Abstract
Eukaryote-derived methioninase, catalyzing the one-step degradation of methionine (Met) to methanethiol (MTL), has received much attention for its low immunogenic potential and use as a therapeutic agent against Met-dependent tumors. Although biological and chemical degradation pathways for Met-MTL conversion are proposed, the concrete molecular mechanism for Met-MTL conversion in eukaryotes is still unclear. Previous studies demonstrated that α-keto-methylthiobutyric acid (KMBA), the intermediate for Met-MTL conversion, was located extracellularly and the demethiolase STR3 possessed no activities towards Met, which rule out the possibility of intracellular Met-MTL conversion pathway inside eukaryotes. We report here that degradation of Met resulted in intracellular accumulation of KMBA in Clonostachys rosea. Addition of Met to culture media led to the production of MTL and downregulation of STR3, while incubation of Met with surrogate substrate α-ketoglutaric acid enhanced the synthesis of MTL and triggered the upregulation of STR3. Subsequent biochemical analysis with recombinant STR3 showed that STR3 directly converted both Met and its transamination product KMBA to MTL. These results indicated that STR3 as rate-limiting enzyme degrades Met and KMBA into MTL. Our findings suggest STR3 is a potential target for therapeutic agents against Met-dependent tumors and aging.
Methionine (Met) is employed at multiple levels in cellular metabolism: as a protein constituent, in the initiation of mRNA translation, and as a regulatory molecule in the form of S-adenosylmethionine. Thus, it can be assumed that Met synthesis, accumulation, and transfer play fundamental roles in cell growth and development1,2,3,4. Despite the importance of Met, Met catabolism is not fully understood especially in eukaryotic cells, most likely because of the undetected transient intermediates and functional redundancy of enzymes within the pathway5,6.
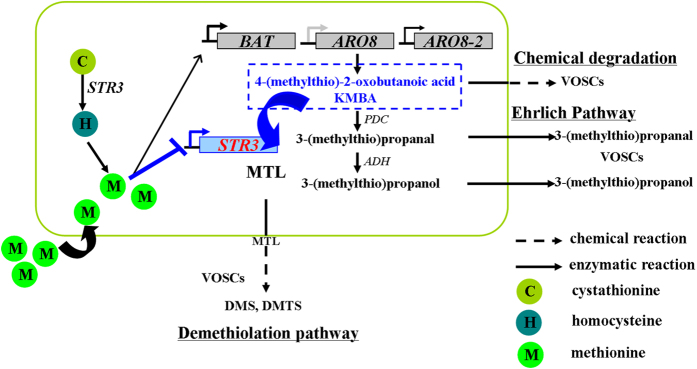
In eukaryotic cells, Met was catabolized to α-keto-methylthiobutyric acid (KMBA) by aminotransferase, KMBA was hypothesized to be converted to methional and methionol via Ehrlich pathway and methanethiol (MTL) via demethiolation pathway, respectively7. However, other studies suggested that KMBA accumulated only extracellularly, proposing that KMBA was highly toxic to cells and actively excreted6,7,8. It is thus currently believed that the degradation of KMBA into volatile organic sulfur-containing compounds (VOSCs) including methional, methanol, MTL and so on is an exclusively chemical reaction dependent on the physicochemical properties of the extracellular medium including pH, redox status, and ionic strength6.
Met degradation pathway genes are highly conserved, Ehrlich pathway genes including aminotransferase genes AAT1, AAT2, ARO8, ARO9, BAT1, BAT2, and α-ketoacid decarboxylase gene PDC are annotated in fungi based on the homology to Saccharomyces cerevisiae genes6,8,9,10. Demethiolation pathway enzymes CYS3 and STR3, annotated as cystathionine β-lyase and γ-lyase, are inferred as the key enzymes directly converting KMBA to MTL for both compounds contain C-S bond11,12,13. While CYS3 only exited in the genome of S. cerevisiae, its homologue in C. rosea were not obtained after searching C. rosea genome13. Recombinant S. cerevisiae CYS3 and STR3 expressed and purified from E. coli displayed a broad specificity toward cysteine-S-conjugates except Met13,14. To date, no gene has been identified for the conversion of Met and its transamination product KMBA into MTL.
Methioninases, catalyzing the one-step degradation of Met to MTL, α-ketobutyrate (α-KB) and ammonium, have received much attention as therapeutic agents against various Met-dependent tumors and aging15,16,17,18,19,20. The therapeutic role is attributed to reducing Met concentration in media deprived tumor cell of dependence on external Met, and trigger an “amino acid response” to allow normal cells to adapt to their environment16,17,19,21. More importantly, methioninase transfected into cancer cells evinced either moderate cell death or severe aggregation18. However, these methioninases are of bacterial origin, and usually associated with high immunogenicity, low substrate specificity, and hazardous effects on the kidney and liver17,22,23,24. The proposed methioninase in fungi needs to be functionally defined for its potential to overcome these defects.
In this study, we located the biodegradation pathway of Met, and then used genetic, biochemical and metabolite–based analysis to prove that STR3 has the ability for one-step degradation of Met and its transamination product KMBA in Clonostachys rosea. Our findings support an effective approach against various Met-dependent tumors and aging.
Results
KMBA, the intermediate for Met-MTL conversion, is located intracellularly
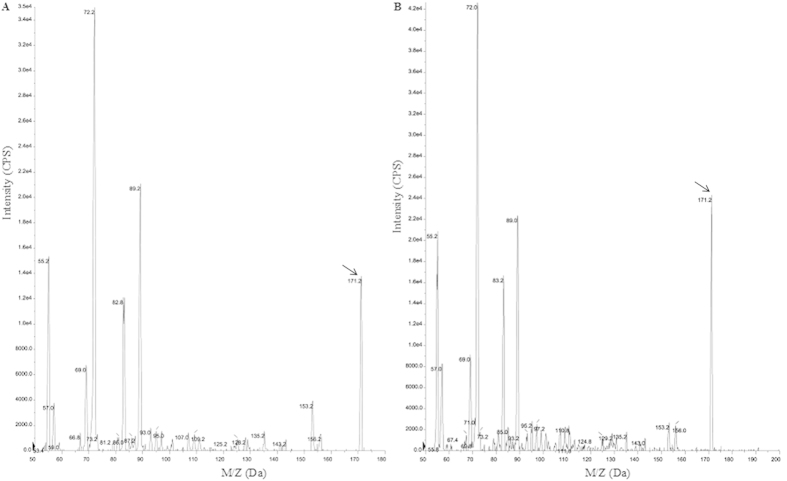
A hypothesis of KMBA-MTL conversion by physicochemical or enzymatic degradation of Met transamination product KMBA was proposed6,7,8. The extracellular or intracellular location of KMBA is a key indicator of KMBA-MTL conversion6. Therefore, we investigated 30 fungal strains in our laboratory to detect KMBA. The possible intracellular KMBA was only found in C. rosea Tang19, liquid chromatography and LTQ-Orbitrap MS analyses demonstrated that the metabolite showed similar molecular weight as KMBA (Fig. 1A,B). Subsequently, intracellular and extracellular KMBA was detected by high-performance liquid chromatography for a period of 7 days. After 1 day, Met degradation resulted in the intracellular accumulation of KMBA. With longer incubation time, KMBA was found to be accumulated intracellularly but was undetectable in extracellular samples (Fig. 2). MTL and its derivatives were also detected during fermentation. Additionally, we did not observe the production of MTL from KMBA after 200 μM KMBA was added into the fermentation broth with mycelia removed (data not shown). These results indicate that KMBA is the intermediate of Met degradation inside cells, suggesting that intracellular Met-MTL conversion pathway existed in C. rosea.
