Summary
Background
Developmental abnormalities observed in Cornelia de Lange syndrome have been genetically linked to mutations in the cohesin machinery. These and other recent experimental findings have led to the suggestion that cohesin, in addition to its canonical function of mediating sister chromatid cohesion, might also be involved in regulating gene expression.
Results
We report that cleavage of cohesin’s kleisin subunit in postmitotic Drosophila salivary glands induces major changes in the transcript levels of many genes. Kinetic analyses of changes in transcript levels upon cohesin cleavage reveal that a subset of genes responds to cohesin cleavage within a few hours. In addition, cohesin binds to most of these loci, suggesting that cohesin is directly regulating their expression. Among these genes are several that are regulated by the steroid hormone ecdysone. Cytological visualization of transcription at selected ecdysone-responsive genes reveals that puffing at Eip74EF ceases within an hour or two of cohesin cleavage, long before any decline in ecdysone receptor could be detected at this locus.
Conclusion
We conclude that cohesin regulates expression of a distinct set of genes, including those mediating the ecdysone response.
Introduction
The regulation of gene expression essential for normal animal development is largely mediated by sequence-specific transcription factors. One of the more mysterious aspects of developmentally regulated transcription concerns how transcription factors bound to remote regulatory sequences modulate transcription of genes many kilobases away while having no effect on neighboring genes. These distant factors must either slide long distances along chromatin fibers or else interact directly with those factors bound close to the start of transcription, with intervening chromatin forming a loop. Because of their proposed roles in chromatin looping, it is suspected that factors that regulate chromatin topology might have key roles in modulating transcription. One such factor is cohesin, a multisubunit complex essential for sister chromatid cohesion and necessary for mitotic chromosome segregation [1]. Cohesin’s Smc1, Smc3, and Rad21/Scc1 subunits form a three-membered ring, within which sister chromatin fibers are entrapped in a process that requires a separate cohesin loading factor composed of the Scc2 and Scc4 proteins. By entrapping unreplicated DNAs, cohesin could, in principle, hold distant sequences of the same chromatid together (in cis) using the same topological principle by which sister DNAs are held together in trans.
Cohesin clearly functions in processes besides sister chromatid cohesion because it is associated with chromatin in most, if not all, quiescent cells [2] and is essential for the pruning of postmitotic neurons, at least partly by regulating levels of ecdysone receptor [3, 4]. Whether or not cohesin regulates transcription has hitherto been investigated mainly by analyzing the effects of its depletion using RNA interference (RNAi). Depletion of its Rad21/Scc1 subunit causes 2-fold changes in expression of the H19 and IGF2 genes in HeLa cells [2] and little or no effect on inducibility of the gene encoding Interferon-γ in T cells, despite destroying a putative loop between its enhancer and promoter sequences [5]. In Drosophila BG3 tissue culture cells, up to 10- to 50-fold changes in the level of transcripts from the enhancer of split and invected-engrailed loci were detected 6 days after RNAi treatment [6]. Intriguingly, substantial changes in mRNA levels for these transcripts were only observed 3 days following RNAi treatment. Though insightful, these experiments have a number of limitations. The effects on transcription are either modest or they are only seen long after cohesin depletion and might therefore be secondary effects due to chromosome missegregation, defective DNA repair, or some other hitherto-uncharacterized state of stress induced by a loss of cohesin activity.
Another line of evidence hinting at a role for cohesin in transcriptional control is the finding that inactivation of one allele of Nipped-B, the Drosophila ortholog of Scc2, alters long-range enhancer-promoter interactions at the homeotic loci cut and Ultrabithorax (Ubx), at least when compromised by a gypsy retrotransposon [7-9]. Moreover, mutating Rad21 in zebrafish reduces expression of the hematopoietic transcription factors RUNX1 and RUNX3 during development [10], whereas mutations in mau-2, the Caenorhabditis elegans Scc4 ortholog, cause defects in axon guidance [11, 12]. Particularly striking is the finding that Cornelia de Lange syndrome (CdLS), a multi-system developmental disorder, is caused (in more than 50% of cases) by haplodeficiency of NIPBL/Delangin, the human Scc2/Nipped-B ortholog [13-15]. Because tissue culture cells derived from CdLS patients have apparently normal sister chromatid cohesion, dysregulated gene expression during embryonic development has been suggested as a potential cause. There are indeed minor changes in the expression of certain genes in NIPBL± mice (up to 2.5-fold) [16] and CdLS patient-derived cell lines (up to 4-fold) [17], but these so far do little to explain the developmental defects associated with CdLS, which could, in principle, be due to defective DNA repair at crucial stages of development.
Ideally, an investigation of cohesin’s role in transcription should aim to observe the immediate consequences of the complex’s inactivation in cells that are neither undergoing mitosis nor replicating their DNA. Sister chromatid cohesion is normally destroyed at the onset of anaphase by separase-mediated cleavage of cohesin’s Rad21/Scc1 α-kleisin subunit, which destroys its topological entrapment of chromatin fibers by opening the cohesin ring [18, 19]. This process can be reproduced in an inducible manner using tobacco etch virus protease (TEV) in strains of Drosophila melanogaster whose α-kleisin Rad21 contains TEV cleavage sites [3, 20]. We describe here the effect on gene expression of TEV-induced Rad21 cleavage in a nonproliferating tissue, which constitutes conclusive evidence that cohesin has a direct role in regulating transcription.
Results
Transcriptional Changes within Salivary Glands Due to Cohesin Cleavage
To analyze cohesin’s role in gene regulation, we used a heat-inducible transgene (hs-TEV) to induce TEV in terminally differentiated third-instar Drosophila salivary glands expressing either wild-type or TEV-cleavable myc10-tagged Rad21 protein (Rad21TEV; see outline in Figure 1A). This tissue undergoes multiple rounds of endoreplication (repeated cycles of S and G phases without intervening mitoses or cell division), giving rise to transcriptionally active giant polytene chromosomes containing ~1000 closely aligned sister DNAs. We have shown previously that heat-shock induction of TEV in late third-instar larvae (at a time when there is no further replication in salivary glands) removes TEV-cleavable Rad21 protein from chromosomes within 4 hr without any obvious change in their morphology [3]. Late third-instar salivary glands are therefore an ideal tissue to study the putative role for cohesin in gene expression, because possible changes in transcript levels cannot be attributed to changes in chromosome morphology, to defective DNA repair during DNA replication, or to side effects caused by chromosome missegregation.
