Abstract
Background:
Endothelin-1 (EDN1) and EDN receptor type A (EDNRA) are implicated in melanocyte functions.
Aim and Objectives:
This study examines the role of EDN1 (G5665T and T-1370G) and EDNRA (C + 70G and G-231A) polymorphisms as a risk factor for vitiligo, and evaluates the relationship between genotypes and clinical characteristics of vitiligo patients.
Materials and Methods:
We analyzed genotype/allele distributions of EDN1 and EDNRA polymorphisms in 100 patients with vitiligo and 185 healthy controls by real-time polymerase chain reaction.
Results:
There was no notable risk for vitiligo afflicted by studied polymorphisms. However, the presence of EDNRA +70 variant G allele was found to be related with decreased risk for development of generalized type of vitiligo (odds ratio [OR]: 0.42, 95% confidence interval [CI] = 0.21–0.86, pcorr = 0.03) and showed protective effect against associated diseases seen in vitiligo (OR: 0.49, 95% CI = 0.27–0.88, pcorr = 0.034). Haplotype analysis demonstrated a strong (disequilibrium coefficient = 0.73, r2 = 0.405) linkage disequilibrium between EDN1 G5665T and T-1370G polymorphisms. The EDN1 5665/-1330 TT haplotype was over represented significantly in controls than in patients (P = 0.04).
Conclusion:
The studied polymorphisms do not seem to be a major risk for vitiligo. Haplotype analysis denoting protective effects against vitiligo may indicate an indirect interaction in the course of vitiligo. In addition, EDNRA + 70 polymorphism is protective against generalized type of vitiligo and associated diseases.
Keywords: Endothelin-1, endothelin receptor A, genetic polymorphism, inflammation, vitiligo
Introduction
What was known?
Research into the pathogenesis of vitiligo has shown a complex of genetic and environmental factors. Endothelin-1 (EDN1) and EDN receptor type A (EDNRA) are implicated in the inflammatory process. The relationships between EDN1 and EDNRA polymorphisms with several diseases have been found.
Vitiligo is an acquired cutaneous disorder with a 0.5–2% incidence worldwide, and is characterized by a loss of melanocytes from epidermis. Although the etiopathogenesis has not been clearly elucidated yet, it was suggested that this common skin disease is a consequence of genetic and environmental factors. In the etiopathogenesis of the disease several mechanisms including autoimmune, autotoxicity, and neural mechanisms induced by biochemical and neurochemical mediators have been proposed.[1] Previously, keratinocyte-derived endothelin-1 (EDN1) has been shown to control melanocyte growth and function.[2,3] It is a potent stimulant on melanocyte proliferation,[4] melanogenesis,[5,6] and migration;[4] implying a link with vitiligo.[5,7,8] EDN1 functions are mediated predominantly by EDN receptor type A (EDNRA). Polymorphisms of EDN1 and EDNRA genes have been investigated in autoimmune diseases such as Hashimoto's thyroiditis (HT),[9] Graves’ disease,[10] scleroderma,[11] primary biliary cirrhosis,[12] and psoriasis[13] as well. There are only two studies in the literature examining the relationship between EDN1 gene polymorphisms and vitiligo with controversial results.[14,15] Kim et al.[14] have found that some haplotypes (GT and AG haplotypes of intron 4 G/A and 5665 G/T polymorphic loci) of EDN1 gene are associated with vitiligo, whereas Lan et al.[15] were unable to show any correlation. To our knowledge, EDNRA polymorphisms were not investigated in vitiligo patients before. Therefore, the aim of this study was to investigate both EDN1 gene polymorphisms (G5665T [or Lys198Asn] and T-1370G) and EDNRA gene polymorphisms (C + 70G and G-231A), in the etiopathogenesis of vitiligo. In addition, genotypes and clinical types of vitiligo found in these patients were also evaluated.
