Abstract
Study Objectives:
Sleep-wake disturbances are often reported in Prader-Willi syndrome (PWS), a rare neurodevelopmental syndrome that is associated with paternally-expressed genomic imprinting defects within the human chromosome region 15q11-13. One of the candidate genes, prevalently expressed in the brain, is the small nucleolar ribonucleic acid-116 (SNORD116). Here we conducted a translational study into the sleep abnormalities of PWS, testing the hypothesis that SNORD116 is responsible for sleep defects that characterize the syndrome.
Methods:
We studied sleep in mutant mice that carry a deletion of Snord116 at the orthologous locus (mouse chromosome 7) of the human PWS critical region (PWScr). In particular, we assessed EEG and temperature profiles, across 24-h, in PWScr m+/p− heterozygous mutants compared to wild-type littermates. High-resolution magnetic resonance imaging (MRI) was performed to explore morphoanatomical differences according to the genotype. Moreover, we complemented the mouse work by presenting two patients with a diagnosis of PWS and characterized by atypical small deletions of SNORD116. We compared the individual EEG parameters of patients with healthy subjects and with a cohort of obese subjects.
Results:
By studying the mouse mutant line PWScrm+/p−, we observed specific rapid eye movement (REM) sleep alterations including abnormal electroencephalograph (EEG) theta waves. Remarkably, we observed identical sleep/EEG defects in the two PWS cases. We report brain morphological abnormalities that are associated with the EEG alterations. In particular, mouse mutants have a bilateral reduction of the gray matter volume in the ventral hippocampus and in the septum areas, which are pivotal structures for maintaining theta rhythms throughout the brain. In PWScrm+/p− mice we also observed increased body temperature that is coherent with REM sleep alterations in mice and human patients.
Conclusions:
Our study indicates that paternally expressed Snord116 is involved in the 24-h regulation of sleep physiological measures, suggesting that it is a candidate gene for the sleep disturbances that most individuals with PWS experience.
Citation:
Lassi G, Priano L, Maggi S, Garcia-Garcia C, Balzani E, El-Assawy N, Pagani M, Tinarelli F, Giardino D, Mauro A, Peters J, Gozzi A, Grugni G, Tucci V. Deletion of the Snord116/SNORD116 alters sleep in mice and patients with Prader-Willi syndrome. SLEEP 2016;39(3):637–644.
Keywords: Prader-Willi, Snord116, sleep, temperature, theta rhythms
Significance.
In this study we identify a novel role of SNORD116/Snord116 in sleep disturbance in Prader-Willi syndrome (PWS). We observed abnormalities in REM sleep both in patients with PWS and in a mouse model of the syndrome, which lacks Snord116. We detected hippocampal alterations in mice that are in agreement with the functional deficit in sleep. PWS is caused by genomic imprinting defects; therefore our findings promote a significant new perspective in the investigation of sleep. This new work, along with previous evidence of parent-of-origin effects on sleep, provides support for a significant role of genomic imprinting in sleep regulatory mechanisms.
INTRODUCTION
Lack of expression of paternally imprinted alleles within the small nuclear ribonucleoprotein N (SNPRN) cluster of the human chromosome region 15q11-13 causes Prader-Willi syndrome (PWS),1 a rare neurodevelopmental disorder that is associated with growth retardation, metabolic abnormalities, hyperphagic behavior, cognitive deficits, and sleep abnormalities. The genetic defect can be quite heterogeneous across patients; it can include various genes and can result from uniparental disomy, deletions, or genomic imprinting deficits. Different patients present with different genetic alterations and recently it has been suggested that the syndrome can be associated with a disruption of the small nucleolar ribonucleic acid (RNA)- 116 (SNORD116, also called HBII-85) gene, a noncoding molecule that participates in the modifications of other small nuclear RNAs and is predominantly expressed in the brain.2 Indeed, a few clinical single cases of PWS with microdeletions within the q11.q13 domain of chromosome 15, including SNORD116, have been reported.3–6 SNORD116 appears to be necessary for PWS to manifest. In fact, within the Prader-Willi genomic region, deletions of MKRN3, MAGEL2, NECDIN,7 or SNORD1158,9 or C15ORF2 and SNURFSNRPN7,10 do not give rise to PWS.
Sleep disturbances represent a significant problem for individuals with PWS and their caregivers; however, this aspect of the syndrome has been poorly investigated to date. Clinical studies have reported hypersomnia, fragmented sleep and apnea,11–14 and abnormal sleep EEG features12,13,15–18 in patients with PWS. More in-depth analyses revealed alterations in the architecture of sleep,13–15 abnormal rapid eye movement (REM) sleep cycles,18 sleep onset REM periods (SOREMPs) and an increased number of REM episodes.12,13,15–17 In the investigations of sleep disturbances in PWS, the genotype of the patients has rarely been taken into account as a variable and, when considered, the details of the extension and type of deletion are not given. Nevertheless, the study of Vgontaz et al.,15 on a small group of patients, reported a higher prevalence of excessive daytime sleep (EDS) and SOREMPs in patients with PWS carrying deletions compared to patients with uniparental disomies.
The creation of several mouse genetic models has provided a valuable set of tools for understanding the genetic mechanisms of the syndrome. Surprisingly, none of the PWS mouse models available has been assessed for the presence of sleep disturbances.19 Therefore, in order to dissect the genetic mechanisms of sleep abnormalities in patients with PWS, investigations that report exact genotypic defects of patients with PWS with sleep disturbances and gene-specific translational models of the syndrome are needed. Here, we present a study in which we report remarkable analogies between the sleep features of the mouse mutant model PWScrm+/p−,2 which lacks Snord116, and two patients with PWS with atypical deletions, one of them carrying a particularly small (372 kb) deletion, inclusive of SNORD116. We further studied the mouse model and we observed a major reduction in the size of the hippocampus of mouse mutants, which may account for the EEG features we report here.
