Abstract
Background: Early-life iron deficiency is a common nutrient deficiency worldwide. Maternal iron deficiency increases the risk of schizophrenia and autism in the offspring. Postnatal iron deficiency in young children results in cognitive and socioemotional abnormalities in adulthood despite iron treatment. The rat model of diet-induced fetal-neonatal iron deficiency recapitulates the observed neurobehavioral deficits.
Objectives: We sought to establish molecular underpinnings for the persistent psychopathologic effects of early-life iron deficiency by determining whether it permanently reprograms the hippocampal transcriptome. We also assessed the effects of maternal dietary choline supplementation on the offspring’s hippocampal transcriptome to identify pathways through which choline mitigates the emergence of long-term cognitive deficits.
Methods: Male rat pups were made iron deficient (ID) by providing pregnant and nursing dams an ID diet (4 g Fe/kg) from gestational day (G) 2 through postnatal day (PND) 7 and an iron-sufficient (IS) diet (200 g Fe/kg) thereafter. Control pups were provided IS diet throughout. Choline (5 g/kg) was given to half the pregnant dams in each group from G11 to G18. PND65 hippocampal transcriptomes were assayed by next generation sequencing (NGS) and analyzed with the use of knowledge-based Ingenuity Pathway Analysis. Real-time polymerase chain reaction was performed to validate a subset of altered genes.
Results: Formerly ID rats had altered hippocampal expression of 619 from >10,000 gene loci sequenced by NGS, many of which map onto molecular networks implicated in psychological disorders, including anxiety, autism, and schizophrenia. There were significant interactions between iron status and prenatal choline treatment in influencing gene expression. Choline supplementation reduced the effects of iron deficiency, including those on gene networks associated with autism and schizophrenia.
Conclusions: Fetal-neonatal iron deficiency reprograms molecular networks associated with the pathogenesis of neurologic and psychological disorders in adult rats. The positive response to prenatal choline represents a potential adjunctive therapeutic supplement to the high-risk group.
Keywords: choline supplementation, fetal iron deficiency, hippocampus, psychological disorders, transcriptome
Introduction
Iron deficiency anemia affects >30% of pregnancies and preschool-age children worldwide (1–5). Iron deficiency during fetal or early postnatal life has been associated with short- and long-term cognitive and behavioral abnormalities in humans, including increased risks for autism, anxiety, depression, and sleep disturbances in childhood (6–9) and anxiety, depression, and schizophrenia in adulthood (10, 11). Iron treatment resolves some acute developmental sensory and motor deficits but fails to mitigate learning and affective behavior (10, 12–16). These persistent effects exact a substantial cost to society in terms of education, jobs, and mental health (17). Similar behavioral abnormalities are evident in rodent models of maternal-fetal iron deficiency (18–22). These effects are accompanied by abnormal neuronal morphology (23, 24) and glutamatergic neurotransmission (25) in the hippocampus that persist into adulthood (22, 23, 25). The molecular underpinnings of the long-term findings need to be elucidated.
Supplementation of choline during late fetal or early neonatal life has been shown to mitigate behavioral abnormalities in models of genetic and early-life environmental insults (26–29). In a rodent model of early-life iron deficiency, prenatal choline supplementation protects against recognition memory deficits and brain-derived neurotrophic factor (Bdnf) dysregulation in formerly iron deficient (FID)9 rats by reversing epigenetic modifications induced by early-life iron deficiency (30, 31). These findings suggest that this adjunct treatment may target molecular mechanisms underlying the long-term sequelae of iron deficiency. The extents to which iron status and choline supplementation affect the entire hippocampal transcriptome and whether the gene networks that are affected relate to the observed behavioral outcomes (22, 30) remain to be determined.
The hippocampus is a component of neural circuitry that regulates cognitive and affective behaviors (32). Perturbations of the hippocampal structure and function have been associated with dysfunction in cognition and social behavior (33), as well as a number of psychopathologies. Reduced hippocampal volume and dysfunction have been correlated with anxiety and schizophrenia (34–37). Hippocampal synaptic impairments have been associated with the pathogenesis of autism spectrum disorders (38). Although these hippocampal sequelae have been demonstrated in adult rats that were iron deficient (ID) during early life (23, 25, 39), the molecular underpinnings remain uncharacterized.
In the present study, we sequenced the hippocampal transcriptome of FID adult rats to identify molecular networks persistently affected by early-life iron deficiency that may underlie the associated cognitive impairments and the elevated risks for the pathogenesis of autism (6, 7) and schizophrenia (11). We also analyzed FID and iron sufficient (IS) rats treated with a choline-fortified diet from gestation day (G) 11 to G18 to determine whether prenatal choline supplementation can prevent changes in gene networks affected by early-life iron deficiency. Collectively, the findings delineate molecular changes underlying the long-term consequences of iron deficiency and provide a potential mechanism by which choline treatment may preserve behavioral function in FID individuals.
Methods
Animals.
G2 pregnant CD1 Sprague-Dawley rats were purchased from Charles Rivers Laboratories. The rats were kept in 12-h:12-h light:dark cycle with ad libitum access to food and water. The induction of fetal-neonatal iron deficiency and supplementation of choline were carried out following a previously described protocol (30) using diets with similar compositions. In brief, pregnant dams were given a purified ID diet (4 mg Fe/kg, 1.1 g choline chloride/kg, TD 80396; Harlan Teklad) from G2 to postnatal day (PND) 7, at which time nursing dams were given a purified IS diet (200 mg Fe/kg, 1.1 g choline chloride/kg, TD 01583; Harlan Teklad) to generate ID pups. Control IS pups were generated from pregnant dams maintained on the purified IS diet. For choline supplementation, pregnant rat dams were fed choline-fortified ID (4 mg Fe/kg, 5.0 g/kg choline chloride) or IS (200 mg Fe/kg, 5.0 g choline chloride/kg) diet from G11 through G18. All litters were culled to 8 pups with 6 males and 2 females at birth to minimize the sex-specific litter biases in maternal nurturing behavior (40). Pups were weaned at PND21, and littermates were housed together with four rats/cage until PND65. Only male offspring were used in the experiments. The University of Minnesota Institutional Animal Care and Use Committee approved all experiments in this study.
Hippocampal dissection.
PND65 rats were killed by injection of pentobarbital (intraperitoneally, 100 mg/kg). Brains were removed from the cranium and bisected along the midline on a metal block cooled in an ice bath. Hippocampi were dissected and immediately flash-frozen in liquid nitrogen and stored at −80°C as previously described (30).
RNA-Seq.