Materials and Methods
The study was approved by the local Ethics Committee. All the patients and controls were recruited from a single center and all provided written informed consent. A total of 100 patients with vitiligo (53 women/47 men) were enrolled in the study. Vitiligo patients older than 18 years, showing any clinical picture except segmentary type and who were not on systemic or topical therapy during previous 2 months were included in the study. Vitiligo was diagnosed on clinical grounds at the Department of Dermatology. The control group consisted of 185, age and sex-matched, dermatology outpatients (95 women/90 men), with no past history of any systemic, infectious, autoimmune, genetic, or atopic disease. A negative family history for vitiligo was also provided. Patients taking any medication including vitamins were excluded. The main diagnoses in the control group were melanocytic nevus and fibroepithelial polyps.
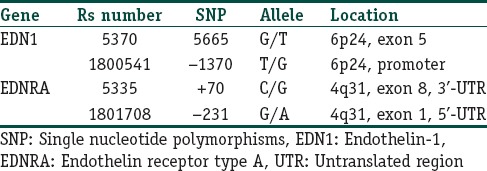
Peripheral venous blood samples were taken in the morning subsequent to an overnight (12 h) fast in EDTA-K3 tubes for genotype analysis. Genomic DNA was isolated from peripheral blood leukocytes by using high pure polymerase chain reaction Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany). We examined the G5665T and T-1370G polymorphisms in EDN1 gene as well as C + 70G and G-231A polymorphisms in EDNRA gene in our DNA samples. Their “rs” numbers and locations are shown in Table 1. These polymorphisms were selected according to the following criteria: Previous association for susceptibility to other diseases,[9,10,11,12,13,14,15] and adequate frequency in Caucasian populations to perform the evaluation. For detection of the mentioned polymorphisms, light SNiP assays were used. Light SNiP assays are based on simple probe melting curve analysis. They consist of premixed primers and probes. They were developed and optimized according to NCBI “rs” numbers of mentioned polymorphisms by Tib MolBiol (Berlin, Germany). The detection of polymorphisms was performed in a LightCycler (Roche Diagnostics, Mannheim, Germany).
Table 1.
SNP in the EDN1 and EDNRA genes
All statistical analyses were performed with SPSS 15.0 for Windows (IBM SPSS Statistics, Chicago, IL, USA). Differences in genotype distributions and allele frequencies in the cases and the controls were compared using the Chi-square test. Bonferroni correction was also applied for the multiple-comparison correction. Bonferroni correction is an adjustment made to P values when several dependent or independent statistical tests are being performed. The statistical significance for deviations from Hardy–Weinberg equilibrium (HWE) was determined using the Pearson Chi-square test. Odds ratios (ORs) were calculated and given with 95% confidence intervals (CIs). The wild-type genotype/allele served as a reference category. Linkage disequilibrium (LD) and haplotype frequencies were estimated using the Haploview 4.2 software (Haploview, MIT/Harvard Broad Institute, MA, USA) and compared between cases and controls using a contingency Chi-square test.[16] In addition, the NCSS 2000 statistical software (NCSS, LCC, Kaysville, Utah, USA) was used to evaluate the power analysis.
