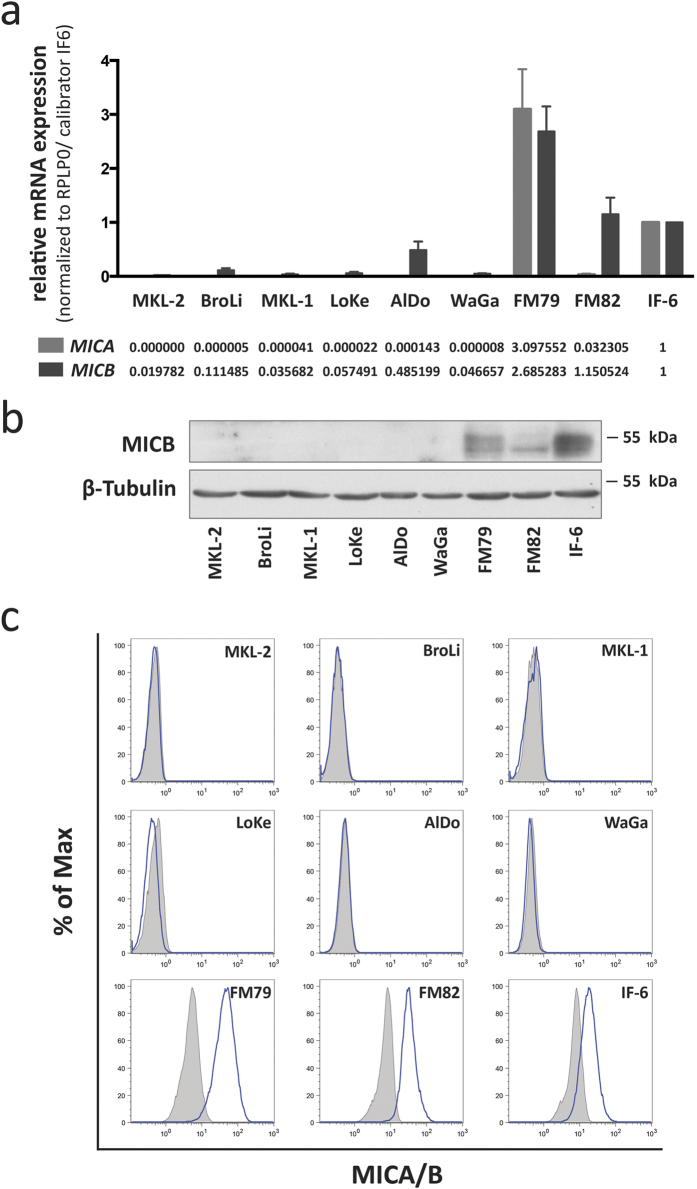
To explore the underlying mechanism, we determined whether the suppression of MICA and MICB expression was robust enough to be maintained when cells were propagated in vitro, in cell culture conditions. This question is of particular relevance since in vitro culture of cells is a well-established factor known to induce MICA and MICB expression23,24. We therefore performed qRT-PCR with MICA and MICB mRNA specific primers using the melanoma cells FM79, FM82 and IF6 as positive controls (Fig. 2a). These analyzes demonstrated that MICA mRNA in MCC cell lines was only expressed in very low amounts, i.e. close to or below the detection threshold; indeed, relative expression values calibrated to IF6 ranged close to zero. Similar to the results from the analyses of the MCC tumor samples, MICB mRNA expression was low, but still higher than MICA mRNA, with relative expression ranging from approximately 0.02 to 0.48 when calibrated to the positive control melanoma cell line IF6’s mRNA expression. Inconsistencies between mRNA and protein expression for MICB have been repeatedly described25, thus, to determine whether the detected MICB mRNA was indeed translated into protein we performed immunoblots of total cell lysates demonstrating that MICB protein was below the detection limit even for the AlDo MCC cell line characterized by the highest MICB mRNA expression (Fig. 2b). Accordingly, flow cytometric analysis for MICA/B membrane expression demonstrated no expression in any of the analyzed MCC cell lines (Fig. 2c). To determine how broadly NKG2D mRNA expression was suppressed, we subjected MCC cells to a number of stress factors known to induce NKG2D ligand expression24,26. These stressors included serum starvation, high concentrations of DMSO, heat shock at 41.5 °C, 1000 U/ml interferon α, and 1000 U/ml interferon γ. These interventions had no or only negligible effects on MICA/B surface expression (Supplementary Fig. 5a–f). Finally, we took advantage of a recently established inducible knock down system for viral T-antigens to stress the cells by withdrawal of the oncogene to which they are addicted; this knock down had no effect on MICB expression (Supplementary Fig. 5g).
Figure 2. MCC cell lines do not express MICA and MICB protein despite low levels of MICA and MICB mRNA.
(a) MICA (light grey) and MICB (dark grey) mRNA expression was determined by qRT-PCR in MCC cells. Relative expression levels were calculated by normalization of CT values to RPLP0 and calibration to the melanoma cell line IF6. (b) MICB protein expression of whole cell lysates was determined by immunoblot; β-tubulin served as a loading control. (c) MICA/B cell surface expression was determined by flow cytometry using an antibody recognizing both MICA and MICB (clone 6D4; blue line); matched isotype control is depicted as grey filled area. Melanoma cell lines FM79, FM82 and IF6, served as positive control for MICA and MICB expression in all assays illustrated in this figure.
The complete lack of MICA and MICB protein expression in MCC cell lines even under diverse cell stress conditions suggests that MICA and MICB silencing in MCC is an active and robust process.
MICA and MICB promoters are silenced by histone hypo-acetylation
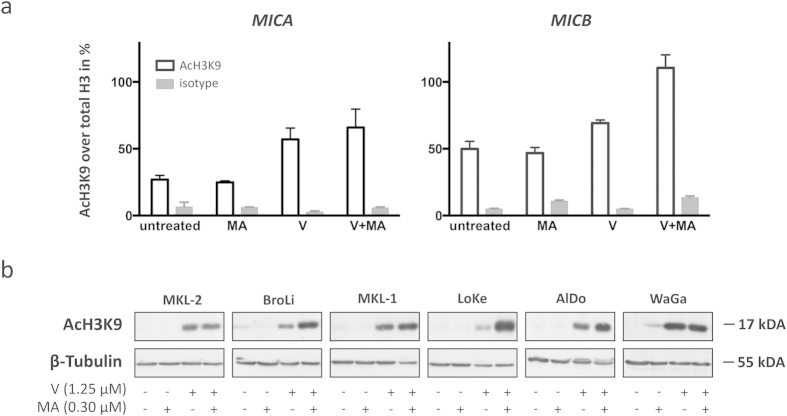
The robust silencing of MICA and MICB expression in MCC observed in vivo and in vitro suggests that this regulation takes place on a transcriptional or epigenetic level rather than post-transcriptionally27. We therefore determined histone H3K9 acetylation at the MICA and MICB promoter region of WaGa cells as a known indicator for transcriptional activity. Chromatin immune precipitation (ChIP) assays clearly demonstrated histone hypo-acetylation at both the MICA and MICB promoter: Only ~20% of histones in the MICA promoter and ~50% of histones in the MICB promoter were acetylated in WaGa cells (Fig. 3a). The lower acetylation levels of histone H3 at the MICA promoter compared to the MICB promoter in WaGa cells is in accordance with a lower expression of MICA compared to MICB mRNA as described above.