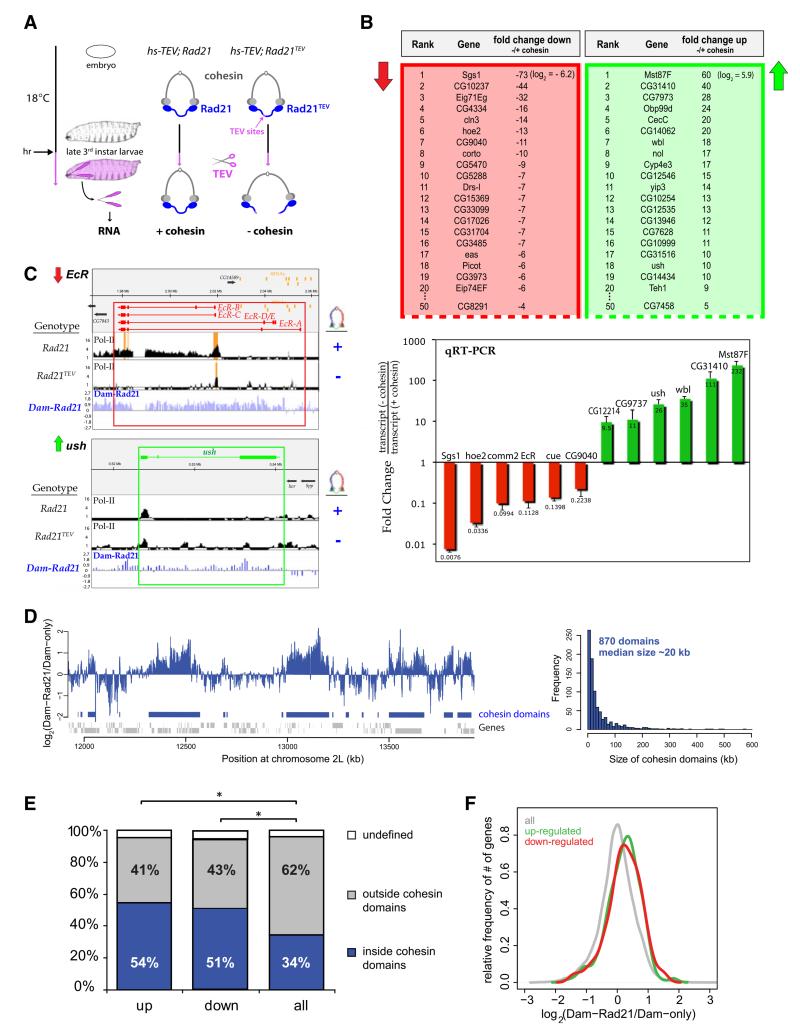
Figure 1. Cleavage of Cohesin Causes Major Transcriptional Changes in Salivary Glands.
Transcriptional changes in salivary glands in the absence versus presence of cohesin were assessed after heat-shock-induced tobacco etch virus protease (TEV) cleavage of cohesin. Green indicates upregulation, red indicates downregulation in the absence of cohesin.
(A) Outline of the heat shock (hs)-TEV system used for differential gene expression profiling in salivary glands. Larvae carrying the hs-TEV construct and containing cohesin complexes with wild-type or TEV-cleavable (purple arrow) Rad21 were raised at 18°C before TEV protease was induced ubiquitously in late third-instar larvae by heat shock (45 min, 37°C). Salivary glands were dissected 10–12 hr after hs, followed by RNA isolation. Rad21 is shown in blue, TEV in purple (see also Figure S1A).
(B) Top: list of the 20 most downregulated and 20 most upregulated genes upon cohesin cleavage in salivary glands identified by microarray analysis (see also Figures S1B and S1C, Table S1, and Table S2). Genes are sorted in descending order based on their average fold change in transcript levels in the absence versus presence of cohesin across seven independent microarrays. A minus indicates fold downregulation. Genes at rank 50 are also given. Bottom: differential expression of selected candidates was confirmed by quantitative real-time PCR (qRT-PCR). Each bar represents the average fold change in the absence versus presence of cohesin of at least three independent experiments (error bars are standard deviations of the mean). Gene names are given above each bar; fold changes are given below/inside each bar.
(C) ChIP-CHIP analysis of the distribution of Pol-II (black plots) in Rad21 (+cohesin) and Rad21TEV (−cohesin) salivary glands 10–12 hr after heat-shock induction of TEV protease. Cohesin binding (blue plots) in salivary glands was assessed by DNA adenine methylase identification (DamID; Dam-Rad21). ChIP-CHIP data is represented as fold enrichment of IP over input (MAT scores; log scale; highly enriched regions (p < 0.0001) are in orange). DamID data are represented as the relative enrichment of methyl-adenine-marked DNA from Dam-Rad21 glands over Dam-only glands (log2 scale). EcR (down-regulated, red box) and ush (upregulated, green box) loci are shown as representative examples (see also Figure S2 for further examples).
(D) Rad21-bound domains (cohesin domains) across a randomly chosen 2 Mb chromosomal region of chromosome 2L. Shown is the relative enrichment of DamRad21 versus Dam-only signal (log2 scale). Rad21 domains are highlighted as blue bars; genes are indicated as gray bars. The size distribution of the total number of 870 Rad21 domains in salivary glands is shown on the right.
(E) Differentially expressed genes are enriched in Rad21 binding. Shown are percentages of transcriptional start sites (TSSs) of cohesin-dependent genes (up- or downregulated) and of all genes that localized inside (blue), outside (gray), or at the border (white) of Rad21-bound regions. The asterisk (*) indicates Fisher’s exact test: p < 0.01.
(F) Average Rad21 binding at the TSSs of upregulated genes (green), downregulated genes (red), and all genes (gray).
RNAs were isolated from salivary glands expressing wild-type (+cohesin) or TEV-cleavable Rad21 (−cohesin) 10–12 hr after heat-shock induction of TEV. This time point was chosen because the heat-shock transcriptional response will have abated, but most Rad21 containing TEV sites remains cleaved, and newly synthesized Rad21TEV does not reaccumulate until about 16 hr after the heat shock (see Figure S1A available online; [3]). Both RNA samples were converted to cDNA, labeled with Cy3 and Cy5, respectively, and hybridized to INDAC FL003 arrays containing 18,240 transcript-specific 70-mer oligonucleotides. Analysis of seven arrays, each hybridized to an independently generated sample pair (Figures S1B and S1C), revealed major differences in the levels of certain transcripts. Cohesin cleavage caused 78 transcripts to increase and 55 to decrease at least 4-fold. Moreover, 419 genes (262 up and 157 down) changed at least 1.5-fold (Figure 1B;Table S1), which suggests that cohesin may function as both an activator and repressor of transcription. Apart from the highly downregulated divergently transcribed Sgs1 and hoe2 gene pair, differentially expressed genes are not clustered in the genome and are implicated in a variety of biological processes (gene ontology [GO] enrichment analysis; Table S2).
Quantitative real-time PCR (qRT-PCR) analysis of selected candidates confirmed differential expression of 16 out of 19 differentially expressed genes tested (Figure 1B and data not shown), revealing up to 100-fold changes. In many cases, changes in transcript levels were accompanied by corresponding changes in association of RNA Polymerase II (Pol-II) with transcription units (Figure 1C; see Figures S2A and S2B for additional examples), as measured by ChIP-CHIP analysis. Thus, TEV cleavage of cohesin depleted Pol-II from the EcR locus, but it increased Pol-II’s association with the ush locus. Consistent with the relatively small set of differentially expressed genes after cleavage of cohesin, major changes in the binding of Pol-II were confined to rare loci.
