Summary
Background
The Cohesin complex that holds sister chromatins together until anaphase is comprised of three core subunits: Smc1 and Smc3, two long-rod-shaped proteins with an ABC-like ATPase head (nucleotide-binding domain [NBD]) and a dimerization domain linked by a 50 nm long intra-molecular antiparallel coiled-coil, and Scc1, an α-kleisin subunit interconnecting the NBD domains of Smc1 and Smc3. Cohesin’s stable association with chromosomes is thought to involve entrapment of chromatin fibers by its tripartite Smc1-Smc3-Scc1 ring via a poorly understood mechanism dependent on a separate Scc2/4 loading complex. A key issue concerns where entrapment initially takes place: at sites where cohesin is found stably associated or at distinct “loading” sites from which it translocates.
Results
In this study, we find transition state mutant versions (Smc1E1158Q and SmcE1155Q) defective in disengagement of their nucleotide binding domains (NBDs), unlike functional cohesin, colocalize with Scc2/4 at core centromeres, sites that catalyze wild-type cohesin’s recruitment to sequences 20 kb or more away. In addition to Scc2/4, the unstable association of transition state complexes with core centromeres requires Scc1’s association with Smc1 and Smc3 NBDs, ATP-driven NBD engagement, cohesin’s Scc3 subunit, and its hinge domain.
Conclusion
We propose that cohesin’s association with chromosomes is driven by two key events. NBD engagement driven by ATP binding produces an unstable association with specific loading sites like core centromeres, whereas subsequent ATP hydrolysis triggers DNA entrapment, which permits translocation along chromatin fibers.
Introduction
Complexes between Smc and kleisin proteins regulate chromosome topology in most living organisms. Smc proteins are rod-shaped proteins with a dimerization domain at one end and an ABC-like nucleotide-binding domain (NBD) at the other with a 50 nm long coiled-coil region in between. Dimerization creates V-shaped molecules whose NBDs bind the winged helical domains of kleisin subunits. One of the best characterized members of this family is the eukaryotic cohesin complex [1-3], which, in addition to its canonical functions in mediating sister chromatid cohesion and double-strand break repair, also regulates transcription, at least in metazoa. Like all eukaryotic Smc-kleisin complexes, cohesin contains a pair of distinct Smc proteins, Smc1 and Smc3, whose NBDs bind C- and N-terminal domains of its Scc1 α-kleisin subunit, respectively, creating a large tripartite ring [4, 5] within which sister DNAs are thought to become entrapped after DNA replication [6-8], a hypothesis known as the ring model. Cohesin has three additional subunits associated with Scc1 [4, 9, 10]: two large α-helical repeat proteins, Scc3 [11] and Pds5 [12, 13], and a protein called Wapl (Rad61 in yeast) that forms a stable complex with Pds5 [14-16].
Preassembled tripartite rings create connections between sister DNAs in a complex and highly regulated process that can in principle be broken into a number of discrete steps. The first of these is the association of cohesin rings with unreplicated chromatin fibers, a process that depends on a separate complex composed of the Scc2 and Scc4 proteins [17, 18]. It has been suggested that this process, like cohesion itself, involves entrapment of chromatin fibers by cohesin rings, in this case individual fibers, by a mechanism that requires transient dissociation of Smc1/Smc3 dimerization domains [19]. There are two possible routes by which sister chromatin fibers become coentrapped during S phase: either rings that have already entrapped unreplicated chromatin fibers are converted during passage of replication forks to ones that entrap sister fibers or the rings that entrap sister fibers are derived from a soluble pool and must reload onto chromatin at the time of replication. Importantly, stable entrapment of sister fibers is accompanied by and dependent on de novo acetylation of Smc3 NBDs by the Eco1 acetyl transferase [16, 20, 21], which has been proposed to lock rings shut [22].
The molecular mechanism by which cohesin loads onto chromosomes is poorly understood. This is partly because the reaction occurs rapidly inside cells and has therefore hitherto defied dissection into discrete steps and partly because it is not known where on chromosomes loading takes place. With a diameter of 30 nm or more, cohesin rings should be capable of sliding along one or more 10 nm chromatin fibers, a process that clearly takes place in vitro [6, 8, 23]. If this also takes place in vivo, then the eventual distribution of cohesin as measured by chromatin immunoprecipitation studies (ChIP) may not in fact reflect where loading took place. The suggestion, either from chromosome spreads [17] or ChIP-chip studies [24, 25], that Scc2/4 and cohesin reside at different genomic locations in yeast has been taken to mean thatcohesin slides from “loading sites” occupied by Scc2/4 to its final chromosomal destination. However, the conclusion that Scc2/4 and cohesin have different genomic distributions has since been disputed [26]. More serious, there is hitherto no evidence that sites occupied by Scc2/4 represent loci at which cohesin is loaded onto chromosomes.
Cohesin is an ABC-like ATPase [27]. Binding of ATP to Smc1 and Smc3 NBDs is thought to induce their engagement and hydrolysis to drive them apart. We reasoned that blocking the ATP binding/hydrolysis cycle at a crucial point might cause cohesin to accumulate at an intermediate stage before the chromatin entrapment process. We show here that Smc1 or Smc3 variants trapped in a transition state with NBDs engaged [27, 28] accumulate to high levels at core centromeres and to a lesser extent within highly transcribed genes. The instability of this association and its colocalization with and dependence on Scc2/4 indicates that transition state cohesin complexes undergo an early step in the loading reaction but fail to entrap chromatin fibers. Our finding that transition state complexes accumulate at core centromeres is consistent with the notion that most pericentric cohesin loads at core centromeres and subsequently translocates considerable distances.
Results
Centromeres Promote Cohesin’s Accumulation 50 Kilobases Away
If specific loci exist for loading cohesin onto chromosomes, then they should increase recruitment of cohesin to neighboring sequences. The best and possibly only current example of such loci in budding yeast are core centromeres, which have been reported to be both necessary and sufficient for the high levels of cohesin throughout a 50 kb pericentric interval [29, 30]. To confirm this remarkable phenomenon, and to determine how far along a chromosome a centromere influences cohesin levels, we used ChIP-SEQ to compare cohesin’s distribution before and after moving the chromosome XIV centromere to a remote cohesin-poor arm region (Figure S1, available online). We chose this technique, which involves high-throughput sequencing to identify and quantify DNA fragments precipitated by antibodies specific for epitopes attached to cohesin subunits, because it is more quantitative and accurate than hybridization to microarrays [31]. To generate reliable binding profiles, we obtained more than 10 million sequence reads for both ChIP and whole-cell extract (WCE) fractions. Sequence coverage was higher than 96% and total redundancy was over 38 times (Table S1). Frequencies of sequence reads within running 500 bp windows (50 bp step size) were normalized by dividing those obtained from immunoprecipitations (IPs) by those obtained from whole-cell extracts (WCEs) and plotted on a linear scale with a baseline corresponding to zero.
If a protein were distributed at random, the ChIP/WCE ratio would equal 1. Crucially, values below 1 do not necessarily imply that the protein in question is not present. They merely show that the protein is present at a level below average. The resulting profiles for Scc1-PK6 reveal pronounced peaks, with enrichment values sometimes above 10, separated by large intervals with enrichment values below average. A key issue, largely ignored by previous ChIP-SEQ or ChIP-CHIP studies, is whether values between peaks represent widespread albeit low-level association or merely “background noise” caused by nonspecific DNA precipitation. To estimate this background, we calculated enrichment values of mitochondrial DNA, which is presumably not associated with either cohesin or its loading complex, and 2 μm DNA, which, though nuclear and packaged into nucleosomes, might lack efficient cohesin loading sites. The profiles of these episomal genomes both lacked discernable peaks and their average enrichment values were 0.04 and 0.11, respectively. The values for 2 μm are remarkably low considering the claim that its STB recruits cohesin [32], and we suggest that they represent an upper bound for background, whereas the lower values for mitochondrial DNA represent a lower bound. Note that if some cohesin really binds 2 μm DNA in vivo, then the background enrichment value could in fact be below 0.11. This novel way of estimating background noise implies that a sizeable fraction of cohesin associated with yeast chromosomes is present within valleys between peaks.