Figure 3. Vorinostat alone or in combination with mithramycin A increases global as well as MICA and MICB promoter-specific histone H3 Lysine 9 (H3K9) acetylation in MCC cell lines.
(a) Chromatin immunoprecipitation (ChIP) assay was performed with untreated, vorinostat (V), mithramycin A (MA), or the combination thereof (V+MA) treated WaGa cells followed by a qRT-PCR using MICA or MICB promoter specific primers. CT values of anti-acetyl-H3K9 (AcH3K9) antibody or rabbit IgG isotype control precipitated DNA were normalized to total histone H3 antibody as described in materials and methods. White bars represent the percentage of acetylated H3K9, grey bars the respective control. Experiments were performed in duplicates and results are expressed as mean ± SEM. (b) Global H3K9 acetylation of untreated, V, MA, or V+MA treated MCC cell lines was determined by immunoblot with the same AcH3K9 antibody used in the ChIP assay; β-tubulin served as loading control.
MICA and MICB promoter acetylation is inducible
Apparently, the expression of MICA and MICB in MCC cell lines is silenced via histone H3 hypo-acetylation at their promoter region. To test whether the silencing of these NKG2D ligands is indeed due to histone hypo-acetylation or if additional mechanisms for MICA and MICB suppression are operative14, we next tested if their expression could be induced by reversal of histone hypo-acetylation. WaGa cells were subjected to a clinically relevant concentration of 1.25 μM (the plasma concentration reached by currently applied treatment regimens) of the histone deacetylase (HDAC) inhibitor vorinostat (SAHA, Zolinza)28. When WaGa cells were cultured for 24 hours in the presence of vorinostat, there was an induction of histone acetylation at the MICA and MICB promoter regions (Fig. 3a). However, with ~56% of histone acetylation in the MICA and ~70% of histone acetylation in the MICB promoter, this induction was rather modest. We therefore combined vorinostat with mithramycin A, a drug that has synergistic effects with HDAC inhibitors by (i) transcriptionally inhibiting the compensatory inductions of certain HDACs29 and (ii) by preventing the formation of SP1/HDAC inhibitory complexes at the promoters’ GC box30. We confirmed these synergistic effects in the MCC cell lines: Treatment with mithramycin A alone already reduced transcription of most class I and II HDAC genes in MCC cell lines and, most importantly, mithramycin A prevented the regulatory induction of HDACs by vorinostat (Supplementary Fig. 6). Subsequently, ChIP assays of accordingly treated MCC cells revealed that the addition of mithramycin A boosted vorinostat induced histone acetylation resulting in an acetylation of ~66% of histones at the MICA promoter and almost complete histone acetylation at the MICB promoter. Treatment with mithramycin A alone had negligible effects on histone acetylation levels in the MICA and MICB promoter of WaGa cells.
To extend this observation to a larger series of MCC cell lines, we performed an immunoblot for global histone acetylation using the same anti-AcH3K9 antibody we employed for the ChIP assay. This analysis confirmed the increased acetylation of global histones upon combined vorinostat and mithramycin A treatment in 4 out of 6 cell lines (Fig. 3b). In MKL-2 and in WaGa cells we did not observe a further increase in global histone H3 acetylation by adding mithramycin A. This observation was unanticipated, since we observed a synergistic effect of the combined drugs on histone H3 acetylation at the MICA and MICB promoter regions of WaGa cells (Fig. 3a). A possible explanation is that both the MICA and MICB promoter regions may be more sensitive to mithramycin A induced histone acetylation due to the presence of a SP1 binding site (GC Box)30.
MICA/B expression can be re-induced on MCC cells in vitro
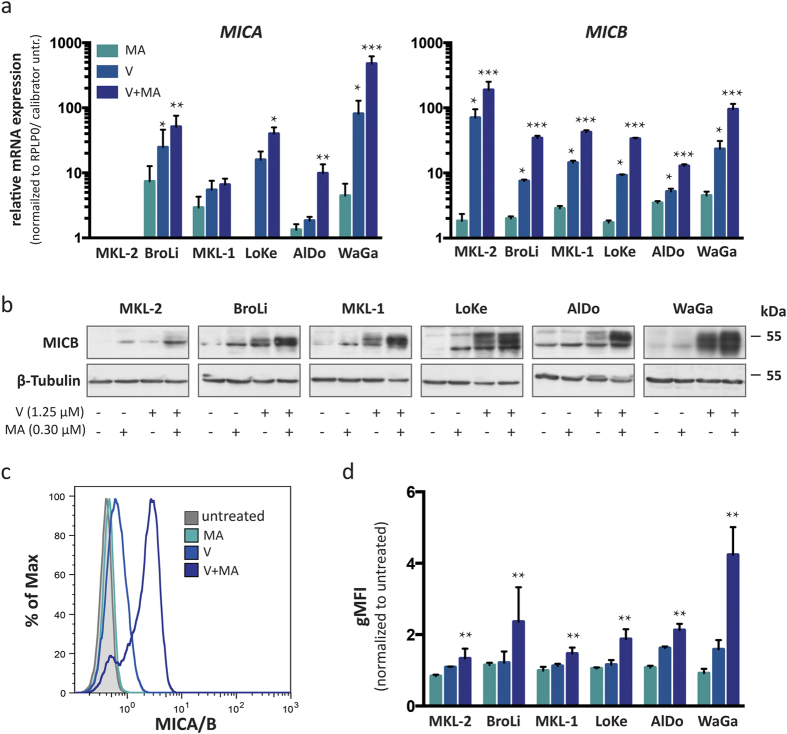
Since the combined treatment of MCC cells with vorinostat and mithramycin A induced strong histone acetylation at the MICA and MICB promoter, we next tested whether this also leads to an increased transcription of MICA and MICB genes. First, we determined the respective mRNA expression with or without treatment by quantitative RT-PCR (Fig. 4a). The combination of vorinostat and mithramycin A led to increased MICA and MICB mRNA expression in all tested MCC cell lines, with the exception of MKL-2 in which no MICA mRNA was detected. Notably, the synergistic effects of vorinostat and mithramycin A were so strong that the relative expression to the respective untreated cell line in Fig. 4a is depicted on a logarithmic (log10) scale. Next, immunoblot and flow cytometry assays confirmed that the increased mRNA expression translated into MICB protein expression in general (Fig. 4b) and more importantly membrane expression (Fig. 4c,d). The degree of induction of membrane bound MICA/B was comparable to that of total MICB protein expression, suggesting that most of the induced MICA/B proteins are transported to the cell surface and no additional post-translational mechanisms interfere with MICA/B surface expression in MCC cells. It should be noted, however, that the combined treatment did not affect all cells of the individual cell lines to the same extent. For a small but distinct subpopulation encompassing between 10 to 15% of the total population, MICA/B surface expression was almost unaltered by this treatment (Fig. 4c).
Figure 4. Induction of MICA and MICB expression by vorinostat alone or in combination with mithramycin A.
