Abstract
Study Objectives:
Narcolepsy with cataplexy is tightly associated with the HLA class II allele DQB1*06:02. Evidence indicates a complex contribution of HLA class II genes to narcolepsy susceptibility with a recent independent association with HLA-DPB1. The cause of narcolepsy is supposed be an autoimmune attack against hypocretin-producing neurons. Despite the strong association with HLA class II, there is no evidence for CD4+ T-cell-mediated mechanism in narcolepsy. Since neurons express class I and not class II molecules, the final effector immune cells involved might include class I-restricted CD8+ T-cells.
Methods:
HLA class I (A, B, and C) and II (DQB1) genotypes were analyzed in 944 European narcolepsy with cataplexy patients and in 4,043 control subjects matched by country of origin. All patients and controls were DQB1*06:02 positive and class I associations were conditioned on DQB1 alleles.
Results:
HLA-A*11:01 (OR = 1.49 [1.18–1.87] P = 7.0*10−4), C*04:01 (OR = 1.34 [1.10–1.63] P = 3.23*10−3), and B*35:01 (OR = 1.46 [1.13–1.89] P = 3.64*10−3) were associated with susceptibility to narcolepsy. Analysis of polymorphic class I amino-acids revealed even stronger associations with key antigen-binding residues HLA-A-Tyr9 (OR = 1.32 [1.15–1.52] P = 6.95*10−5) and HLA-C-Ser11 (OR = 1.34 [1.15–1.57] P = 2.43*10−4).
Conclusions:
Our findings provide a genetic basis for increased susceptibility to infectious factors or an immune cytotoxic mechanism in narcolepsy, potentially targeting hypocretin neurons.
Citation:
Tafti M, Lammers GJ, Dauvilliers Y, Overeem S, Mayer G, Nowak J, Pfister C, Dubois V, Eliaou JF, Eberhard HP, Liblau R, Wierzbicka A, Geisler P, Bassetti CL, Mathis J, Lecendreux M, Khatami R, Heinzer R, Haba-Rubio J, Feketeova E, Baumann CR, Kutalik Z, Tiercy JM. Narcolepsy-associated HLA class I alleles implicate cell-mediated cytotoxicity. SLEEP 2016;39(3):581–587.
Keywords: autoimmunity, hypocretin/orexin, CD4, CD8, cytotoxicity
Significance.
Although evidence strongly indicates that immune-related mechanisms are involved in the pathogenesis of narcolepsy with hypocretin deficiency, functional data are missing. The strong association with HLA class II genes is not corroborated by any CD4+ T Cell implication and inflammation. Additionally, these genes are not normally expressed in the brain. Here, we show that several HLA class I variants are associated with narcolepsy. Since these molecules are expressed in the brain, our results suggest that a cytotoxic (CD8+ T Cell or NK cell) mechanism might be the final step in the disease, leading to hypocretin neuronal destruction.
INTRODUCTION
Narcolepsy is a primary disorder of vigilance states characterized by episodes of unwanted sleep and sudden loss of muscle tone triggered by strong emotions (cataplexy). Although the HLA association found in 1984 led to the autoimmune hypothesis of narcolepsy, it was only after the discovery of hypocretin (orexin) deficiency in 2000, due to the loss of hypocretin-producing neurons, that the autoimmune hypothesis gained substantial basis.1–3 Nevertheless, convincing evidence of an autoimmune attack against hypocretin neurons is still lacking. Also, the nature of such autoimmunity remains elusive. The discovery of auto-reactive antibodies against TRIB2 in a limited number of narcolepsy patients suggested a humoral immune mechanism.4 As opposed to established and well-documented autoimmune disorders, T-cell-mediated autoimmunity has not yet been found in narcolepsy,5 except for rare cases of symptomatic narcolepsy in which CD8+ T-cells may be pathogenic.6
By far major genetic susceptibility determinants of narcolepsy, as in other autoimmune disorders, are found within the HLA region.7,8 Among HLA class II genes, the DRB1*15:01-DQA1*01:02-DQB1*06:02 is the disease-associated haplotype in narcolepsy. In European populations, the three alleles of this haplotype are in complete linkage disequilibrium (LD), so that any single allele is representative of the haplotype. Early serologic typings in narcolepsy reported associations with HLA-B7 and A3 in Caucasian and B35 in Japanese patients.9–12 However, these original findings were subsequently interpreted as secondary (through LD) to HLA DRB1-DQB1 association. Studies including molecular genotyping of HLA class I genes (A, B, and C) in large populations with narcolepsy are lacking. Nevertheless, a recent study used molecular HLA class I typing in 304 cases and 304 controls and imputed class I genotypes from previous genome-wide association studies (from HLA region SNP data) in White and Chinese populations.13 This study found (in addition to DPB1) significant associations between narcolepsy and HLA-A*11:01, B*35:03, and B*51:01.