Figure 1. Intracellular KMBA was identified by LC-MS.
(A) LC-MS analysis of standard KMBA, (B) LC-MS analysis of intracellular KMBA.
Figure 2. KMBA was located exclusively intracellularly in C. rosea.
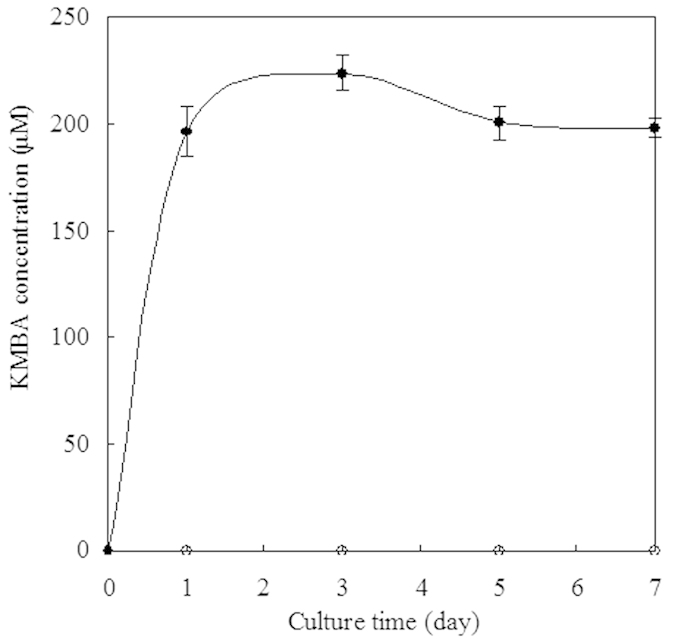
(●intracellular KMBA, ○extracellular KMBA).
ARO8-2, BAT and STR3 are putative enzymes for Met-MTL conversion
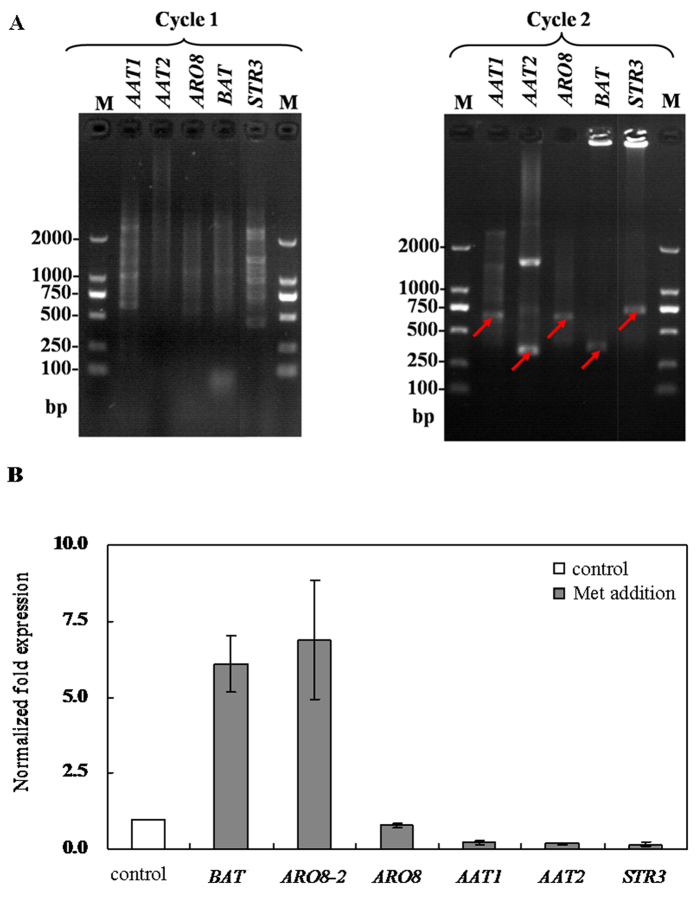
Aminotransferase and demethiolase are involved in Met-MTL conversion in Ascomycetes especially Saccharomycetes. To explore the intracellular conversion pathway in C. rosea, we mined Kyoto Encyclopedia of Genes and Genomes (KEGG) to retrieve the sets of aminotransferase and demethiolase, and conducted multiple alignments of amino acid sequences by ClustalX. Based on the analysis of the highly conserved regions, degenerate PCR primers were designed for cloning Met-MTL pathway gene homologs in C. rosea (Table 1). After two rounds of degenerate PCR, we found three groups of orthologous genes encoding aminotransferase, including aspartate amino acid aminotransferase genes AAT1 and AAT2, aromatic acid aminotransferase genes ARO8 and ARO8-2, and branched-chain amino acid aminotransferase genes BAT (Fig. 3A). By using NCBI blastX, the coding DNA sequences were aligned and the analysis results showed that AAT1, AAT2, ARO8, ARO8-2 and BAT were 71%, 81%, 88%, 77% and 78% identical to the previously reported homologous genes of Sordariomycetes at amino acid level. Another gene ortholog encoding demethiolase was also successfully amplified, which showed 87% identity to STR3 at the amino acid level (Fig. 3A). Subsequently, full-length cDNAs containing these Met-MTL pathway genes were isolated from cDNA library by RACE (rapid-amplification of cDNA ends) technology.