METHODS
Animal Husbandry
We investigated mice carrying a deletion (Figure 1) of the PWS critical region (PWScr) including Snord116 and IPW exons A1/A2, B, and C.2 In the Istituto Italiano di Tecnologia (IIT), mice were bred and maintained through paternal inheritance on a C57BL/6J background. All experiments were conducted in their home cages, the environmental temperature was 23° C. Mice were 8 to 9 mo old when tested because the phenotype is reported to be more evident with age both in patients20 and in mice with a deletion of Snord116.21 All animal procedures were approved by the ethical national committee in Italy, for IIT Genova. Mouse husbandry followed ARRIVE guidelines (http://www.nc3rs.org.uk/arrive-guidelines). For the genotyping assay see the supplemental material.
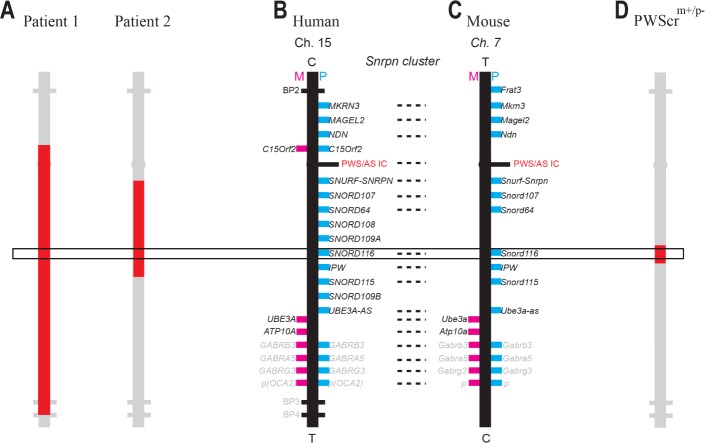
Figure 1.
Human SNRPN cluster and mouse orthologous Snrpn cluster (not to scale). (A) Representation of the deletion of Patient 1 and Patient 2. Patient 1 carries an atypical deletion with the proximal breakpoint at RP11-484P15 and the distal breakpoint located in the common BP4. The size of the deletion is 6, 27 Mb. Patient 2 exhibits a deletion with the proximal breakpoint at base 22,648,348 and the distal one at base 23,020,695 of chromosome 15. The size of the deletion is 372 Kb. (B) Human SNRPN cluster of chromosome 15 and (C) the orthologues Snrpn cluster of chromosome 7C in mice. Pink rectangles are maternally expressed genes, blue rectangles are paternally expressed genes; when both pink and blue rectangles are present, genes are expressed biallelically. Gene expression/imprinting profiles are reported only for the brain. C, centromere; T, telomere; note that genes are in the same order in human and mice chromosomes but in reverse orientation. Dashed line links homologues. (D) Representation of the deletion of the Prader-Willi syndrome critical region including Snord116 and IPW exons A1/A2, B, and C carried by the murine model PWScrm+/p− studied in this investigation.
In vivo EEG Recordings in Mice
Sleep and temperature profiles were investigated in 7 PWScrm+/p− mice and 7 PWScrm+/p+ littermate controls, all males. A wireless implant, as described in the study by Lassi et al.,22 was used (see supplemental material).
However, the transmitter receiving the EEGs was implanted subcutaneously and contained a sensor that detected peripheral body temperature. EEG measures allow defining three major states of mammalian life: wakefulness, rapid eye movement (REM), and nonrapid eye movement (NREM) sleep (Figure 2A). For each animal and then for each genotype we analyzed the percentage of time spent in sleep, NREM, and REM, and the number of NREM sleep episodes and REM sleep per hour, during 24 h, across light (L) and dark (D) phases (LD: 12:12). Unusual interruptions of wakefulness by REM sleep appeared evident while visually inspecting the physiological signals of PWScrm+/p− mice; we scored these REM sleep intrusions and quantified them. Moreover, we confirmed these wake-to-sleep transitions by video-scoring random episodes. The spectral characteristics of the EEG were further analyzed. EEG power density for delta (0, 5–4 Hz) and theta (5–9 Hz) frequencies in NREM and REM sleep, across 12 h of light and across 12 h of dark were calculated. Temperature values were collapsed in 3-h bins and compared between geno-types. See supplemental material for further details.
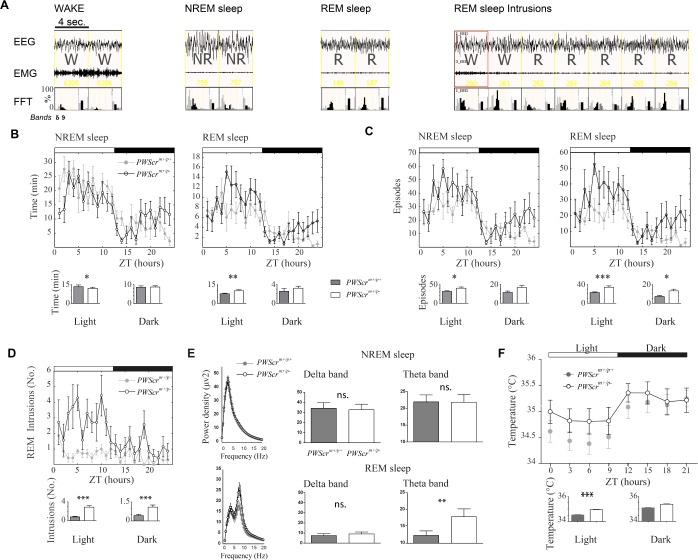
Figure 2.
