Abstract
The exposome concept promotes use of omic tools for discovering biomarkers of exposure and biomarkers of disease in studies of diseased and healthy populations. A two-stage scheme is presented for profiling omic features in serum to discover molecular biomarkers and then for applying these biomarkers in follow-up studies. The initial component, referred to as an exposome-wide-association study (EWAS), employs metabolomics and proteomics to interrogate the serum exposome and, ultimately, to identify, validate and differentiate biomarkers of exposure and biomarkers of disease. Follow-up studies employ knowledge-driven designs to explore disease causality, prevention, diagnosis, prognosis and treatment.
Keywords: Biomarker, exposome, metabolomics, proteomics, EWAS, discovery
Introduction
Although the term ‘biomarker’ refers to any measurable state in a living organism, a useful biomarker can differentiate between biological states, particularly those represented by diseased and healthy populations. Useful biomarkers are diverse, ranging from simple physiological parameters, such as blood pressure, to complex patterns in genome scans. Discovery of new biomarkers is important for epidemiology, which seeks causes of diseases (NRC 1987) (hereafter ‘biomarkers of exposure’), as well as for diagnosis and treatment of diseases (BDWG 2001) (hereafter ‘biomarkers of disease’). Both avenues of inquiry have benefited from recognition that diseases represent aberrations in molecular processes involving the genome and subsequent omic phenomena (Gerszten & Wang 2008; Griffin et al. 2011), features of which can potentially serve as biomarkers (Holmes et al. 2008b). This paradigm has motivated movement away from knowledge-based (reductionist) approaches for discovering biomarkers and towards data-driven (omic) strategies (Zhang et al. 2011). Since phenotypic variation is caused by a combination of genetic (G) factors and environmental (E) exposures (representing all non-genetic influences) (Gibson 2008; Smith 2011), biomarkers are needed to classify both dimensions of the GxE space. Certainly genome-wide association studies (GWAS) offer superb descriptions of the G matrix (Goldstein 2009; Hindorff et al. 2009). However, elaboration of the E matrix has been impeded by conceptual and methodological hurdles (Vineis 2004; Thomas 2010; Rappaport 2011; Smith 2011). The imbalance in coverage of G- and E-related risks spawned the concept of the ‘exposome’, representing all environmental exposures received by an individual during life (Wild 2005). By functionally defining the exposome as the set of all biologically-active chemicals in a person's blood – from both exogenous and endogenous processes – it is possible to harness omic tools for filling in much of the E matrix (Rappaport & Smith 2010). Thus, biomarker discovery should be an essential element in characterizing health-impairing exposures. Subsequent knowledge-based investigations can link biomarkers with exposure sources, establish causality, and investigate the systems biology of important toxicants and disease processes, thereby offering avenues for prevention, diagnosis and treatment of diseases (Nicholson 2006).
Whereas the genome gives rise to a programmed set of molecules (Zhu et al. 2012) in the blood, the exposome is functionally represented by the complementary set of chemicals derived from sources outside of genetic control, including the diet (Holmes et al. 2008a), pathogens (Tsai & Chung 2010), the microbiome (Nicholson et al. 2005), smoking (Smith et al. 2003), psychosocial stress (Epel et al. 2006), drugs (Hiemke 2008) and pollution (Vineis & Perera 2007). By conducting exposome-wide association studies, or EWAS, it should be possible to discover components of the exposome that, over time, cause complex chronic diseases. Since only about one third of these disease risks have been attributed to known risk factors (primarily smoking, diet and exercise) (Lopez et al. 2006), EWAS are essential for establishing a firm molecular basis for environmental causes of disease. Carefully conducted EWAS can also differentiate biomarkers of exposure from molecular signatures of disease pathology, referred to here as biomarkers of disease. This commentary will develop a structured approach for joining EWAS, designed for biomarker discovery, with subsequent inquiries focusing upon disease causality, diagnosis and treatment.