MICA and MICB mRNA and protein expression of untreated MCC cell lines were compared to the respective expression after treatment with vorinostat (V, light blue), mithramycin A (MA, turquoise), or the combination thereof (V+MA, dark blue). (a) mRNA expression of MICA and MICB was determined by qRT-PCR in duplicates in three independent experiments; CT values were normalized to RPLP0 and calibrated to the ΔCT value of the respective untreated cell line; relative mRNA expression is depicted on a logarithmic scale (log 10) ± SEM. (b) MICB expression in whole cell lysates of MCC cell lines was detected by immunoblot using a MICB specific antibody; β-tubulin served as loading control. (c,d) MICA/B cell surface expression was determined by flow cytometry using an antibody recognizing both MICA and MICB (clone 6D4), which is exemplified for WaGa (c); the results for all cell lines are depicted as the geometric mean fluorescence intensity (gMFI) of MICA/B staining, normalized to the respective untreated cell lines ± SEM in three independent experiments (d). Statistical analysis was performed using the Friedman test as indicated.
When we subjected MCC cells to increasing concentrations of vorinostat starting at 1.25 μM used throughout the previous experiments to 10 μM we observed that vorinostat alone is capable of inducing strong MICA/B surface expression in the majority of AlDo, BroLi and WaGa cells if present at high concentrations, whereas in LoKe, MKL-1 and MKL-2 cells the magnitude of vorinostat inducible MICA/B expression is lower and the plateau of expression is reached already at intermediate concentrations (Supplementary Fig. 7). This observation together with the observation that a subpopulation of cells did not respond to the synergistic effect of mithramycin A suggests that histone hypo-acetylation at the MICA and MICB promoter is maintained by different mechanisms depending on both the cell line per se as well as the functional state of the cell.
An alternative HDAC inhibitor, i.e. the “classical” HDAC inhibitor trichostatin A (TSA), alone or in combination with mithramycin A produced essentially the same results as observed with the “second-generation” HDAC inhibitor vorinostat suggesting that only class I and II HDACs are involved in silencing MICA and MICB (Supplementary Fig. 8).
Induction of MICA/B enhances LAK cell mediated lysis of MCC cells
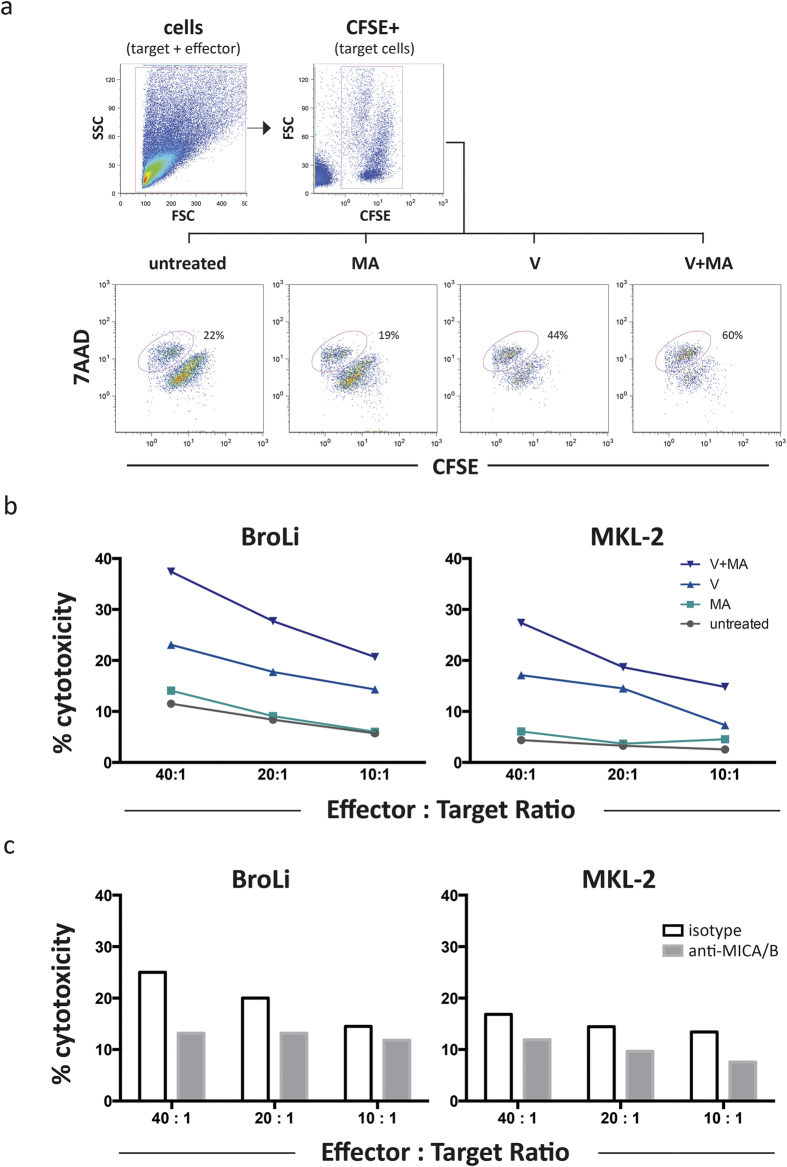
The expression of NKG2D ligands such as MICA and MICB render tumor cells more susceptible to being killed by NK and T cells. Notably, T-cell activation can be mediated by NKG2D even without contribution of TCR-recognition31. Thus, we addressed whether induction of MICA and MICB on the surface of MCC cells results in increased killing by cytotoxic cells. The circumstance that MCC cell lines grow in spheroids necessitated the use of a flow cytometry based cytotoxic assay. Lymphokine- activated killer (LAK) cells were chosen as effector cells because they are a clinically applicable heterogeneous population of NKG2D+, interleukin 2 (IL-2) activated NK, NKT and T cells32 and we are currently conducting a clinical trial based on the antibody targeted delivery of IL-2 to the MCC tumor microenvironment (www.immomec.com). The gating strategy to differentiate dead from living cells is illustrated in Fig. 5a for untreated or treated MCC cells (BroLi) at an effector to target ratio of 40:1. In line with the magnitude of the induced membrane expression of MICA/B, the highest LAK cell mediated cytotoxicity was observed for MCC cells treated with the combination of vorinostat and mithramycin A, whereas mithramycin A alone had no and vorinostat alone only an intermediate effect (Fig. 5b). Notably, the LAK cell mediated cytotoxicity correlated with the surface expression of MICA/B. A blocking experiment using saturating amounts of an anti-MICA/B antibody confirmed that increased lysis of MCC cells was indeed dependent on the induction of MICA/B molecules (Fig. 5c). Hence, vorinostat and mithramycin A mediated induction of MICA/B molecules is responsible for the augmented sensitivity of treated MCC cells towards LAK cell mediated lysis.
Figure 5. Inhibition of HDACs in MCC cell lines increased their susceptibility to LAK cell mediated lysis, which is is subdued by MICA/B blockade.