Sleep profiling in PWScrm+/p− mutant mice versus controls. (A) Example of electroencephalographic traces (i.e. electroencephalography and electromyography) scored as wakefulness, nonrapid eye movement (NREM) sleep, rapid eye movement (REM) sleep, and REM intrusions. Fast Fourier transforms (FFTs) are represented for each epoch in which delta (light gray bars) and theta (black bars) are plotted. (B) Total time spent asleep per hour in NREM and REM sleep across the 24 h in PWScrm+/p− mice versus controls. (C) Amount of episodes of sleep, NREM and REM sleep, per hour, across the 24 h in PWScrm+/p− mice versus controls. An episode is intended as one or more consecutive epochs of the same stage (i.e. wake [W], NREM sleep or REM sleep). (D) REM sleep intrusions during wakefulness in mutants versus control mice. (E) Power densities of the whole spectrum of frequencies and detailed histograms for delta and theta EEG frequencies (μV2) in NREM and REM sleep in mutants versus control mice in the light phase. (F) Peripheral temperature of PWScrm+/p− mice and controls across 24 h. Histograms show the sleep and temperature means ± standard error of the mean (SEM), in the light and dark phases, of the two genotypes. All graphs are presented as mean ± SEM across 24 h. Light and dark phases are indicated by the white and black strips on top of each graph. ZT, Zeitgeber time. Statistical significance is represented as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Morphology Assessment: Magnetic Resonance Imaging
High-resolution morphoanatomical magnetic resonance imaging (MRI) was performed on brains of 12 PWScrm+/p− (32–45 w old) and 12 PWScrm+/p+ (30–47 w old) male mice. Intergroup differences in local gray-matter volume were mapped using voxel-based morphometry (VBM) as previously described.23 Gray matter of spatially normalized subjects was then segmented using a Markov random field model using a six-class segmentation of the study-based template as a prior to initialize the process. See supplemental material for further details.
Patients with PWS
Here we present two patients with a diagnosis of PWS and atypical deletion that included SNORD116. The human study protocol was approved by the local ethical committee of the IRCCS, Istituto Auxologico Italiano, Piancavallo, Verbania, (Italy). Written informed consent was obtained from parents and patients. The severity of illness was determined by Holm diagnostic criteria.24 Both patients underwent growth hormone (GH) therapy. Because of the metabolic characteristics that accompany the syndrome, we compared the individual EEG parameters of patients with healthy subjects but also with a cohort of obese subjects. Then, 14 healthy individuals and 10 obese subjects, half males and half females, mean age: 39.3 ± 4.1 and 41.3 ± 3.7 y, respectively, were recruited as control groups. Obese individuals matched the patients for their body mass index (BMI, 34 ± 3.4) and suffered from apnea (apnea-hypopnea index [AHI] = 16.7 ± 2.06). Healthy subjects had no sleep disturbances according to the Pittsburgh Sleep Quality Index (PSQI). For the genotyping assay and sleep assessment, see the supplemental material.
Patient 1
Patient 1 is a 39-y-old man. The deletion size in Patient 1 (Figure 1) is approximately 6,27 Mb, with a distal breakpoint mapping at the common BP4 of chromosome 15 and an atypical proximal breakpoint located approximately 575 kb downstream of BP2 of chromosome 15. Therefore, MKRN3, MAGEL2, and NDN genes, which are usually deleted as a consequence of the typical PWS deletions, are preserved. The patient was born at term with hypogenitalism, muscle hypotonia, feeding difficulties, failure to thrive, and facial features. Overall, he presented with a global developmental delay and incomplete sexual development. Hyperphagia appeared when the patient was 2 y with concomitant weight gain leading to severe obesity in adolescence. He underwent biliopancreatic diversion at 19 y and now weighs 86.3 kg; he is 161 cm tall and his BMI is 33.3 kg/m2. Polysomnography (PSG) revealed irregular breathing with sleep apnea events (AHI = 3.9).
Patient 2
Patient 2 is a 43-y-old woman. She carries a very small, 372-kb deletion of the region defined by the SNRPN probe on chromosome 15 (Figure 1). The deletion comprises part of SNURF-SNRPN and the following C/D box snoRNA genes: SNORD107, SNORD108, SNORD109A, SNORD64 (also known as HBII-436, HBII-437, HBII-438A, HBII-13, respectively), the multiple copy cluster of SNORD116 (HBII-85), and 21 copies of SNORD115.
The patient was born after 42 w of gestation and exhibited severe muscle hypotonia and feeding difficulties. She presented with delayed motor development and failure to gain weight in the first years of life. Primary amenorrhea and mild mental retardation are present. Obesity started at the age of 6 y and hyperphagia appeared later. Her actual weight is 66.9 kg, and her height is 141.3 cm, for a BMI of 33.5 kg/m2. PSG evaluation showed hypoventilation and sleep apnea (AHI = 4.1).
We elaborated the percentage of REM and NREM sleep over the total sleep time and the number of REM-NREM sleep cycles in 1 night and of REM sleep episodes per hour (REM sleep fragmentation) for each subject. Furthermore we compared the spectral characteristics of the EEG by analyzing the EEG power density for delta (0, 5–4 Hz) and theta (5–9 Hz) frequencies in NREM and REM sleep, between patients and controls. Wakefulness was not assessed because human subjects EEG was recorded only at night.
Statistics
The statistical tests were performed with MATLAB and GraphPad Prism 5.0.