Disease pathways
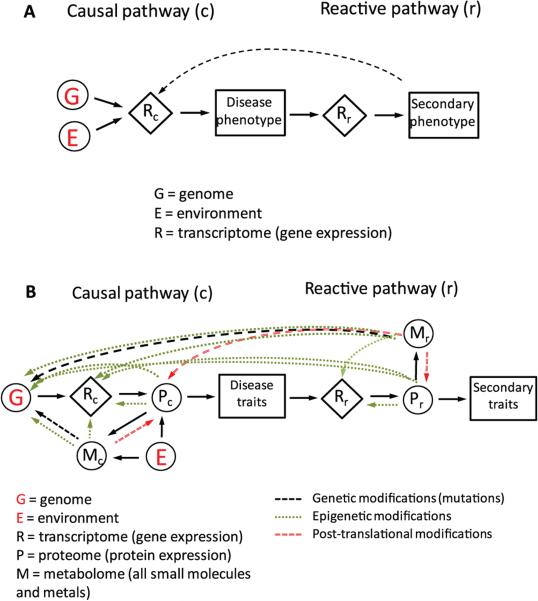
Although omic processes are conceptualized as a linear series, i.e. genome (DNA) → transcriptome (RNA) → proteome (large molecules) → metabolome (small molecules) (Wenk 2005; Gerszten & Wang 2008; Zhu et al. 2012), there is considerable cross talk among them, often involving epigenetic events. For example, some proteins can activate or repress transcription processes (MacQuarrie et al. 2011) and small reactive molecules can modify DNA, RNA and functional proteins to produce mutations and epigenetic changes (Sturla 2007; Liebler 2008). Thus it is reasonable to ask which ‘omes’ offers the most promise for discovering useful biomarkers? Before attempting to answer this question, it is helpful to consider the molecular nature of the exposome and its position in the GxE space that gives rise to disease phenotypes. Here, I will extend the informative construct of Schadt et al. (2005) that modeled complex disease traits by interactions between the genome (G) and transcriptome (R). As shown in Figure 1A, Schadt et al. differentiated gene pathways that initiate disease traits (‘causal pathway’) from those representing secondary traits resulting from disease progression (‘reactive pathway’). Although the influence of the environment (E) is depicted in Schadt's model by an ethereal link to the causal transcriptome, the nature and molecular basis for environmental effects were not elaborated. To include functional measures of cellular status and regulatory processes, Figure 1B extends Schadt's model to incorporate the proteome (P) and metabolome (M). This extended model, posits that the primary biologic mechanism for implementing molecular signals of E is via M and P. In this depiction, M represents all small molecules and metals in the systemic circulation, regardless of their origins. Since it includes many reactive species, M can affect G (via mutation and epigenetic modifications), R (via epigenetic modifications) and P (via post-translational modifications). Of course, R is a major contributor to P, which can also affect G and R through epigenetic mechanisms. Thus, M, P, and R are all linked with each other, and with E generally, leading to correlated features in subject's omic profiles and the associated networks at a given time. Since connections among G, R, P and M exist in both the causal and reactive pathways in Figure 1B, the full transcriptome is represented by R = Rc + Rr, the proteome by P = Pc + Pr and the metabolome by M = Mc + Mr. As will be shown, this mingling of causal and reactive omes complicates differentiation between biomarkers of exposure and biomarkers of disease.
Figure 1.
Pathways showing influences of genetic (G) and environmental (E) factors on chronic diseases. A) Model of causal and reactive pathways due to effects of G and E on gene expression (R), as reported by Shadt et al. (Schadt et al. 2005). B) Model of causal and reactive pathways from (A) extended to include the proteome (P) and metabolome (M).
EWAS and applications of biomarkers
Figure 1B clarifies the role that EWAS can play in differentiating between biomarkers of environmental exposures (causal pathway) and biomarkers of disease (reactive pathway). Since biologically-active molecules are transported to and from cells and tissues by serum, the serum represents a reservoir of chemicals derived from both xenobiotic processes and all cellular genomes. In addition, serum is archived in many epidemiologic studies, including large prospective cohorts that offer particular advantages for comparisons of incident disease cases and controls (Collins 2004; Ollier et al. 2005; Pischon et al. 2008). Thus, it makes sense to focus on serum for characterizing exposomes and for linking exposures with disease. Indeed, one can envision a serum exposome, elaborated by measurements of M and P in serum or plasma. Prominent components of the serum exposome include lipids, sugars, nucleotides, amino acids and metabolites, reactive electrophiles (notably, reactive oxygen, nitrogen, chlorine and carbonyl species), drugs, metals, micronutrients, receptor-binding agents (e.g. hormones and xenoestrogens) and inflammatory markers (cytokines, chemokines, eicosanoids, vasoactive amines and growth factors) (Rappaport & Smith 2010; Li et al. 2011). Although the transcriptome (R) reflects effects of environmental factors (Gibson 2008), and with sufficient cataloguing of reference chemicals might offer clues regarding important exposures (Lamb 2007), it would generally be of secondary interest in identifying molecular biomarkers.