Results
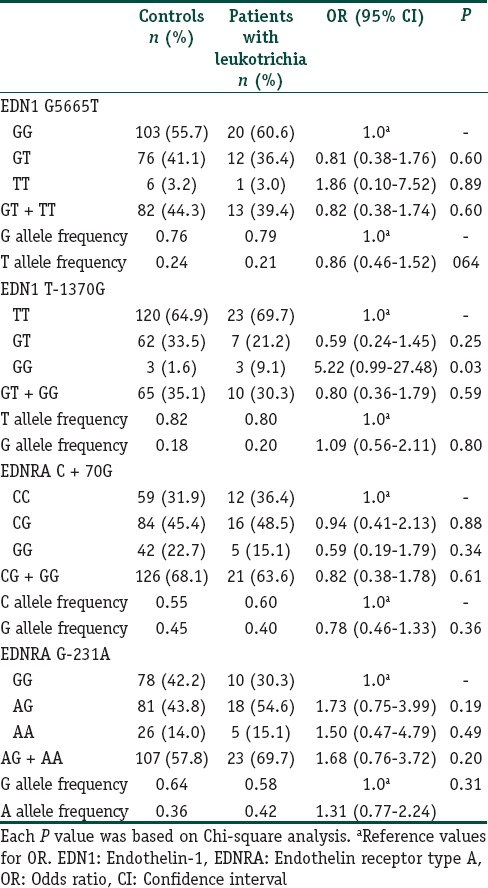
A total of 285 subjects (100 vitiligo and 185 controls) were included in this case control study. Table 2 depicts the clinical characteristics of the vitiligo patients including the clinical type of disease, duration, family history, leukotrichia, stability within 1 year, and associated diseases. The mean ages were 38.6 ± 12.0 years (range 18–76) and 38.2 ± 9.7 years (range 18–72 years) for patients and controls, respectively. There was no significant difference between the study and control groups in terms of mean age and sex distribution. We had 87% power to detect an effect size (W) of 0.20 using a 2 degrees of freedom (α = 0.05). The genotypic and allelic distributions of EDN1 (G5665T and T-1370G) and EDNRA (C + 70G and G-231A) polymorphisms for patients and controls are shown in Table 3. All genotype distributions were in accordance with the HWE between controls and patients. The allelic frequencies of the studied polymorphisms found in our control population were similar to the results in Turkish,[9,10] English,[11,17] and German[18] populations. Statistical analyses showed no direct association between vitiligo and the four individual polymorphisms studied. However, when the relationship between these polymorphisms and clinical characteristics of patients were evaluated, we found that EDNRA + 70 variant G allele and genotypes containing G (CG, GG and CG + GG) showed protective effects from presence of associated disease in patients with vitiligo [Table 4]. When the Bonferroni correction was performed, the significance still existed. In our vitiligo group, we designated a large spectrum of autoimmune diseases as follows: HT, type 1 diabetes mellitus, rheumatoid arthritis, alopecia areata, idiopathic thrombocytopenic purpura, and multiple sclerosis. In addition, G allele seems to protect against generalized type of vitiligo when compared to focal type (OR: 0.42, 95% CI = 0.21–0.86, pcorr = 0.03 [Table 5]). In addition, frequency of GG genotype was marginally lower in generalized type of vitiligo when compared to focal type (OR: 0.18, pcorr = 0.057 [Table 5]). However, there is no significant difference in the distribution of this allele between generalized and acrofacial types [Table 5]. EDN1-1370 GG genotype was 5-fold more in vitiligo patients with leukotrichia in comparison with healthy controls which may be seen in Table 6. This significant evidence did not survive the Bonferroni correction for multiple testing.
Table 2.
Characteristics of the patients with vitiligo
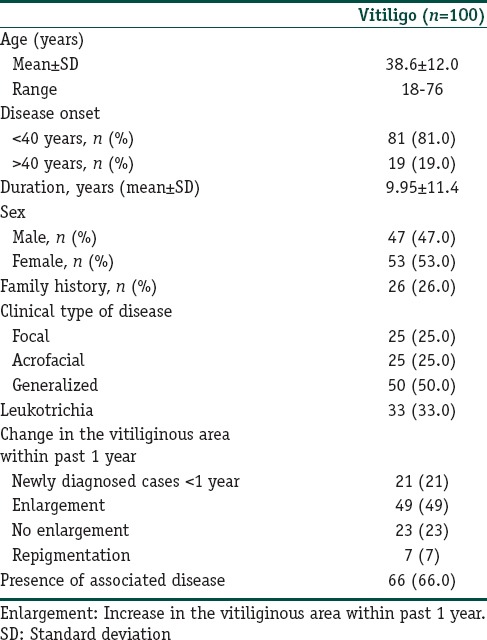
Table 3.