Two-way mixed analyses of variance (ANOVAs) (Genotype × Time) were run for all sleep parameters in order to compare PWScrm+/p− with PWScrm+/p+ mice across light and dark phases. For the power spectral analysis of delta and theta frequencies we run paired two-tailed t-tests. Comparisons of the mean temperature in the light phase and the mean temperature in the dark phase, between genotypes, was done with paired two-tailed t-tests. For the MRI analysis, voxelwise cross-subject statistic was performed using a nonparametric permutation test with 5,000 permutations and a cluster-based threshold of 0.05. To compare patients with PWS with healthy controls and obese subjects, we run one-way repeated-measures ANOVAs for the power spectral analysis of delta and theta frequencies of NREM and REM sleep.
The following P values were considered significant differences: < 0.05, < 0.01, and < 0.001.
RESULTS
REM/NREM Sleep Differs in PWScrm+/p− Mice Compared with Controls during Light Phase
Although the total sleep time across 24 h was similar between genotypes, the architecture of REM and NREM sleep changed in the two groups of mice. During the light phase of the light-dark schedule, when mice are mostly asleep, the amount of REM sleep increased (P < 0.01; Figure 2B) whereas the amount of NREM sleep decreased (P < 0.02) in PWScrm+/p− mutants compared to their littermate controls. No differences emerged during the dark phase for these parameters. We assessed the quality of sleep by looking at sleep fragmentation, measured as the number of either REM or NREM sleep episodes per hour. Indeed, the increased REM sleep amount in mutants was due to an increased number of REM sleep episodes per hour both in light (P < 0.001) and dark (P < 0.02) conditions (Figure 2C). Yet, mutants had significantly higher NREM sleep episodes (P < 0.01) compared to controls during the light phase. In addition, mutants presented REM sleep intrusions (Figure 2A and 2D) that are rare in controls (light: P < 0.001; dark: P < 0.001). Finally, spectral analyses of EEG revealed that the amplitude of REM sleep theta power was increased in PWScrm+/p− mice compared to wild-type mice (P < 0.001). Interestingly, this alteration affected specifically theta frequencies in the light phase of mutants (Figure 2E; P < 0.001). No differences were found for the spectral power analysis of wakefulness and NREM sleep either in dark or light phases (Figure 2E, only NREM sleep in the light phase shown).
PWScrm+/p− Mice Show an Increased Peripheral Temperature during Light
In assessing physiological parameters during long-term recording we monitored the circadian profile of peripheral body temperature in mice by means of subcutaneous implants (see Table S1, supplemental material). It emerged that PWScrm+/p− mice showed notable alterations of their peripheral temperature throughout 24 h. In particular, mutants presented higher peripheral temperature in the light phase (P < 0.001) and presented a trend to elevation of temperature in the dark phase (P = 0.08) of the light-dark cycle (Figure 2F).
PWS Patients with Atypical Deletions Show an Abnormal REM Sleep Compared with Healthy Controls
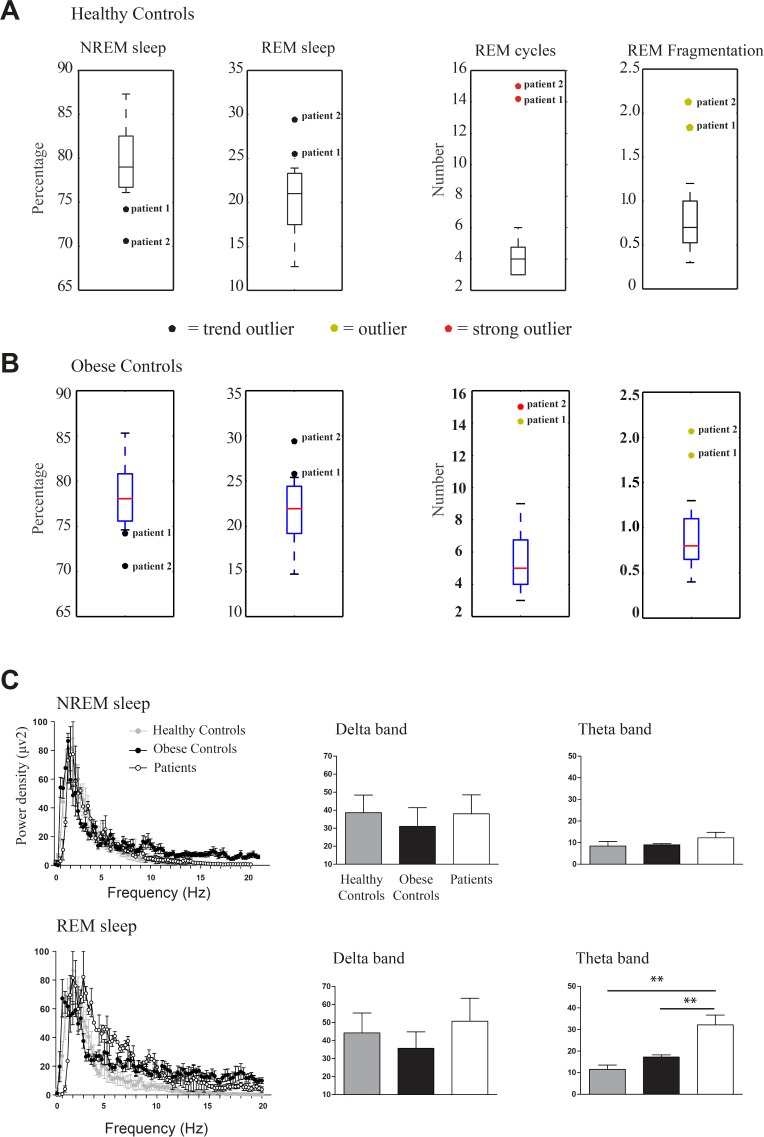
Genetic tests confirmed a small deletion in both patients embracing the SNORD116. A QFQ-banded chromosome analysis was performed to exclude chromosomal abnormalities, and it showed a normal karyotype for both patients. According to Holm diagnostic criteria,24 8 points are needed for a diagnosis of PWS; patients 1 and 2 scored 11 and 10, respectively (see Table S1). Thus, diagnoses of PWS were confirmed genetically and clinically (see Figure 1 and Table S1). We compared the individual EEG parameters of these two case studies within larger and representative cohorts of obese and healthy subjects. Patients with PWS presented with a decreased NREM and an increased REM sleep compared to control subjects. The percentages of NREM and REM sleep in PWS patients were outside the lower and upper limits, respectively, of the distribution of values obtained from controls (Figure 3A and 3B); patients were outliers for the number of REM sleep cycles and for REM sleep episodes during nocturnal sleep. Moreover, several REM sleep intrusions occurred in nocturnal sleep in patients with PWS, making them also outliers for this parameter. About five REM sleep periods occurred during a normal, nondisturbed, nocturnal sleep of 8 h; therefore, values higher than 1 in the fragmentation index (right panel of Figure 3A and 3B) indicate a REM sleep disruption. Patient 1 also presented REM sleep intrusions in wakefulness. Only theta, and not delta, power was significantly different, in REM sleep in the light phase (P < 0.001; Figure 3C). In particular, REM sleep theta power was higher compared to that of healthy controls (P < 0.01) and obese controls (P < 0.01) whereas obese subjects did not differ from healthy controls.