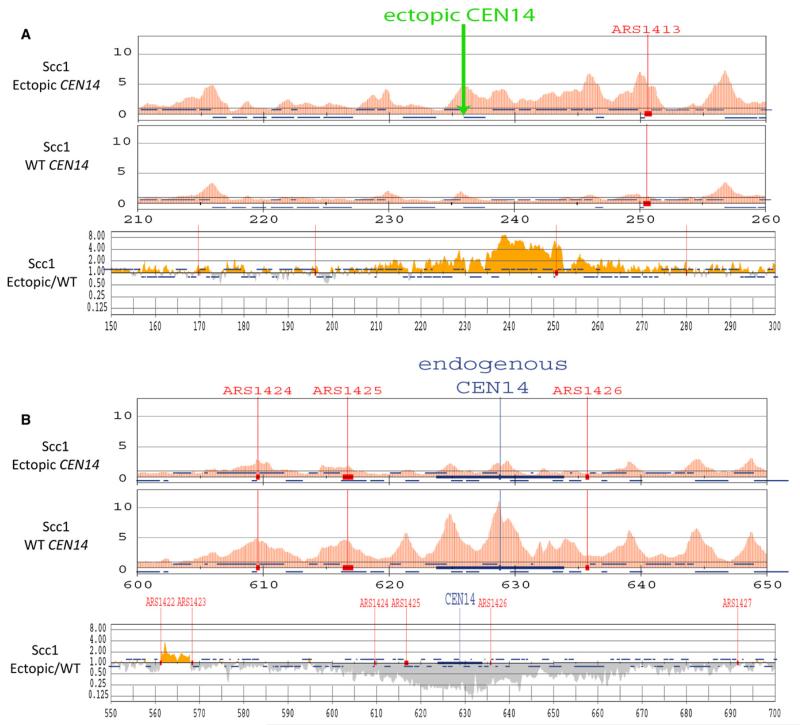
Translocation of CEN14 reduced cohesin’s association with sequences within a 100 kb interval around the deleted centromere (Figure 1A) and increased it within a similar sized interval around the insertion site (Figure 1B). The effect was quantified by plotting the ratio between enrichment values within running 500 bp intervals along the genome before and after CEN14 translocation. There was little systematic effect on unaltered chromosomes or on the bulk of chromosome XIV itself. In contrast, there were 2- to 8-fold decreases and/or increases within a 20 kb window and more modest, but nevertheless detectable, changes within a 100 kb window surrounding the deletion and/or insertion sites. Inactivation of the nonessential Chl4 subunit of the kinetochore-specific Ctf19 complex reduced cohesin levels within a similarly wide window around core centromeres (Figure S1C), confirming that proteins associated with core kinetochores are necessary for cohesin’s recruitment to distant pericentric sequences [29, 33].
Figure 1. Centromeres and Kinetochore Proteins Promote Recruitment of Scc2/4 and Cohesin.
(A and B) An ectopic centromere promotes cohesin’s accumulation 20 kb away. The distribution of Scc1-PK9 was measured by ChIP-SEQ in exponentially grown cells from strain K16670 in which CEN14 was moved to a site between ADE12 and ALG9 (top panels) and compared to that of Scc1 in wild-type (WT) cells (K16586) (top and middle panels). The ratios of Scc1 ChIP-SEQ signals between K16670 and wild-type were mapped to chromosome XIV (bottom panels). Yellow bars indicate the ratio is more than 1.0 and gray bars indicate the ratio is less than 1.0. The Scc1 distribution within a 50 kb region along ectopic CEN14 (A) or endogenous CEN14 (B) is shown. See also Figure S1.
Cohesin Blocked in a Transition State Accumulates at Core Centromeres
One explanation for the kinetochore effect is that most pericentric cohesin loads onto chromosomes at core centromeres and subsequently translocates into neighboring sequences. According to the ring model, centromeres and their associated proteins catalyze entrapment of chromatin fibers by cohesin rings that subsequently slide large distances along the chromatin fiber, away from the initial site of entrapment. An alternative is that core centromeres by some mysterious means enhance loading of cohesin throughout pericentric regions; in other words, cohesin loads at sites close to where it eventually resides. To distinguish these, it is necessary to observe cohesin complexes in the act of loading. According to the sliding model, they will be found at core centromeres, whereas according to the local loading model, they will be found throughout pericentric sequences.
One way of addressing where cohesin initially entraps chromatin fibers would be to analyze cohesin complexes blocked at various stages in the binding and hydrolysis of ATP. This was initially performed with quantitative PCR to measure co-precipitation of core centromere, inner and outer pericentric, and chromosomal arm DNAs with epitope-tagged wild-type or mutant Smc proteins (ChIP-qPCR). Because the mutations are lethal [28], experiments were performed with cells expressing untagged wild-type protein alongside mutant versions. Mutations in Walker A motifs that abolish binding of ATP to Smc1 (Smc1K39I) or Smc3 (Smc3K38I) abolished association with all DNAs tested (Figure 2A). In contrast, the Smc1E1158Q and Smc3E1155Q mutations, which prevent hydrolysis of ATP bound to Smc1 and Smc3 NBDs, respectively [27, 28], and are predicted to block or at least delay cohesin in a transition state with nucleotide bound between engaged NBDs, actually increased association with core centromeres while decreasing it within pericentric or arm sequences (Figure 2A). The reduction at most sequences other than core centromeres is consistent with the previous observation that Smc1E1158Q and Smc3E1155Q reduce cohesin’s overall chromatin association in chromosome spreads [28]. The striking increase at core centromeres had been overlooked in the above study. This is not a general property of Smc1/3 proteins defective in ATP hydrolysis because cohesin complexes that bind but cannot hydrolyze ATP because signature motif mutations Smc1S1130R or Smc3S1128R that prevent NBD engagement do not associate with centromeres (Figure S2A).
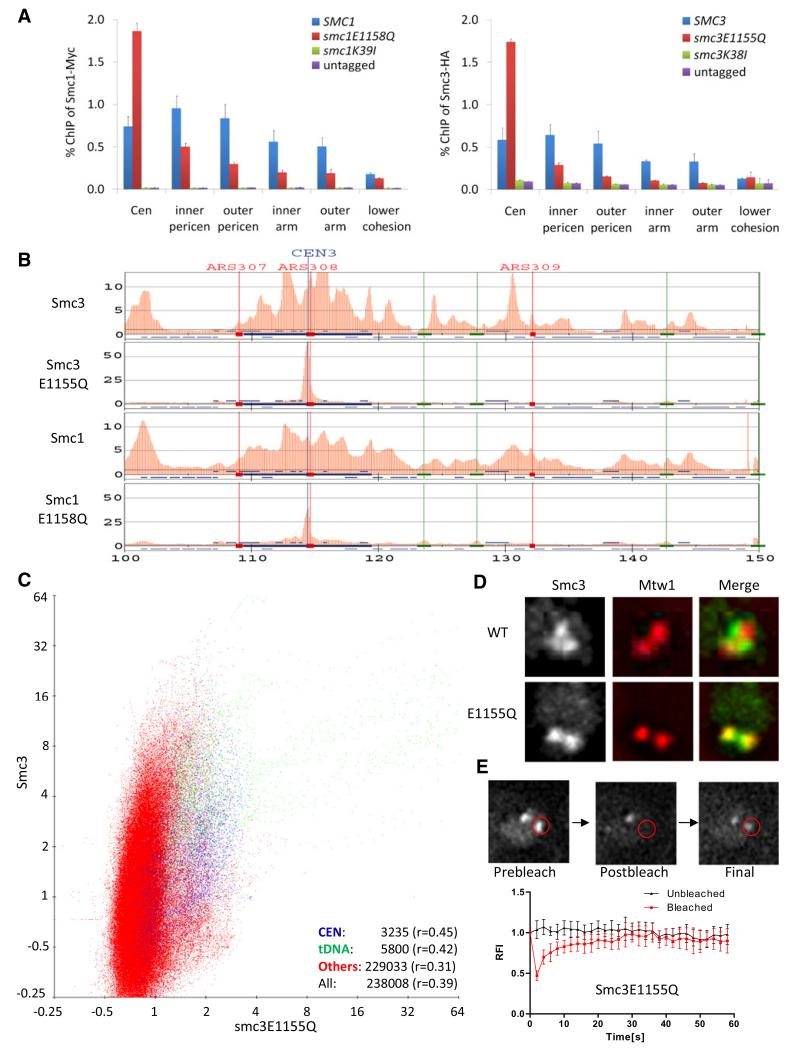
Figure 2. Unstable Association of Hydrolysis-Defective Cohesin Complexes with Centromeres.