The transcriptional program within salivary glands changes both during the third-instar larval stage and at the transition to the pupal stage [21]. It was therefore possible that the observed differences in gene expression were related to differences in the developmental stage, especially when comparing fly stocks carrying different transgenes (Rad21TEV animals develop slightly slower at 18°C than Rad21 animals). To exclude the possibility that the changes in gene expression after TEV cleavage of cohesin were due to any minor developmental difference caused by the presence of TEV sites within Rad21, we analyzed Pol-II profiles from salivary glands of Rad21TEV larvae in the absence of TEV protease induction (+cohesin) and performed qRT-PCR analysis to compare transcript levels of selected differentially expressed candidates in larvae of different ages with those in the absence versus presence of cohesin. The results of these experiments implied that the majority of changes in gene expression after TEV cleavage of cohesin were indeed due to loss of cohesin (Figures S3A and S3B).
Differentially Expressed Genes Are Preferentially Bound by Cohesin
To analyze cohesin’s distribution in salivary glands, we used DNA adenine methylase identification (DamID) [22, 23], which involves detecting sites of adenine methylation in transgenic strains expressing bacterial DNA adenine methyltransferase (Dam) fused to a protein of interest, in this case Rad21. DamID was chosen because we were not successful in obtaining sufficiently high quality Rad21myc-ChIPed starting material, most likely because of poor efficiency of myc antibodies for ChIP. Because Dam fusion proteins must be expressed at very low levels to ensure specificity of methylation, it is not possible to assess their functionality directly. We therefore measured the ability of mRNAs, encoding Rad21 or Dam-Rad21, to rescue mitotic defects associated with TEV-induced cleavage of cohesin during syncytial divisions in Rad21TEV embryos [20]. mRNAs encoding Dam-Rad21 were, similarly to those encoding wild-type Rad21, able to rescue precocious sister chromatid separation caused by TEV injection (Movie S1, Movie S2, and Movie S3), suggesting that the Dam-Rad21 fusion protein is functional, at least in conferring sister chromatid cohesion. The Dam-Rad21 binding profile, namely the relative enrichment of methyl-adenine-marked DNA fragments from Dam-Rad21 third-instar salivary glands over a Dam-only control, shows Rad21 enrichment at large regions of the genome containing one or more transcription units (Figure 1D; see Figures S2A and S2B for additional examples). A domain detection algorithm (see Experimental Procedures for details) identified a total of 870 Rad21-bound regions (so-called cohesin domains) varying in size from ~2 to ~650 kb, with a median size of ~20 kb (Figure 1D). Cohesin domains cover 33% of the genome and contain 34% of all the transcription start sites (TSSs, defined as the 1 kb region downstream of the transcriptional start). These cohesin domains identified in salivary glands substantially overlap with the Smc1-bound domains identified by ChIP-CHIP in cultured cells [24]. The TSSs of genes defined as cohesin-bound by Misulovin and colleagues [24] are significantly enriched inside our cohesin domains (p < 10−5, data not shown), confirming the validity of our approach.
Notably, cohesin domains are significantly enriched in genes that are differentially expressed upon loss of cohesin. Fifty-four percent of TSSs of upregulated genes and 51% of the TSSs of downregulated genes localize within a cohesin domain, which is a significantly (Fisher’s exact test: up, p = 2.36e−11; down, p = 1.12e−05) larger number compared to only 34% of all TSSs (Figure 1E). Calculation of the average Rad21 binding at the TSS of each gene confirmed that TSSs of cohesin-dependent genes are significantly enriched for Rad21 binding (Wilcoxon test: up, p = 4.3e−11; down, p = 3.19e−08;Figure 1F). Together, these results are consistent with previous reports [6, 17] and suggest that cohesin may indeed be the primary cause of the transcriptional changes observed for more than half of the genes whose expression changes after cohesin cleavage.
Cohesin Is an Essential Regulator of the Transcriptional Response to Ecdysone
We noticed that several of the differentially expressed genes had previously been implicated in the 20-hydroxyecdysone (ecdysone) response, including the ecdysone receptor (EcR) itself, whose protein level is reduced by cohesin cleavage in postmitotic neurons [3, 4]. Encouraged by these findings, we addressed whether cohesin may have a general role in the transcriptional regulation of ecdysone-responsive genes. Comparison of the published list of 555 genes whose expression levels change upon ecdysone treatment in cultured larval organs [25] to our list of differentially expressed genes after cohesin cleavage revealed that out of the 424 ecdysone-responsive genes for which we had expression data, 33 (7.7%) were differentially expressed (18 up and 15 down) after TEV cleavage of cohesin, a significantly larger number than expected by chance (2.7%, Fisher’s exact test: p = 1.19−07; Figure 2A). Plotting the changes in gene expression after cohesin cleavage for ecdysone-responsive genes versus all genes confirmed that ecdysone-responsive genes are preferentially up- or downregulated after cohesin removal (Siegel-Tukey test: p = 1.04e−28; Figure 2B), suggesting that cohesin plays a so-far-unrecognized role as mediator of the transcriptional response to ecdysone in larval salivary glands.
Figure 2. Cohesin Regulates the Expression of Ecdysone-Responsive Genes.
Differential gene expression in salivary glands after cohesin cleavage was assessed at ecdysone-responsive genes [25].
(A) Percentages of ecdysone-responsive and of all genes that are upregulated (green), downregulated (red), and unchanged (white) after TEV cleavage of cohesin. The asterisk (*) indicates Fisher’s exact test: p < 0.01.
(B) Log2-fold changes in gene expression after TEV cleavage of cohesin for ecdysone-responsive genes (yellow) versus all genes (gray).
(C) Pol-II- and Rad21-binding profiles at Eip74EF and Eip75B are shown as representative examples for ecdysone-responsive loci (see Figure 1C for further details; see also Figure S2).
(D) Ecdysone-responsive genes that are differentially expressed upon cohesin cleavage are enriched in Rad21 binding. Shown are percentages of TSSs of ecdysone-responsive cohesin-dependent genes, of all ecdysone-responsive genes, and of all genes that localized inside (blue), outside (gray), or at the border (white) of Rad21-bound regions. The asterisk (*) indicates Fisher’s exact test: p < 0.01.
(E) Average Rad21 binding at the TSSs of four different categories of genes: ecdysone-responsive genes that are upregulated (green) or downregulated (red) after cohesin cleavage, all ecdysone-reponsive genes (yellow), and all genes (gray).
In support of a direct (versus indirect) regulatory role for cohesin in the ecdysone response, our statistical analysis revealed that the TSSs of 23 out of the 33 ecdysone-responsive genes whose expression changed following cohesin cleavage localized within a cohesin domain (see Figure 2C and Figures S2A and S2B for examples). This is a significantly (Fisher’s exact test: p = 2.03e−03) larger number than expected by chance, namely 69.7% versus 46.6% of all ecdysone-responsive genes versus 34.5% of all genes of which differential expression was measured (Figure 2D). In addition, calculation of the average Rad21 binding at TSSs confirmed that the TSSs of ecdysone-responsive genes—and especially of the subset of genes that are differentially expressed after cohesin cleavage—are preferentially bound by Rad21 (Figure 2E).