Table 1. Primers for gene cloning and expression.
| Genes | Primers | Sequence |
|---|---|---|
| Homology-based cloning | ||
| AAT1 | AAT1-5F | 5′-ATHAAYYTNGGNGTNGGNGCNTA-3′ |
| AAT1-3R | 5′-NARNCCNGTRTANGCRAACAT-3′ | |
| AAT1-3R-22 | 5′-CCCATRTTYTTNGCRAANSWYTG-3′ | |
| AAT2 | AAT2-5F | 5′-YTNGGNATHGGNGCNTAGMGNGA-3′ |
| AAT2-3R | 5′-DATNCKNCCNSWCATHGTNC-3′ | |
| AAT2-5F-22 | 5′-CAYGCNTGYGCNCAYAAYCCNAC-3′ | |
| AAT2-3R-22 | 5′-GGNGGRTTNSWDATYTCNSWNC-3′ | |
| ARO8 | ARO8-5F2 | 5′-ATHWSNYTNGGNGGNGGNYTNCC-3′ |
| ARO8-3R-23 | 5′-RAACCANSWNCCNCKNGCNAC-3′ | |
| ARO8-4R | 5′-RTARTANGGYTCRTCYTCDATDAT-3′ | |
| BAT | BAT-5F1 | 5′-TAYGCNACNGARTGYTTYGARGG-3′ |
| BAT-3R1 | 5′-RTCNARNGGNGCNGTDATNA-3′ | |
| BAT-5F2 | 5′-GNAARYTNMGNYTNTTYMGNCC-3′ | |
| BAT-3R2 | 5′-RTARTTNGCNCCNACYTT-3′ | |
| STR3 | STR3-5F | 5′-MGNWSNGGNAAYCCNACNMG |
| STR3-3R | 5′-ARNSWRTTNACRCANCCRAA-3′ | |
| STR3-3R2 | 5′-NCCNGTYTCRAANSWNARNACNGC-3′ | |
| Full-length cDNA cloning | ||
| AAT1 | AAT1-3RA1 | 5′-CCCTCGACAGGCTCGCCATTACCCAG-3′ |
| AAT1-3NRA2 | 5′-GTGCTGAGTTCCTTCAGCGCTGGTACAG-3′ | |
| AAT1-5RA1 | 5′-CACTGGTCCTGGCTGGGGTCAACA-3′ | |
| AAT1-5NRA2 | 5′-GAGGAGTGTTCTTGAGGTCCTCAACCAG-3′ | |
| AAT2 | AAT2-3RA1 | 5′-GAGCTGCTCATCGCCCAGAGCTTC-3′ |
| AAT2-3NRA2 | 5′-GTGCTGGCTGCTTCCACGCCATCACCTC-3′ | |
| AAT2-5RA1 | 5′-CTCACTCATTAGGCACCCCAGGC-3′ | |
| AAT2-5NRA2 | 5′-CACAGGAAACAGCTATGACCATGATTAC-3′ | |
| ARO8 | ARO8-3RA1 | 5′-TGACCTCTCCATCGCCCTCAACTAC-3′ |
| ARO8-3NRA2 | 5′-GAACACACCGAGCTGGTCTGCCGCCCTC-3′ | |
| ARO8-5RA1 | 5′-TCGTCCATTGCCTCGGGGATGAGTC-3′ | |
| ARO8-5NRA2 | 5′-GAACATGTTGATTCCAAGAGGGGCGAATG-3′ | |
| ARO8-2 | ARO8-23RA1 | 5′-AGCTCCATGCAGCAGCGAGACCTTC-3′ |
| ARO8-23NRA2 | 5′-CCAGCCGGACTGGCTCATCCCTCAAGGGTG-3′ | |
| ARO8-25RA1 | 5′-AGCGCAGAAGCTGGGCTGAGCCAC-3′ | |
| ARO8-25NRA2 | 5′-CACTCTGGCCGTCTCGGAGATCGTGCTTG-3′ | |
| BAT | BAT-3RA1 | 5′-GAGCAGCGCGGATACTCCCTCTAC-3′ |
| BAT-3NRA2 | 5′-CCAGGCTCAGCCCTCCTCTATGTTATTG-3′ | |
| BAT-5RA1 | 5′-GTGACGTACTGCTCCTCGCCGAAG-3′ | |
| BAT-5NRA2 | 5′-CAGGTTCTGCTGGTGGCCCCTGGAGCTG-3′ | |
| STR3 | STR3-3RA1 | 5′-TCCTCGAAACACCTACAAACCCTC-3′ |
| STR3-3NRA2 | 5′-CCAACAATCGCCCGGAATGCCCACGAAG-3′ | |
| STR3-5RA1 | 5′-GAATGGCAAGAGTCTTGACTCCTC-3′ | |
| STR3-5NRA2 | 5′-TCATTGGGTGACAGACCGCAGCCAGTTG-3′ | |
| STR3 expression in S. cerevisiae | ||
| STR3 | YSTR3-5H | 5′-CCCAAGCTTAACACAATGTCTGCCCCGCCTCCGCCAAATG-3′ |
| YSTR3-3BH | 5′-CGCGGATCCTTAGTGGTGGTGGTGGTGGTGTTTGGCTGTGCGTGGAGTTCGTC-3′ | |
| F5 | 5′-AGTATCAACAAAAAATTGTTA-3′ | |
| R3 | 5′-GAATGTAAGCGTGACATAACTAA-3′ | |
The restriction enzyme sites are underlined, and the His-tag encoding sequences are in bold.
Figure 3. Role of ARO8-2, BAT and STR3 in Met-MTL conversion.
(A) Met-MTL conversion pathway genes obtained by homology-based cloning, (B) Transcriptional analysis of putative pathway genes (Normalized fold expression values for synthase genes were relative to the control without Met addition. The error type was standard deviation).
To better understand the role of these putative genes involved in Met-MTL conversion in C. rosea, we assessed the changes in gene transcriptional expression patterns after addition of 5 g/L Met into the culture media. The addition of Met resulted in upregulation of aminotransferase genes BAT and ARO8-2 by 6.09 and 6.89 folds, while downregulation of aspartate amino acid aminotransferase genes AAT1 and AAT2, and demethiolase gene STR3 by 4.55, 5.88 and 6.25 folds, respectively (Fig. 3B). No obvious change was found in aromatic acid aminotransferase gene ARO8 (Fig. 3B). These results were consistent with the observations in Yarrowia lipolytica and Kluyveromyces lactis that MTL biosynthesis was initiated by the upregulation of branched-chain amino acid aminotransferase genes and aromatic acid aminotransferase genes9,25. However, in contrast to the studies showing the effects of Met addition on transcriptional expression of STR3 in Kluyveromyces lactis and Yarrowia lipolytica8,9, we found that MTL production induced by Met addition was accompanied with reduction of STR3, suggesting that STR3 may be a rate-limiting enzyme controlling Met-MTL conversion in C. rosea.