Distribution of genotypes and allele frequencies for patients with vitiligo and control group
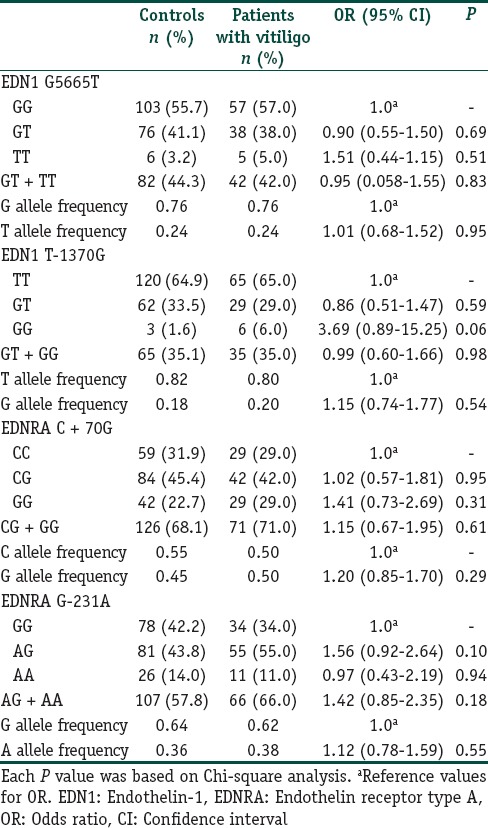
Table 4.
Distribution of EDNRA C + 70G genotypes and allele frequencies for patients with vitiligo: Impact of presence of associated disease
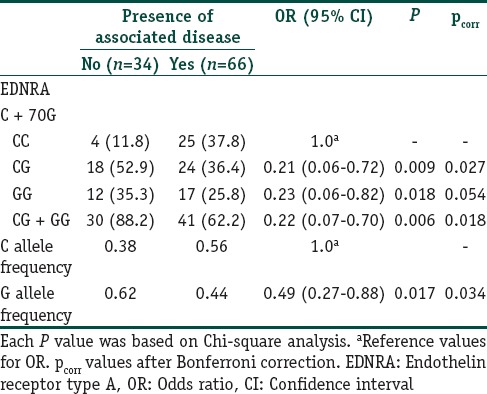
Table 5.
Distribution of EDNRA C + 70G genotypes and allele frequencies for patients with focal, acrofacial and generalized type of vitiligo
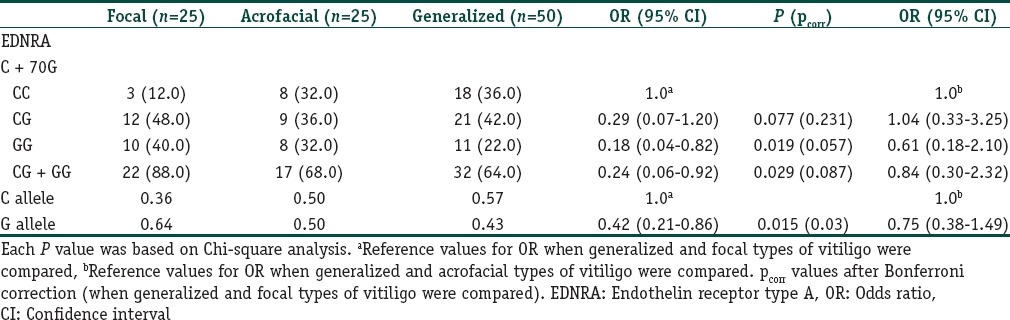
Table 6.
Distribution of genotypes and allele frequencies for vitiligo patients with leukotrichia and control group
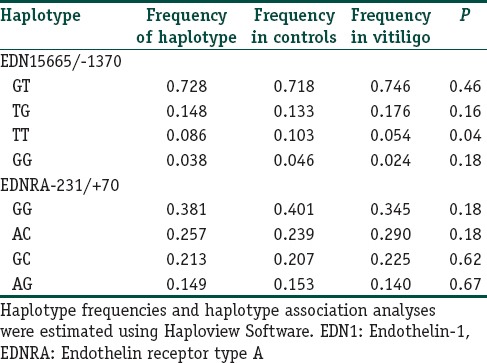
Levontin's standardized disequilibrium coefficient (D’) was calculated as a measure for LD between the studied polymorphisms in the EDN1 and EDNRA genes [Figure 1]. EDN1 5665 and -1370 were found to be in strong LD (D’ = 0.73, r2 = 0.405). In addition, a weak LD between EDNRA − 231 and + 70 was found (D’ = 0.22, r2 = 0.032). Haplotype frequencies are shown in Table 7. The most frequent haplotype among the patients and controls were EDN1 5665/-1370 GT and EDNRA − 231/+70 GG. The EDN1 5665/-1330 TT haplotype was significantly overrepresented in controls than in patients (P = 0.04).