Timed Cohesin Cleavage Specifically in Salivary Glands
Changes in transcript levels 10 hr after cohesin cleavage do not necessarily imply that cohesin directly regulates transcription, even if cohesin is present at the locus in question. For example, members of the ecdysone signaling gene family are induced sequentially by a pulse of the steroid hormone ecdysone that initiates the larval-to-pupal transition. Because transcription of EcR is also reduced upon cohesin cleavage, it is possible that the effect of cohesin cleavage on the aforementioned genes is merely due to reduced levels of EcR protein. To distinguish primary from secondary effects, it was therefore essential to evaluate the kinetics of changes in transcript levels that occur upon cohesin cleavage.
The heat-shock system used to induce TEV in our original screen has a number of limitations for this purpose. First, early effects could be missed because strong heat shocks have a drastic effect on transcription. Second, cohesin reappears on chromosomes between 15 and 20 hr after hs-TEV induction because of resynthesis of Rad21TEV and degradation of TEV protease (Figure S1A; Figure 3B; data not shown), which precludes evaluation of long-term effects of cohesin cleavage. Third, the heat-shock promoter is transcribed in all larval tissues, and effects on transcription in one tissue (in this case salivary glands) might, in principle, be caused by changes that had occurred in another. For example, cohesin cleavage would have drastic and pleiotropic effects on proliferating neuroblasts, muscle cell precursors, and imaginal disc cells.
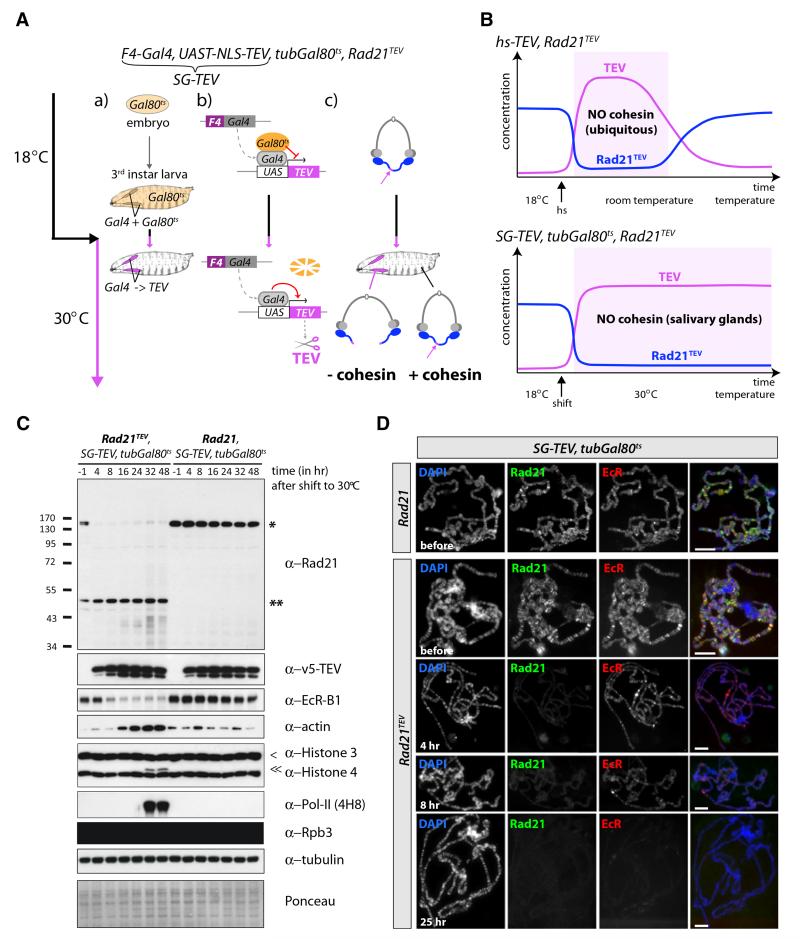
Figure 3. Timed Salivary Gland-Specific Cleavage of Cohesin.
(Aa–Ac) Outline of the tubGal80ts/SG-TEV system that enables control of timing and salivary gland specificity of cohesin cleavage. Larvae surviving on Rad21TEV and encoding tubGal80ts and SG-TEV (F4-Gal4, UAST-NLS-TEV) were raised at 18°C, the permissive temperature for the ubiquitously expressed Gal4 inhibitor Gal80ts. By shifting late third-instar larvae to the restrictive temperature (30°C), Gal80ts is degraded, which enables the salivary gland-specific Gal4 driver F4-Gal4 to induce TEV protease expression specifically in salivary glands (Aa). (Ab) and (Ac) summarize the states of transcription and cohesin, respectively, in salivary glands at 18°C and 30°C. Rad21 is shown in blue, TEV in purple, Gal80ts in orange.
(B) Schematic comparison of the effects of the hs-TEV versus SG-TEV/tubGal80ts systems on the level of functional cohesin complexes (presence of un-cleaved Rad21TEV) after induction of TEV protease. Concentrations of TEV (purple) and Rad21TEV (blue) are plotted against time. Rad21TEV starts to reaccumulate in the hs-TEV system about 15 hr after TEV induction because of degradation of TEV and resynthesis of Rad21TEV (top), whereas continuous expression of TEV in salivary glands at 30°C prevents reappearance of full-length Rad21TEV in this tissue in the SG-TEV/tubGal80ts system (bottom).
(C) Western blot analysis of salivary gland extracts from SG-TEV/tubGal80ts larvae surviving either on Rad21TEV or Rad21. Extracts were prepared before (t = −1; TEV off) and at different time points after shifting third-instar larvae to 30°C (TEV on). Blots were probed with the indicated antibodies. Full-length Rad21 (*) and the C-terminal TEV cleavage fragment (**), as well as full-length Histone H3 (<) and N-terminally clipped Histone H3 (< <), are indicated. Tubulin and Ponceau stainings were used as loading controls.
(D) Representative polytene chromosome spreads from SG-TEV/tubGal80ts crawling third-instar larvae surviving either on Rad21 or Rad21TEV were prepared before (TEV off) and at various times after shifting third-instar larvae to 30°C (TEV on). Polytene chromosomes were coimmunostained with antibodies against Rad21 and EcR-B1. The chromosome morphology was visualized by DAPI staining. In the overlay (right panels), DAPI is shown in blue, Rad21 in green, and EcR-B1 in red. Scale bars are 50 μm (top two rows) and 100 μm (bottom three rows).
To control both the timing and tissue specificity of cohesin cleavage, we expressed TEV protease with a nuclear localization signal from a transgene (UAST-NLS-TEV) whose promoter contained multiple Gal4 binding sites [3]. Tissue specificity was conferred by a second transgene, F4-Gal4 [26], that produces the Gal4 transcriptional activator protein from a salivary gland-specific driver. For clarity, we will refer to the combination of UAST-NLS-TEV and F4-Gal as SG-TEV. Lastly, temporal control of transcription was conferred by a third transgene, tubGal80ts, that expresses (ubiquitously) the temperature-sensitive Gal80 protein (Gal80ts), which binds to and inhibits Gal4 at 18°C (the permissive temperature) but not at 30°C (the restrictive temperature) [27]. Though complex, this system ensures that TEV is only expressed in salivary glands upon transfer of larvae to the restrictive temperature (see Figure 3A). Importantly, normal development and the transcriptional programs that underlie it continue after larvae are shifted permanently to 30°C, and it should therefore be possible to evaluate both short- and long-term effects of tissue-specific cohesin cleavage (see Figure 3B).