(A) Hydrolysis-defective cohesin is enriched at centromeres. Association with defined loci of wild-type and mutant Smc proteins tagged with myc9 (Smc1) or HA3 (Smc3) was measured by ChIP-qPCR. Chromatin was immunoprecipitated from extracts prepared from exponentially grown cells of strains K699, K11850, K11852, K11857, K11872, K13560, and K13561. The following abbreviations are used: Cen, centromere; inner pericen, inner pericentromere; outer pericen, outer pericentromere. Error bars represent standard deviation (SD); n = 3.
(B) Genome-wide distribution of Smc3-HA3 and Smc3E1155Q-HA3. Crude extracts prepared from exponentially grown yeast cells (K16586 and K17458) were used for ChIP-SEQ. Red bars represent binding ratios within running 500 bp windows (50 bp step size) showing enrichment in the ChIP fraction. Positions of the centromere (CEN) and autonomously replicating sequences (ARSs) are shown. The horizontal axis represents kilobases along chromosome III.
(C) Correlations between Smc3 and Smc3E1155Q. ChIP-SEQ signals were pooled from running 500 bp windows along each chromosome every 50 bp. Smc3E1155Q-HA3 signals from each window were plotted against those of Smc3-HA3. ChIP signals within 500 bp of tDNAs are marked as green dots, those within 5000 bp of centromeres are marked as blue dots, and the rest are marked as red dots. The correlation coefficients for centromeres, tDNA, the others, and the total are shown bottom right.
(D) Localization of Smc3 and Smc3E1155Q in live diploid cells. Smc3 and Smc3E1155Q were fused with GFP (K18232 and K16715). Mtw1-RFP was used as a kinetochore marker. Smc3 forms pericentromeric barrels between sister kinetochore clusters at metaphase, whereas Smc3E1155Q colocalizes with Mtw1.
(E) Association of Smc3E1155Q with centromeres is unstable. FRAP was measured in diploid yeast cells expressing Smc3E1155Q-GFP. One of two Smc3E1155Q fluorescent foci was bleached by exposing the area marked by a red circle to an argon laser for 200 ms. Relative fluorescence intensities of unbleached (black) and bleached (red) signals are plotted over time. Smc3E1155Q -GFP recovered with t1/2 = 3.4 s; n = 6; the error bars represent SD. The signal detected just after photobleaching (around 45% of that before photobleaching) was due to rapid turnover of soluble pool of Smc3E1158Q. See also Figure S2.
To confirm and extend these findings, we compared the distributions of Smc3 and Smc3E1155Q using ChIP-SEQ. Figure 2B shows that the distributions of Smc3 or Smc3E1155Q around CEN3 are very different. Smc3 accumulates in a number of above average peaks within a 50 kb pericentric interval, whereas Smc3E1155Q accumulates to very high levels in a narrow zone surrounding the core centromere itself and at much lower levels at several neighboring loci. Similar distributions and differences between Smc3 and Smc3E1155Q were found around the centromeres of all chromosomes. A scatter plot of Smc3 versus Smc3E1155Q revealed little colocalization (Figure 2C). The correlation coefficients at tDNAs, around centromeres, and at noncentromeric non-tDNA sequences were 0.42, 0.45, and 0.31, respectively. ChIP-SEQ revealed that the genomic distribution of Smc1E1158Q was similar to that of Smc3E1155Q and likewise differed greatly from its wild-type cognate (Figure 2B). The most striking aspect about the distribution of cohesin complexes containing either Smc1E1158Q or Smc3E1155Q is their accumulation to very high levels around core centromeres and their reduced association with most other sites occupied by wild-type complexes.
Unstable Association with Centromeres of Transition State Cohesin
To visualize wild-type and mutant Smc proteins in live cells, we tagged the proteins with GFP. Both Smc1E1158Q and Smc3E1155Q accumulated within nuclei and within them formed distinct foci colocalizing throughout most of the cell cycle with the kinetochore protein Mtw1 tagged with RFP (data not shown). During metaphase, both Mtw1 and the mutant Smc proteins formed two distinct foci corresponding to sister kinetochore clusters that have been partially pulled apart by microtubules (biorientation) (Figure 2D and Figure S2B), a pattern that is distinct from that of pericentric cohesin marked by Smc3-GFP (Figure 2D), which forms a barrel of fluorescence between bioriented Mtw1 foci [16, 34]. Thus, the accumulation of Smc1E1158Q and Smc3E1155Q at core centromeres observed by ChIP-SEQ is also seen in live cells. The Smc1 ATP-binding mutant (Smc1K39I) failed to accumulate within nuclei (Figure S2B), presumably because it cannot bind Scc1 [28]. In contrast, Smc3K38I, which forms cohesin rings, accumulated within nuclei but did not form either pericentric barrels or foci in the vicinity of kinetochores (Figure S2B).
To address whether the association of Smc1E1158Q- or Smc3E1155Q-GFP with core centromeres is a stable one, we selectively bleached one of the sister clusters within metaphase cells and measured its rate of recovery (FRAP) (Figure 2E and Figure S2C). Remarkably, fluorescence of bleached and unbleached Smc1E1158Q- or Smc3E1155Q-GFP clusters converged within 20 s (half-time for recovery: 5–7 s). In contrast, similar experiments bleaching one side of Smc1-GFP barrels in metaphase cells indicated that cohesin associates stably with pericentric chromatin at this stage of the cell cycle (Figure S2D and [35]). The rapid turnover of Smc1E1158Q- or Smc3E1155Q-GFP is not a general property of proteins associated with kinetochores because FRAP demonstrated the Mtw1-GFP turns over slowly, if at all (Figure S2E). The dynamic behavior of the mutant proteins suggests that they are unlikely to confer sister chromatid cohesion even at centromeres. To test this, we used a physical assay (sedimentation velocity combined with gel electrophoresis [6]) to ascertain whether the mutant proteins generate sister minichromosome cohesion in cells depleted (with a degron system) of cognate wild-type protein. As expected, wild-type but not hydrolysis-defective Smc proteins supported minichromosome cohesion (Figures S2F–S2H).
Scc2/4 Is Necessary but Not Sufficient for Association of Transition State Complexes with Chromatin
The finding that Smc1E1158Q- and Smc3E1155Q-containing cohesin accumulate at core centromeres raises the possibility that, though dysfunctional, these complexes nevertheless undergo an early step in the loading process. If so, they might be expected to colocalize with Scc2/4 complexes. Because it is difficult to compare our current ChIP-SEQ data with previous ChIP-CHIP data on Scc2 [24, 26] and because of conflicting results concerning the genomic distribution of Scc2/4 [24, 26], we reinvestigated its distribution using ChIP-SEQ. This revealed that both proteins are enriched at core centromeres and specific chromosome arm loci (Figure 3A and Figure S3A). Average enrichment values for mitochondrial and 2 μm DNAs, which give lower and upper estimates of the background noise, were 0.06 and 0.275, respectively. As expected, scatter plots of Scc2 versus Scc4 (Figure S3B) revealed a striking correlation around tDNAs (r = 0.98), within a 10 kb interval around centromeres (r = 0.98), and throughout the rest of the genome (r = 0.91). This analysis also revealed that Scc4 associates with a number of nontranscribed loci in the absence of Scc2 (red points above the line), suggesting that the former might have a second, Scc2-independent function.