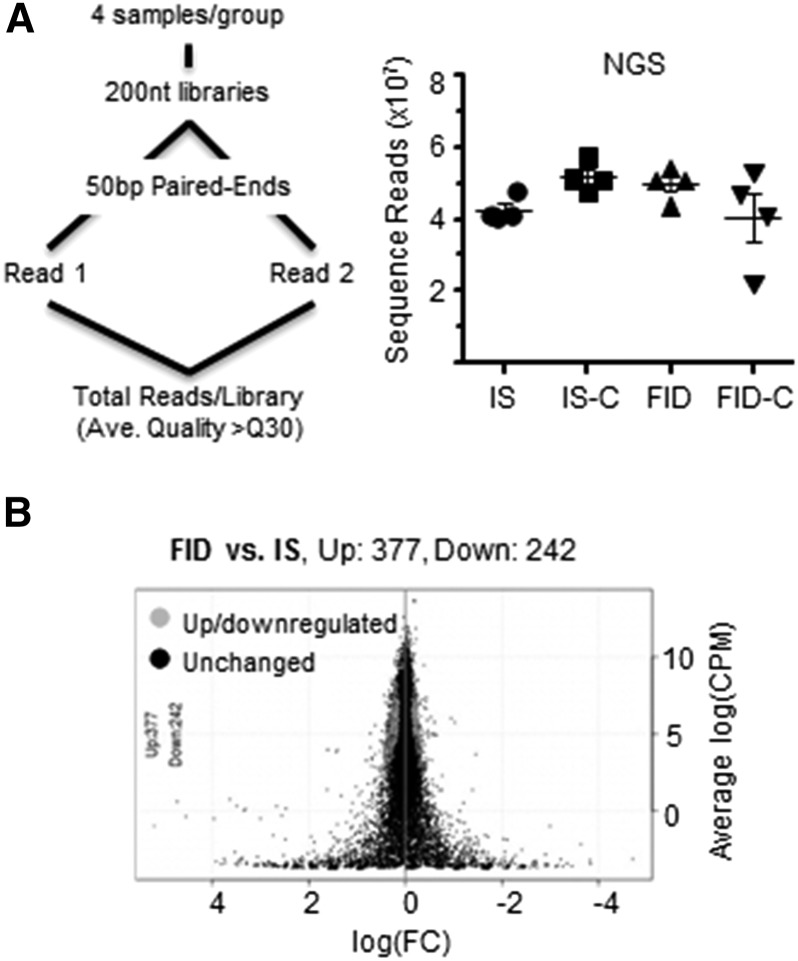
Hippocampal RNA (n = 4/group) was isolated by using RNAqueous RNA isolation kit (Ambion Inc.), quantified, and validated by RiboGreen RNA quantification (Invitrogen) and an Agilent 2100 Bioanalyzer (Agilent Inc.). The 16 pups for this analysis came from 12 dams (3 dams/group). Sample size was estimated with the use of power analysis described previously (41) to achieve optimal NGS data with high quality reads and alignments. Barcoded libraries were created for 4 biological replicates/group [IS, iron sufficient and choline supplemented (IS-C), FID, and formerly iron deficient and choline supplemented (FID-C)] with the use of a TruSeq RNA v2 kit (Illumina). Libraries were size-selected to have ∼200-bp inserts. Each library was sequenced as a 50-bp pair end across 2 lanes on the flow cell by using the HiSeq2000 (Agilent Inc.). Except for 1 FID-C library, the total depth of sequence reads were >4.0 × 107/library with mean quality scores above Q30 (Figure 1A). NGS data are deposited and stored at the Minnesota Super Computing Institute and can be made available upon request.
FIGURE 1.
RNA-Seq data generated from PND65 male rat hippocampus following gestational iron deficiency with or without choline supplementation. (A) Flow chart of RNA sample preparation and sequencing by Illumina HiSeq. The right panel shows total NGS reads from RNA libraries for each diet group. (B) Log(FC) plot shows differentially expressed genes between FID and IS data sets. Ave., average; bp, base pairs; CPM, count per million; Down, downregulated. FC, fold change; FID, formerly iron-deficient; FID-C, FID choline; IS, iron sufficient; IS-C, IS choline; NGS, next generation sequencing; nt, nucleotides; PND, postnatal day; Up, upregulated.
qRT-PCR.
The qRT-PCR experiments were carried out as described previously (31) with 5 biological replicates (n = 5/group). The sample size was chosen based on our previous reports (30, 31). The 20 pups for this analysis came from 16 dams, which were different from the dams used in the RNA-Seq experiments. In brief, 1 μg of total RNA was used to generate cDNA (High Capacity cDNA reverse transcription kit; Applied Biosystems) and diluted 10-fold to give a 200-μL final volume. Gene expression was quantified with the use of TaqMan Gene Expression Assay probes (Applied Biosystems) and a MX3000P instrument (Stratagene). Samples were analyzed in technical duplicates. Fold changes are means of duplicates, normalized to β-actin (internal loading control).
Bioinformatic Analysis.
RNA-Seq reads were qualified by using Galaxy FastQC and aligned with the use of the TopHat package (42) and the rat genome (rn4). Differential gene expression analyses were performed by using the EgdeR package (43). Differentially expressed genes between groups (FID compared with IS, IS-C compared with IS, FID-C compared with FID, and FID-C compared with IS-C) were identified with a log2 fold change of 0.20 and a false discovery rate (FDR) of < 5%. Hierarchic clustering analysis was performed by using the differentially expressed genes observed at least once. Ward’s criterion for genes with 1 − (correlation coefficient) was used as a distance measure. A clustering heat map was generated by using a z score scaled across all replicates for each gene.
Ingenuity Pathway Analysis.
The knowledge-based Ingenuity Pathway Analysis (IPA; Qiagen) was employed to identify networks, canonic pathways, molecular and cellular functions, and behavioral and neurologic disorders as previously described (44). The “core” analyses were performed employing a high-stringency filter, direct interactions, experimentally observed confidence, and cutoff settings at log2 (fold change): 0.2 and P < 0.05. IPA comparison analyses were performed to identify the effects of early-life iron deficiency or choline treatment among tested groups.
Statistics.
For qRT-PCR validation, interactions between iron status and choline treatment were analyzed by 2-factor ANOVA with post hoc Bonferroni-corrected t test for differences between groups. α was set at < 0.05. Values are means ± SEMs. There were no significant differences in variance between the groups that were compared. For IPA comparison analysis, Fisher’s exact test was used to calculate a P value for the association among genes in the data sets, pathways, and functions.
Results
Fetal-neonatal iron deficiency reprogrammed the adult hippocampal transcriptome.
Fetal-neonatal iron deficiency persistently altered the PND65 FID hippocampal transcriptome. NGS sequenced 10,787 loci of IS and FID transcriptomes. Differential gene expression analysis identified 377 upregulated and 242 downregulated genes (Figure 1B, FDR < 0.05), which represents 5.74% of the mapped loci. These genes were categorized as follows: 41–53% other (e.g., structural, transmembrane, nuclear factor, and unknown), 14–20% enzyme, 8% transcription regulator, 7–8% transporter, 5–6% kinase, 2–4% ion channel, 3–4% peptidase, 3% phosphatase, 2–4% G-protein–coupled receptor, 1% transmembrane receptor, and <1% of translation regulator, growth factor, and nuclear hormone receptor. To independently verify the RNA-Seq data, we used qPCR to analyze selected up- and downregulated genes implicated in neural plasticity (e.g., ApoE, Gda, Pld5, and Mbp) and behavior (e.g., Drd1a, Cartpt, and Igf1). The qPCR data confirmed the RNA-Seq results (Table 1).
TABLE 1.
Real-time (TaqMan) PCR validation of RNA-Seq data of PND65 rat hippocampal transcriptome following gestational iron deficiency with or without choline supplementation1
| Fold of IS |
||||||||
| RNA-Seq |
Real-time PCR |
|||||||
| Iron status × choline2 |
||||||||
| Gene | IS-C | FID | FID-C | IS-C | FID | FID-C | P value | F value |
| ApoE | 0.91 | 0.86* | 0.93 | 0.76*** | 0.73*** | 0.99 | <0.0001 | 42.2 |
| Cartpt | 0.58** | 0.50*** | 0.45*** | 0.94 | 0.58 | 1.15 | 0.08 | 3.58 |
| Drd1a | 1.08 | 1.22 | 1.00 | 1.18 | 1.39** | 1.07 | 0.0005 | 16.6 |
| Gas5 | 0.50** | 0.64 | 0.80 | 0.35*** | 0.44** | 0.66*** | <0.0001 | 114 |
| Gda | 1.19** | 1.24*** | 1.06 | 1.33*** | 1.58*** | 1.14 | <0.0001 | 139 |
| Hcrt | 0.74 | 0.57* | 1.00 | 0.75 | 0.44** | 1.07 | 0.06 | 4.17 |
| Igf1 | 1.03 | 1.33* | 1.06 | 1.04 | 1.20** | 0.96 | 0.0066 | 8.98 |
| Mbp | 0.94 | 0.86* | 1.04 | 0.64** | 0.61*** | 0.79 | 0.0002 | 22.2 |
| Pld5 | 1.27 | 2.57*** | 1.89*** | 1.39** | 2.35*** | 1.11 | <0.0001 | 40.6 |
| Pmf1 | 0.95 | 0.47* | 0.96 | 0.61*** | 0.62*** | 0.74** | <0.0001 | 90.5 |
Values are means (n = 4 for RNA-Seq and n = 5 for real-time PCR). *, **, ***Different from IS: *P < 0.05, ** P < 0.01, ***P < 0.001. FID, formerly iron deficient; FID-C, FID-choline; IS, iron sufficient; IS-C, IS choline; PND, postnatal day. A list of definitions of gene and protein names used in this table is included in Supplemental Material.