Salivary gland development occurs normally at 18°C in both Rad21TEV and control (Rad21) larvae containing SG-TEV/tubGal80ts, demonstrating efficacy of Gal80ts in repressing SG-TEV at this temperature. In contrast, transfer of animals to 30°C during early larval stages blocks salivary gland growth in Rad21TEV but not Rad21 larvae (data not shown), confirming that cohesin has an essential role during salivary gland endocycles [3] despite the lack of chromosome segregation. To address cohesin’s role in salivary glands that have completed their endocycles, we shifted late third-instar larvae from 18°C to 30°C. Western blots of salivary gland extracts showed that TEV is undetectable prior to the temperature shift, accumulates within 4 hr of the shift, and remains at high levels thereafter (Figure 3C). Accumulation of TEV was accompanied by a permanent decline in TEV-cleavable Rad21 but not wild-type Rad21. Importantly, neither TEV protease nor Rad21TEV cleavage fragments could be detected in Rad21TEV larvae from which salivary glands had been removed (data not shown), confirming the tissue specificity of F4-Gal4.
Chromosome spreads showed that Rad21TEV but not wild-type Rad21 disappeared from polytene chromosomes within 2 to 4 hr of the temperature shift (Figure 3D; Figure 5A). The sustained removal of cohesin, only possible using the SG-TEV/tubGal80ts system, revealed a number of posttranscriptional, most likely stress-induced, alterations at late (>24 hr) time points, e.g., increase in actin protein levels, clipping of histone H3 [28], and the dramatic appearance of what appears to be a posttranslationally modified version of the large subunit of Pol-II (Figure 3C).
Controlled Salivary Gland-Specific Cohesin Cleavage Reveals Both Rapid and Slow Changes in Gene Expression
To identify genes whose expression is altered before such pleiotropic changes in cell physiology take place and that are therefore good candidates for being directly regulated by cohesin, we used the SG-TEV/tubGal80ts system combined with qRT-PCR analysis to measure the transcript levels of a subset of cohesin-dependent genes in salivary glands of late third-instar larvae (staged upon collection) over a 48 hr period following induction of cohesin cleavage. All mRNA levels were normalized using tubulin mRNA, which did not alter upon cohesin cleavage using the heat-shock system. Figure 4 plots the ratios of Rad21TEV and Rad21 mRNA levels following TEV induction. Importantly, the ratio for a cohesin-independent gene, Rpll215, which encodes the large subunit of Pol-II, remained close to 1 at all time points (Figure 4). Of six transcripts downregulated by cohesin cleavage, three (EcR-B1, Eip74EF, and comm2) declined to minimal levels within 4 hr of the temperature shift and three (Sgs1, Eip75B, and CG31698) declined more gradually, reaching minimal levels only after 16 hr (Figure 4, black lines). Upregulated genes could also be divided into early- and late-response categories. Transcript levels of wbl and CG12214 showed negligible changes during the first 4 hr and increased to maximal levels only after 16 hr, whereas ush (and, to a lesser extent, Mst87F) increased within 4 hr. Time courses of mRNA levels following heat-shock-induced cohesin cleavage confirmed the pattern of early and late responses (Figure 4, gray lines), at least over the first 16 hr (using this system analysis beyond 16 hr was not possible because of recovery of full-length Rad21TEV protein;Figure S1A). Notably, the TSSs of all five early-response genes (3 down and 2 up) that were identified by our time course analysis are located inside a Rad21-bound domain (data not shown), which strongly suggests that they are indeed directly regulated by cohesin.
Figure 4. Loss of Cohesin in Salivary Glands Causes Both Rapid and Slow Changes in Transcript Levels.
The kinetics of transcriptional changes upon cleavage of cohesin in salivary glands were assessed by qRT-PCR analysis using the SG-TEV/tubGal80ts (black lines) and hs-TEV (gray lines) systems. Plots show tubulin-normalized, averaged fold differences in transcript levels in the absence (Rad21TEV) versus presence (Rad21) of cohesin over time (in hours; t = −1 indicates time point before TEV induction), obtained from three independent time courses per TEV-expression system (error bars show standard deviations). Genes were classified as early and late response genes based on rapid or gradual changes in transcript levels within the first 4 or 16 hr, respectively. Green indicates upregulation, red indicates downregulation, gray indicates no change in the absence of cohesin.
Rapid Reduction of Eip74EF mRNA Is Not Due to Loss of Ecdysone Receptor
The decline of ecdysone-regulated genes could be partly or wholly an indirect effect caused by the rapid decline of EcR-B1 mRNA levels following cohesin cleavage. To address this, we used western blots to measure the level of ecdysone receptor protein and polytene chromosome spreads to measure its association with specific loci after shifting SG-TEV/tubGal80ts larvae to 30°C. This revealed little or no change in protein levels or chromosomal association of EcR-B1 during the first 4 hr (Figures 3C and 3D). The 10-fold decrease in Ei-p74EF transcript levels within this period (Figure 4) cannot therefore be attributed to a lack of the hormone receptor. A lack of receptor could be responsible for the steep decline between 8 and 16 hr of mRNAs from the Sgs1 locus, which encodes an ecdysone-inducible salivary gland-specific glue protein and at which we detect only background levels of cohesin (Figure 4; Figure S3A). A decline in EcR might also contribute to the gradual decrease of Eip75B between 4 and 16 hr (Figure 4).
Cohesin Is Required for Puffing at Eip74EF
Eip74EF and Eip75B belong to a group of genes whose activity can be visualized cytologically on polytene chromosomes in late third-instar larvae [29]. High rates of transcription induced by the late third-instar peak of ecdysone cause a characteristic decondensation or “puffing” of these loci, which happen to be located at adjacent bands, namely 74 and 75. Because puffing at band 75 occurs within 5 to 10 min of puffing at band 74, creating highly characteristic twin puffs at this stage of development [21], the 74-75 region is particularly easy to locate on polytene spreads (Figure 5A; see also model in Figure 5D) and enabled us to reliably monitor puffing at both loci within the same spread.