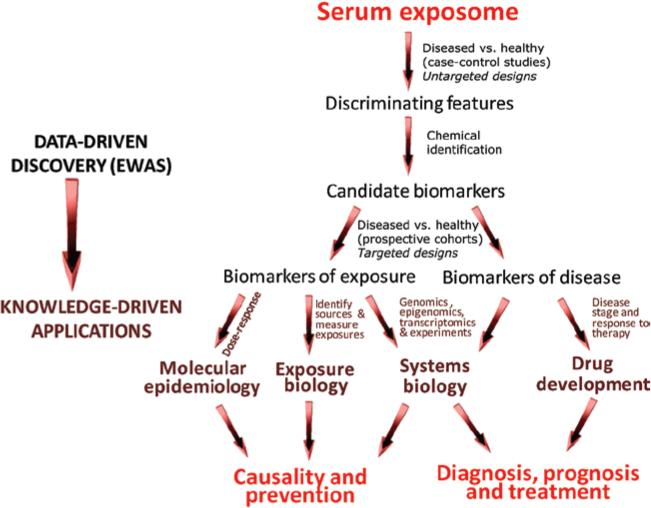
Figure 2 offers a protocol for conducting EWAS and follow-up studies, which borrows some characteristics from previous work (Hanash et al. 2008; Koulman et al. 2009; Kitteringham et al. 2009; Patel et al. 2010; Zhang et al. 2011). The protocol includes an initial data-driven phase to discover biomarkers, followed by a knowledge-driven phase to address hypotheses regarding disease causality and progression. Referring first to biomarker discovery, untargeted omic profiles (or semi-targeted profiles based upon large numbers of known analytes) would be compared between healthy and diseased subjects to seek features that consistently discriminate according to disease status. Because the serum exposome is staggeringly complex, a given EWAS would focus upon a class of omic features, such as, small polar molecules (Wang et al. 2011b), N-linked glycoproteins (Bones et al. 2011) or reactive electrophiles stabilized as adducts of human serum albumin (Li et al. 2011). Also, several EWAS could investigate different classes of omic features with the same serum samples.
Figure 2.
Scheme for conducting exposome-wide-association studies (EWAS) to discover serum biomarkers of exposure and disease and for applying biomarkers to investigate disease causality, prevention, diagnosis, prognosis and treatment.
During the last decade, liquid-chromatography tandem mass spectrometry (LC-MS/MS) and nuclear magnetic resonance (NMR) spectroscopy have been increasingly used – along with multivariate statistics – to perform global analyses of small molecules and proteins in human serum (Hanash et al. 2008; Nordstrom & Lewensohn 2010; Roux et al. 2011; Calligaris et al. 2011). Although the scheme shown in Figure 2 anticipates that cross-sectional (case-control) studies would provide serum for initial comparisons, samples from prospective cohorts would be preferred if they are available. In any case, given the high-dimensional data and associated false discovery rates, the strength of associations is enhanced when the same discriminating features are observed in independent studies.
The second step in the discovery phase involves unambiguous chemical identification of the discriminating features (Koulman et al. 2009). When mass spectral and chromatographic properties of these features match those in available databases, the identities can sometimes be determined relatively quickly by comparing accurate masses and retention times with reference standards. In other cases, identification requires laborious experiments involving high-resolution mass spectrometry, NMR, and synthesis of reference compounds. In any event, the identified molecules serve as candidate biomarkers for subsequent studies.