Figure 1.
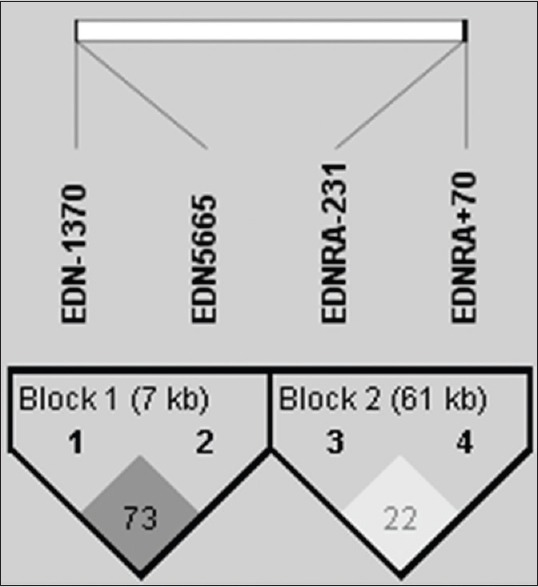
Haplotype analysis and linkage disequilibrium patterns of studied polymorphisms. There was strong linkage disequilibrium between endothelin-1 5665/-1370 (D’ = 0.73, r2 = 0.405); and weak linkage disequilibrium between endothelin receptor type A − 231/+70 (D’ = 0.22, r2 = 0.032)
Table 7.
Haplotype analysis of the EDN1 and EDNRA polymorphisms in patients with vitiligo and control subjects
Discussion
EDN1 is a well-known peptide for its role in epidermal hyperpigmentation by signaling mechanisms of mitogenesis and melanogenesis.[4,5,6] Previously, the keratinocyte-derived EDN1 has been shown to be decreased in vitiligo.[8] Furthermore, EDN1 was found to be increased in patients undergoing PUVA treatment indicating a role of this peptide in vitiligo pathogenesis.[19] Besides, EDN1 is a strong vasoconstrictive agent which may contribute to controlling the inflammatory process seen in vitiligo. Indeed, blocking the EDN1/EDNRA signaling resulted in the loss of pigmentation in vivo.[5] However, the exact role of EDN1 in vitiligo has not been clearly elucidated yet.
EDN1 gene polymorphisms have been investigated and incriminated in the etiopathogenesis of various diseases before. The mutant T allele of a G5665T polymorphism in EDN1 gene was found as a risk factor for some diseases[18,20,21,22] and responsible for disease progression in others.[12] Some studies were shown that the presence of T allele was associated with higher EDN1 levels, raised systolic blood pressure, and predisposed patients to preeclampsia, suggesting the functional importance of this polymorphism in pregnancy.[21,22] Iglarz et al.[23] confirmed the impact of the G5665T polymorphism on vascular reactivity. In the present study, EDN1 5665 T allele was not found to have any impact on vitiligo. The lack of association between G5665T with HT[9] is also noteworthy since HT is a close counterpart of vitiligo.
Two former studies investigating the role of G5665T polymorphism in vitiligo did not show a significant relationship as well.[14,15] Considering the potential regulatory effects of promoter region polymorphisms on gene expression and plasma protein levels, we also studied T-1370G polymorphism which is located in the promoter region of EDN1 gene. Recently, it has been shown that the EDN1 mRNA and the secreted EDN1 levels in primary osteosarcoma hcell culture obtained from TT homozygotes were significantly higher than those from GG homozygotes.[24] Interestingly, the GG genotype frequency of T-1370G polymorphism, related with the low production of EDN1,[17,21] was increased 5-fold in vitiligo patients with leukotrichia in comparison to controls. Leukotrichia does not correlate with disease activity. However, this subset of patients is relatively therapy resistant, and this information may be valuable in determining therapy options. However, in the present study we did not detect a relationship between this polymorphism and vitiligo in general. Lack of association between G5665T and T-1370G polymorphisms and vitiligo indicates that these polymorphisms alone may not play a major role in the pathogenesis of this autoimmune disease.
