Abstract
Embryogenesis is an important component in the life cycle of most plant species. Due to the difficulty in embryo isolation, the global gene expression involved in plant embryogenesis, especially the early events following fertilization are largely unknown in radish. In this study, three cDNA libraries from ovules of radish before and after fertilization were sequenced using the Digital Gene Expression (DGE) tag profiling strategy. A total of 5,777 differentially expressed transcripts were detected based on pairwise comparison in the three libraries (0_DAP, 7_DAP and 15_DAP). Results from Gene Ontology (GO) and pathway enrichment analysis revealed that these differentially expressed genes (DEGs) were implicated in numerous life processes including embryo development and phytohormones biosynthesis. Notably, some genes encoding auxin response factor (ARF ), Leafy cotyledon1 (LEC1) and somatic embryogenesis receptor-like kinase (SERK ) known to be involved in radish embryogenesis were differentially expressed. The expression patterns of 30 genes including LEC1-2, AGL9, LRR, PKL and ARF8-1 were validated by qRT-PCR. Furthermore, the cooperation between miRNA and mRNA may play a pivotal role in the radish embryogenesis process. This is the first report on identification of DEGs profiles related to radish embryogenesis and seed development. These results could facilitate further dissection of the molecular mechanisms underlying embryogenesis and seed development in radish.
Although plant embryogenesis is usually studied in the context of seed development, there are many alternative routes to embryo initiation, including somatic embryogenesis, microspore embryogenesis and zygotic embryo initiation1. These modes of embryogenesis are a crucial developmental period in life cycle of flowering plants2,3. During embryogenesis, the single-celled zygote, the product of fertilization of female gamete with male gamete, follows a defined pattern of cell division and differentiation to the mature embryo4. The developmental events are highly conserved among diverse angiosperms, playing an important role in defining key aspects of seed formation and diversity5,6. Embryo development consisted of two distinct phases including early morphological events and late maturation7. Recent studies demonstrated that early embryogenesis could be considered as a miniature model of plant development based on hallmarks of several key embryonic developmental processes8,9. However, the molecular events related to plant early embryogenesis are largely unknown compared with other biological processes, which is partially due to poor accessibility and tiny size of embryos in many higher plants2.
The morphogenesis phase of embryogenesis is well described in model plants such as Arabidopsis thaliana10,11. In addition, embryogenesis-related genes have been identified in Arabidopsis2,3, Brassica napus12, Oryza sativa9, Gossypium hirsutum13 and Cucumis sativus14. For example, Tzafrir et al. identified 244 candidate EMB genes in Arabidopsis required for normal embryo development using embryo-defective mutants15. Among these EMB genes, the BABY BOOM (BBM) gene, a transcription factor of AP2/ERF family, was originally isolated as a marker for embryogenic cells in tissue culture16. Ectopic expression of BBM can induce spontaneous somatic embryos and cotyledon-like structure formation from the vegetative tissues of young seedlings17.
Similar to the phenotypes observed in the transgenic lines with overexpression of BBM, ectopic or spontaneous embryogenesis phenotypes have been reported in lines with overexpression of the LEAFY COTYLEDON1 (LEC1), LEC2, FUSCA3 (FUS3), ABA INSENSITIVE3 (ABI3) and WUSCHEL (WUS) genes in Arabidopsis12. Stone et al. reported that LEC1 and LEC2 are required genetically for the completion of normal seed development and may have complementary but partially redundant functions in embryo formation in Arabidopsis18, while ABI3 is involved in embryo maturation. For instance, it could promote transcription of LEC2, FUS3 and ABI3, as well as regulated transcription of a member of subfamily B-3 of the Apetala2/Ethylene Response Factor (AP2/ERF) transcription factors3,19. Moreover, it was reported that the Agamous-like15 (AGL15) was expressed in embryogenic cultures, which was further integrated into different signaling processes during embryogenesis20. Schmidt et al. originally identified somatic embryogenesis receptor-like kinases (SERKs)21, which expressed from the eight-celled stage through to the globular stage, while no transcript was detected in unfertilized flowers.
Radish, Raphanus sativus L. (2n = 2x = 18), is a major root vegetable crop of the Brassicaceae family. The thick roots and fresh sprouts are harvested as vegetables. While in North America, radish seeds are produced for oil production. Seeds contain the embryo that develops into an adult plant1,22. Thus, the embryogenesis is crucial for seed production, quality and yield of seed reproduction in elite seed breeding program for the root crops and the oilseeds in radish22. To enhance the selfing seeding rate, it is necessary to understand the molecular processes leading to the early embryonic process in radish. However, little is known about the genetic basis of radish embryogenesis.
The complex physiological and molecular processes during plant embryogenesis result from an intricate program of gene expression23. Thus, gene expression study can provide an entry point to analyze the plant embryogenesis processes. With the advent in sequencing technology, an ever increasing number of studies have been conducted to obtain systemic transcript profiles during plant embryogenesis using RNA-seq24. Nevertheless, there is no published study on identification of differentially expressed transcripts during radish embryogenesis. In this study, a comprehensive radish transcriptomes and microRNA (miRNA) expression profiles were obtained during embryogenesis and the early development of seed development using RNA-seq. Genes associated with transcriptional regulation, signaling pathways and metabolic pathways were over-represented among differentially expressed genes (DEGs) in radish embryogenesis and seed development. Based on an integrative analysis of embryogenesis related miRNAs and DEGs, a schematic model of radish embryogenesis was proposed.
Results
Properties of the sequenced cDNA libraries
To identify genes involved in radish embryogenesis, three cDNA libraries were constructed from tissues of ovules at 0, 4–7 and 10–15 day after pollination (DAP). A total of 7,191,808, 7,235,080 and 6,963,130 reads were generated from these three libraries (0_DAP, 7_DAP and 15_DAP, respectively) (Table 1), from which 73.79%, 73.59% and 70.84% reads could be successfully mapped to the reference genome (Fig. 1). The perfect match reads were 3,874,571 (0_DAP), 3,871,479 (7_DAP) and 3,557,884 (15_DAP). To evaluate the quality of the RNA-seq data, the distribution of unigene coverage in each sample was analyzed, which was the number of clean reads that aligned to the reference genes. As showed in Supplementary Fig. S1, the distributions of the transcripts obtained in the three libraries (0_DAP, 7_DAP and 15_DAP) were uniform, and most of which covered more than 50% reference genes (0_DAP, 57% of all genes; 7_DAP, 52% of all genes; 15_DAP, 51% of all genes; Supplementary Table S1), implying that the transcript sequences detected by RNA-seq technology are reliable.
Table 1. Summary of alignment statistics of RNA-seq in three libraries (0_DAP, 7_DAP and 15_DAP) mapped to reference genome.
| 0_DAP |
7_DAP |
15_DAP |
||||
|---|---|---|---|---|---|---|
| reads number | percentage | reads number | percentage | reads number | percentage | |
| Total Reads | 7,191,808 | 100.00% | 7,235,080 | 100.00% | 6,963,130 | 100.00% |
| Total BasePairs | 352,398,592 | 100.00% | 354,518,920 | 100.00% | 341,193,370 | 100.00% |
| perfect match | 3,874,571 | 71.05% | 3,871,479 | 71.44% | 3,557,884 | 70.03% |
| <=2bp mismatch | 1,578,680 | 28.95% | 1,547,650 | 28.56% | 1,522,565 | 29.97% |
| unique match | 4,307,081 | 78.98% | 4,253,014 | 78.48% | 4,048,952 | 79.70% |
| multi-position match | 1,146,170 | 21.02% | 1,166,115 | 21.52% | 1,031,497 | 20.30% |
Figure 1. The quality assessment of sequencing data from 0_DAP, 7_DAP and 15_DAP libraries.