STR3 is inferred as rate-limiting enzyme controlling Met-MTL conversion
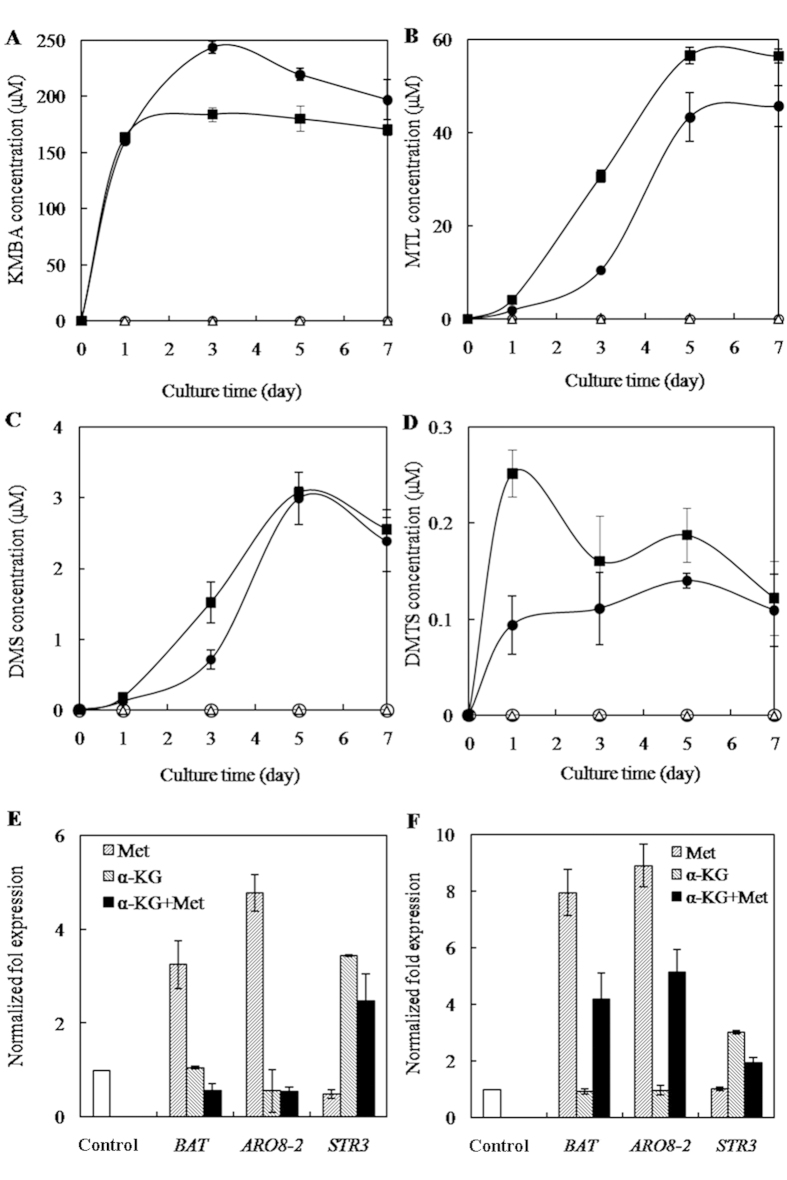
MTL production was closely positively correlated with the transcriptional expression of Met-MTL pathway genes6,7,8,9,26. In addition, a previous study showed that addition of surrogate substrate α-ketoglutaric acid (α-KG) stimulated MTL production28. To further identify synthase genes involved in Met-MTL conversion in C. rosea, we added α-KG into fermentation media and analyzed the mRNA expression changes of possible pathway genes. The addition of α-KG alone induced the upregulation of STR3, but MTL and its derivatives were not detected in the fermentation media, which is due to the limited intracellular Met. Relative to L-Met addition alone, the incubation of L-Met with α-KG for 3 day reduced the production of KMBA (Met aminotransferation product) by 24.46%, and the mRNA levels of aminotransferase genes BAT and ARO8-2 by 5.80 and 8.85 folds (Fig. 4A,E). However, the production of MTL and its derivatives DMS and DMTS was increased by 193.58%, 114.08% and 45.45%, and the demethiolase gene STR3 was correspondingly upregulated by 5.06 fold (Fig. 4B–E). Similar effects of α-KG addition on the transcriptional expression of BAT, ARO8-2 and STR3, and MTL production were observed after 5 days of incubation (Fig. 4F). These results demonstrated that the addition of α-KG inhibited the transaminase activity but stimulated the demethiolase activity. Correlation of the upregulation of STR3 with the increased production of MTL and its derivatives even with the downregulation of BAT and ARO8-2 indicated that STR3 may be the key enzyme controlling Met-MTL conversion.
Figure 4. Functional analyses of STR3 in Met-MTL conversion.
(A–D) Effect of α-KG addition on the production of KMBA (A), MTL (B), DMS (C) and DMTS (D) (■The addition of α-KG with Met, ●The addition of Met, □The addition of α-KG, ○Control), (E,F) Effect of α-KG addition on transcriptional expression of Met-MTL conversion pathway genes on days 3 (E) and 5 (F) (Normalized fold expression values for synthase genes were relative to the control without Met addition. The error type was standard deviation).
STR3 catalyzes direct conversion of both Met and KMBA into MTL
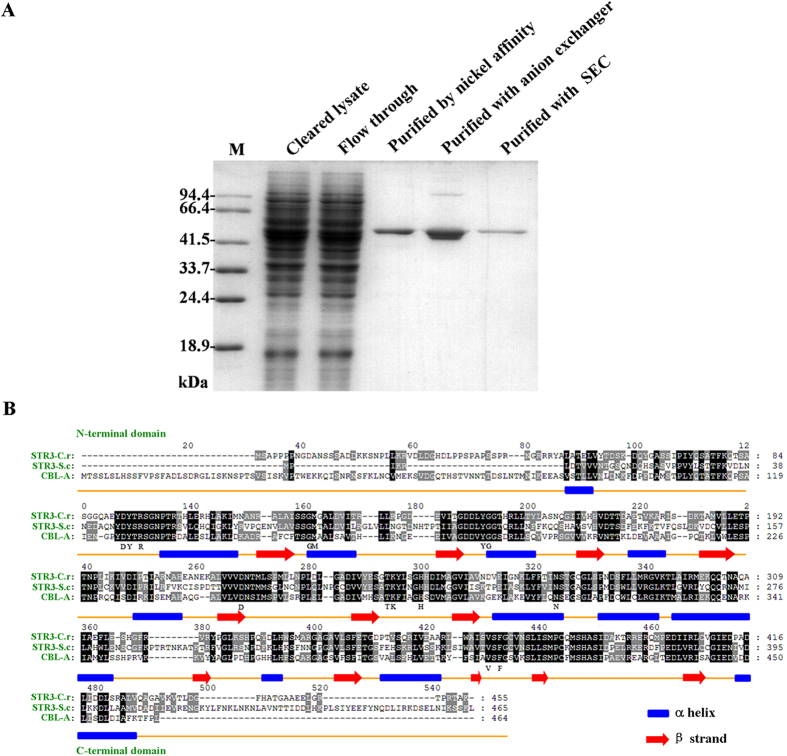
Recombinant STR3 was expressed in S. cerevisiae and purified using Ni-nitrilotriacetic acid chromatography to capture the C-terminal six-histidine-tagged protein27. Subsequently, nearly pure STR3 was obtained by anion-exchange column and size-exclusion chromatography (Fig. 5A). Monomeric recombinant STR3 has a predicted molecular weight of 49 kDa, as determined by migration on an SDS-PAGE gel (Fig. 5A). The identity of the purified protein as the STR3-encoded protein was confirmed by MALDI-TOF MS (E value of 2.3e−43, sequence coverage 56%).