On the other hand, our haplotype analysis revealed that there was strong LD between G5665T and T-1370G polymorphisms. The EDN1 5665/-1370 TT haplotype was significantly less presented in vitiligo patients in comparison with controls. T alleles in both 5665 and -1370 are known to be related with the high production of EDN1 protein[17,21] which causes melanocyte proliferation and melanogenesis. Actually, many studies have shown that vitiligo skin expresses low levels of EDN1.[5,8] Consequently, our haplotype results are in line with what is expected; namely, high levels of EDN1 means increased amount of melanin protection against vitiligo. Lan et al.[15] and Kim et al.[14] also did haplotype analyses between G5665T and two different intronic polymorphisms in EDN1 gene (intron 2 A/G and intron 4 G/A, respectively). Lan et al.[15] could not find a significant difference between cases and controls in haplotype frequencies of G5665T and intron 2 A/G. However, Kim et al.[14] reported that GT and AG haplotype frequencies at intron 4G/A and G5665T loci of EDN1 gene in focal and segmental clinical types differed significantly from healthy controls. These findings suggest that EDN1 polymorphisms may not be a direct risk factor alone, but their presence, especially with G5665T may contribute as additional/cumulative factors in the development of vitiligo.
EDNRA mediates main functions of EDN1. Regarding polymorphisms of EDNRA gene, the functional consequence of EDNRA G-231A and C + 70G polymorphisms are unknown, since the functional studies are not yet available. The C + 70G and G-231A polymorphisms of EDRA gene are within the 3’-untranslated region (UTR) and the 5’- UTR of the gene, respectively. 3’- UTRs of eukaryotic mRNAs are usually implicated in the posttranslational regulation and have a pivotal role in message stability.[25] The functional consequences of 5’- UTR polymorphisms might be related to the regulatory sequences of gene transcriptions and associated with altered gene expression.[14] In addition, such polymorphisms may create novel splice sites influencing the function of the receptor and receptor-ligand interactions. Therefore, these polymorphisms could have significant effects on mRNA expression and message stability, and thus affecting the total amount and activity of the functional receptor. On the other hand, receptor-ligand interactions may change. Our study showed the variant G allele of EDNRA +70 polymorphism was protective against the generalized type of vitiligo. It was previously demonstrated that that carrying variant allele protects also from early onset disease of HT.[9] We think it may be possible that +70 wild C allele carrying receptors bind their ligands with decreased affinity, resulting in at least weakening of signals at the membrane level, or slowing internalization of signal in the melanocytes with subsequent ceasing of melanocyte proliferation or melanocyte destruction. In another study, examining the differences of EDN1 and EDNRA expressions, Aly et al.[26] did not find any difference between keratinocytes of vitiligo lesions and perilesional normal epidermis. However, in that study[26] the patients were few in numbers (n = 10) and we think a larger number of cases and advanced methods of evaluation could show different results. Altogether, it can be concluded that EDNRA does not contribute to vitiligo process, rather its +70 G polymorphism is protective against both generalized type of vitiligo and associated diseases. Although most vitiligo patients are otherwise healthy, vitiligo may be associated with a large spectrum of autoimmune diseases.
Conclusion
This is one of the few studies examining the EDN1 and EDNRA polymorphisms in vitiligo. Our results suggest that EDN1 (G5665T and T-1370G) and EDNRA (C+70G and G-231A) polymorphisms may not have a causative effect in vitiligo. In contrast, haplotype analyses of these polymorphisms denote protective effects against vitiligo. Furthermore, EDNRA +70 polymorphism which is protective against generalized type of vitiligo and associated diseases, may indicate that there should be other causes, yet unknown, disrupting the normal function of keratinocyte-melanocyte interaction, in the course of vitiligo.
Financial support and sponsorship
This study was supported by the Research Fund of the University of Istanbul.
Conflicts of interest
There are no conflicts of interest.
What is new?
The studied Endothelin.1 (EDN1) (G5665T and T.1370G) and EDN receptor type A (C+70G and G-231A) polymorphisms may not have a causative effect in vitiligo. In fact, haplotype analyses denoting protective effects against generalized vitiligo and associated diseases suggest that there should be other causes disrupting melanogenesis in the course of vitiligo.
Acknowledgment
This study was supported by the Research Fund of the University of Istanbul (Project No: UDP-53896).