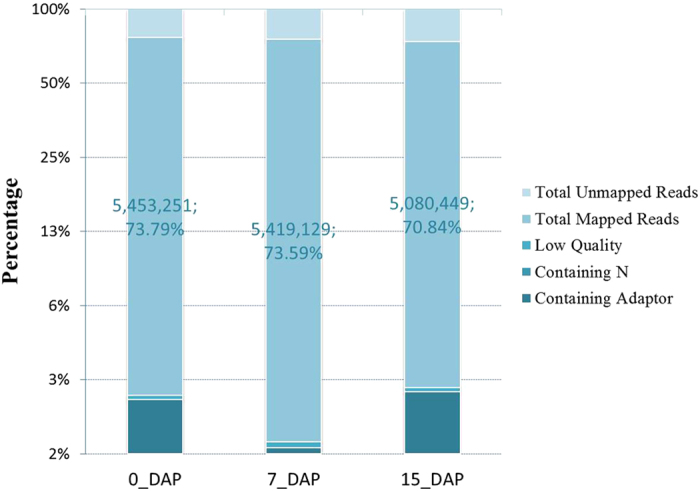
The large portion per bar showed total number and percentage of reads mapped to the reference sequences.
Identification of differentially expressed transcripts
By comparing our three Illumina libraries, a great number of differentially expressed transcripts were identified that were likely involved in radish embryogenesis. The expression levels of genes in three libraries were analyzed by determining the number of reads per kb per million (RPKM) of clean tags. Using the false discovery rate (FDR) ≤ 0.001 and |log2Ratio| ≥ 1 as the threshold values, a total of 5,777 transcripts which accounted for approximately 11.17% of the total transcripts present in three libraries were differentially expressed during radish embryogenesis based on pairwise comparison between the three libraries (Fig. 2A and Supplementary Fig. S2). Between library 0_DAP and 15_DAP, 2,990 and 4,918 transcripts were up- and down- regulated, respectively. A total of 2,301 transcripts were up-regulated and 4,146 transcripts were down-regulated between 0_DAP and 7_DAP library. The complete list of the DEGs during embryogenesis in radish was shown in Supplementary Table S2. Commonly up- and down-regulated transcripts were identified among embryogenesis and seed development stages to determine the degree of overlap (Fig. 2B, C). Maximum number of commonly up- (848) and down- (1197) regulated transcripts were observed between 0_DAP-vs-7_DAP and between 0_DAP-vs-15_DAP comparisons, respectively (Fig. 2B, C).
Figure 2. Differentially expressed transcripts during radish embryogenesis (0_DAP, 7_DAP and 15_DAP).
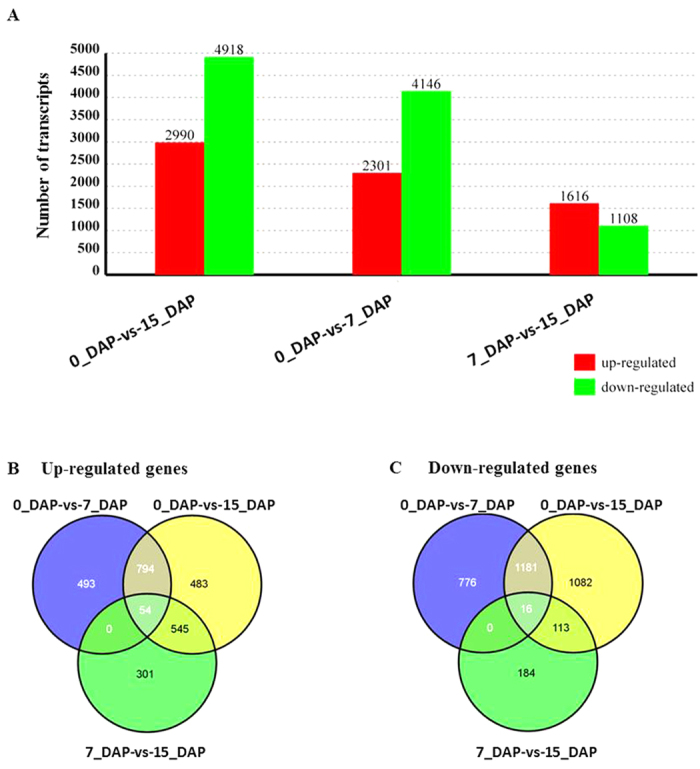
(A) Number of differentially expressed transcripts in three libraries (0_DAP, 7_DAP and 15_DAP) based on pairwise comparison. Numbers of up- and down-regulated genes were summarized; (B, C) Number of commonly differentially expressed transcripts among the three libraries (0_DAP, 7_DAP and 15_DAP). (B, C) showed the number distribution of up- and down-regulated transcripts based on pairwise comparison, respectively.
Cluster analysis of differentially expressed transcripts
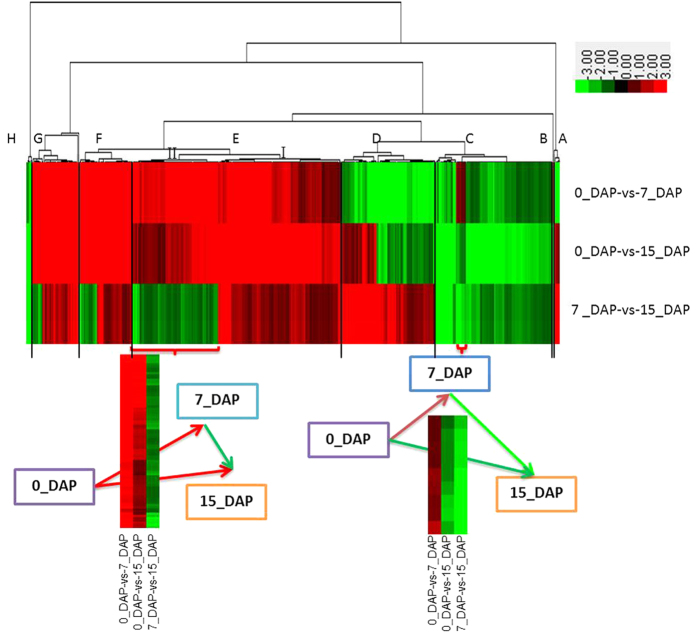
To explore patterns of co-regulation of the DEGs during embryogenesis and seed development in radish, expression profiles of these genes were clustered using the hierarchical clustering algorithm and visualized in a heat map. The expression patterns were separated into eight major clusters (A–H) (Fig. 3). Transcripts present in each of the three libraries within each cluster were shown in Supplementary Table S3. Among these major clusters, the up-regulated transcripts were enriched in cluster A, D, E, F and G, while the down-regulated transcripts were enriched in cluster B and C. All the six transcripts in cluster A, 85 out of 87 in cluster D, 112 out of 191 in cluster E, 32 out of 49 in cluster F were up-regulated at the late stage (15_DAP) of radish embryogenesis. Moreover, among these transcripts, the differentially expressed genes in cluster E (II) including WRKY22, zinc finger homeodomain protein 1, efflux carrier of polar auxin transport, lipoxygenase 2 and beta glucosidase 18, were expressed at a higher level during early (7_DAP) embryogenesis, implying that these genes may play a key role in initial embryo development and morphogenesis phase in radish. Only one transcript in cluster B, and nine out of 107 transcripts in cluster C were up-regulated in 0_DAP-vs-7_DAP, whereas all the genes in cluster H were down-regulated in any pairwise comparison of libraries (Fig. 3).