The flow cytometry based cytotoxicity assay was performed as described in Material and Methods. (a) The gating strategy is illustrated for untreated and treated BroLi cells used at an effector to target ratio of 40:1; target cells were gated as CFSE positive cells in an FSC/CFSE plot, lysed target cells were defined as 7AAD/CFSE double positive cells and are quantified as percentage of all target cells. (b) Untreated (grey), vorinostat (V, light blue), mithramycin A (MA, turquoise), or the combination thereof (V+MA, dark blue) treated BroLi and MKL-2 cells served as target cells for LAK cells at the indicated effector to target ratios in a 4h cytotoxicity assay. The lysis of the respective target is given as average of three independent experiments. (c) Vorinostat plus mithramycin A treated BroLi and MKL-2 cells served as target cells for LAK cells at the indicated effector to target ratios in a 4h cytotoxicity assay in the presence of saturating amounts of a MICA/B specific blocking antibody (grey bars) or an isotype control antibody (white bars); Fc receptors of effector cells were blocked by saturating amounts of F(ab)2 fragments.
MICB expression can be re-induced on MCC cells in vivo
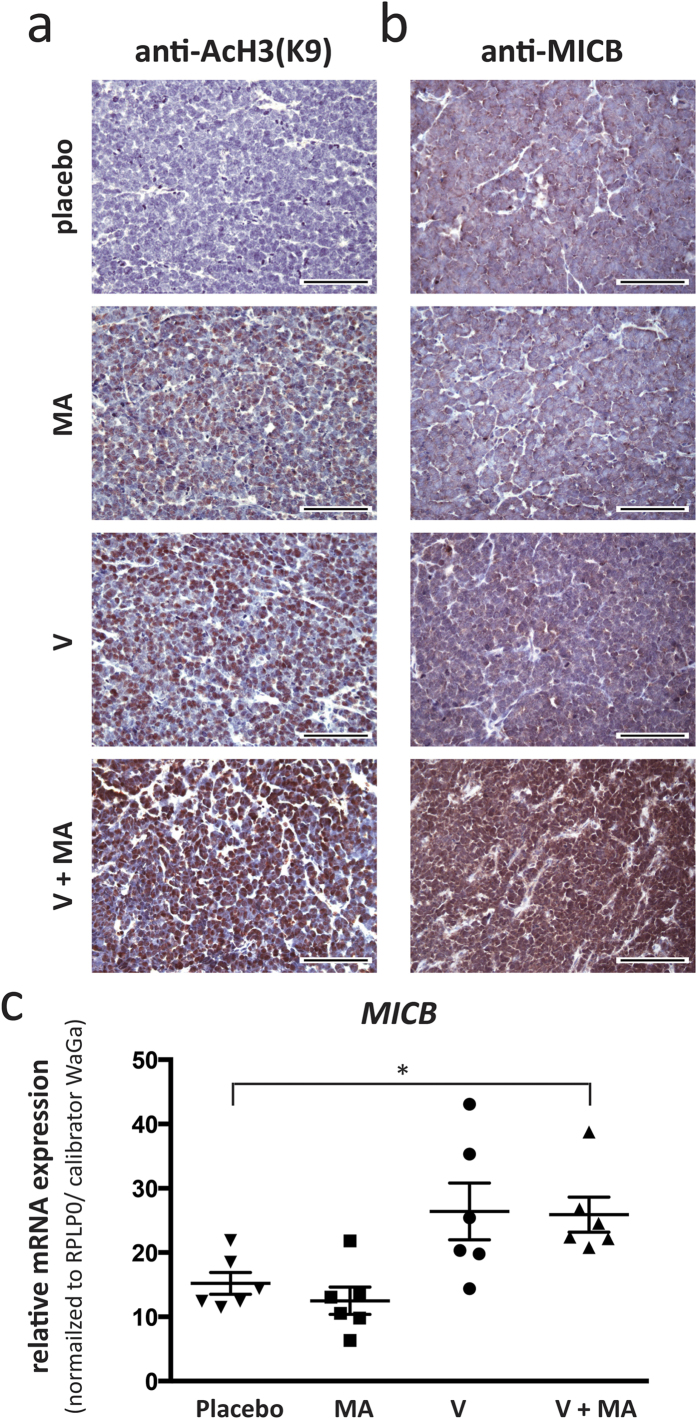
To translate these in vitro observations into a preclinical in vivo setting, we took advantage of a recently established MCC xenotransplantation model in which MCC tumors are induced by subcutaneous injection of WaGa cells in NOD/SCID mice33. Once tumors reached a volume of approximately 100 mm3, mice were treated for two weeks by intraperitoneal injections of placebo, vorinostat, mithramycin A or the combination thereof at concentrations resulting in serum levels corresponding to those in the clinical setting in humans. After the last dosage animals were sacrificed, and the xenotransplants subjected to detailed characterization. By means of immunohistochemistry, we confirmed both the induction of histone H3 acetylation as well as MICB protein expression (Fig. 6a). In xenotransplanted tumors of mice treated with placebo, histones were hypo-acetylated and inhibition of HDACs induced histone H3K9 acetylation in vivo. A substantial induction of histone acetylation, however, was already achieved by vorinostat or mithramycin A alone; still this induced histone acetylation was further enhanced by the combined therapy. The more pronounced effects of the single agents may be due to either the prolonged exposure of either drug in the in vivo experiments (14 days with 10 days of drug administration versus 24h) and/or by other mechanisms such as inflammatory responses, interaction of MCC cells with components of the microenvironment or hypoxia. In accordance with the higher level of histone acetylation in MCC xenotransplants, MICB expression was already present in tumors of untreated mice. Still, the induced histone H3K9 acetylation also resulted in an increased expression of MICB protein (Fig. 6b). Quantification of MICB mRNA expression clearly demonstrated the synergism of the drug combination (p < 0.05) (Fig. 6c); however, it was not as evident as in the in vitro experiments.
Figure 6. Vorinostat and mithramycin A treatment induces histone H3K9 acetylation and MICB expression in MCC cells in vivo.
NOD.CB17/Prkdcscid mice (n = 6 for each treatment group) bearing subcutaneous xenotransplants of WaGa cells were treated with placebo, vorinostat (V) mithramycin A (MA), or the combination thereof (V+MA) as described in materials and methods. Immunohistochemistry on FFPE fixed tumor samples obtained after two weeks of treatment was performed using antibodies specific against AcH3K9 (a) or MICB (b). Representative examples are depicted at 40× magnification, scale bar is 100 μm. (c) mRNA was isolated from cryopreserved tumors and qRT-PCR was performed using primers specific for MICB. CT values were normalized to RPLP0 and calibrated to in vitro cultured WaGa cells. Statistical analysis was performed using the Kruskal-Wallis test.
Discussion
Virally associated cancers are characterized by a pronounced immunogenicity. To this end, the higher prevalence of Merkel cell carcinoma (MCC) in immune compromised patients, the high rate of spontaneous regression and the improved prognosis for patients whose tumor is infiltrated by T lymphocytes prompted the discovery of the Merkel cell polyomavirus (MCPyV). It has been shown that most MCC patients mount specific T-cell responses against epitopes derived from proteins encoded by the transforming MCPyV early genes10,11, which are persistently expressed in MCC lesions in patients and are required for ongoing growth of MCC cell lines7,8. Despite this immunogenicity, MCC is a very aggressive cancer causing disease-specific death in more than 40% of the patients after primary diagnosis and of almost all patients once distant metastases occur3. It should be further noted, that more than 90% of MCCs arise in fully immune competent patients12, suggesting tumor specific immune escape mechanisms must be employed. The two main pathways that allow tumors to escape the immune system are loss of immunogenic determinants and the tumor-driven suppression or desensitization of the immune response. Here, we present strong evidence that epigenetic silencing of the stress induced non-classical MHC class I molecules MICA and MICB is one of the major immune escape mechanisms of MCC. These molecules are ligands of the immune regulatory receptor NKG2D expressed on a variety of cytotoxic effector cells of both the innate and adaptive immune system and, most importantly, the NKG2D-NKG2D ligand system has been identified as being essential for the immune surveillance of cancer41.