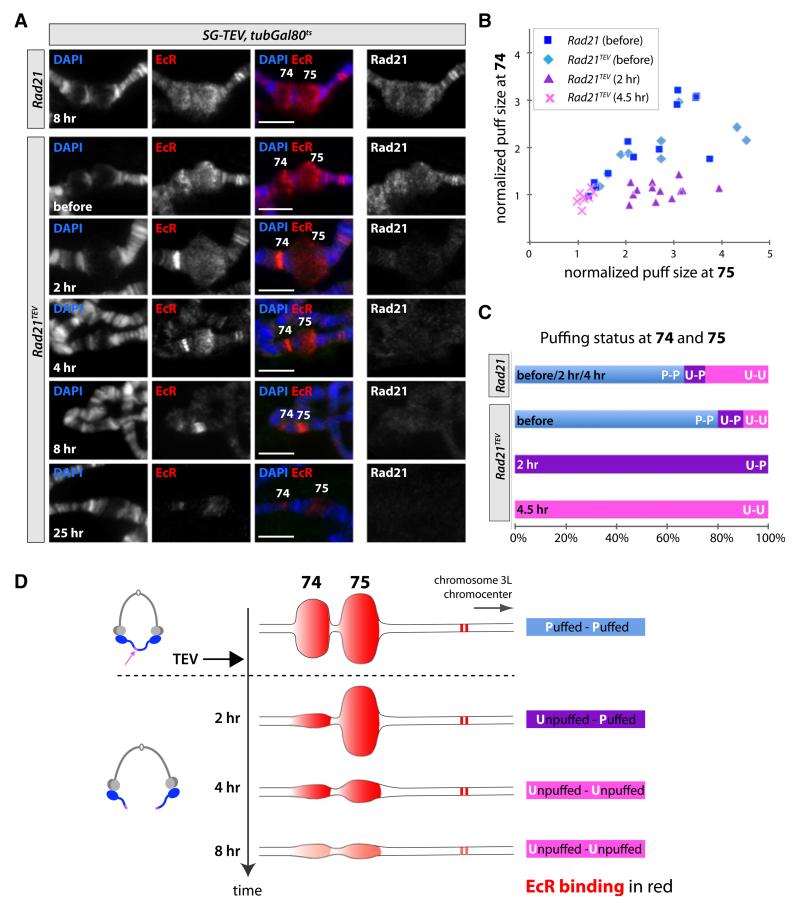
Figure 5. Cohesin Is Required for Puffing at Eip74EF and Eip75B.
The ecdysone-induced late third-instar puffing response at the neighboring chromosomal bands 74 and 75 (harboring Eip74EF and Eip75B, respectively) was analyzed by polytene chromosome spreads before and after salivary gland-specific TEV protease induction in SG-TEV/tubGal80ts late third-instar larvae surviving either on Rad21TEV or Rad21.
(A) Representative polytene chromosome spreads from salivary glands in the presence (Rad218h and Rad21TEVbefore) and absence of cohesin (Rad21TEV, later time points) were coimmunostained with antibodies against Rad21 and EcR-B1. The chromosome morphology was visualized by DAPI staining. Positions of bands 74 and 75 are indicated in overlays (DAPI in blue, EcR-B1 in red). The two adjacent sharp EcR-stained bands at the right side of band 75 (toward the chromocenter) were used to identify the 74-75 locus. Scale bars are 50 μm.
(B) Quantification of normalized puff sizes (see Experimental Procedures) at bands 74 and 75 in the presence of cohesin (blue squares and light blue diamonds) and at 2 hr (purple triangles) and 4.5 hr (pink crosses) after cohesin cleavage. Each data point represents the normalized puff size at band 74 plotted against the normalized puff size at band 75 from the same polytene chromosome spread.
(C) Changes in puffing status at bands 74 and 75 upon cleavage of cohesin. Based on the ratio and absolute sizes of the 74 and 75 puffs, each twin locus was classified as either puffed-puffed (P-P, blue), with both 74 and 75 decondensed, unpuffed-puffed (U-P, purple), with 74 condensed and 75 decondensed, or unpuffed-unpuffed (U-U, pink; puffed loci: normalized width > 1.5). The graph plots the percentage of spreads belonging to each category for each experimental condition.
(D) Scheme illustrating the distinct puffing stages at 74-75 observed before and after TEV cleavage of cohesin. EcR binding to 74 and 75, as well as to the two adjacent bands toward the chromocenter, is highlighted in red.
Polytene chromosome spreads were prepared from Rad21 (control, +cohesin) and Rad21TEV (−cohesin) late third-instar larvae before and after salivary gland-specific TEV cleavage of cohesin. To monitor association of cohesin and ecdysone receptor with particular loci along chromosomes, we coimmunostained spreads with antibodies against Rad21 and EcR-B1. Plotting of normalized puff sizes at bands 74 and 75 (see Experimental Procedures) along the y and x axes, respectively, confirmed that there was no significant difference in the puffing pattern of Rad21 and Rad21TEV salivary glands before TEV protease induction (Figure 5B, blue symbols). Even though there was considerable variation in the size of puffs between spreads (presumably because of small differences in the developmental state between larvae), puff size at 74 correlated with puff size at 75 within individual chromosomes. The constant ratio of normalized puff sizes at 74 and 75 is consistent with simultaneous induction and regression of these twin puffs [21]. Immunostaining showed that both puffs were enriched for EcR-B1 (Figure 5A), as has been previously described [30]. Both puffs were also enriched for Rad21, which is consistent with our DamID binding profile for cohesin at these loci (Figure 2C; Figure S2B).
Neither puffing nor EcR-B1- and Rad21-binding patterns changed upon shifting Rad21 (control) animals to 30°C (data not shown). In contrast, TEV protease induction caused rapid and major alterations to the 74-75 chromosomal region in Rad21TEV salivary glands. Thus, Rad21 staining at both loci (as well as at all chromosomal loci, data not shown) declined to background levels within 2 hr, implying complete cleavage and dissociation of cohesin within this short time period (Figure 5A). Strikingly, cohesin cleavage was accompanied by a dramatic reduction of puffing at band 74. Because high levels of EcR-B1 persisted at this locus during this period, a sharp, intense band of EcR-B1 staining was created at 74 (Figure 5A, “2 hr” panels). Puffs at band 75, on the other hand, were little affected 2 hr after the temperature shift, resulting in a dramatic reduction in the ratio of puff sizes at 74 and 75 within chromosomes sampled at the 2 hr time point (Figures 5A and 5B, purple triangles; see also Figure 5C for averaged quantification of the puffing status at 74 and 75). The contraction of band 74, but not band 75, so soon after cohesin’s removal, without any detectable loss of EcR, suggests that cohesin plays a direct role in transcribing the former. Though band 75 remained in a puffed state in the absence of cohesin for a longer period than band 74, it too declined by 4 hr, notably before any noticeable decline in EcR-B1 associated with the locus (Figures 5A and 5B, pink crosses; see also quantification in Figure 5C). Transcription from band 75 might therefore require cohesin activity at this locus, as well as EcR protein.
Discussion
Development of a method to cleave Rad21 with TEV protease in a time- and tissue-specific manner has enabled us to assess the immediate and long-term consequences of cohesin inactivation on transcription in third-instar salivary glands from D. melanogaster. We chose this postmitotic tissue to ensure that any effects of cohesin inactivation on the transcriptional apparatus could not be attributed to indirect or knock on effects of chromosome missegregation or defective DNA repair due to the absence of cohesin’s canonical function, namely sister chromatid cohesion. Despite this precaution, cohesin cleavage causes, from 24 hr onward, major changes in cellular physiology, some of which most likely reflect a general stress-related response (see Figure 3C). We cannot at this stage ascertain whether these highly pleiotropic events are triggered by changes in gene expression that precede them or by the loss of a novel cohesin function that we are currently unaware of. In either case, our observations demonstrate that it is very difficult to attribute functions to cohesin in regulating gene expression merely by observing the long-term consequences of its inactivation. Changes in gene expression that only occur 24 hr or more after cohesin’s removal from chromosomes could be secondary or tertiary events triggered by fundamental changes in cell physiology.