Two-factor ANOVA.
The FID adult hippocampal transcriptome showed altered gene networks associated with nervous system development and psychological disorders.
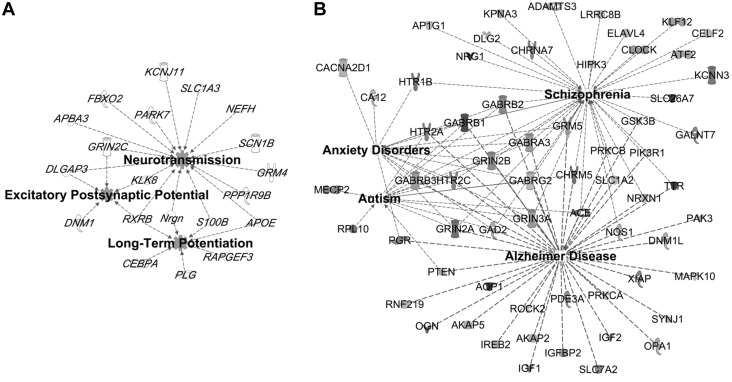
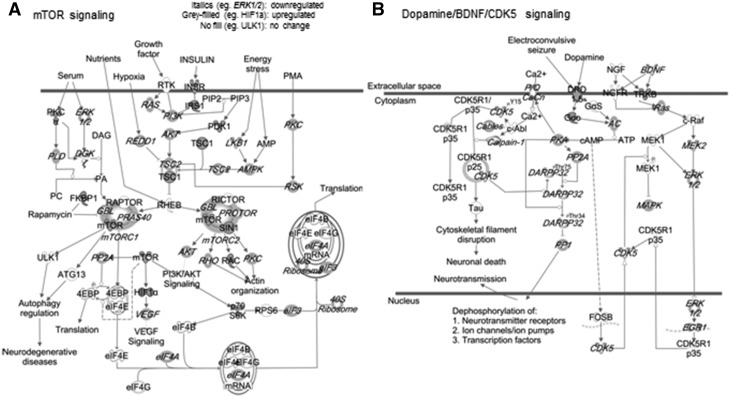
Fetal-neonatal iron deficiency downregulated genes that map onto functional networks previously implicated following early-life iron deficiency (19, 25, 45, 46), including neurotransmission and neuroplasticity (Figure 2A). Networks that were not previously known to be sensitive to iron deficiency were also identified (Table 2), including genes within a large interactive network that are regulated by transcription factors important for nervous system development (Supplemental Figure 1A). Similar to previous findings in the ID hippocampus (45), a number of upregulated genes in the FID hippocampus mapped onto the pathogenesis of Alzheimer disease (Figure 2B). Many of the substantially upregulated genes are involved in the etiology of anxiety disorders, autism, and schizophrenia (Figure 2B, Table 3). The upregulated genes clustered to an interactive network centering on RNA polymerase II, histone H3, and Akt, which are important regulators of gene expression and cellular growth (Supplemental Figure 1B). Canonic pathway analysis identified signaling mechanisms that have previously been shown to be disrupted by fetal-neonatal iron deficiency, including mechanistic target of rapamycin (mTOR)/p70S6K, dopamine, and BDNF signaling pathways (Figure 3) (46–48). Gene dysregulation was bidirectional (up and down) in some pathways (Figure 3) and unidirectional in others (Supplemental Table 1). The dysregulated genes in the FID transcriptome predicted alterations in the activity of upstream regulators, which are implicated in mediating neural plasticity and transcriptional regulation (Supplemental Figure 2).
FIGURE 2.
Annotated functions associated with dysregulated genes in the hippocampus on PND65 of FID rats. A number of downregulated factors (A) are known to be involved in neurotransmission and long-term potentiation, whereas a greater number of upregulated factors (B) are implicated in the pathogenesis of Alzheimer disease, autism, schizophrenia, and anxiety disorders. FID, formerly iron deficient; PND, postnatal day. A list of definitions of gene and protein names used in this figure is included in Supplemental Material.
TABLE 2.
Top 5 decreased diseases and functions in the hippocampus on PND65 of FID rats annotated by Ingenuity Pathway Analysis1
| Category | Disease or function annotation | P value | Molecules | Molecules, n |
| Cell-to-cell signaling | Neurotransmission | 4.33 × 10−5 | Apba3, Apoe, Dlgap3, Fbxo2, Grin2c, Grm4, Kcnj11, Klk8, Nefh, Nrgn, Park7, Ppp1r9b, Rxrb, S100b, Scn1b, Slc1a3 | 16 |
| Release and reuptake of neurotransmitter | 2.19 × 10−2 | Gsn, Nr2f6, Park7, Plg | 4 | |
| Long-term potentiation | 3.04 × 10−2 | Apoe, Cebpa, Nrgn, Plg, Rapgef3, Rxrb, S100b | 7 | |
| Nervous system development and function | Development of neurons | 3.24 × 10−5 | Apoe, Arhgef25, Bcan, Brsk1, Ccdc64, Clu, Cyth2, Dlgap3, Fam107a, Grm4, Gsn, Klk8, Mbp, Ncdn, Nefh, Nr2f6, Numbl, Plg, Ppp1r9b, Prkcsh, S100b, Slc1a3, Stk11, Ubb | 24 |
| Abnormal morphology of neurons | 9.38 × 10−4 | Aadat, Apoe, Brsk1, Cdk5r2, Dnm1, Fbxo2, Glb1, Gm2a, Klk8, Nefh, Ppp1r9b, Pvalb, Scn1b, Slc1a3 | 14 | |
| Outgrowth of neurites | 1.12 × 10−3 | Apoe, Arhgef25, Bcan, Brsk1, Cdk5r2, Cnpy2, Fam107a, Nefh, Pcsk1n, Plg, Ralgds, Scn1b | 12 | |
| Abnormal morphology of hippocampus | 1.27 × 10−3 | Apoe, Brsk1, Cdk5r2, Klk8, Lhx2, Ppp1r9b | 6 | |
| Behavior | Cognition | 1.38 × 10−3 | Apoe, Cebpa, Deaf1, Fam107a, Fbxo2, Gm2a, Grm4, Kcnj11, Klk8, Ncdn, Park7, Rapgef3, Rxrb, S100b | 14 |
| Locomotion | 1.05 × 10−3 | Aadat, Apoe, Dnm1, Kcnj11, Lgi4, Park7, Pcsk1n, Plg, Ptger1, Rxrb, Scn1b, Slc27a4 | 12 | |
| Lipid metabolism | Concentration of lipid | 1.81 × 10−3 | Alb, Apoe, Aqp3, Cbs/Loc102724560, Cebpa, Clu, Cpe, Ctss, Cxcl14, Glb1, Gm12253, Mif, Neu1, Pck2, Pcyt2, Plg, Pvalb, Rapgef3, Sec14l2, Slc27a4, Thrsp | 21 |
| Quantity of steroid | 2.25 × 10−3 | Alb, Apoe, Cbs/Loc102724560, Cebpa, Clu, Ctss, Cxcl14, Mif, Neu1, Plg, Pvalb, Rapgef3, Sec14l2, Slc27a4 | 14 | |
| Synthesis of lipid | 1.09 × 10−2 | Alb, Ang, Apoe, Cebpa, Clu, Gsta3, Igfbp7, Mif, Pcyt2, Plg, Sec14l2, Slc1a3, Slc27a4, Slc9a3r2, Stk11, Thrsp | 16 | |
| Molecular transport | Transport of molecule | 6.94 × 10−3 | Afm, Agap3, Ak1, Alb, Ang, Apoe, Aqp3, Brsk1, Cpe, Erp29, Fxyd7, Gm2a, Grin2c, Grm4, Gsn, Hgs, Ifi27, Kcnj11, Lin7b, Mif, Mlc1, Nmb, Pnp, Psd, Ptger1, Rtn2, Scn1b, Slc1a3, Slc27a4, Slc9a3r2, Stk11, Stx4 | 32 |
| Release of catecholamine | 7.54 × 10−3 | Gsn, Nr2f6, Park7, Plg | 4 | |
| Efflux of cholesterol | 1.12 × 10−2 | Apoe, Clu, Ctss, Rxrb | 4 |
FID, formerly iron deficient; PND, postnatal day. A list of definitions of gene and protein names used in this table is included in Supplemental Material.