Figure 3.
Sleep in patients with Prader-Willi syndrome compared to healthy and obese control groups. Boxplots of percentages of nonrapid eye movement (NREM) and rapid eye movement (REM) sleep, number of REM sleep cycles and REM sleep episodes for healthy controls (A) and obese subjects (B); colored circles indicate the outlier position of the two patients respect to the control distributions. (C) Power densities of the whole spectrum of frequencies and detailed histograms for delta and theta electroencephalographic frequencies (μV2) in NREM and REM sleep in patients, healthy and obese individuals. Graphs are presented as mean ± standard error of the mean; **P < 0.01.
Snord116 Deletion Leads to Brain Morphological Changes
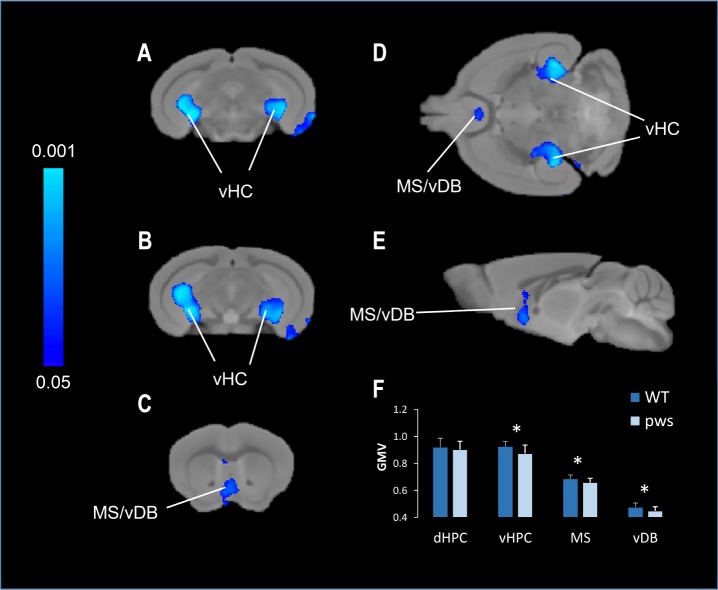
To investigate whether the physiological abnormalities that we observed in mice and humans carrying the Snord116 deletion are linked to specific brain morphological changes, we investigated MRI changes in mice. This was done by using VBM (see supplemental material). VBM highlighted bilateral foci of reduced gray-matter volume in ventral hippo-campal areas and in regions of the septum and diagonal band of PWScrm+/p− mice, with respect to controls (Figure 4). The effect was significant both at the voxel level (P < 0.01, TFCE corrected) and in terms of anatomical regions of interest (P < 0.05). In particular, we observed a major reduction in the size of the hippocampus (Figure 4). The hippocampal involvement appeared to be specific with lack of gray-matter alterations in its dorsal or more posterior portions.
Figure 4.
Magnetic resonance imaging analyses in PWScrm+/p− mice compared to wild-type littermate controls. Representative coronal (A–C), horizontal (D), and sagittal (E) slice reconstruction of the areas showing a statistically significant decrease in gray-matter volume (GMV) in PWScrm+/p− mice compared with control littermates. Decrease of gray-matter volume was evident in the vHPC, DG (A,B), vDB (C) and MS (D,E). The statistical significance is indicated by blue color coding (P < 0.05).The arrows indicate the placement of region of interest for post hoc analyses in the vHPC, MS, and vDB areas. Bar graphs (F) illustrate the mean and standard deviation of GMV of dHPC, vHPC, MS, and vDB plotted as a function of genotype. vHPC, ventral hippocampus; dHPC, dorsal hippocampus; MS, medial septal nuclei; vDB, ventral nuclei of the diagonal band; GMV, gray matter volume. Statistical significance is represented as *P < 0.05.