Since biomarkers can reflect either causal exposures or manifestations of disease progression (see Figure 1B), the discovery process concludes with targeted measurements of candidate biomarkers in archived serum from prospective-cohort studies, where specimens had been collected prior to diagnosis. Targeted analyses are amenable to high-throughput methods, appropriate for assays involving small quantities of serum from many subjects (Dieterle & Marrer 2008; Griffiths et al. 2010; Hanash et al. 2011). As shown in Figure 2, candidate biomarkers that remain associated with disease status would be regarded as biomarkers of exposure while those that don’t would be better suited for evaluating stages of disease progression and therapy (biomarkers of disease).
Biomarkers of exposure and disease that emerge from the discovery phase are worthy of additional inquiries, which harness knowledge-driven designs to investigate causality, prevention, diagnosis, prognosis and treatment of diseases (shown in the lower portion of Figure 2). Evidence that biomarkers of exposure are on the causal pathway (Hill 1965) would include a dose-response relationship (molecular epidemiology), correlation with identifiable sources of exposure and biotransformation (exposure biology) and links with additional omic and experimental models that provide mechanistic insights (systems biology). Likewise, the diagnostic and prognostic value of biomarkers of disease would be enhanced by knowledge of the systems biology, and drug development would benefit when biomarkers are measured in individuals at various stages of disease and under alternative therapeutic regimens (Baker 2005; LaBaer 2005; Dieterle & Marrer 2008).
Proof of concept studies
With the widespread availability of LC-MS/MS and NMR platforms, metabolomics and proteomics have been increasingly used to discover new biomarkers (Griffiths et al. 2010; Gerszten et al. 2011). Because metabolomic methods currently enjoy a more mature state of standardization than proteomic methods, the avenues of research illustrated in Figure 2 will be illustrated with some recent studies of small molecules in serum or plasma from diseased and healthy subjects. To illustrate the breadth of diseases that have been investigated, Table 1 reproduces a portion of a recent review by Nordstrom and Lewensohn (2010), which catalogued scores of metabolomic studies involving biofluids and tissue specimens. The 11 publications summarized in Table 1 compare serum metabolomic profiles between cases and controls in investigations of neurologic, immunological and cardiovascular diseases and cancer. Despite the modest numbers of subjects (31–220 total case and control subjects), multiple discriminating features were observed in each of the studies. Because of high false-discovery rates, this is to be expected and underscores the need for independent validation of discriminating features. Also, chemical identities were assigned to several discriminating features in 10 of the 11 studies and thus would serve as candidate biomarkers.
Table 1.
Summary of results from metabolomic investigations of serum/plasma from case-control studies, showing numbers of subjects, discriminating features and identified features, as reported by (Nordstrom & Lewensohn 2010).
| Disease | Disease class | No. of subjects | Discrim. features | Ident. features | Reference |
|---|---|---|---|---|---|
| Huntington's disease | Neurologic | 50 | 15 | 15 | (Underwood et al. 2006) |
| Parkinson's disease | Neurologic | 88 | 17 | 3 | (Bogdanov et al. 2008) |
| Motor neuron disease | Neurologic | 58 | 76 | 0 | (Rozen et al. 2005) |
| Celiac disease | Immunologic | 68 | 16 | 16 | (Bertini et al. 2009) |
| Ischemia | Cardiovascular | 31 | 5 | 5 | (Barba et al. 2008) |
| Myocardial injury | Cardiovascular | 72 | 13 | 13 | (Lewis et al. 2008) |
| Myocardial ischemia | Cardiovascular | 36 | 23 | 6 | (Sabatine et al. 2005) |
| Myocardial ischemia | Cardiovascular | 39 | 4 | 4 | (Lin et al. 2009) |
| Renal cell carcinoma | Cancer | 129 | 14 | 14 | (Gao et al. 2008) |
| Pancreatic cancer | Cancer | 190 | 3 | 3 | (Beger et al. 2006) |
| Prostate cancer | Cancer | 220 | 10 | 10 | (Osl et al. 2008) |
The metabolomic studies shown in Table 1 represent the first phase of EWAS, where discriminating features were sought in one or more cross-sectional studies of a human disease. To illustrate metabolomic applications that have proceeded further in the context of Figure 2, three additional investigations will be mentioned that validated candidate biomarkers in independent cross-sectional and/or prospective studies. The first, by Hazen and coworkers, began with untargeted metabolomics of polar molecules in plasma from two cross-sectional studies of cardiovascular disease (CVD) (Wang et al. 2011b). Of more than 2000 features, 18 were associated with disease status in both sets of samples. Three of these were identified as choline and its two metabolites, betaine and trimethylamine N-oxide (TMAO). Targeted analyses of these candidate biomarkers in plasma from an independent sample of CVD patients showed atherosclerosis risks increasing with serum concentrations, particularly for TMAO, which displayed about a two-fold risk for the highest quartile of plasma levels compared to the lowest quartile. Experiments with mice showed that in vivo production of TMAO involved an intermediate step requiring gut microbes to metabolize choline to trimethylamine. This study indicates that TMAO is a potentially causal biomarker of dietary exposure to choline – which is derived from eggs, milk, red meat, poultry, seafood and lecithin (a prominent food additive) – and its interactions with the gut microbiome. Since both the diet and gut microbes vary significantly across human populations (Holmes et al. 2008a), TMAO could be an important biomarker for knowledge-driven studies of CVD.