TABLE 3.
Top 5 increased diseases and functions in the hippocampus on PND65 of FID rats annotated by Ingenuity Pathway Analysis1
| Category | Disease or function annotation | P value | Molecules | Molecules, n |
| Neurologic disease | Alzheimer disease | 1.05 × 10−5 | Ace, Adam10, Akap2, Akap5, Aqp1, Chrm5, Dhx9, Dnm1l, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Grin3a, Gsk3b, Hdac9, Htr2c, Lphn2, Mapk10, Opa1, Pcdh11×, Pgr, Pten, Rab14, Rnf219, Soat1, Synj1, Tph1, Ttr | 29 |
| Movement disorders | 1.86 × 10−4 | Abcb7, Anxa2, Aqp1, Ca13, Chrm5, Chrna7, Clcn3, Cntn1, Dgkb, Esrrg, Etv1, Faslg, Fgf12, Fgf14, Fmod, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Gnaq, Gnat2, Grin3a, Gsk3b, Htr1b, Htr2c, Igfbp5, Kcnab1, Lrrn1, Mpz, Oprl1, Pde10a, Pdpk1, Pkia, Plcb4, Ppargc1a, Ptpn9, Rnf2, Scn3a, St8sia3, Synj1, Top1, Traf6, Usp13 | 44 | |
| Huntington disease | 1.83 × 10−3 | Aqp1, Chrm5, Dgkb, Esrrg, Fgf12, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Gnat2, Grin3a, Htr1b, Htr2c, Kcnab1, Lrrn1, Pde10a, Pkia, Ppargc1a, Scn3a, St8sia3, Synj1, Top1, Traf6, Usp13 | 25 | |
| Psychological disorders | Mood disorders | 3.37 × 10−8 | Akap10, Ankrd6, Ca13, Chrm5, Chrna7, Dip2c, Dpp10, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Galr1, Grin3a, Gsk3b, Hdac9, Htr1b, Htr2c, Htr4, Pde10a, Pde11a, Pde1a, Pgr, Scn3a, Synj1, Tacr1, Tph1, Ttr, Ubr1, Zkscan1 | 30 |
| Autism | 6.15 × 10−4 | Gad2, Grin3a, Hdac9, Htr2c, Pten, Smad4 | 6 | |
| Schizophrenia | 1.14 × 10−3 | Ace, Ap1g1, Atf2, Chrm5, Chrna7, Clint1, Elavl4, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Grin3a, Gsk3b, Hdac9, Htr1b, Htr2c, Htr4, Kpna3, Sp4, Syt1, Tph1, Ttr | 23 | |
| Behavior | Anxiety | 8.05 × 10−7 | Akap5, Bmpr1a, Chrna7, Elavl4, Gabrg2, Gad2, Galr1, Htr1b, Htr4, Lrrn1, Nampt, Npy2r, Plcb4, Slc4a10, Tacr1, Trpc4 | 16 |
| Exploratory behavior | 5.97 × 10−3 | Gsk3b, Htr4, Kcnb2, Lrrn1, Lsamp, Pde10a | 6 | |
| Emotional behavior | 1.05 × 10−2 | Arid5b, Gabrg2, Gad2, Galr1, Gsk3b, Htr1b, Htr2c, Npy2r, Rab3c, Tacr1, Tph1 | 11 | |
| Nutritional disease | Eating disorders | 1.67 × 10−6 | Ca13, Chrm5, Gabra2, Gabrb2, Gabrb3, Gabrg2, Grin3a, Htr1b, Htr2c, Htr4, Npy2r, Pank1, Pgr, Pten | 14 |
| Obesity | 1.84 × 10−5 | Ace, Arid5b, Ca13, Chm, Crbn, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Grin3a, Hdac9, Htr1b, Htr2c, Htr4, Jak2, Med1, Nampt, Npy2r, Nr2c2, Pde10a, Pde11a, Ppargc1a, Pten, Scn3a | 25 | |
| Metabolic disease | Amyloidosis | 1.06 × 10−5 | Ace, Adam10, Akap2, Akap5, Aqp1, Chrm5, Dhx9, Dnm1l, Gabra2, Gabrb2, Gabrb3, Gabrg2, Gad2, Grin3a, Gsk3b, Hdac9, Htr2c, Igfbp5, Lphn2, Mapk10, Opa1, Pcdh11×, Pgr, Pten, Rab14, Rnf219, Soat1, Synj1, Tph1, Ttr | 30 |
FID, formerly iron deficient; PND, postnatal day. A list of definitions of gene and protein names used in this table is included in Supplemental Material.
FIGURE 3.
Altered mTOR/p70S6K (A) and dopamine/BDNF/CDK5 (B) signaling pathways in the hippocampus on PND65 of FID rats. BDNF, brain-derived neurotrophic factor; CDK5, cyclin-dependent kinase 5; FID, formerly iron deficient; mTOR, mechanistic target of rapamycin; PND, postnatal day. A list of definitions of gene and protein names used in this figure is included in Supplemental Material.
Choline supplementation during late gestation reduced the effects of fetal-neonatal iron deficiency on the adult hippocampal transcriptome.
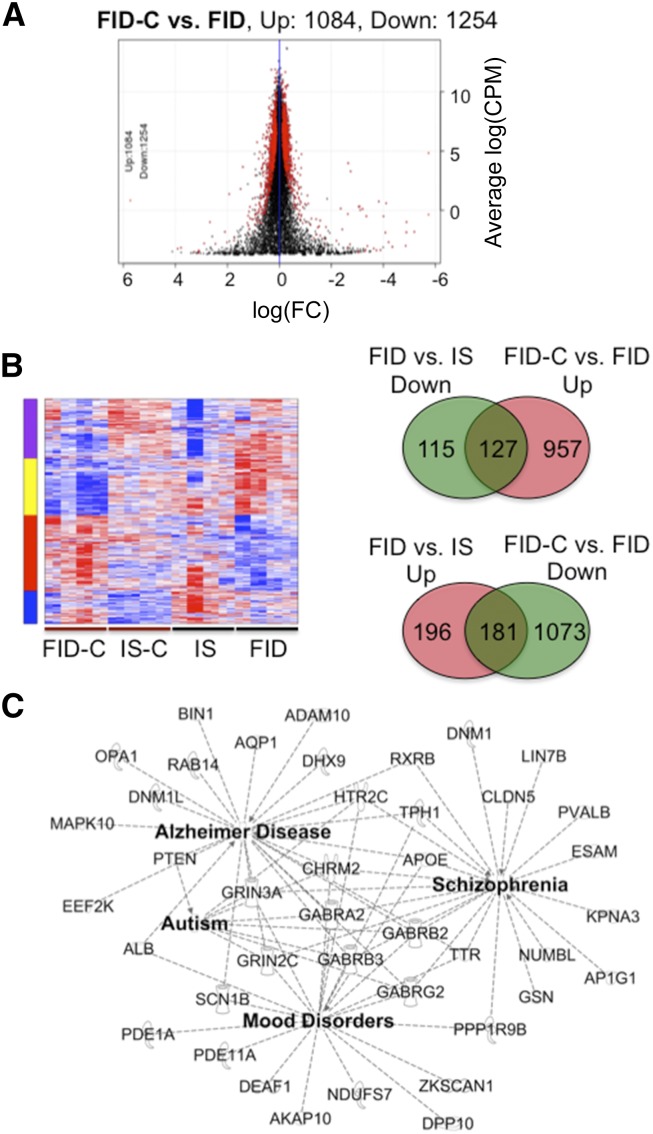
In the FID and FID-C transcriptomes, 10,839 loci were sequenced, of which 1084 upregulated and 1254 downregulated genes were altered, representing 21.6% of the mapped loci (Figure 4A). Hierarchic gene clustering identified a set of genes dysregulated by iron deficiency that responded to choline treatment (Figure 4B). These include a normalization of iron deficiency–induced downregulation of 127 genes and upregulation of 181 genes (Figure 4B, Venn diagrams). Functional annotation by IPA showed a partial normalization of specific molecular and cellular functions altered by early-life iron deficiency (Supplemental Table 2). The greatest recovery of iron deficiency-impaired functions occurred in gene networks associated with neurologic disorders including autism, schizophrenia, and Alzheimer disease (Figure 4C), as well as in neurotransmission and cellular microtubule dynamics (data not shown). In total, 13 of 18 autism-associated genes and 21 of 51 schizophrenia-associated genes were recovered by prenatal choline treatment (Supplemental Table 3). A large number of the normalized genes mapped onto interactive networks that mediate transcriptional regulation and neuronal growth and differentiation (Supplemental Figure 3).