DISCUSSION
In our study we report, for the first time, evidence that genetic deletion within the SNORD116/Snord116 region affects sleep physiology. The dysregulation of REM sleep, which is proved by its increased amount, fragmentation and misplacement emerged in both the PWScrm+/p− mice and the patients with PWS, suggesting common regulatory mechanisms between human and mouse REM sleep. Remarkably, in both species we observed spectral defects of the theta power of the EEG despite cortical differences such as thickness and gyrification.25
In mammals the REM/theta trait is organized and maintained within specific brain circuits that involve the hippocampus and the septum diagonal band complex.26,27 Many studies in rodents have shown that the hippocampus is the main generator of theta, which through the septum distributes the rhythm across the brain.28–30 Thus, the electrophysiological defects, due to Snord116 deletion, are coherent with the bilateral alterations of septum and hippocampus that we identified in PWScrm+/p− mice.
Human and rodent REM sleep changes are also influenced by metabolic variations such as those induced by the thermo-regulatory demand.31 The drop of body temperature during sleep is linked to heat dissipation, which produces an increase of skin temperature. Warm skin promotes the heat dissipation from the core to the environment.32 We reasoned that, although both body and proximal skin temperature decrease while sleeping and exhibit a similar circadian phase, the mutants' gradient of core-to-peripheral temperature might be increased. The fact that REM sleep is particularly vulnerable to thermo-regulatory demands33–35 would explain our observation of an alteration of REM sleep.
Our previous work on the imprinted gene Gnas revealed a reduction of REM sleep following core temperature increase in mice.22 Therefore, the temperature and sleep phenotypes of PWScrm+/p− mice reinforce the idea that REM sleep and body temperature share common metabolic mechanisms depending on genomic imprinting.
Nevertheless, we cannot exclude that brain temperature also may be altered and have an effect on REM sleep. Deboer and Tobler36 have shown in the Djungarian hamster that, by lowering the cortical temperature, the frequency of REM sleep episodes increased. The REM sleep alteration we report here could also be affected by the temperature of the hypothalamus; the latter affects also REM sleep occurrence37 and sleep propensity.38
In our study the sleep phenotype is accounted for by both brain morphological differences and metabolic temperature variations. Thermoregulation instability in patients with PWS has been reported but, as often happens for this syndrome, the complexity of genetic causes corresponds to variable symptoms severity: from high fever39 and sudden death40 to no obvious issues. Temperature problems are considered supportive findings among the criteria for a diagnosis of PWS (based on consensus criteria24); they are not scored but they increase the certainty of a diagnosis of PWS. Nevertheless, no systematic study on body temperature, PWS, and sleep has been conducted so far.
Skryabin et al.,2 when engineering the PWScrm+/p− mouse, investigated the expression of different targets of the Snrpn cluster in the brain, in particular whether the Snord116 deletion affected Necdin, Magel2, Mkrn3, Frat3, other snoRNAs, and Ipw exons. The authors confirmed the complete absence of MBII-85 snoRNA though all other genes and snoRNAs of the locus were unaffected except for a slight decrease in the expression of exons F and G of Ipw. To the purpose of our study this is particularly important for refining the role of specific gene regulatory processes in mediating circadian clock and sleep. Although the circadian clock is organized by self-sustained ∼24-h cycles of transcriptional and translational positive/negative feedback loops,41 sleep is subjected to an independent homeo-static control.42 Consistent evidence has shown that at a genetic level circadian clock and sleep homeostasis are linked and often are both compromised in diseases.43 The Prader-Willi genomic region appears to contain genetic regulatory mechanisms that account for both circadian clock and sleep. Indeed, of the paternally expressed protein-coding genes of the Prader-Willi region, Magel2 was found to modulate circadian rhythms.44 In our PWScrm+/p− mouse model circadian rhythms were normally regulated as this model presents intact Magel2 expression.2 The experimental isolation of the role of MBII-85 snoRNA indicates that abnormal sleep is an endophenotype of this deletion. It is improbable that the Ipw exons are directly responsible for the sleep phenotype as it was shown that lack of Ipw transcripts do not lead to PWS phenotypes both in humans8 and mice.45 However, a recent study on parthenogenesis and PWS induced pluripotent stem cells (iPSCs) has demonstrated a role of non-coding RNA IPW in upregulating maternally expressed genes in the imprinted DLK1-DIO3 cluster on chromosome 1446; the latter has been reported to play a role in thermogenesis.1 Thus, although Ipw deletion may not give rise to the full sleep phenotype in our PWScrm+/p− mouse model, it may indirectly contribute to thermoregulatory processes that influence sleep.
Our data further advocate a role of genomic imprinting in sleep-wake regulatory processes, consistently with other clinical observations, such as Angelman syndrome,47–49 and with animal studies, such as studies in Ube3am−/p+,50 +/Ex1a mice,22 and hybrids that show parent-of-origin sleep regulation.51 To date, accumulating evidence assigns a primary role of genomic imprinting in the regulation of REM sleep. REM sleep is mainly present in mammals and characterizes the phylogenetic transition from the reptile lineage to the monotremes line.52
Interestingly, both in the PWScrm+/p− mice and in the previously studied +/Ex1a model,22 we have found sleep and temperature alterations. REM sleep is the predominant form of sleep among mammals early in development,53 and genomic imprinting exerts a fundamental role in brain developmental functions.1 The concomitant effects that imprinting genes exert on REM sleep and thermoregulations suggests that this epigenetic mechanism, which controls allelic-specific expression mainly in developing brain, may then affect sleep structure in adulthood.
In conclusion, in our study we annotate REM/NREM sleep abnormalities as direct and indirect endophenotypes of Snord116 and Ipw A-C deletion within the Prader-Willi genomic region. This report adds translational validity of the PWScrm+/p− murine model to the investigation of PWS.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank Prof. Jürgen Brosius and Dr. Boris Skryabin for making the PWScrm+/p− mice available. We also thank Dr. Alberto Galbusera for technical assistance in performing the MRI experiments. Thank you also to Monica Morini and the animal facility staff at IIT.