The second study, by Gerszten and coworkers, investigated metabolite profiles as predictors of incident diabetes in serum from two large prospective cohort studies (Wang et al. 2011a). Based upon results from earlier cross-sectional studies, the investigators targeted 61 small polar molecules, notably amino acids and urea-cycle and nucleotide metabolites. Elevated levels of four branched-chain and aromatic amino acids (leucine, valine, phenylalanine and tyrosine) were significantly associated with incident diabetes in both cohorts up to 12 y before elevation of fasting glucose levels. Also, a three-amino acid panel (isoleucine, phenylalanine and tyrosine) predicted fourfold to sevenfold diabetes risks for the highest quartiles of measurements compared to the lowest quartiles in the two populations. This study strongly suggests that elevation of some amino acids in serum represents an early predictor of insulin resistance. Because these biomarkers of disease preceded clinical symptoms of diabetes by several years, they could be useful for investigating disease mechanisms, diagnosis and therapy.
The final study highlights work by Ritchie and coworkers to discover biomarkers of colorectal cancer (CRC) (Ritchie et al. 2010a,b; Ritchie et al. 2011). Using untargeted measurements of several thousand small non-polar molecules in serum from three independent case-control studies, the investigators reported a set of 13 common features that were strongly associated with CRC. Although not unambiguously identified, these 13 chemicals displayed mass spectral characteristics suggestive of a single class of gastrointestinal tract acids (GTAs) containing between 28 and 36 carbon atoms (Ritchie et al. 2010a). Because targeted follow-up analyses found lower levels of GTAs in CRC cases than in controls, the authors concluded that GTAs were protective of CRC, probably because they possess anti-inflammatory properties similar to those of the structurally similar resolvins and protectins that are products of omega-3 fatty acid metabolism (Ritchie et al. 2010a; Ritchie et al. 2011). Results from additional cross-sectional studies of CRC patients showed that levels of GTAs were independent of tumor stage and did not return to normal values after surgical or drug interventions, while parallel measurements in controls showed significant reductions in GTA levels with age (Ritchie et al. 2010b). These results indicate that GTAs are potentially causal biomarkers of (protective) exposure that should be used in future investigations of causality and prevention of CRC and inflammatory bowel diseases.
Discussion
Biomarker discovery has grown dramatically over the last decade, mostly in the quest for biomarkers of disease that can guide development of drugs and diagnostic products. Although this research has been disappointing, in the sense that it has ushered relatively few commercial products through the validation pipeline (Baker 2005), it has vigorously embraced the omics revolution and thereby offers hope that whole new classes of biomarkers of disease will be found. Parallel developments of biomarkers of exposure have been more modest, not only because these biomarkers lack clear commercial interests, but also because knowledge-driven designs are still favored over omic tools for characterizing exposures. This is unfortunate because our current superficial understanding of environmental (i.e. non-genetic) causes of chronic diseases (Lopez et al. 2006) is unlikely to improve by investigating known or suspected risk factors with greater vigor. Given recent promotion of the exposome concept (Wild 2012) and its functional analog as the totality of bioactive chemicals in the blood (Rappaport & Smith 2010), it is now possible for epidemiologists to join the omics revolution, and thus for these two lines of biomarker discovery to converge. Models of disease pathways (Figure 1) provide insight into omic connections and also permit differentiation between bio-markers of exposure (causal pathway) and biomarkers of disease (reactive pathway).