FIGURE 4.
Prenatal choline supplementation partially normalized dysregulated genes in the hippocampus on PND65 of FID rats. (A) Scatter plot showing differentially expressed genes between the FID-C and FID groups. (B) Heat map showing gene clusters from all treatment groups. The key indicates differential gene expression by diet. Overlapped regions of Venn diagrams depict genes (127 downregulated and 181 upregulated) altered in the FID and normalized in the FID-C group. (C) Interactive gene networks implicated in the pathogenesis of Alzheimer disease, schizophrenia, autism, and mood disorders that were normalized by prenatal choline treatment of the FID group. CPM, counts per million; Down, downregulated; FC, fold change; FID, formerly iron deficient; FID-C, FID-choline; IS, iron sufficient; IS-C, IS choline; PND, postnatal day; Up, upregulated. A list of definitions of gene and protein names used in this figure is included in Supplemental Material.
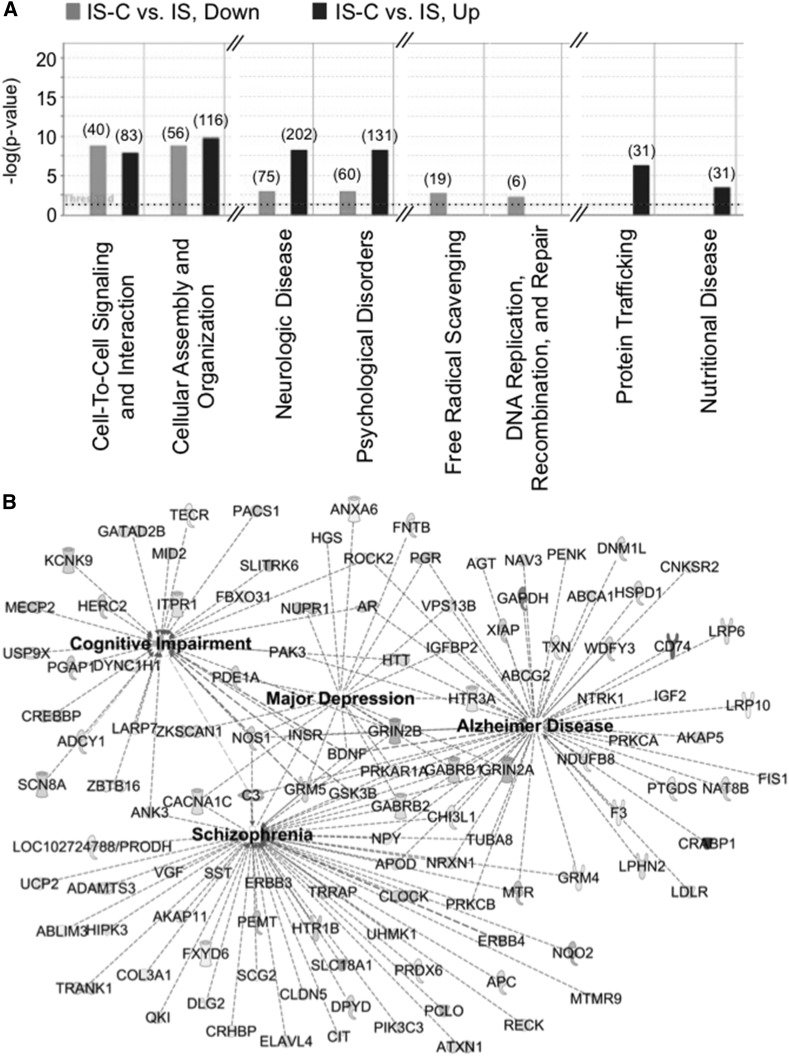
Late gestational dietary choline supplementation reprograms the hippocampal transcriptome of the adult IS rats.
The long-term effects of dietary choline supplementation during pregnancy on the adult hippocampal transcriptome in the always IS animals are unknown. Thus, we examined the effects of prenatal choline supplementation on the adult hippocampal transcriptome. NGS data of IS and IS-C transcriptomes mapped onto 10,799 loci. Analysis of differentially expressed genes identified 636 upregulated and 563 downregulated genes (Figure 5A, FDR < 0.05), representing 11.1% of the mapped loci. Dysregulated genes mapped onto functional networks that are important for cell-to-cell signaling, cellular organization, neurologic disease, and psychological disorders (Figure 5A). The substantially dysregulated genes were also involved in the pathogenesis of Alzheimer disease, depression, and schizophrenia (Figure 5B).
FIGURE 5.
Prenatal choline supplementation reprogrammed the hippocampal transcriptome of PND65 IS-C rats. (A) Bar graphs showing functions that are associated with dysregulated genes in the transcriptome of IS-C rats. The key above the graph explains the number of altered genes and direction of change relative to IS rats. (B) Gene networks altered in the IS-C hippocampus that are implicated in the cognitive impairment and the pathogenesis of schizophrenia, major depression, and Alzheimer disease. Down, downregulated; IS, iron sufficient; IS-C, IS choline; PND, postnatal day; Up, upregulated. A list of definitions of gene and protein names used in this figure is included in Supplemental Material.
Discussion
Characterizing the functional networks of genes persistently altered by early-life iron deficiency is an important step in ultimately developing improved preventive and remedial strategies, particularly in light of the failure of iron treatment alone to diminish long-term dysfunction. To identify such networks and pathways, we examined the adult hippocampal transcriptome. Our results demonstrate the wide-ranging effects of early-life iron deficiency by identifying over 600 genes and their relevant molecular networks that are permanently altered by this nutrient deficiency, including disruptions in known pathways such as mTOR and Bdnf.
The alterations that were identified in the molecular networks are consistent with observed long-term behavioral deficits, including cognitive deficits and anxiety, found both in FID humans (20, 21, 49, 50) and in rodent models (6–17, 51). The changes within specific gene networks may constitute key components of the neurobiological mechanisms underlying the effects of iron deficiency. For example, altered expression of genes associated with neuronal transmission and abnormal neuronal morphology may underlie disruptions in long-term potentiation (22, 25) and dendritic branching (23) seen in FID animals. Thus, the altered interactive gene networks identified in this study help establish the molecular mechanisms through which iron deficiency leads to these long-term behavioral deficits in FID individuals (17). We also identified pathways, which, to our knowledge, are novel, that were altered in the FID hippocampal transcriptome, including glucocorticoid receptor signaling and the oxidative stress response pathway (Supplemental Figures 4 and 5). Coupled with the change in dopamine receptor signaling (Figure 2B), altered glucocorticoid receptor signaling is consistent with increased anxiety behavior and altered stress responses observed in both FID rats (18, 19, 21, 52) and humans (8, 53). The altered NRF2-mediated oxdative stress response pathway is of interest given its role in maintaining cellular homeostasis in responses to diverse extracellular stressors (54, 55). Moreover, emerging evidence suggests a link between NRF2-mediated signaling and the etiology of autism spectrum disorders (56) and schizophrenia (57, 58), both of which have been associated with the offspring of maternal iron deficiency in humans (6, 7, 11).