MICA and MICB are not or only slightly expressed by MCC tumors in situ and completely absent on MCC cell lines in vitro. We demonstrate that the lack of MICA and MICB mRNA and protein expression in MCC is largely due to epigenetic silencing via histone hypo-acetylation in their promoter region. This epigenetic silencing is very robust even in the presence of several well-established stress factors known to induce their expression. However, this silencing can be abrogated by treatment with HDAC inhibitors both in vitro and in vivo. Since the ultimate goal of our studies was to establish a therapeutic approach for MCC, we used a clinically relevant concentration, i.e. concentrations attained in patients treated with the FDA approved HDAC inhibitor vorinostat (SAHA, ZolinzaTM)28. Although this concentration of vorinostat increased histone acetylation at the MICA and MICB promoter as well as subsequent mRNA and protein surface expression in MCC cell lines, the effects were not very strong. This outcome was not entirely surprising, given the fact that HDAC inhibitors are very potent treating hematological malignancies, but have so far failed to achieve significant effects as mono-therapy for solid tumors in vivo34. Classical MCC cell lines grow as 3D-cultures in large spheroids and therefore represent the in vivo situation of a solid tumor much closer than other cancer cell lines35. To increase the susceptibility of cancers to HDAC inhibitors, they are frequently combined with other drugs36. Mithramycin A is a gene selective Sp1 inhibitor, which has been reported to potentiate HDAC inhibitor induced transcriptional activation37. To this end, promoter acetylation of MICA and MICB genes and subsequent mRNA and protein expression were markedly enhanced in MCC cells upon this combined treatment. This, however, is not in line with observations in other cell types where Sp1 appears to be necessary for MICA and MICB transcription and thus mithramycin A inhibits their expression38,39. Treatment of MCC cell lines with vorinostat or mithramycin A markedly reduced (and the combination almost fully eliminated) Sp1 protein expression. This finding was in line with our observation of decreased Sp1 binding at the MICA and MICB promoter (Supplementary Fig. 9). Thus, we assume that in MCC cells, Sp1 is not necessary (or when in a complex with certain HDAC molecules even inhibitory) for the transcription of MICA and MICB. This altered Sp1 function may be caused by the presence of MCPyV. Likewise, Venkataraman et al.38 reported that in cytomegalovirus-infected cells, unlike in non-infected cells, MICA and MICB expression was independent of Sp1.
In a small sub-population of MCC cells, induction of MICA/B surface expression by treatment with HDAC inhibitors in combination with mithramycin A was not as pronounced as in the main population (Fig. 4c and Supplementary Fig. 8a). These cells may exploit resistance mechanisms such as increased drug efflux or represent a slow cycling subpopulation. Since we could detect this subpopulation only by flow cytometry analysis for MICA/B surface expression, other mechanisms to counteract MICA and MICB surface expression, e.g. cytoplasmic retention or shedding from the cell surface, may be operative14,20.
The principal effect of NKG2D signaling is an enhanced cytotoxic activity of lymphocytes towards the NKG2D ligand-expressing cells. In line with this notion, we observed that HDAC inhibitor treatment of MCC cells resulted in an increased susceptibility to LAK cell-mediated killing. Importantly, this increased cytotoxicity was decreased by a MICA/B blocking antibody. However, this blockade only partially diminished LAK cell mediated cytotoxicity, suggesting that besides MICA and MICB, other immunogenic molecules are induced by the combined treatment with vorinostat and mithramycin A. Indeed, HDAC inhibitors are known to induce the expression of other NKG2D ligands such as ULBPs40. It should be further noted that LAK cells include several possible effector cells. NKG2D is expressed as an activating or co-activating receptor not only on NK cells and CD8+ T cells, but also on γδ T cells which are important for surveillance of virally associated cancers19,41. In murine models, γδ T cells are strongly protective against polyomavirus induced tumors; this protection critically depends on their activation via NKG2D42.
We recently reported that MCPyV-specific CD8+ T cells are present in peripheral blood of more than half of MCC patients10, and that intra-tumoral infiltration of CD8+ lymphocytes is a positive prognostic marker for these patients15. Unfortunately, MCPyV-reactive CD8+ T cells are not fully functional in most MCC patients43. This exhausted phenotype was associated with expression of PD-1 on the MCPyV-reactive T cells; notably, PD-L1 expression has been reported for both MCC cells and myeloid cells infiltrating the tumor microenvironment10,22. Signaling via NKG2D may prevent the exhaustion of MCPyV-reactive CD8+ T cells. In addition to restoring preexisting T-cell responses, induction of NKG2D ligand expression on MCC cells is likely to trigger new T-cell responses. Activation of NK and γδ T cells via NKG2D increases tumor cell killing and thus cross-presentation of antigens, as well as production of chemokines and cytokines attracting and activating CD8+ T cells44,45. Furthermore, naïve CD8+ T cells express NKG2D as a co-activating receptor and binding to NKG2D ligands boosts their activation46. In preclinical models it is well established that NKG2D ligand over-expression on tumor cells results in an increased priming and activation of tumor-specific CD8+ T cells and long lasting T-cell memory responses even against NKG2D-negative tumor cells47: (i) Induction of NKG2D ligands on carcinoma cells boosts anti-tumor effects of CTLA-4 blockade48, and (ii) treatment with immune stimulating cytokines such as IL-2 and IL-12 is more effective against NKG2D ligand expressing tumors49.
Thus, HDAC inhibitor mediated MICA and MICB induction in MCC is likely to enhance the effects of immune therapeutic approaches currently tested in the clinic: (i) autologous MCPyV specific CD8+ T cell transfer (NCT01758458), (ii) CTLA-4 blocking antibody ipilimumab (NCT02196961), (iii) PD-L1 blocking antibody MSB0010718C (NCT02155647), or cytokine based therapies using (iv) tumor-stroma targeting antibody-IL2 fusion proteins (NCT02054884) or (v) IL12-encoding plasmids delivered by electroporation (NCT01440816).
A limitation of our study is the use of allogeneic LAK cells as effector cells in the cytotoxicity experiments. Unfortunately, autologous peripheral blood or tumor infiltrating lymphocytes from the same patients the respective MCC cell lines were derived from, are not available. LAK cells are a heterogeneous population of highly activated T, NK and NKT cells; hence, it was not possible to further scrutinize the detailed mechanisms by which HDAC inhibition in MCC cells boosts their susceptibility to immune recognition. Furthermore, we focused in this study on the transcription and membrane expression of only MICA and MICB. The expression of other NKG2D ligands is likely to be regulated by histone acetylation as well; however, the fact that the increased susceptibility of MCC cells after inhibition of histone acetylation to LAK cell mediated cytotoxicity is abrogated by an MICA/B blocking antibody strongly argues that induced MICA and MICB expression is the dominating effect.