Figure 3. Cluster diagram of DEGs based on pairwise comparison among the three libraries (0_DAP, 7_DAP and 15_DAP) in radish.
Expression levels for DEGs for the three groups of pairwise comparisons were hierarchically clustered and shown in a heat-map. Level of expression was represented by color scale from green (down-regulated expression) to red (up-regulated expression), as indicated by a scale bar in the upper right corner. The dendrogram of distances were also shown for genes, names of which were presented in Supplementary Table S3. The two small heat-maps placed lower showed the enlarged view of the two sections in red braces.
GO classification and enrichment analysis of the DEGs
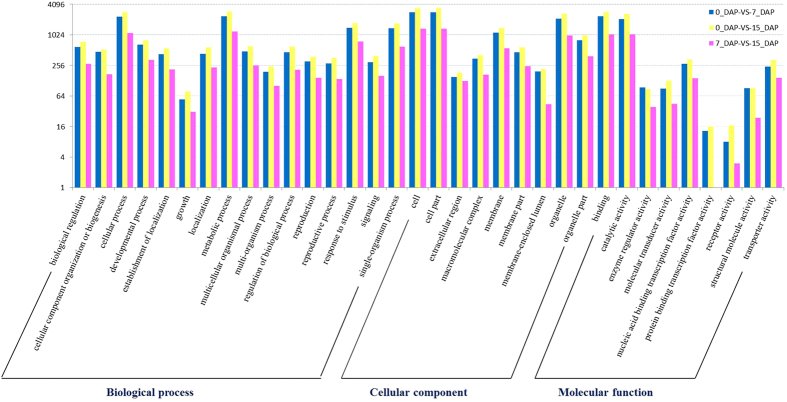
To get better understanding of the biological functions of these transcripts during radish embryogenesis, DEGs were annotated with Gene Ontology (GO) categories based on GO assignments of top matches in the NCBI non-redundant (nr) database. Based on sequence similarities, 4,513 (0_DAP-vs-7_DAP), 5,576 (0_DAP-vs-15_DAP) and 2,091 (7_DAP-vs-15_DAP) DEGs were categorized into 34 GO terms, respectively (Fig. 4). Among GO term, the ‘metabolic process’, ‘cell’, ‘cell part’, ‘organelle’, ‘binding’ and ‘catalytic activity’ terms were dominant among DEGs. Only a few of the DEGs between 0_DAP and 7_DAP as well as 0_DAP and 15_DAP were classified in the categories ‘protein binding transcription factor activity’, whereas no DEG between 7_DAP and 15_DAP was annotated.
Figure 4. Histogram of level 2 Gene ontology classification of DEGs during embryogenesis in radish.
Results are summarized for three main GO categories: biological process (P), molecular function (F), and cellular component (C). The x-axis and y-axis indicate the names and the number of each GO term, respectively.
According to GO functional enrichment analysis at early stage of embryogenesis, a total of 6 (cellular component), 16 (molecular function) and 54 (biological process) terms were significantly enriched (Bonferroni-corrected P-value ≤ 0.05; Supplementary Table S4). The enrichment of the genes participated in various metabolic and reproductive processes including cell wall (GO:0005618), post-embryonic morphogenesis (GO:0009886), embryonic axis specification (GO:0000578), embryo development (GO:0009790), regulation of post-embryonic development (GO:0009791), embryonic pattern specification (GO:0009880), embryonic morphogenesis (GO:0048598) and floral organ development (GO:0048437).
Functional classification using the KEGG database
To further identify the functions of DEGs during embryogenesis and seed development, functional classification and pathway assignment of DEGs were performed using the KEGG database with a hypergeometric test and Bonferroni Correction25. In total, 127 KEGG pathways had at least one DEG. As shown in Table 2, the annotated DEGs were predominantly enriched in 20 KEGG pathways (Supplementary Table S5), particularly plant hormone signal transduction [ko04075], starch and sucrose metabolism [ko00500], brassinosteroid biosynthesis [ko00905] and phenylpropanoid biosynthesis [ko00940], indicating that these were the active pathways during radish embryogenesis and seed development. Plant hormone signal transduction contains several biosynthesis and metabolic patterns, including Tryptophan metabolism (ko00380, seven DEGs), Zeatin biosynthesis (ko00908, four DEGs), Brassinosteroid biosynthesis (ko00905, six DEGs), Diterpenoid biosynthesis (ko00904, two DEGs), Carotenoid biosynthesis (ko00906, four DEGs), Cysteine and methionine metabolism (ko00270, three DEGs), α-Linolenic acid metabolism (ko00592, three DEGs) and Phenylalanine metabolism (ko00360, two DEGs) (Supplementary Fig. S3). Some DEGs involved in these plant hormone signal transduction pathways were shown in Table 3.
Table 2. The enriched pathways of significantly differentially-regulated DEGs in three libraries (0_DAP, 7_DAP and 15_DAP) based on pairwise comparison.
| Pathways | ID | 0_DAP-vs-7_DAP |
0_DAP-vs-15_DAP |
7_DAP-vs-15_DAP |
|||
|---|---|---|---|---|---|---|---|
| No. of DEG | p-Value | No. of DEG | p-Value | No. of DEG | p-Value | ||
| Starch and sucrose metabolism | ko00500 | 150 | 4.21E-18 | 155 | 3.17E-11 | 77 | 2.54E-11 |
| Pentose and glucuronate interconversions | ko00040 | 84 | 2.29E-11 | 78 | 3.50E-05 | 42 | 7.81E-07 |
| Plant hormone signal transduction | ko04075 | 216 | 7.96E-11 | 256 | 1.76E-09 | 118 | 1.34E-09 |
| Ribosome biogenesis in eukaryotes | ko03008 | 72 | 1.09E-05 | 79 | 0.000538 | 8 | 0.999389 |
| DNA replication | ko03030 | 33 | 0.000254 | 37 | 0.001201 | 2 | 0.997389 |
| Brassinosteroid biosynthesis | ko00905 | 13 | 0.003174 | 9 | 0.252944 | 10 | 0.000297 |
| Glycosphingolipid biosynthesis-globo series | ko00603 | 9 | 0.007287 | 8 | 0.070432 | 3 | 0.223588 |
| Thiamine metabolism | ko00730 | 6 | 0.01125 | 3 | 0.434719 | 2 | 0.227954 |
| Indole alkaloid biosynthesis | ko00901 | 12 | 0.020144 | 17 | 0.002195 | 10 | 0.000767 |
| Zeatin biosynthesis | ko00908 | 18 | 0.037568 | 18 | 0.201559 | 16 | 0.000138 |
| Vitamin B6 metabolism | ko00750 | 7 | 0.04039 | 8 | 0.05051 | 4 | 0.066465 |
| Folate biosynthesis | ko00790 | 7 | 0.161538 | 3 | 0.927808 | 1 | 0.884103 |
| Phenylpropanoid biosynthesis | ko00940 | 67 | 0.166402 | 82 | 0.23998 | 56 | 1.77E-06 |
| Biotin metabolism | ko00780 | 2 | 0.20004 | 3 | 0.086867 | 2 | 0.059061 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | ko00400 | 16 | 0.805192 | 22 | 0.729426 | 15 | 0.046203 |
| Glycerophospholipid metabolism | ko00564 | 25 | 0.833352 | 35 | 0.70619 | 9 | 0.945405 |
| Glycine, serine and threonine metabolism | ko00260 | 24 | 0.90091 | 34 | 0.802886 | 26 | 0.003606 |
| Alanine, aspartate and glutamate metabolism | ko00250 | 22 | 0.940543 | 33 | 0.807366 | 24 | 0.009459 |
| Fatty acid biosynthesis | ko00061 | 8 | 0.96842 | 35 | 0.000121 | − | − |
| Glutathione metabolism | ko00480 | 25 | 0.997484 | 38 | 0.985319 | 12 | 0.975054 |
Table 3. The identified genes involved in plant hormone signal transduction pathway.