Mechanisms underlying the risk for psychiatric disorders such as schizophrenia (11) that are associated with early-life iron deficiency remains uncharacterized. Adult FID rats exhibit a deficit in sensorimotor gating during prepulse inhibition (22, 59) and reduced social interaction (30), suggesting that a rodent model of early-life iron deficiency displays a schizophrenia-like phenotype in adulthood. Here we report that about 51 genes were altered in the FID hippocampal transcriptome, which are mapped onto the pathogenesis of schizophrenia (Supplemental Figure 6, FID compared with IS), providing a potential mechanism for the association between schizophrenia and early-life iron deficiency. The findings also highlight the consistency between dysregulated gene networks and functional outcomes in FID rats (22).
Prenatal choline supplementation substantially reprogrammed the adult hippocampal transcriptome of both FID and always IS animals. This effect was much more pronounced in the FID hippocampus, in which choline supplementation modified expression of nearly twice the number of genes as in the IS-C group. Our major finding was that choline supplementation normalized expression levels of more than half of all genes dysregulated by iron deficiency and multiple molecular networks, including those implicated in neurotransmission, autism, and schizophrenia. Interestingly, although choline treatment facilitates the normalization of a subset of dysregulated genes induced by early-life iron deficiency that are associated with schizophrenia (21 of 51), expression of a larger number of genes within the interactive network were also modified by choline treatment (Supplemental Table 3). Many of these genes were functional analogues of the non-recovered genes, suggesting that choline promotes a corrective action by influencing the regulation of alternative substrates. The beneficial effects of prenatal choline treatment are consistent with previous studies that show the increased expression of synaptic plasticity genes in animal models of early-life environmental insults (30, 60, 61). Collectively, these findings provide new insights into the molecular mechanisms by which early-life iron deficiency and prenatal choline treatment modify gene regulation that ultimately lead to long-term behavioral abnormalities. Despite the incomplete recovery of the transcriptome in the FID-C hippocampus, our findings suggest that choline supplementation could be effective as an adjunct therapy in humans to prevent some long-term behavioral consequences of fetal or early postnatal iron deficiency.
The extensive overlap between the negative early-life iron deficiency effects and the positive effects of prenatal choline treatment on gene regulation suggests a common underlying mechanism. One potential mechanism is stable epigenetic modifications across the genome (31, 62, 63). We have previously demonstrated that early-life iron deficiency alters RNA polymerase II activity and modifies histone H3 of the Bdnf promoter (31), which could potentially serve as a mechanism for the long-term downregulation of hippocampal Bdnf. In the present study, we found alterations in gene networks centered on RNA polymerase II and histone H3 in the FID hippocampus, which were normalized with concurrent choline supplementation. This finding provides additional evidence that choline supplementation may be targeting epigenetic mechanisms underlying the long-term cellular and behavioral effects of iron deficiency. Given the evidence that choline supplementation influences DNA methylation (62, 63), future studies could determine whether these epigenetic changes involved DNA methylation or histone modification and whether they occurred at gene regulatory regions (e.g., promoters and enhancers).
Consistent with previous studies, prenatal choline supplementation produced beneficial effects on gene expression in FID hippocampus. However, it appears to have some negative effects on the hippocampal transcriptome of IS rats. These findings suggest that the effects of choline on hippocampal gene expression may be dependent in part on iron status. In the IS rats, prenatal choline supplementation altered expression of genes within molecular networks associated with neuropathologies and psychological disorders. One major finding is that dysregulated genes existed within functional networks associated with schizophrenia (Figure 5C). Although more functional analysis is needed to corroborate these molecular changes, our preliminary finding of reduced sensorimotor gating (data not shown) in IS-C rats is consistent with the prediction based on gene expression analysis. Our negative findings were somewhat unexpected with respect to previous studies that report the beneficial effects of prenatal choline supplementation in normal animals with improved hippocampal expression of Bdnf (60), neurotransmission (64, 65), dendritic structure (66), and spatial memory (67). The discrepant outcomes in these studies may be due to the differences in the routes of choline delivery (feed pellets compared with drinking water), dosages, treatment durations, and the postnatal ages at which analyses were performed. In particular, timing of maternal choline supplementation appears to be a key factor. Supplementation throughout gestation and lactation (G2 to PND7) shows a downregulation of hippocampal Bdnf expression in both FID and IS rats, whereas choline supplementation given on G11–18 results in upregulation of hippocampal Bdnf in the FID offspring (30, 68). Williams and colleagues (60, 61) reported that prenatal choline treatment induces bidirectional changes in expression of synaptic plasticity genes (e.g., increased Bdnf and double cortin, lower glial fibrillary acidic protein); however, these changes were not accompanied by improved long-term potentiation in normal adult rats. Likewise, no change in hippocampal expression of nerve growth factors was observed between choline-treated and untreated normal male rats at 24 mo of age (69). Collectively, the effects of choline supplementation on gene expression in the brain appear largely positive but may also be dependent on the design of the study, where the dosage, treatment duration, and type of early-life insult need to be considered. Clearly, there is a need for additional research, particularly on the timing and dosage of choline supplementation, to clarify how this supplementation can benefit hippocampal development and, ultimately, cognitive and socioemotional behaviors in the FID and IS animals.
In conclusion, early-life iron deficiency is a major clinical condition with long-lasting cognitive, emotional, and behavioral effects even after iron repletion. Accumulating evidence shows an increased risk for autism (6, 7) and schizophrenia (11) following maternal and fetal iron deficiency. These long-term effects constitute the true cost to society (17). Our findings highlight molecular pathways disrupted in the FID hippocampus that have been found to be associated with cognitive impairments and the development of psychopathologies (e.g., autism and schizophrenia). The findings underscore the need to identify populations at risk for brain iron deficiency before the detection of systemic iron deficiency anemia, by which time the brain has already become ID (3, 15, 70). In order to do this, the identification of novel molecular markers that can index brain iron status is needed. This would advance the field beyond the current methods of screening for iron deficiency anemia (e.g., hematocrit, ferritin, and hepcidin), which are inadequate to detect brain iron deficiency. The disruptions of specific molecular networks were diminished by choline treatment during late gestation, supporting the use of choline as an accessible and effective adjunct therapy for those at risk for fetal and early postnatal iron deficiency (71–73).
Acknowledgments
We thank Diana Wallin for editorial assistance. PVT designed, conducted, analyzed, and wrote the paper and had primary responsibility for deciding its final content; BCK conducted, analyzed, and wrote the paper; MTP performed and analyzed prepulse inhibition data; K-JW performed bioinformatics analysis; JCG reviewed and edited the paper; RAS and MKG designed, reviewed, and edited the paper. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: BDNF, brain-derived neurotrophic factor; FDR, false discovery rate; FID, formerly iron deficient; FID-C, formerly iron deficient and choline supplemented; G, gestational day; ID, iron deficient; IPA, Ingenuity Pathway Analysis; IS, iron sufficient; IS-C, iron sufficient and choline supplemented; mTOR, mechanistic target of rapamycin; NGS, next generation sequencing; PND, postnatal day.