HLA class II molecules are constitutively expressed only by antigen-presenting cells such as dendritic cells, macrophages, or B cells, while HLA class I molecules are ubiquitously expressed by nearly all nucleated cells. Accordingly, HLA class II molecules are not expressed by neurons while HLA class I molecules can be.14 Therefore, the ultimate autoimmune reaction leading to neuronal cell death could involve HLA class I-restricted CD8+ reactive T-cells. Since HLA class I and II molecules are expressed by different cell types and shape different T-cell-mediated immune reactions, genetic associations to each HLA class bring different immunological information on the disease process, although the involvement of both is possible as exemplified in multiple sclerosis and type 1 diabetes.15,16 Thus the aim of the present study was to generate HLA class I genotyping data in narcolepsy with cataplexy and their matched controls and by controlling for the LD between HLA class I and the most important class II alleles, search for independent associations with HLA class I loci.
METHODS
Patients and Controls
Patients from the Netherlands (n = 334), France (n = 258), Germany (n = 223), Switzerland (n = 67), and Poland (n = 62) were included in the analysis. Detailed description of these patients was recently published.7,17 All patients were HLA DQB1*06:02 positive. Our populations did not include familial forms, subjects with mixed non-European origin, or carrying rare (< 1%) DQB1 alleles (DQB1*03:04, DQB1*06:01, and DQB1*06:09). In 406 patients, genotypings were also generated for DRB1, DQA1, and DQB1 loci. Among these, DQB1*06:02 was in complete LD with DQA1*01:02 and with almost complete LD with DRB1*15:01 (403 out of 406, or 99.26%), confirming that DQB1*06:02 tags perfectly the narcolepsy-associated class II haplotype. Four-digit HLA-DQB1, A, B, and C genotyping was generated for all patients by PCR-Luminex. HLA DQB1*06:02 positive controls were taken from panels of randomly selected, healthy, representative, and unrelated Dutch (n = 2,158), French (n = 583), German (n = 559), Swiss (n = 570), and Polish/Slovak (n = 173) subjects.
Linkage Disequilibrium and Haplotype Construction
To evaluate the LD between DQB1 and A, B, and C loci, 736 French and 1,239 Dutch control subjects from the general population (unselected by DQB1 genotype) were used. Haplotype frequencies and phase segregations of four-loci alleles (HLA-A, B, C and DQB1) were inferred from merged population data set of unphased genotypes observed in the patients and controls as described previously.18 Briefly, four-loci unphased genotypes were automatically encoded (and haplotypes were decoded afterwards) using PHASE_KEY v.1.0 (software available at http://www.ihit.waw.pl/zaklad-immunogenetyki.html). The most probable four-loci haplotype segregations were established for all patients and controls using Bayesian implementation of expectation maximization (EM) algorithm provided by PHASE v. 2.1.19 The iterative processing involved burn-in iterations set to 200 and 1,000 final iterations. Informed consent was obtained from all subjects and the experimental protocols were approved by each country's ethics institution.
Statistical Analysis
Associations between HLA class I alleles and narcolepsy, independent of HLA class II (DQB1) alleles were tested by the Cochran-Mantel-Haenszel test.20 To ensure the robustness of our findings, two additional tests were performed: (1) for each class I allele we performed logistic regression including all DQB1 alleles as covariates; (2) we also performed logistic regression for each class I allele, stratifying our samples according to DQB1 alleles. These stratified results were then meta-analyzed using fixed effect inverse variance weighting. We also applied Cochran's heterogeneity test for the results of all countries to ensure that our findings hold irrespective of the origin of the samples. Our major findings were confirmed by these additional analyses and no cross-cohort heterogeneity was detected. Thus we present the results for the Cochran-Mantel-Haenszel test only. The analysis was restricted to Class I alleles with allele frequencies > 0.5% in at least one control sample and to alleles detected in all countries. We estimated the effective number of tests (i.e., the equivalent number of independent alleles by taking into account the LD between alleles) as proposed by Gao et al.21 The 45 A, B, and C alleles tested are equivalent to 29 independent tests, thus we used the modified Benjamini-Hochberg step-up procedure for 29 tests to control the false discovery rate (FDR) at 5%.