Recently, many exciting developments have led to new, effective cancer immunotherapies50. Immune checkpoint blockade, cytokines with and without tumor targeting, as well as adoptive T cell transfer with and without chimeric antigen receptors results in objective, long lasting clinical responses with response rates, speed and depth even in advanced tumor stages51. However, a majority of patients still do not benefit from therapy. Predictive biomarkers for response to immunotherapy are immune response gene signatures or the presence of clonally expanded CD8+ T cells within the tumor52. Unfortunately, only 20% of the patients’ MCC lesions are characterized by such a favorable immune signature15. The lack of MICA and MICB expression of MCC cells is likely to contribute to this immunological state as re-induction of these NKG2D ligands by HDAC inhibition restores the susceptibility of MCC cells to cytotoxic lymphocytes. Thus, “epigenetic priming” of cancer cells for immune recognition appears to be a valuable addition to current immune therapeutic interventions for MCC53.
Material and Methods
Patients
A total of 50 archived paraffin-embedded tumor samples from 34 MCC patients were selected from the Department of Dermatology, Medical University of Graz. All tumor samples were histologically confirmed MCC lesions, i.e. primary tumors, local recurrences as well as skin and nodal metastasis. Utilization of the tumor specimens for this study was approved by the institutional review board of the Medical University of Graz (24-295 ex 11/12) and the methods were carried out in accordance with the approved guidelines.
Immunohistochemistry (IHC)
IHC was performed on formalin fixed and paraffin embedded (FFPE) tissue using the Autostainer Link 48 (Dako, Glostrup, Denmark). After deparaffinization in xylene, sections were rehydrated with 100%, 96%, 70%, and 50% ethanol for 5 min each and finally rinsed with demineralized water. Antigen retrieval for the anti-MICA antibody was performed with EDTA (1mM EDTA, 0.05% Tween 20, pH 8.0) and for the anti-MICB antibody with citrate (Dako retrival solution, cat. no. S1699, pH 6) in a steamer at 100 °C for 30 minutes. After cooling for 20 minutes and two additional washing steps, sections were blocked with peroxidase blocking solution (Dako) followed by incubation over night at 4 °C with antibodies to MICA (AF130, R&D Systems, MN, USA) or MICB (orb 1241, Biorbyte, Cambridge, UK) diluted in antibody diluent (Dako) to 1:200 or 1:100, respectively. After washing steps, incubation with a biotinylated secondary antibody, further washing steps, addition of streptavidin peroxidase, detection was obtained using ImmPACT NovaRED Peroxidase Substrate (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions. After counterstaining of nuclei with haematoxylin (Dako), sections were dehydrated and mounted in Tissue Tek glass mounting medium (Sakura Finetek, Torrance, CA, USA). Three independent investigators (CR, DS, JCB) classified the tumors as positive or negative for expression of MICA and MICB.
Cell culture
The MCC cell lines WaGa, MKL-1, MKL-2, BroLi, AlDo, LoKe54,55 and melanoma cell lines FM79, FM82, IF656 have been described before. All cell lines were maintained in RPMI-1640 (PAN Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO, USA) and 1% penicillin/streptomycin (Biochrome, Berlin, Germany). For the cell line AlDo the medium was additionally supplemented with 30% fibroblast conditioned medium. For treatment with specific inhibitors, cells were cultured at a concentration of 1 × 106 cells/ml in 6 well plates. Inhibitors were dissolved according to the manufacturers’ guidelines and used at 1.25 μM vorinostat (Selleckchem, Munich, Germany), 0.3 μM mithramycin A (Sigma) and 0.3 μM trichostatin A (Selleckchem) for 24 hours if not otherwise stated.
Quantitative real time-PCR
RNA of in vitro propagated cells or cryopreserved xenotransplants was isolated using PeqGOLD total RNA Kit (Peqlab, Erlangen, Germany) and transcribed into cDNA with the Transcriptor First Strand cDNA Synthesis Kit (Roche Life Science, Indianapolis, IN, USA) according to the manufacturer’s instructions. Quantitative real time polymerase chain reactions (qRT-PCR) were performed using SYBR green or TaqMan PCR master mix (Sigma) on the StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). RPLP0 was used as endogenous control, and detected with the sense-primer: CCA TCA GCA CCA CAG CCT TA, the antisense-primer: GGC GAC CTG GAA GTC CAA CT, and the probe ATC TGC TGC ATC TGC TTG GAG CCC A. MICA and MICB mRNA was detected using the SYBR green primers: MICA/B-sense: CAC CTG CTA CAT GGA ACA CAG C, MICA-antisense: TAT GGA AAG TCT GTC CGT TGA CTC T, and MICB-antisense: ACA TGG AAT GTC TGC CAA TGA TC. Relative quantification was calculated by the ΔΔCT method using the melanoma cell line IF6 as calibrator.
Immunoblotting
Cell lysates were generated by lysing 3 × 106 cells per sample in protein extraction buffer supplemented with a proteinase inhibitor cocktail as described before57. Lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-Page), samples were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA), blocked for 1 hour in a blocking buffer according to the respective antibody’s data sheet and then incubated overnight at 4 °C with primary antibodies diluted in phosphate buffered saline (PBS) with 0.1% Tween 20 (PBST) or tris buffered saline with 0.1% Tween-20 (TBST) according to data sheet: MICB (R&D Systems, Minneapolis, MN, USA) 1:1000 in PBST, acetyl-histone H3 (Lys9) (Cell Signaling Technology, Danvers, MA, USA) 1:1000 in TBST, Sp1 (Cell Signaling) 1:1000 in TBST or β-tubulin (Sigma) 1:1000 in PBST. After membranes were washed trice for 10 minutes each in the respective buffer, they were incubated for 1h with the appropriate peroxidase-coupled secondary antibodies (Dako), followed visualization using the ECL Western Blotting Substrate (Pierce, Rockford, IL, USA).
Flow Cytometry
Cell surface expression of MICA and MICB was determined by flow cytometry. 1 × 106 cells were washed with ice cold PBS and incubated with the PE-conjugated anti-human MICA/B antibody (6D4; Biolegend, San Diego, CA, USA) in PBS with 0.1% bovine serum albumin (BSA) for 90 minutes at 4 °C in the dark. After washing steps, cells were stained with 10 μg/ml 7-aminoactinomycin (7AAD, Sigma) to exclude non-viable and measured on a FC500 Flow Cytometer (Beckman Coulter, Brea, CA, USA). Flow cytometry data were analyzed with FlowJo Version 8.7 software (TreeStar, Sunnyvale, CA, USA).