| Entry | Gene Name | Definition | E. C. Number |
|---|---|---|---|
| K13946 | AUX1 | auxin influx carrier (AUX1 LAX family) | |
| K14485 | TIR1 | transport inhibitor response 1 | |
| K14484 | AUX/IAA | auxin-responsive protein IAA | |
| K14486 | ARF | auxin response factor | |
| K14487 | GH3 | auxin responsive GH3 gene family | |
| K14488 | SAUR | SAUR family protein | |
| K14489 | CRE1 | Arabidopsis histidine kinase 2/3/4 (cytokinin receptor) | EC:2.7.13.3 |
| K14490 | AHP | histidine-containing phosphotransfer peotein | |
| K14491 | B-ARR | two-component response regulator ARR-B family | |
| K14492 | A-ARR | two-component response regulator ARR-A family | |
| K1449 | GID1 | gibberellin receptor GID1 | EC:3.-.-.- |
| K14495 | GID2 | F-box protein GID2 | |
| K14494 | DELLA | DELLA protein | |
| K1451 | CTR1 | serine/threonine-protein kinase CTR1 | EC:2.7.11.1 |
| K14512 | MPK6 | mitogen-activated protein kinase 6 | EC:2.7.11.24 |
| K14513 | EIN2 | ethylene-insensitive protein 2 | |
| K14515 | EBF1/2 | EIN3-binding F-box protein | |
| K14516 | ERF1/2 | ethylene-responsive transcription factor 1 | |
| K13416 | BAK1 | brassinosteroid insensitive 1-associated receptor kinase 1 | EC:2.7.10.12.7.11.1 |
| K13415 | BRI1 | protein brassinosteroid insensitive 1 | EC:2.7.10.1 2.7.11. |
| K1450 | BSK | BR-signaling kinase | EC:2.7.11. |
| K14503 | BZR1/2 | brassinosteroid resistant 1/2 | |
| K14504 | TCH4 | xyloglucan:xyloglucosyl transferase TCH4 | EC:2.4.1.207 |
| K14505 | CYCD3 | cyclin D3, plant |
Experimental validation of DEGs through qRT-PCR
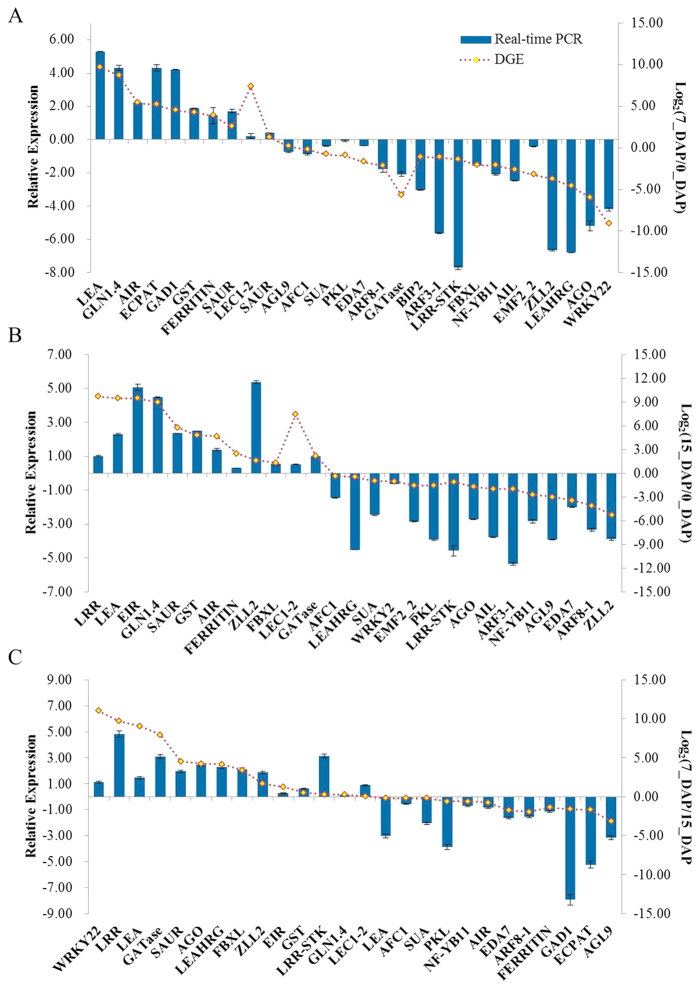
To evaluate the validity of Illumina sequencing analysis and further assess the patterns of DEGs, 28, 27 and 26 genes were selected from the three pairwise comparisons (0_DAP-vs-7_DAP, 0_DAP-vs-15_DAP, 7_DAP-vs-15_DAP) for qRT-PCR analysis, respectively (Fig. 5). For all of the selected genes, the expression patterns determined in real-time RT-PCR were in agreement with those based on RNA-seq. Nonetheless, there were discrepancies with respect to degrees of differential expression between the two methods. This might be attributed to the differences in the algorithms used to determine expression levels. The high-throughput sequencing technique generated absolute rather than relative expression measurements. Furthermore, to visualize the correlations intuitively, real-time RT-PCR was performed on 17 DEGs in three comparative groups (0_DAP-vs-7_DAP, 0_DAP-vs-15_DAP and 7_DAP-vs-15_DAP), which showed different expression profiles during early embryonic process, with high or low expression levels at one or more time points. The Pearson correlation coefficient was calculated by SPSS to assess the correlation between different platforms (Supplementary Fig. S4). Overall, we observed a good concordance in the expression patterns of DEGs obtained by both the RNA-seq and qRT-PCR method as indicated by the overall correlation coefficient (0.84), implying the reliability of the transcriptomic profiling data.
Figure 5. RT-qPCR validation of some DGEs during radish embryogenesis.
The relative expression of DGEs between 0_DAP and 7_DAP libraries (A), 0_DAP and 15_DAP libraries (B) and 7_DAP and 15_DAP libraries (C) were analyzed by the 2−ΔΔCT method.
DEGs involved in embryo induction, formation and maturation stages
To understand molecular mechanism of embryogenic competence acquisition embryogenesis and seed development, the DEGs with a higher degree of differential expression (|log2Ratio| ≥ 2) were further characterized (Supplementary Table S6). Several differentially expressed transcription factors, transporters, hormone response/transduction genes and transcripts encoding embryo defective protein etc. were identified, including LEA (late embryogenesis abundant protein), SERK (somatic embryogenesis receptor-like kinase), EIR (auxin efflux carrier family proteins), ARFs (auxin response factor), ANT (aintegumenta), GST (glutathione S-transferase), WRKYs (WRKY transcription factors), FERRITIN, AG/ZLL2 (argonaute/Zwille-like proteins) and AGLs (Agamous-like MADS-box proteins).