RESULTS
Single HLA Class I Allele Association
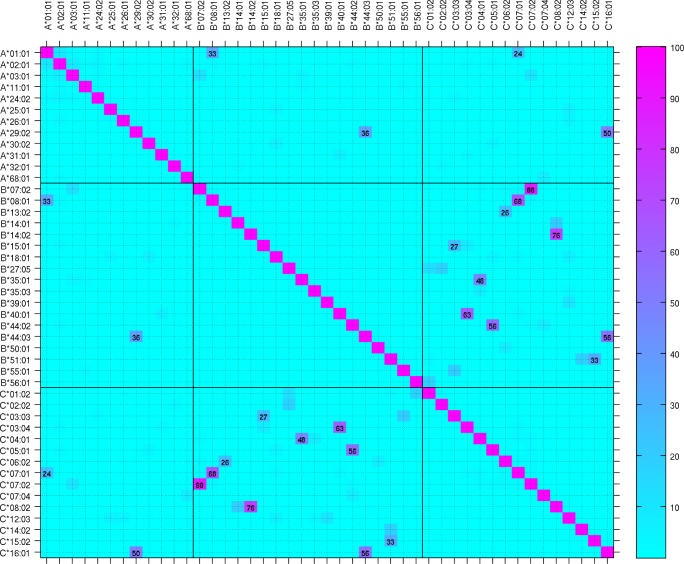
The frequency of the 45 class I alleles tested is reported in Table S1 (supplemental material). Although in many studies, control subjects are just matched for ethnicity (e.g., of European descent), substantial differences in HLA allele frequencies are commonly observed between sub-populations and even within the same country. Allele frequency heterogeneity test indicated that 15 of the 45 alleles show significant between-country heterogeneity (Table S1). We therefore tested for allelic associations independently in each country with their matched controls. Since we have matched our control populations for DQB1*06:02, we first tested HLA class I associations with narcolepsy without correcting for LD with HLA class II (DQB1) alleles (Table S2, supplemental material). As opposed to DQB1*06:02 unmatched controls, associations with known alleles with strong LD with DQB1*06:02 (e.g., B*07:02 and C*07:02; Figure 1) were strongly attenuated. Nevertheless, other narcolepsy-associated DQB1 alleles (DQB1*02, *03:01, *05:01, and *06:03) with potential LD with class I alleles might bias these results.
Figure 1.
Linkage disequilibrium between HLA-DQB1 and HLA class I alleles in the general population. Heat map of linkage disequilibrium between HLA-DQB1 and HLA-A, B, C alleles analyzed in combined Dutch and French general population unselected for DQB1*06:02. Major significant LD values are indicated.
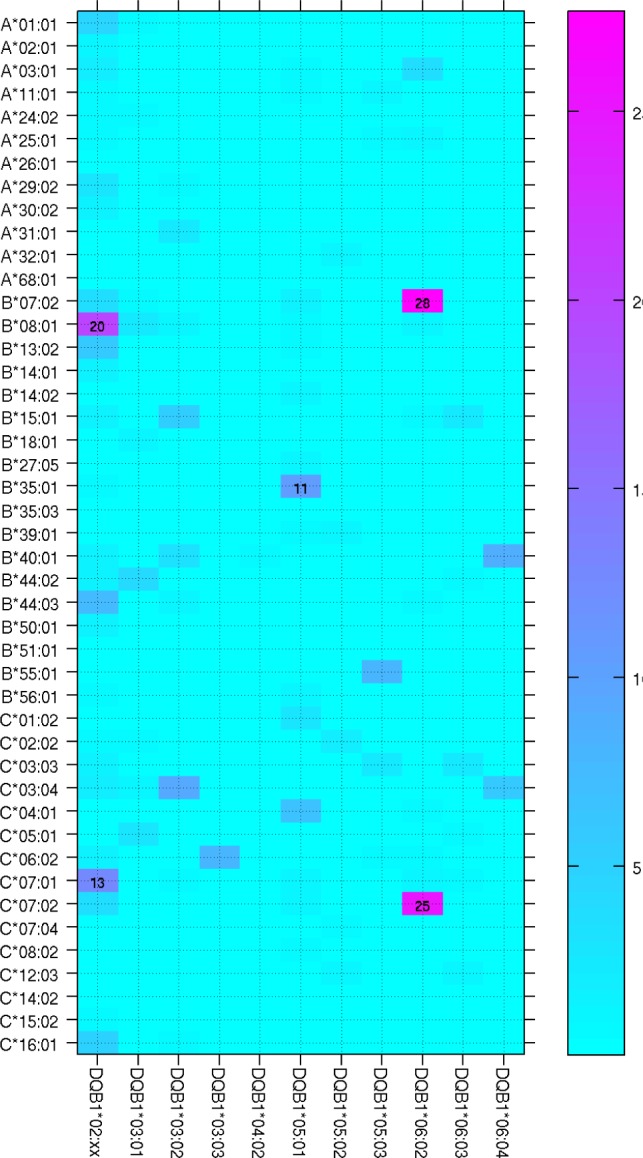
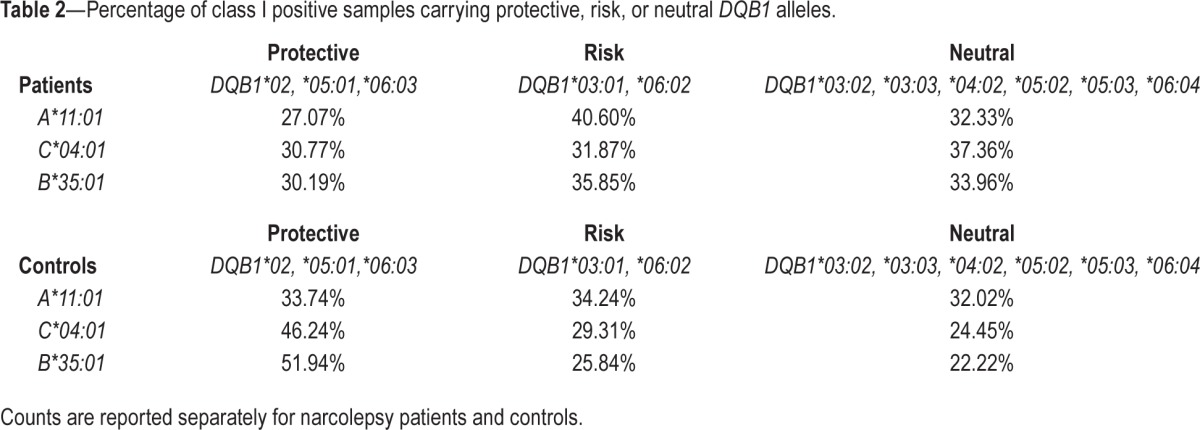