Our observations reveal an additional complication in interpreting gene expression changes. Several of the genes whose expression is affected by cohesin cleavage are genes regulated by the ecdysone receptor, whose abundance declines after 8 hr, presumably because of an almost immediate, cohesin cleavage-dependent decline in its mRNA. Thus, the precipitous decline in Sgs1 mRNAs that takes place between 8 and 16 hr could be caused by the lack of ecdysone receptor and not by the lack of cohesin per se. Such phenomena could explain many late responses to cohesin inactivation.
Given these considerations, it is clear that in order to attribute a role for cohesin in regulating a gene on the basis of changes in its expression upon cohesin inactivation, it is necessary to demonstrate a change in transcription as soon as cohesin has been removed from chromosomes and, crucially, long before any major change in cell physiology or in the concentration of other transcription regulators. Two genes stand out in this regard, namely EcR encoding the ecdysone receptor and Eip74EF encoding an ecdysone-dependent transcription factor. Eip74EF is a particularly good candidate, because heavy transcription of this gene in third-instar larvae gives rise to a cytologically visible puff. Cohesin is associated with this puff, and its removal by Rad21 cleavage is accompanied by an immediate cessation of puffing. Crucially, contraction of band 74 caused by Rad21’s removal takes place several hours before any decline in ecdysone receptor associated with it. We therefore suggest that cohesin present at Eip74EF has a direct role in maintaining transcription of the gene. We have no reason to believe that the same is not also true for EcR, though we have not observed it at a cytological level. Because transcription of most genes is unaffected by cohesin cleavage, it is striking that transcription of EcR, as well as of a direct target gene, Eip74EF, appears to be directly regulated by cohesin. Our finding that ecdysone-responsive genes in general are enriched in cohesin domains and preferentially misregulated following cohesin cleavage suggests a common aspect of the transcription process at these loci that renders them particularly dependent on cohesin. It is conceivable that the interplay between the core set of gene regulatory mechanisms (transcription factors, enhancers, promoters, etc.) was insufficient to achieve the precise control that was required to orchestrate the dramatic ecdysone-induced changes that occur during the larval-to-pupal metamorphosis. It is also conceivable that cohesin, because of its ability to encircle chromatin strands, was particularly suited to fulfill this role, either by facilitating interactions between distant DNA elements in cis or by its ability to slide along DNA (see below).
Although Eip74EF may be the best example of a gene directly regulated by cohesin, it is by no means the only candidate. Reduced puffing at its twin, the adjacent Eip75B, also occurs before any obvious decline in ecdysone receptor at this locus. Although the drop in Eip75B mRNAs that occurs 8 hr after induction of Rad21 cleavage may be due to a decline in ecdysone receptor, the more modest decrease that occurs earlier may be due to a direct effect of cohesin’s dissociation from the locus. There are other genes, for example comm2, whose mRNAs decline rapidly upon cohesin cleavage, and these may also be directly regulated by cohesin. Interestingly, transcripts from at least two genes, namely ush and Mst87F, rise rapidly after cohesin cleavage, suggesting that although cohesin promotes transcription at certain genes, it exerts repression at others.
Cohesin’s canonical function is to mediate sister chromatid cohesion. It is currently thought to perform this by entrapping sister DNAs inside a tripartite ring formed by its Smc1, Smc3, and Rad21/Scc1 subunits [1]. This raises the important question of whether cohesin regulates gene expression using a similar topological principle. With this in mind, it has been repeatedly proposed that cohesin might regulate gene expression by facilitating the formation or maintenance of loops between remote regulatory elements and promoter regions. Such loops have not been visualized directly but have instead been inferred from coprecipitation of remote DNA sequences following formaldehyde fixation. According to this somewhat indirect assay, long-term cohesin depletion reduces interaction between an enhancer at the 3′ end of the H19 gene with a remote CCCTC-binding factor (CTCF) binding site that controls imprinting of the IGF2-H19 locus [31]. Loss of the putative loop between the CTCF binding site and the H19 enhancer is thought to enable the enhancer to activate the neighboring IGF2 gene. Cohesin depletion also disrupts a similar type of long-range interaction between distant (cohesin-associated) CTCF sites at the INFG locus, though in this case, cohesin depletion has little effect on inducibility of the locus by cytokine [5]. The observation that cohesin in Drosophila, unlike its enrichment at CTCF binding sites in human cells [2, 32], is associated with large domains raises the possibility that it can also regulate transcription by means other than the formation of loops between remote regulatory elements. By entrapping DNAs inside rings capable of sliding along chromatin, cohesin complexes may provide a potentially mobile platform for the stable association of other factors necessary for regulating (positively or negatively) the movement of polymerases through transcription units. Cohesin’s intriguing potential to modulate chromatin, together with its binding to regions covering several transcription units, is seemingly at odds with our finding that differentially expressed genes are not clustered in the genome. Whatever the activity is that cohesin brings along, our data suggest that its absence affects only a subset of genes that are normally exposed to it. Our identification of ecdysone-responsive genes as a class of cohesin-dependent genes highlights that there might exist still-unknown common determinants or gene-specific regulators that render a gene susceptible to changes in cohesin binding.
Experimental Procedures
Fly Strains
Flies surviving on myc10-tagged wild-type Rad21 (Rad21: w1118; +/+; Rad21ex3, P[w+, tubpr < Rad21-myc10 < SV40]) or myc10-tagged TEV-cleavable Rad21 (Rad21TEV: w1118; +/+; Rad21ex15, P[w+, tubpr < Rad21(550-3TEV)-myc10 < SV40]), as well as heat-inducible TEV-protease-expressing flies (hs-TEV: w1118; hs-NLS-v5-TEV-NLS2; Rad21ex3/TM6B Tb ubiquitin-GFP), have been described previously [3]. For salivary gland-specific TEV cleavage, the tubGal80ts transgene [33] was recombined with the Rad21 null mutant Rad21ex15 and the nuclear UAST-NLS-TEV transgene [3] and crossed to the F4-Gal4 driver line [26] to generate w1118; F4-Gal4; tubGal80ts Rad21ex15 UAST-NLS-TEV/Tm6B Tb ubiquitin-GFP flies. For DamID, flies carrying 5xUAST-Dam-myc-Rad21 (Dam-Rad21) were generated by standard P element-mediated transgenesis (BestGene). Flies carrying 5xUAST-Dam (Dam-only) have been published before [34]. For details on the cloning, see Supplemental Experimental Procedures. To test for the functionality of Dam-myc-Rad21 constructs, we performed rescue experiments after TEV cleavage of Rad21TEV in syncytial embryos. Embryo preparation, synthesis of mRNA coding for wild-type and Dam-myc-tagged Rad21, and mRNA/TEV protease injections were performed as previously described [20]. A complete list of genotypes of all fly strains used in this study can be found in the Supplemental Experimental Procedures.