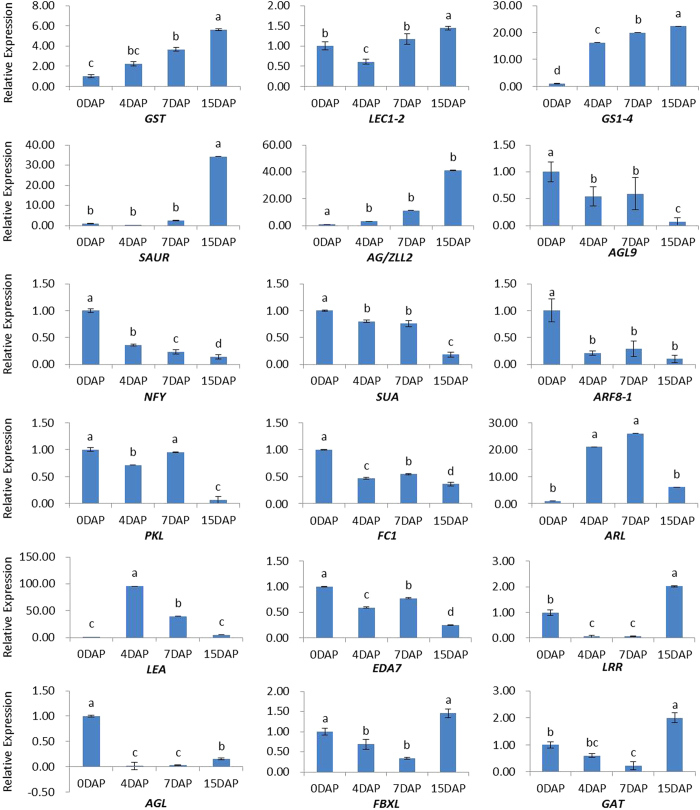
To further investigate and verify the expression variations of the DEGs, transcriptional qRT-PCR analysis of 18 selected genes including AGL9, ARF8-1, LEC1-2, LEA protein, NF-Y (nuclear factor Y) and GST, were performed for a more detailed analysis of ovule gene expression after pollination in different days (0 DAP, 4 DAP, 7 DAP and 15 DAP). Among these genes associated with embryo induction and formation, they can be classified into four groups. The first group including GST, LEC1-2 transcription factor, GS1-4 (glutamine synthetase cytosolic isozyme 1–4), SAUR (small auxin up RNA protein), AG/ZLL2, showed an up-regulated expression during the embryo induction and formation stages (Fig. 6). DEGs in the second group were mostly transcription factors where some were reported previously as important regulators of plant developmental process, including AGL9, NF-Y, SUA (suppressor of ABI3-5), ARF8-1, PKL (PICKLE) and FC1 (fus3-complementing gene 1). In addition, the expression of ARL (auxin-responsive-like protein) and LEA reached a peak at 4–7 DAP ovule, suggesting that these genes may play an important role in morphogenesis and maturation of embryos. Some other genes in group 4 including EDA7 (embryo sac development arrest 7 protein), LRR (leucine-rich repeat-containing protein), AGL, FBXL (F-box/LRR-repeat protein) and GAT (glutamine amidotransferase) were found to be down-regulated during early embryogenesis and up-regulated slightly in the late stage. These observations showed that some transcription factors and genes, such as LEC1-2, ARFs and AGLs, were responsive to phytohormone, which could be involved in normal morphogenesis phase of zygotic embryogenesis in radish.
Figure 6. Relative expression levels of DEGs in four developmental stages of radish embryogenesis.
The normalized expression levels at 0 DAP were arbitrarily set to 1. Different letters indicate significant differences at P < 0.05 according to Duncan’s multiple range tests. Each bar shows the mean ± SE of triplicate assays.
Integrative analysis of DEGs and miRNAs during embryogenesis
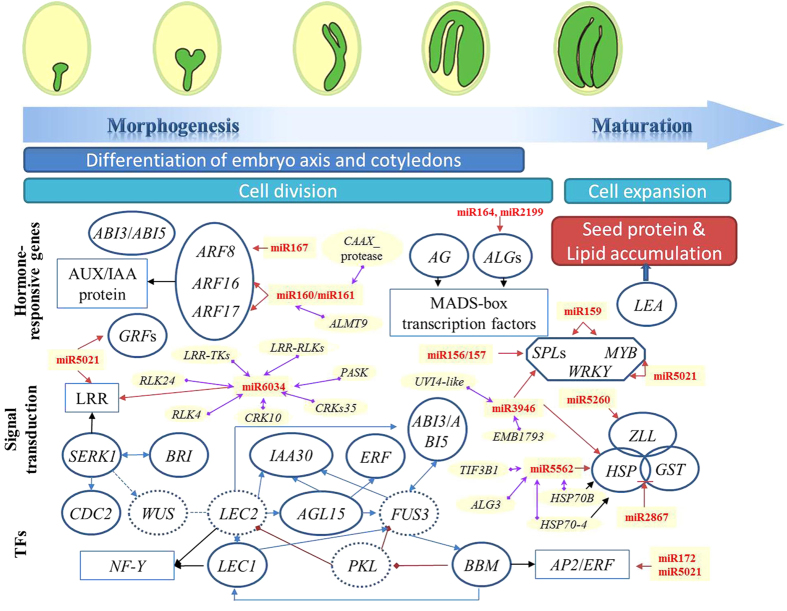
Previous studies have extensively showed that many miRNA-mRNA pairs were involved in embryogenesis, such as miR160/167-ARFs, miR156/157-SPLs and miR172-AP226,27,28. Based on the previously identified radish embryogenesis-related miRNAs29, and the DGE data in this study, we found that the differential expression of several genes likely involved in developmental and physiological events during radish development were modulated, to some extent, by the activity of miRNAs (Fig. 7). Our data sets revealed that 57 candidate genes were involved in regulation of phytohormones (Supplementary Table S7). These genes including ARFs, GH3, IAAs, ARP (auxin-repressed protein), EIR, ERF (ethylene-responsive transcription factor) and SAUR were modulated during different stages of embryogenesis, and their expression profiles were correlated with several miRNAs such as miR160/161 and miR167 (Fig. 7), indicating their potential key roles and regulatory relationships during embryogenesis and seed development in radish. Moreover, one BBM gene (targeted by miR172 and miR5021), 32 LRR genes (targeted by miR5021 and miR6034), 12 LEA genes and 3 AGLs genes (targeted by miR164 and miR2199) were significantly differentially expressed at different development stages of radish embryo (Fig. 7 and Supplementary Table S7). During embryogenesis, a series of developmental transition steps were orchestrated by transcription factors including those in the WRKY gene family30. A total of 28 WRKY gene transcripts representing 17 family members were identified (Supplementary Table S7).
Figure 7. A proposed model of genetic and molecular interactions in the regulatory network during embryogenesis in radish.
Embryo development consists of two phases, morphogenesis and maturation. Arrows in blue and black indicate regulation and affiliation, respectively, whereas red lines between genes indicate transcriptional repression. The circle with solid and dotted line represented genes identified or not in radish and the corresponding miRNAs are shown in red. Genes in yellow boxes were connected with miRNAs by purple arrows using Cytoscape, which were visualized as miRNA-mRNA interactions.
To gain in-depth insight into the potential relationship between DEGs and embryogenesis-related miRNAs29, a co-expression analysis was performed based on the mRNA and miRNA data to identify DEGs that were likely co-regulated with differentially expressed miRNAs. Among them, 75 pairs of miRNA/mRNA were found as co-expressed (Supplementary Table S8) and a partial regulatory network was visualized in Fig. 7. Further analysis showed that 33 pairs containing 19 known miRNAs and 28 unique transcripts showed a negative correlation in expression trends during early embryogenesis, providing valuable information for the regulation of miRNA in the regulatory network of radish embryogenesis.