Figure 5. Functional analyses of STR3 in vitro.
(A) SDS-PAGE of purified STR3, SEC: size exclusion chromatography; (B) Partial sequence alignment of STR3 with other demethiolases. Number is provided for aCBL, the catalytic residues (Y181, D253, K278), binding sites related to PLP (Y127, R129, G157, M158, T277), substrate binding sites (D126, G182, N307, V404, F406) in aCBL are indicated, STR3-C.r: C. rosea STR3(KT157524), STR3-S.c: S. cerevisiae STR3 (EDZ72299), CBL-A: Arabidopsis CBL (NP_191264).
To further support our conclusion that STR3 is a rate-limiting enzyme controlling Met-MTL conversion, Met and its transamination product KMBA were used as substrates for STR3 in biochemical reactions. Except the controls, MTL was detected in reaction systems either with Met or KMBA, indicating that STR3 has the ability to convert Met and KMBA to MTL. STR3 had a Km of 196.37 mM for Met (Vmax = 0.034 μM·min−1) and 16.68 mM for KMBA (Vmax = 1.56 μM min−1), showing that STR3 can degrade Met and KMBA, but preferred the substrate KMBA to its precursor Met. This suggests that one-step degradation of Met into MTL exists, but the conversion of KMBA to MTL is the primary pathway in C. rosea.
STR3 protein contains 455 amino acids with 49% and 51% identity to Arabidopsis CBL (cystathionine β-lyase) and S. cerevisiae STR3, respectively. Both CBL and STR3 are PLP-dependent and selectively cleave the covalent bond at β- or γ-position of sulfur-containing amino acid substrates29. Based on Arabidopsis CBL structure30, STR3 is predicted to have an N-terminal PLP binding domain and similar binding or catalytic site residues as CBL (Fig. 5B).
PLP dependence and cystathionine specificity
To validate that PLP is a cofactor in STR3 activity, different molar ratios between PLP and STR3 in enzymatic reactions were investigated to determine the demethiolation activity of STR3. Demethiolation activity was detected even without exogenous PLP addition, but the addition of exogenous PLP significantly enhanced STR3 activity (Table 2), which indicated that purified STR3 may carry intracellular PLP. This result was confirmed by detecting the characteristic absorbance of PLP at 420 nm31.
Table 2. Effect of cofactor PLP levels on STR3 activity.
| Molar ratio (PLP/STR3) | Relative activity (%) |
|---|---|
| 0 | 100 |
| 0.5 | 132.6 ± 1.7 |
| 1 | 137.4 ± 2.5 |
| 2 | 140.7 ± 2.8 |
Specific activity toward KMBA was 0.53 ± 0.0030 μM MTL·h−1·mg protein−1. The relative activity of demethiolation of KMBA to MTL was set at 100%.
The other sulfur-containing amino acids with Cβ, γ-S bonds were further assayed as potential substrates for the STR3 enzyme (Table 3). Thionitrobenzoic acid derivatives produced by the reaction between thiol and 5, 5′-dithiobis-2-nitrobenzoic acid were detected in all reactions, suggesting that STR3 has β, γ-lyase activity with a broad range of substrate specificity. Furthermore, in all tested sulfur-containing amino acids, L-cystathionine was the most effective physiological substrate for STR3 (Table 3). It indicated that STR3 catalyzed the conversion of cystathionine to homocysteine, the precursor of Met.
Table 3. Substrate specificity of purified STR3.
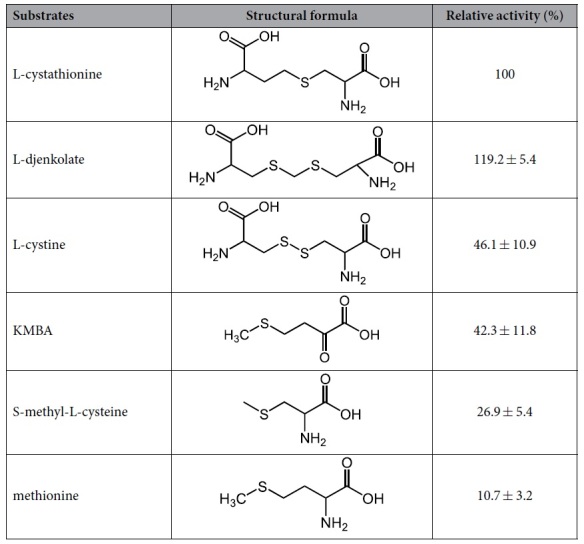
Demethiolation activities with different substrates were determined in the standard enzyme assay system. Specific activity toward L-cystathionine was 22.04 ± 2.40 mM MTL·min−1·mg protein−1. The relative activity of demethiolation of L-cystathionine to thiol was set at 100%.
Discussion
In this study, we demonstrated that KMBA, the intermediate of Met-MTL conversion, was located intracellularly and STR3 is the key enzyme controlling Met-MTL conversion in C. rosea. Importantly, biochemical analysis with purified recombinant STR3 protein showed that STR3 catalyzed the direct conversion of both Met and KMBA to MTL (Fig. 6). Our findings indicate that STR3 is the key demethiolase controlling Met-MTL conversion in C. rosea.
Figure 6. The Met-MTL conversion pathway proposed in the present study.
The blue marks indicate proposed functions of STR3 in the present study. After Met addition, KMBA as a Met degradation intermediate was localized intracellularly; BAT and ARO8-2 are activated, but STR3 was repressed accompanied with MTL production; STR3 catalyzes direct conversion of Met and its transaminated product KMBA into MTL. VOSCs: volatile organic sulfur-containing compounds including 3-(methylthio)propanal, 3-(methylthio)propanol and MTL, DMS, DMTS.
The MTL branch of Met degradation pathways represents the basic metabolism in eukaryotes based on gene annotation studies11,26,32. Previous studies demonstrated that KMBA was located extracellularly and STR3 possessed no activities towards Met, which rule out the possibility of intracellular Met-MTL conversion pathway inside eukaryotes6,8,14,26. Therefore, the precise Met-MTL conversion pathway remains unclear. Our genetic, biochemical and metabolic studies demonstrated that KMBA is located intracellularly and STR3 is a key demethiolase in Met-MTL conversion in Clonostachys rosea, unraveling the complex metabolic pathway of Met in eukaryotes.