By taking into account all DQB1 alleles, Cochran-Mantel-Haenszel test revealed significant DQB1-independent associations with HLA-A*11:01, C*04:01, and B*35:01. All three alleles increased the risk of narcolepsy (Table 1). The two other analyses (conditional and stratified) confirmed these associations and additionally indicated a protective association conferred by B*08:01 (Tables S3 and S4, supplemental material). The associations with C*04:01, and B*35:01 are consistent with high LD between these 2 alleles (Figure 2). Accordingly, conditional analysis indicated 2 independent associations, one with A*11:01 and the other with C*04:01 or B*35:01. Also, associations with B*08:01 and potentially with A*01:01 might be due to LD between these 2 alleles (Figure 2), which are part of the extended conserved haplotype known as “A1-B8-DR3,” or “8.1.” The complete class II-independent associations with HLA-A*11:01, C*04:01, and B*35:01 are demonstrated by the fact that these alleles occurred in all DQB1 haplotypes and even less frequently in at risk DQB1 alleles carriers (Table 2).
Table 1.
Odds ratios for top 10 HLA class I alleles associated with narcolepsy independent of HLA-DQB1 alleles.
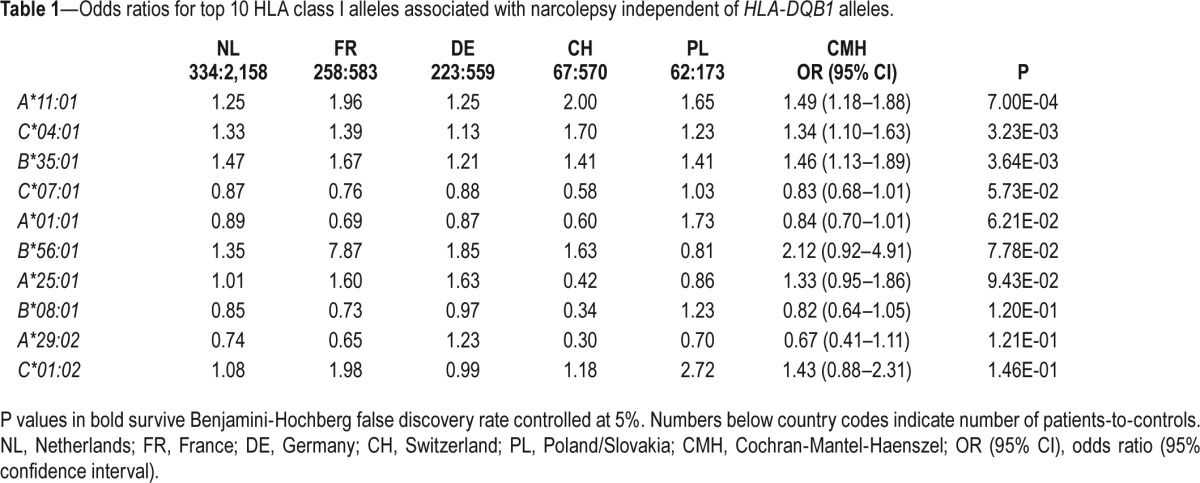
Figure 2.
Linkage disequilibrium between HLA class I alleles in the general population. Heat map of linkage disequilibrium between HLA-A, B, and C alleles analyzed in combined Dutch and French general population unselected for DQB1*06:02. Major significant LD values are indicated.
Table 2.
Percentage of class I positive samples carrying protective, risk, or neutral DQB1 alleles.