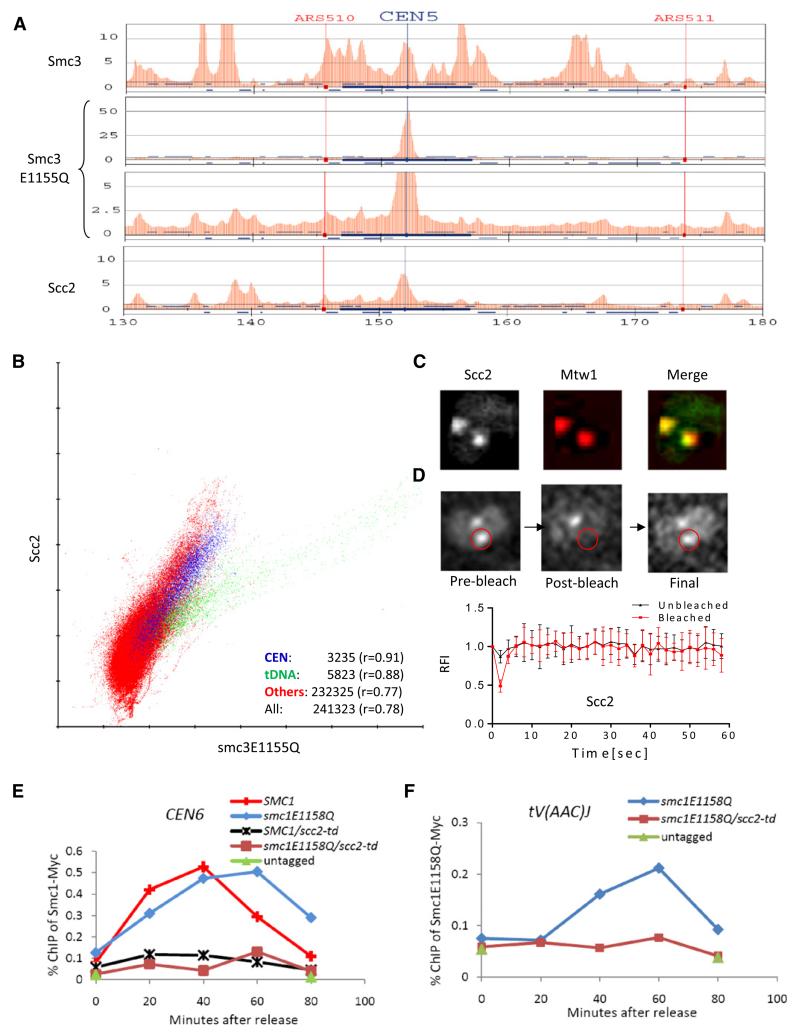
Figure 3. Hydrolysis-Defective Cohesin Colocalizes with and Depends on Scc2/4.
(A) ChIP-SEQ distributions of Smc3-HA3, Smc3E1155Q-HA3, and Scc2-FLAG6 at selected regions of chromosome V. Yeast strains K13560, K13561, and K17458 were used.
(B) Correlations of Smc3E1155Q with Scc2 performed as described in Figure 2C.
(C) Localization of Scc2-GFP in live diploid cells (K16442). Kinetochores are marked by Mtw1-RFP.
(D) Association of Scc2-GFP with centromeres is unstable. FRAP performed as in Figure 2E showed that fluorescence recovered after photobleaching with t1/2 = 0.8 s; n = 6.
(E and F) Association of Smc1E1158Q with chromatin is Scc2-dependent. Exponential phase cultures of strains K16799, K16800, K16811, K16812, and K16331 growing in YEPraff at 25°C were arrested in G1 with α-factor. Degradation of td-Scc2 was triggered by shifting cultures to YEPgal, 20 μg/ml doxycycline, and 37°C for 1 hr before transferring cells to pheromone-free YEPgal media containing 20 μg/ml doxycycline. Chromatin was immunoprecipitated with myc9 tags, and association with indicated loci of myc9-tagged Smc1 and Smc1E1158Q was measured with ChIP-qPCR. See also Figure S3.
Scatter plots between Scc2 and Smc3 revealed a very different picture (Figure S3C). The correlation coefficients at tDNAs, around centromeres, and at noncentromeric non-tDNA sequences were 0.48, 0.54, and 0.2, respectively. These data confirm previous suggestions [17, 24] that the distributions of Scc2/4 and cohesin are distinct and are inconsistent with the recent claim that Scc2/4 and cohesin largely colocalize throughout pericentric and arm regions [26]. Interestingly, scatter plots demonstrated that the distributions of Smc3E1155Q and Scc2 are, in contrast, broadly similar (Figure 3B), with striking correlations at centromeres (r = 0.91), at tDNAs (r = 0.88), and at all other sequences (r = 0.78). Importantly, there is much greater colocalization between Smc3E1155Q and Scc2 than between Smc3E1155Q and wild-type Smc3. Colocalization at centromeres was confirmed by live-cell imaging. Scc2-GFP colocalized with Mtw1-RFP in metaphase cells, a pattern resembling that of Smc1E1158Q or Smc3E1155Q but not that of wild-type Smc1/3 proteins (Figure 3C). Indeed both Scc2- and Scc4-GFP colocalized with Smc1E1158Q-RFP (Figure S3D). FRAP demonstrated that Scc2/4 complexes turn over very rapidly in the vicinity of kinetochores, with a half life of 2–4 s (Figure 3D). Interestingly, Scc2-GFP’s half life was noticeably shorter than that of Smc1E1158Q- or Smc3E1155Q-GFP whether or not cells expressed Smc1E1158Q.
If Smc1E1158Q- and Smc3E1155Q-containing cohesin complexes participate in a key early event during the loading process, then their association with chromosomes might be expected to depend on Scc2/4 activity. To test this, we used ChIP-qPCR to compare association of Smc1 and Smc1E1158Q with core centromeres as wild-type (SCC2) or Scc2 depleted cells (scc2-td; Figure S3E and S3F) re-enter the cell cycle after pheromone-induced G1 arrest (Figure 3E). This revealed that association with Smc1E1158Q was as dependent on Scc2 as wild-type Smc1. Association of Smc1E1158Q with selected arm sites, namely the tV(AAC)J tDNA (Figure 3F), SNR10, and SSB2 genes (data not shown), was similarly affected by Scc2 depletion. However, association of Scc2/4 with a chromosomal locus is insufficient to recruit Smc1E1158Q because artificial tethering of a functional Scc2-TetR fusion to a cluster of Tet operators 38 Kb from CEN5 fails to recruit either Smc1-GFP or Smc1E1158Q-GFP to this location, despite efficiently recruiting Scc4-GFP (Figure S3G). This also demonstrates that association of Smc1E1158Q and Smc3E1155Q with core centromeres does not arise merely because the mutant complexes still bind Scc2/4 despite failing to associate stably with chromosomes.
Scc2/4 and Smc3E1155Q Accumulate on Highly Transcribed Genes
Visual inspection revealed that Scc2, Scc4, and Smc3E1155Q frequently colocalized along chromosome arms with tRNA genes (Figure 4A), as recently found for condensin [36]. ChIP-SEQ revealed a strong correlation (r = 0.87) between Scc2 in the vicinity of tDNAs (marked in green) and Pol III occupancy (Figure 4B). ChIP-SEQ also revealed that Scc2/4 levels were invariably high throughout the transcription units of genes with high Pol II occupancy. When Pol II levels are plotted versus Scc2, the majority of points lie on a steep arc (Figure 4C). As expected, tDNA loci (green) and sequences around CENs (blue), which have an ability to recruit Scc2 independent of PolII, lie to the right of this arc. For sequences (marked in red) that are neither in the vicinity of tDNAs nor centromeres, the correlation coefficient between Pol II and Scc2 is 0.78. The correlation between Scc2 and Pol II is even more striking when the analysis is restricted to ORFs (Figure S4A).