REFERENCES
- 1.Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15:517–30. doi: 10.1038/nrg3766. [DOI] [PubMed] [Google Scholar]
- 2.Skryabin BV, Gubar LV, Seeger B, et al. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahoo T, del Gaudio D, German JR, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duker AL, Ballif BC, Bawle EV, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196–201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieth E, Eddiry S, Gaston V, et al. Highly restricted deletion of the SNORD116 region is implicated in Prader-Willi syndrome. Eur J Hum Genet. 2015;23:252–5. doi: 10.1038/ejhg.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Smith AJ, Purmann C, Walters RG, et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet. 2009;18:3257–65. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanber D, Giltay J, Wieczorek D, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–90. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Runte M, Varon R, Horn D, Horsthemke B, Buiting K. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum Genet. 2005;116:228–30. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- 9.Schule B, Albalwi M, Northrop E, et al. Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome. BMC Med Genet. 2005;6:18. doi: 10.1186/1471-2350-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson WP, Bottani A, Xie YG, et al. Molecular, cytogenetic, and clinical investigations of Prader-Willi syndrome patients. Am J Hum Genet. 1991;49:1219–34. [PMC free article] [PubMed] [Google Scholar]
- 11.Harris JC, Allen RP. Is excessive daytime sleepiness characteristic of Prader-Willi syndrome? The effects of weight change. Arch Pediatr Adolesc Med. 1996;150:1288–93. doi: 10.1001/archpedi.1996.02170370066011. [DOI] [PubMed] [Google Scholar]
- 12.Manni R, Politini L, Nobili L, et al. Hypersomnia in the Prader Willi syndrome: clinical-electrophysiological features and underlying factors. Clin Neurophysiol. 2001;112:800–5. doi: 10.1016/s1388-2457(01)00483-7. [DOI] [PubMed] [Google Scholar]
- 13.Priano L, Grugni G, Miscio G, et al. Sleep cycling alternating pattern (CAP) expression is associated with hypersomnia and GH secretory pattern in Prader-Willi syndrome. Sleep Med. 2006;7:627–33. doi: 10.1016/j.sleep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Camfferman D, McEvoy RD, O'Donoghue F, Lushington K. Prader Willi Syndrome and excessive daytime sleepiness. Sleep Med Rev. 2008;12:65–75. doi: 10.1016/j.smrv.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Vgontzas AN, Bixler EO, Kales A, et al. Daytime sleepiness and REM abnormalities in Prader-Willi syndrome: evidence of generalized hypoarousal. Int J Neurosci. 1996;87:127–39. doi: 10.3109/00207459609070832. [DOI] [PubMed] [Google Scholar]
- 16.Vgontzas AN, Kales A, Seip J, et al. Relationship of sleep abnormalities to patient genotypes in Prader-Willi syndrome. Am J Med Genet. 1996;67:478–82. doi: 10.1002/(SICI)1096-8628(19960920)67:5<478::AID-AJMG7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 17.Vela-Bueno A, Kales A, Soldatos CR, et al. Sleep in the Prader-Willi syndrome. Clinical and polygraphic findings. Arch Neurol. 1984;41:294–6. doi: 10.1001/archneur.1984.04050150072020. [DOI] [PubMed] [Google Scholar]
- 18.Hertz G, Cataletto M, Feinsilver SH, Angulo M. Sleep and breathing patterns in patients with Prader Willi syndrome (PWS): effects of age and gender. Sleep. 1993;16:366–71. doi: 10.1093/sleep/16.4.366. [DOI] [PubMed] [Google Scholar]
- 19.Resnick JL, Nicholls RD, Wevrick R. Recommendations for the investigation of animal models of Prader-Willi syndrome. Mamm Genome. 2013;24:165–78. doi: 10.1007/s00335-013-9454-2. [DOI] [PubMed] [Google Scholar]
- 20.Sinnema M, Einfeld SL, Schrander-Stumpel CT, Maaskant MA, Boer H, Curfs LM. Behavioral phenotype in adults with Prader-Willi syndrome. Res Dev Disabil. 2011;32:604–12. doi: 10.1016/j.ridd.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Ding F, Li HH, Li J, Myers RM, Francke U. Neonatal maternal deprivation response and developmental changes in gene expression revealed by hypothalamic gene expression profiling in mice. PLoS One. 2010;5:e9402. doi: 10.1371/journal.pone.0009402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassi G, Ball ST, Maggi S, et al. Loss of Gnas imprinting differentially affects REM/NREM sleep and cognition in mice. PLoS Genet. 2012;8:e1002706. doi: 10.1371/journal.pgen.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dodero L, Damiano M, Galbusera A, et al. Neuroimaging evidence of major morpho-anatomical and functional abnormalities in the BTBR T+TF/J mouse model of autism. PLoS One. 2013;8:e76655. doi: 10.1371/journal.pone.0076655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm VA, Cassidy SB, Butler MG, et al. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics. 1993;91:398–402. [PMC free article] [PubMed] [Google Scholar]
- 25.Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 2014;15:217–32. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andy OJ, Stephan H. The septum in the human brain. J Comp Neurol. 1968;133:383–410. doi: 10.1002/cne.901330308. [DOI] [PubMed] [Google Scholar]
- 27.Kitchigina V, Popova I, Sinelnikova V, et al. Disturbances of septohippocampal theta oscillations in the epileptic brain: reasons and consequences. Exp Neurol. 2013;247:314–27. doi: 10.1016/j.expneurol.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Buzsaki G. Theta oscillations in the hippocampus. Neuron. 2002;33:325–40. doi: 10.1016/s0896-6273(02)00586-x. [DOI] [PubMed] [Google Scholar]