Discussion
Embryogenesis represents an elaborate and complex phase in plant life cycles, which condenses the fundamental processes underlying plant development into a short sequence of predictable steps5,8. Previous studies have identified several genes involved in regulating plant embryogenesis mainly by mRNA differential display (mRNADD)31 or hybridization-based approaches32. In contrast to the above-mentioned approaches, the RNA-seq technology has clear advantages for transcriptome profiling, which can generate millions of sequence reads with high reproducibility, and create a comprehensive view of the participation of several multi-gene families in plant embryogenesis33,34. However, no study on comprehensive identification of the DEGs during embryogenesis has yet been conducted in radish. This study was undertaken with the goal to investigate the genes regulating radish embryogenesis and seed development. By pairwise comparisons of the data between three libraries (0_DAP, 7_DAP and 15_DAP), differential transcript abundance and regulation were demonstrated. Genes associated with secondary metabolism, signal transduction, hormone response/transduction and development were identified, and their expression profiles were characterized in detail.
Transcription regulation underlying embryogenesis in radish
Large-scale transcription analyses of embryogenesis had been demonstrated in several species12,35. Because multiple transcription factors (TFs) were differentially expressed durng embryogenesis and a number of them have known roles during embryogenesis in other plant models, our study contributes to a better understanding of embryogenesis process10,36. In this study, at least two groups of TFs (AP2/ERF Domain Protein and MADS-Domain Protein AGAMOUS-LIKE) were present with complex expression profiles. Several members in the AP2/ERF family had also been reported to be involved in microspore-, somatic- and zygotic embryogenesis32. In this study, the radish orthologs of APETALA2 (AP2, negatively controlling cell proliferation during seed development), AINTEGUMENTA (ANT, expressing in the primordia of cotyledons) and BABY BOOM (BBM, promoting cell proliferation and morphogenesis during embryogenesis) were differentially expressed during radish embryogenesis and seed development, consistent with their actions in Arabidopsis thaliana17, Brassica napus16, Capsicum annuum37 and Elaeisguineensis38. These findings suggested that these TFs potentially participate in the specific process embryogenesis and early seedling development in radish9.
The AINTEGUMENTA-LIKE (AIL) transcription factor genes including BBM, ANT and PLETHORA (PLT) genes in the AP2/ERF family and were expressed in young tissues. These ANT genes play key roles in many developmental processes, e.g. embryogenesis, meristem maintenance, organ positioning and growth39,40. Previous studies showed that ANT had a central role in ovule development through the activation of HD-ZIP III genes that promote apical fate during early embryogenesis39, in accordance with the results obtained in this study. Currently, the BBM gene had been regarded as a transcriptional activator or repressor in a wide range of developmental pathways, such as plant embryogenesis, and its targets support a role for AIL proteins in the regulation of cell proliferation and differentiation39.
Although some regulators can directly influence embryo identity in somatic tissue, other factors act in a more indirect manner. The latter might involve an increase in the capacity to induce somatic embryos in response to other triggers rather than in inducing embryogenesis directly1. The AGL genes, belonging to MADS-domain protein family, were detected during radish embryogenesis, which were also reported to express in young developing embryos, promote somatic embryogenesis and enhance production of secondary embryonic tissue in soybean41, rice33 and Arabidopsis20. In the present study, four AGL genes were down-regulated, suggesting that they preferentially accumulated during the early stages of radish embryo development and played an important role in the beginning of embryogenesis42.
In addition to the AP2s and AGLs, the LEC2 and WUS homeodomain protein genes were identified but not differentially expressed under the threshold value in our data. The LEC1 (At-NF-YB9), a member of NF-Y family homologous with LEC1-LIKE (L1L; At-NF-YB6), was also identified in this study. LEC1 was a general integrator of light and hormone signaling during embryogenesis43 and its action included processes beyond the embryo, e.g. etiolation responses in young seedlings44. In the present study, the expression levels of LEC1-2 analyzed by qRT-PCR were low at 4 DAP and gradually increased in later stages. PKL, a putative chromatin remodeling factor, showed an opposite expression trend, leading to the hypothesis that PKL repressed LEC1-2 TF during radish embryogenesis. Consistent with LEC overexpression phenotypes, pkl mutants accumulated storage macromolecules and give rise to embryogenic calli in culture45.
Crucial roles of hormone in embryogenesis and seed development
In dicotyledonous plant, the embryogenesis begins with fertilization of egg and subsequently the zygote undergoes cell elongations and divisions4. Plant growth regulators (PGRs), ARFs, Aux/IAAs, SUA and ARL were involved in plant embryogenesis4,36. In Arabidopsis, Xiang et al. demonstrated that the auxin signaling events associated with gene activity were more prevalent in early developmental stages when the post-fertilization sporophytic program is initiated2. In this study, genes related to auxin biosynthetic and metabolic processes, polar auxin transport, homeostasis and auxin-mediated signaling, were highly expressed during radish embryogenesis and seed development, which was consistent with the observations in rice46, cotton36 and Arabidopsis30,34. Auxin is considered to be a critical PGR in cell division and differentiation, and its level surges occurred during embryogenesis with consequent accumulation of numerous mRNAs9,36. Some highly expressed auxin-related genes, including auxin-responsive family protein (ARL), EIR, GH3s, PINs and small auxin-up RNAs (SAURs) were differentially expressed, suggesting that they might play important roles in development process of radish embryogenesis9.
Moreover, the brassinosteroids (BRs) are an important class of signaling hormones involved in plant growth and development47. In this study, the plant hormone signal transduction “super-pathway” was amongst the most predominant one. This super-pathway includes a total of eight pathways (Supplementary Fig. S3). Among them, BR biosynthesis pathway has been widely reported to interact with other phytohormones influencing plant development48,49. BRs are a class of polyhydroxylated steroidphyto hormones essential for plant growth and development, including cell division47, cell elongation50, root development48, flowering time51 and embryogenesis52. The BRs could regulate gene expression through a receptor kinase-mediated signal transduction pathway53. In particular, the main BR-perceiving receptor, BR-insensitive1 (BRI1) and BAK1/SERK3 are co-receptors for brassinosteroids, and this SERKs of the leucine-rich repeat receptor-like kinase subfamily II (LRR-RLK II) that are associated with the process of somatic embryogenesis9. BRI1, was also important for cell fate specification in the stem vasculature and seedling root epidermis54,55. Further studies were required to unravel the functional effect of brassinosteroids in regulating radish embryogenesis.
Signal transduction pathway in embryogenesis
The process of embryogenesis involves a series of dramatic transitions due to differential gene expression as well as various signal transduction pathways for activating or repressing gene sets9,12. A number of studies had reported that the Receptor-Like Kinases (RLKs) and Receptor-Like Cytoplasmic Kinases (RLCKs) are involved in many different signaling processes such as hormone signaling, growth and development and have important functions during embryonic intercellular signaling event55,56, among which, the SERK gene, a member of LRR-RLKs, was one of the most important genes regulating a successive downstream signal transducers in the signal transduction pathway (Fig. 7). Some SERK genes were recognized as markers for cell competent to form somatic embryos in culture, but specific functions for the SERKs in zygotic embryogenesis were elusive55. In the present study, we found a great number of SERKs [EC: 2.7.10.1, 2.7.11.1] as a leucine-rich repeat (LRR) transmembrane protein kinase in the KEGG pathway (Supplementary Table S6), which may play a crucial role in triggering embryogenesis in plants3,9. Consistent with our results, Somleva et al. demonstrated that the SERK gene was expressed in a subpopulation of cells competent to form somatic embryos during the induction of embryogenic cell formation in D.glomerata57. Hecht et al. confirmed the SERK1 could enhance the ability of suspension cells to undergo embryogenesis by ectopic overexpression in Arabidopsis58.