References
- 1.Le Poole IC, Wañkowicz-Kaliñska A, van den Wijngaard RM, Nickoloff BJ, Das PK. Autoimmune aspects of depigmentation in vitiligo. J Investig Dermatol Symp Proc. 2004;9:68–72. doi: 10.1111/j.1087-0024.2004.00825.x. [DOI] [PubMed] [Google Scholar]
- 2.Falabella R. Vitiligo and the melanocyte reservoir. Indian J Dermatol. 2009;54:313–8. doi: 10.4103/0019-5154.57604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi Y, Hearing VJ. Physiological factors that regulate skin pigmentation. Biofactors. 2009;35:193–9. doi: 10.1002/biof.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KY, Jeon SY, Hong JW, Choi KW, Lee CY, Choi SJ, et al. Endothelin-1 enhances the proliferation of normal human melanocytes in a paradoxical manner from the TNF-α-inhibited condition, but tacrolimus promotes exclusively the cellular migration without proliferation: A proposed action mechanism for combination therapy of phototherapy and topical tacrolimus in vitiligo treatment. J Eur Acad Dermatol Venereol. 2013;27:609–16. doi: 10.1111/j.1468-3083.2012.04498.x. [DOI] [PubMed] [Google Scholar]
- 5.Imokawa G, Kobayashi T, Miyagishi M, Higashi K, Yada Y. The role of endothelin-1 in epidermal hyperpigmentation and signaling mechanisms of mitogenesis and melanogenesis. Pigment Cell Res. 1997;10:218–28. doi: 10.1111/j.1600-0749.1997.tb00488.x. [DOI] [PubMed] [Google Scholar]
- 6.Mou KH, Zhang XQ, Yu B, Zhang ZL, Feng J. Effects of endothelin-1 on melanocyte adhesion and migration. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2004;29:247–51. [PubMed] [Google Scholar]
- 7.Singh S, Singh U, Pandey SS. Serum concentration of IL-6, IL-2, TNF-α, and IFNg in vitiligo patients. Indian J Dermatol. 2012;57:12–4. doi: 10.4103/0019-5154.92668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moretti S, Spallanzani A, Amato L, Hautmann G, Gallerani I, Fabbri P. Vitiligo and epidermal microenvironment: Possible involvement of keratinocyte-derived cytokines. Arch Dermatol. 2002;138:273–4. doi: 10.1001/archderm.138.2.273. [DOI] [PubMed] [Google Scholar]
- 9.Aydin AF, Vural P, Oruç ÇU, Dogru-Abbasoglu S, Özderya A, Karadag B, et al. The evaluation of endothelin 1 (EDN1) and endothelin receptor type A (EDNRA) gene polymorphisms in Hashimoto's thyroiditis. Int Immunopharmacol. 2014;21:181–5. doi: 10.1016/j.intimp.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Aydin AF, Develi-Is S, Dogru-Abbasoglu S, Vural P, Ozderya A, Karadag B, et al. Polymorphisms of endothelin 1 (G5665T and T-1370G) and endothelin receptor type A (C 70G and G-231A) in Graves’ disease. Int Immunopharmacol. 2014;18:198–202. doi: 10.1016/j.intimp.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca C, Renzoni E, Sestini P, Pantelidis P, Lagan A, Bunn C, et al. Endothelin axis polymorphisms in patients with scleroderma. Arthritis Rheum. 2006;54:3034–42. doi: 10.1002/art.22036. [DOI] [PubMed] [Google Scholar]
- 12.Mantaka A, Goulielmos GN, Koulentaki M, Tsagournis O, Voumvouraki A, Kouroumalis EA. Polymorphisms of genes related to endothelial cells are associated with primary biliary cirrhosis patients of Cretan origin. Hum Immunol. 2012;73:829–35. doi: 10.1016/j.humimm.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Vasku V, Vasku A, Tschöplová S, Izakovicová Hollá L, Semrádová V, Vácha J. Genotype association of C(-735) T polymorphism in matrix metalloproteinase 2 gene with G (8002) A endothelin 1 gene with plaque psoriasis. Dermatology. 2002;204:262–5. doi: 10.1159/000063355. [DOI] [PubMed] [Google Scholar]