Chromatin immune precipitation (ChIP)
ChIP assays were performed using the SimpleChiP® Enzymatic Chromatin IP Kit with Agarose beads (Cell Signaling). In brief, proteins were cross-linked to DNA with 1.5% formaldehyde for 10 minutes. Nuclear membranes were broken up using the UP50H Sonicator (Hielscher, Teltow, Germany) set to 100%, 0.9 output for 20 seconds, 6 times in a row with incubation on ice for 30 seconds between sonication pulses. Afterwards, antibodies against histone H3 (cat. no. 6420), acetyl-histone H3 (Lys9) (AcH3K9) (cat. no. 9671), or normal rabbit IgG (Cell Signaling) were used for immune precipitation. The immune precipitated DNA was subsequently analyzed by qRT-PCR, using SYBR green primers specific to the MICA or MICB promoter region: MICA promoter sense CGG ATC CTG GAA TAC GTG GG, antisense ACT CAC ACC TGC CCG TTA TG; MICB promoter sense GCG ACA GGG TCC AGG TCG TGC TC, antisense CCC TAC GTC GCC ACC TTC TCA GCT. The percentage of acetylated histones (AcH3K9) was normalized to total Histones H3 and calculated using following equation:
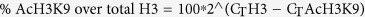 |
Flow cytometry based cytotoxicity assay
Peripheral blood mononuclear cells (PBMCs) where isolated via gradient centrifugation with LymphoprepTM (Stemcell Technologies, Vancouver, BC, Canada), and cultured for 3 days in CellGro®SCGM (CellGenix, Freiburg, Germany) supplemented with 10% FBS (Invitrogen, Grand Island, NY, USA) and 500 IU interleukin 2 (IL-2) per ml (Miltenyi Biotec, Bergisch Gladbach, Germany) to generate lymphokine-activated killer (LAK) cells. MCC cell lines served as target cells either without or with inhibitor (Vorinostat, mithramycin A, or combination thereof) treatment for 12 hours at the concentrations described above; the shortened incubation time was chosen to assure that cells are indeed vital and target cell membranes are fully intact for the cytotoxicity assay. After target cells were washed 3 times in RPMI with 10% FBS, they were labeled by incubation in 3 μM CFSE (Sigma) in pre-warmed RPMI with 10% FBS for 10 minutes at 37 °C, followed by another round of 3 washing steps to remove any excessive CFSE.
2 × 104 CFSE labeled target cells were incubated alone to establish spontaneous cell death, or co-incubated at varying effector:target ratios, i.e. 40:1, 20:1, 10:1, for 4h at 37 °C in a total volume of 100 μl. Before flow cytometry, cells incubated in 10 μg/ml 7AAD (Sigma). Dead target cells were defined as CFSE+/7AAD+, and the percentage of cytotoxicity was calculated as following:
 |
For blocking experiments target cells were incubated with saturating concentrations of blocking antibodies against MICA/B (clone 6D4; Biolegend) or isotype control for 2h at 37 °C. before incubation with LAK cells. Pre incubation with F(ab’)2 fragments for 30 minutes (Life Technologies) was performed to avoid Fc-receptor mediated antibody-dependent cell-mediated cytotoxicity.
Xenotransplantation experiments
Six-week-old female NOD.CB17/Prkdcscid mice were obtained from Charles River Laboratories, and housed under specific pathogen-free conditions. Tumors were induced by s.c. injection as described before30. Twenty-four days after tumor cell inoculation, when the tumors reached a volume of approximately 100 mm3, treatment was started. Mice were divided into four groups of six mice each ensuring an equal overall tumor burden. Immediately before injection a 1 M (264.3 mg/ml) vorinostat stock solution was diluted in 45% Polyethylene glycol (PEG-400, Sigma) to 10 mg/ml and 60 mg/kg bodyweight were administered i.p. per mouse. For mithramycin A, a 5 mg/ml stock solution was diluted in H2O to 0.33 mg/ml and 0.2 mg/kg bodyweight were injected i.p. per mouse. The placebo group received the same volume of the respective solvents. Mice were treated five consecutive days a week for two weeks. Afterwards tumor tissue was formalin fixed and paraffin embedded for IHC or cryo-preserved for RNA isolation. All animal studies had been approved by the Austrian ministry of education and science according to the regulations for animal experimentation (BMWF-66.010/0151-II/3b/2012).
Statistical Analyses
Statistical analyses were performed using Graphpad Prism 6.0 Software (Graphpad Software Inc., San Diego, CA, USA). Cell culture experiments were analyzed using Friedman test, a paired non-parametric ANOVA. The xenotransplantation experiments were analyzed using Kruskal-Wallis test, an unpaired non-parametric ANOVA. A p-value smaller than 0.05 was considered significant; the respective p-values are indicated in the figures as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
Additional Information
How to cite this article: Ritter, C. et al. Reversal of epigenetic silencing of MHC class I chain-related protein A and B improves immune recognition of Merkel cell carcinoma. Sci. Rep. 6, 21678; doi: 10.1038/srep21678 (2016).
Supplementary Material
Acknowledgments
We thank Gerlinde Mayer and Ulrike Schmidbauer for assistance with immunohistochemistry; Lorenzo Cerroni and Isabella Fried for their help analyzing the histological slides; Ulrike Fackelmann for assistance with the animal experiment and Roland Houben for providing us with the inducible MCPyV T-antigen knockdown cell lines. This Study was supported by FP7 grant IMMOMEC provided by the European Commission.
Footnotes
Author Contributions C.R., D.S. and J.C.B. designed the study, wrote the manuscript and evaluated the immunohistochemistry stainings; C.R. and K.F. performed the experiments and analyzed the data. K.G.P. and P.N. provided tissue microarrays, cDNA array and clinical data. All authors reviewed the manuscript.