References
- 1.Siddappa AM, Rao R, Long JD, Widness JA, Georgieff MK. The assessment of newborn iron stores at birth: A review of the literature and standards for ferritin concentrations. Neonatology 2007;92:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georgieff MK, Wewerka SW, Nelson CA, Deregnier RA. Iron status at 9 months of infants with low iron stores at birth. J Pediatr 2002;141:405–9. [DOI] [PubMed] [Google Scholar]
- 3.Petry CD, Eaton MA, Wobken JD, Mills MM, Johnson DE, Georgieff MK. Iron deficiency of liver, heart, and brain in newborn infants of diabetic mothers. J Pediatr 1992;121:109–14. [DOI] [PubMed] [Google Scholar]
- 4.Chockalingam UM, Murphy E, Ophoven JC, Weisdorf SA, Georgieff MK. Cord transferrin and ferritin values in newborn infants at risk for prenatal uteroplacental insufficiency and chronic hypoxia. J Pediatr 1987;111:283–6. [DOI] [PubMed] [Google Scholar]
- 5.Sweet DG, Savage G, Tubman TR, Lappin TR, Halliday HL. Study of maternal influences on fetal iron status at term using cord blood transferrin receptors. Arch Dis Child Fetal Neonatal Ed 2001;84:F40–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt RJ, Tancredi DJ, Krakowiak P, Hansen RL, Ozonoff S. Maternal intake of supplemental iron and risk of autism spectrum disorder. Am J Epidemiol 2014;180:890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidrak S, Yoong T, Woolfenden S. Iron deficiency in children with global developmental delay and autism spectrum disorder. J Paediatr Child Health 2014;50:356–61. [DOI] [PubMed] [Google Scholar]
- 8.Lozoff B, Jimenez E, Hagen J, Mollen E, Wolf AW. Poorer behavioral and developmental outcome more than 10 years after treatment for iron deficiency in infancy. Pediatrics 2000;105:E51. [DOI] [PubMed] [Google Scholar]
- 9.Peirano PD, Algarin CR, Garrido MI, Lozoff B. Iron deficiency anemia in infancy is associated with altered temporal organization of sleep states in childhood. Pediatr Res 2007;62:715–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozoff B, Beard J, Connor J, Barbara F, Georgieff M, Schallert T. Long-lasting neural and behavioral effects of iron deficiency in infancy. Nutr Rev 2006;64:S34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Insel BJ, Schaefer CA, McKeague IW, Susser ES, Brown AS. Maternal iron deficiency and the risk of schizophrenia in offspring. Arch Gen Psychiatry 2008;65:1136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian P, Murray-Kolb LE, Khatry SK, Katz J, Schaefer BA, Cole PM, Leclerq SC, Tielsch JM. Prenatal micronutrient supplementation and intellectual and motor function in early school-aged children in Nepal. JAMA 2010;304:2716–23. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Yan H, Zeng L, Cheng Y, Liang W, Dang S, Wang Q, Tsuji I. Effects of maternal multimicronutrient supplementation on the mental development of infants in rural western China: Follow-up evaluation of a double-blind, randomized, controlled trial. Pediatrics 2009;123:e685–92. [DOI] [PubMed] [Google Scholar]
- 14.Chang S, Zeng L, Brouwer ID, Kok FJ, Yan H. Effect of iron deficiency anemia in pregnancy on child mental development in rural China. Pediatrics 2013;131:e755–63. [DOI] [PubMed] [Google Scholar]
- 15.Siddappa AM, Georgieff MK, Wewerka S, Worwa C, Nelson CA, Deregnier RA. Iron deficiency alters auditory recognition memory in newborn infants of diabetic mothers. Pediatr Res 2004;55:1034–41. [DOI] [PubMed] [Google Scholar]
- 16.Lukowski AF, Koss M, Burden MJ, Jonides J, Nelson CA, Kaciroti N, Jimenez E, Lozoff B. Iron deficiency in infancy and neurocognitive functioning at 19 years: Evidence of long-term deficits in executive function and recognition memory. Nutr Neurosci 2010;13:54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lozoff B, Smith JB, Kaciroti N, Clark KM, Guevara S, Jimenez E. Functional significance of early-life iron deficiency: Outcomes at 25 years. J Pediatr 2013;163:1260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eseh R, Zimmerberg B. Age-dependent effects of gestational and lactational iron deficiency on anxiety behavior in rats. Behav Brain Res 2005;164:214–21. [DOI] [PubMed] [Google Scholar]
- 19.Beard J, Erikson KM, Jones BC. Neonatal iron deficiency results in irreversible changes in dopamine function in rats. J Nutr 2003;133:1174–9. [DOI] [PubMed] [Google Scholar]
- 20.Carlson ES, Tkac I, Magid R, O’Connor MB, Andrews NC, Schallert T, Gunshin H, Georgieff MK, Petryk A. Iron is essential for neuron development and memory function in mouse hippocampus. J Nutr 2009;139:672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felt BT, Beard JL, Schallert T, Shao J, Aldridge JW, Connor JR, Georgieff MK, Lozoff B. Persistent neurochemical and behavioral abnormalities in adulthood despite early iron supplementation for perinatal iron deficiency anemia in rats. Behav Brain Res 2006;171:261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pisansky MT, Wickham RJ, Su J, Fretham S, Yuan LL, Sun M, Gewirtz JC, Georgieff MK. Iron deficiency with or without anemia impairs prepulse inhibition of the startle reflex. Hippocampus 2013;23:952–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunette KE, Tran PV, Wobken JD, Carlson ES, Georgieff MK. Gestational and neonatal iron deficiency alters apical dendrite structure of CA1 pyramidal neurons in adult rat hippocampus. Dev Neurosci 2010;32:238–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgenson LA, Wobken JD, Georgieff MK. Perinatal iron deficiency alters apical dendritic growth in hippocampal CA1 pyramidal neurons. Dev Neurosci 2003;25:412–20. [DOI] [PubMed] [Google Scholar]
- 25.Jorgenson LA, Sun M, O’Connor M, Georgieff MK. Fetal iron deficiency disrupts the maturation of synaptic function and efficacy in area CA1 of the developing rat hippocampus. Hippocampus 2005;15:1094–102. [DOI] [PubMed] [Google Scholar]
- 26.Thomas JD, Abou EJ, Dominguez HD. Prenatal choline supplementation mitigates the adverse effects of prenatal alcohol exposure on development in rats. Neurotoxicol Teratol 2009;31:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas JD, La Fiette MH, Quinn VR, Riley EP. Neonatal choline supplementation ameliorates the effects of prenatal alcohol exposure on a discrimination learning task in rats. Neurotoxicol Teratol 2000;22:703–11. [DOI] [PubMed] [Google Scholar]
- 28.Nag N, Mellott TJ, Berger-Sweeney JE. Effects of postnatal dietary choline supplementation on motor regional brain volume and growth factor expression in a mouse model of Rett syndrome. Brain Res 2008;1237:101–9. [DOI] [PubMed] [Google Scholar]
- 29.Schaevitz L, Berger-Sweeney J, Ricceri L. One-carbon metabolism in neurodevelopmental disorders: Using broad-based nutraceutics to treat cognitive deficits in complex spectrum disorders. Neurosci Biobehav Rev 2014;46:270–84. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy BC, Dimova JG, Siddappa AJ, Tran PV, Gewirtz JC, Georgieff MK. Prenatal choline supplementation ameliorates the long-term neurobehavioral effects of fetal-neonatal iron deficiency in rats. J Nutr 2014;144:1858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran PV, Kennedy BC, Lien YC, Simmons RA, Georgieff MK. Fetal iron deficiency induces chromatin remodeling at the Bdnf locus in adult rat hippocampus. Am J Physiol Regul Integr Comp Physiol 2015;308:R276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 2010;65:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubin RD, Watson PD, Duff MC, Cohen NJ. The role of the hippocampus in flexible cognition and social behavior. Front Hum Neurosci 2014;8:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, Agartz I, Westlye LT, Haukvik UK, Dale AM, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 2015. Aug 18 (Epub ahead of print; DOI: 10.1038/mp.2015.118). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrisberger F, Smieskova R, Schmidt A, Lenz C, Walter A, Wittfeld K, Grabe HJ, Lang UE, Fusar-Poli P, Borgwardt S. BDNF Val66Met polymorphism and hippocampal volume in neuropsychiatric disorders: A systematic review and meta-analysis. Neurosci Biobehav Rev 2015;55:107–18. [DOI] [PubMed] [Google Scholar]