Aminotransferases involved in the conversion of Met into MTL are highly conserved in fungi5,6,8,10,26. Upregulation of branched-chain amino acid aminotransferase gene BAT1 and aromatic acid aminotransferase gene ARO8 led to higher MTL production6,8,9,27,33. However, the functional role of demethiolase STR3 in the process of MTL biosynthesis is still controversial as different studies reported contrasting results when fermentation strains were inoculated into different meida9,14,33. In this study, addition of Met triggered the downregulation of demethiolase gene STR3, which is consistent with the result in Yarrowia lipolytica9,33. In contrast, the addition of Met together with α-KG reversed the effect of Met alone, and the elevated STR3 expression was accompanied with increased MTL production, suggesting that STR3 is a rate-limiting enzyme controlling the conversion of Met into MTL.
Biochemical analyses indicated that STR3 exhibited the ability for one-step degradation of Met and its transamination product KMBA, revealing a novel enzymatic role distinct from its previously identified features as a eukaryotic family member. Met restriction suppressed cancer cell metabolism and extended lifespan of normal cell15,19,20,21. Due to the low immunogenicity of C. rosea STR3, enzymatic or dietary depletion of Met in vivo makes STR3 an ideal therapeutic candidate against various types of Met-dependent tumors and aging17,18,34,35,36. These PLP-dependent enzymes share an appreciable degree of sequence homology and exhibit similarities in their three-dimensional structures37,38. Amino acid residues at the enzyme active site rather than the structural fold control substrate specificity and reaction type29,39. C. rosea STR3 has similar amino acid residues at the enzyme active site, substrate binding site and cofactor binding site as Arabidopsis CBL. Further analysis of the different amino acid sites of STR3 is required to understand the substrate specificity for Met. Additionally, STR3 is a cystathionine β-lyase (CBL) with a higher catalytic efficiency for L-cystathionine than other physiological substrates. In this catalytic reaction, cystathionine is cleaved by STR3 to produce homo-Cys, the direct precursor of Met in eukaryotes2,30. STR3 expression repressed by Met addition may be related with feedback effects. Thus, STR3 is a potential key target for the biosynthesis of Met and MTL in eukaryotes by metabolic engineering40,41.
In conclusion, STR3 displayed demethiolase activities to Met and its transamination product KMBA. Together with genetic and metabolite data, we demonstrated that the direct conversion of Met and its transamination product KMBA into MTL controlled by STR3 occurs in C. rosea. Our findings provide a rationale for Met depletion in vivo or dietary supplementation of methioninase in cancer therapy and anti-aging, and the regulation of essential amino acid Met content and volatile flavor constituent MTL in plants and yeast, respectively.
Methods
Chemicals
Chemical standards of MTL and its derivatives DMS and DMTS with over 98% purity were purchased from J&K Scientific Ltd (Beijing, China). All analytical grade reagents for STR3 enzymatic assay were purchased from Biosharp (Shenzhen, China), Tokyo Chemical Industry (Shanghai, China) and Sigma-Aldrich (St. Louis, MO).
Microbial strains, media, and culture conditions
Chemically competent E. coli DH5α cells (TransGen Biotech, Beijing, China) were used for amplification of pYES2 plasmid (Invitrogen). Growth and selection were carried out in Luria-Bertani medium supplemented with 100 mg/L ampicillin. Commercial S. cerevisiae strain INVSc1 (Invitrogen) was used as a host strain for the expression of STR3. Yeasts were cultivated at 30 °C in either a rich medium YPD (Yeast Extract Peptone Dextrose medium, containing 1% (w/v) yeast extract, 2% peptone, and 2% glucose), or a synthetic minimal defined medium SC (synthetic complete medium, containing 2% glucose or raffinose, 0.67% yeast nitrogen base without amino acids (Biosharp)). Uracil (SC-U) was omitted to obtain selective plates for growing pYES2 transformants.
Thirty isolated fungal strains were grown on potato-agar-dextrose slants, precultured in fermentation medium (sucrose, 35 g/L; peptone, 2.5 g/L; yeast extract, 2.5 g/L; KH2PO4 ·H2O, 1 g/L; MgSO4·7H2O 0.5 g/L; Vitamin B1, 0.05 g/L), and then their ability to degrade Met was tested by inoculating them in fermentation medium added with 5 g/L Met. Met transamination product KMBA was only detected in the culture inoculated with Clonostachys rosea Tang 19, which was identified by ITS sequence (Genbank accession number: KT007105). This strain was selected for further study.
Extraction and identification of KMBA by LC-MS
Intracellular KMBA was extracted as follows6. Fungal mycelia cultured in fermentation medium with 5 g/L Met were collected by filtration, washed with 20 mL of cold ultrapure water, and then filtered again. Mycelial pellets were stored at −80 °C. The pellets were resuspended with 1 mL of 1% (v/v) formic acid, incubated for 10 min at 95 °C and centrifuged for 30 min at 4 °C. The supernatant was lyophilized and stored at −80 °C. Before injection, samples were resuspended in water containing 0.1% formic acid and stored at 4 °C. Chromatographic separation was performed on a Reprosil-Pur Basic C18 column (4.6 mm × 250 mm × 5 μm) from Dr. Maisch GmbH (Germany) using a liquid chromatography 20AD system (Shimadzu Corporation, Japan). The optimized mobile phase was 0.1% formic acid water solution. The column oven temperature was set to 30 °C, and the flow rate was 1 mL/min. The detection wavelength was 197 nm. The retention time for KMBA was 12.5 min. For the determination of extracellular KMBA, 1 mL fermentation broth was centrifuged at 13,000 g for 30 min and then filtered through a 0.22 μm filter prior to detection.
Chromatographic purification of KMBA was carried on Amethyst C18-H column (10 mm × 250 mm × 5 μm) from Sepax Technologies (Newark, DE). Targeted samples were identified by comparison with the standard compounds, followed by tandem mass spectrometry (MS/MS) on a Finnigan TSQ Quantum Ultra AM instrument (Thermal, USA). The metabolites were confirmed by standard MS using positive electrospray ionization and scanned in the normal mass range from 50 m/z to 180 m/z. Characteristic fragment ions were detected by second-order MS.
Cloning of Met-MTL pathway genes
The coding sequences of Met-MTL pathway genes including aminotransferase genes AAT1, AAT2, BAT and ARO8, and demethiolase gene STR3 of C. rosea Tang 19 were amplified by PCR using degenerated forward/reverse primers (Table 1) designed on the basis of conserved regions of fungi in the previously reported sequences5,6,9. The full-length cDNAs encoding Met-MTL pathway genes were obtained by 5′ and 3′ RACE using nested primers (Table 1). The prediction of open reading frames (ORF) of these genes was conducted using the program ORF finder at the NCBI web site. Genbank numbers of AAT1, AAT2, BAT, ARO8, ARO8-2 and STR3 were KT157525, KT157520, KT157521, KT157522, KT157523, KT157524, respectively.