HLA Class I Haplotype Association
To verify if our allelic associations may result from extended haplotypes, the class I haplotypes (HLA-A, B, C) and extended four-loci haplotypes (HLA-A, B, C and DQB1) were compared between cases and controls from each country. The class I A*11:01-C*04:01-B*35:01 haplotype had no statistically different incidence in cases and controls (2.7% and 2.2%, respectively, OR = 1.25, [0.77–2.03], P = 0.36), con -firming the independent allelic associations. Three extended haplotypes were significantly associated: A*03:01-C*04:01-B*35:01-DQB1*06:02 (OR = 2.39 [1.35–4.22], P = 2.80*10 −3), A*03:01-C*04:01-B*35:01-DQB1*05:01 (OR = 0.02 [0.00– 0.16], P = 9.41*10−4), A*03:01-C*07:02-B*07:02-DQB1*06:02 (OR = 0.72 [0.58–0.90], P = 3.11*10 −3). The increased susceptibility to narcolepsy in carriers of A*03:01-C*04:01-B*35:01-DQB1*06:02 haplotype can be attributed to its allele components due to independent associations of C*04:01, B*35:01 and DQB1*06:02. Interestingly, the protective A*03:01-C*04:01-B*35:01-DQB1*05:01 haplotype including the risk alleles C*04:01 and B*35:01 was not found in any patient. The protective property of A*03:01-C*07:02-B*07:02-DQB1*06:02 haplotype is not straightforward. It may protect from narcolepsy by the involvement of class III genes of ancestral haplotype or DPB1 alleles (not tested here), discordant peptide presentation repertoires between class I and class II alleles and clonal deletion of some naïve thymocytes and/or involvement of protective DQB1 alleles in trans.
HLA Class I Epitope Association
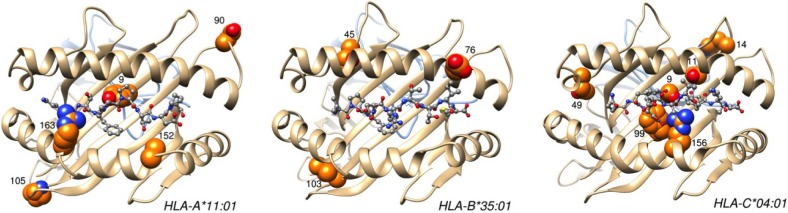
One major limitation of HLA class I allelic association is the large number of alleles (here 190 alleles) that decreases the statistical power, therefore limiting the analysis to the most frequent alleles (here 45 alleles). Additionally, this analysis does not take into account shared epitopes among alleles, which are actually responsible for antigen presentation. To overcome these limitations, we analyzed HLA class I polymorphic amino acids associated with susceptibility and resistance to narcolepsy with cataplexy. Several HLA class I amino acids were found associated with narcolepsy, independently of HLA class II (Table 3). These associations were stronger than allelic associations. By searching for alleles that best tag significant amino-acid associations, again A*11:01, B*35:01, and C*04:01 were identified, confirming our allele frequency analysis (Table 3). Interestingly, based on crystal structure of HLA-A, B, and C, several of these associated amino acids are of notable functional importance (Figure 3). HLA-A-Tyr9, Ala152, Arg163, HLA-B-Glu76, HLA-C-Ser9, Ser11, Phe99, and Arg156 are found in the peptide-binding grove. HLA-A-Arg163 and HLA-B-Glu76 can affect T cell receptor (TCR) recognition and HLA-B-Glu76 can interact with killer immunoglobulin receptors. Among these amino acids several are found within the same HLA alleles and therefore closely associated. Accordingly, conditional analysis revealed strong associations only with HLA-ATyr9 (OR = 1.32 [1.15–1.52], P = 6.95*10 −5) and HLA-C-Ser11 (OR = 1.34 [1.15–1.57] P = 2.43*10−4).
Table 3.
Odds ratios for HLA class I amino-acids significantly associated with narcolepsy independent of HLA-DQB1 alleles.