Additionally, several genes encoding enzymes involved in embryo development have been validated by qRT-PCR in the present study, including GST, GAT and GS1-4 with high expression at 15 DAP (Fig. 6), which were involved in controlling seed development59,60. Similarly, in triticale, Grabowska et al. demonstrated that during the early phase of seed development, the activity of glutamine synthetase with a high level was observed as early as 3 day after flowering (DAF) and increased and reached its maximum at 15 DAF59, consisting with our result. GSTs induced by various exogenous factors such as ethylene and auxins have also been shown to act as modulators of signal transduction pathways that control cell proliferation, which were up-regulated during the early events of somatic embryogenesis in a variety of plant species31,60 and zygotic embryogenesis in our study. However, the regulation mechanism and functions of these genes during the developmental process of embryos needs to be further explored.
In conclusion, this is the first study to characterize the DEGs expression profiling during radish embryogenesis and seed development with RNA-seq technology. A total of 5,777 DEGs were detected by pairwise comparisons among three cDNA libraries, including genes involved in secondary metabolism, signal transduction, hormone response/transduction and development in plants. Several identified pathways such as plant hormone signal transduction, metabolism, embryonic axis specification, embryo development and morphogenesis were involved in radish embryogenesis. Furthermore, a model of regulatory relationship between DEGs and miRNAs was proposed that might be central to radish embryogenesis and seed development. Our findings not only provide putative components and regulatory network associated with radish embryogenesis, but also facilitate further dissection of the molecular mechanisms underlying embryo morphogenesis and seed development in root vegetable crops.
Methods
Plant materials
Immature ovules and zygotes (0, 4, 6, 7, 10, 12, 13 and 15 DAP) from a radish (Raphanus sativus L.) advanced inbred line ‘NAU-DY13’ (early bolting and flowering) were randomly collected from self-pollinated ovaries growing in a growth chamber with a 16 h light at 26 °C/8 h dark at 20 °C cycle and immediately frozen in liquid nitrogen and stored at −80 °C for further use.
DGE library construction and Illumina sequencing
The tissue samples from zygotes of 0 day after pollination, 4–7 days after pollination and 10–15 days after pollination were separately pooled into three collections for RNA preparation. Total RNA from each collection was extracted using Trizol® Reagent (Invitrogen) following the manufacturer’s instructions. Library construction was carried out at Beijing Genomics Institute (BGI, Shenzhen, China) using Illumina’s DGE tag profiling technology. In brief, 6 μg of total RNA was used for mRNA capture with magnetic oligo (dT) beads, and then the first and second strand cDNA was synthesized. After purification and adaptor ligation, PCR amplification was performed for library construction. Finally, the qualified and quantified sample libraries were sequenced via Illumina HiSeqTM 2500 platform.
Data processing and digital tag profiling
The clean tags were obtained by removing 3′ adaptor fragments, low-quality tags, and several types of impurities from the raw reads. Then, the clean tags were mapped to the radish reference sequences containing genomic survey sequences (GSS), EST sequences and our mRNA transcriptome sequences by SOAP2. No more than two mismatches were allowed in the alignment61. Statistical analysis was performed to identify differentially expressed genes (DEGs) among different libraries using a rigorous algorithm described previously62. The expression level for each gene was determined by the reads number uniquely mapped to the specific gene and the total number of uniquely mapped reads in the library. Gene expression was normalized using the Reads Per kb per Million (RPKM) method63.
Differentially expressed gene analysis
Regarding the significance of digital gene expression profiles62, a rigorous algorithm supplied from BGI was used to compare the differences in gene expression between each two DGE libraries. The threshold P-values were adjusted by the multiple testing procedures described by Benjamini and Yekutieli64 by controlling FDR (false discovery rate). In this study, FDR ≤ 0.001 and the absolute value of |log2Ratio| ≥ 1 were used as the threshold for judging significance difference of the gene expression.
Functional annotation of DEGs
The DEGs were subjected to Gene Ontology (GO) database (http://www.geneontology.org/) and mapped to the reference canonical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG). The target sequences were allocated to the corresponding functional categories on the basis of the BLAST searches by GO annotation using default parameters. The gene expression patterns of each pairwise comparison (0_DAP-vs-7_DAP, 0_DAP-vs-15_DAP, 7_DAP-vs-15_DAP) was analyzed and genes were clustered according to their expression level using self-organizing map using Cluster 3.0 with all the default parameters except the Euclidean distance of similarity metric. Additionally, the expression values were log2-transformed. Heat-maps with cluster data were then constructed using Java Tree View (http://jtreeview.sourceforge.net/) for visualization of the hierarchical clustering results65.
Quantitative real-time RT-PCR validation
Quantitative real-time PCR (qRT-PCR) was employed to validate the results from high-throughput sequencing of the differentially expressed genes. Total RNAs were obtained from five radish samples (0, 4, 7 and 15 DAP) as described above. All reactions were performed on an iCycler iQ real-time PCR detection system (BIO-RAD) with three biological and three technological replications, respectively, which were carried out in a total volume of 20 μl including 0.2 μM primer pairs, 2 μl diluted cDNA and 10 μl 2×SYBR Green PCR Master Mix (TaKaRa Bio Inc., Dalian, China). The amplification reactions were incubated at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 58 °C for 15 s, and 72 °C for 20 s. The Actin gene was selected as the internal control. Relative gene expression levels were calculated using the 2−△△Ct method. The primers for the selected transcripts were shown in Supplementary Table S9.
Correlation analysis between radish embryogenesis-related miRNAs and DEGs
The correlation analysis between mRNA and miRNA expression during embryogenesis in radish was performed using Cytoscape_v3.2.1 software66. The radish embryogenesis-associated miRNA data were obtained from our previous study27. Briefly, to identify the co-regulated differentially expressed miRNAs and mRNAs, the target genes of miRNA were compared with the normalized DEGs. The relationship between miRNA and mRNA was assessed according to the log2 value. For instance, the log2 value of miRNA and its corresponding gene were both above 0, and then they were up-up regulation. After establishing the contingency table, a visualization of nodes and edges as a two-dimensional network layout was constructed67.
Additional Information
How to cite this article: Zhai, L. et al. Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.). Sci. Rep. 6, 21652; doi: 10.1038/srep21652 (2016).
Supplementary Material
Acknowledgments
This work was in part supported by grants from the National Natural Science Foundation of China (31372064, 31501759), the National Key Technologies R & D Program of China (2012BAD02B01) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). We thank Dr. Shiu Shin-Han at Michigan State University for his critical review and helpful comments.
Footnotes
Author Contributions L.Z. and L.L. conceived and designed the experiments. L.Z. and H.F. performed the sample collection. L.Z., C.L., X.L. and Y.W. were responsible for high-throughput deep sequencing analysis and expression analysis. L.Z. drafted the manuscript. L.X., M.M.E., X.Z. and L.L. reviewed and edited the manuscript. All authors read and approved the final manuscript.