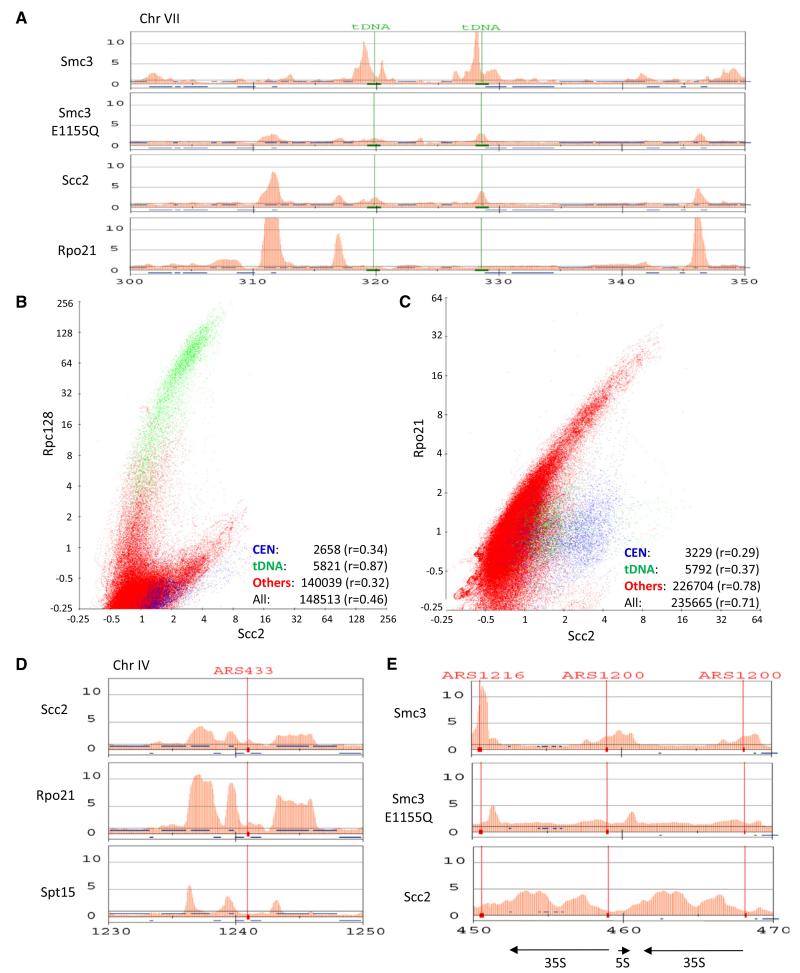
Figure 4. Scc2/4 and Hydrolysis-Defective Cohesin Accumulate on Highly Transcribed Genes.
(A) Scc2 and Smc3E1155Q but not Smc3 colocalize with PolII subunit Rpo21 a PolII. Yeast cells expressing Rpo21-Flag3 (K17460) were used for ChIP-SEQ. The distributions of Smc3-HA3, Smc3E1155Q-HA3, and Scc2-FLAG6 are compared to Rpo21-FLAG3 within a selected region of chromosome VII. tDNAs are indicated with green lines.
(B) Correlation of Rpc128 (K18393) with Scc2 performed as described in Figure 2C.
(C) Correlation of Rpo21 (K18394) with Scc2 performed as described in Figure 2C.
(D) The distributions of Scc2, Rpo21, and Spt15 within a selected region of chromosome IV.
(E) The distributions of Smc3, Smc3E1155Q, and Scc2 at rDNA loci with transcription units of 35S and 5S shown at the bottom. See also Figure S4.
To confirm that ChIP-SEQ has sufficient resolution to distinguish transcription units from their promoters, we analyzed the distribution of the TATA-box binding protein (TBP) encoded by SPT15. Though most Spt15 is associated with Pol III transcription units, it was also enriched at the 5′ ends of highly transcribed Pol II genes. Importantly, its distribution at such genes was distinct from that of PolII and Scc2 (Figure 4D). Spt15 is concentrated at promoters, whereas PolII and Scc2 are found throughout ORFs. ChIP-SEQ confirmed that Scc2/4 as well as PolII are recruited to galactose-inducible genes when cells are grown in galactose instead of glucose (Figure S4B). Interestingly, Scc2/4, Smc1E1158Q, and Smc3E1155Q are preferentially associated with the Pol I transcription units within rDNA and not, as in the case of wild-type cohesin, with the nontranscribed spacer (Figure 4E). This raises the possibility that in addition to a specialized kinetochore-driven reaction, Scc2/4 and cohesin recruitment along chromosome arms may be associated with transcription by all three polymerases.
Cohesin Completely Blocked with NBDs Stably Engaged Associates with Centromeres
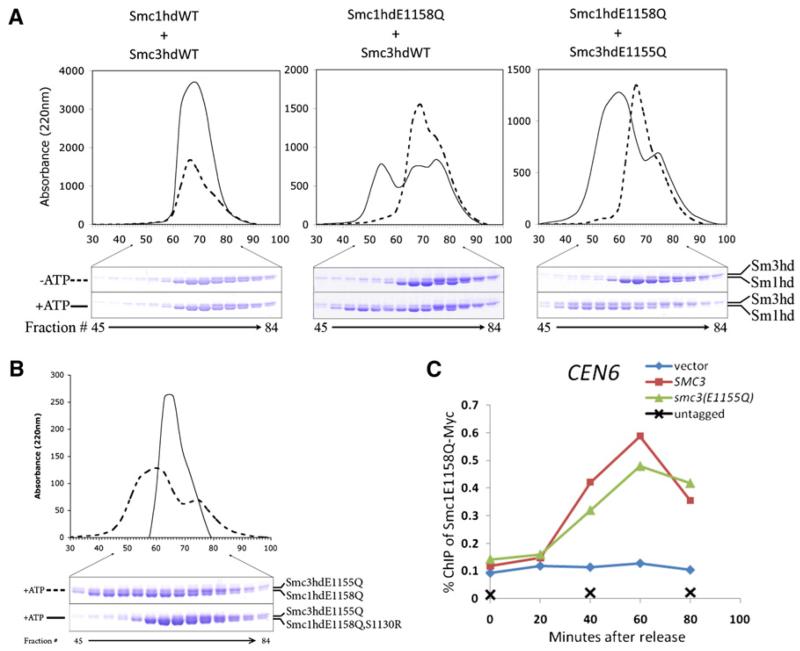
Current models presume that ATP induces engagement of Smc1 and Smc3 NBDs, which is a prerequisite for ATP hydrolysis. However, the observation that isolated Smc1 NBDs associated with Scc1’s C-terminal winged helical domain can alone hydrolyse ATP [27] raises the possibility that NBD engagement in fact involves association of like heads from different Smc1/3 heterodimers. To investigate the specificity of NBD engagement, we used gel filtration to measure association of wild-type and ATP hydrolysis-defective Smc1 and Smc3 NBDs (Figure 5A). Smc1E1158Q but not wild-type Smc1 formed homodimers in the presence of ATP, but neither Smc3E1155Q nor wild-type Smc3 did so. Importantly, ATP-induced heterodimers between Smc3E1155Q (but not Smc3) and Smc1E1158Q but not between Smc3E1155Q and wild-type Smc1 (Figure 5A). Signature motif mutations (either Smc1S1130R or Smc3S1128R) abolished formation of stable complexes between Smc1E1158Q and Smc3E1155Q (Figure 5B and data not shown), demonstrating the key role of these residues in NBD engagement. The lack of stable complexes involving wild-type NBDs is presumably due to ATP hydrolysis. Given that Smc1 and Smc3 NBDs are attached to each other via Smc1/3 hinges, these data suggest that Smc1NBDs engage with Smc3 NBDs rather than with themselves in vivo.
Figure 5. Cohesin Completely Blocked with NBDs Stably Engaged Associates with Centromeres.
(A) ATP-dependent engagement of Smc1 and Smc3 NBDs. Equal amounts (3 nmol) of recombinant WT or ATP hydrolysis-defective Smc NBDs (Smc1 was associated with Scc1’s C-terminal fragment [F451–A566]) were subjected to size exclusion chromatography in the presence or absence of ATP. No dimer formation was observed with ATPγS or AMPPNP (data not shown).
(B) Disruption of ATP-dependent engagement of Smc1 and Smc3 NBDs by mutation in signature motif. Equal amounts (3 nmol) of recombinant indicated Smc NBDs were subjected to size exclusion.