Effect of Met and α-KG addition on the conversion of Met to MTL
The C. rosea culture was initiated by the addition of 0.5% (w/v) L-Met or 0.098% (w/v) α-KG (Sigma-Aldrich) with respective controls included. After culturing for 3 to 5 days in the fermentation medium, C. rosea mycelia were collected and the transcriptional expression of Met-MTL pathway genes was analyzed by quantitative real-time PCR. RNA was isolated using the MasterPure Fungal RNA Purification Kit (Biozym Scientific GmbH). RNA concentration and purity (A260/A280 ratio) were measured by using the Take3 micro-volume plate of BioTek Epoch Microplate Spectrophotometer. Complementary DNA (cDNA) was synthesized using a PrimeScriptTM 1st Strand cDNA Synthesis Kit (TaKaRa Biotechnology (Dalian) Co., Ltd) with 0.5 μg of total RNA as the template. The 18S rRNA served as the reference gene for normalization of gene expression. Quantitative real-time PCR was performed on a StepOnePlus Real-Time PCR system in duplicate for at least three independent experiments. SYBR dye (TaKaRa Biotechnology (Dalian) Co., Ltd) was used to visualize gene amplification. The internal primers for RACE cloning were used to amplify the target sequence from different Met-MTL pathway genes (Table 1).
Met catabolic products including KMBA, MTL and its derivatives DMS and DMTS were determined after the cultures were incubated for 1, 3, 5 and 7 days. KMBA was extracted and detected following the above mentioned method. MTL and its derivatives were extracted and detected as follows. A 5-mL volume of fermentation culture was added into a 15-mL vial, the vial was tightly capped with a silicon septum and pre-equilibrated at 40 °C for 5 min, and then solid-phase microextraction (SPME) device was inserted into the headspace of vial to extracte MTL and its derivatives for 20 min at 40 °C. MTL and its derivatives were analyzed by gas chromatography (GC-2010 with a flame ionization detector (FID) from Shimadzu Technologies Inc., Tokyo, Japan) with an oven temperature program. First, the temperature was maintained at 35 °C for 3 min. Subsequently, the temperature reached 100 °C, with an increment of 8 °C/min. Then, the temperature was raised to 220 °C, with an increment of 10 °C for 1 min. Three independent cultures were analyzed for each condition.
Expression and purification of STR3
The isolation and manipulation of DNA from C. rosea were performed using E.Z.N.A Fungal DNA kit (Omega) according to the manufacturer’s protocol. Primer pairs YSTR3-5H and YSTR3-3BH were designed to amplify the complete ORF of STR3 from chromosomal DNA of C. rosea (Table 1). HindIII (in the forward primer) and BamHI (in the reverse primer) restriction sites were introduced to ensure that STR3 was correctly inserted into yeast expression plasmid pYES2. For purification of STR3, a His-tag encoding fragment was included before the stop codon (TTA). After the PCR product was cloned into pYES2, the resulting plasmid, pYES2-STR3, was transformed into S. cerevisiae strain INVSc1 as described24. Transformants were selected in SC medium lacking uracil (SC-U) and further confirmed by colony PCR with universal primers of F5 and R3.
After 20 h of galactose induction, yeast cultures were pelleted by centrifugation at 2,000 g for 5 min at 4 °C. Intracellular proteins were extracted by vigorously grinding the pelleted yeast cells cooled in liquid nitrogen. The lysates were centrifuged at 13,000 g for 1 h at 4 °C and filtered through a 0.22 μm (Millipore) filter. The resulting clear lysate was then exposed to nickel affinity chromatography using a 1-mL Ni-Sepharose 6 Fast Flow matrix according to the manufacturer’s instructions (GE Lifesciences). STR3 was purified by washing with 3 column volumes of NiA buffer containing 50 mM Tris, pH 8.5, 250 mM imidazole.
The collected sample was loaded on a Mono Q 10/100 GL (GE) equilibrated with buffer A (pH 7.5, 30 mM Tris-HCl, 15 mM NaCl, 1 mM EDTA, and 2 mM dithiothreitol). Only one peak was detected by using a linear gradient method, and was eluted with 516 mM NaCl. Subsequently, Size exclusion chromatography was carried out with a Superdex 200HR 10/30 analytical column (GE Lifesciences) using an AKTATM Pure 25 (FPLC, Pharmacia/GE Lifesciences). The flow rate was 0.4 mL/min. Proteins were separated by electrophoresis on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Identification of STR3
STR3 resolved by SDS-PAGE was subjected to a trypsin digestion and analyzed by the 4700 Proteomics Analyzer MALDI-TOF/TOF (Applied Biosystems, Framingham, MA). Based on the combined MS and MS/MS spectra, STR3 was identified based on a 95% or higher confidence interval of their scores in the MASCOT V2.0 search engine (Matrix Science, London, U.K.). Functional roles of STR3 as demethiolase involved in the conversion of Met to MTL were measured using Met and its transamination product KMBA as substrates. Enzymatic reaction was conducted in a total reaction volume of 5 mL containing a final concentration of 1 mg/mL of STR3, 50 mM Tris-HCl (pH8.0), 5 μM PLP and 20 mM of Met or KMBA. Reaction mixtures were incubated for 1 h at pH 8.0. MTL production via demethiolation of Met and KMBA was determined by GC according to the method mentioned above. Control experiments referred to the complete assay with either omission of STR3 or with denatured STR3 (boiled for 10 min) and MTL and its derivatives were not detected after enzymatic reaction. Meanwhile, different concentrations of cofactor PLP (pyridoxal-5′-phosphate) were added to the standard enzyme assay mixture to investigate the effect of cofactor on STR3 activity toward KMBA. Demethiolating activities of STR3 were measured using the other sulfur-containing amino acids including S-methyl-L-cysteine, L-cystine, L-cystathionine and L-djenkolate as the substrates, as previously described13. After reactions were conducted in a total volume of 1 mL at pH 8.0 and 30 °C for 15 min, thionitrobenzoic acid derivatives produced by the reaction between thiol and 5, 5′-dithiobis-2-nitrobenzoic acid were measured spectrophotometrically at 412 nm. All the experiments were performed in triplicate.
Additional Information
How to cite this article: Jia, K.-Z. et al. Clonostachys rosea demethiolase STR3 controls the conversion of methionine into methanethiol. Sci. Rep. 6, 21920; doi: 10.1038/srep21920 (2016).
Acknowledgments
Financial supports from the National Natural Science Foundation of China (NSFC, Project Nos. 21376066, 81503112, 21506049 and 31570054), and Hubei Provincial Natural Science Foundation for Innovative Research Team (2015CFA013) are gratefully acknowledged. Prof. Ya-Jie Tang also thanks the National High Level Talents Special Support Plan (“Million People Plan”) by the Organization Department of the CPC Central Committee (2014), Training Program for Top Talents in Hubei Province (2013), and Training Program for Huanghe Talents in Wuhan Municipality (2014).