References
- Radoeva T. & Weijers D. A roadmap to embryo identity in plants. Trends Plant Sci. 19, 709–716 (2014). [DOI] [PubMed] [Google Scholar]
- Xiang D. et al. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol. 156, 346–356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A. & Bozhkov P. V. Somatic embryogenesis: life and death processes during apical-basal patterning. J. Exp. Bot. 65, 1343–1360 (2014). [DOI] [PubMed] [Google Scholar]
- ten Hove C. A., Lu K. J. & Weijers D. Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142, 420–430 (2015). [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Paiva G. d. & Yadegari R. Plant embryogenesis: zygote to seed science. Science 266, 605–614 (1994). [DOI] [PubMed] [Google Scholar]
- Braybrook S. A. & Harada J. J. LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 236, 647–657 (2012). [DOI] [PubMed] [Google Scholar]
- Palovaara J., Saiga S. & Weijers D. Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci. 18, 514–521 (2013). [DOI] [PubMed] [Google Scholar]
- Yang X. & Zhang X. Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 29, 36–57 (2010). [Google Scholar]
- Jenik P. D., Gillmor C. S. & Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 23, 207–236 (2007). [DOI] [PubMed] [Google Scholar]
- Jeong S., Bayer M. & Lukowitz W. Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo. J. Exp. Bot. 62, 1687–1697 (2011). [DOI] [PubMed] [Google Scholar]
- Malik M. R. et al. Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol. 144, 134–154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Yang X., Yuan D., Zeng F. & Zhang X. GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton. Plant Biotechnol. J. 9, 1038–1048 (2011). [DOI] [PubMed] [Google Scholar]
- Wisniewska A. et al. Identification of genes up-regulated during somatic embryogenesis of cucumber. Plant Physiol. Biochem. 50, 54–64 (2012). [DOI] [PubMed] [Google Scholar]
- Tzafrir I. et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulinska-Lukaszek K., Tobojka M., Adamiok A. & Kurczynska E. Expression of the BBM gene during somatic embryogenesis of Arabidopsis thaliana. Biol. Plant. 56, 389–394 (2012). [Google Scholar]
- Stone S. L. et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98, 11806–11811 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R. et al. The Arabidopsis somatic embryogenesis receptor-like kinase1 protein complex includes brassinosteroid-insensitive1. Plant Cell 18, 626–638 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare D., Tang W., Hill K. & Perry S. E. The MADS-domain transcriptional regulator AGAMOUS-LIKE 15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol. 146, 1663–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E., Guzzo F., Toonen M. & De Vries S. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124, 2049–2062 (1997). [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N. & Wobus U. Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol. 64, 189–217 (2013). [DOI] [PubMed] [Google Scholar]
- Ge X. X. et al. Transcriptional profiling of genes involved in embryogenic, non-embryogenic calluses and somatic embryogenesis of Valencia sweet orange by SSH-based microarray. Planta 236, 1107–1124 (2012). [DOI] [PubMed] [Google Scholar]
- Dang Z. et al. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genomics 14, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. & Lai Z. Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in Longan. PloS One 8, e60337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Hernandez E. C. et al. Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front Plant Sci. 6, 555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J Exp Bot. 64, 1521–1536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L. et al. Genome-wide identification of embryogenesis-associated microRNAs in radish (Raphanus sativus L.) by high-throughput sequencing. Plant Mol. Biol. Rep. 32, 900–915 (2014). [Google Scholar]
- Sun X., Shantharaj D., Kang X. & Ni M. Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 13, 611–620 (2010). [DOI] [PubMed] [Google Scholar]
- Galland R., Randoux B., Vasseur J. & Hilbert J. L. A glutathione S-transferase cDNA identified by mRNA differential display is upregulated during somatic embryogenesis in Cichorium. Biochim. Biophys. Acta. 1522, 212–216 (2001). [DOI] [PubMed] [Google Scholar]
- Tsuwamoto R., Fukuoka H. & Takahata Y. Identification and characterization of genes expressed in early embryogenesis from microspores of Brassica napus. Planta 225, 641–652 (2007). [DOI] [PubMed] [Google Scholar]
- Xu H., Gao Y. & Wang J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PloS One 7, e30646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. W., Casson S. A. & Lindsey K. Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 143, 924–940 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N., Goffard N., Nizamidin M. & Rolfe B. G. Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol. 8, 110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 12, 110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irikova T., Grozeva S. & Denev I. Identification of BABY BOOM and LEAFY COTYLEDON genes in sweet pepper (Capsicum annuum L.) genome by their partial gene sequences. Plant Growth Regul. 67, 191–198 (2012). [Google Scholar]
- Morcillo F. et al. EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta 226, 1353–1362 (2007). [DOI] [PubMed] [Google Scholar]
- Horstman A., Willemsen V., Boutilier K. & Heidstra R. AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci. 19, 146–157 (2014). [DOI] [PubMed] [Google Scholar]
- Duan C. et al. Identification of the Hevea brasiliensis AP2/ERF superfamily by RNA sequencing. BMC Genomics 14, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding E. W., Tang W., Nichols K. W., Fernandez D. E. & Perry S. E. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 133, 653–663 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Zheng Y. & Perry S. E. AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 161, 2113–2127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K. et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24, 4777–4792 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A. et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71, 427–442 (2012). [DOI] [PubMed] [Google Scholar]
- Braybrook S. A. & Harada J. J. LECs go crazy in embryo development. Trends Plant Sci. 13, 624–30 (2008). [DOI] [PubMed] [Google Scholar]
- Xue L. J., Zhang J. J. & Xue H. W. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PloS One 7, e31081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse W., van Mourik S., Albrecht C., van Leeuwen J. & de Vries S. A mathematical model for the coreceptors somatic embryogenesis receptor-like kinase 1 and somatic embryogenesis receptor-like kinase 3 in brassinosteroid insensitive1-mediated signaling. Plant Physiol. 163, 1472–1481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel C. F., Osmont K. S. & Hardtke C. S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461 (2006). [DOI] [PubMed] [Google Scholar]
- Li J., Li Y., Chen S. & An L. Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot. 61, 4221–4230 (2010). [DOI] [PubMed] [Google Scholar]
- Yang X. H., Xu Z. H. & Xue H. W. Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 17, 116–131 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D. The molecular intersection of brassinosteroid-regulated growth and flowering in Arabidopsis. Proc. Natl. Acad. Sci. USA 105, 7345–7346 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullman G., Zhang Y. & Phan B. Brassinolide improves embryogenic tissue initiation in conifers and rice. Plant Cell Rep. 22, 96–104 (2003). [DOI] [PubMed] [Google Scholar]
- Lin W. et al. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110, 12114–12119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K. T., Chen A. Y. & Nemhauser J. L. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 106, 8073–8076 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M. D., Bryan A. C., Racolta A., Jerosky K. V. & Tax F. E. A few standing for many: embryo receptor-like kinases. Trends Plant Sci. 16, 211–217 (2011). [DOI] [PubMed] [Google Scholar]
- Aan den Toorn M., Albrecht C. & de Vries S. On the Origin of SERKs: Bioinformatics Analysis of the Somatic Embryogenesis Receptor Kinases. Mol. Plant. 8, 762–782 (2015). [DOI] [PubMed] [Google Scholar]
- Somleva M. N., Schmidt E. D. L. & de Vries S. C. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep. 19, 718–726 (2000). [DOI] [PubMed] [Google Scholar]
- Hecht V. et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816 (2001). [PMC free article] [PubMed] [Google Scholar]
- Grabowska A., Kwinta J. & Bielawski W. Glutamine synthetase and glutamate dehydrogenase in triticale seeds: molecular cloning and genes expression. Acta Physiol. Plant. 34, 2393–2406 (2012). [Google Scholar]
- Stasolla C. Glutathione redox regulation of in vitro embryogenesis. Plant Physiol. Biochem. 48, 319–327 (2010). [DOI] [PubMed] [Google Scholar]
- Li R. Q. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
- Audic S. & Claverie J. M. The significance of digital gene expression profiles. Genome Res. 7, 986–995 (1997). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001). [Google Scholar]
- Saldanha A. J. Java Treeview-extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- He C. Y., Cui K., Zhang J. G., Duan A. G. & Zeng Y. F. Next-generation sequencing-based mRNA and microRNA expression profiling analysis revealed pathways involved in the rapid growth of developing culms in Moso bamboo. BMC Plant Biol. 13, 119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.