Gene Expression Profiling
Virgin female flies expressing Rad21 with (Rad21TEV) or without (transgenic or endogenous Rad21) TEV cleavage sites as their sole source of Rad21 were crossed to male flies carrying heat-shock-inducible TEV protease in a Rad21-null background (hs-TEV flies). Crosses were kept at 18°C under noncrowded conditions. TEV protease expression was induced in late third-instar Tb− GFP− larvae by heat shock (45 min in a 36.5°C water bath, followed by 10–12 hr incubation at room temperature). For each of the seven microarray experiments, 10–20 salivary gland pairs of Rad21TEV (−cohesin) and Rad21 (+cohesin) crawling third-instar larvae (staged upon collection) were dissected, and total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. cDNA preparation, Cy3 and Cy5 labeling of the sample pairs, hybridization to Drosophila long oligonucleotide cDNA arrays (FL002), array scanning, normalization, and basic statistical analysis (Bioconductor package, Limma) were performed at the FlyChip facility in Cambridge, UK. Data are presented as vsn-normalized log2 ratio (log2[−cohesin]/[+cohesin]).
qRT-PCR analysis of selected candidate genes was performed according to standard procedures. For details, see Supplemental Experimental Procedures.
DamID Analysis of Cohesin Binding in Salivary Glands
Genomic DNA was isolated from salivary glands of homozygous Dam-Rad21 or Dam-only crawling third-instar larvae. In vivo methylated DNA was amplified as described before [22]. Differentially labeled fragments of both samples were pooled and hybridized to microarrays carrying 380,000 60-mer DNA oligonucleotides [35] (Roche-NimbleGen), with a median probe spacing of 300 bp. Probes were mapped to D. melanogaster Release 5 genome. Microarray data analysis was performed with R (http://www.r-project.org). Raw data was LOESS normalized and median centered, and dye swap arrays were averaged. Rad21 domains were defined using a two-state hidden Markov model. Further details are available in the Supplemental Experimental Procedures. All downstream analyses were performed using custom made R scripts, which are available upon request.
ChIP-CHIP Analysis
Pol-II chromatin immunoprecipitations (chromatin-IPs) of third-instar larval salivary glands, using the CTD4H8 mouse anti-Pol-II antibody (Upstate), were performed according to [36] and [37], with minor modifications (see Supplemental Experimental Procedures for details).
Immunolabeling and Analysis of Polytene Chromosome Squashes
Polytene chromosome spreads were prepared according to standard procedures and stained overnight at 4°C with primary antibodies (gp-anti-Rad21 [1:500], mouse-anti-EcR-B1 [1:200]). Immunocomplexes were detected with Alexa-conjugated secondary antibodies and were mounted using Vectashield mounting medium containing DAPI (Vector Laboratories). Fluorescent images were acquired with an AXIO Imager.Z1 microscope (Zeiss) equipped with 40× and 63× EC Plan-Neofluar oil objectives and a CoolSNAP HQ charge-coupled device camera (Photometrics) using Meta-Morph software (Universal Imaging).
To analyze puffing of bands 74 and 75, we took only chromosome spreads in which those chromosome loci could be identified unambiguously (based on the characteristic twin puffed morphology and neighboring EcR double bands; see Figure 5D) into consideration. The width of each puff was measured and normalized to the average width of three neighboring bands.
Western Blot Analysis
Western blot analysis was performed from dissected third-instar larval salivary glands and whole larvae, according to standard protocols. All antibodies used are listed in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank B. Edgar, J. Mellor, and J. Mummery-Widmer (Knoblich laboratory) for reagents; Guillaume Filion for helping with the cohesin domain definition; B. Fischer (Fly-CHIP Cambridge) and Y. Katou for help with microarray analysis and ChIP-CHIP experiments, respectively; the central microarray facility from the Netherlands Cancer Institute for DamID-array hybridization; J. Metson, P. Guna, and W. Talhout for technical assistance; and all members of the B.v.S. and K.N. laboratories for discussions and comments on the manuscript. A.P. currently holds an EMBO Long-Term Fellowship. R.A.O. holds a postdoctoral fellowship from the Fundaç ão para a Ciência e a Tecnologia of Portugal. T.I. is supported by a Grant-in-Aid for Young Scientists (A) from the Japanese Society for the Promotion of Science (JSPS). Work in the laboratory of K.S. is supported by a grant of the Cell Innovation Program from Japan’s Ministry of Education, Culture, Sports, Science and Technology and a Grant-in-Aid for Scientific Research (S) from the JSPS. Work in the laboratory of B.v.S. is supported by a Netherlands Genomics Initiative grant. Work in the laboratory of K.N. is supported by grants from the Medical Research Council and Wellcome Trust.
Footnotes
Accession Numbers
Gene expression, DamID, and Pol-II ChIP-CHIP data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE24063 (gene expression and Pol-II) and GSE21874 (DamID).
Supplemental Information includes Supplemental Experimental Procedures, three figures, two tables, and three movies and can be found with this article online at doi:10.1016/j.cub.2010.09.006.
References
- 1.Nasmyth K, Haering CH. Cohesin: Its roles and mechaisms. Annu. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 2.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 3.Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS ONE. 2009;4:e6202. doi: 10.1371/journal.pone.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 11.Bénard CY, Kébir H, Takagi S, Hekimi S. mau-2 acts cell-autonomously to guide axonal migrations in Caenorhabditis elegans. Development. 2004;131:5947–5958. doi: 10.1242/dev.01433. [DOI] [PubMed] [Google Scholar]
- 12.Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 15.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 16.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, et al. Transcriptional dys-regulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 19.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 20.Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI. Induction by ecdysone in salivary glands of D. melanogaster cultured in vitro. Chromosoma. 1972;38:255–281. doi: 10.1007/BF00290925. [DOI] [PubMed] [Google Scholar]
- 22.Greil F, Moorman C, van Steensel B. DamID: Mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol. 2006;410:342–359. doi: 10.1016/S0076-6879(06)10016-6. [DOI] [PubMed] [Google Scholar]
- 23.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 24.Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckstead RB, Lam G, Thummel CS. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss A, Herzig A, Jacobs H, Lehner CF. Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol. 1998;8:239–242. doi: 10.1016/s0960-9822(98)70090-9. [DOI] [PubMed] [Google Scholar]
- 27.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 28.Santos-Rosa H, Kirmizis A, Nelson C, Bartke T, Saksouk N, Cote J, Kouzarides T. Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 2009;16:17–22. doi: 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhimulev IF, Belyaeva ES, Semeshin VF, Koryakov DE, Demakov SA, Demakova OV, Pokholkova GV, Andreyeva EN. Polytene chromosomes: 70 years of genetic research. Int. Rev. Cytol. 2004;241:203–275. doi: 10.1016/S0074-7696(04)41004-3. [DOI] [PubMed] [Google Scholar]
- 30.Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 31.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Buttitta LA, Katzaroff AJ, Perez CL, de la Cruz A, Edgar BA. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell. 2007;12:631–643. doi: 10.1016/j.devcel.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Vogel MJ, Pagie L, Talhout W, Nieuwland M, Kerkhoven RM, van Steensel B. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenetics Chromatin. 2009;2:1. doi: 10.1186/1756-8935-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 36.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 37.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.