(C) Cohesin incapable of hydrolyzing ATP associated with both NBDs also associates with centromeres. Exponential phase cells of strains K16331, K17240, K17241, and K17242 growing at 25°C were arrested in G1 with α-factor. Degradation of Smc3-td was induced and cells released from pheromone as in Figure 2E. The association of Smc1E1158Q-MYC9 with centromere was measured by ChIP-qPCR. See also Figure S5.
To address whether association with core centromeres of cohesin complexes containing either Smc1E1158Q or Smc3E1155Q is also a property of complexes that hydrolyse neither ATP molecule and therefore accumulate in a state with Smc1 and Smc3 NBDs stably engaged, we created a strain expressing Smc1E1158Q tagged with Myc, wild-type Smc1, either Smc3E1155Q or wild-type Smc3, and a temperature sensitive degron-degradable Smc3 expressed under tetracycline control (smc3-td). Cells were arrested in G1 as a result of incubation in the presence of pheromone, Smc3 expression turned off by addition of doxycycline, and Smc3 degradation induced by shift to 37°C and addition of galactose to induce the degron-specific ubiquitin protein ligase Ubr1 (Figure S5A and S5B). Cells were subsequently released into the cell cycle by pheromone removal under circumstances in which Smc1E1158Q-Myc binds Smc3E1155Q in one strain and wild-type Smc3 in the other. ChIP-qPCR revealed that Smc1E1158Q-Myc associated with core centromeres to similar levels in both strains (Figure 5C) upon resynthesis of Scc1 in late G1. Crucially, association depended on expression of Smc3 or Smc3E1155Q, proving that Smc3-td had indeed been depleted. We conclude that complexes in which Smc1 and Smc3 NBDs are stably trapped in an engaged state (so stable that it can be detected by gel filtration) also associate with core centromeres, a finding that contrasts with the observation that mutations predicted to block bacterial SMC proteins in an equivalent state reduce DNA binding in vitro [37].
Ring Formation and Scc3 Are Required for Association of Transition State Cohesin with Sites Occupied by Scc2/4
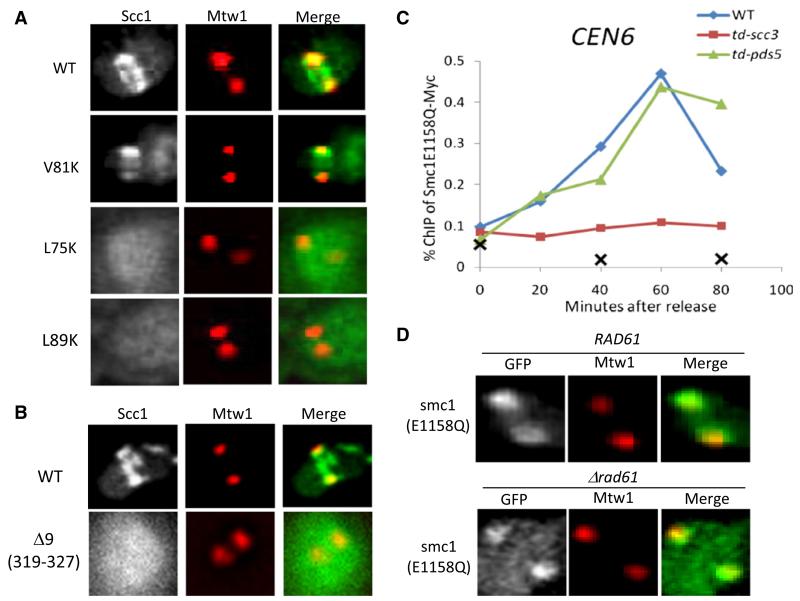
To test the role of cohesin ring formation, we analyzed the effect of Scc1 mutations (Figure S6A). When coexpressed with Smc1E1158Q, wild-type Scc1-GFP colocalized with Mtw1 foci as well as forming pericentric barrels in between (Figure 6A). Interestingly, the Mtw1-associated foci persisted after anaphase onset, suggesting that Scc1 associated with Smc1E1158Q is not cleaved by separase (Figure S6C). Both live imaging (Figure 6A) and ChIP-qPCR (Figure S6B) showed that L75K and L89K, which disrupt interaction between Smc3’s NBD and Scc1’s N-terminal domain [27], abolish association of Scc1-GFP with Mtw1 as well as pericentric barrels. V81K, which is also lethal but still permits association with Smc3 NBDs, resembled wild-type (Figure 6A and Figures S6A and S6B), though pericentric barrels were weaker. We conclude that ring formation as well as NBD engagement is necessary for centromere/kinetochore association.
Figure 6. Ring Formation and Scc3 Are Required for Cohesin Loading.
(A) Ring formation is required for cohesin loading. Wild-type or mutant Scc1-GFP was coexpressed with Smc1E1158Q (K16764, K16765, K16766, and K16767) and imaged in live cells. Wild-type Scc1-GFP and Scc1V81K-GFP colocalize with Mtw1-RFP foci and, in addition, form pericentric barrels in between. L75K and L89K mutations abolish both association with Mtw1 and barrel formation.
(B) Scc3 is required for cohesin loading. Wild-type or mutant Scc1-GFP (K16915) was coexpressed with Smc1E1158Q and visualized by fluorescence microscopy. Wild-type Scc1-GFP colocalizes with Mtw1 foci as well as forming pericentric barrels in between. Scc1Δ319-327-GFP merely accumulates within nuclei.
(C) Association of Smc1E1158Q with chromatin depends on Scc3 but not Pds5. Exponential phase cells of strains K16331, K16812, K17299, and K17300 were arrested at G1 phase with α-factor at 25°C. Degradation of Scc3-td or Pds5-td was induced and cells released from pheromone as in Figure 2E. Association of myc9-tagged Smc1E1158Q with centromeres was measured by ChIP-qPCR.
(D) Smc1E1158Q-GFP was expressed in wild-type or rad61Δ strains (K16445 and K16796) and visualized together with Mtw1-RFP by fluorescence microscopy. See also Figure S6.
A similar approach enabled us to address the role of cohesin’s Scc3 subunit. Deletion of Scc1 amino acids 319-327 (Δ9) disrupts association with Scc3 (Figure S6D) but has no effect on ring formation (data not shown). It also abolishes Scc1-GFP pericentric barrels and foci associated with Mtw1 when coexpressed with Smc1E1158Q (Figure 6B). To confirm this and to address the role of Pds5, we created strains in which either Scc3 or Pds5 are depleted with degron alleles (scc3-td and pds5-td). ChIP-qPCR revealed that association of Smc1E1158Q myc9 with core centromeres as G1 cells enter S phase is greatly reduced by Scc3 depletion but unaffected by that of Pds5 (Figure 6C and Figures S6E and S6F). ChIP-qPCR as well as live-cell imaging showed that loss of Pds5’s partner, namely Rad61/Wapl, also had little or no effect (Figure 6D and Figure S6G). We conclude that Scc3 but not Pds5/Wapl is required for association of Smc1E1158Q with core centromeres.