This commentary offers a unified, structured scheme for using omics to discover biomarkers of both exposure and disease and then for exploiting these bio-markers in follow-up studies of disease causality, prevention, diagnosis, prognosis and treatment (Figure 2). The data-driven component of the scheme, referred to as an EWAS, employs metabolomics and proteomics to interrogate components of the serum exposome in diseased and healthy subjects and, ultimately, to identify and validate biomarkers of exposure and biomarkers of disease. Metabolomics and proteomics are highlighted because they represent identifiable molecular entities that reflect the interplay between genetic and environmental factors. Proof-of-concept EWAS have already identified promising new biomarkers of major chronic diseases, including cardiovascular disease, diabetes and cancer. Given the relentless advances in analytical platforms that are amenable to either untargeted profiling or high-throughput, targeted analyses of small molecules and proteins, it is reasonable to anticipate a host of new biomarkers in the next decade. It should be exciting.
Acknowledgments
Declaration of interest
This work has been supported by grant U54ES016115 from the U.S. National Institute for Environmental Health Sciences (NIEHS) through the trans-National Institutes of Health (NIH) Genes, Environment, and Health Initiative.
References
- Baker M. In biomarkers we trust? Nat Biotechnol. 2005;23:297–304. doi: 10.1038/nbt0305-297. [DOI] [PubMed] [Google Scholar]
- Barba I, de León G, Martín E, Cuevas A, Aguade S, Candell-Riera J, Barrabés JA, Garcia-Dorado D. Nuclear magnetic resonance-based metabolomics predicts exercise-induced ischemia in patients with suspected coronary artery disease. Magn Reson Med. 2008;60:27–32. doi: 10.1002/mrm.21632. [DOI] [PubMed] [Google Scholar]
- BDWG Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- Beger RD, Schnackenberg LK, Holland RD, LI D, Dragan Y. Metabonomic models of human pancreatic cancer using 1D proton NMR spectra of lipids in plasma. Metabolomics. 2006;2:125–134. [Google Scholar]
- Bertini I, Calabrò A, De Carli V, Luchinat C, Nepi S, Porfirio B, Renzi D, Saccenti E, Tenori L. The metabonomic signature of celiac disease. J Proteome Res. 2009;8:170–177. doi: 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, Flint Beal M. Metabolomic profiling to develop blood biomarkers for Parkinson's disease. Brain. 2008;131:389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- Bones J, Byrne JC, O'Donoghue N, McManus C, Scaife C, Boissin H, Nastase A, Rudd PM. Glycomic and glycoproteomic analysis of serum from patients with stomach cancer reveals potential markers arising from host defense response mechanisms. J Proteome Res. 2011;10:1246–1265. doi: 10.1021/pr101036b. [DOI] [PubMed] [Google Scholar]
- Calligaris D, Villard C, Lafitte D. Advances in top-down proteomics for disease biomarker discovery. J Proteomics. 2011;74:920–934. doi: 10.1016/j.jprot.2011.03.030. [DOI] [PubMed] [Google Scholar]
- Collins FS. The case for a US prospective cohort study of genes and environment. Nature. 2004;429:475–477. doi: 10.1038/nature02628. [DOI] [PubMed] [Google Scholar]
- Dieterle F, Marrer E. New technologies around biomarkers and their interplay with drug development. Anal Bioanal Chem. 2008;390:141–154. doi: 10.1007/s00216-007-1688-y. [DOI] [PubMed] [Google Scholar]
- Epel ES, Lin J, Wilhelm FH, Wolkowitz OM, Cawthon R, Adler NE, Dolbier C, Mendes WB, Blackburn EH. Cell aging in relation to stress arousal and cardiovascular disease risk factors. Psychoneuroendocrinology. 2006;31:277–287. doi: 10.1016/j.psyneuen.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Gao H, Dong B, Liu X, Xuan H, Huang Y, Lin D. Metabonomic profiling of renal cell carcinoma: high-resolution proton nuclear magnetic resonance spectroscopy of human serum with multivariate data analysis. Anal Chim Acta. 2008;624:269–277. doi: 10.1016/j.aca.2008.06.051. [DOI] [PubMed] [Google Scholar]