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.J.T. and K.Z.J. conceived the project. Y.J.T. and K.Z.J. designed the experiments. K.Z.J., Q.Z., L.Y.S. and Y.H.X. implemented the analysis workflow and conducted the experiments. Y.J.T., K.Z.J. and H.M.L. analysed and interpreted the results. K.Z.J. and Q.Z. prepared all figures and Tables. Y.J.T. and K.Z.J. prepared and wrote the manuscript. All authors reviewed, commented on and approved the final manuscript.
References
- Hesse H. et al. Current understanding of the regulation of methionine biosynthesis in plants. J Exp Bot 55, 1799–1808 (2004). [DOI] [PubMed] [Google Scholar]
- Thomas D. & Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61, 503–532 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N. et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metabol 19, 780–794 (2014). [DOI] [PubMed] [Google Scholar]
- Binet R., Fernandez R. E., Fisher D. J. & Maurelli A. T. Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter. Mbio 2, e00051–00011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpete P. et al. Methionine catabolism in Saccharomyces cerevisiae. FEMS Yeast Res 6, 48–56 (2006). [DOI] [PubMed] [Google Scholar]
- Hebert A. et al. New insights into sulfur metabolism in yeasts as revealed by studies of Yarrowia lipolytica. Appl Environ Microbiol 79, 1200–1211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfi K., Landaud S. & Bonnarme P. Evidence for distinct L-methionine catabolic pathways in the yeast Geotrichum candidum and the bacterium Brevibacterium linens. Appl Environ Microbiol 72, 2155–2162 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert A. et al. Exploration of sulfur metabolism in the yeast Kluyveromyces lactis. Appl Microbiol Biotechnol 91, 1409–1423 (2011). [DOI] [PubMed] [Google Scholar]
- Cholet O., Henaut A., Hebert A. & Bonnarme P. Transcriptional analysis of L-methionine catabolism in the cheese-ripening yeast Yarrowia lipolytica in relation to volatile sulfur compound biosynthesis. Appl Environ Microbiol 74, 3356–3367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. et al. Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464, 1033–1038 (2010). [DOI] [PubMed] [Google Scholar]
- Hansen J. & Johannesen P. F. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol Gen Genet 263, 535–542 (2000). [DOI] [PubMed] [Google Scholar]
- Splivallo R., Ottonello S., Mello A. & Karlovsky P. Truffle volatiles: from chemical ecology to aroma biosynthesis. New Phytol 189, 688–699 (2011). [DOI] [PubMed] [Google Scholar]
- Yamagata S. et al. Cloning and bacterial expression of the CYS3 gene encoding cystathionine γ-lyase of Saccharomyces cerevisiae and the physicochemical and enzymatic properties of the protein. J Bacteriol 175, 4800–4808 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. et al. Engineering Saccharomyces cerevisiae to release 3-mercaptohexan-1-ol during fermentation through overexpression of an S. cerevisiae gene, STR3, for improvement of wine aroma. Appl Environ Microbiol 77, 3626–3632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strekalova E., Malin D., Good D. M. & Cryns V. L. Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing TRAIL receptor-2 expression. Clin Cancer Res (2015), 10.1158/1078-0432.CCR-14-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X. et al. Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet 11, e1005158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. et al. In vivo efficacy of recombinant methioninase is enhanced by the combination of polyethylene glycol conjugation and pyridoxal 5′-phosphate supplementation. Cancer Res 63, 8377–8383 (2003). [PubMed] [Google Scholar]
- Ventachalam K. V. Novel cancer therapy: Targeting methionine metabolism. FASEB J 29, supplement 897.832 (2015). [Google Scholar]
- Lee B. C. et al. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat Commun 5, 3592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandison R. C., Piper M. D. & Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckenstuhl C. et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet 10, e1004347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed A. S. Microbial L-methioninase: production, molecular characterization, and therapeutic applications. Appl Microbiol Biotechnol 86, 445–467 (2010). [DOI] [PubMed] [Google Scholar]
- Cantor J. R. et al. Therapeutic enzyme deimmunization by combinatorial T-cell epitope removal using neutral drift. Proc Natl Acad Sci USA 108, 1272–1277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov N. A. et al. Pre-steady-state kinetic and structural analysis of interaction of methionine gamma-lyase from Citrobacter freundii with inhibitors. J Biol Chem 290, 671–681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagkli D. M. et al. Kluyveromyces lactis and Saccharomyces cerevisiae, two potent deacidifying and volatile-sulphur-aroma-producing microorganisms of the cheese ecosystem. Appl Microbiol Biotechnol 73, 434–442 (2006). [DOI] [PubMed] [Google Scholar]
- Bondar D. C., Beckerich J. M. & Bonnarme P. Involvement of a branched-chain aminotransferase in production of volatile sulfur compounds in Yarrowia lipolytica. Appl Environ Microbiol 71, 4585–4591 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D. St Jean A., Woods R. A. & Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20, 1425 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. W. et al. Volatile sulphur compounds and pathways of L-methionine catabolism in Williopsis yeasts. Appl Microbiol Biotechnol 95, 1011–1020 (2012). [DOI] [PubMed] [Google Scholar]
- Eliot A. C. & Kirsch J. F. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem 73, 383–415 (2004). [DOI] [PubMed] [Google Scholar]
- Breitinger U. et al. The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity. Plant Physiol 126, 631–642 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry-Weeks C. R., Keith J. M. & Thompson J. Toxicity of Bordetella avium beta-cystathionase toward MC3T3-E1 osteogenic cells. J Biol Chem 268, 7298–7314 (1993). [PubMed] [Google Scholar]
- Howell K. S. et al. Genetic determinants of volatile-thiol release by Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol 71, 5420–5426 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholet O., Henaut A., Casaregola S. & Bonnarme P. Gene expression and biochemical analysis of cheese-ripening yeasts: focus on catabolism of L-methionine, lactate, and lactose. Appl Environ Microbiol 73, 2561–2570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K. et al. Methioninase gene therapy of human cancer cells is synergistic with recombinant methioninase treatment. Cancer Res 60, 2696–2702 (2000). [PubMed] [Google Scholar]
- Vander Heiden M. G. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 10, 671–684 (2011). [DOI] [PubMed] [Google Scholar]
- Maddocks O. D. et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansonius J. N. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol 8, 759–769 (1998). [DOI] [PubMed] [Google Scholar]
- Schneider G., Kack H. & Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure 8, R1–6 (2000). [DOI] [PubMed] [Google Scholar]
- Song H. G., Xu R. & Guo Z. H. Identification and characterization of a methionine gamma-Lyase in the calicheamicin biosynthetic cluster of Micromonospora echinospora. Chembiochem 16, 100–109 (2015). [DOI] [PubMed] [Google Scholar]
- Dharmendra Kumar J. G. Methionine production by fermentation. Biotechnol Adv 23, 41–61 (2005). [DOI] [PubMed] [Google Scholar]
- Galili G. & Amir R. Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol J 11, 211–222 (2013). [DOI] [PubMed] [Google Scholar]