A Role for Smc1/3 Hinges in Association of Transition State Cohesin with Sites Occupied by Scc2/4
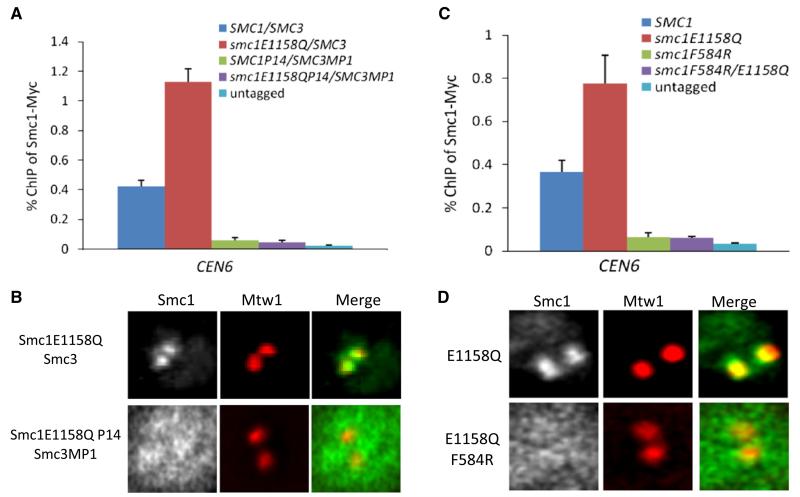
The proposal that entrapment of chromatin by cohesin requires transient dissociation of its Smc1/3 hinge [19] suggests that this domain might also be involved at an early stage in chromatin engagement. We therefore used ChIP-qPCR and live-cell imaging to address whether a Smc1E1158Q protein that forms heterodimers with Smc3 via an MP1-p14 dimerization module also associates with core centromeres. Both assays demonstrated that replacement of Smc1/3 hinges by MP1-p14 abolishes centromere association (Figures 7A and 7B and Figure S7A) despite permitting the formation of cohesin rings whose NBDs can engage [19]. Thus, engagement of Smc1 and Smc3 NBDs, their interconnection by Scc1, and the binding of Scc3 are together insufficient to drive the early loading reaction undergone by complexes containing Smc1E1158Q. A cohesin hinge dimerization domain is also required.
Figure 7. Smc1/3 Hinges Are Required for Cohesin Loading.
(A and B) Hinge-substituted Smc1/3 heterodimers are not loaded onto chromosomes. (A) Association with CEN6 of myc9-tagged Smc1 or Smc1E1158Q with wild-type or p14/MP1 hinge domains measured by ChIP-qPCR in asynchronous yeast extract/peptone/dextrose (YPD) cultures of strains K699, K11587, K13585, K14133, and K16874. The error bars represent SD; n = 3.
(B) Smc1E1158Q-GFP or Smc1P14/E1155Q-GFP was coexpressed with an extra copy of Smc3 or with Smc3MP1 (K16936 and K17070). GFP fusion proteins were visualized by fluorescence microscopy. Hinge substitution abolished association of Smc1E1158Q-GFP with Mtw1-RFP foci in metaphase cells.
(C and D) The F584R Smc1 hinge mutation abolishes cohesin’s association with centromeres. (C) Association with CEN6 of myc9-tagged Smc1 or Smc1E1158Q proteins with wild-type or Smc1F584R hinge domains measured by ChIP-qPCR in asynchronous YPD cultures of strains K699, K11857, K14133, K14134, and K17000. The error bars represent SD; n = 3. (D) Smc1E1158Q-GFP or Smc1F584R/E1155Q-GFP (K16895 and K17070) was coexpressed with an extra copy of Smc3 and visualized together with Mtw1-RFP by fluorescence microscopy. See also Figure S7.
We recently discovered that Smc1 F584R, which largely destroys the hinge’s south Smc1/3 interface and possibly causes a rearrangement of its torroidal structure, reduces cohesin’s association with chromosomes without greatly reducing association (in vivo) with either Smc3 or Scc1 [35]. A trivial explanation for the lowered chromatin association is that the mutation so weakens the hinge interface that chromatin fibers rapidly escape entrapment. However, it is possible that the mutation also directly affects an early step in cohesin’s engagement with chromatin. To investigate this, we used ChIP-qPCR and live-cell imaging to compare association with centromeres of single mutant Smc1E1158Q and double mutant Smc1F584R/E1158Q proteins in cells expressing two copies of SMC3 to ensure that the mutant proteins do not have to compete with wild-type Smc1 for binding to Smc3. Both assays revealed that Smc1F584R abolishes association of Smc1E1158Q with centromeres (Figures 7C and 7D). These data confirm that association with centromeres of cohesin whose NBDs are trapped in a transition state involves cohesin’s hinge. If, as is likely, Smc1E1158Q-containing cohesin complexes never entrap chromatin fibers, the effect of Smc1F584R cannot be attributed to chromatin fiber escape.
Discussion
Evidence that Core Centromeres Are Cohesin Loading Sites
To address the mechanism by which cohesin loads onto chromatin fibers, it is necessary to identify loci at which loading/entrapment takes place, in particular genomic addresses where it occurs at high rates. The notion that loading involves creation of a topological linkage mediated by entrapment of DNA by cohesin rings suggests that loci at which cohesin rings initially entrap DNA need not correspond to those at which they subsequently reside as rings may be capable of sliding along chromatin fibers while remaining stably associated with them. Indeed, there is little or no evidence that the majority of sites at which cohesin resides are sufficient to load cohesin onto chromosomes. Clear exceptions in budding yeast are core centromeres that are both necessary and sufficient for recruitment of cohesin to neighboring pericentric sequences [29, 30]. Using ChIP-SEQ, we have confirmed and extended this important conclusion, observing that core centromeres affect the density of cohesin up to 50 kb away.
One can envision two types of mechanism by which core centromeres, via their kinetochore associated proteins, recruit cohesin to much wider pericentric domains. According to one, they affect the structure and/or modification of pericentric chromatin in a manner that facilitates loading of cohesin locally throughout the entire domain [29]. An alternative is that they catalyze the loading of cohesin strictly in the vicinity of core centromeres from whence it slides to neighboring sequences, presumably by virtue of the topological engagement that is the product of the loading reaction. According to the local loading model, early intermediates in the loading reaction should be found throughout pericentric sequences normally associated with high densities of cohesin, whereas according to the sliding model, such intermediates will be found concentrated at core centromeres. Because loading clearly occurs along chromosome arms in the absence of centromeres, the sliding model envisions that loading intermediates will be also associated with sequences along chromosome arms, albeit at much lower levels than at core centromeres.
It is not yet possible to turn on and off the loading reaction in vivo in a manner rapid enough to detect wild-type complexes engaging with and then moving away from loading sites, but we reasoned that it might be possible to trap an early intermediate in the loading reaction by introducing mutations that abort cohesin’s ATP hydrolysis cycle. We found that cohesin complexes that engage Smc1 and Smc3 NBDs in the presence of ATP, but are defective in hydrolyzing nucleotide bound either to Smc1 (Smc1E1158Q) or Smc3 (Smc3E1155Q), do indeed associate with chromosomes. Crucially, they do so in a pattern that is distinct from that of wild-type complexes but very similar to that of cohesin’s Scc2/4 loading complex. The most striking aspect is a massive accumulation at all 16 core centromeres. The instability of the association suggests that it precedes topological entrapment of chromatin fibers. Importantly, association of Smc1E1158Q- and Smc3E1155Q-containing cohesin rings with centromeres is abolished by signature motif mutations that prevent NBD engagement and depends on the activity of Scc2/4 complexes, on formation of tripartite cohesin rings, on binding of Scc3 to cohesin’s Scc1 subunit, and on stable binding of Smc1 to Smc3 via their hinge domains, all properties essential for the recruitment of wild-type cohesin rings to pericentric regions.
Without observing wild-type complexes in the act of loading, we cannot yet be certain that the state entered into by Smc1E1158Q- and Smc3E1155Q-containing cohesin complexes resembles an intermediate during a normal loading reaction. It is nevertheless unlikely that accumulation of Smc1E1158Q or Smc3E1155Q with core centromeres arises merely from the binding to Scc2/4 at this location of an otherwise dead cohesin complex. Smc1E1158Q and Smc3E1155Q associated with kinetochores are twice as abundant as Scc2/4 (data not shown), persist for longer, and are not recruited to tandem Tet operators bound by Scc2/4 complexes. This last observation demonstrates that Scc2/4 is insufficient to recruit Smc1E1158Q and Smc3E1155Q. Their association with core centromeres requires, in addition, specific kinetochore proteins (data not shown). Furthermore, the dependence on a wide variety of distant domains and interactions within the cohesin complex implies that the reaction is far more complex than simple binding.