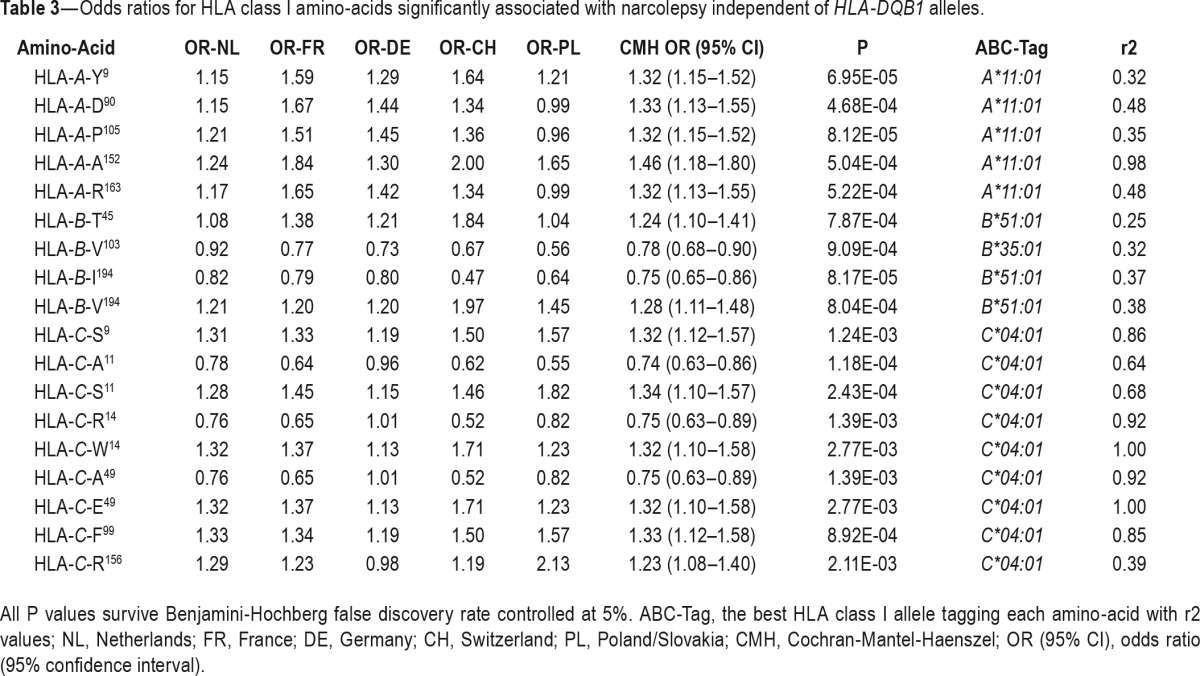
Figure 3.
Key narcolepsy-associated HLA class I residues positioned on crystal structures. HLA and β2-microglobulin are shown in ribbon representation, colored in brown and blue, respectively. Epitopes are displayed in ball and stick representation, colored according to the atom types. Important HLA residues are displayed as spheres, colored according to the atom types except for orange carbon atoms. The figure was prepared with UCSF Chimera28, using the Protein Databank29 entries 1X7Q30 (HLA-A*11:01), 3LKO31 (HLA-B*35:01), and 1IM932 (HLA-C*04:01).
DISCUSSION
The tight association between the HLA class II DQB1*06:02 allele and narcolepsy with cataplexy is well established and the original associations with class I genes, such as B7 specificity, are definitely due to high LD between DQB1*06:02 and class I alleles such as B*07:02 (Figure 1). Nevertheless, the association is much stronger with DQB1 locus since B*07:02 does not occur in all DQB1*06:02-associated haplotypes (association with DQB1*06:02 is found in over 98% while B*07:02 is found in less than 65% of narcolepsy patients). In addition to LD between class I and II genes, finding associations with class I genes is hampered by a large number of class I alleles, requiring large patient populations, unavailable in rare disorders like narcolepsy. By controlling for LD, we identified HLA-A*11:01, B*35:01, and C*04:01 as new narcolepsy susceptibility alleles. Conditioned and unconditioned results very strongly agreed for A*11:01. However, the signal from B*35:01 and C*04:01 could not be resolved: conditioning on each other gave nonsignificant results both ways (P > 0.3). Since B*35:01-C*04:01 haplotype is the fourth most common haplotype in our control population we cannot decide which allele is actually causal.
In our previous paper,7 we examined the association of class II alleles with narcolepsy in largely overlapping samples and (except for the DQB1*06:02) found that several class II alleles have strong protective effects (OR = 0.19–0.76) while the class I alleles described here are markers of susceptibility (ORs ranging 1.34–1.49). Note that class I associations not conditioned on DQB1 alleles show much larger effect sizes (see Table S2).
Although our study is so far one of the largest ones, including 944 patients, we obviously lack statistical power to detect all potential narcolepsy-associated HLA class I alleles. We therefore speculate that the HLA class I alleles found here to be suggestively associated with narcolepsy might turn out to be significant in larger samples. Association with HLA-A*11:01 was also reported recently, together with suggestive association with C*04:01.13 Nevertheless, associations with HLA-B*35:03 and B*51:01 found previously are not replicated by our results. While both alleles have similar frequencies in our populations and those of Ollila et al.,13 we only found them marginally significant (with similar ORs) in the unconditioned analysis. Hence we suspect that these associations are not robust to conditioning on DQB1 alleles or they may have different LD in our populations. On the other hand, our associations with B*35:01 and B*08:01 were not reported before. The former had similar frequencies in the imputed genotypes of Caucasians in Ollila et al.,13 but different ones in their typed samples. The latter showed opposite behavior, following our trend in the directly typed samples and discrepant frequencies in the imputed ones. Thus, Ollila et al.,13 may have missed some of our findings due to their possibly poor allele imputation, or different allelic frequencies between controls in the two studies.