- 14.Kim HJ, Choi CP, Uhm YK, Kim YI, Lee JW, Yoon SH, et al. The association between endothelin-1 gene polymorphisms and susceptibility to vitiligo in a Korean population. Exp Dermatol. 2007;16:561–6. doi: 10.1111/j.1600-0625.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 15.Lan CC, Ko YC, Tu HP, Wu CS, Lee CH, Wu CS, et al. Association study between keratinocyte-derived growth factor gene polymorphisms and susceptibility to vitiligo vulgaris in a Taiwanese population: Potential involvement of stem cell factor. Br J Dermatol. 2009;160:1180–7. doi: 10.1111/j.1365-2133.2009.09064.x. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Panoulas VF, Douglas KM, Smith JP, Taffé P, Stavropoulos-Kalinoglou A, Toms TE, et al. Polymorphisms of the endothelin-1 gene associate with hypertension in patients with rheumatoid arthritis. Endothelium. 2008;15:203–12. doi: 10.1080/10623320802228708. [DOI] [PubMed] [Google Scholar]
- 18.Bühler K, Ufer M, Müller-Marbach A, Brinkmann U, Laule M, Stangl V, et al. Risk of coronary artery disease as influenced by variants of the human endothelin and endothelin-converting enzyme genes. Pharmacogenet Genomics. 2007;17:77–83. doi: 10.1097/01.fpc.0000230118.26581.40. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Naser MB, El-Khateeb EA, Sallam TH, Habib MA. Endothelin-1 is significantly elevated in plasma of patients with vitiligo treated with psoralen plus ultraviolet A. Clin Exp Dermatol. 2006;31:571–5. doi: 10.1111/j.1365-2230.2006.02148.x. [DOI] [PubMed] [Google Scholar]
- 20.Baráth A, Endreffy E, Bereczki C, Gellén B, Szücs B, Németh I, et al. Endothelin-1 gene and endothelial nitric oxide synthase gene polymorphisms in adolescents with juvenile and obesity-associated hypertension. Acta Physiol Hung. 2007;94:49–66. doi: 10.1556/APhysiol.94.2007.1-2.6. [DOI] [PubMed] [Google Scholar]
- 21.Barden AE, Herbison CE, Beilin LJ, Michael CA, Walters BN, Van Bockxmeer FM. Association between the endothelin-1 gene Lys198Asn polymorphism blood pressure and plasma endothelin-1 levels in normal and pre-eclamptic pregnancy. J Hypertens. 2001;19:1775–82. doi: 10.1097/00004872-200110000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal PK, Jain V, Srinivasan R, Jha V. Maternal EDN1 G5665T polymorphism influences circulating endothelin-1 levels and plays a role in determination of preeclampsia phenotype. J Hypertens. 2009;27:2044–50. doi: 10.1097/HJH.0b013e32832f7f3f. [DOI] [PubMed] [Google Scholar]
- 23.Iglarz M, Benessiano J, Philip I, Vuillaumier-Barrot S, Lasocki S, Hvass U, et al. Preproendothelin-1 gene polymorphism is related to a change in vascular reactivity in the human mammary artery in vitro. Hypertension. 2002;39:209–13. doi: 10.1161/hy0202.103442. [DOI] [PubMed] [Google Scholar]
- 24.Zang X, Zhou Y, Huang Z, Zhang C. Endothelin-1 single nucleotide polymorphisms and risk of pulmonary metastatic osteosarcoma. PLoS One. 2013;8:e73349. doi: 10.1371/journal.pone.0073349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonenberg N. mRNA translation: Influence of the 5’ and 3’ untranslated regions. Curr Opin Genet Dev. 1994;4:310–5. doi: 10.1016/s0959-437x(05)80059-0. [DOI] [PubMed] [Google Scholar]
- 26.Aly DG, Salem SA, Abdel-Hamid MF, Youssef NS, El Shaer MA. Endothelin-1 and its A and B receptors: Are they possibly involved in vitiligo? Acta Dermatovenerol Croat. 2013;21:12–8. [PubMed] [Google Scholar]