We therefore suggest that despite their ultimate dysfunction, Smc1E1158Q and Smc3E1155Q complexes undergo an early step in the loading process. More specificially, we propose that the mutations block the transition from a state in which cohesin has associated with loading sites in a Scc2/4-dependent manner to one in which the ring entraps the chromatin fiber and, as a consequence, is capable of sliding into neighboring sequences. The rapid turnover of Smc1E1158Q and Smc3E1155Q complexes at centromeres suggests that association is an unstable one and that the mutant complexes undergo a series of futile cycles, a property that would cause them to interfere with the loading of wild-type complexes and explain why overexpression of either Smc1E1158Q or Smc3E1155Q is lethal [38].
If this interpretation is correct, then our finding that Smc1E1158Q and Smc3E1155Q accumulate at core centromeres represents the first direct evidence that the latter recruit cohesin to pericentric sequences by acting not as long-range enhancers of loading locally within pericentric sequences [29] but as loading and/or entrapment sites from which cohesin slides into neighboring sequences. Because core centromeres affect the density of cohesin up to 50 kb away, our data suggest that cohesin may be capable of sliding considerable distances along chromatin fibers, which would explain why wild-type cohesin and its Scc2/4 loading complex rarely colocalize in yeast. Our data are consistent with the suggestion [24] that Scc2/4 complexes accumulate (albeit not stably as we show here) at loading sites, whereas cohesin slides to neighboring sequences, which are frequently though not exclusively regions of convergent transcription.
Our finding that both Scc2/4 and transition state cohesin complexes colocalize with Pol I, II, and III transcription units along chromosome arms raises the possibility that transcribing polymerases may catalyze, albeit at a much slower rate than core centromeres, the loading process along chromosome arms. A connection between transcription and Smc-kleisin complex loading may widespread as Bacillus subtilis Smc-kleisin complexes colocalize with highly active transcription units as well as ParS sites (the bacterial equivalent to yeast centromeres) [39]. Interestingly, both Nipped B (Scc2) and cohesin are frequently associated with actively transcribed regions in Drosophila melanogaster tissue culture cells [40]. It is also intriguing that Scc2 (Nipbl), more so even than cohesin itself, colocalizes with the mediator complex in ES cells [41]. It is conceivable therefore that the loading of cohesin along chromosome arms might be promoted by the transcription apparatus in a wide variety of eukaryotic cells. Loading during transcription is not the sole mechanism by which cohesin associates with chromosome arms because there is little or no transcription during cleavage divisions in amphibians. This might explain why Scc2/4 and cohesin loading have been linked to the Drf1 subunit of the Cdc7 kinase [42] and formation of prereplication complexes [43, 44] in Xenopus egg extracts.
The Mechanism of Chromatin Fiber Entrapment
If we are correct in thinking that the state entered into by Smc1E1158Q- and Smc3E1155Q-containing cohesin complexes resembles an intermediate during a normal loading reaction and that the end product is chromatin fiber entrapment, then the behavior of the mutant complexes ought to reveal the normal entrapment mechanism. We presume that entrapment can be broken down into three steps, namely, ring opening, strand passage (be it DNA or protein), and ring closure. We suspect that ring opening is linked either to NBD engagement or disengagement. Closure on the other hand could in principle be a spontaneous process because Smc1 and Smc3 hinges bind spontaneously with a low nanomolar association constant [35]. More complicated models, according to which hydrolysis of ATP bound to different heads catalyzes very different steps, is hard to reconcile with the rather similar phenotypes of Smc1E1158Q and Smc3E1155Q. Our finding that association of hydrolysis-defective complexes with centromeres is an unstable one indicates that rings have not yet closed around chromatin. This raises the possibility that in the presence of Scc2/4 and kinetochore proteins, NBD engagement, which is considered the power stroke of ABC-like transporters, triggers opening, whereas disengagement, which is blocked by Smc1E1158Q and Smc3E1155Q, triggers closure and thereby chromatin fiber trapping. However, if opening occurs at Smc1/3 hinges, [19] and this is induced by NBD engagement, why does Smc1F584R, which reduces the stability of Smc1/3 hinges, greatly reduce association of hydrolysis-defective complexes with kinetochores? This conundrum cannot be avoided by supposing that rings open instead through dissociation of Scc1 from Smc3 NBDs because mutations favoring dissociation at this interface also abolish association of hydrolysis-defective complexes with kinetochores. This lead us to suggest a rather different model (Figure S7B) in which Scc2/4, Scc3, and kinetochore proteins associated with a chromatin substrate facilitate interaction of closed Smc1/3 hinges with engaged NBDs. Hinges are subsequently opened by disengagement of NBDs driven by ATP hydrolysis. According to this scenario, hinge reassociation leading to chromatin entrapment is driven by the free energy of hinge-dimerization not by ATP binding or hydrolysis. Irrespective of which interpretation is correct, our observations stress that Smc1/3 hinges have a crucial role at an early stage in the process by which chromatin fibers are entrapped by cohesin rings. The less rapid turnover of hydrolysis-defective cohesin complexes than their Scc2/4 loading factors at kinetochores suggests that the transition state adopted by the former does not necessarily include a Scc2/4 complex stably bound to it.
In conclusion, we provide evidence that an early step in the process by which cohesin is loaded onto chromosomes involves a nontopological association of cohesin complexes whose Smc1 and Smc3 have engaged in the presence of ATP. The identification of this transition state has enabled us to distinguish the roles of Scc2/4 and non-Smc cohesin subunits. Although Scc2/4 and Scc3 are essential for forming transition complexes at loading sites, the Pds5/Wapl complex is not and must have a function later in the process by which sister chromatid cohesion is established.
Experimental Procedures
Yeast strains were generated by standard protocol and the detailed genotypes were described in the Supplemental Information. Live-cell imaging and FRAP assay were performed as described [35] with diploid yeast cells homozygous for GFP-tagged protein. ChIP-qPCR was performed as described [35]. Briefly, qPCR was run with Corbett Rotorgene cycler. Primers pairs used for chromosome VI were listed in Table S3. For depletion of degron-fused protein, see the Supplemental Information. ChIP-SEQ assay was performed as described in Supplemental Information. Head domains of Smc1 and Smc3 were expressed and purified as described in [4] with some modifications (see Supplemental Information). Analytical gel filtration for binding analysis were performed as described in the Supplemental Information.
Supplementary Material
Acknowledgments
We are grateful to Nasmyth lab members for useful discussions, R. Parton for help with microscopy, K. Nakagawa for ChIP-SEQ, B. Panaretou and K. Bloom for providing useful yeast strains and plasmids, and S. Gruber for critical reading of the manuscript. T.I. and Y.K. were supported by Grant-in-Aid for Young Scientists (A) from MEXT. K.S. was supported by a grant of the Cell Innovation Program and Grant-in-Aid for Scientific Research (S) from the MEXT, Japan. K.N. was supported by Cancer Research UK and the Wellcome Trust.
Footnotes
Accession Numbers
All data have been deposited in the National Center for Biotechnology Information Sequence Read Archive database (http://trace.ncbi.nlm.nih.gov/Traces/sra/) with the accession number SRP004703.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and three tables and can be found with this article online at doi:10.1016/j.cub.2010.12.004.
References
- 1.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 3.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 4.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 5.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol. Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh SK, Huang C-C, Hajra S, Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 2010;38:570–584. doi: 10.1093/nar/gkp993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mc Intyre J, Muller EG, Weitzer S, Snydsman BE, Davis TN, Uhlmann F. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: Opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009;23:2224–2236. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 14.Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 15.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 18.Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr. Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 19.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 21.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 22.Beckouët F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt CK, Brookes N, Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009;10:R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arumugam P, Nishino T, Haering CH, Gruber S, Nasmyth K. Cohesin’s ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr. Biol. 2006;16:1998–2008. doi: 10.1016/j.cub.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 29.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 31.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh SK, Hajra S, Paek A, Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 33.Fernius J, Marston AL. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009;5:e1000629. doi: 10.1371/journal.pgen.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra A, Hu B, Kurze A, Beckouët F, Farcas AM, Dixon SE, Katou Y, Khalid S, Shirahige K, Nasmyth K. Both interaction surfaces within cohesin’s hinge domain are essential for its stable chromosomal association. Curr. Biol. 2010;20:279–289. doi: 10.1016/j.cub.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirano M, Hirano T. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 2004;23:2664–2673. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 40.Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 44.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.