- 36.Heckers S, Rauch SL, Goff D, Savage CR, Schacter DL, Fischman AJ, Alpert NM. Impaired recruitment of the hippocampus during conscious recollection in schizophrenia. Nat Neurosci 1998;1:318–23. [DOI] [PubMed] [Google Scholar]
- 37.Heckers S, Konradi C. GABAergic mechanisms of hippocampal hyperactivity in schizophrenia. Schizophr Res 2015;167:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Won H, Mah W, Kim E. Autism spectrum disorder causes, mechanisms, and treatments: Focus on neuronal synapses. Front Mol Neurosci 2013;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao R, Tkac I, Schmidt AT, Georgieff MK. Fetal and neonatal iron deficiency causes volume loss and alters the neurochemical profile of the adult rat hippocampus. Nutr Neurosci 2011;14:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosten TA, Huang W, Nielsen DA. Sex and litter effects on anxiety and DNA methylation levels of stress and neurotrophin genes in adolescent rats. Dev Psychobiol 2014;56:392–406. [DOI] [PubMed] [Google Scholar]
- 41.Ching T, Huang S, Garmire LX. Power analysis and sample size estimation for RNA-Seq differential expression. RNA 2014;20:1684–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran PV, Dakoji S, Reise KH, Storey KK, Georgieff MK. Fetal iron deficiency alters the proteome of adult rat hippocampal synaptosomes. Am J Physiol Regul Integr Comp Physiol 2013;305:R1297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlson ES, Stead JD, Neal CR, Petryk A, Georgieff MK. Perinatal iron deficiency results in altered developmental expression of genes mediating energy metabolism and neuronal morphogenesis in hippocampus. Hippocampus 2007;17:679–91. [DOI] [PubMed] [Google Scholar]
- 46.Tran PV, Fretham SJ, Carlson ES, Georgieff MK. Long-term reduction of hippocampal brain-derived neurotrophic factor activity after fetal-neonatal iron deficiency in adult rats. Pediatr Res 2009;65:493–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fretham SJ, Carlson ES, Georgieff MK. Neuronal-specific iron deficiency dysregulates mammalian target of rapamycin signaling during hippocampal development in nonanemic genetic mouse models. J Nutr 2013;143:260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McEchron MD, Goletiani CJ, Alexander DN. Perinatal nutritional iron deficiency impairs noradrenergic-mediated synaptic efficacy in the CA1 area of rat hippocampus. J Nutr 2010;140:642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corapci F, Radan AE, Lozoff B. Iron deficiency in infancy and mother-child interaction at 5 years. J Dev Behav Pediatr 2006;27:371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lozoff B, Beard J, Connor J, Barbara F, Georgieff M, Schallert T. Long-lasting neural and behavioral effects of iron deficiency in infancy. Nutr Rev 2006;64:S34–43; discussion S72–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt MK, Muslimatun S, West CE, Schultink W, Hautvast JG. Mental and psychomotor development in Indonesian infants of mothers supplemented with vitamin A in addition to iron during pregnancy. Br J Nutr 2004;91:279–86. [DOI] [PubMed] [Google Scholar]
- 52.Weinberg J, Levine S, Dallman PR. Long-term consequences of early iron deficiency in the rat. Pharmacol Biochem Behav 1979;11:631–8. [DOI] [PubMed] [Google Scholar]
- 53.Felt BT, Peirano P, Algarin C, Chamorro R, Sir T, Kaciroti N, Lozoff B. Long-term neuroendocrine effects of iron-deficiency anemia in infancy. Pediatr Res 2012;71:707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Tan J, Miao Y, Lei P, Zhang Q. ROS and autophagy: Interactions and molecular regulatory mechanisms. Cell Mol Neurobiol 2015;35:615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bakunina N, Pariante CM, Zunszain PA. Immune mechanisms linked to depression via oxidative stress and neuroprogression. Immunology 2015. Jan 10 (Epub ahead of print; DOI: 10.1111/imm.12443). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Napoli E, Wong S, Hertz-Picciotto I, Giulivi C. Deficits in bioenergetics and impaired immune response in granulocytes from children with autism. Pediatrics 2014;133:e1405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bitanihirwe BK, Woo TU. Oxidative stress in schizophrenia: An integrated approach. Neurosci Biobehav Rev 2011;35:878–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martins-de-Souza D, Harris LW, Guest PC, Bahn S. The role of energy metabolism dysfunction and oxidative stress in schizophrenia revealed by proteomics. Antioxid Redox Signal 2011;15:2067–79. [DOI] [PubMed] [Google Scholar]
- 59.Harvey L, Boksa P. Additive effects of maternal iron deficiency and prenatal immune activation on adult behaviors in rat offspring. Brain Behav Immun 2014;40:27–37. [DOI] [PubMed] [Google Scholar]
- 60.Wong-Goodrich SJ, Mellott TJ, Glenn MJ, Blusztajn JK, Williams CL. Prenatal choline supplementation attenuates neuropathological response to status epilepticus in the adult rat hippocampus. Neurobiol Dis 2008;30:255–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong-Goodrich SJ, Glenn MJ, Mellott TJ, Liu YB, Blusztajn JK, Williams CL. Water maze experience and prenatal choline supplementation differentially promote long-term hippocampal recovery from seizures in adulthood. Hippocampus 2011;21:584–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J 2006;20:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003;23:5293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montoya D, Swartzwelder HS. Prenatal choline supplementation alters hippocampal N-methyl-D-aspartate receptor-mediated neurotransmission in adult rats. Neurosci Lett 2000;296:85–8. [DOI] [PubMed] [Google Scholar]
- 65.Pyapali GK, Turner DA, Williams CL, Meck WH, Swartzwelder HS. Prenatal dietary choline supplementation decreases the threshold for induction of long-term potentiation in young adult rats. J Neurophysiol 1998;79:1790–6. [DOI] [PubMed] [Google Scholar]
- 66.Li Q, Guo-Ross S, Lewis DV, Turner D, White AM, Wilson WA, Swartzwelder HS. Dietary prenatal choline supplementation alters postnatal hippocampal structure and function. J Neurophysiol 2004;91:1545–55. [DOI] [PubMed] [Google Scholar]
- 67.Williams CL, Meck WH, Heyer DD, Loy R. Hypertrophy of basal forebrain neurons and enhanced visuospatial memory in perinatally choline-supplemented rats. Brain Res 1998;794:225–38. [DOI] [PubMed] [Google Scholar]
- 68.Georgieff MK, Brunette KE, Tran PV. Early life nutrition and neural plasticity. Dev Psychopathol 2015;27:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glenn MJ, Kirby ED, Gibson EM, Wong-Goodrich SJ, Mellott TJ, Blusztajn JK, Williams CL. Age-related declines in exploratory behavior and markers of hippocampal plasticity are attenuated by prenatal choline supplementation in rats. Brain Res 2008;1237:110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lozoff B, Clark KM, Jing Y, Armony-Sivan R, Angelilli ML, Jacobson SW. Dose-response relationships between iron deficiency with or without anemia and infant social-emotional behavior. J Pediatr 2008;152:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bastian TW, Prohaska JR, Georgieff MK, Anderson GW. Perinatal iron and copper deficiencies alter neonatal rat circulating and brain thyroid hormone concentrations. Endocrinology 2010;151:4055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bastian TW, Prohaska JR, Georgieff MK, Anderson GW. Fetal and neonatal iron deficiency exacerbates mild thyroid hormone insufficiency effects on male thyroid hormone levels and brain thyroid hormone-responsive gene expression. Endocrinology 2014;155:1157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Georgieff MK, Mills MM, Gordon K, Wobken JD. Reduced neonatal liver iron concentrations after uteroplacental insufficiency. J Pediatr 1995;127:308–11. [DOI] [PubMed] [Google Scholar]