- Gerszten RE, Asnani A, Carr SA. Status and prospects for discovery and verification of new biomarkers of cardiovascular disease by proteomics. Circ Res. 2011;109:463–474. doi: 10.1161/CIRCRESAHA.110.225003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature. 2008;451:949–952. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]
- Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Atherton H, Shockcor J, Atzori L. Metabolomics as a tool for cardiac research. Nat Rev Cardiol. 2011;8:630–643. doi: 10.1038/nrcardio.2011.138. [DOI] [PubMed] [Google Scholar]
- Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP. Targeted metabolomics for biomarker discovery. Angew Chem Int Ed Engl. 2010;49:5426–5445. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers–blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- Hiemke C. Clinical utility of drug measurement and pharmacokinetics: therapeutic drug monitoring in psychiatry. Eur J Clin Pharmacol. 2008;64:159–166. doi: 10.1007/s00228-007-0430-1. [DOI] [PubMed] [Google Scholar]
- Hill AB. The environment and disease: association or causation? Proc R Soc Med. 1965;58:295–300. [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008a;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008b;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Koulman A, Lane GA, Harrison SJ, Volmer DA. From differentiating metabolites to biomarkers. Anal Bioanal Chem. 2009;394:663–670. doi: 10.1007/s00216-009-2690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer J. So, you want to look for biomarkers (introduction to the special biomarkers issue). J Proteome Res. 2005;4:1053–1059. doi: 10.1021/pr0501259. [DOI] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M, Farrell L, Asnani A, Cyrille M, Ramanathan A, Shaham O, Berriz G, Lowry PA, Palacios IF, Tasan M, Roth FP, Min J, Baumgartner C, Keshishian H, Addona T, Mootha VK, Rosenzweig A, Carr SA, Fifer MA, Sabatine MS, Gerszten RE. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–3512. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, Rappaport SM. Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Mol Cell Proteomics. 2011;10:M110 004606. doi: 10.1074/mcp.M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem Res Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhang J, Gao P. Silent myocardial ischemia is associated with altered plasma phospholipids. J Clin Lab Anal. 2009;23:45–50. doi: 10.1002/jcla.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 2011;27:141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol Syst Biol. 2006;2:52. doi: 10.1038/msb4100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- Nordström A, Lewensohn R. Metabolomics: moving to the clinic. J Neuroimmune Pharmacol. 2010;5:4–17. doi: 10.1007/s11481-009-9156-4. [DOI] [PubMed] [Google Scholar]
- NRC. Biological markers in environmental health research. Committee on Biological Markers of the National Research Council. Environ Health Perspect. 1987;74:3–9. doi: 10.1289/ehp.74-1474499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollier W, Sprosen T, Peakman T. UK Biobank: from concept to reality. Pharmacogenomics. 2005;6:639–646. doi: 10.2217/14622416.6.6.639. [DOI] [PubMed] [Google Scholar]
- Osl M, Dreiseitl S, Pfeifer B, Weinberger K, Klocker H, Bartsch G, Schäfer G, Tilg B, Graber A, Baumgartner C. A new rule-based algorithm for identifying metabolic markers in prostate cancer using tandem mass spectrometry. Bioinformatics. 2008;24:2908–2914. doi: 10.1093/bioinformatics/btn506. [DOI] [PubMed] [Google Scholar]
- Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS ONE. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K, van der Schouw YT, Spencer E, Moons KG, Tjønneland A, Halkjaer J, Jensen MK, Stegger J, Clavel-Chapelon F, Boutron-Ruault MC, Chajes V, Linseisen J, Kaaks R, Trichopoulou A, Trichopoulos D, Bamia C, Sieri S, Palli D, Tumino R, Vineis P, Panico S, Peeters PH, May AM, Bueno-de-Mesquita HB, van Duijnhoven FJ, Hallmans G, Weinehall L, Manjer J, Hedblad B, Lund E, Agudo A, Arriola L, Barricarte A, Navarro C, Martinez C, Quirós JR, Key T, Bingham S, Khaw KT, Boffetta P, Jenab M, Ferrari P, Riboli E. General and abdominal adiposity and risk of death in Europe. N Engl J Med. 2008;359:2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2011;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SA, Ahiahonu PW, Jayasinghe D, Heath D, Liu J, Lu Y, Jin W, Kavianpour A, Yamazaki Y, Khan AM, Hossain M, Su-Myat KK, Wood PL, Krenitsky K, Takemasa I, Miyake M, Sekimoto M, Monden M, Matsubara H, Nomura F, Goodenowe DB. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010a;8:13. doi: 10.1186/1741-7015-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SA, Heath D, Yamazaki Y, Grimmalt B, Kavianpour A, Krenitsky K, Elshoni H, Takemasa I, Miyake M, Sekimoto M, Monden M, Tomonaga T, Matsubara H, Sogawa K, Matsushita K, Nomura F, Goodenowe DB. Reduction of novel circulating long-chain fatty acids in colorectal cancer patients is independent of tumor burden and correlates with age. BMC Gastroenterol. 2010b;10:140. doi: 10.1186/1471-230X-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SA, Jayasinghe D, Davies GF, Ahiahonu P, Ma H, Goodenowe DB. Human serum-derived hydroxy long-chain fatty acids exhibit anti-inflammatory and anti-proliferative activity. J Exp Clin Cancer Res. 2011;30:59. doi: 10.1186/1756-9966-30-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Lison D, Junot C, Heilier JF. Applications of liquid chromatography coupled to mass spectrometry-based metabolomics in clinical chemistry and toxicology: A review. Clin Biochem. 2011;44:119–135. doi: 10.1016/j.clinbiochem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Rozen S, Cudkowicz ME, Bogdanov M, Matson WR, Kristal BS, Beecher C, Harrison S, Vouros P, Flarakos J, Vigneau-Callahan K, Matson TD, Newhall KM, Beal MF, Brown RH, Jr, Kaddurah-Daouk R. Metabolomic analysis and signatures in motor neuron disease. Metabolomics. 2005;1:101–108. doi: 10.1007/s11306-005-4810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, Berriz GF, Roth FP, Gerszten RE. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Perfetti TA, Garg R, Hansch C. IARC carcinogens reported in cigarette mainstream smoke and their calculated log P values. Food Chem Toxicol. 2003;41:807–817. doi: 10.1016/s0278-6915(03)00021-8. [DOI] [PubMed] [Google Scholar]
- Smith GD. Epidemiology, epigenetics and the ‘Gloomy Prospect’: embracing randomness in population health research and practice. Int J Epidemiol. 2011;40:537–562. doi: 10.1093/ije/dyr117. [DOI] [PubMed] [Google Scholar]
- Sturla SJ. DNA adduct profiles: chemical approaches to addressing the biological impact of DNA damage from small molecules. Curr Opin Chem Biol. 2007;11:293–299. doi: 10.1016/j.cbpa.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309–2324. doi: 10.1038/onc.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood BR, Broadhurst D, Dunn WB, Ellis DI, Michell AW, Vacher C, Mosedale DE, Kell DB, Barker RA, Grainger DJ, Rubinsztein DC. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain. 2006;129:877–886. doi: 10.1093/brain/awl027. [DOI] [PubMed] [Google Scholar]
- Vineis P. A self-fulfilling prophecy: are we underestimating the role of the environment in gene-environment interaction research? Int J Epidemiol. 2004;33:945–946. doi: 10.1093/ije/dyh277. [DOI] [PubMed] [Google Scholar]
- Vineis P, Perera F. Molecular epidemiology and biomarkers in etiologic cancer research: the new in light of the old. Cancer Epidemiol Biomarkers Prev. 2007;16:1954–1965. doi: 10.1158/1055-9965.EPI-07-0457. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011a;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011b;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- Zhang GF, Sadhukhan S, Tochtrop GP, Brunengraber H. Metabolomics, pathway regulation, and pathway discovery. J Biol Chem. 2011;286:23631–23635. doi: 10.1074/jbc.R110.171405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. Stitching together Multiple Data Dimensions Reveals Interacting Metabolomic and Transcriptomic Networks That Modulate Cell Regulation. PLoS Biol. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]