The strongest genetic association with autoimmune diseases is found with HLA class II alleles. Nevertheless, recent studies reported convincing evidence for independent associations with class I alleles in several autoimmune disorders such as type 1 diabetes and multiple sclerosis.15,16 The implication of class I genes in autoimmune diseases may help to understand the underlying immune mechanisms. CD4+ T-cells are HLA class II-restricted and mediate the production of cytokines such as interleukins and TNF-α, resulting in inflammation and promotion of B- and T- cell activation and differentiation, while CD8+ T-cells are HLA class I-restricted and are responsible for killing of virally infected or tumor cells, as well as transplant rejection. Although the HLA class II associations led to extensive work on CD4+ T-cell autoreactivity in autoimmune disorders, evidence strongly supports also a critical role for CD8+ T-cells that can modulate the immune response either directly through cytotoxic killing of cells or by regulatory (protective) mechanisms.22
The most specific narcolepsy allele DQB1*06:02 is the strongest protective allele in type 1 diabetes. Interestingly, A*11:01, and C*04:01 found here as risk alleles for narcolepsy are protective in type 1 diabetes,15 suggesting an exclusive HLA contribution to these two conditions. The susceptibility association with single class I alleles found here points to the main HLA function, the better presentation of tissue specific peptides within peptide presentation repertoires of class I-associated alleles. Physiologically, HLA class I and II molecules present mostly self-peptides that are recognized by naive T cells with low/intermediate affinity of their TCR for the MHC-peptide complex.23 The process is regulated by peptide presentation repertoires partly overlapping between HLA molecules24 and tight selection of thymocytes directed to promiscuous tissue-specific peptides expressed in the thymus.25 Presentation of slightly modified peptides and generation of CD8+ T cell clones with increased affinity of TCR for MHC-peptide (above the activation threshold) is needed to induce a cytotoxic attack.26 These observations indicate a crucial role of HLA peptide presentation repertoires in both the scope of thymic selection and the induction of T-cell cytotoxicity. Class I allelic associations in DQB1*06:02 positive populations may point to the relation between class I and class II peptide repertoires. This suggests that peptide presentation repertoires of associated class I molecules fall within the framework of primarily associated class II molecule repertoire, thus shaping the cytotoxic attack, presumably specific to hypocretin-producing neurons. A T-cell clone with such a narrow specificity can be prone to clonal deletion in certain class I/II combinations.
Also, HLA class I-encoded molecules interact with killer immunoglobulin-like receptors, mostly expressed on natural killer cells and a subset of memory T cells to provide inhibitory signals. Our finding that A*11:01 and C*04:01 increase the risk of narcolepsy with cataplexy not only provides a genetic basis for a cell-mediated mechanism but also suggests a potential immune pathway critically involving natural killer cells. Also, leukocyte Ig-like receptors (LILRs) bind class I molecules and can modify the antigen presentation by macrophages and dendritic cells. The activating LILRA1 and LILRA3 receptors (CD85) bind preferentially the HLA-C heavy chain27 and modulate T-cell proliferation. Whether C*04:01-restricted CD8+ T-cells occur in narcolepsy patients needs further investigations.
In conclusion, the discovery of independent HLA class I associations with narcolepsy provides the first genetic evidence that a direct CD8 and/or NK-mediated immune reaction, potentially targeting hypothalamic hypocretin neurons, can occur in this disease.
DISCLOSURE STATEMENT
This was not an industry supported study. This work was supported by the University of Lausanne and the European Narcolepsy Network (EU-NN). ZK received financial support from the Leenaards Foundation and the Swiss National Science Foundation. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank Vincent Zoete from the SIB Molecular Modeling Group and the Protein Modeling Facility of the Lausanne University for molecular visualization and Stanislaw Nowak for expansion of PHASE 2.1 by source code modification.