- 29.Sirota A, Montgomery S, Fujisawa S, Isomura Y, Zugaro M, Buzsaki G. Entrainment of neocortical neurons and gamma oscillations by the hippocampal theta rhythm. Neuron. 2008;60:683–97. doi: 10.1016/j.neuron.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubenov EV, Siapas AG. Hippocampal theta oscillations are travelling waves. Nature. 2009;459:534–9. doi: 10.1038/nature08010. [DOI] [PubMed] [Google Scholar]
- 31.Krauchi K, Deboer T. The interrelationship between sleep regulation and thermoregulation. Front Biosci (Landmark Ed) 2010;15:604–25. doi: 10.2741/3636. [DOI] [PubMed] [Google Scholar]
- 32.Van Someren EJ. Mechanisms and functions of coupling between sleep and temperature rhythms. Prog Brain Res. 2006;153:306–24. doi: 10.1016/S0079-6123(06)53018-3. [DOI] [PubMed] [Google Scholar]
- 33.Krauchi K, Deboer T. The interrelationship between sleep regulation and thermoregulation. Front Biosci. 2010;15:604–25. doi: 10.2741/3636. [DOI] [PubMed] [Google Scholar]
- 34.Valatx JL, Roussel B, Cure M. [Sleep and cerebral temperature in rat during chronic heat exposure] Brain Res. 1973;55:107–22. doi: 10.1016/0006-8993(73)90491-5. [DOI] [PubMed] [Google Scholar]
- 35.Amici R, Cerri M, Ocampo-Garces A, et al. Cold exposure and sleep in the rat: REM sleep homeostasis and body size. Sleep. 2008;31:708–15. doi: 10.1093/sleep/31.5.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deboer T, Tobler I. Shortening of the photoperiod affects sleep distribution, EEG and cortical temperature in the Djungarian hamster. J Comp Physiol A. 1996;179:483–92. doi: 10.1007/BF00192315. [DOI] [PubMed] [Google Scholar]
- 37.Parmeggiani PL, Agnati LF, Zamboni G, Cianci T. Hypothalamic temperature during the sleep cycle at different ambient temperatures. Electroencephalogr Clin Neurophysiol. 1975;38:589–96. doi: 10.1016/0013-4694(75)90159-5. [DOI] [PubMed] [Google Scholar]
- 38.Gong H, Szymusiak R, King J, Steininger T, McGinty D. Sleep-related c-Fos protein expression in the preoptic hypothalamus: effects of ambient warming. Am J Physiol Regul Integr Comp Physiol. 2000;279:R2079–88. doi: 10.1152/ajpregu.2000.279.6.R2079. [DOI] [PubMed] [Google Scholar]
- 39.Ince E, Ciftci E, Tekin M, et al. Characteristics of hyperthermia and its complications in patients with Prader Willi syndrome. Pediatr Int. 2005;47:550–3. doi: 10.1111/j.1442-200x.2005.02124.x. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson DA, Anaya TM, Clayton-Smith J, et al. Unexpected death and critical illness in Prader-Willi syndrome: report of ten individuals. Am J Med Genet A. 2004;124A:158–64. doi: 10.1002/ajmg.a.20370. [DOI] [PubMed] [Google Scholar]
- 41.Barnard AR, Nolan PM. When clocks go bad: neurobehavioural consequences of disrupted circadian timing. PLoS Genet. 2008;4:e1000040. doi: 10.1371/journal.pgen.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tucci V. Sleep, circadian rhythms, and interval timing: evolutionary strategies to time information. Front Integr Neurosci. 2011;5:92. doi: 10.3389/fnint.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franken P. A role for clock genes in sleep homeostasis. Curr Opin Neurobiol. 2013;23:864–72. doi: 10.1016/j.conb.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Kozlov SV, Bogenpohl JW, Howell MP, et al. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–72. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- 45.Ding F, Prints Y, Dhar MS, et al. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm Genome. 2005;16:424–31. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- 46.Stelzer Y, Ronen D, Bock C, Boyle P, Meissner A, Benvenisty N. Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells. Stem Cell Reports. 2013;1:79–89. doi: 10.1016/j.stemcr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dan B, Boyd SG. Angelman syndrome reviewed from a neurophysiological perspective. The UBE3A-GABRB3 hypothesis. Neuropediatrics. 2003;34:169–76. doi: 10.1055/s-2003-42213. [DOI] [PubMed] [Google Scholar]
- 48.Miano S, Bruni O, Leuzzi V, Elia M, Verrillo E, Ferri R. Sleep polygraphy in Angelman syndrome. Clin Neurophysiol. 2004;115:938–45. doi: 10.1016/j.clinph.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Valente KD, Andrade JQ, Grossmann RM, et al. Angelman syndrome: difficulties in EEG pattern recognition and possible misinterpretations. Epilepsia. 2003;44:1051–63. doi: 10.1046/j.1528-1157.2003.66502.x. [DOI] [PubMed] [Google Scholar]
- 50.Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol Dis. 2005;20:471–8. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 51.Tinarelli F, Garcia-Garcia C, Nicassio F, Tucci V. Parent-of-origin genetic background affects the transcriptional levels of circadian and neuronal plasticity genes following sleep loss. Philos Trans R Soc Lond B Biol Sci. 2014;369:20120471. doi: 10.1098/rstb.2012.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel JM, Manger PR, Nienhuis R, Fahringer HM, Pettigrew JD. Monotremes and the evolution of rapid eye movement sleep. Philos Trans R Soc Lond B Biol Sci. 1998;353:1147–57. doi: 10.1098/rstb.1998.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005;437:1264–71. doi: 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.