References
- Becker J. C. & zur Hausen A. Cells of origin in skin cancer. J Invest Dermatol 134, 2491–2493 (2014). [DOI] [PubMed] [Google Scholar]
- Lemos B. D. et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. J Am Acad Dermatol 63, 751–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson N. C. Merkel cell carcinoma: changing incidence trends. J Surg Oncol. 89, 1–4 (2004). [DOI] [PubMed] [Google Scholar]
- Arora R., Chang Y. & Moore P. S. MCV and Merkel cell carcinoma: a molecular success story. Cure Opin Virol 2, 489–498 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P. S. & Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer 10, 878–889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J. A. & Garcea R. L. A cornucopia of human polyomaviruses. Nat Rev Microbiol 11, 264–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R. et al. Merkel Cell Polyomavirus-Infected Merkel Cell Carcinoma Cells Require Expression of Viral T Antigens Merkel Cell Polyomavirus-Infected Merkel Cell Carcinoma Cells Require Expression of Viral T Antigens. J Virol 84, 7064–7072 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M., Kwun H. J., Feng H., Chang Y. & Moore P. S. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest 121, 3623–3634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugamata A., Goya K. & Yoshizawa N. A case of complete spontaneous regression of extremely advanced Merkel cell carcinoma. J Surg Case Rep 10, doi: 10.1093/jscr/2011.10.7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afanasiev O. K. et al. Merkel polyomavirus-specific T cells fluctuate with merkel cell carcinoma burden and express therapeutically targetable PD-1 and Tim-3 exhaustion markers. Clin Cancer Res 19, 5351–5360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngaa R. et al. T-cell Responses to Oncogenic Merkel Cell Polyomavirus Proteins Distinguish Patients with Merkel Cell Carcinoma from Healthy Donors. Clin Cancer Res 20, 1768–1778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M. et al. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol 58, 375–381 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S., Afanasiev O. & Nghiem P. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Curr Oncol Rep 13, 1420–1421 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet D. H., Gasser S., Gowen B. G., Deng W. & Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol 31, 413–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson K. G. et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol 29, 1539–1546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Messina L., Reyburn H. T. & Vales-Gomez M. Human NKG2D-ligands: cell biology strategies to ensure immune recognition. Front Immunol 3, 299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci USA 96, 6879–6884 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter C. S. et al. Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol 118, 600–605 (2002). [DOI] [PubMed] [Google Scholar]
- Bauer S. Activation of NK Cells and T Cells by NKG2D, a Receptor for Stress-Inducible MICA. Science 285, 727–729 (1999). [DOI] [PubMed] [Google Scholar]
- Marcus A. et al. Recognition of Tumors by the Innate Immune System and Natural Killer Cells. Advances in Immunology. Adv Immunol 122, 91–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms P. W. et al. Distinct Gene Expression Profiles of Viral- and Nonviral-Associated Merkel Cell Carcinoma Revealed by Transcriptome Analysis. J Invest Dermatol 133, 936–945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson E. J. et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: Association with inflammation, Merkel cell polyomavirus and overall survival. Cancer Immunol Res 1, 54–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V., Steinle A., Bauer S. & Spies T. Recognition of Stress-Induced MHC Molecules by Intestinal Epithelial T Cells. Science 279, 1737–40 (1998). [DOI] [PubMed] [Google Scholar]
- Groh V. et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA 93, 12445–12450 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol 9, 1065–1073 (2008) [DOI] [PubMed] [Google Scholar]
- Hong J., Shao T., Sun X., Li G. & Xu J. Interferon gamma up-regulates major-histocompatibility-complex class I-related chain A expression and enhances major-histocompatibility-complex class I-related chain A-mediated cytolysis of human corneal epithelium by natural killer cells in vitro. J Interferon Cytokine Res 32, 115–120 (2012). [DOI] [PubMed] [Google Scholar]
- Steffen P. A. & Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nature 15, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- Mann B. S., Johnson J. R., Cohen M. H., Justice R. & Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 12, 1247–1252 (2007). [DOI] [PubMed] [Google Scholar]
- Sleiman S. F. et al. Histone Deacetylase Inhibitors and Mithramycin A Impact a Similar Neuroprotective Pathway at a Crossroad between Cancer and Neurodegeneration Sama. Pharmaceuticals 4, 1183–1195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. & Davie J. R. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat 192, 275–283 (2010). [DOI] [PubMed] [Google Scholar]
- Meresse B. et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21, 357–366 (2004). [DOI] [PubMed] [Google Scholar]
- West E. J., Scott K. J., Jennings V. A. & Melcher A. A. Immune activation by combination human lymphokine-activated killer and dendritic cell therapy. Br J Cancer 105, 787–795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmes C. et al. Type I and II IFNs inhibit Merkel cell carcinoma via modulation of the Merkel cell polyomavirus T antigens. Cancer Res 72, 2120–2128 (2012). [DOI] [PubMed] [Google Scholar]
- Qiu T. et al. Effects of treatment with histone deacetylase inhibitors in solid tumors: a review based on 30 clinical trials. Future Oncol 9, 255–269 (2013). [DOI] [PubMed] [Google Scholar]
- Friedrich J., Seidel C., Ebner R. & Kunz-Schughart L. A. Spheroid-based drug screen: considerations and practical approach. Nat Protoc 4, 309–324 (2009). [DOI] [PubMed] [Google Scholar]
- Dokmanovic M., Clarke C. & Marks P. A. Histone Deacetylase Inhibitors: Overview and Perspectives. Mol Cancer Res 5, 981–989 (2007). [DOI] [PubMed] [Google Scholar]
- Silva G., Cardoso B. A., Belo H. E. L. & Almeida A. O. N. M. Vorinostat induces apoptosis and differentiation in myeloid malignancies: genetic and molecular mechanisms. Plos One 8, e53766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman G. M., Suciu D., Groh V., Boss J. M. & Spies T. Promoter region architecture and transcriptional regulation of the genes for the MHC class I-related chain A and B ligands of NKG2D. J Immunol 178, 961–969 (2007). [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rodero S. et al. Transcriptional regulation of MICA and MICB: a novel polymorphism in MICB promoter alters transcriptional regulation by Sp1. Eur J Immunol 37, 1938–1953 (2007). [DOI] [PubMed] [Google Scholar]
- López-Soto A., Folgueras A. R., Seto E. & Gonzalez S. HDAC3 represses the expression of NKG2D ligands ULBPs in epithelial tumour cells: potential implications for the immunosurveillance of cancer. Oncogene 28, 2370–2382 (2009). [DOI] [PubMed] [Google Scholar]
- Raulet D. H. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol 3, 781–790 (2003). [DOI] [PubMed] [Google Scholar]
- Mishra R. et al. NK cells and gamma delta T cells mediate resistance to polyomavirus-induced tumors. Plos Pathog 6, e1000924 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowlatshahi M. et al. Tumor-specific T cells in human Merkel cell carcinomas: a possible role for Tregs and T-cell exhaustion in reducing T-cell responses. J Invest Dermatol 133, 1879–1889 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Chen Y., Xiao W., Sun R. & Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol 10, 230–252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.-L. et al. γδ T Cells and Their Potential for Immunotherapy. Int. J. Biol. Sci. 10, 119–135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maasho K., Opoku-Anane J., Marusina A. I., Coligan J. E. & Borrego F. Cutting Edge: NKG2D Is a Costimulatory Receptor for Human Naive CD8+ T Cells. J Immunol 174, 4480–4484 (2005). [DOI] [PubMed] [Google Scholar]
- Diefenbach A., Jensen E. R., Jamieson A. M. & Raulet D. H. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature 413, 165–171 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruocco M. G. et al. Suppressing T cell motility induced by anti–CTLA-4 monotherapy improves antitumor effects. J Clin Invest. 122, 3718–3730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth M. J. NKG2D Recognition and Perforin Effector Function Mediate Effective Cytokine Immunotherapy of Cancer. J Exp Med 200, 1325–1335 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin-Frankel J. Cancer Immunotherapy-Breakthrough of the year 2013. Science 342, 1432–3 (2013). [DOI] [PubMed] [Google Scholar]
- Wargo J. A., Cooper Z. A. & Flaherty K. T. Universes Collide: Combining Immunotherapy with Targeted Therapy for Cancer. Cancer Discov, doi: 10.1158/2159-8290.CD-14-0477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Galon J., Fuchs C. S. & Dranoff G. Cancer immunology-analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol 279, 1737–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. P. et al. Merkel Cell Carcinoma: Epidemiology, Target, and Therapy. Curr Derm Rep 3, 46–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R. et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer 130, 847–856 (2011). [DOI] [PubMed] [Google Scholar]
- Houben R. et al. Characterization of functional domains in the Merkel cell polyoma virus Large T antigen. Int J Cancer, doi: 10.1002/ijc.29200 (2014). [DOI] [PubMed] [Google Scholar]
- Kirkin A. F. et al. Generation of human-melanoma-specific T lymphocyte clones defining novel cytolytic targets with panels of newly established melanoma cell lines. Cancer Immunol Immunother 41, 71–81 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov S. et al. Cancer cells become susceptible to natural killer cell killing after exposure to histone deacetylase inhibitors due to glycogen synthase kinase-3-dependent expression of MHC class I-related chain A and B. Cancer Res 65, 11136–11145 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.