REFERENCES
- 1.Juji T, Satake M, Honda Y, Doi Y. HLA antigens in Japanese patients with narcolepsy. All the patients were DR2 positive. Tissue Antigens. 1984;24:316–9. doi: 10.1111/j.1399-0039.1984.tb02144.x. [DOI] [PubMed] [Google Scholar]
- 2.Nishino S, Ripley B, Overeem S, Lammers GJ, Mignot E. Hypocretin (orexin) deficiency in human narcolepsy. Lancet. 2000;355:39–40. doi: 10.1016/S0140-6736(99)05582-8. [DOI] [PubMed] [Google Scholar]
- 3.Peyron C, Faraco J, Rogers W, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–7. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 4.Cvetkovic-Lopes V, Bayer L, Dorsaz S, et al. Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J Clin Invest. 2010;120:713–9. doi: 10.1172/JCI41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liblau RS, Vassalli A, Seifinejad A, Tafti M. Hypocretin (orexin) biology and the pathophysiology of narcolepsy with cataplexy. Lancet Neurol. 2015;14:318–28. doi: 10.1016/S1474-4422(14)70218-2. [DOI] [PubMed] [Google Scholar]
- 6.Dauvilliers Y, Bauer J, Rigau V, et al. Hypothalamic immunopathology in anti-Ma-associated diencephalitis with narcolepsy-cataplexy. JAMA Neurol. 2013;70:1305–10. doi: 10.1001/jamaneurol.2013.2831. [DOI] [PubMed] [Google Scholar]
- 7.Tafti M, Hor H, Dauvilliers Y, et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2014;37:19–25. doi: 10.5665/sleep.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hor H, Kutalik Z, Dauvilliers Y, et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat Genet. 2010;42:786–9. doi: 10.1038/ng.647. [DOI] [PubMed] [Google Scholar]
- 9.Seignalet J, Billiard M. Possible association between HLA-B7 and narcolepsy. Tissue Antigens. 1984;23:188–9. doi: 10.1111/j.1399-0039.1984.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 10.Mueller-Eckhardt G, Meier-Ewert K, Schendel DJ, Reinecker FB, Multhoff G, Mueller-Eckhardt C. HLA and narcolepsy in a German population. Tissue Antigens. 1986;28:163–9. doi: 10.1111/j.1399-0039.1986.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 11.Poirier G, Montplaisir J, Decary F, Momege D, Lebrun A. HLA antigens in narcolepsy and idiopathic central nervous system hypersomnolence. Sleep. 1986;9:153–8. doi: 10.1093/sleep/9.1.153. [DOI] [PubMed] [Google Scholar]
- 12.Matsuki K, Juji T, Tokunaga K, Naohara T, Satake M, Honda Y. Human histocompatibility leukocyte antigen (HLA) haplotype frequencies estimated from the data on HLA class I, II, and III antigens in 111 Japanese narcoleptics. J Clin Invest. 1985;76:2078–83. doi: 10.1172/JCI112211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ollila HM, Ravel JM, Han F, et al. HLA-DPB1 and HLA Class I confer risk of and protection from narcolepsy. Am J Hum Genet. 2015;96:136–46. doi: 10.1016/j.ajhg.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cebrian C, Zucca FA, Mauri P, et al. MHC-I expression renders catecholaminergic neurons susceptible to T-cell-mediated degeneration. Nat Commun. 2014;5:3633. doi: 10.1038/ncomms4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble JA, Valdes AM, Varney MD, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59:2972–9. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luca G, Haba-Rubio J, Dauvilliers Y, et al. Clinical, polysomnographic and genome-wide association analyses of narcolepsy with cataplexy: a European Narcolepsy Network study. J Sleep Res. 2013;22:482–95. doi: 10.1111/jsr.12044. [DOI] [PubMed] [Google Scholar]
- 18.Nowak J, Mika-Witkowska R, Mendek-Czajkowska E, et al. Association of HLA haplotypes with paroxysmal nocturnal hemoglobinuria. Transplant Proc. 2010;42:3266–70. doi: 10.1016/j.transproceed.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nathan M. Chi-square tests with one degree of freedom; extensions of the Mantel-Haenszel Procedure. J Am Statistical Assoc. 1963;58:90–700. [Google Scholar]
- 21.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–9. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 22.Saxena A, Martin-Blondel G, Mars LT, Liblau RS. Role of CD8 T cell subsets in the pathogenesis of multiple sclerosis. FEBS Lett. 2011;585:3758–63. doi: 10.1016/j.febslet.2011.08.047. [DOI] [PubMed] [Google Scholar]
- 23.Hickman HD, Luis AD, Buchli R, et al. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172:2944–52. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 24.Barber LD, Gillece-Castro B, Percival L, Li X, Clayberger C, Parham P. Overlap in the repertoires of peptides bound in vivo by a group of related class I HLA-B allotypes. Curr Biol. 1995;5:179–90. doi: 10.1016/s0960-9822(95)00039-x. [DOI] [PubMed] [Google Scholar]
- 25.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H, Wu Y, Liang B, et al. An affinity/avidity model of peripheral T cell regulation. J Clin Invest. 2005;115:302–12. doi: 10.1172/JCI23879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DC, Kosmoliaptsis V, Apps R, et al. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J Immunol. 2011;186:2990–7. doi: 10.4049/jimmunol.1003078. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blicher T, Kastrup JS, Buus S, Gajhede M. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr D Biol Crystallogr. 2005;61:1031–40. doi: 10.1107/S0907444905013090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gras S, Kedzierski L, Valkenburg SA, et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc Natl Acad Sci U S A. 2010;107:12599–604. doi: 10.1073/pnas.1007270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–60. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.